- Institute of Engineering in Life Sciences, Section IV: Biomolecular Separation Engineering, Karlsruhe Institute of Technology (KIT), Karlsruhe, Germany
Non-enveloped virus-like particles (VLPs) are versatile protein nanoparticles with great potential for biopharmaceutical applications. However, conventional protein downstream processing (DSP) and platform processes are often not easily applicable due to the large size of VLPs and virus particles (VPs) in general. The application of size-selective separation techniques offers to exploit the size difference between VPs and common host-cell impurities. Moreover, size-selective separation techniques offer the potential for wide applicability across different VPs. In this work, basic principles and applications of size-selective separation techniques are reviewed to highlight their potential in DSP of VPs. Finally, specific DSP steps for non-enveloped VLPs and their subunits are reviewed as well as the potential applications and benefits of size-selective separation techniques are shown.
1 Introduction
Virus-like particles (VLPs) are multimeric structures that resemble viruses but lack the viral genome which makes them non-infectious (Chackerian, 2007). The dense and repetitive structure of antigens leads to a high immunogenicity of VLPs. The high immunogenicity in combination with their non-infectivity makes VLPs potent vaccine candidates (Chackerian, 2007). Non-enveloped VLPs consist of at least one structural protein, several of which assemble into capsids of one or more layers (Roy & Noad, 2009). Non-enveloped VLP-based vaccines licensed for human use protect against hepatitis B virus, human papillomavirus (HPV), hepatitis E virus, and one chimeric VLP-based vaccine protects against malaria (Lua et al., 2014; Nooraei et al., 2021). Another application of non-enveloped VLPs is their utilization as viral vectors for the delivery of nucleic acids, peptides, or drugs (Garcea & Gissmann, 2004; Le & Müller, 2021). In the case of enveloped VLPs, an additional lipid membrane layer is formed by the budding of capsids from the host cell (Fuenmayor et al., 2017). Enveloped VLPs can also consist of a layer of host cell lipid membrane containing viral proteins (Lua et al., 2014). The insertion of antigens or epitopes into the subunit proteins (of non-enveloped and enveloped VLPs), for example by genetic fusion or chemical conjugation, leads to chimeric VLPs (Grgacic & Anderson, 2006; Roldão et al., 2010). Chimeric VLPs were developed as prophylactic and therapeutic vaccines against several diseases including infectious diseases and cancers (Roldão et al., 2010; Mohsen et al., 2020). Illustrations and further information on the different VLP types can be found in Lua et al. (2014).
VLPs can be produced in bacteria, yeast, insect, plant, or mammalian cells (Kushnir et al., 2012; Fuenmayor et al., 2017). VLP properties influence the choice of the host cell for recombinant production of VLPs, for example, eukaryotic cells for lipid envelopes or post-translational protein modifications. Enveloped VLPs are released from cells by budding while viral proteins may also be released by cell lysis. Non-enveloped VLPs can spontaneously self-assemble in host cells (in vivo), even in prokaryotic ones such as Escherichia coli (E. coli) (Kushnir et al., 2012). Therefore, non-enveloped VLPs are often produced in microbial cells which usually requires cell lysis, such as for hepatitis B core antigen (HBcAg) in E. coli (Schumacher et al., 2018) or HPV in Saccharomyces cerevisiae (Cook et al., 1999). The necessity of a lysis step or low cell viability leads to a release of host cell impurities into the VLP-containing process solution impacting the downstream processing (DSP) (Moleirinho et al., 2020).
DSP is divided into recovery and purification in this review. The recovery of non-enveloped VLPs is similar to the one in conventional protein DSP. The purification of non-enveloped VLPs can be subdivided into two parts (Pattenden et al., 2005). The first part is the purification of the particles as a whole by removing process-related impurities, for example host cell impurities. This initial purification is referred to as VLP capture and shown as the first step in Figure 1A. It is similar to or overlapping with the purification of non-enveloped viruses and to some extent also with enveloped VLPs and viruses. These different virus-based or viral particles are referred to as virus particles (VPs) in this review. The second part of purification is more specific to non-enveloped VLPs and is optional depending on the VLP and its application. It aims to improve the particle quality since the in vivo assembly of VLPs may lead to malformed particles (Roldão et al., 2012), thus product-related impurities. The in vitro disassembly of VLPs into subunit proteins and subsequent reassembly (Figure 1A) were shown to improve VLP properties such as immunogenicity and stability (Mach et al., 2006; Zhao, et al., 2012a; Zhao, et al., 2012b). Furthermore, this process sequence enables to remove entrapped impurities (Vicente, et al., 2011a; Link et al., 2012; Strods et al., 2015; Mohsen et al., 2018). For gene therapy applications, bound nucleic acids need to be removed to ensure product safety and free binding sites for target nucleic acids (Strods et al., 2015; Petrovskis et al., 2021). Another approach is performing purification of VLP subunits in the disassembled state, followed by in vitro assembly (Liew et al., 2012; Gerstweiler et al., 2021) as shown in Figure 1B. This approach offers the possibility to use purification techniques applied in conventional protein purification without adaptions due to the particulate structure of VLPs. However, prevention of self-assembly during processing is required, for example by supplementing reducing, chelating, or chaotropic agents. The presence of these substances may impact or limit the unit operations available. Furthermore, VLPs consisting of different subunit proteins would require multiple downstream processes. This processing route also prevents exploiting the size difference between particles and impurities during purification (Morenweiser, 2005). Using size-selective separation techniques offers the possibility to process different variants of a VP or VLP subunit similarly. For example, different virus strains, chimeric VLP candidates, or therapeutic cargo within the particle could be purified using the same platform process. However, compared to monoclonal antibodies, platform processes for VPs are not well established (Moleirinho et al., 2020).
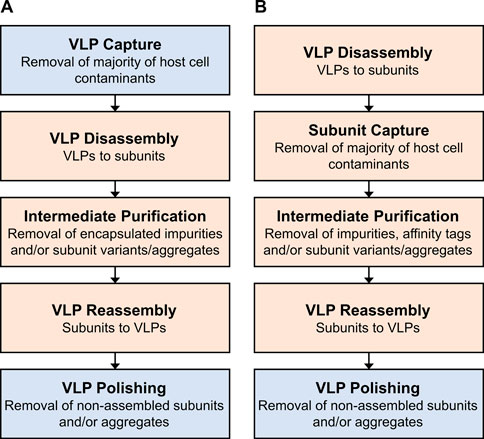
FIGURE 1. Overview of purification steps and approaches for non-enveloped VLPs. (A) Initial purification of assembled VLPs followed by dis- and reassembly. (B) Purification of VLP subunits followed by reassembly. The purification techniques presented in Section 2 and Section 3 can be applied in the process steps highlighted in blue and orange, respectively. Note, depending on the impurity profile and purity demands, process steps might require more than one unit operation.
