
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 21 June 2023
Sec. Nanobiotechnology
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1201300
 Yunze Sun1,2†
Yunze Sun1,2† Yiran Wu1,2†
Yiran Wu1,2† Dachuan Ma3
Dachuan Ma3 Jian-Jun Li2*
Jian-Jun Li2* Xianming Liu3*
Xianming Liu3* Yuanjiang You2
Yuanjiang You2 Jun Lu4
Jun Lu4 Zhen Liu5
Zhen Liu5 Xin Cheng5
Xin Cheng5 Yuguang Du2*
Yuguang Du2*Glycans are an important group of natural biopolymers, which not only play the role of a major biological energy resource but also as signaling molecules. As a result, structural characterization or sequencing of glycans, as well as targeted synthesis of glycans, is of great interest for understanding their structure–function relationship. However, this generally involves tedious manual operations and high reagent consumptions, which are the main technical bottlenecks retarding the advances of both automatic glycan sequencing and synthesis. Until now, automated enzymatic glycan sequencers or synthesizers are still not available on the market. In this study, to promote the development of automation in glycan sequencing or synthesis, first, programmed degradation and synthesis of glycans catalyzed by enzymes were successfully conducted on a digital microfluidic (DMF) device by using microdroplets as microreactors. In order to develop automatic glycan synthesizers and sequencers, a strategy integrating enzymatic oligosaccharide degradation or synthesis and magnetic manipulation to realize the separation and purification process after enzymatic reactions was designed and performed on DMF. An automatic process for enzymatic degradation of tetra-N-acetyl chitotetraose was achieved. Furthermore, the two-step enzymatic synthesis of lacto-N-tetraose was successfully and efficiently completed on the DMF platform. This work demonstrated here would open the door to further develop automatic enzymatic glycan synthesizers or sequencers based on DMF.
Glycans, nucleic acids, and proteins are three major classes of natural biopolymers (Wen et al., 2018). In addition to their traditionally recognized roles as energy sources for living organisms, it is now well known that glycans play important signaling roles in a variety of physiological and pathological processes, including cell growth and proliferation (Varki, 2017), immune responses (Crocker et al., 2007), angiogenesis and tumor cell metastasis (Lou et al., 2014), protein folding and degradation (Parodi, 2000), cell–cell communications (Cannon and Helenius, 1999), and cell–pathogen interactions (Kato and Ishiwa, 2015). Identifying links between glycan structures and functions (Dennis et al., 2009), monitoring glycosylation in disease diagnosis and prognosis (Peracaula et al., 2008), and elucidating molecular mechanisms of glycans with structural differences involved in pathogenesis (Freeze and Aebi, 2005) are highlighted in current glycan-related studies. Therefore, developing precise, robust, and sensitive methodologies for the analysis and synthesis of glycans with definite structures is critical.
However, the extremely high diversity of isomerization makes structural elucidation of glycans a challenging job. The most frequently used methods for the structural analysis of glycans include capillary electrophoresis (CE) (Guttman, 1997), nuclear magnetic resonance spectroscopy (NMR) (Wyss et al., 1995), mass spectrometry (MS) (Morelle and Michalski, 2007), and liquid chromatography (LC) (Royle et al., 2008), which are often combined when being applied (Houel et al., 2014). In order to identify glycosidic linkages, an exoglycosidase-based digestion technique by highly specific non-reducing end exoglycosidase is routinely used for glycan sequencing coupled with the aforementioned methods (Tzur et al., 2008). However, this approach often involves tedious and time-consuming manual processes (Szigeti and Guttman, 2017).
The availability of pure glycans is a prerequisite for understanding the molecular details of their functions (Rillahan and Paulson, 2011) and producing homogeneous glycoconjugates (Kosik et al., 2010). The chemical synthesis of oligosaccharides generally consists of a glycosyl donor, a glycosyl acceptor, and an activator (Wen et al., 2018). Numerous chemical strategies have been explored for the assembly of complex glycans, such as Schmidt’s imidate (Hummel and Schmidt, 1997), Nicolaou’s two-stage activation (Nicolaou and Mitchell, 2001), Danishefsky’s glycal assembly (Danishefsky and Bilodeau, 1996), Kahne’s sulfoxide glycosylation (Yan and Kahne, 1996), Boons’ polymer-supported solution-phase synthesis (Zhu and Boons, 2000), Ye and Huang’s preactivation glycosylation (Miermont et al., 2007), and Wong’s reactivity-based one-pot synthesis (Koeller and Wong, 2000). Compared with the chemical synthesis of glycans, enzymatic synthesis has attracted more attention due to high stereoselectivity, high regioselectivity, non-protection, mild reaction conditions, and environmental friendliness (Chen, 2015). In recent years, one-pot multienzyme (OPME) systems for oligosaccharide synthesis have been well established (Yu and Chen, 2016), which allow the use of designated enzymatic modules for extending specific monosaccharides with specific configurations and linkages on the glycan (Ye et al., 2019).
Tedious procedures and high labor costs are common technical limitations to rapid advances in both glycan sequencing and synthesis. Thus, automatic strategies have been developed for glycan synthesis. The first automated glycan synthesizer was set up by modifying a peptide synthesizer based on a solid-phase synthetic method (Plante et al., 2001). Later, the enzymatic synthesis of oligosaccharides was achieved in an automated manner using a commercially available peptide synthesizer (Zhang et al., 2018). More recently, based on an ISYNTH AI SWING workstation (a liquid handling system), an automated platform for enzyme-mediated oligosaccharide synthesis was built (Li et al., 2019). By contrast, automated glycan sequencers have been not reported yet, even though some explorations have been made for automatic glycan sequencing. For instance, a novel automated approach for N-glycan sequencing of biopharmaceuticals by combining capillary electrophoresis and exoglycosidases has been reported (Szigeti and Guttman, 2017). Miniaturized platforms for implementing enzyme-promoted synthesis and degradation of oligosaccharides with high automation and reliability are strongly needed for processing a small number of samples. Chemists have long been interested in miniaturizing chemical reactions to take advantage of favorable scaling of diffusion and heat exchange (Davies, 2019; Hardwick and Ahmed, 2020).
Digital microfluidics (DMF) is a novel microscale liquid-handling platform on which picoliter- and microliter-sized droplets are manipulated on arrays of electrodes coated with a hydrophobic insulator (Samiei et al., 2016). On a DMF chip, droplets can be manipulated to move, dispense, mix, split, and merge with ease of automation. DMF is being extensively used to miniaturize a wide range of biochemical applications, such as PCR (Sanders et al., 2011), single cell analysis and immunoassay (He et al., 2015), and chemical synthesis of peptides (Jebrail et al., 2012; Xing et al., 2021), with the advantages of much less amount of sample consumptions, faster heat and mass transfer and reaction rates, and higher integration capacity. To our knowledge, there has been no precedence for enzymatic degradation and synthesis of oligosaccharides reported on the DMF platform. In this report, the viability of using the DMF platform for enzymatic degradation and synthesis of oligosaccharides was investigated.
