- 1Clostridia Research Group, BBSRC/EPSRC Synthetic Biology Research Centre (SBRC), School of Life Sciences, Biodiscovery Institute, The University of Nottingham, Nottingham, United Kingdom
- 2Department of Microbiology, Faculty of Life Sciences, Ahmadu Bello University, Zaria, Nigeria
- 3NIHR Nottingham Biomedical Research Centre, Nottingham University Hospitals NHS Trust, The University of Nottingham, Nottingham, United Kingdom
The current climate crisis has emphasised the need to achieve global net-zero by 2050, with countries being urged to set considerable emission reduction targets by 2030. Exploitation of a fermentative process that uses a thermophilic chassis can represent a way to manufacture chemicals and fuels through more environmentally friendly routes with a net reduction in greenhouse gas emissions. In this study, the industrially relevant thermophile Parageobacillus thermoglucosidasius NCIMB 11955 was engineered to produce 3-hydroxybutanone (acetoin) and 2,3-butanediol (2,3-BDO), organic compounds with commercial applications. Using heterologous acetolactate synthase (ALS) and acetolactate decarboxylase (ALD) enzymes, a functional 2,3-BDO biosynthetic pathway was constructed. The formation of by-products was minimized by the deletion of competing pathways surrounding the pyruvate node. Redox imbalance was addressed through autonomous overexpression of the butanediol dehydrogenase and by investigating appropriate aeration levels. Through this, we were able to produce 2,3-BDO as the predominant fermentation metabolite, with up to 6.6 g/L 2,3-BDO (0.33 g/g glucose) representing 66% of the theoretical maximum at 50°C. In addition, the identification and subsequent deletion of a previously unreported thermophilic acetoin degradation gene (acoB1) resulted in enhanced acetoin production under aerobic conditions, producing 7.6 g/L (0.38 g/g glucose) representing 78% of the theoretical maximum. Furthermore, through the generation of a ΔacoB1 mutant and by testing the effect of glucose concentration on 2,3-BDO production, we were able to produce 15.6 g/L of 2,3-BDO in media supplemented with 5% glucose, the highest titre of 2,3-BDO produced in Parageobacillus and Geobacillus species to date.
Highlights
• Acetoin and 2,3-butanediol production engineered in a thermophilic Gram-positive chassis.
• Formation of by-products was minimized by deleting competing pathways.
• Identification and subsequent deletion of a previously unreported thermophilic acetoin degradation gene.
• Acetoin produced at 7.6 g/L (0.38 g/g glucose), 78% of the theoretical maximum.
• 2,3-butanediol was the predominant fermentation metabolite, up to 6.6 g/L (0.33 g/g glucose), 65% of the theoretical maximum.
1 Introduction
2,3-butanediol (2,3-BDO) and 3-hydroxybutanone (acetoin) are important chemicals with numerous applications in a number of different sectors. 2,3-BDO and its derivatives are involved in the manufacture of food additives and flavorings, pharmaceuticals, printing inks, synthetic rubber and plastics, fuel additives and antifreeze agents in addition to many more (Biswas et al., 2012; Cho et al., 2015; Maina et al., 2022). Its global market value is predicted to be $300 million by 2030 (Chemicals and Materials Market Research Report, 2019). Likewise, acetoin serves as a building block for the synthesis of cosmetics and pharmaceuticals and is also widely used as an additive in food flavoring and electronic cigarettes (Bao et al., 2014; Allen et al., 2016; Kandasamy et al., 2016). In recognition of its versatility, acetoin was named as one of the 30 platform chemicals produced from biomass by the United States Department of Energy (Werpy and Petersen, 2004).
Currently, both chemicals are primarily derived from petrochemicals by chemical-synthesis. However, concerns over the finite nature of fossil fuels and the current climate crisis, coupled with increasing demands, have provided a greater incentive to produce 2,3-BDO and acetoin via sustainable and renewable approaches. Microbial-based fermentations using lignocellulose, a renewable feedstock, has begun to attracted considerable attention in recent years (Xiao and Lu, 2014; Yang et al., 2017; Zhang et al., 2019). This is especially true for acetoin, as using fermentation is substantially more favourable in the food industry due to stringent safety regulations (Bao et al., 2014).
Previous efforts directed at producing 2,3-BDO relied upon using bacteria such as Klebsiella penumoniae, Klebsiella oxytoca and Paenibacillus polymyxa, which were known to produce significant amounts (De Mas et al., 1988; Ramachandran and Goma, 1988; Qureshi and Cheryan, 1989; Häßler et al., 2012). Further increases in the productivity of these native producers were sought through genetic modification (Supplementary Table S1), through which yields of up to 150 g L−1 were achieved (Song et al., 2019). However, Klebsiella species are pathogenic, compromising their use in industrial production (Yang and Zhang, 2018). Acetoin production was primarily achieved using Bacillus species. The best reported yield was obtained using Bacillus amyloliquefaciens (Luo et al., 2014), whereby 0.37 g g−1 glucose was achieved following an adaptive evolution strategy. More recently, thanks to advances in genetic modification tools, 2,3-BDO and acetoin production has been successfully achieved in a range of non-native, mostly mesophilic hosts, including Saccharomyces cerevisiae; Escherichia coli; Corynebacterium and Pichia pastoris (Yang and Zhang, 2018; Zhang et al., 2019) (Supplementary Table S2).
Meanwhile, the use of thermophiles as industrial platforms has been gaining increasing attention due to their many favourable features. These include, lower contamination risks, higher feed conversion rates and reduced cooling costs. They also confer desirable properties on the growth medium, such as reduced viscosity, reduced energy requirements for mixing and increased diffusion rates and substrate solubility (Chang and Yao, 2011; Keller et al., 2014). Temperatures above 50°C also favour consolidated bioprocessing (CBP), a process that allows the cost-effective, simultaneous enzyme-mediated saccharification and fermentation of lignocellulosic biomass into desired products by a single microorganism (Chinn and Mbaneme, 2015).
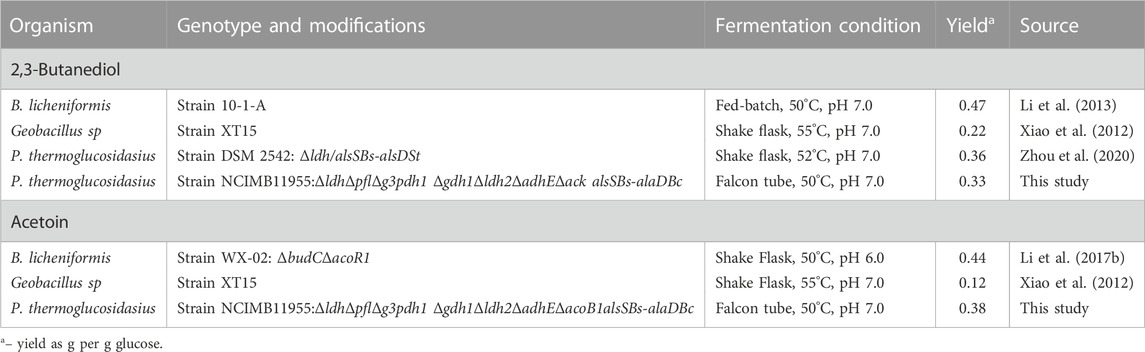
Thus far, a very limited number of thermophilic organisms have been exploited for the production of 2,3-BDO and acetoin (Table 1). High titre 2,3-BDO/acetoin yields have been documented using B. licheniformis strains (Li et al., 2013) with 94% and 84% of theoretical maximum yield (TMY), respectively. Although encouraging, B. licheniformis mainly produces optical inactive meso-2,3-BDO and is difficult to genetically manipulate, which impedes further improvements (Li et al., 2017a; Schilling et al., 2020; Zhou et al., 2020). Parageobacillus species also have potential for 2,3-BDO and acetoin production under elevated temperatures. Wild isolate Geobacillus sp XT15, (Xiao et al., 2012), produced both acetoin and 2,3-BDO, though at a much lower yield (0.22 and 0.12 g/g glucose). More recently up to 7.2 g L−1, representing 72% theoretical maximum pure R,R-2,3-BDO production, was achieved in Parageobacillus thermoglucosidasius 95A1 (Zhou et al., 2020) using shake-flask fermentation at 55°C.
Here, Parageobacillus thermoglucosidasius NCIMB 11955 is used as the single chassis to produce both 2,3-BDO and acetoin at elevated temperatures through metabolic engineering. This particular strain is versatile both in term of growth condition and genetic accessibility. It can ferment a wide range of sugars typical of those found in lignocellulosic substrates and has been previously been modified to create an industrially relevant strain, TM242, for ethanol production (Cripps et al., 2009; Hussein et al., 2015). Lately, more sophisticated genetic methods are been developed for the strain, including gene knockout/in using allele coupled exchange (ACE) and CRISPR mediated gene-deletion (Reeve et al., 2016; Sheng et al., 2017; Lau et al., 2021) that together allow for more precise and targeted metabolic engineering. These methods have additionally been used to extend substrate range through the successful expression of the genomically integrated Caldicellulosiruptor bescii celA gene enabling direct utilization of lignocellulosic substrate by the organism (Bashir et al., 2019).
To achieve high titre 2,3-BDO and acetoin production, three interrelated approaches were taken by: 1) implementing an efficient acetoin/2,3-BDO pathway through the assembly and expression of genes encoding acetolactate synthase [EC 2.2.1.6] (ALS), acetolactate decarboxylase [EC 4.1.1.5] (ALD) and acetoin reductase/2,3-BDO-dehydrogenase [EC 1.1.1.4] or AR/BDH (Xiao and Lu, 2014); 2) inactivation of genes encoding enzymes participating in competing pathways, and; 3) alleviating redox issues through decoupling of the expression of the native bdh using a constitutive promoter and different aeration conditions. The outcome of this strategy were engineered strains that produced 2,3-BDO and acetoin at 50°C at levels close to the theoretical maximum yield (TMY). In both cases, with all major competing pathways disrupted, including ethanol production, up to 90% of the carbon flux entered the acetoin/2,3-BDO pathway, giving scope for further improvement.
2 Materials and methods
2.1 Chemicals and reagents
All chemicals and reagents were obtained from Sigma-Aldrich Ltd., (Poole, United Kingdom). DNA polymerases, restriction enzymes, T4 ligase and DNA ladders were purchased from New England Biolabs or Promega (Southampton, United Kingdom). EDTA-Free Protease Inhibitor Cocktail Tablets were purchased from Roche (Mannheim, Germany)
2.2 Strains and cultivation
All bacterial strains used in this study are listed in Table 2. Unless otherwise stated, E. coli Top 10 was used as a host for plasmid construction and E. coli B21 (DE3) was used for protein expression. P. thermoglucosidasius NCIMB 11955 was used for 2,3-BDO and acetoin production. All E. coli strains were grown in Luria-Bertani (LB) agar or LB broth containing appropriate antibiotics (kanamycin: 50 μg/mL; ampicillin: 100 μg/mL) at 30°C or 37°C with agitation of 200 rpm. P. thermoglucosidasius strains were routinely grown on TSA agar or in 2SPYNG broth (Sheng et al., 2017), with kanamycin when necessary at 12.5 μg/mL, at 50°C–60°C, with agitation of 250 rpm.
To assess growth on acetoin and 2,3-BDO, WT P. thermoglucosidasius was grown in CBM (Morris, 1971) (consisted of, per litre of deionized water, 200 mg MgSO4·7H2O, 7.58 mg MnSO4·H2O, 10 mg FeSO4·7H2O, 1 mg p-aminobenzoic acid, 2 μg biotin, 1 mg thiamine, 4 g acid hydrolysed casein hydrolysate, 0.5 g K2HPO4 and 0.5 g KH2PO4) with appropriate carbon sources at 1%.
For acetoin and 2,3-BDO production, typically, appropriate strains from a 10 mL overnight (O/N) preculture were inoculated to prewarmed modified ASYE medium (2 mM MgSO4, 0.1 mM CaCl2, 2 mM citric acid, 100 μM FeSO4·7H2O, 16.85 mM NiCl3·6H2O, 12.5 mM biotin, 0.1 M HEPES buffer, 0.01 g L−1 thiamine, ×5 dilution of Sigma-Aldrich Ltd., M9 minimal salts, ×1,000 dilution of Trace Metal Mix A5, 2% yeast extract, 1%–7% glucose) at a 10% (v/v). Fermentation was carried out on a rotatory shaker at 250 rpm at 50°C for 48 h in sealed 50 mL Falcon tubes with various volumes as indicated before analysis.
