
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 04 May 2023
Sec. Synthetic Biology
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1183354
Synthetic biology combines the disciplines of biology, chemistry, information science, and engineering, and has multiple applications in biomedicine, bioenergy, environmental studies, and other fields. Synthetic genomics is an important area of synthetic biology, and mainly includes genome design, synthesis, assembly, and transfer. Genome transfer technology has played an enormous role in the development of synthetic genomics, allowing the transfer of natural or synthetic genomes into cellular environments where the genome can be easily modified. A more comprehensive understanding of genome transfer technology can help to extend its applications to other microorganisms. Here, we summarize the three host platforms for microbial genome transfer, review the recent advances that have been made in genome transfer technology, and discuss the obstacles and prospects for the development of genome transfer.
Synthetic biology is an area that emerged in the early 21st century, and is based on elucidating and simulating the basic laws of biosynthesis (Kiga and Yamamura, 2008; Zhang et al., 2023). Its main application is the artificial design and construction of new biological systems, such as the establishment of bio-manufacturing pathways for drugs, functional materials, and energy substitutes (Bibi and Ahmed, 2020; Clarke and Kitney, 2020; Burgos-Morales et al., 2021). Genome synthesis is an essential part of this. It enables us to create living cells with fully controllable biological properties by de novo synthesis and assembly of rationally designed genomes (Baby et al., 2019; Labroussaa et al., 2019; Venetz et al., 2019; Zhang et al., 2020; Venter et al., 2022). Advances in synthetic genomics have facilitated the development of new tools and methods for synthesizing, assembling, modifying, and transferring complete microbial genomes (Lu et al., 2018; Liu et al., 2019). However, due to the slow growth rate, insufficient DNA recombination ability, and low transformation efficiency of the organisms, the tools and methods cannot always be performed directly in the original species. Therefore, it is necessary to transfer the genome into model organisms such as Saccharomyces cerevisiae, Escherichia coli, or Bacillus subtilis (Blount, 2015; Nielsen, 2019; Errington and Aart, 2020; Koster et al., 2022; Malci et al., 2022). Model organisms have the advantages of a short life cycle, a clear genetic background, easy cultivation, and a simple experimental procedure, making them very suitable as a platform for genome synthesis. The increasing size of artificially synthesized genomes and species poses a challenge not only for large genome synthesis, but also for genome transfer.
Whole genome transfer is the direct way to obtain complete genomes or eukaryotic chromosomes in other species, but in the case of unsuccessful genome transfer, a stepwise method can also be used to transfer the target genome (Karas et al., 2015). Genome transfer is divided into whole genome transfer and genomic fragment transfer. Whole genome transfer usually involves direct transfer into recipient cells (Karas et al., 2013a; Karas et al., 2014; Baby et al., 2018). For example, the whole genome of Mycoplasma mycoides was transferred into the related species Mycoplasma capricolum, thereby transforming one species into another (Lartigue et al., 2007). In contrast, genomic fragment transfer usually requires model organisms to act as platforms (Gibson et al., 2008b). A whole genome transfer process is divided into three parts: the first part involves the transfer of the entire genome or large genomic fragments into a suitable model organism. The second part is editing and modification, which is carried out using well-established genetic systems in the model organism. The final part is the transfer of the manipulated genome into the recipient cells of interest. The combination of genome transfer technology and genome engineering of model organisms is a powerful approach for manipulating both synthetic and natural microbial genomes (Gibson et al., 2010). If genome transfer technology can be applied to more microbial species, it could revolutionize microbial genetics and produce a new generation of artificially designed microorganisms.
In recent years, whole genome assembly technologies have flourished, with genome transfer playing an important role in this. With this aim in mind, this review sets out to describe the background of microbial genome transfer, especially cross-species transfer, focusing on the genome transfer using three different model organisms as platforms. In addition, we discuss the factors that influence genome transfer and examine its future prospects.