2 Virus particles
This section focuses on the removal of process-related impurities which is usually performed early in DSP. Typical process-related impurities resulting from host cells are proteins, nucleic acids, and cell debris. Product-related impurities are aggregated VPs, fragmented VPs, or empty viral vectors (Gagnon, 2009). Co-packaged host cell deoxyribonucleic acid (DNA) and non-infectious or empty particles are considered product-related impurities in the case of viral vectors (European Medicines Agency Committee for Advanced Therapies CAT, 2018). Further considerations for product-related impurities of non-enveloped VLPs are reviewed in Section 3.
Table 1 lists recently published DSP approaches for a variety of VPs. It provides an overview of state-of-the-art strategies with the potential for DSP of non-enveloped VLP, even though not all listed VPs are non-enveloped VPs or VLPs. The capture step, at the beginning of the purification train, aims to remove most of the process-related impurities. For chromatographic separation, highly selective affinity ligands are available, for example, protein A for monoclonal antibodies. However, for VPs, the number of commercially available affinity ligands is small or non-existent requiring costly and custom-made solutions. Alternatives for VPs are separations based on size promising universal processing independent of strain, construct, or candidate. Additionally, size-selective separation is beneficial due to the large size differences between VPs and common impurities. This is also reflected in Table 1 in which the size-selective separation techniques filtration, steric exclusion chromatography (SXC), size-exclusion chromatography (SEC), multimodal SEC (mmSEC), and precipitation are frequently used. These techniques are highlighted in the following paragraphs.
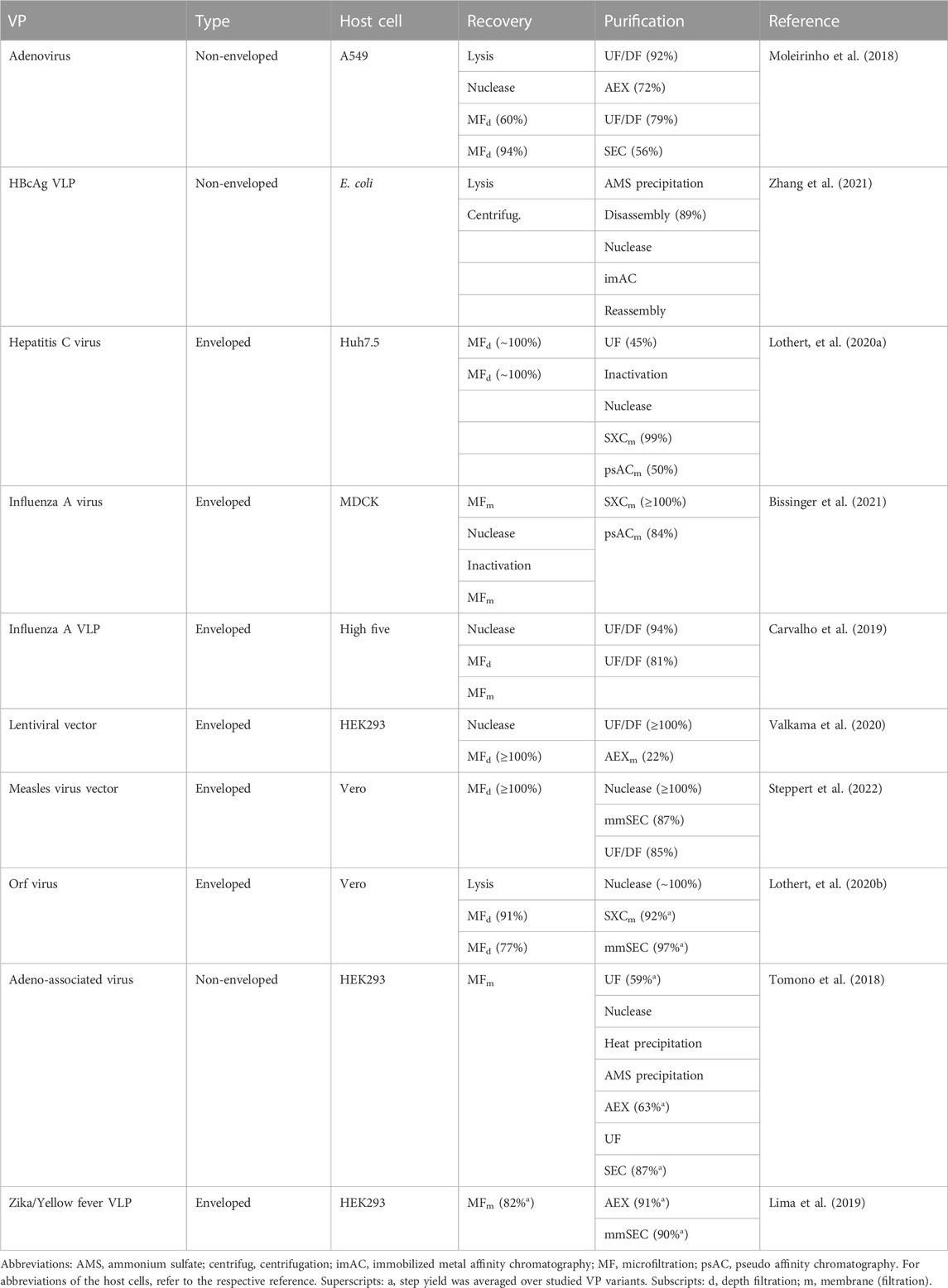
TABLE 1. Examples of recently published downstream processes (recovery and purification with step yield in parentheses when available) for VPs. If the recovery steps do not contain a lysis step, culture supernatants were processed.