Unless specified, all chemicals were of analytical grade and purchased from Sigma (St. Louis, Missouri, United States), Aladdin (Shanghai, China), or Beijing Solarbio Science and Technology Co. Ltd (Beijing, China). Tetra-N-acetyl chitotetraose was purchased from Mega (Bray, Ireland). Chitosan oligosaccharide RX14 was obtained from the company China RongXin Biotechnology Co., Ltd. (Suzhou, China). 7-Aminonaphthalene-1,3-disulfonic acid (Tag1) was purchased from TCI Development Co., Ltd. (Shanghai, China). Tag2 (7-(2-(2-(N-methyl-aminooxy)ethoxy)ethoxy)naphthalene-1,3-disulfonic acid) was synthesized by Shanghai Nafu Biotechnology Co. Ltd. according to the published procedure (Li et al., 2019) (Shanghai, China).
The DMF platform was set up according to the published procedure (Supplementary Figure S1) (Li et al., 2022).
On the two-plate DMF device used for enzymatic reactions, 80 actuation electrodes were fabricated on the bottom plate, which were covered with 8 μm thick SU-8 as the dielectric layer and 50 nm thick Teflon as the hydrophobic layer to prevent damage to the electrodes, while the top plate was also covered with the same hydrophobic layer. Four semicircular notches and four through holes were fabricated on the top plate with a laser scriber for droplet loading and collecting with a pipette. The device was assembled by joining a top and bottom plate with a spacer formed by three layers of double-sided tape with a thickness of approximately 0.15 mm. Accordingly, the volume of one droplet on one electrode was around 2–2.5 μL.
Streptomyces alfalfae β-1,4-N-acetylglucosaminidase (GlcNAcase) (Lv et al., 2019), Trichoderma reesei β-1,4-glucosaminidase (GlcNase) (Ike et al., 2006), Bifidobacterium longum N-acetylhexosamine-1-kinase (NahK) (Li et al., 2011), Pasteurella multocida N-acetylglucosamine uridylyltransferase (GlmU) (Chen et al., 2011), Helicobacter pylori β-1,3-N-acetylglucosaminyltransferase (LgtA) (Li et al., 2016), Escherichia coli K-12 pyrophosphatase (PpA) (Li et al., 2013), Escherichia coli K-12 galactokinase (GalK) (Chen et al., 2002), Bifidobacterium longum UDP-sugar pyrophosphorylase (USP) (Muthana et al., 2012), and Escherichia coli O55:H7 β-1,3-galactosyltransferase (WbgO) (Liu et al., 2009) were expressed and purified as reported in the literature.
In general, all enzymes were overexpressed in E. coli BL21(DE3) and induced by isopropyl 1-thio-D-galactopyranoside (IPTG) according to the aforementioned published procedures. The cells were collected by centrifugation and suspended in the equilibration buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0). Cells were disrupted by sonication and centrifuged at 15,000 g for 20 min at 4°C to remove insoluble cell debris. The supernatants were applied to a Ni2+-NTA agarose column and washed with equilibration buffer and elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 50/200 mM imidazole, pH 8.0) in turn to remove protein impurities. The fractions showing enzyme activities were pooled and concentrated. SDS-PAGE was carried out on a 12% separation gel. Protein concentration was measured by using the Bradford method with bovine serum albumin as the standard.
The droplets were actuated with a high-voltage alternative wave generator (QFC 2020D, Dalian Quantum Fluid Control Technology, China) in a sinusoid waveform (300 V RMS with a frequency of 1 kHz). The actuation of droplets started from loading deionized water to test the actuation versatility of the DMF device. The reagents used on the DMF device included saccharides, salts, enzymes, and magnetic beads. For saccharides, 100 mM glucose, 100 mM galactose, 100 mM lactose, and 10 mg/mL RX14 were tested, respectively. For salts, 50 mM NaOAc buffer (pH = 5.5), 1 M Tris-HCl buffer (pH = 8.0), 60 mM NH4HCO3, and 100 mM MgCl2 were tested, respectively. For enzymes, each enzyme mentioned previously was tested. Before loading, Pluronic F127 surfactant was added at a concentration of 0.05% (v/v).
Tetra-N-acetyl chitotetraose (A4) at a concentration of 5 mg/mL was loaded in one droplet, and GlcNAcase at a concentration of 0.1 mg/mL in NaOAc buffer (50 mM, pH = 5.5) was loaded in another one.
As for another reaction, chitosan oligosaccharide RX14 at a concentration of 5 mg/mL was loaded in one droplet, and GlcNAcase and GlcNase at a concentration of 0.1 mg/mL in NaOAc buffer (50 mM, pH = 5.5) was loaded in another one.
On the DMF platform, the two droplets were mixed evenly and left for 1 h for enzymatic degradation. After the reaction, the droplet containing the degradation product was taken out through a hole on the top plate and characterized by MS.
Substrates and enzymatic modules were loaded in two droplets. One droplet contained 3 mM lactose, 3 mM N-acetylglucosamine, 3.6 mM ATP, and 3.6 mM UTP, while another one contained enzymatic module 1 (Supplementary Table S1), 10 mM MgCl2, and 50 mM NH4HCO3.
Like the process for oligosaccharide degradation, the two droplets were mixed evenly and left for 1 h for enzymatic synthesis. After the reaction, the droplet containing the synthesized product was taken out through a hole on the top plate and characterized by mass spectrometry.
Tag1 (0.03 mmol) and tetra-N-acetyl chitotetraose (A4, 0.0024 mmol) were dissolved in 1 mL 15% CH3COOH solution. Next, sodium cyanoborohydride (NaBH3CN, 0.03 mmol) was added. The reaction mixture was incubated at 50°C for 15 h (Chiesa and Oneill, 1994). The reaction mixture was purified on a Bio-Gel P-2 column from Bio-Rad (Hercules, California, United States). The column was eluted with water. The fractions containing the product were combined and lyophilized. The product, A4-Tag1, was confirmed by MS (Supplementary Figure S2).
Tag2 (0.008 mmol) was dissolved in NaOAc buffer (100 mM, pH 4.2, 100 µL). Next, lactose (Lac, 0.08 mmol) was added. The reaction mixture was incubated at 37°C for 24 h (Li et al., 2019). The reaction mixture was purified on a Bio-Gel P-2 column. The column was eluted with water. The fractions containing the product were combined and lyophilized. The product, Lac-Tag2, was confirmed by MS (Supplementary Figure S3).
Tetra-N-acetyl chitotetraose modified with Tag1 (A4-Tag1) at a concentration of 1 mg/mL was loaded in one droplet, while GlcNAcase at a concentration of 0.02 mg/mL in NaOAc buffer (50 mM, pH = 5.5) was loaded in another droplet.
On the DMF platform, the two droplets were mixed evenly and left for 1 h for enzymatic degradation. Then reaction mixture was mixed with DEAE (diethylaminoethyl) magnetic beads in one droplet, which were suspended in the droplet to capture products modified with Tag1. The DEAE magnetic beads and supernatant were separated by a magnet, and the supernatant containing the degradation products was taken out through a hole on the top plate for MS analysis. Subsequently, DEAE magnetic beads were washed in turn with water and 60 mM NH4HCO3. Finally, the captured products on DEAE magnetic beads were released by washing with 0.3 M NH4HCO3, and the eluent obtained was transferred and characterized by MS.