2.3 General cloning and transformation
Plasmids and primers used in this study are listed in Supplementary Tables S3,S4. All primers were designed using ApE (University of Utah) and analysed with OligoAnalyzer Tool (IdtDNA). For the amplification of the desired DNA products, Phusion High-fidelity DNA polymerase was used for cloning purpose and dream Tag Green PCR (Thermo Fisher Scientific, United Kingdom) master mix was used for strain verification. DNA fragments that had undergone restriction digestions were purified using Zymoclean™ Gel DNA Recovery Kit (ZymoResearch, United Kingdom) before ligation using LigaFast™ Rapid DNA Ligation System (Promega). Chemically competent E. coli Top10 cells produced in-house were used to propagate the constructed plasmids and plasmids were harvested using Monarch® Plasmid Miniprep Kit (New England BioLabs). All cloning procedures were conducted following the manufacturer’s instructions. Sanger sequencing (Source Biosciences; EurofinsDNA) were routinely performed to confirm the authenticity of plasmid constructs. Preparation of P. thermoglucosidasius electro-competent cells and electroporation procedure using Genepulser electroporator (BioRad, United Kingdom) was undertaken in accordance with the method previously described (Sheng et al., 2017).
2.4 Construction of 2,3-BDO biosynthesis pathway
All of the plasmid constructs carrying the genes of the 2,3-BDO biosynthesis pathway are based on pMTL61110 (Sheng et al., 2017), which carries the ColE1 and pUB110 origins of replication, a thermostable kanamycin resistance gene and multiple cloning sites (MCS). Parts included the T1T2 double terminator of the E. coli rrnB gene, the Pldh promoter of the Bacillus stearothermophilus ldh gene and the ALS and ALD genes synthesized by Genewiz Ltd., and codon optimized for P. thermoglucosidasius NICMB 11955. Operons were constructed in a stepwise manner. First, Pldh was fused with the ALS gene using SOEing PCR and primers N_pLDH_F_AvrII, N_pLDH_BALS_R, N_BALS_LDH_F and N_BALS_R_NdeI, which generated a DNA fragment flanked by AvrII and NdeI restriction sites. Following successful ligation into LS-ALS1, the P. thermoglucosidasius GPAD gene promoter (Pgpad) was fused with the ALD(n) gene concomitant with the introduction of a synthetic RBS (ribosome binding site). Primers used were GPAD_F_NotI, GPAD_R_Bot9, ALD(n)_Bot9_F and ALD(n)_R_XhoI. The resultant DNA fragment, flanked by NotI and XhoI restriction sites, was combined with the previous fragment carrying ALS to give plasmid pMTL-BD(n) (where n = 1, 3 and 5.) Plasmid pMTL-BD3b could not be propagated in E. coli due to cell toxicity, instead, inserts containing the promoter of the rplS gene fused to the native bdh gene (SOEing PCR with primers BDH1_F_PheB, BDH1_R_NheI, Rpls_F_XhoI, Rpls_R_PheB) were ligated with pMTL-BD3 after restriction digest with XhoI and NheI and electroporated directly into NICMB 11955 following dialysis treatment. Colony PCR and Sanger sequencing were used to verify that each plasmid was correct.
2.5 Strain construction using CRISPR
All constructs for various targeted gene knockouts were based on pMTL67555 (Lau et al., 2021) which is based on the Streptococcus thermophilus Cas9-3. Synthetic knockout cassettes contained target specific sgRNA, identified using Benchling CRISPR Guide Design software (www.benchling.com), and homology arms (HA) synthesized by Genewiz Ltd. HA consists of 501 bp regions of DNA from up- (5′) and down- (3′) stream of the gene to be inactivated. Ligation of the fragments between the BamHI and AscI restriction sites of pMTL67555 yielded the knockout plasmids. Deletion cassettes were designed to generated an in-frame deletion of the targeted gene comprising just 3 codons, including the start and stop codons. Following the successful transformation of the knockout plasmids into NICMB 11955, transformants are passaged on TSA-kanamycin agar for 3 days before verification of gene knockout by colony PCR. Once verified, the mutants were passaged for at least 6 times in 2SPYNG without selection (at 12 h intervals) for plasmid curing.
2.6 Enzyme assays
Crude cell extracts were prepared using BugBuster 10X protein extraction reagent (Merck). The protein extraction buffer (1X) consisted of BugBuster 10X (2 mL), sodium phosphate buffer 50 mM (18 mL), EDTA-free protease inhibitor tablet (Roche), and lysozyme (20 mg/mL). Total protein concentrations of the soluble fraction of cell lysates were determined using BCA-kit (Thermo Fisher Scientific, United Kingdom) following the manufacturer’s instructions and a standard curve generated from BSA (200–1,000 μg/mL) was used to calculate the protein concentrations. Assays for acetoin reductase activity were carried out at 60°C, initiated by adding 10 μL crude cell lysate to a solution containing 50 mM acetoin, 0.5 mM NADH or NADPH and 1 mM TCEP (Nicholson, 2008). Oxidation of NADH to NAD+ was monitored spectrophotometrically by the AR dependent rate of A340 decrease. Acetolactate decarboxylase activity was assayed as previously described (Dulieu and Poncelet, 1999). In short, substrate solution was prepared by diluting 0.05 mL of ethyl 2-acetoxy-2-methylacetoacetate with 1.5 mL of H2O and 1.5 mL of 1 M NaOH. After stirring for 20 min, it was made up to 25 mL with 50 mM MES, adjusting to pH 6.0 using 0.5 M HCL. The reaction was initiated following incubation of cell lysates with substrate at 60°C for 20 min before adding Voges-Proskauer Reagent A and Reagent B and 280 µL 0.5% (w/v) creatinine. After incubation at 40 min at room temperature, A522 was used for quantification of the final acetoin concentration using an acetoin standard curve. Negative controls included reaction with and without substrate in the absence of ALD. One Unit of ALD activity is defined as the concentration of acetoin by ALD (absorbance/a constant molar coefficient ε) per 1 mg of protein.
2.7 Analytical methods
Growth of all bacterial cultures was monitored by measuring optical density at 600 nm (OD600) using a Pharmacia Novaspec II. Metabolites were analysed using high-performance liquid chromatography (HPLC-UV and RI), performed using the Dionex UltiMate 3,000 System and 5 mM H2SO4 as the mobile phase. Valeric acid at 50 mM was used as an internal standard. The sample supernatants were filtered with 0.2 μm filters and mixed in 1:1 ratio with diluent. The Bio-Rad Aminex HPX-87H 300 mm × 7.8 mm × 9 μm column was used for sample elucidation, running at a flow rate of 0.5 mL/min, at 35°C for 55 min. All experiments were performed in triplicate and data analysed using PRISM (GraphPad Software, La Jolla, United States) and Excel (Microsoft).
3 Results
3.1 Verification of the native acetoin/2,3-BDO pathway
The route to 2,3-BDO from pyruvate involves the intermediate metabolites acetolactate and acetoin, catalyzed by ALS, ALD and BDH. The P. thermoglucosidasius NICMB 11955 genome possesses genes encoding homologues of ALS (BCV53_05415) and BDH (BCV53_03685) but not ALD. Although it may still be able to produce 2,3-BDO via the oxygen-dependent, spontaneous oxidation of acetolactate to di-acetyl, no trace was evident in culture supernatants following growth of the WT in ASYE medium. The major metabolites (Supplementary Figure S1) were lactate (74%), acetate (16%) and ethanol (8%).
In previous work (Sheng et al., 2017), various P. thermoglucosidasius mutants were constructed by allelic exchange using a heterologous pyrE gene as a counter selection marker and the requisite ΔpyrE derivative of NICMB 11955. These included a ΔldhΔpfl double mutant in which the major fermentative pathways to lactate and formate had been eliminated. In such mutants, pyruvate is accumulated to high (37 mM) concentrations (Extance et al., 2013). We speculated that the acetyl-CoA pathway in this mutant would be overloaded and carbon flux would overflow into the acetoin/2,3-BDO pathway. To test this hypothesis, we first repaired the pyrE mutant allele to wildtype using ACE. The strain generated, LS100, was shown to produce up to 5 mM 2,3-BDO under the same conditions as when the WT produced none (Supplementary Table S5).
The data obtained with LS100 provided indirect evidence that the bdh gene identified in the P. thermoglucosidasius NICMB 11955 genome encodes a BDH with activity against acetoin. To confirm this, we PCR amplified, cloned and expressed the gene in E. coli using the PET expression system (pET17b). Enzyme assays with acetoin as the substrate were conducted at 60°C using crude cell extracts of the E. coli clone generated. These showed (Supplementary Figure S2) that the enzyme is an NADH dependent alcohol dehydrogenase with significant activity towards acetoin at thermophilic temperatures.
3.2 Selection and characterization of pathway components
The native P. thermoglucosidasius ALS is not an ideal candidate for acetolactate production as it is anabolic, belonging to the branched chain amino acid biosynthetic pathway. Enzymes of this type are generally highly regulated by product inhibition (Weinstock et al., 1992; Elisáková et al., 2005). Essential to acetolactate formation is a catabolic ALS not subject to product inhibition. Previous work indicates that the Bacillus subtilis ALS (ALS1) is a suitable candidate (Lin et al., 2014; Zhou et al., 2020). Interestingly, we found that ALS1 shares near identical (99%) AA sequence homology to the ALS of B. licheniformis. In addition, we also identified another ALS, from Bacillus coagulans 2–6 (ALS2) (Wang et al., 2014), a species shown to produce acetoin and 2,3-BDO as the primary fermentation products under aerobic conditions (Su and Xu, 2014). ALS2 is annotated as being catabolic and shares 65% AA homology with ALS1.
The conversion of acetolactate to acetoin also needs to be efficiently catalyzed. An effective ALD is, therefore, essential. Furthermore, it is desirable to have a wide selection of pathway components to give greater flexibility for a more integrated approach to metabolic engineering, as sometimes the introduction of high enzyme activities throughout a pathway, especially in heterologous hosts, can lead to depletion of intermediates, causing diminished growth, cell toxicity, and reduced product yields (Lechner et al., 2016).
Genome sequences of the two thermophilic producers (Xiao et al., 2012; Li et al., 2017b) were unavailable and reports of thermophilic ALDs are rare. Hence, six novel ALDs, from a wide range of organisms able to grow at 50°C and above, were selected as potential candidates. The only mesophile source in this selection was the ALD of Exiguobacterium acetylicum (formerly Brevibacterium acetylicum). However, previous reports have indicated that its ALD (ALD1) possessed promising properties including stability at 60°C, high activity at 50°C and a broad pH range of 5–8 (Ohshiro et al., 1989). Others include two from bacilli, ALD2 (B. coagulans) (Wang et al., 2014) and ALD3 (Bacillus cereus) (Finlay et al., 2000); one from an anaerobe, ALD4 (P. thermopropionicum) (Imachi et al., 2002) and two fungal ALDs, ALD5 (Chaetomium thermophilum) (Kellner et al., 2016) and ALD6 (Thielavia terrestris) (Berka et al., 2011).
The synthetic genes (Genewiz Ltd.) encoding the ALD candidates were codon optimized and cloned into the P. thermoglucosidasius expression vector pMTL61110_pldh between its XbaI and KpnI restriction sites. This positioned the ALD genes between the strong Pldh promoter of the G. stearothermophilus ldh (lactate dehydrogenase) gene (Bartosiak-Jentys et al., 2013) and the T1T2 double terminator of the E. coli rrnB gene (Figure 1). The 3’ 6 bp spacer sequence of the native Pldh RBS were replaced with the palindromic XbaI recognition sequence for convenience of cloning. The plasmids were transformed into WT P. thermoglucosidasius NICMB 11955 and the ALDs were allowed to express before crude cell lysates were prepared and assayed for acetolactate dehydrogenase activity (Materials and Methods). Significant activities were observed in extracts made from cells producing three of the ALDs. ALD2 had the highest activity (0.81 ± 0.00 U/mg), followed by ALD6 and ALD4, which exhibited 73% and 70%, respectively, of the activity of ALD1 (Figure 1).