Filtration, such as (cross-flow) microfiltration and cross-flow ultrafiltration/diafiltration (UF/DF), is commonly used during DSP of VPs as indicated by Wolff & Reichl (2011) and in Table 1. Microfiltration, mostly in normal-flow mode, is used for clarification during recovery whereas both cross-flow mirco- and UF/DF are used during purification. VPs are either retained by the membrane and smaller impurities (for example host cell proteins and DNA) are depleted or larger particles (for example cell debris or aggregates) are retained and VPs permeate through the membrane (Wickramasinghe et al., 2005; Grzenia et al., 2008). Cross-flow UF/DF has several advantages compared to other commonly used purification techniques for VPs. The small pore size of UF membranes makes them an alternative to ultracentrifugation (Reiser, 2000; Peixoto et al., 2007). Compared to UF/DF, ultracentrifugation, both density gradient and continuous, is generally considered to be poorly scalable, costly, time-consuming, and requires an additional buffer exchange in case of density gradients (Morenweiser, 2005; Vicente, et al., 2011b; Wolff & Reichl, 2011; Ladd Effio & Hubbuch, 2015; Nestola et al., 2015; Moleirinho et al., 2020). Carvalho et al. (2019) showed that the purification of Influenza A VLPs using two UF/DF steps is more efficient in terms of process duration and buffer consumption when compared to ion exchange chromatography and SEC, however with the drawback of low DNA removal. The use of UF/DF during purification offers potential integration of a formulation step which can also be performed by UF/DF (Liew et al., 2012; Wagner et al., 2014; Carvalho et al., 2019; Valkama et al., 2020; Moreira et al., 2021).
SEC separates molecules according to their ability to penetrate the pores of porous resin beads within a column. Therefore, larger particles are excluded from either all or some of the pores depending on the pore and particle size. With decreasing size, solutes diffuse into an increasing fraction of the pores leading to a longer residence time in the column. SEC thus has a high selectivity for the separation of VPs and host cell impurities, and is mostly independent of the liquid phase conditions (Gagnon, 2009). SEC with pore sizes that exclude VPs enables their elution in the void volume of the column while impurities elute later. This enables higher flow rates, shorter columns, and higher loads, reducing some of the drawbacks in conventional protein SEC (Gagnon, 2009). However, SEC leads to dilution and is limited by restricted loading volumes, especially for separation problems with similar size magnitudes, such as VPs and their aggregates. SEC thus benefits from reduced process volumes and is therefore often performed toward the end of purification as shown in Table 1 and other studies (Peixoto et al., 2007; Rodrigues et al., 2007; Tomono et al., 2016). Similarly to UF/DF mentioned above, the polishing of VPs by SEC is often performed in combination with a formulation step (Peixoto et al., 2007; Dormond et al., 2010; Merten et al., 2011; Moleirinho et al., 2018).
Next to SEC as a conventional chromatographic technique for VP purification (Gagnon, 2009), modern mmSEC (commercially available as Capto Core 400/700 by Cytiva) is increasingly applied for purification in recent publications [Table 1 and (Weigel et al., 2014; Reiter et al., 2019)]. mmSEC is based on the core-shell bead technology where the core with multimodal ligands is surrounded by an inert size-restricting shell. This technology enables the binding of smaller impurities and recovery VPs in the flow-through. The drawback of mmSEC is that the required capacity is determined by the impurity content, not the target species content. This property makes it more attractive in later process stages with a lower impurity burden, for example during polishing.
Another chromatographic technique that is beneficial for the purification of VPs and larger proteins is SXC (J. Lee et al., 2012). Here, the addition of polyethylene glycol (PEG) to the VP solution increases the free energy of the system. The increase in free energy is attributed to the steric exclusion of PEG molecules from the VP surface and other surfaces leading to an energetically unfavorable discontinuity in PEG concentration. This excluded volume effect is highly correlated to the VP or protein hydrodynamic radius but is also affected by other solute and solvent properties (J. Lee et al., 2012). The association of VPs with each other and hydrophilic surfaces reduces the free energy, which is thermodynamically more favorable. Using monolith columns (J. Lee et al., 2012) or membranes (Marichal-Gallardo et al., 2017) as stationary phases allows for the binding and elution of VPs by increasing and decreasing the PEG concentration in the mobile phase, respectively. In addition to the examples in Table 1, SXC is applied as a capture step for adeno-associated viruses achieving a high recovery (Marichal-Gallardo et al., 2021). The advantages of SXC are binding and elution under practically any conditions only requiring the addition and removal of PEG, respectively, as well as low costs and good scalability using membranes (Marichal-Gallardo et al., 2017).
Purification of VPs using PEG precipitation underlies similar principles as SXC (J. Lee et al., 2012). As for SXC, larger proteins or particles tend to precipitate earlier than smaller species at identical conditions (Rothstein, 1994). Besides using non-ionic polymers such as PEG, protein precipitation is performed by increasing the salt concentration (salting-out), for example using ammonium sulfate as a precipitant (Wingfield, 1998). Salting-out is dominated by a preferential exclusion of the precipitant from the hydrated protein surface leading to self-association and precipitation (Timasheff & Arakawa, 1988; Wingfield, 1998). Precipitation is usually applied for capturing at the beginning of purification. Examples are the precipitation of norovirus VLPs using PEG (Koho et al., 2012) or precipitation of different HBcAg VLP candidates (Schumacher et al., 2018; Zhang et al., 2021) and adeno-associated virus strains using ammonium sulfate (Tomono et al., 2016; 2018). For the screening of VLP precipitation conditions, advanced analytical and predictive tools were recently developed (Vormittag et al., 2020; Wegner & Hubbuch, 2022). Integration of precipitation, wash, and re-dissolution with cross-flow filtration led to synergies improving process performance (Hillebrandt et al., 2020).
2.1 Discussion of non-size-selective separation techniques
A benchmark for host cell DNA of continuous cell lines according to the European Pharmacopoeia is a reduction to a maximum of 10 ng per vaccine dose to minimize tumorigenic potential (Council of Europe, 2017). The reduction of host cell nucleic acids in DSP of VPs is often performed enzymatically by nucleases at the end of upstream processing or early in DSP. Nuclease application is present in almost all processes listed in Table 1 and in older downstream processes such as for HPV VLPs (Cook et al., 1999). However, nucleases require specific conditions for optimal activity and thus temperature, pH, or ion composition may have to be adapted. Especially for high nucleic acid burden after cell lysis, efficient processing may require a trade-off between incubation time and product loss due to instability at the chosen conditions (Kawka et al., 2021). However, also short processing within 30 min, at room temperature, and without product loss was demonstrated (Lothert, et al., 2020a). Nevertheless, even for long incubation times, nucleic acid removal was observed to be incomplete (Weigel et al., 2014; Lothert, et al., 2020b; Kawka et al., 2021). At a large production scale, the use of nuclease and required conditioning steps may considerably increase production costs. Nuclease removal during further DSP is another important consideration for patient safety. For gene therapy applications, the removal of nucleases is a crucial step as it prevents the digestion of target nucleic acids.