Lactose modified with Tag2 (Lac-Tag2) at a concentration of 3 mM was loaded in the first droplet. Enzymatic module 1, 10 mM MgCl2, 50 mM NH4HCO3, 3 mM N-acetylglucosamine, 3.6 mM ATP, and 3.6 mM UTP were loaded in the second droplet. Enzymatic module 2, 10 mM MgCl2, 50 mM NH4HCO3, 3 mM galactose, 3.6 mM ATP, and 3.6 mM UTP were loaded in the third droplet. The components of the enzymatic modules are shown in Supplementary Table S2.
On the DMF platform, the first two droplets were mixed evenly and left for 1 h for enzymatic synthesis. Then the reaction mixture was mixed with DEAE magnetic beads in one droplet. The DEAE magnetic beads were suspended in the droplet to capture the products modified with Tag2. The DEAE magnetic beads and supernatants were separated by a magnet, and the supernatant, including enzymes, excess (sugar) nucleotides, MgCl2, and NH4HCO3, was taken out and discarded. Subsequently, DEAE magnetic beads were washed in turn with water and 60 mM NH4HCO3. Next, the captured products with Tag2 on DEAE magnetic beads were released by washing with 0.3 M NH4HCO3, and the eluent obtained was mixed evenly with the third droplet and left for 1 h for the next enzymatic synthesis reaction. Like the first cycle, DEAE magnetic beads were used for capturing the synthesized products. Finally, the eluent washed by 0.3 M NH4HCO3 was transferred and characterized by MS.
The eluent from 2.9 containing lacto-N-tetraose modified with Tag2 (LNT-Tag2) was mixed with a droplet (2 μL) of trifluoroacetic acid (0.25%, v/v) on the DMF platform. The reaction mixture was agitated at room temperature for 2 h (Prudden et al., 2014). After the reaction, the product was mixed with a droplet of 50 mM NH4HCO3 to adjust the pH. Then, DEAE magnetic beads were used for capturing Tag2 removed from LNT-Tag2, and the supernatant including lacto-N-tetraose (LNT) was taken out and characterized by MS.
Analysis of the products was performed on the UltiMate-3000-ISQ-EM (Thermo Fisher Scientific). The droplets were diluted to 100 μL before analysis. Samples were loaded directly without columns. The mobile phase was water, and the flow rate was 0.1 mL/min.
The two-plate DMF device was established according to a published procedure (Li et al., 2022).
A droplet on a digital microfluidic chip was actuated under the electrowetting mechanism (Wheeler, 2008), by which the wettability of its static hydrophobic surface can be tuned to hydrophilic under an applied electric field, providing additional charges into the triple contact line of the droplet, the chip surface, and the surrounding oil. Under the two-plate format, as in our DMF chip, when the adjacent electrode of a droplet is charged, the electrocapillary force on the triple line breaks the horizontal force balance and pulls the droplet onto the activated electrode. This is the basic working mechanism of droplet actuation. When two droplets are transported onto the same electrode, they are fused into one bigger droplet. Therefore, when the two droplets carrying two reactants, respectively, are merged, a reaction takes place in the merged droplet.
Initially, droplets containing different substances, including saccharides, enzymes, and salts, involved in enzymatic synthesis and degradation of oligosaccharides were manipulated on the DMF platform to test the feasibility of our methodology. Droplets containing salts or saccharides can be actuated smoothly in both air and silicone oil (cSt 1.5), while those containing enzymes can only be actuated in silicone oil after being doped with 0.05% Pluronic F127 (Luk et al., 2008). In addition, evaporation of droplets on the DMF chip is an issue. In the air, a droplet with a volume of 2 μL can only be kept for around 1–2 h at room temperature. In order to avoid evaporation of droplets, the droplets can be immersed in silicone oil so that they can be kept for at least 10 h at room temperature. In order to be compatible with the DMF platform, the droplets of the enzymatic reactions were conducted in the presence of 0.05% Pluronic F127 in silicone oil in the following experiments unless indicated otherwise.
We tried degradation of oligosaccharides by specific non-reducing end exoglycosidases on the DMF platform. Specific non-reducing end GlcNAcase (β-1,4-N-acetylglucosaminidase) (EC 3.2.1.52) was used to hydrolyze tetra-N-acetyl chitotetraose (A4) into N-acetylglucosamine (GlcNAc) (Figure 1A). A4 was loaded in a droplet, and GlcNAcase and a buffer were carried in another droplet. The two droplets were mixed evenly and incubated for enzymatic degradation of A4. After 1 h, the droplet containing the degradation product was taken out through a hole on the top plate and characterized by mass spectrometry (Supplementary Video S1). According to MS characterization of GlcNAcase-catalyzed degradation of tetra-N-acetyl chitotetraose (A4), ESI-MS m/z was calculated for the degradation product GlcNAc (C8H16NO6) as [M + H]+ 222.22 and found to be 222.13. The peak corresponding to A4 (calculated as [M + H]+ 831.80) was not detected based on MS. MS analysis indicated that the degradation product GlcNAc was detected and enzymatic activities were not affected by the electric field (Figure 1B). Similarly, specific non-reducing end GlcNase (β-1,4-glucosaminidase) (EC 3.2.1.165) was used to degrade chitosan oligosaccharide RX14, and the product glucosamine (GlcN) was observed (Supplementary Figure S4). These experiments demonstrated that enzymatic degradation of oligosaccharides in one droplet was successfully achieved on our DMF platform.
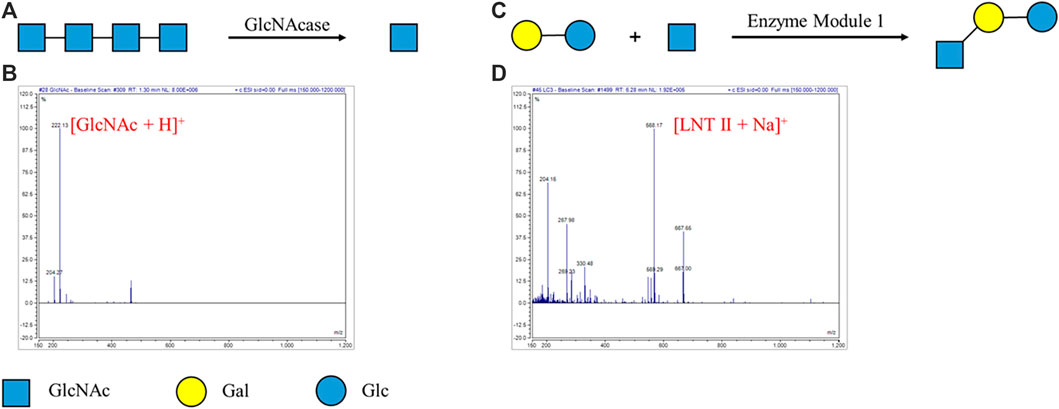
FIGURE 1. Enzymatic reaction for degradation and synthesis of oligosaccharides on the DMF device. (A) GlcNAcase-catalyzed degradation of tetra-N-acetyl chitotetraose (A4). (B) MS characterization of GlcNAcase-catalyzed degradation of tetra-N-acetyl chitotetraose (A4). ESI-MS m/z calculated for C8H16NO6 [M + H]+ 222.22 and found to be 222.13. The peak corresponding to A4 (calculated as [M + H]+ 831.80) was not detected. (C) Enzymatic module-catalyzed synthesis of lacto-N-triose II (LNT II). (D) MS characterization of enzymatic module-catalyzed synthesis of lacto-N-triose II (LNT II). ESI-MS m/z calculated for C20H35NO16Na [M + Na]+ 568.48 and found to be 568.17. The peak corresponding to lactose (calculated as [M + Na]+ 365.29) was not detected.