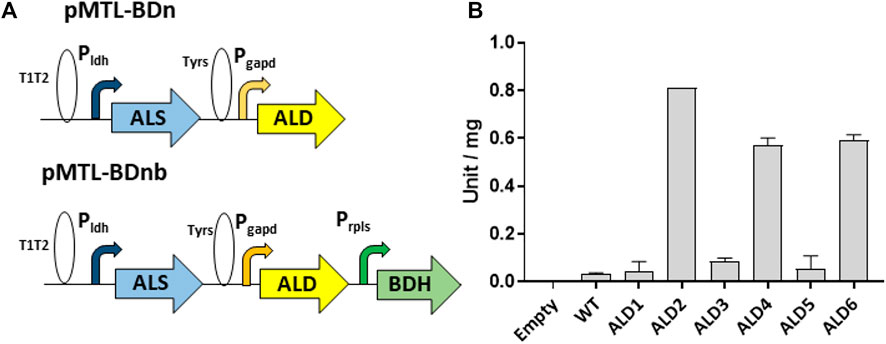
FIGURE 1. Assembly of enzyme variants in the acetoin/2,3-BDO pathway and assay of acetolactate decarboxylase activities. (A) Gene configuration of the constructed acetoin/2,3-BDO pathway. The ALS gene is expressed from the G. stereothermophilus lactate dehydrogenase promoter (Pldh) with the native RBS, the ALD gene is expressed via PT glyceraldehyde 3-phosphate dehydrogenase promoter and RBS9 (TAGATAAAGGAGGTAGAACT, taken from the RBS of the locus tag CLB_1812 of Clostridium botulinum ATCC 3502, accession no. CP000726) while the BDH gene is expressed from the native rplS promoter and G. stereothermophilus pheB RBS. T1T2 and TyrS are transcriptional terminators from the E. coli rrnB and B. subtilis tyrS genes, respectively. (B) Assay of α-acetolactate decarboxylases expressed in PT. Y-axis represents unit per mg of protein in the crude cell lysate. Error bars represent standard deviation, n = 3.
3.3 Implementation of acetoin/2,3-BDO pathway in P. thermoglucosidasius
The components of the acetoin/2,3-BDO operons were assembled such that the ALS and ALD genes were placed under the control of the Pldh and Pgapd promoters, respectively (Figure 1). To prevent the possibility of recombination between identical sequences, the native ldh RBS preceded ALS genes, while ALD genes relied on the Bot9 RBS (Lau et al., 2021). Each promoter-gene cassette was separated by two unique restriction sites (SbfI/KpnI for ALS and NotI/XhoI for ALD) for the convenience of replacing/swapping cassettes.
During the assembly of the various combinations of ALS and ALD genes, a major obstacle observed was cell toxicity both when cloning, in E. coli and when the shuttle vectors were transferred into the P. thermoglucosidasius host. Four E. coli strains (Top10, JM109, DH5α and NEB F−) were tested and only JM109 and NEB F− allowed propagation of the desired plasmids. No growth was evident in the case of DH5α, while although Top10 transformants grew, the plasmids were found to have acquired mutations effecting the integrity of the cloned genes. Insertions/deletions and nucleotide substitutions that respectively caused frameshifts and nonsense mutations in the ALD gene proved to be a frequent occurrence when plasmids were introduced into P. thermoglucosidasius. In total, just three cassettes (BD2, BD3 and BD5) were obtained that were cloneable in E.coli and which maintained structural integrity (verified by Sanger sequencing) for at least three passages following transformation into P. thermoglucosidasius. Plasmids carrying these cassettes (encoded enzymes in brackets), were pMTL-BD2 (ALS2-ALD1), pMTL-BD3 (ALS1-ALD3) and pMTL-BD5 (ALS1-ALD5).
Acetoin and 2,3-BDO levels were characterized in transformants of both the P. thermoglucosidasius WT and the LS100 mutant carrying plasmids pMTL-BD2, pMTL-BD3 and pMTL-BD5 to identify which combination of ALS and ALD was the most productive (Figure 2). Levels of 2,3-BDO were increased in all the strains. For the WT, cells harbouring pMTL-BD3 produced up to 29 mM 2,3-BDO, whereas cells carrying pMTL-BD2 and pMTL-BD5 produced much lower levels of 3.34 mM and 2.81 mM, respectively. For LS100, up to 31 mM 2,3-BDO was produced by cells carrying either pMTL-BD2 or pMTL-BD3, with a lower 26 mM for those cells harbouring pMTL-BD5, representing a 5.9-, 6- and 4.8-fold increase, respectively, compared to the same strain without the ALS-ALD operons. For acetoin, a low level (4–10 mM) was evident in all WT strains carrying all three operons whilst insignificant quantities were evident in all LS100 strains. Taking the amount of glucose consumed into account, LS100 cells carrying pMTL-BD3 had the highest 2,3-BDO yield at 0.2 G/G glucose (g/g) This equates to 40% of theoretical maximum yield (TMY). Remarkably, most of the 2,3-BDO produced was the RR-stereoisomer with up to 95% optical purity. This is in contrast to the ratio observed between the meso- and RR-2,3-BDO forms in the plasmid-free LS100 strain which is 1:1. The formation of the different 2,3-BDO isomers is dependent upon acetoin stereoisomerism, where the 3 S form arises through the spontaneous di-acetyl route and the 3R form results from the active ALD route. We concluded that ALD contributed to all of the increase in 2,3-BDO level, and that the native BDH is chiral-specific. This has industrial significance as pure chiral compounds are more valuable and highly sought after (Fu et al., 2016; Kandasamy et al., 2016).
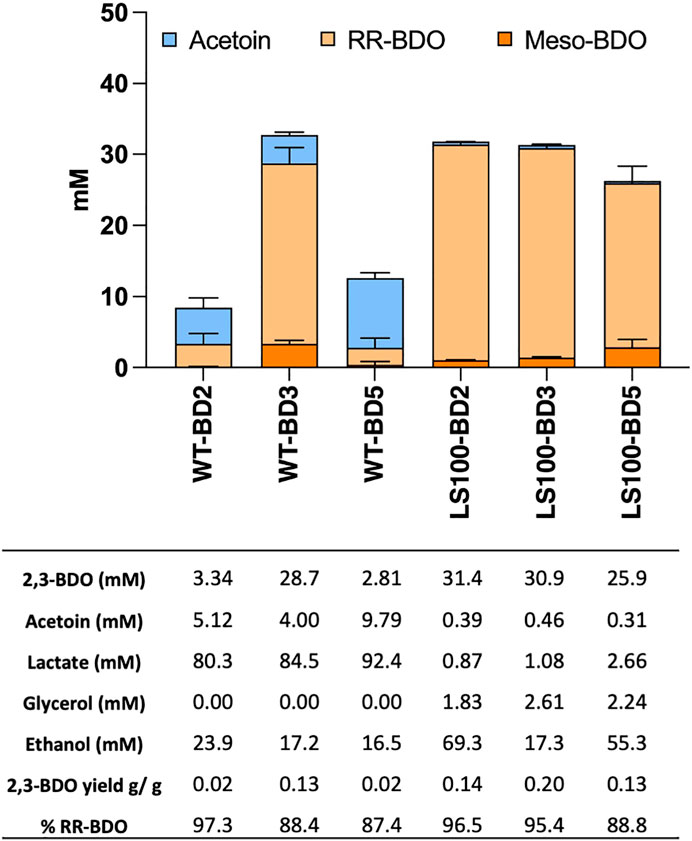
FIGURE 2. Product profiles of the P. thermoglucosidasius WT and LS100 (ΔldhΔpfl) double mutant strain carrying the assembled, plasmid-based, heterologous acetoin/2,3-BDO pathways. Strains carried either plasmids pMTL-BD2, pMTL-BD3 or pMTL-BD5. Cultures were grown for 48 h in 10 mL of M-ASYE medium containing 2% (w/v) yeast extract and 2% (w/v) glucose in 50 mL falcon tubes at 50°C with shaking (250 rpm). Concentrations are calculated as the average three biological replicates. Error bars represent the standard deviation. BDO = 2,3- butanediol, RR-BDO = RR-2,3-butanediol, meso-BDO = meso-2,3- butanediol.
3.4 Deletion of competing pathways
The Embden-Meyerhof-Parnas (EMP) pathway requires two NAD + for the production of ATP. In aerobes, NAD+ is recycled by the oxidation of NADH by molecular oxygen. Under oxygen-limiting conditions, a number of fermentative routes in P. thermoglucosidasius are utilized for redox recycling. While they help to maintain redox balance, carbons diverted into various chemicals severely impact the yield of the desired product. A number of studies have shown that deleting genes (ldh and adhE) of competing pathways (lactate and ethanol formation) are essential for elevated production of 2,3-BDO (Jantama et al., 2015; Radoš et al., 2015; Erian et al., 2018). Although the major fermentative routes to lactate and formate have been deleted in strain LS100, its metabolite profile indicated that lactate and glycerol were still being made as well as significant quantities of ethanol. Accordingly, the inactivation of genes encoding the enzymes involved in their production could be beneficial to 2,3-BDO production.
Prevention of ethanol formation could be most simply achieved (Figure 3) by knocking out the adhE gene (BCV53_03330) as reported previously (Extance et al., 2013). Although the ldh gene is already deleted in strain LS100, analysis of the NICMB 11955 genome revealed the presence of another gene encoding a secondary LDH, annotated as L-LDH (BCV53_05985) and sharing no sequence similarities to other non-P. thermoglucosidasius strains. The gene was designated ldh2. In the case of glycerol, biosynthesis can occur either via glycerol-3-phosphate dehydrogenase (G3PDH) (Albers et al., 1996) or via glycerol dehydrogenase (GDH) (Piattoni et al., 2013; Wang et al., 2016). Two copies of G3PDH exist in S. cerevisiae, and deletion of either reduced the production of glycerol while elimination of both entirely blocked glycerol biosynthesis (Medina et al., 2010; Hubmann et al., 2011). When coupled with the 2,3-BDO pathway, increased production was observed (Albers et al., 1996). Two G3PDHs (1,2) (BCV53_15365, BCV53_15685) and two GDHs (1,2) (gldA, BCV53_10775) are present in P. thermoglucosidasius NICMB 11955. G3PDH1, also annotated as a FAD dependent oxidoreductase, encodes a 550 AA protein sharing 67% and 30% sequence identity to B. subtilis and S. cerevisiae, respectively. G3PDH2 codes for a 348 AA enzyme that is highly conserved across species (97% in bacilli and 87% in S. cerevisiae). GDH1 encodes a 363 AA protein sharing 61% identity to the GDH of K. pneumoniae and 50% to that of B. subtilis while GDH2 is 358 AA with no significant sequence identity to proteins in any other genus.
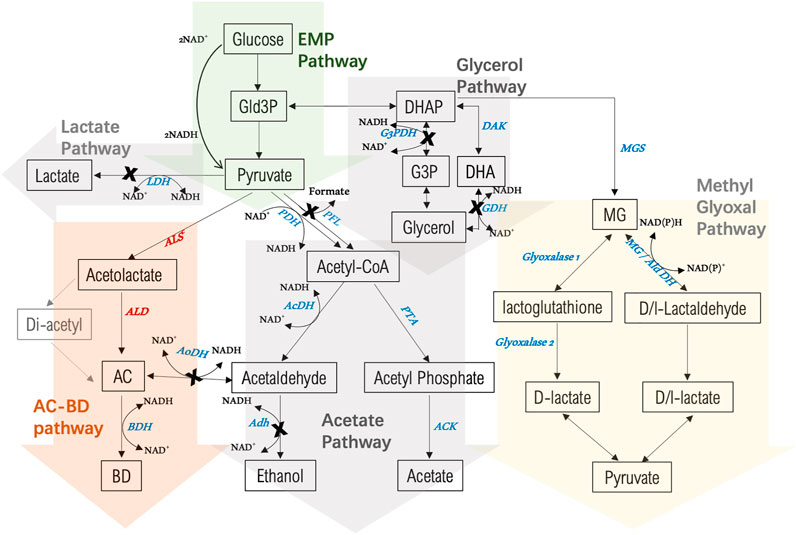
FIGURE 3. Overview of 2,3-BDO biosynthesis and by-product pathways in P. thermoglucosidasius NCIMB 11955 starting from glucose as a substrate. Enzymes involved in 2,3-BDO biosynthesis are in red: ALS: acetolactate synthase; ALD: acetolactate decarboxylase; BDH: 2,3-butanediol dehydrogenase. Enzymes of competing pathways are in blue: LDH: lactate dehydrogenase; G3PDH: glycerol-3-phospahte dehydrogenase; DAK: dihydroxyacetone kinase; GDH: glycerol dehydrogenase; AcDH: Acetaldehdyde dehydrogenase; AoDH: Acetoin dehydrogenase; Adh: alcohol dehydrogenase; PTA: phosphate acetyltransferase; ACK: acetate kinase; MGS: methylglyoxal synthase; AldDH: aldehyde dehydrogenase. A cross indicates where the encoding gene has been deletion in the present study. AC = acetoin, BD = 2,3-butanediol.