Apart from size-selective separations, heparin and sulfated cellulose are applied as adsorbers for (pseudo) affinity chromatography. Both have a similar molecular structure and were shown to preferentially bind certain VPs (Opitz et al., 2007; 2009). Heparin ligands were applied for the separation of human immunodeficiency virus-1 gag VLPs and extracellular vesicles (Reiter et al., 2019) as well as HPV VLP purification (Kim et al., 2010). Sulfated cellulose membrane adsorbers were used for pseudo affinity capture of influenza virus (Opitz et al., 2009; Weigel et al., 2016) and VLPs (Carvalho et al., 2018) as well as for polishing of hepatitis C and influenza virus (Table 1). Further chromatography types are also used where anion exchange chromatography (AEX) is often applied for purification of enveloped (Table 1) and non-enveloped VPs such as adenovirus (Kawka et al., 2021), norovirus VLPs (Koho et al., 2012), and HPV VLPs (Cook et al., 1999). A potential disadvantage of ion exchange chromatography is the elution at a high ionic strength or a different pH, which might lead to VP aggregation or instability and thus requires optimization for each new strain or candidate. Overall, bead-based bind-elute chromatography of VPs, such as ion-exchange or affinity, often suffers from low dynamic binding capacities due to size-exclusion and low diffusion coefficients of VPs (Gagnon, 2009). These disadvantages can be mitigated using stationary phases for which convective mass transfer of VPs to the ligands is dominant, for example membranes or monoliths (Gagnon, 2009).
3 Non-enveloped VLPs
As described above, non-enveloped VLPs can be produced as subunits, purified, and assembled (Figure 1B) or first purified as a whole, optionally disassembled into subunits, and subsequently reassembled (Figure 1A). With regards to the latter approach, Section 2 elaborates on the purification of VPs as a whole which enables to exploit their size for purification. This section reviews subsequent purification steps including VLP dis- and reassembly which are specific for non-enveloped VLP and their subunits. For each process step, one or more size-selective purification approaches are presented.
The disassembly of non-enveloped VLPs can be achieved by changing the liquid phase conditions, for example by the addition of dithiothreitol in combination with a pH increase for HPV VLPs (Mach et al., 2006), the addition of urea or guanidine hydrochloride at low ionic strength for HBcAg VLPs (K. W. Lee & Tan, 2008; Singh & Zlotnick, 2003), or a pH increase at low ionic strength for Norwalk VLPs (Ausar et al., 2006). Liquid phase conditions for disassembly at a laboratory scale are achieved by the direct addition of substances (Zlotnick et al., 1996), by the addition of stock solutions (Hillebrandt et al., 2021; K. W; Lee & Tan, 2008; Valentic et al., 2022), or by dialysis (Ausar et al., 2006; Porterfield et al., 2010; Holmes et al., 2015; Strods et al., 2015). At a larger scale, DF was shown to achieve efficient disassembly processing and purification while avoiding target protein loss due to local urea concentration or pH peaks (Hillebrandt et al., 2021). Another option is pelleting the target protein in form of inclusion bodies (Suffian et al., 2017) or by precipitation (Zhang et al., 2021) with subsequent re-solubilization at disassembly conditions. However, pelleting the target protein by centrifugation was shown to entrap impurities and leads to longer re-solubilization times due to floc compaction when compared to DF-based processing (Hammerschmidt et al., 2016; Hillebrandt et al., 2020).
At a laboratory scale, the purification of the disassembled VLP subunits is often performed by SEC (Strods et al., 2015; Schumacher et al., 2018) or poly-histidine tags with corresponding affinity chromatography (Middelberg et al., 2011; Zhang et al., 2021). However, SEC has limited scalability and affinity tags may lead to undesired alterations in the protein structure or require subsequent removal of the tag. Alternatively, disassembled VLPs were separated from nucleic acids by cross-flow DF and from higher molecular weight species by dead-end UF (Hillebrandt et al., 2021). Other (chromatographic) separations, for example ion exchange, are also conceivable, when compatible with disassembly agents as mentioned above. Strods et al. (2015) suggested and applied alkaline hydrolysis of nucleic acid impurities which, however, requires sufficient stability of the subunit proteins.
Reassembly is usually initiated by reversing the disassembly conditions, hence by increasing ionic strength, decreasing pH to the neutral range, or removing chaotropic or reducing agents (Wingfield et al., 1995; Mach et al., 2006; Ren et al., 2006; Porterfield et al., 2010; Zhang et al., 2021). For gene therapy applications, reassembly can be induced by the addition of nucleic acids which are subsequently encapsulated similarly as in natural virus assembly (Porterfield et al., 2010; Strods et al., 2015; Petrovskis et al., 2021). It is worth mentioning, that different molecular variants of a VLP show different dis- and (re-) assembly behavior, which also affects process development (Rüdt et al., 2019; Hillebrandt et al., 2021; Valentic et al., 2022). VLP dis- and reassembly were investigated in the frame of DF providing a scalable process and the opportunity to improve VLP yield by controlled buffer exchange (Liew et al., 2012; Hillebrandt et al., 2021). Furthermore, process monitoring approaches were developed to monitor product and process properties during DF-based VLP dis- and reassembly (Rüdt et al., 2019; Hillebrandt et al., 2022).
4 Conclusion
This review summarizes the fundamentals and applications of several different size-selective separation techniques for the DSP of VPs. Recently published processing routes and advantages of size-selective separation for VPs and non-enveloped VLPs are highlighted. In the future, the presented size-selective approaches can serve as building blocks for platform downstream processes. A combination of different size-selective techniques may lead to synergistic effects in process performance while promising wide applicability.
Author contributions
NH initiated the work, evolved the concepts and structure of the manuscript, performed the literature review and selection, created the table and figure, and drafted the manuscript. JH supervised the work and reviewed the final manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors would like to thank Annabelle Dietrich and Philipp Vormittag for reviewing the manuscript. We acknowledge support by the KIT-Publication Fund of the Karlsruhe Institute of Technology.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AEX, anion exchange chromatography; DF, diafiltration; DNA, deoxyribonucleic acid; DSP, downstream processing; E. coli, Escherichia coli; HBcAg, hepatitis B core antigen; HPV, human papillomavirus; mmSEC, multimodal size-exclusion chromatography; PEG, polyethylene glycol; SEC, size-exclusion chromatography; SXC. steric exclusion chromatography; UF, ultrafiltration; VLP, virus-like particle; VP, virus particle.