Encouraged by these results, enzymatic module-catalyzed synthesis of oligosaccharide lacto-N-triose II (LNT II) was also attempted (Figure 1C). The substrates, including lactose, GlcNAc, ATP, and UTP, were loaded in a droplet, while the enzymatic module 1 (including NahK, GlmU, LgtA, PpA), MgCl2, and buffer were put in another one. The ratio of different enzymes in the enzymatic modules was optimized for efficient enzymatic synthesis. The two droplets were mixed and incubated for 1 h and characterized by mass spectrometry. According to MS characterization of enzymatic module-catalyzed synthesis of lacto-N-triose II (LNT II), ESI-MS m/z was calculated for LNT II (C20H35NO16Na) as [M + Na]+ 568.48 and found to be 568.17. The peak corresponding to lactose (calculated as [M + Na]+ 365.29) was not detected based on MS (Figure 1D). MS analysis indicated that nearly all substrates were converted into the product LNT II.
In order to develop automatic glycan synthesizers and sequencers, a strategy integrating enzymatic oligosaccharide degradation or synthesis and magnetic manipulation to realize the separation and heterogeneous purification process after enzymatic reactions on the DMF platform was designed (Figure 2). The sulfonate tags bear two sulfonate groups, which can be captured onto anion exchange resin and released by 0.3 M NH4HCO3. The sulfonated tags can be efficiently attached to the reducing ends of the starting saccharides (Figure 3), which have been demonstrated not to interfere with enzymatic reactions (Chiesa and Oneill, 1994; Li et al., 2019).
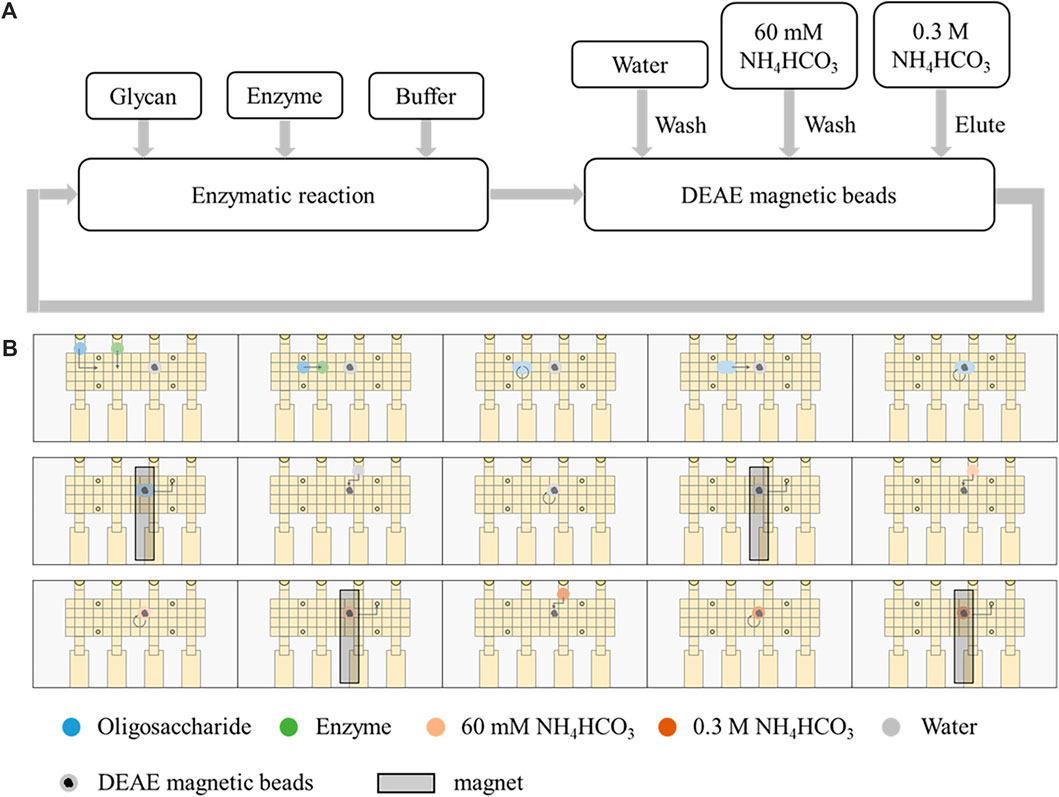
FIGURE 2. Automatic strategy coupling enzymatic reactions and magnetic manipulation on the DMF device. (A) Schematic processes of enzymatic reactions and separation by DEAE magnetic beads. (B) Schematic of the processes on the DMF device.
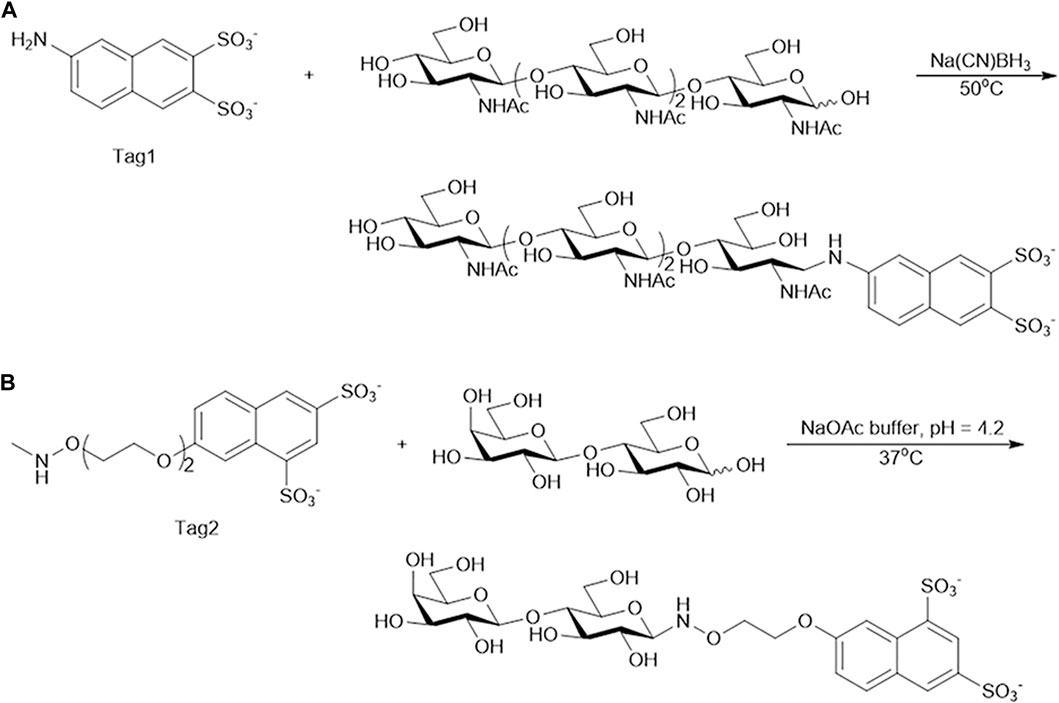
FIGURE 3. Synthesis of oligosaccharide modified with Tag. (A) Tetra-N-acetyl chitotetraose (A4) modified with Tag1. (B) Lactose modified with Tag2.