Using LS100 as the parental strain, the following deletion mutants were created: LS101 (ΔldhΔpflΔg3pdh1); LS102 (ΔldhΔpflΔgdh1); LS103 (ΔldhΔpflΔldh2), LS104 (ΔldhΔpflΔg3pdh1Δgdh1); LS105 (ΔldhΔpflΔg3pdh1Δgdh1Δldh2); LS200 (LS100ΔadhE); LS201 (LS101ΔadhE); LS202 (LS102ΔadhE); LS204(LS104ΔadhE) and LS205 (LS105ΔadhE). Despite numerous attempts, the two genes g3pdh2 and gdh2 could not be deleted. Temporary deletions were observed and confirmed by PCR, however, curing the deletion vector proved problematic and when the plasmid was lost reversion back to the parental strain was always observed. It is likely that those are essential genes, for example, the highly conserved g3pdh2 could be used for the biosynthesis of phospholipid membrane (Coleman et al., 2019).
The various mutant strains obtained were transformed with pMTL-BD3 and the fermentation profiles of the resultant transformants analyzed (Figure 4). Other than the adhE knockouts, improvements were observed for all the additional mutants compared to LS100. Mutants carrying Δldh2 had the highest 2,3-BDO level, with 44 mM and 50 mM (0.22 and 0.23 g/g) for LS103 and LS105, respectively. No differences were observed for the single and double glycerol pathway mutants. Ethanol levels were also increased, with up to a 3-fold increase with LS105. For the AdhE mutants, ethanol production was largely eliminated, but 2,3-BDO levels were much reduced to levels of between 13 and 19 mM. Lactate levels increased slightly (1.5–2.0 mM) with single g3pdh1 deletions and were reduced in the single ldh2 mutant (0.8 mM). However, with both g3pdh1 and gdh1 deleted, the lactate level increased significantly (up to 9 mM) in strains with or without Δldh, indicating the presence of alternative routes. Other studies (Loftie-Eaton et al., 2013) suggest that the methylglyoxal pathway most likely contributes to this increase. P. thermoglucosidasius NICMB 11955 appears to possess this pathway: methylglyoxal synthase (BCV53_15505), three glyoxylase enzymes (BCV53_06495, BCV53_09900, BVC53_15250) and methylglyoxal reductase (BCV53_06315).
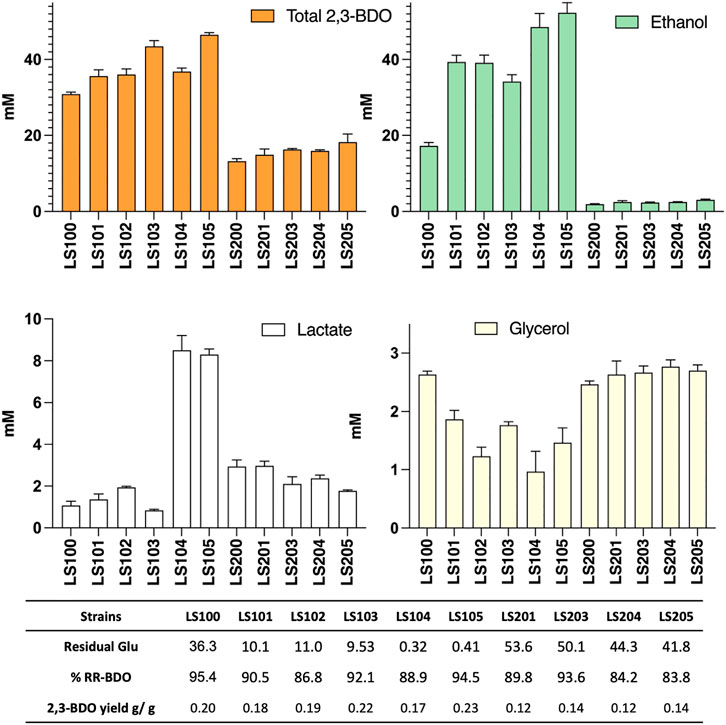
FIGURE 4. The effect on acetoin and 2,3-BDO yields of targeting various competing pathways in strains with (LS100—LS105) and without (LS200—LS205) a functional adhE gene. All strains carry plasmid pMTL-DB3 and were grown at 50°C for 48 h in 10 mL of M-ASYE medium containing 2% (w/v) glucose and yeast extract in a 50 mL falcon tube with shaking (250 rpm). 2,3-BDO level represents the sum of meso-2,3-butanediol and (R,R)-2,3-butanediol. The presence of an adhE deletion almost eliminated ethanol production but inhibited growth and 2,3-BDO production. Acetoin is not detected under this condition. Concentrations are calculated as the average of three biological replicates. Error bars represent standard deviation. BDO = 2,3- butanediol, RR-BDO = RR-2,3-butanediol. Strains with genes deleted are indicated as follows: LS100 (ΔldhΔpfl), LS101 (ΔldhΔpflΔg3pdh1), LS102 (ΔldhΔpflΔgdh1), LS103 (ΔldhΔpflΔldh2), LS104 (ΔldhΔpflΔg3pdh1Δgdh1), LS105 (ΔldhΔpflΔg3pdh1Δgdh1Δldh2), LS200 (LS100ΔadhE), LS201 (LS101ΔadhE); LS202 (LS102ΔadhE); LS204(LS104ΔadhE) and LS205 (LS105ΔadhE).
Glycerol levels were reduced in all non-ΔadhE strains, with the LS102 and LS104 producing only 1.1 and 0.6 mM, respectively, most likely a consequence of the redox imbalance caused by the loss of the capacity to regenerate NAD+ with the loss of the ethanol pathway. Thus, in the ΔadhE strains, both lactate and glycerol levels were significantly increased, despite almost double the amount of residual glucose being left in the media compare to the corresponding non-adhE mutants.
3.5 Addressing redox constraints in ΔadhE mutants
With the disruption of lactate production, the acetyl-CoA pathway plays a major role in maintaining redox balance under oxygen limitation. It follows that ethanol is a major by-product during 2,3-BDO fermentation and deletion of adhE reduces growth and productivity. Whereas Zhou et al. (2020) reported a decrease in titre and an increase in yield for 2,3-BDO in the ADH mutants in P. thermoglucosidasius strain DSM 2542, we observed both a decrease in titre and yield. However, it is still desirable to maximize carbon flux to the acetoin/2,3-BDO pathway necessitating alternative ways of co-factor regeneration.
It has been shown that both increasing oxygen supply and over-expression of BDH can enhance 2,3-BDO production (Kim et al., 2016). Accordingly, we explored the effect of aeration by varying fermentation volume, using 2 mL, 3 mL and 5 mL. Data obtained indicated a general increase in carbon flux of the acetoin/2,3-BDO pathway with decreasing fermentation volume. More than 60 mM (acetoin + 2,3-BDO) was obtained from 3 mL cultures compared to less than 20 mM across all five ΔadhE strains. In terms of 2,3-BDO production, the highest titre achieved was 34 mM, using strain LS205 habouring pMTL-BD3. With more aeration, production switched to acetoin, and in 2 mL cultures up to 52 mM was produced (Figure 5A).
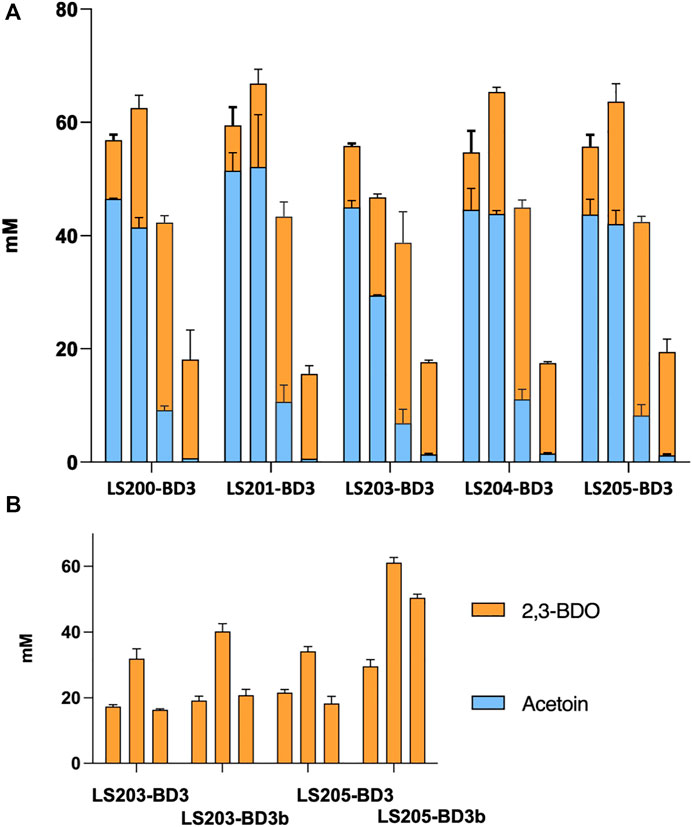
FIGURE 5. The effect of aeration and BDH overexpression on yields of acetoin and 2,3-BDO. (A) Combined acetoin and 2,3-BDO concentrations in ΔadhE strains (LS200-LS205) habouring plasmid pMTL-BD3 under different aeration conditions. The media employed was M-ASYE medium containing 2% (w/v) yeast extract and glucose. All fermentations were undertaken at 50°C for 48 h and in 50 mL falcon tubes with shaking (250 rpm). High to low aeration conditions were imposed by the use of either 2 mL, 3 mL, 5 mL or 10 mL culture volumes, represented by column 1–4, left to right. Increasing aeration increases carbon flux to the acetone/2,3-BDO pathway. Concentrations are calculated as an average of three biological replicates. (B) 2,3-BDO concentrations in strain LS203 and LS205 with (pMTL-BD3) and without (pMTL-BD3b) overexpression of the native bdh. Overexpression of bdh increased 2,3-BDO concentrations across three aeration conditions (3 mL, 5 mL and 10 mL fermentation volumes: bar 1–3, left to right, in each strain). Concentrations are calculated as average of three biological replicates.
To assess the effect of overexpressing bdh we placed it under the control of the native rplS promoter and the G. stearothermophilus pheB RBS (ribosome binding site), which is reported to give strong, constitutive expression levels under varied growth conditions (Reeve et al., 2016). The bdh gene was firstly fused with the promoter-RBS and then ligated onto pMTL-BD3 between the XhoI and NheI sites to generate pMTL-BD3b. To circumvent toxicity issues in E. coli, the ligation product was transformed directly into the appropriate P. thermoglucosidasius NICMB 11955 strain following de-ionizing dialysis treatment and correct transformants were verified by colony PCR and Sanger sequencing (Supplementary Figure S3). Increases in 2,3-BDO production were observed under the three fermentation conditions tested. The highest increase occurred in a 5 mL fermentation volume, with up to 40 and 61 mM being were obtained with LS203 and LS205 carrying pMTL-BD3, respectively. This represented a 1.3-fold (LS203) and 2-fold (LS205) increase (Figure 5B). However, when tested in strain LS105 (which does not carry the adhE deletion) under limited aeration, the levels of 2,3-BDO decreased. There was also a 2-fold decrease in ethanol produced and glucose consumed (Data not shown). We concluded that this is because increased bdh expression shifts the carbon flux towards the acetoin/2,3-BDO pathway, but without sufficient aeration, NAD+ deficiency brings about growth termination. Clearly, 2,3-BDO levels are determined by the intricate balance of oxygen supply and acetoin conversion. To optimize further, a wider range of fermentation volumes were analyzed, and at 6 mL, the highest amount of 2,3-BDO was produced at 63.4 mM (Table 3).