References
Ausar, S. F., Foubert, T. R., Hudson, M. H., Vedvick, T. S., and Middaugh, C. R. (2006). Conformational stability and disassembly of Norwalk virus-like particles. J. Biol. Chem. 281 (28), 19478–19488. doi:10.1074/jbc.M603313200
Bissinger, T., Wu, Y., Marichal-Gallardo, P., Riedel, D., Liu, X., Genzel, Y., et al. (2021). Towards integrated production of an influenza A vaccine candidate with MDCK suspension cells. Biotechnol. Bioeng. 118 (10), 3996–4013. doi:10.1002/bit.27876
Carvalho, S. B., Fortuna, A. R., Wolff, M. W., Peixoto, C., M Alves, P., Reichl, U., et al. (2018). Purification of influenza virus-like particles using sulfated cellulose membrane adsorbers. J. Chem. Technol. Biotechnol. 93 (7), 1988–1996. doi:10.1002/jctb.5474
Carvalho, S. B., Silva, R. J. S., Moleirinho, M. G., Cunha, B., Moreira, A. S., Xenopoulos, A., et al. (2019). Membrane-based approach for the downstream processing of influenza virus-like particles. Biotechnol. J. 14 (8), 1800570. doi:10.1002/biot.201800570
Chackerian, B. (2007). Virus-like particles: Flexible platforms for vaccine development. Expert Rev. Vaccines 6 (3), 381–390. doi:10.1586/14760584.6.3.381
Cook, J. C., Joyce, J. G., George, H. A., Schultz, L. D., Hurni, W. M., Ja323nsen, K. U., et al. (1999). Purification of virus-like particles of recombinant human papillomavirus type 11 major capsid protein L1 from Saccharomyces cerevisiae. Protein Expr. Purif. 17 (3), 477–484. doi:10.1006/prep.1999.1155
Council of Europe (2017). “Cell substrates for the production of vaccines for human use,” in European Pharmacopoeia. 9th ed. Strasbourg, France: Council of Europe, 602–606.
Dormond, E., Chahal, P., Bernier, A., Tran, R., Perrier, M., and Kamen, A. (2010). An efficient process for the purification of helper-dependent adenoviral vector and removal of helper virus by iodixanol ultracentrifugation. J. Virological Methods 165 (1), 83–89. doi:10.1016/j.jviromet.2010.01.008
European Medicines Agency Committee for Advanced Therapies (CAT) (2018). Guideline on the quality, non-clinical and clinical aspects of gene therapy medicinal products. Eur. Med. Agency Guidel. 44, 1–41.
Fuenmayor, J., Gòdia, F., and Cervera, L. (2017). Production of virus-like particles for vaccines. New Biotechnol. 39, 174–180. doi:10.1016/j.nbt.2017.07.010
Gagnon, P. (2009). “Chromatographic purification of virus particles,” in Encyclopedia of industrial Biotechnology. Editor M. C. Flickinger (New Jersey, United States: John Wiley & Sons, Inc), 1–21. doi:10.1002/9780470054581.eib583
Garcea, R. L., and Gissmann, L. (2004). Virus-like particles as vaccines and vessels for the delivery of small molecules. Curr. Opin. Biotechnol. 15 (6), 513–517. doi:10.1016/j.copbio.2004.10.002
Gerstweiler, L., Billakanti, J., Bi, J., and Middelberg, A. (2021). Comparative evaluation of integrated purification pathways for bacterial modular polyomavirus major capsid protein VP1 to produce virus-like particles using high throughput process technologies. J. Chromatogr. A 1639, 461924. doi:10.1016/j.chroma.2021.461924
Grgacic, E. V. L., and Anderson, D. A. (2006). Virus-like particles: Passport to immune recognition. Methods 40 (1), 60–65. doi:10.1016/j.ymeth.2006.07.018
Grzenia, D. L., Carlson, J. O., and Wickramasinghe, S. R. (2008). Tangential flow filtration for virus purification. J. Membr. Sci. 321 (2), 373–380. doi:10.1016/j.memsci.2008.05.020
Hammerschmidt, N., Hobiger, S., and Jungbauer, A. (2016). Continuous polyethylene glycol precipitation of recombinant antibodies: Sequential precipitation and resolubilization. Process Biochem. 51 (2), 325–332. doi:10.1016/j.procbio.2015.11.032
Hillebrandt, N., Vormittag, P., Bluthardt, N., Dietrich, A., and Hubbuch, J. (2020). Integrated process for capture and purification of virus-like particles: Enhancing process performance by cross-flow filtration. Front. Bioeng. Biotechnol. 8, 489. doi:10.3389/fbioe.2020.00489
Hillebrandt, N., Vormittag, P., Dietrich, A., and Hubbuch, J. (2022). Process monitoring framework for cross-flow diafiltration-based virus-like particle disassembly: Tracing product properties and filtration performance. Biotechnol. Bioeng. 119 (6), 1522–1538. doi:10.1002/bit.28063
Hillebrandt, N., Vormittag, P., Dietrich, A., Wegner, C. H., and Hubbuch, J. (2021). Process development for cross-flow diafiltration-based VLP disassembly: A novel high-throughput screening approach. Biotechnol. Bioeng. 118 (10), 3926–3940. doi:10.1002/bit.27868
Holmes, K., Shepherd, D. A., Ashcroft, A. E., Whelan, M., Rowlands, D. J., and Stonehouse, N. J. (2015). Assembly pathway of Hepatitis B core virus-like particles from genetically fused dimers. J. Biol. Chem. 290 (26), 16238–16245. doi:10.1074/jbc.M114.622035
Kawka, K., Wilton, A. N., Madadkar, P., Medina, M. F. C., Lichty, B. D., Ghosh, R., et al. (2021). Integrated development of enzymatic DNA digestion and membrane chromatography processes for the purification of therapeutic adenoviruses. Sep. Purif. Technol. 254, 117503. doi:10.1016/j.seppur.2020.117503
Kim, H. J., Kim, S. Y., Lim, S. J., Kim, J. Y., Lee, S. J., and Kim, H.-J. (2010). One-step chromatographic purification of human papillomavirus type 16 L1 protein from Saccharomyces cerevisiae. Protein Expr. Purif. 70 (1), 68–74. doi:10.1016/j.pep.2009.08.005