In 2005, it was demonstrated that whole genomes from other organisms could be transferred into B. subtilis (Itaya et al., 2005). The research involved transferring the Synechocystis PCC6803 genome into the genome of B. subtilis cells, resulting in chimeric chromosomes. Subsequent research extended this approach to develop genome transfer methods using B. subtilis as a platform. Then, in 2007, Carole Lartigue et al. achieved the first complete genome transfer from M. mycoides to M. capricolum (Lartigue et al., 2007). In this experiment, the recipient genome was completely replaced by the donor genome. Based on this, the Venter research group achieved the complete chemical synthesis of the Mycoplasma genitalium genome in 2008 (Gibson et al., 2008a; Gibson et al., 2008b). Then, in 2010, the synthetic M. mycoides genome was transferred into M. capricolum cells, producing new Mycoplasma cells that could function normally (Gibson et al., 2010). The researchers used yeast as a temporary and modified platform for the synthetic genome (Gibson et al., 2008a; Gibson et al., 2008b). The cloning of the entire bacterial genome as centromeric plasmids in yeast was a breakthrough, allowing one-step genome transfer. Several extensions of this method have been developed in order to transfer whole prokaryotic genomes or eukaryotic chromosomes (Table 1). Escherichia coli is a commonly used model organism, and has the advantages of a short generation time, combined with simple and well-understood genetic manipulation methods (Ruiz and Silhavy Thomas, 2022). Although no studies have demonstrated the transfer of whole genomes into E. coli, megabase-sized plasmids can nonetheless be stably maintained in E. coli (Mukai et al., 2020). The development of methods to clone and maintain large genomic fragments in E. coli would greatly facilitate genome assembly and transfer technology. As mentioned above, both B. subtilis and S. cerevisiae are useful platforms for genome and chromosome transfer, and E. coli is also an important platform for maintaining the assembly of large DNA fragments.
Bacillus subtilis is a typical platform used in many biotechnology and synthetic biology applications, and has proven itself to be an essential system for genome transfer (Johnston et al., 2014). Bacillus subtilis has the ability to take up exogenous DNA, and the exogenous DNA is usually integrated into the B. subtilis chromosome by RecA-mediated homologous recombination (Yadav et al., 2012; Yadav et al., 2014). Itaya et al. first proposed the use of the B. subtilis genome as a vector (BGM vector) for genomic sequence transfer (Itaya, 1995). They transferred a 48.5 kb length of E. coli prophage λ-DNA into B. subtilis by iterative assembly. The BGM vector is a new cloning and transfer system, and in order to test its ability to clone and transfer large genomic DNA, the same team cloned approximately 120 kb of mouse genomic DNA into BGM. The results showed that the stability of the mouse DNA could be maintained, proving that the BGM vector could at least carry DNA fragments up to 120 kb (Itaya et al., 2000; Itaya et al., 2003). Later, based on the BGM vector, the inchworm elongation method was proposed, in which the positioning and orientation of two DNAs will form an LPS (Landing Pad Sequences, LPS) array (LPA), and as the LPA slides, it leads to elongation of the adjacent target DNA (Itaya et al., 2005). To demonstrate the feasibility of this approach, the whole 3.5 Mb genome of Synechocystis PCC6803 has been completely transferred into the B. subtilis genome. However, the inchworm extension method requires long, contiguous DNA as a template, which limits its application.
To overcome this limitation, Itaya et al. proposed the domino method, which connects DNA sequences in BGM vectors by homologous recombination between overlapping sequences to assemble large genomic fragments (Itaya et al., 2008) (Figure 1). The domino method has several advantages. For example, it does not require the preparation of large, high-purity DNA molecules, the structure of the final recombinant genome can be designed as desired, and the cloned DNA will maintain its structural stability. However, the domino method is usually used to transfer DNA smaller than 100 kb, and the transfer efficiency decreases significantly when the size of DNA increases to 100 kb. Therefore, researchers have developed a new conjugation transfer system that eliminates the domino method’s restrictions on the size of cloned DNA and thereby improves the transfer efficiency, achieving rapid transfer of 875 kb DNA (Itaya et al., 2018). Another study, aimed at simplifying the domino method, was conducted by Juhas et al., who combined Gibson Assembly and λ-red recombination in E. coli with RecA-mediated homologous recombination in B. subtilis (Juhas and Ajioka, 2016). The aim was to transfer bacterial artificial chromosome (BAC)-mediated DNA into the B. subtilis chromosome. Ultimately, they integrated the 10 kb DNA fragment from E. coli K12 MG1655 into the B. subtilis chromosome. To avoid irrational restructuring problems, Ogawa et al. developed an inducible recA expression BGM vector (iREX), which improved the stability of the inserted fragment by deleting endogenous recA and introducing a xylose-inducible recA expression cassette (Ogawa et al., 2015). Thus, the expression of recA was controlled by xylose in the medium.