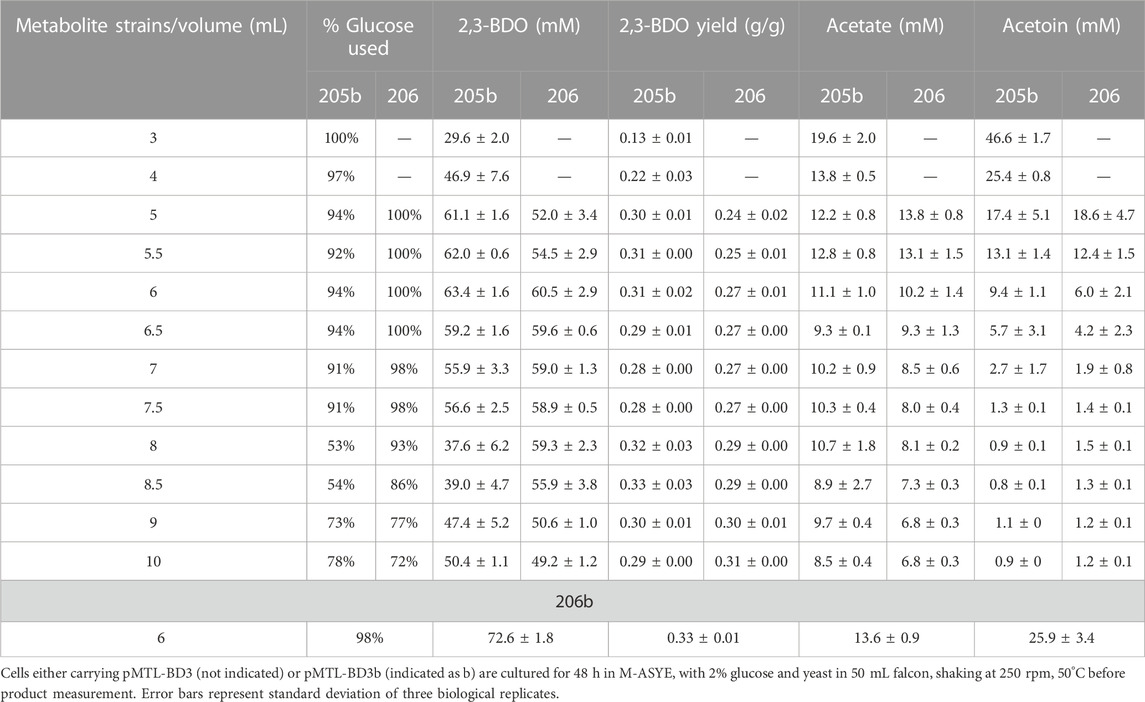
TABLE 3. Key metabolite profiles of engineered strains with (LS205) and without (LS206) an ack deletion under different fermentation volumes.
3.6 Identification of an acetoin utilization gene
In acetoin-excreting bacteria, acetoin catabolism can occur via the lipoic acid-mediated conversion of acetoin to acetaldehyde, allowing the re-utilization of the 4 carbon compound for energy (Xiao and Xu, 2007). Acetoin utilization depletes carbon flux from the acetoin/2,3-BDO pathway. It follows that measures that disrupt acetoin utilization, either through deletion of catalytic component or transcriptional regulators, can lead to increased acetoin levels (Zhang et al., 2016; Li et al., 2017a).
To test whether P. thermoglucosidasius NICMB 11955 can catabolise acetoin, we grew the WT strain in defined medium containing 1% (w/v) acetoin or 2,3-BDO as the sole carbon source in baffled-flasks at 60°C. NICMB 11955 utilized both acetoin and 2,3-BDO rapidly, reaching a maximum OD600 of >8.0 in 12 h. A protein BLAST (BLASTp) search of the P. thermoglucosidasius genome identified three potential candidates involved in catabolism. The first candidate AcuABC (locus BCV53_18410-18420) is annotated as acetoin utilization gene ABC and comprises a polycistronic operon with AcuA separated by 34 bp from AcuB and AcuB overlapped by 4 bp with AcuC. The further two candidates were annotated as (acetyl-transferring) pyruvate dehydrogenase complex (PDHC) E1 component subunits (BCV53_10285 and 10935).
The AcuABC proteins share 80%, 55% and 70% sequence identity with the B. subtilis acetoin utilization proteins, AcuA (ACDH), AcuB, and AcuC (histone deacetylase), one of two separate operons (AcuABC and AcoABCL) responsible for acetoin consumption in this bacillus (Grundy et al., 1993; Ali et al., 2001). Interestingly, 88 bp 3’ of the P. thermoglucosidasius NICMB 11955 acuC gene is the a gene coding for the catabolite control protein A (CCPA), which in B. subtilis was reported to negatively regulate acetoin utilization by repression of AcoR, the transcriptional activator of AcuABC (Ali et al., 2001; Peng et al., 2020).
The two identified PDHCs belong to the 2-oxoacid hydrogenase complex (OADHCs.) family, a super-family containing many different dehydrogenase complexes, including the acetoin dehydrogenase system (ADHC) (Perham, 2003). The NCBI annotation pipeline is based upon sequence identity and domain predictions (Tatusova et al., 2016), therefore annotation can be misleading. On closer inspection, both PDCs showed sequence identity (75% and 46%) to B. subtilis Acetoin:2,6-dichlorophenolindophenol (DCPLP) oxidoreductase subunit alpha. A study by Oppermann et al. (1991), suggests that the alpha subunit is likely the catalytic subunit responsible for the cleavage of acetoin into acetate and acetaldehyde. Hence, we decided to test this hypothesis by deleting the encoding genes, using CRISPR/Cas, and assessing the effect on acetoin utilization. For the purpose of this study we renamed the two PDHC (BCV53_10285 and 10935) as AcoB1 and AcoB2.
To counter acetoin utilization the mutant strains generated were LS106 (ΔacoB1), LS107 (ΔacoB2), LS108 (ΔacuA), LS109 (ΔacuB), LS110 (ΔacuC) and LS111 (ΔacuA-C). Deletion of acoB1 had a dramatic effect, with no growth evident on media containing either acetoin or 2,3-BDO as the sole carbon source. In contrast, all other mutants had a similar growth profile to the WT. Using glucose, slower growth was observed with all mutants, and the maximum OD600 of LS106 (ΔacoB1) and LS111 (ΔacuC) were slightly reduced compared to WT. For further validation, we expressed the AcoB1 gene in the ΔacoB1 strain under the control of the Pldh promoter using the expression plasmid pMTL61110_pldh. Complementation of the WT growth profile were observed for both acetoin and 2,3-BDO (Figure 6).
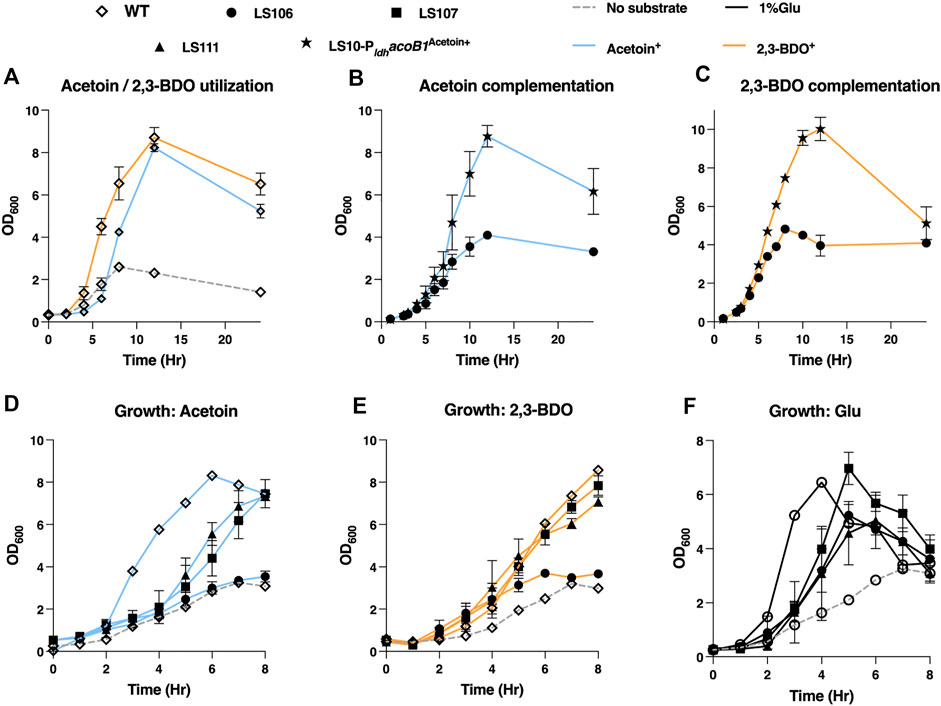
FIGURE 6. Testing the ability of P. thermoglusidasius NCIMB 119 to consume acetoin and/or 2,3-BDO. (A) Growth curve of P. thermoglusidasius NCIMB 11955 on acetoin (AC) and 2,3-BDO (BDO) as the sole source of carbon. (B) Growth on acetoin of NCIMB 11955 deletion mutants ΔacoB1 (LS106), ΔacoB2 (LS107) and ΔacuA-C (LS111). (C) Growth on 2,3-BDO of LS106, LS107 and LS111. (D) Growth of LS106, LS107 and LS111 on glucose. (E) Complementation of LS106 growth on acetoin through plasmid-based overexpression the acoB1 gene. (F) Complementation of LS106 growth on 2,3-BDO through plasmid-based overexpression the acoB1 gene. In all cases, time zero = inoculation of overnight liquid culture to OD600 of 0.25–0.5. Cells were propagated at 60°C in CBM with a 1% (w/v) carbon source and shaking (250 rpm) in 250 mL baffled flasks. CBM without a carbon source was used as negative control. Error bars represent the standard deviation of three biological replicates.
3.7 Enhanced acetoin production using ΔacoB1 strains
Following the identification of the acetoin utilization gene acoB1, it was knocked out in the five ΔadhE strains generating five replicate, double knock-out mutants designated LS300, LS301, LS303, LS304 and LS305 (Table 2). These were transformed with plasmid pMTL-BD3, grown in 2 mL fermentation volumes and the acetoin and 2,3-BDO levels measured. A significant increase was observed, with up to 97 mM (acetoin + 2,3-BDO) produced after 48 h, representing 87% of all carbon flux. While 2,3-BDO levels remained at around 10 mM in all five strains, acetoin production was greatly enhanced with the highest titre of 7.7 g/L (87 mM) in strain LS305, equating to a yield of 0.38 g/g glucose (78% TMY) (Table 4).
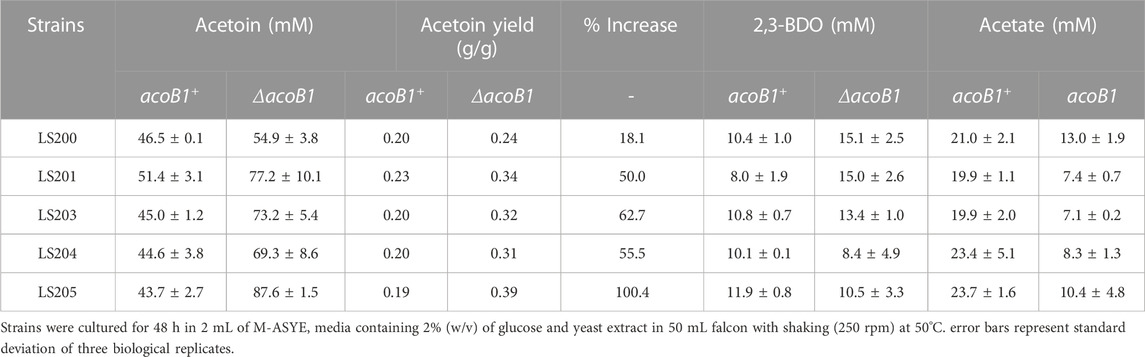
TABLE 4. Comparison of key metabolite profiles of LS200 (ΔadhE) strains carrying pMTL-BD3 with and without deletion of acoB1 under aerobic conditions.
With nearly 90% of the carbon entering the acetoin/2,3-BDO pathway, we hypothesized that abolishing the production of 2,3-BDO by deletion of bdh would further enhance acetoin production. The bdh gene was therefore knocked in the five ΔacoB1/ΔadhE double mutant strains generating strains LS400, LS401, LS403, LS404 and LS405 (Table 2). Production of 2,3-BDO in these strains was entirely abolished, however, acetoin titres also decreased. To investigate whether oxygen supply was the limiting factor, baffled shake-flasks were used in place of falcon-tube concentration (Supplementary Figure S2). Acetoin titres, however, remained essentially the same.
3.8 Disruption of acetate kinase
Thus far, for both acetoin and 2,3-BDO production, acetate was the only by-product measured that was still produced in significant amounts. In other studies, disruption of acetate production has been shown to enhanced 2,3-BDO yields (Jantama et al., 2015). Accordingly, the genes encoding phosphate acetyltransferase (pta) (BCV53_03505) and acetate kinase (ack) (BCV53_18310) were targeted for deletion in strains LS205 and LS305. Only ack mutants, LS206 and LS306, were obtained. Both strains were transformed with pMTL-BD3 to allow investigation of the effect of the ack mutation on acetoin/2,3-BDO production.