To verify this automatic process, GlcNAcase was still used to degrade A4 modified at the reducing end with the sulfonate Tag1 (A4-Tag1) on the DMF platform (Figure 4A). The process on the DMF platform was shown in Supplementary Video S2. First, the droplet containing A4-Tag1 and another one containing GlcNAcase and buffer were mixed and incubated for 1 h. Then, DEAE (diethylaminoethyl) magnetic beads were suspended in the droplet to capture the products modified with Tag1. The DEAE magnetic beads and supernatants were separated by a magnet, and the supernatant droplet ① containing GlcNAc cleaved off from the non-reducing end of A4-Tag1 by GlcNAcase was taken out through a hole on the top plate for MS analysis. Subsequently, DEAE magnetic beads were washed in turn with water and 60 mM NH4HCO3. Finally, the captured products with Tag1 on DEAE magnetic beads were released by washing with a small volume of 0.3 M NH4HCO3, and the eluted droplet ② obtained was transferred and characterized by MS. According to MS results (Figure 4B; Supplementary Figure S5), GlcNAc (calculated as [M + H]+ 222.22 and found to be 222.05) was found in the droplet ① and GlcNAc-Tag1 (calculated for C18H20N8O11S2 as [M - 2H]2− 252.25 and found to be 252.13) was also detected in droplet ②. In addition, the peak corresponding to A4-Tag1 (calculated for C42H59N5O26S2 as [M - 2H]2− 557.03) was not detected based on MS in droplet ②. These MS results proved that A4-Tag1 in a single droplet was broken down into GlcNAc and GlcNAc-Tag1, and the degradation products were completely separated by DEAE magnetic beads through an automatic process on the DMF platform.
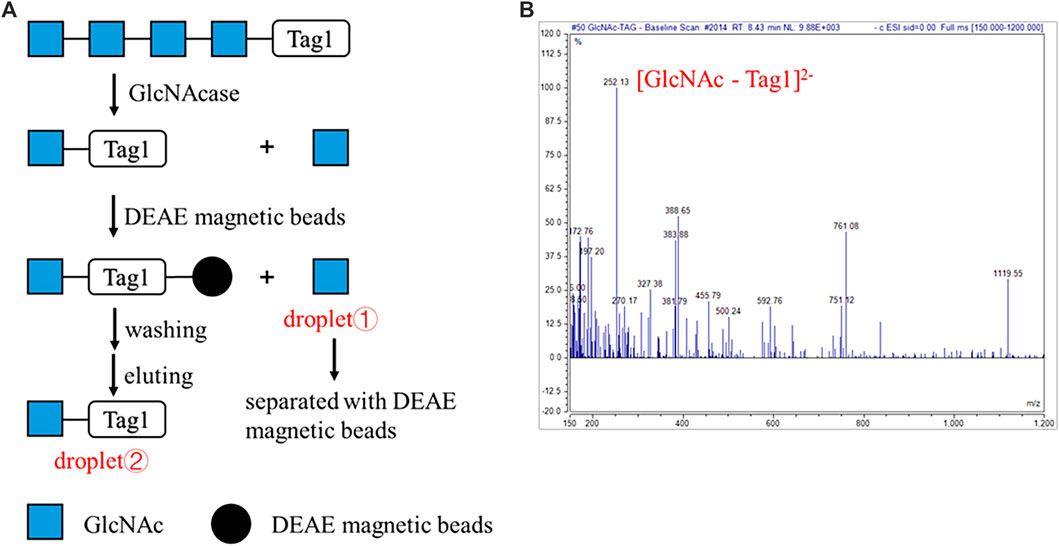
FIGURE 4. Automatic strategy of enzymatic reaction for oligosaccharides degradation on the DMF device. (A) Schematic processes of the automatic strategy of GlcNAcase-catalyzed degradation of tetra-N-acetyl chitotetraose (A4). (B) MS characterization of the product GlcNAc-Tag1. ESI-MS m/z calculated for C18H20N8O11S2 [M - 2H]2− 252.25 and found to be 252.13. The peak corresponding to A4-Tag1 (calculated for C42H59N5O26S2 as [M - 2H]2− 557.03) was not detected.
The automatic process for enzymatic synthesis of oligosaccharides on the DMF platform was similar to that of degradation of oligosaccharides. Two enzymatic modules were used to convert lactose modified at the reducing end with another sulfonate tag (Lac-Tag2) into Lacto-N-tetraose-Tag2 (LNT-Tag2) and, finally, into LNT (Figure 5A). As aforementioned, the droplet containing Lac-Tag2, ATP, and UTP and another one containing enzymatic module 1 (including NahK, GlmU, LgtA, PpA), MgCl2, buffer, and GlcNAc were mixed and incubated for 1 h. Then, lacto-N-triose II-Tag2 (LNT II-Tag2), the formed product with the sulfonate Tag2, was quantitatively captured onto DEAE magnetic beads, and the rest of the reaction components including enzymes and excess monosaccharides, and NTPs were separated with magnetic beads by a magnet. Then, the captured intermediate was washed in turn with H2O and 60 mM NH4HCO3 and was finally released by washing with 0.3 M NH4HCO3. Similarly, as the enzymatic synthesis of LNT II-Tag2 by enzymatic module 1, LNT II-Tag2 was further transformed into LNT-Tag2 by the enzymatic module 2 (including GalK, USP, WbgO, PpA). Finally, the Tag2 was removed under mild acidic conditions to give LNT as the final product having a free reducing end. During each step, the intermediates and the final product were characterized by MS. LNT II-Tag2 (calculated for C35H50N2O24S2 as [M - 2H]2− 473.46 and found to be 473.01), LNT-Tag2 (calculated for C41H60N2O29S2 as [M - 2H]2− 554.52 and found to be 554.14), and LNT (calculated for C26H45NO21Na as [M + Na]+ 730.62 and found to be 730.23) were observed (Figures 5B, C; Supplementary Figure S6). The extent of the reaction in DMF was not measured since it was difficult to quantify by HPLC for the substrates or products in a droplet. Nevertheless, the peak corresponding to Lac-Tag2 (calculated for C27H37NO19S2 [M - 2H]2− 371.86) was not detected based on MS. Therefore, it was speculated that nearly all substrates were converted into the product lacto-N-tetraose. These results proved that LNT was successfully synthesized on the DMF platform through an automatic process.