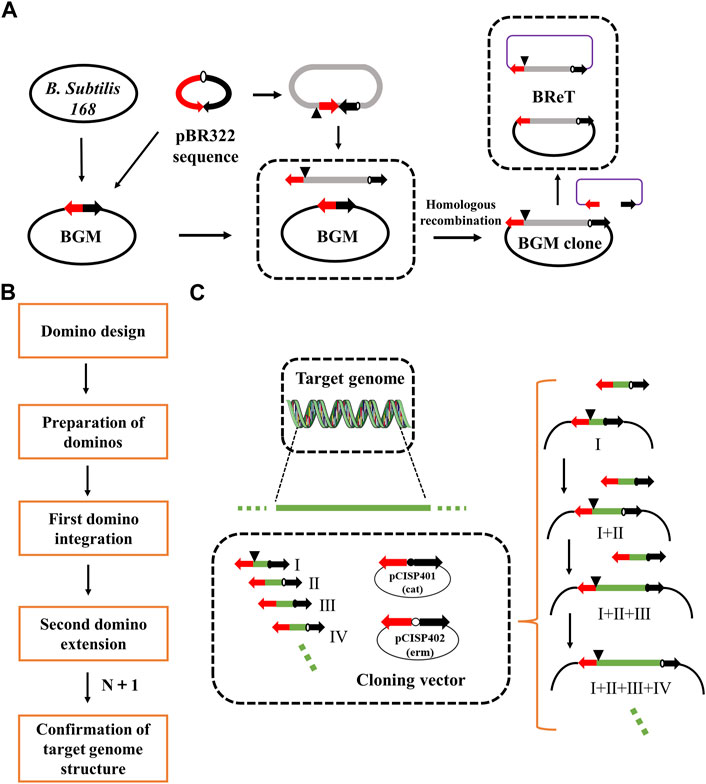
FIGURE 1. Domino cloning and BReT (Bacillus recombinational transfer retrieval). (A) Domino elongated DNA in the BGM vector by cloning between GpBR sequences, indicated by two arrows. BReT-mediated transfer occurs by homologous recombination between the pBR322 sequence of GpBR and the incoming linearized BReT plasmid; (B) the domino process; and (C) key steps of the domino process. DNA fragments are assembled into the BGM vector by homologous recombination between overlapping sequences. The dominoes were prepared in two plasmids, pCISP401 and pCISP402, with the alternating use of the two antibiotic selection markers allowing for multiple rounds of domino extension (cat, chloramphenicol; erm, erythromycin).
As a model organism, S. cerevisiae was the first eukaryote to be sequenced and has long been used as a platform to transfer DNA molecules from a variety of donor organisms in the form of yeast centromeric plasmids and yeast artificial chromosomes (Cherry et al., 2012; Vashee et al., 2020). Using yeast to transfer bacterial genomes or eukaryotic chromosomes requires the insertion of genetic elements from the yeast, including an autonomously replicating sequence (ARS), a centromere (CEN), and a selection marker to ensure that the cloned DNA can replicate and maintain itself. Genomes with low G + C% do not usually require an ARS, as the AT-rich consensus motif (ARS-like function) can occur naturally within their own sequence (Lartigue et al., 2009; Tan et al., 2021). To date, typical bacterial genomes and eukaryotic chromosomes have been transferred into yeast, with genome sizes ranging from 0.204 Mb to 1.8 Mb and GC content ranging from 24% to 55% (O'Neill et al., 2012; Karas et al., 2013a; Labroussaa et al., 2016).