LS306 (pMTL-BD3) failed to grow under the conditions tested. This may be partly due the level of acetyl-CoA in this strain being insufficient. Acetyl-CoA can be reassimilated through the AcDH-mediated conversion of acetoin to acetaldehyde using cofactor CoA and NAD+. Acetaldehyde is further oxidised to acetyl-CoA through the activity of acetaldehyde dehydrogenase (AldDH) (Wolfe, 2005; Extance et al., 2016). Analysis of the P. thermoglucosidasius NICMB 11955 genome identified four putative genes encoding an AldDH, namely, BCV53_01645, BVC53_06570, BVC53_ 12995 and BVC53_14470. Alternatively, acetyl-CoA re-assimilation can occur either via the combined action of PtA and AckA or through acetyl-CoA synthetase (ACS) (Pinhal et al., 2019; Meng et al., 2021). Since most routes from pyruvate to acetyl-CoA are disrupted in our strain, and with carbon fluxes diverted towards the acetoin/2,3-BDO pathway, the re-assimilation pathways might be of vital importance to provide cells with sufficient acetyl-CoA for the TCA cycle and biosynthesis.
In LS206 (pMTL-BD3), 2,3-BDO the titre was comparable to that of LS205 habouring pMTL-BD3b (LS205b) when using 5–7.5 mL fermentation volumes, and much higher under more oxygen-limited conditions (Table 3). However, 2,3-BDO yields were slightly lower in LS206 (pMTL-BD3) compared to LS205b and more glucose was consumed for similar amounts of 2,3-BDO produced. Despite disruption of ack, acetate was still present, though at a slightly lower level compared to LS205b, probably due to the presence of alternative routes to its formation, involving, for instance, acetyl-CoA ligase (Pineda et al., 2016) and acetyl-CoA hydrolase (Lee et al., 1996), both of which are present in the P. thermoglucosidasius NICMB 11955 genome (BCV53_18295; BCV53_18405 and BCV53_05785). Next, 2,3-BDO level was measured in LS206b. Again, the decoupling of bdh expression positively affected 2,3-BDO production, with up to 73 mm obtained, representing 0.33 g/g yield, the highest yield obtained in this study and 66% of the TMY. In addition, up to 25.9 mM acetoin was obtained, indicating that up to 89% of total carbon flux went into the acetoin/2,3-BDO pathway (Figure 7C).
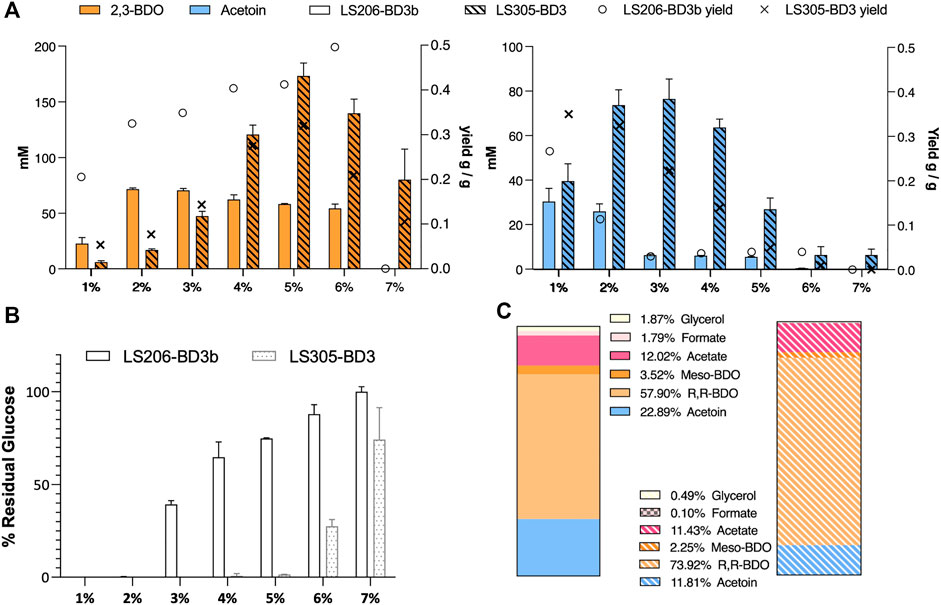
FIGURE 7. The effect of glucose concentration on acetoin and 2,3-BDO production. (A) Acetoin and 2,3-BDO levels under different glucose concentrations. Yield (right Y-axis) is gram of product produced per gram of glucose consumed. 2,3-BDO yields increase while acetoin yields decrease with higher glucose concentrations. More glucose was consumed under more aerated condition (2 mL fermentation volume, strain LS305 habouring pMTL-BD3), and less under oxygen limitation (6 mL fermentation volume, LS206 habouring pMTL-BD3b) (B) At 7% glucose and above, cells became unviable. Error bars represent the standard deviation of three biological replicates. (C) Percentage product profiles of strain LS206 [pMTL-BD3b] grown at 50°C for 48 h, with shaking (250 rpm), in 6 mL M-ASYE with 2% (w/v) glucose (plain) and LS305 [pMTL-BD3] in 2 mL M-ASYE with 5% (w/v) glucose (stripped). The majority of carbon flux was in the acetoin/2,3-BDO pathway (84% and 88%) with 2,3-BDO being the predominant product (61.4% and 76%). Values are the means of three biological replicates. BDO = 2,3-butanediol and AC = Acetoin.E.
3.9 Effect of glucose concentration on acetoin and 2,3-BDO production
Finally, we sought to investigate the effect of different concentrations of glucose on our engineered strains. Not only it is important that the strains can tolerate a range of glucose concentrations for industrial flexibility, different glucose concentrations tend to introduce significant changes in the cell physiology such as increased osmotic stress (Gonzalez et al., 2016), inhibition and activation of certain metabolic pathways (Guidi et al., 2010). In general, glucose concentration positively affected 2,3-BDO titres but hampered acetoin production. In LS206 (pMTL-BD3b), for instance, using 6 mL fermentation volumes, a near linear relationship in 2,3-BDO yield was observed with increasing glucose concentration, with up to 0.5 g/g in 6% glucose, representing near 100% TMY. However, 2,3-BDO titres plateaued at 2% glucose, with less glucose being utilized above this concentration. It seems that excessive glucose negatively affects catabolism. In LS305 (pMTL-BD3), the use of small (2 mL) culture volume alleviated the constraints on glucose utilization, which was completely utilized at concentrations up to 5% glucose. Increases in 2,3-BDO yield mirrored the trend seen with LS206 (pMTL-BD3b), with titres peaking at 0.32 ± 0.2 g/g when the glucose concentration was 5%, representing a titre of 15.6 ± 1.4 g/L, the highest reported in Geobacillus (Figure 7).
4 Discussion
Acetoin and 2,3-BDO are biologically related chemicals of significant commercial interest. Their bioproduction using microorganisms may provide a more carbon neutral alternative to the traditional petrochemical routes. Here, through rational metabolic engineering, the thermophilic chassis P. thermoglucosidasius NCIMB 11955 has been established as a platform organism for bioproduction of both acetoin and 2,3-BDO.
Although previous work has demonstrated the production of 2,3-BDO in P. thermoglucosidasius DSM 2542 (Zhou et al., 2020), the authors reported inherent problems with redox imbalance and failed to solve the issue of by-product (ethanol) formation. It has been proposed that the physiological role of the 2,3-BDO pathway is to supply NADH for elimination of the toxic acetate produced during overflow metabolism, channelling reducing power to ethanol production (Meng et al., 2021). This suggests the formation of 2,3-BDO and ethanol occurs at a 1:1 ratio, which is what we observed in all the adhE mutants carrying the 2,3-BDO pathway. Disruption of the ethanol pathway resulted in increased acetate titres and decreased 2,3-BDO production. It is important to minimize by-product formation, as not only is it wasteful of substrate, it may also complicate downstream extraction adding to processing costs. We confronted the redox issue through adjusted aeration and decoupling the expression of the native bdh, which we identified and characterized. Along with disruptions of genes targeting metabolites of the mixed fermentation pathway (including lactate, glycerol, formate and acetate) 2,3-BDO is produced as the major product at 6.6 g/L, representing a yield of 0.33 g/g and 66% of TMY. Although this failed to exceed the 7.2 g/L titre in P. thermoglucosidasius DSM 2542 reported previously (Zhou et al., 2020), ethanol production is completed eliminated, and only acetate and acetoin levels exceeded 10 mM. Amongst all the metabolites measured, 61% was 2,3-BDO, of which 94% was the R,R-enantiomer. Along with 23% acetoin, carbon flux to the acetoin/2,3-BDO route altogether represents 84% of total metabolites, with 89% carbon from glucose converted, giving scope for further improvement.
Product utilization was also investigated. Many bacterial species, including B. subtilis, Micrococcus urea, Alcaligenes eutrophus, Enterococcus faecalis, Pelobacter carbinolicus, Klebsiella pneumoniae and Clostridium magnum, are able to degrade acetoin (Ali et al., 2001). Acetoin catabolism in P. thermoglucosidasius, and thermophilic microorganisms in general, has not been described. Within any bioprocesses, it is important to ensure that the generated compounds are not re-metabolised by the producing organisms (Arenas-López et al., 2019). Unfortunately, NCIMB 11955 was found to utilize both acetoin and 2,3-BDO as sole carbon source for growth, a property that required elimination.
Two independent pathways of acetoin catabolism have been proposed in B. subtilis, one involving the AcuABC operon (Grundy et al., 1993) and the other involving the Aco operon encoding the multicomponent acetoin dehydrogenase complex (Huang et al., 1999). Putative genes for both pathways are present in the NCIMB 11955 strain, but acetoin appears to be catabolized solely via the acetoin dehydrogenase (AcDH) route, as deletion of the AcuABC operon in failed to prevent acetoin utilization, while a single deletion of the AcoB1 gene abolished growth on both acetoin and 2,3-BDO.
AcDH belongs to the family of 2-oxo acid dehydrogenase complexes which uses the reducing power of NAD+ to convert 2-oxo acids to the corresponding acyl-CoA derivatives and produce NADH and CO2 (De Kok et al., 1998). During 2,3-BDO fermentation, when oxygen is limiting and NAD+ are not recycled due to only one produced via the acetoin/2,3-BDO route, AcDH is likely inactive. However, with more aeration, NAD+ availability would be sufficient to drive acetoin catabolism, which can affect product titre. Therefore, we hypothesized that deletion of the AcoB1 gene would have profound implications in more aerobic conditions and less or no effect under oxygen limitation, hence more relevant for acetoin production. Data using the acoB1 mutant supports this: at both 5 mL and 10 mL fermentation volumes, 2,3-BDO produced was comparable in strains LS205 (without acoB1 deletion) and LS305 (with acoB1 deletion). However, when using 2 mL, acetoin production increased from 43.7 ± 2.7 mM to 86.0 ± 0.1 mM, respectively, for the same strains, representing a near 100% increase in titre.
Further strategies to enhance acetoin production, including deletion of bdh, and varied glucose concentration were unsuccessful. Deletion of the native bdh gene resulted in lower acetoin production levels, while its production decreased proportionally to glucose concentration, suggesting that unknown mechanisms such as osmotic stress can regulate 2,3-BDO and acetoin biosynthesis. Remarkably, under aerobic conditions, the predominant metabolite in LS305 shifted from acetoin to 2,3-BDO with increasing glucose concentration, amounting to 15.6 g/L at 5% glucose, representing 64% TMY. The yield obtained here is comparable to what was achieved with LS206 (pMTL-BD3b), (0.32 g/g vs. 0.33 g/g), with the benefit of being able to metabolize more glucose. Moreover, carbon fluxes to the acetoin/2,3-BDO pathway is also comparable, if not better, compared to LS206 (pMTL-BD3b), (88% vs. 84% total metabolites), with 76% being 2,3-BDO, of which 97% is the R-R enantiomer (Figure 7C). These data underline the benefit of incorporating a acoB1 deletion mutation in strains to support high 2,3-BDO titres under more aerated conditions.