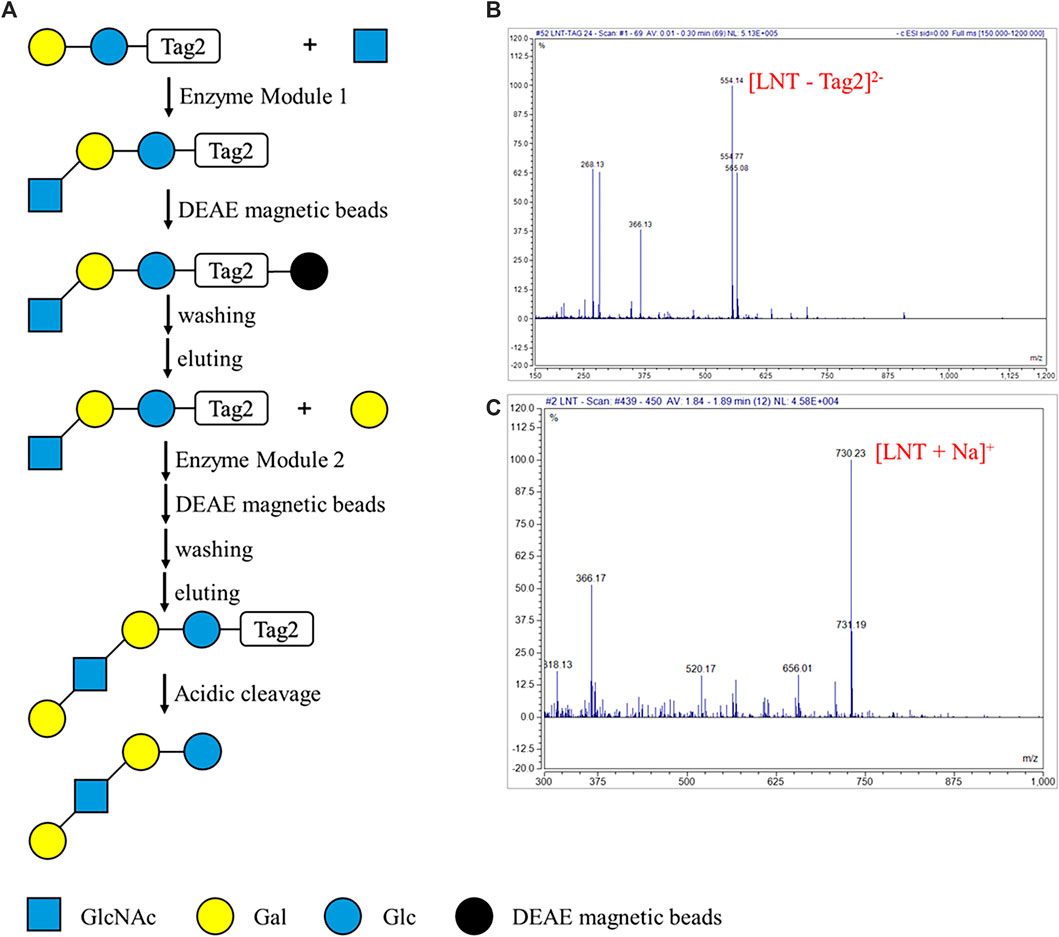
FIGURE 5. Automatic strategy of enzymatic reaction for oligosaccharides synthesis on the DMF device. (A) Schematic processes of the automatic strategy of enzymatic module-catalyzed synthesis of lacto-N-tetraose (LNT). (B) MS characterization of the key intermediate product LNT-Tag2. ESI-MS m/z calculated for C41H60N2O29S2 [M - 2H]2− 554.52 and found to be 554.14. The peak corresponding to Lac-Tag2 (calculated for C27H37NO19S2 [M - 2H]2− 371.86) was not detected. (C) MS characterization of the LNT. ESI-MS m/z calculated for C26H45NO21Na [M + Na]+ 730.62 and found to be 730.23.
Enzymatic degradation of tetra-N-acetyl chitotetraose (A4) and enzymatic synthesis of lacto-N-triose II (LNT II) were achieved successfully on our DMF platform. Moreover, a strategy integrating enzymatic oligosaccharide degradation or synthesis and magnetic manipulation to realize the separation and heterogeneous purification process after enzymatic reactions on the DMF platform was designed. Using this strategy, automatic enzymatic degradation of tetra-N-acetyl chitotetraose modified with Tag1 (A4-Tag1) and automatic enzymatic synthesis of lacto-N-tetraose (LNT) were realized. In conclusion, we proved the feasibility of enzymatic degradation and synthesis of oligosaccharides and developed an automatic process on the DMF platform by combining with magnetic beads-based separation and purification. Here, the DMF platform exhibits the potential capabilities of versatile, multiplexed, and automatable biochemical operations like oligosaccharide degradation and synthesis. In the future, our DMF platform would be integrated with micropumps for pipelined reactions and instruments for in situ detection so that the whole automatic process can be realized under the control of integrated programs. Above all, this work would lay the foundation for the development of automatic enzymatic glycan synthesizers and sequencers based on DMF.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
YD and J-JL conceptualized the project. YS, J-JL, and XL wrote the manuscript with input from all authors. YS and YW performed the experiments. YD and J-JL supervised the project. YD and XL acquired funding. All authors contributed to the article and approved the submitted version.
YD is grateful to the Natural Science Foundation of China (no. 21877114). XL is grateful to the Institute of Process Engineering Open Fund (no. 2014KF-03) and the National Natural Science Foundation of China (no. 31927802).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1201300/full#supplementary-material
Cannon, K. S., and Helenius, A. (1999). Trimming and readdition of glucose to N-linked oligosaccharides determines calnexin association of a substrate glycoprotein in living cells. J. Biol. Chem. 274 (11), 7537–7544. doi:10.1074/jbc.274.11.7537
Chen, X. (2015). “Human milk oligosaccharides (HMOS): Structure, function, and enzyme-catalyzed synthesis,” in Advances in carbohydrate chemistry and biochemistry. Editors D. C. Baker, and D. Horton (San Diego: Elsevier Academic Press Inc), Vol. 72, 113–190.