The whole genome transfer into yeast can be carried out by means of centromeric plasmids. The first approach is to insert the yeast vector (ARS, CEN, and selection marker) into the genome prior to yeast transformation. After this, the newly marked genome can be transferred to yeast in two ways: one is completely isolated from donor cells and then transferred to the yeast, the other is transferred to the yeast by way of cell fusion (Figure 2A). Before cell fusion, cells need to be treated with enzymes, ultrasound, etc. to remove yeast cell walls and produce spheroplasted cells. Yeast spheroplasts can not only be transformed with purified DNA, but can also be fused with other yeast strains or bacterial cells to allow DNA transfer (Zhou et al., 2009; Benders et al., 2010; Tagwerker et al., 2012). The advantage of this method is that the yeast vector insertion site can be selected without affecting the viability of the donor cells. Mycoplasma have been successfully cloned in yeast, including M. genitalium (0.6 Mb), Mycoplasma pneumoniae (0.8 Mb), and M. mycoides subspecies capri (1.1 Mb) (Benders et al., 2010). These organisms were initially selected for genome cloning because of their small genome size and special genetic code (the UGA encoding tryptophan instead of a stop codon), which avoids toxicity to the host yeast cells. Subsequently, other bacterial genomes with standard genetic codes have also been successfully transferred into yeast, including the 1.8 Mb genome of Haemophilus influenzae and the 1.66 Mb genome of cyanobacterium Prochlorococcus marinus MED4 (Tagwerker et al., 2012; Karas et al., 2013a). However, in another study, the genome of Acholplasma laylawii PG-8A failed to transfer into yeast using this method (Karas et al., 2012). The researchers found that a gene encoding an extracellular endonuclease was toxic to yeast; after inactivating this gene, its genome was found to be stable in yeast. The second approach is transformation-associated recombination (TAR) cloning, which exploits yeast’s ability to efficiently recombine DNA fragments (Lee et al., 2015). In this approach, the genome was isolated, then linearized in vitro, and finally co-transformed into yeast with a linear yeast vector containing homology sequences (Figure 2B) (Kouprina and Larionov, 2003; 2016; Rideau et al., 2017). For example, the genome of Mycoplasma hominis was transferred into yeast in a single step, and successfully modified using the CRISPR/Cas9 editing tool. A variation of this approach is the CReasPy cloning (Figure 2B). It combines CRISPR/Cas9 gene editing technology with the efficient homologous recombination of yeast to simultaneously transfer and edit the bacterial genomes. Using this approach, the 0.816 Mb genome of M. pneumoniae was successfully transferred into yeast (Ruiz et al., 2019). Another method is CasHRA, which combines CRISPR/Cas9, yeast homologous recombination, and yeast protoplast fusion (Zhou et al., 2016). This involves the co-introduction of multiple large circular DNAs into yeast by protoplast fusion, followed by linearisation by gRNA-guided Cas9 protein cleavage, and finally DNA assembly using the yeast homologous recombination system. Using CasHRA, Zhou et al. successfully assembled and transferred the 1.03 Mb minimal E. coli genome into yeast.