5 Conclusion
In conclusion, we have created platform strains for the production of acetoin and 2,3-BDO at thermophilic temperatures. By addressing issues of redox imbalance and product utilization, substantial 2,3-BDO was produced with minimal by-product formation. In parallel, for the first time in the industrially relevant strain P. thermoglucosidasius NCIMB 11955 we demonstrated high yields of acetoin, on par with the highest reported in the literature. This work also serves as a foundation for the optimization of fermentation at a larger scale, with much potential for further improvement.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
NM, YZ, and LS conceptualized the study. LS designed and carried out the experiments. AM contributed in performing the experimental work. ML provided tools and guidance in their use. Funding acquisition was by NM and YZ. The manuscript was drafted by LS and revised by AM, ML, and NM. All authors approved the final version.
Funding
This work was supported by the UK Biotechnology and Biological Sciences Research Council (grant number BB/N022718/1), the IB Catalyst grant MAXIBIO, and through the award of a BBSRC CASE studentship [grant number BB/L016478/1] to ML. supported by ATUM (formerly DNA2.0). AM was supported by the Petroleum Technology Development Fund, Nigeria [PTDF/ED/PHD/MA/1334/18] and by MAXIBIO.
Acknowledgments
We thank David Tooth and Matthew Abbott for assistance with HPLC–UV analysis, and all members of SBRC who helped to carry out this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1191079/full#supplementary-material
References
Albers, E., Larsson, C., Lidén, G., Niklasson, C., and Gustafsson, L. (1996). Influence of the nitrogen source on Saccharomyces cerevisiae anaerobic growth and product formation. Appl. Environ. Microbiol. 62, 3187–3195. doi:10.1128/aem.62.9.3187-3195.1996
Ali, N. O., Bignon, J., Rapoport, G., and Debarbouille, M. (2001). Regulation of the acetoin catabolic pathway is controlled by Sigma L in Bacillus subtilis. Bacillus subtilis J. Bacteriol. 183, 2497–2504. doi:10.1128/jb.183.8.2497-2504.2001
Allen, J. G., Flanigan, S. S., LeBlanc, M., Vallarino, J., MacNaughton, P., Stewart, J. H., et al. (2016). Flavoring chemicals in e-cigarettes: Diacetyl, 2,3-pentanedione, and acetoin in a sample of 51 products, including fruit-candy-and cocktail-flavored e-cigarettes. Environ. Health Perspect. 124, 733–739. doi:10.1289/ehp.1510185
Arenas-López, C., Locker, J., Orol, D., Walter, F., Busche, T., Kalinowski, J., et al. (2019). The genetic basis of 3-hydroxypropanoate metabolism in Cupriavidus necator H16. Biotechnol. Biofuels 12, 150–216. doi:10.1186/s13068-019-1489-5
Bao, T., Zhang, X., Rao, Z., Zhao, X., Zhang, R., Yang, T., et al. (2014). Efficient whole-cell biocatalyst for acetoin production with NAD+ regeneration system through homologous Co-expression of 2,3-butanediol dehydrogenase and NADH oxidase in engineered Bacillus subtilis. PLoS One 9, e102951. doi:10.1371/journal.pone.0102951
Bartosiak-Jentys, J., Hussein, A. H., Lewis, C. J., and Leak, D. J. (2013). Modular system for assessment of glycosyl hydrolase secretion in Geobacillus thermoglucosidasius. Microbiol 159, 1267–1275. doi:10.1099/mic.0.066332-0
Bashir, Z., Sheng, L., Anil, A., Lali, A., Minton, N. P., and Zhang, Y. (2019). Engineering Geobacillus thermoglucosidasius for direct utilisation of holocellulose from wheat straw. Biotechnol. Biofuels 12, 199. doi:10.1186/s13068-019-1540-6
Berka, R. M., Grigoriev, I. V., Otillar, R., Salamov, A., Grimwood, J., Reid, I., et al. (2011). Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat. Biotechnol. 29, 922–927. doi:10.1038/nbt.1976
Biswas, R., Yamaoka, M., Nakayama, H., Kondo, T., Yoshida, K. I., Bisaria, V. S., et al. (2012). Enhanced production of 2,3-butanediol by engineered Bacillus subtilis. Appl. Microbiol. Biotechnol. 94, 651–658. doi:10.1007/s00253-011-3774-5
Chang, T., and Yao, S. (2011). Thermophilic, lignocellulolytic bacteria for ethanol production: Current state and perspectives. Appl. Microbiol. Biotechnol. 92, 13–27. doi:10.1007/s00253-011-3456-3
Chemicals and Materials Market Research Report, 2019. 2, 3-butanediol market (application: Intermediate chemicals, plastics, food additives, cosmetics and others)-global industry analysis, size, share, growth, trends and forecast, 2019- 2027. Available at: https://www.transparencymarketresearch.com/2-3-butanediol-market.html (Accessed: February 10, 2020).
Chinn, M., and Mbaneme, V. (2015). Consolidated bioprocessing for biofuel production: Recent advances. Energy Emiss. Control Technol. 3, 23–44. doi:10.2147/eect.s63000
Cho, S., Kim, T., Woo, H. M., Lee, J., Kim, Y., and Um, Y. (2015). Enhanced 2,3-butanediol production by optimizing fermentation conditions and engineering Klebsiella oxytoca M1 through overexpression of acetoin reductase. PLoS One 10, e0138109. doi:10.1371/journal.pone.0138109
Coleman, G. A., Pancost, R. D., Williams, T. A., and Dagan, T. (2019). Investigating the origins of membrane phospholipid biosynthesis genes using outgroup-free rooting. Genome Biol. Evol. 11, 883–898. doi:10.1093/gbe/evz034
Cripps, R. E., Eley, K., Leak, D. J., Rudd, B., Taylor, M., Todd, M., et al. (2009). Metabolic engineering of Geobacillus thermoglucosidasius for high yield ethanol production. Metab. Eng. 11, 398–408. doi:10.1016/j.ymben.2009.08.005
De Kok, A., Hengeveld, A. F., Martin, A., and Westphal, A. H. (1998). The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria. Biochim. Biophys. Acta 1385, 353–366. doi:10.1016/s0167-4838(98)00079-x
De Mas, C., Jansen, N. B., and Tsao, G. T. (1988). Production of optically active 2, 3-butanediol byBacillus polymyxa. Bacillus polymyxa Biotechnol. Bioeng. 31, 366–377. doi:10.1002/bit.260310413
Dulieu, C., and Poncelet, D. (1999). Spectrophotometric assay of α-acetolactate decarboxylase. Enzyme Microb. Technol. 25, 537–542. doi:10.1016/s0141-0229(99)00079-4
Elisáková, V., Pátek, M., Holátko, J., Nesvera, J., Leyval, D., Goergen, J.-L., et al. (2005). Feedback-resistant acetohydroxy acid synthase increases valine production in Corynebacterium glutamicum. Appl. Environ. Microbiol. 71, 207–213. doi:10.1128/aem.71.1.207-213.2005
Erian, A. M., Gibisch, M., and Pflügl, S. (2018). Engineered E. coli W enables efficient 2,3-butanediol production from glucose and sugar beet molasses using defined minimal medium as economic basis. Microb. Cell Fact. 17, 190–217. doi:10.1186/s12934-018-1038-0
Extance, J., Crennell, S. J., Eley, K., Cripps, R., Hough, D. W., and Danson, M. J. (2013). Structure of a bifunctional alcohol dehydrogenase involved in bioethanol generation in Geobacillus thermoglucosidasius. Acta Crystallogr. Sect. D. Biol. Crystallogr. 69, 2104–2115. doi:10.1107/s0907444913020349
Extance, J., Danson, M. J., and Crennell, S. J. (2016). Structure of an acetylating aldehyde dehydrogenase from the thermophilic ethanologen Geobacillus thermoglucosidasius. Protein Sci. 25, 2045–2053. doi:10.1002/pro.3027
Finlay, W. J. J., Logan, N. A., and Sutherland, A. D. (2000). Bacillus cereus produces most emetic toxin at lower temperatures. Lett. Appl. Microbiol. 31, 385–389. doi:10.1046/j.1472-765x.2000.00835.x
Fu, J., Huo, G., Feng, L., Mao, Y., Wang, Z., Ma, H., et al. (2016). Metabolic engineering of Bacillus subtilis for chiral pure meso-2,3-butanediol production. Biotechnol. Biofuels 9, 90. doi:10.1186/s13068-016-0502-5
Gonzalez, R., Morales, P., Tronchoni, J., Cordero-Bueso, G., Vaudano, E., Quirós, M., et al. (2016). New genes involved in osmotic stress tolerance in Saccharomyces cerevisiae. Front. Microbiol. 7, 1545–1612. doi:10.3389/fmicb.2016.01545
Grundy, F. J., Waters, D. A., Takova, T. Y., and Henkin, T. M. (1993). Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol. Microbiol. 10, 259–271. doi:10.1111/j.1365-2958.1993.tb01952.x
Guidi, F., Magherini, F., Gamberi, T., Borro, M., Simmaco, M., Modesti, A., et al. (2010). Effect of different glucose concentrations on proteome of Saccharomyces cerevisiae. Biochim. Biophys. Acta - Proteins Proteomics 1804, 1516–1525. doi:10.1016/j.bbapap.2010.03.008
Häßler, T., Schieder, D., Pfaller, R., Faulstich, M., and Sieber, V. (2012). Enhanced fed-batch fermentation of 2,3-butanediol by Paenibacillus polymyxa DSM 365. Bioresour. Technol. 124, 237–244. doi:10.1016/j.biortech.2012.08.047
Huang, M., Oppermann-Sanio, F. B., and Steinbüchel, A. (1999). Biochemical and molecular characterization of the Bacillus subtilis acetoin catabolic pathway. J. Bacteriol. 181, 3837–3841. doi:10.1128/jb.181.12.3837-3841.1999
Hubmann, G., Guillouet, S., and Nevoigt, E. (2011). Gpd1 and Gpd2 fine-tuning for sustainable reduction of glycerol formation in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 77, 5857–5867. doi:10.1128/aem.05338-11
Hussein, A. H., Lisowska, B. K., and Leak, D. J. (2015). The genus Geobacillus and their biotechnological potential. Adv. Appl. Microbiol. 92, 1–48. doi:10.1016/bs.aambs.2015.03.001
Imachi, H., Sekiguchi, Y., Kamagata, Y., Hanada, S., Ohashi, A., and Harada, H. (2002). Pelotomaculum thermopropionicum gen. nov., sp. nov., an anaerobic, thermophilic, syntrophic propionate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 52, 1729–1735. doi:10.1099/00207713-52-5-1729
Jantama, K., Polyiam, P., Khunnonkwao, P., Chan, S., Sangproo, M., Khor, K., et al. (2015). Efficient reduction of the formation of by-products and improvement of production yield of 2,3-butanediol by a combined deletion of alcohol dehydrogenase, acetate kinase-phosphotransacetylase, and lactate dehydrogenase genes in metabolically engineered Klebsiella oxytoca in mineral salts medium. Metab. Eng. 30, 16–26. doi:10.1016/j.ymben.2015.04.004
Kandasamy, V., Liu, J., Dantoft, S. H., Solem, C., and Jensen, P. R. (2016). Synthesis of (3R)-acetoin and 2,3-butanediol isomers by metabolically engineered Lactococcus lactis. Sci. Rep. 6, 36769–9. doi:10.1038/srep36769
Keller, M., Loder, A., Basen, M., Izquierdo, J., Kelly, R. M., and Adams, M. W. W. (2014). Production of lignofuels and electrofuels by extremely thermophilic microbes. Biofuels 5, 499–515. doi:10.1080/17597269.2014.996729
Kellner, N., Schwarz, J., Sturm, M., Fernandez-Martinez, J., Griesel, S., Zhang, W., et al. (2016). Developing genetic tools to exploit Chaetomium thermophilum for biochemical analyses of eukaryotic macromolecular assemblies. Sci. Rep. 6, 20937–21010. doi:10.1038/srep20937
Kim, T., Cho, S., Lee, S. M., Woo, H. M., Lee, J., Um, Y., et al. (2016). High production of 2,3-Butanediol (2,3-BD) by Raoultella ornithinolytica B6 via optimizing fermentation conditions and overexpressing 2,3-BD Synthesis Genes. PLoS One 11, 01650766–e165115. doi:10.1371/journal.pone.0165076