Koho, T., Mäntylä, T., Laurinmäki, P., Huhti, L., Butcher, S. J., Vesikari, T., et al. (2012). Purification of norovirus-like particles (VLPs) by ion exchange chromatography. J. Virological Methods 181 (1), 6–11. doi:10.1016/j.jviromet.2012.01.003
Kushnir, N., Streatfield, S. J., and Yusibov, V. (2012). Virus-like particles as a highly efficient vaccine platform: Diversity of targets and production systems and advances in clinical development. Vaccine 31 (1), 58–83. doi:10.1016/j.vaccine.2012.10.083
Ladd Effio, C., and Hubbuch, J. (2015). Next generation vaccines and vectors: Designing downstream processes for recombinant protein-based virus-like particles. Biotechnol. J. 10 (5), 715–727. doi:10.1002/biot.201400392
Le, D. T., and Müller, K. M. (2021). In vitro assembly of virus-like particles and their applications. Life 11 (4), 334. doi:10.3390/life11040334
Lee, J., Gan, H. T., Latiff, S. M. A., Chuah, C., Lee, W. Y., Yang, Y. S., et al. (2012). Principles and applications of steric exclusion chromatography. J. Chromatogr. A 1270, 162–170. doi:10.1016/j.chroma.2012.10.062
Lee, K. W., and Tan, W. S. (2008). Recombinant Hepatitis B virus core particles: Association, dissociation and encapsidation of green fluorescent protein. J. Virological Methods 151 (2), 172–180. doi:10.1016/j.jviromet.2008.05.025
Liew, M. W. O., Chuan, Y. P., and Middelberg, A. P. J. (2012). Reactive diafiltration for assembly and formulation of virus-like particles. Biochem. Eng. J. 68, 120–128. doi:10.1016/j.bej.2012.07.009
Lima, T. M., Souza, M. O., and Castilho, L. R. (2019). Purification of flavivirus VLPs by a two-step chomatographic process. Vaccine 37 (47), 7061–7069. doi:10.1016/j.vaccine.2019.05.066
Link, A., Zabel, F., Schnetzler, Y., Titz, A., Brombacher, F., and Bachmann, M. F. (2012). Innate immunity mediates follicular transport of particulate but not soluble protein antigen. J. Immunol. 188 (8), 3724–3733. doi:10.4049/jimmunol.1103312
Lothert, K., Offersgaard, A. F., Pihl, A. F., Mathiesen, C. K., Jensen, T. B., Alzua, G. P., et al. (2020a). Development of a downstream process for the production of an inactivated whole hepatitis C virus vaccine. Sci. Rep. 10 (1), 16261. doi:10.1038/s41598-020-72328-5
Lothert, K., Pagallies, F., Eilts, F., Sivanesapillai, A., Hardt, M., Moebus, A., et al. (2020b). A scalable downstream process for the purification of the cell culture-derived Orf virus for human or veterinary applications. J. Biotechnol. 323, 221–230. doi:10.1016/j.jbiotec.2020.08.014
Lua, L. H. L., Connors, N. K., Sainsbury, F., Chuan, Y. P., Wibowo, N., and Middelberg, A. P. J. (2014). Bioengineering virus-like particles as vaccines. Biotechnol. Bioeng. 111 (3), 425–440. doi:10.1002/bit.25159
Mach, H., Volkin, D. B., Troutman, R. D., Wang, B., Luo, Z., Jansen, K. U., et al. (2006). Disassembly and reassembly of yeast-derived recombinant human papillomavirus virus-like particles (HPV VLPs). J. Pharm. Sci. 95 (10), 2195–2206. doi:10.1002/jps.20696
Marichal-Gallardo, P., Börner, K., Pieler, M. M., Sonntag-Buck, V., Obr, M., Bejarano, D., et al. (2021). Single-use capture purification of adeno-associated viral gene transfer vectors by membrane-based steric exclusion chromatography. Hum. Gene Ther. 32 (17–18), 959–974. doi:10.1089/hum.2019.284
Marichal-Gallardo, P., Pieler, M. M., Wolff, M. W., and Reichl, U. (2017). Steric exclusion chromatography for purification of cell culture-derived influenza A virus using regenerated cellulose membranes and polyethylene glycol. J. Chromatogr. A 1483, 110–119. doi:10.1016/j.chroma.2016.12.076
Merten, O.-W., Charrier, S., Laroudie, N., Fauchille, S., Dugué, C., Jenny, C., et al. (2011). Large-scale manufacture and characterization of a lentiviral vector produced for clinical ex vivo gene therapy application. Hum. Gene Ther. 22 (3), 343–356. doi:10.1089/hum.2010.060
Middelberg, A. P. J., Rivera-Hernandez, T., Wibowo, N., Lua, L. H. L., Fan, Y., Magor, G., et al. (2011). A microbial platform for rapid and low-cost virus-like particle and capsomere vaccines. Vaccine 29 (41), 7154–7162. doi:10.1016/j.vaccine.2011.05.075
Mohsen, M. O., Gomes, A., Vogel, M., and Bachmann, M. (2018). Interaction of viral capsid-derived virus-like particles (VLPs) with the innate immune system. Vaccines 6 (3), 37. doi:10.3390/vaccines6030037
Mohsen, M. O., Speiser, D. E., Knuth, A., and Bachmann, M. F. (2020). Virus-like particles for vaccination against cancer. Wiley Interdiscip. Rev. Nanomedicine Nanobiotechnology 12 (1), 15799–e1617. doi:10.1002/wnan.1579
Moleirinho, M. G., Rosa, S., Carrondo, M. J. T., Silva, R. J. S., Hagner-McWhirter, Å., Ahlén, G., et al. (2018). Clinical-grade oncolytic adenovirus purification using polysorbate 20 as an alternative for cell lysis. Curr. Gene Ther. 18 (6), 366–374. doi:10.2174/1566523218666181109141257
Moleirinho, M. G., Silva, R. J. S., Alves, P. M., Carrondo, M. J. T., and Peixoto, C. (2020). Current challenges in biotherapeutic particles manufacturing. Expert Opin. Biol. Ther. 20 (5), 451–465. doi:10.1080/14712598.2020.1693541
Moreira, A. S., Faria, T. Q., Oliveira, J. G., Kavara, A., Schofield, M., Sanderson, T., et al. (2021). Enhancing the purification of Lentiviral vectors for clinical applications. Sep. Purif. Technol. 274, 118598. doi:10.1016/j.seppur.2021.118598
Morenweiser, R. (2005). Downstream processing of viral vectors and vaccines. Gene Ther. 12 (S1), S103–S110. doi:10.1038/sj.gt.3302624