Chen, X., Liu, Z. Y., Zhang, J. B., Zhang, W., Kowal, P., and Wang, P. G. (2002). Reassembled biosynthetic pathway for large-scale carbohydrate synthesis: Alpha-gal epitope producing "superbug. Chembiochem 3 (1), 47–53. doi:10.1002/1439-7633(20020104)3:1<47:aid-cbic47>3.0.co;2-n
Chen, Y., Thon, V., Li, Y. H., Yu, H., Ding, L., Lau, K., et al. (2011). One-pot three-enzyme synthesis of UDP-GlcNAc derivatives. Chem. Commun. 47 (38), 10815–10817. doi:10.1039/c1cc14034e
Chiesa, C., and Oneill, R. A. (1994). Capillary zone electrophoresis of oligosaccharides derivatized with various aminonaphthalene sulfonic-acids. Electrophoresis 15 (8-9), 1132–1140. doi:10.1002/elps.11501501171
Crocker, P. R., Paulson, J. C., and Varki, A. (2007). Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7 (4), 255–266. doi:10.1038/nri2056
Danishefsky, S. J., and Bilodeau, M. T. (1996). Glycals in organic synthesis: The evolution of comprehensive strategies for the assembly of oligosaccharides and glycoconjugates of biological consequence. Angewandte Chemie-International Ed. Engl. 35 (13-14), 1380–1419. doi:10.1002/anie.199613801
Davies, I. W. (2019). The digitization of organic synthesis. Nature 570 (7760), 175–181. doi:10.1038/s41586-019-1288-y
Dennis, J. W., Nabi, I. R., and Demetriou, M. (2009). Metabolism, cell surface organization, and disease. Cell 139 (7), 1229–1241. doi:10.1016/j.cell.2009.12.008
Freeze, H. H., and Aebi, M. (2005). Altered glycan structures: The molecular basis of congenital disorders of glycosylation. Curr. Opin. Struct. Biol. 15 (5), 490–498. doi:10.1016/j.sbi.2005.08.010
Guttman, A. (1997). Multistructure sequencing of N-linked fetuin glycans by capillary gel electrophoresis and enzyme matrix digestion. Electrophoresis 18 (7), 1136–1141. doi:10.1002/elps.1150180719
Hardwick, T., and Ahmed, N. (2020). Digitising chemical synthesis in automated and robotic flow. Chem. Sci. 11 (44), 11973–11988. doi:10.1039/d0sc04250a
He, J. L., Chen, A. T., Lee, J. H., and Fan, S. K. (2015). Digital microfluidics for manipulation and analysis of a single cell. Int. J. Mol. Sci. 16 (9), 22319–22332. doi:10.3390/ijms160922319
Houel, S., Hilliard, M., Yu, Y. Q., McLoughlin, N., Martin, S. M., Rudd, P. M., et al. (2014). N- and O-glycosylation analysis of etanercept using liquid chromatography and quadrupole time-of-flight mass spectrometry equipped with electron-transfer dissociation functionality. Anal. Chem. 86 (1), 576–584. doi:10.1021/ac402726h
Hummel, G., and Schmidt, R. R. (1997). A versatile synthesis of the lactoneo-series antigens — Synthesis of sialyl dimer lewis X and of dimer lewis Y. Tetrahedron Lett. 38 (7), 1173–1176. doi:10.1016/s0040-4039(97)00006-3
Ike, M., Isami, K., Tanabe, Y., Nogawa, M., Ogasawara, W., Okada, H., et al. (2006). Cloning and heterologous expression of the exo-beta-D-glucosaminidase-encoding gene (gls93) from a filamentous fungus, Trichoderma reesei PC-3-7. Appl. Microbiol. Biotechnol. 72 (4), 687–695. doi:10.1007/s00253-006-0320-y
Jebrail, M. J., Assem, N., Mudrik, J. M., Dryden, M. D. M., Lin, K., Yudin, A. K., et al. (2012). Combinatorial synthesis of peptidomimetics using digital microfluidics. J. Flow Chem. 2 (3), 103–107. doi:10.1556/jfc-d-12-00012
Kato, K., and Ishiwa, A. (2015). The role of carbohydrates in infection strategies of enteric pathogens. Trop. Med. health 43 (1), 41–52. doi:10.2149/tmh.2014-25
Koeller, K. M., and Wong, C. H. (2000). Synthesis of complex carbohydrates and glycoconjugates: Enzyme-based and programmable one-pot strategies. Chem. Rev. 100 (12), 4465–4494. doi:10.1021/cr990297n
Kosik, O., Auburn, R. P., Russell, S., Stratilova, E., Garajova, S., Hrmova, M., et al. (2010). Polysaccharide microarrays for high-throughput screening of transglycosylase activities in plant extracts. Glycoconj. J. 27 (1), 79–87. doi:10.1007/s10719-009-9271-8
Li, H. B., Liu, X. M., Zhu, F. J., Ma, D. C., Miao, C. Y., Su, H. R., et al. (2022). Spatial barcoding-enabled highly multiplexed immunoassay with digital microfluidics. Biosens. Bioelectron. 215, 114557. doi:10.1016/j.bios.2022.114557
Li, L., Liu, Y. H., Wan, Y., Li, Y. H., Chen, X., Zhao, W., et al. (2013). Efficient enzymatic synthesis of guanosine 5 '-Diphosphate-Sugars and derivatives. Org. Lett. 15 (21), 5528–5530. doi:10.1021/ol402585c
Li, T., Liu, L., Wei, N., Yang, J. Y., Chapla, D. G., Moremen, K. W., et al. (2019). An automated platform for the enzyme-mediated assembly of complex oligosaccharides. Nat. Chem. 11 (3), 229–236. doi:10.1038/s41557-019-0219-8
Li, Y. H., Xue, M. Y., Sheng, X., Yu, H., Zeng, J., Thon, V., et al. (2016). Donor substrate promiscuity of bacterial beta 1-3-N-acetylglucosaminyltransferases and acceptor substrate flexibility of beta 1-4-galactosyltransferases. Bioorg. Med. Chem. 24 (8), 1696–1705. doi:10.1016/j.bmc.2016.02.043
Li, Y. H., Yu, H., Chen, Y., Lau, K., Cai, L., Cao, H. Z., et al. (2011). Substrate promiscuity of N-acetylhexosamine 1-kinases. Molecules 16 (8), 6396–6407. doi:10.3390/molecules16086396
Liu, X. W., Xia, C. F., Li, L., Guan, W. Y., Pettit, N., Zhang, H. C., et al. (2009). Characterization and synthetic application of a novel beta 1,3-galactosyltransferase from Escherichia coli O55:H7. Bioorg. Med. Chem. 17 (14), 4910–4915. doi:10.1016/j.bmc.2009.06.005
Lou, Y. W., Wang, P. Y., Yeh, S. C., Chuang, P. K., Li, S. T., Wu, C. Y., et al. (2014). Stage-specific embryonic antigen-4 as a potential therapeutic target in glioblastoma multiforme and other cancers. Proc. Natl. Acad. Sci. U. S. A. 111 (7), 2482–2487. doi:10.1073/pnas.1400283111
Luk, V. N., Mo, G. C. H., and Wheeler, A. R. (2008). Pluronic additives: A solution to sticky problems in digital microfluidics. Langmuir 24 (12), 6382–6389. doi:10.1021/la7039509