Lau, M. S. H., Sheng, L., Zhang, Y., and Minton, N. P. (2021). Development of a suite of tools for genome editing in Parageobacillus thermoglucosidasius and their use to identify the potential of a native plasmid in the generation of stable engineered strains. ACS Synth. Biol. 10, 1739–1749. doi:10.1021/acssynbio.1c00138
Lechner, A., Brunk, E., and Keasling, J. D. (2016). The need for integrated approaches in metabolic engineering. Cold Spring Harb. Perspect. Biol. 8, a023903–a023918. doi:10.1101/cshperspect.a023903
Lee, F. J. S., Lin, L. W., and Smith, J. A. (1996). Acetyl-CoA hydrolase involved in acetate utilization in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1297, 105–109. doi:10.1016/0167-4838(96)00109-4
Li, C., Gai, Z., Wang, K., and Jin, L. (2017a). Engineering Bacillus licheniformis as a thermophilic platform for the production of l-lactic acid from lignocellulose-derived sugars. Biotechnol. Biofuels 10, 235. doi:10.1186/s13068-017-0920-z
Li, C., Wei, X., Yu, W., Wen, Z., and Chen, S. (2017b). Enhancement of acetoin production from Bacillus licheniformis by 2,3-butanediol conversion strategy: Metabolic engineering and fermentation control. Process Biochem. 57, 35–42. doi:10.1016/j.procbio.2017.03.027
Li, L., Zhang, L., Li, K., Wang, Y., Gao, C., Han, B., et al. (2013). A newly isolated Bacillus licheniformis strain thermophilically produces 2,3-butanediol, a platform and fuel bio-chemical. Biotechnol. Biofuels 6, 123. doi:10.1186/1754-6834-6-123
Lin, P. P., Rabe, K. S., Takasumi, J. L., Kadisch, M., Arnold, F. H., and Liao, J. C. (2014). Isobutanol production at elevated temperatures in thermophilic Geobacillus thermoglucosidasius. Metab. Eng. 24, 1–8. doi:10.1016/j.ymben.2014.03.006
Loftie-Eaton, W., Taylor, M., Horne, K., Tuffin, M. I., Burton, S. G., and Cowan, D. A. (2013). Balancing redox cofactor generation and ATP synthesis: Key microaerobic responses in thermophilic fermentations. Biotechnol. Bioeng. 110, 1057–1065. doi:10.1002/bit.24774
Luo, Q., Wu, J., and Wu, M. (2014). Enhanced acetoin production by Bacillus amyloliquefaciens through improved acetoin tolerance. Process Biochem. 49, 1223–1230. doi:10.1016/j.procbio.2014.05.005
Maina, S., Prabhu, A. A., Vivek, N., Vlysidis, A., Koutinas, A., and Kumar, V. (2022). Prospects on bio-based 2,3-butanediol and acetoin production: Recent progress and advances. Biotechnol. Adv. 54, 107783. doi:10.1016/j.biotechadv.2021.107783
Medina, V. G., Almering, M. J. H., Van Maris, A. J. A., and Pronk, J. T. (2010). Elimination of glycerol production in anaerobic cultures of a Saccharomyces cerevisiae strain engineered to use acetic acid as an electron acceptor. Appl. Environ. Microbiol. 76, 190–195. doi:10.1128/aem.01772-09
Meng, W., Zhang, L., Cao, M., Zhang, Y., Zhang, Y., Li, P., et al. (2021). 2,3-Butanediol synthesis from glucose supplies NADH for elimination of toxic acetate produced during overflow metabolism. Cell Discov. 7, 43. doi:10.1038/s41421-021-00273-2
Nicholson, W. L. (2008). The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Appl. Environ. Microbiol. 74, 6832–6838. doi:10.1128/aem.00881-08
Ohshiro, T., Aisaka, K., and Uwajima, T. (1989). Purification and characterization of α-acetolactate decarboxylase from Brevibacterium acetylicum. Agric. Biol. Chem. 53, 1913–1918. doi:10.1271/bbb1961.53.1913
Oppermann, F. B., Schmidt, B., and Steinbuchel, A. (1991). Purification and characterization of acetoin:2,6-dichlorophenolindophenol oxidoreductase, dihydrolipoamide dehydrogenase, and dihydrolipoamide acetyltransferase of the Pelobacter carbinolicus acetoin dehydrogenase enzyme system. J. Bacteriol. 173, 757–767. doi:10.1128/jb.173.2.757-767.1991
Peng, Q., Zhao, X., Wen, J., Huang, M., Zhang, J., and Song, F. (2020). Transcription in the acetoin catabolic pathway is regulated by AcoR and CcpA in Bacillus thuringiensis. Microbiol. Res. 235, 126438. doi:10.1016/j.micres.2020.126438
Perham, R. N. (2003). Swinging arms and swinging domains in multifunctional enzymes: Catalytic machines for multistep reactions. Ann. Rev. Biochem. 69, 961–1004. doi:10.1146/annurev.biochem.69.1.961
Piattoni, C. V., Figueroa, C. M., Asención Diez, M. D., Parcerisa, I. L., Antuña, S., Comelli, R. A., et al. (2013). Production and characterization of Escherichia coli glycerol dehydrogenase as a tool for glycerol recycling. Process Biochem. 48, 406–412. doi:10.1016/j.procbio.2013.01.011
Pineda, E., Vázquez, C., Encalada, R., Nozaki, T., Sato, E., Hanadate, Y., et al. (2016). Roles of acetyl-CoA synthetase (ADP-forming) and acetate kinase (PPi-forming) in ATP and PPi supply in Entamoeba histolytica. Biochim. Biophys. Acta 1860, 1163–1172. doi:10.1016/j.bbagen.2016.02.010
Pinhal, S., Ropers, D., Geiselmann, J., and De Jong, H. (2019). Acetate metabolism and the inhibition of bacterial growth by acetate. J. Bacteriol. 201, 001477–e219. doi:10.1128/jb.00147-19
Qureshi, N., and Cheryan, M. (1989). Production of 2,3-butanediol by Klebsiella oxytoca. Appl. Microbiol. Biotechnol. 30, 440–443. doi:10.1007/bf00263847
Radoš, D., Carvalho, A. L., Wieschalka, S., Neves, A. R., Blombach, B., Eikmanns, B. J., et al. (2015). Engineering Corynebacterium glutamicum for the production of 2,3-butanediol. Microb. Cell Fact. 14, 171. doi:10.1186/s12934-015-0362-x
Ramachandran, K. B., and Goma, G. (1988). 2,3-Butanediol production from glucose by Klebsiella pneumoniae in a cell recycle system. J. Biotechnol. 9, 39–46. doi:10.1016/0168-1656(88)90013-2
Reeve, B., Martinez-Klimova, E., de Jonghe, J., Leak, D. J., and Ellis, T. (2016). The Geobacillus plasmid set: A modular toolkit for thermophile engineering. ACS Synth. Biol. 5, 1342–1347. doi:10.1021/acssynbio.5b00298
Schilling, C., Ciccone, R., Sieber, V., and Schmid, J. (2020). Engineering of the 2,3-butanediol pathway of Paenibacillus polymyxa DSM 365. Metab. Eng. 61, 381–388. doi:10.1016/j.ymben.2020.07.009
Sheng, L., Kovács, K., Winzer, K., Zhang, Y., and Minton, N. P. (2017). Development and implementation of rapid metabolic engineering tools for chemical and fuel production in Geobacillus thermoglucosidasius NCIMB 11955. Biotechnol. Biofuels 10, 5. doi:10.1186/s13068-016-0692-x
Song, C. W., Park, J. M., Chung, S. C., Lee, S. Y., and Song, H. (2019). Microbial production of 2,3-butanediol for industrial applications. J. Ind. Microbiol. Biotechnol. 46, 1583–1601. doi:10.1007/s10295-019-02231-0
Su, F., and Xu, P. (2014). Genomic analysis of thermophilic Bacillus coagulans strains: Efficient producers for platform bio-chemicals. Sci. Rep. 4, 3926–4010. doi:10.1038/srep03926
Tatusova, T., Dicuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucl. Acids Res. 44, 6614–6624. doi:10.1093/nar/gkw569
Wang, L., Cai, Y., Zhu, L., Guo, H., and Yu, B. (2014). Major role of NAD-dependent lactate dehydrogenases in the production of L-lactic acid with high optical purity by the thermophile Bacillus coagulans. Appl. Environ. Microbiol. 80, 7134–7141. doi:10.1128/aem.01864-14
Wang, L., Wang, J., Shi, H., Gu, H., Zhang, Y., Li, X., et al. (2016). Characterization of glycerol dehydrogenase from Thermoanaerobacterium thermosaccharolyticum DSM 571 and GGG motif identification. J. Microbiol. Biotechnol. 26, 1077–1086. doi:10.4014/jmb.1512.12051
Weinstock, O., Sella, C., Chipman, D. M., and Barak, Z. (1992). Properties of subcloned subunits of bacterial acetohydroxy acid synthases. J. Bacteriol. 174, 5560–5566. doi:10.1128/jb.174.17.5560-5566.1992
T. A. Werpy, and G. Petersen (Editors) (2004). Top value added chemicals from biomass, volume I: Results of screening for potential candidates from sugars and synthesis gas (United States: Pacific Northwest National Laboratory and National Renewable Energy Laboratory).
Wolfe, A. J. (2005). The acetate switch. Microbiol. Mol. Biol. Rev. 69, 12–50. doi:10.1128/mmbr.69.1.12-50.2005
Xiao, Z., and Lu, J. R. (2014). Strategies for enhancing fermentative production of acetoin: A review. Biotechnol. Adv. 32, 492–503. doi:10.1016/j.biotechadv.2014.01.002
Xiao, Z., Wang, X., Huang, Y., Huo, F., Zhu, X., Xi, L., et al. (2012). Thermophilic fermentation of acetoin and 2,3-butanediol by a novel Geobacillus strain. Biotechnol. Biofuels 5, 88. doi:10.1186/1754-6834-5-88
Xiao, Z., and Xu, P. (2007). Acetoin metabolism in bacteria. Crit. Rev. Microbiol. 33, 127–140. doi:10.1080/10408410701364604
Yang, T., Rao, Z., Zhang, X., Xu, M., Xu, Z., and Yang, S.-T. (2017). Metabolic engineering strategies for acetoin and 2,3-butanediol production: Advances and prospects. Crit. Rev. Biotechnol. 37, 990–1005. doi:10.1080/07388551.2017.1299680
Yang, Z., and Zhang, Z. (2018). Recent advances on production of 2, 3-butanediol using engineered microbes. Biotechnol. Adv. 37, 569–578. doi:10.1016/j.biotechadv.2018.03.019
Zhang, B., Li, X. L., Fu, J., Li, N., Wang, Z., Tang, Y. J., et al. (2016). Production of acetoin through simultaneous utilization of glucose, xylose, and arabinose by engineered Bacillus subtilis. PLoS One 11, 01592988–e159314. doi:10.1371/journal.pone.0159298
Zhang, X., Han, R., Bao, T., Zhao, X., Li, X., Zhu, M., et al. (2019). Synthetic engineering of Corynebacterium crenatum to selectively produce acetoin or 2,3-butanediol by one step bioconversion method. Microb. Cell Fact. 18, 128–212. doi:10.1186/s12934-019-1183-0
Keywords: P. thermoglucosidasius NCIMB 11955, acetoin, 2,3-butanediol, thermophile, CRISPR CAS
Citation: Sheng L, Madika A, Lau MSH, Zhang Y and Minton NP (2023) Metabolic engineering for the production of acetoin and 2,3-butanediol at elevated temperature in Parageobacillus thermoglucosidasius NCIMB 11955. Front. Bioeng. Biotechnol. 11:1191079. doi: 10.3389/fbioe.2023.1191079
Received: 21 March 2023; Accepted: 20 April 2023;
Published: 02 May 2023.
Edited by:
Jeong Chan Joo, Catholic University of Korea, Republic of KoreaReviewed by:
Ji-Sook Hahn, Seoul National University, Republic of KoreaBo Yu, Chinese Academy of Sciences (CAS), China
Eun Ju Eun, Jeonbuk National University, Republic of Korea
Copyright © 2023 Sheng, Madika, Lau, Zhang and Minton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nigel P. Minton, bmlnZWwubWludG9uQG5vdHRpbmdoYW0uYWMudWs=
†Present address: Lili Sheng, Zhejiang Huarui Biotechnology Ltd., Huzhou, China
‡ORCID: Abubakar Madika, https://orcid.org/0000-0002-6224-8998; Matthew S. H. Lau, https://orcid.org/0009-0000-5962-7228; Ying Zhang, https://orcid.org/0000-0003-3074-9998; Nigel P. Minton, https://orcid.org/0000-0002-9277-1261
 Lili Sheng1†
Lili Sheng1† Abubakar Madika
Abubakar Madika Matthew S. H. Lau
Matthew S. H. Lau Ying Zhang
Ying Zhang Nigel P. Minton
Nigel P. Minton