Nestola, P., Peixoto, C., Silva, R. R. J. S., Alves, P. M., Mota, J. P. B., and Carrondo, M. J. T. (2015). Improved virus purification processes for vaccines and gene therapy. Biotechnol. Bioeng. 112 (5), 843–857. doi:10.1002/bit.25545
Nooraei, S., Bahrulolum, H., Hoseini, Z. S., Katalani, C., Hajizade, A., Easton, A. J., et al. (2021). Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnology 19 (1), 59. doi:10.1186/s12951-021-00806-7
Opitz, L., Lehmann, S., Reichl, U., and Wolff, M. W. (2009). Sulfated membrane adsorbers for economic pseudo-affinity capture of influenza virus particles. Biotechnol. Bioeng. 103 (6), 1144–1154. doi:10.1002/bit.22345
Opitz, L., Lehmann, S., Zimmermann, A., Reichl, U., and Wolff, M. W. (2007). Impact of adsorbents selection on capture efficiency of cell culture derived human influenza viruses. J. Biotechnol. 131 (3), 309–317. doi:10.1016/j.jbiotec.2007.07.723
Pattenden, L. K., Middelberg, A. P. J., Niebert, M., and Lipin, D. I. (2005). Towards the preparative and large-scale precision manufacture of virus-like particles. Trends Biotechnol. 23 (10), 523–529. doi:10.1016/j.tibtech.2005.07.011
Peixoto, C., Sousa, M. F. Q., Silva, A. C., Carrondo, M. J. T., and Alves, P. M. (2007). Downstream processing of triple layered rotavirus like particles. J. Biotechnol. 127 (3), 452–461. doi:10.1016/j.jbiotec.2006.08.002
Petrovskis, I., Lieknina, I., Dislers, A., Jansons, J., Bogans, J., Akopjana, I., et al. (2021). Production of the HBc protein from different HBV genotypes in E. coli. Use of reassociated HBc VLPs for packaging of ss- and dsRNA. Microorganisms 9 (2), 283. doi:10.3390/microorganisms9020283
Porterfield, J. Z., Dhason, M. S., Loeb, D. D., Nassal, M., Stray, S. J., and Zlotnick, A. (2010). Full-length hepatitis B virus core protein packages viral and heterologous RNA with similarly high levels of cooperativity. J. Virology 84 (14), 7174–7184. doi:10.1128/JVI.00586-10
Reiser, J. (2000). Production and concentration of pseudotyped HIV-1-based gene transfer vectors. Gene Ther. 7 (11), 910–913. doi:10.1038/sj.gt.3301188
Reiter, K., Aguilar, P. P., Wetter, V., Steppert, P., Tover, A., and Jungbauer, A. (2019). Separation of virus-like particles and extracellular vesicles by flow-through and heparin affinity chromatography. J. Chromatogr. A 1588, 77–84. doi:10.1016/j.chroma.2018.12.035
Ren, Y., Wong, S.-M., and Lim, L.-Y. (2006). In vitro-reassembled plant virus-like particles for loading of polyacids. J. General Virology 87 (9), 2749–2754. doi:10.1099/vir.0.81944-0
Rodrigues, T., Carvalho, A., Carmo, M., Carrondo, M. J. T., Alves, P. M., and Cruz, P. E. (2007). Scaleable purification process for gene therapy retroviral vectors. J. Gene Med. 9 (4), 233–243. doi:10.1002/jgm.1021
Roldão, A., Mellado, M. C. M., Castilho, L. R., Carrondo, M. J. T., and Alves, P. M. (2010). Virus-like particles in vaccine development. Expert Rev. Vaccines 9 (10), 1149–1176. doi:10.1586/erv.10.115
Roldão, A., Mellado, M. C. M., Lima, J. C., Carrondo, M. J. T., Alves, P. M., and Oliveira, R. (2012). On the effect of thermodynamic equilibrium on the assembly efficiency of complex multi-layered virus-like particles (VLP): The case of rotavirus VLP. PLoS Comput. Biol. 8 (2), e1002367. doi:10.1371/journal.pcbi.1002367
Rothstein, F. (1994). “Differential precipitation of proteins,” in Protein purification process engineering. Editor R. G. Harrison (New York City: Marcel Dekker), 18, 115–208.
Roy, P., and Noad, R. (2009). Virus-like particles as a vaccine delivery system: Myths and facts. Adv. Exp. Med. Biol. 655, 145–158. doi:10.1007/978-1-4419-1132-2_11
Rüdt, M., Vormittag, P., Hillebrandt, N., and Hubbuch, J. (2019). Process monitoring of virus-like particle reassembly by diafiltration with UV/Vis spectroscopy and light scattering. Biotechnol. Bioeng. 116 (6), 1366–1379. doi:10.1002/bit.26935
Schumacher, J., Bacic, T., Staritzbichler, R., Daneschdar, M., Klamp, T., Arnold, P., et al. (2018). Enhanced stability of a chimeric Hepatitis B core antigen virus-like-particle (HBcAg-VLP) by a C-terminal linker-hexahistidine-peptide. J. Nanobiotechnology 16 (1), 39. doi:10.1186/s12951-018-0363-0
Singh, S., and Zlotnick, A. (2003). Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. J. Biol. Chem. 278 (20), 18249–18255. doi:10.1074/jbc.M211408200
Steppert, P., Mosor, M., Stanek, L., Burgstaller, D., Palmberger, D., Preinsperger, S., et al. (2022). A scalable, integrated downstream process for production of a recombinant measles virus-vectored vaccine. Vaccine 40 (9), 1323–1333. doi:10.1016/j.vaccine.2022.01.004
Strods, A., Ose, V., Bogans, J., Cielens, I., Kalnins, G., Radovica, I., et al. (2015). Preparation by alkaline treatment and detailed characterisation of empty Hepatitis B virus core particles for vaccine and gene therapy applications. Sci. Rep. 5 (1), 11639. doi:10.1038/srep11639
Suffian, I. F. B. M., Garcia-Maya, M., Brown, P., Bui, T., Nishimura, Y., Palermo, A. R. B. M. J., et al. (2017). Yield optimisation of hepatitis B virus core particles in E. coli expression system for drug delivery applications. Sci. Rep. 7 (1), 43160. doi:10.1038/srep43160
Timasheff, S. N., and Arakawa, T. (1988). Mechanism of protein precipitation and stabilization by co-solvents. J. Cryst. Growth 90 (1–3), 39–46. doi:10.1016/0022-0248(88)90296-5