Lv, C., Gu, T., Xu, K., Gu, J., Li, L., Liu, X., et al. (2019). Biochemical characterization of a beta-N-acetylhexosaminidase from Streptomyces alfalfae and its application in the production of N-acetyl-d-glucosamine. J. Biosci. Bioeng. 128 (2), 135–141. doi:10.1016/j.jbiosc.2019.01.017
Miermont, A., Zeng, Y. L., Jing, Y. Q., Ye, X. S., and Huang, X. F. (2007). Syntheses of lewis(x) and dimeric lewis(x): Construction of branched oligosaccharides by a combination of preactivation and reactivity based chemoselective one-pot Glycosylations. J. Org. Chem. 72 (23), 8958–8961. doi:10.1021/jo701694k
Morelle, W., and Michalski, J. C. (2007). Analysis of protein glycosylation by mass spectrometry. Nat. Protoc. 2 (7), 1585–1602. doi:10.1038/nprot.2007.227
Muthana, M. M., Qu, J. Y., Li, Y. H., Zhang, L., Yu, H., Ding, L., et al. (2012). Efficient one-pot multienzyme synthesis of UDP-sugars using a promiscuous UDP-sugar pyrophosphorylase from Bifidobacterium longum (BLUSP). Chem. Commun. 48 (21), 2728–2730. doi:10.1039/c2cc17577k
Nicolaou, K. C., and Mitchell, H. J. (2001). Adventures in carbohydrate chemistry: New synthetic technologies, chemical synthesis, molecular design, and chemical biology. Angew. Chemie-International Ed. 40 (9), 1576–1624. doi:10.1002/1521-3773(20010504)40:9<1576:Aid-anie15760>3.0.Co;2-g
Parodi, A. J. (2000). Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem. J. 348, 1–13. doi:10.1042/bj3480001
Peracaula, R., Barrabes, S., Sarrats, A., Rudd, P. M., and de Llorens, R. (2008). Altered glycosylation in tumours focused to cancer diagnosis. Dis. Markers 25 (4-5), 207–218. doi:10.1155/2008/797629
Plante, O. J., Palmacci, E. R., and Seeberger, P. H. (2001). Automated solid-phase synthesis of oligosaccharides. Science 291 (5508), 1523–1527. doi:10.1126/science.1057324
Prudden, A. R., Chinoy, Z. S., Wolfert, M. A., and Boons, G. J. (2014). A multifunctional anomeric linker for the chemoenzymatic synthesis of complex oligosaccharides. Chem. Commun. (Camb) 50 (54), 7132–7135. doi:10.1039/c4cc02222j
Rillahan, C. D., and Paulson, J. C. (2011). “Glycan microarrays for decoding the glycome,” in Annual review of biochemistry. Editors R. D. Kornberg, C. R. H. Raetz, J. E. Rothman, and J. W. Thorner (Palo Alto: Annual Reviews), Vol. 80, 797–823.
Royle, L., Campbell, M. P., Radcliffe, C. M., White, D. M., Harvey, D. J., Abrahams, J. L., et al. (2008). HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal. Biochem. 376 (1), 1–12. doi:10.1016/j.ab.2007.12.012
Samiei, E., Tabrizian, M., and Hoorfar, M. (2016). A review of digital microfluidics as portable platforms for lab-on a-chip applications. Lab. Chip 16 (13), 2376–2396. doi:10.1039/c6lc00387g
Sanders, R., Huggett, J. F., Bushell, C. A., Cowen, S., Scott, D. J., and Foy, C. A. (2011). Evaluation of digital PCR for absolute DNA quantification. Anal. Chem. 83 (17), 6474–6484. doi:10.1021/ac103230c
Szigeti, M., and Guttman, A. (2017). Automated N-glycosylation sequencing of biopharmaceuticals by capillary electrophoresis. Sci. Rep. 7, 11663. doi:10.1038/s41598-017-11493-6
Tzur, Y., Markovich, A., and Lichtenstein, R. G. (2008). A two-dimensional array for simultaneous sequencing of N- and O-glycans and their glycoforms on specific glycosylation sites. J. Proteome Res. 7 (3), 1188–1198. doi:10.1021/pr070392+
Wen, L., Edmunds, G., Gibbons, C., Zhang, J., Gadi, M. R., Zhu, H., et al. (2018). Toward automated enzymatic synthesis of oligosaccharides. Chem. Rev. 118 (17), 8151–8187. doi:10.1021/acs.chemrev.8b00066
Wheeler, A. R. (2008). Putting electrowetting to work. Science 322 (5901), 539–540. doi:10.1126/science.1165719
Wyss, D. F., Choi, J. S., Li, J., Knoppers, M. H., Willis, K. J., Arulanandam, A. R. N., et al. (1995). Conformation and function of the N-linked glycan in the adhesion domain of human CD2. Science 269 (5228), 1273–1278. doi:10.1126/science.7544493
Xing, Y., Liu, Y., Chen, R., Li, Y., Zhang, C., Jiang, Y., et al. (2021). A robust and scalable active-matrix driven digital microfluidic platform based on printed-circuit board technology. Lab. Chip 21 (10), 1886–1896. doi:10.1039/d1lc00101a
Yan, L., and Kahne, D. (1996). Generalizing glycosylation: Synthesis of the blood group antigens Le(a), Le(b), and Le(X) using a standard set of reaction conditions. J. Am. Chem. Soc. 118 (39), 9239–9248. doi:10.1021/ja9608555
Ye, J., Xia, H., Sun, N., Liu, C.-C., Sheng, A., Chi, L., et al. (2019). Reprogramming the enzymatic assembly line for site-specific fucosylation. Nat. Catal. 2 (6), 514–522. doi:10.1038/s41929-019-0281-z
Yu, H., and Chen, X. (2016). One-pot multienzyme (OPME) systems for chemoenzymatic synthesis of carbohydrates. Org. Biomol. Chem. 14 (10), 2809–2818. doi:10.1039/c6ob00058d
Zhang, J., Chen, C., Gadi, M. R., Gibbons, C., Guo, Y., Cao, X., et al. (2018). Machine-Driven enzymatic oligosaccharide synthesis by using a peptide synthesizer. Angew. Chem. Int. Ed. Engl. 57 (51), 16638–16642. doi:10.1002/anie.201810661
Keywords: digital microfluidics (DMF), oligosaccharides synthesis, oligosaccharides degradation, exoglycosidase digestion, glycosyltransferase (GT), enzymatic module
Citation: Sun Y, Wu Y, Ma D, Li J-J, Liu X, You Y, Lu J, Liu Z, Cheng X and Du Y (2023) Digital microfluidics-engaged automated enzymatic degradation and synthesis of oligosaccharides. Front. Bioeng. Biotechnol. 11:1201300. doi: 10.3389/fbioe.2023.1201300
Received: 06 April 2023; Accepted: 08 June 2023;
Published: 21 June 2023.
Edited by:
Hongxu Lu, Shanghai Institute of Ceramics, Chinese Academy of Science, ChinaReviewed by:
Yuan Liu, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences (CAS), ChinaCopyright © 2023 Sun, Wu, Ma, Li, Liu, You, Lu, Liu, Cheng and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Jun Li, ampsaUBpcGUuYWMuY24=; Xianming Liu, bGl1eGlhbm1pbmdAZGljcC5hYy5jbg==; Yuguang Du, eWdkdUBpcGUuYWMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.