Tomono, T., Hirai, Y., Okada, H., Adachi, K., Ishii, A., Shimada, T., et al. (2016). Ultracentrifugation-free chromatography-mediated large-scale purification of recombinant adeno-associated virus serotype 1 (rAAV1). Mol. Ther. - Methods & Clin. Dev. 3, 15058. doi:10.1038/mtm.2015.58
Tomono, T., Hirai, Y., Okada, H., Miyagawa, Y., Adachi, K., Sakamoto, S., et al. (2018). Highly efficient ultracentrifugation-free chromatographic purification of recombinant AAV serotype 9. Mol. Ther. - Methods & Clin. Dev. 11, 180–190. doi:10.1016/j.omtm.2018.10.015
Valentic, A., Müller, J., and Hubbuch, J. (2022). Effects of different lengths of a nucleic acid binding region and bound nucleic acids on the phase behavior and purification process of HBcAg virus-like particles. Front. Bioeng. Biotechnol. 10, 929243–929311. doi:10.3389/fbioe.2022.929243
Valkama, A. J., Oruetxebarria, I., Lipponen, E. M., Leinonen, H. M., Käyhty, P., Hynynen, H., et al. (2020). Development of large-scale downstream processing for lentiviral vectors. Mol. Ther. - Methods & Clin. Dev. 17, 717–730. doi:10.1016/j.omtm.2020.03.025
Vicente, T., Mota, J. P. B., Peixoto, C., Alves, P. M., and Carrondo, M. J. T. (2011a). Rational design and optimization of downstream processes of virus particles for biopharmaceutical applications: Current advances. Biotechnol. Adv. 29 (6), 869–878. doi:10.1016/j.biotechadv.2011.07.004
Vicente, T., Roldão, A., Peixoto, C., Carrondo, M. J. T., and Alves, P. M. (2011b). Large-scale production and purification of VLP-based vaccines. J. Invertebr. Pathology 107, S42–S48. doi:10.1016/j.jip.2011.05.004
Vormittag, P., Klamp, T., and Hubbuch, J. (2020). Optimization of a soft ensemble vote classifier for the prediction of chimeric virus-like particle solubility and other biophysical properties. Front. Bioeng. Biotechnol. 8, 881–917. doi:10.3389/fbioe.2020.00881
Wagner, J. M., Pajerowski, J. D., Daniels, C. L., McHugh, P. M., Flynn, J. A., Balliet, J. W., et al. (2014). Enhanced production of chikungunya virus-like particles using a high-pH adapted spodoptera frugiperda insect cell line. PLoS ONE 9 (4), e94401. doi:10.1371/journal.pone.0094401
Wegner, C. H., and Hubbuch, J. (2022). Calibration-free PAT: Locating selective crystallization or precipitation sweet spot in screenings with multi-way PARAFAC models. Front. Bioeng. Biotechnol. 10, 1051129–1051218. doi:10.3389/fbioe.2022.1051129
Weigel, T., Solomaier, T., Peuker, A., Pathapati, T., Wolff, M. W., and Reichl, U. (2014). A flow-through chromatography process for influenza A and B virus purification. J. Virological Methods 207, 45–53. doi:10.1016/j.jviromet.2014.06.019
Weigel, T., Solomaier, T., Wehmeyer, S., Peuker, A., Wolff, M. W., and Reichl, U. (2016). A membrane-based purification process for cell culture-derived influenza A virus. J. Biotechnol. 220, 12–20. doi:10.1016/j.jbiotec.2015.12.022
Wickramasinghe, S. R., Kalbfuß, B., Zimmermann, A., Thom, V., and Reichl, U. (2005). Tangential flow microfiltration and ultrafiltration for human influenza A virus concentration and purification. Biotechnol. Bioeng. 92 (2), 199–208. doi:10.1002/bit.20599
Wingfield, P. T. (1998). “Protein precipitation using ammonium sulfate,” in Current protocols in protein science (New Jersey, United States: John Wiley & Sons, Inc), 3. doi:10.1002/0471140864.psa03fs13
Wingfield, P. T., Stahl, S. J., Williams, R. W., and Steven, A. C. (1995). Hepatitis core antigen produced in Escherichia coli: Subunit composition, conformation analysis, and in vitro capsid assembly. Biochemistry 34 (15), 4919–4932. doi:10.1021/bi00015a003
Wolff, M. W., and Reichl, U. (2011). Downstream processing of cell culture-derived virus particles. Expert Rev. Vaccines 10 (10), 1451–1475. doi:10.1586/erv.11.111
Zhang, Y., Liu, Y., Zhang, B., Yin, S., Li, X., Zhao, D., et al. (2021). In vitro preparation of uniform and nucleic acid free Hepatitis B core particles through an optimized disassembly-purification-reassembly process. Protein Expr. Purif. 178, 105747. doi:10.1016/j.pep.2020.105747
Zhao, Q., Allen, M. J., Wang, Y., Wang, B., Wang, N., Shi, L., et al. (2012a). Disassembly and reassembly improves morphology and thermal stability of human papillomavirus type 16 virus-like particles. Nanomedicine Nanotechnol. Biol. Med. 8 (7), 1182–1189. doi:10.1016/j.nano.2012.01.007
Zhao, Q., Modis, Y., High, K., Towne, V., Meng, Y., Wang, Y., et al. (2012b). Disassembly and reassembly of human papillomavirus virus-like particles produces more virion-like antibody reactivity. Virology J. 9 (1), 52. doi:10.1186/1743-422X-9-52
Keywords: virus particles, virus-like particles, vaccines, downstream processing, purification, size-selective
Citation: Hillebrandt N and Hubbuch J (2023) Size-selective downstream processing of virus particles and non-enveloped virus-like particles. Front. Bioeng. Biotechnol. 11:1192050. doi: 10.3389/fbioe.2023.1192050
Received: 22 March 2023; Accepted: 08 May 2023;
Published: 25 May 2023.
Edited by:
Michel Eppink, Wageningen University and Research, NetherlandsReviewed by:
Patricia Vázquez-Villegas, Tecnologico de Monterrey, MexicoCopyright © 2023 Hillebrandt and Hubbuch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jürgen Hubbuch, anVlcmdlbi5odWJidWNoQGtpdC5lZHU=
 Nils Hillebrandt
Nils Hillebrandt Jürgen Hubbuch
Jürgen Hubbuch