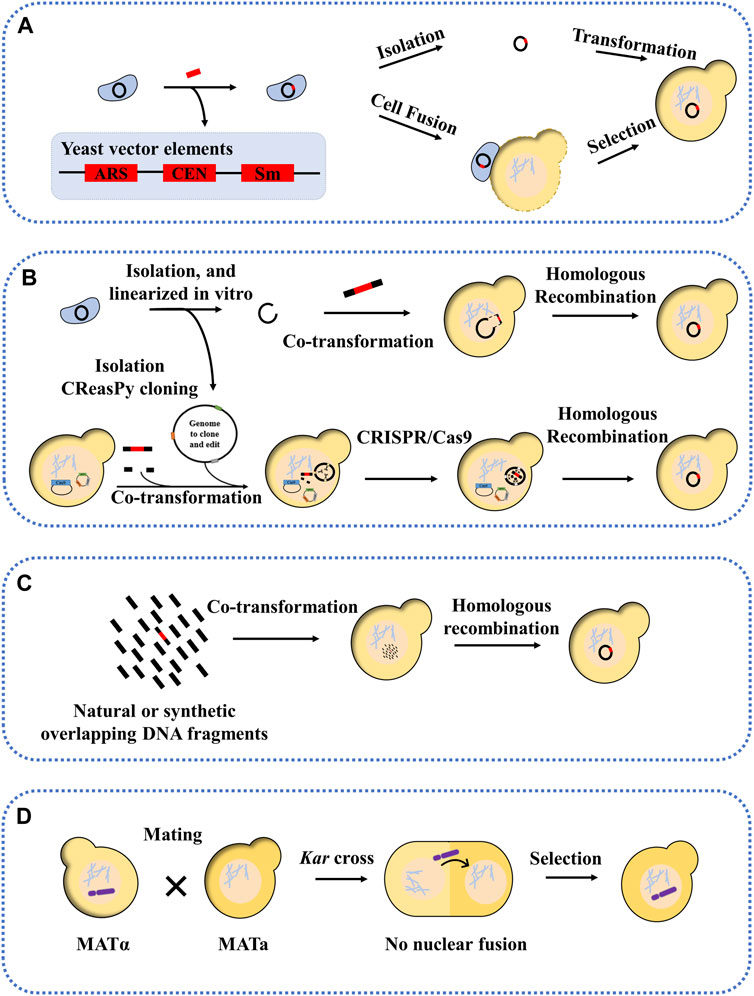
FIGURE 2. Methods for transferring natural or synthetic genomes into yeast. (A) Yeast sequence, necessary conditions for the propagation of foreign genomes in yeast (called the yeast vector), includes an autonomously replicating sequence (ARS), a centromere (CEN), a selection marker is inserted into the genome by transformation, after which the entire genome is isolated or cloned into the yeast by induced cell fusion; (B) the genome to be transferred is linearized and co-transformed into the yeast with the yeast vector, showing overlapping sequences and CReasPy cloning; (C) Cloning into yeast by assembling multiple overlapping fragments; (D) Kar cross transfer YAC.
Genomic segment transfer methods, an extension of the TAR cloning technique, can be performed by using multiple overlapping fragments of yeast transformation (Figure 2C) (Gibson et al., 2008b; Karas et al., 2013b). For example, the 2.7 Mb genome of Synechococcus elongatus with 55% GC content was divided into 30 overlapping fragments with homologous arms, each approximately 112 kb in length. Similarly, for eukaryotic chromosomes, Karas et al. successfully assembled and transferred the chromosomes 25 and 26 of Phaeodactylum tricornutum in yeast using the TAR cloning technique, starting from a DNA fragment of approximately 100 kb. And in another study, the mitochondrial genome of P. tricornutum was also successfully cloned and transferred into yeast, with a size of 60 kb–95 kb (Cochrane et al., 2020). The above studies have shown that the transferred genomic fragments cannot exceed 200 kb, but the addition of ARS can significantly increase the transferred and assembled fragment length (454 kb) and increase the stability of larger genomic DNA in yeast.
The yeast artificial chromosome (YAC) is an efficient tool for transferring large genomic fragments (Coulson et al., 1988; Larin et al., 1991). Most YAC libraries are constructed in haploid yeast strains, and it is necessary to transfer the YAC from the host strain to the target yeast strain. Researchers have developed a new approach to efficiently transfer YAC into target yeast strains, a method known as kar cross (Figure 2D) (Spencer et al., 1994). This approach is based on the fact that yeast chromosomes can be transferred from one nucleus to another between kar1 mutants and wild strains (Georgieva and Rothstein, 2002). The principle is based on the fact that, when yeast cells mate, nuclear fusion occurs immediately after cell fusion, with no intervening cell or nuclear division, resulting in a diploid. If the nuclear fusion gene (such as kar1, kar2, etc.) is mutated in one of the mating partners, nuclear fusion cannot occur, resulting in a heterokaryon containing two haploid nuclei (Dutcher, 1981; Yang and Kuang, 1996). In this case, it is possible that the target chromosome, such as YAC, could be transferred from one nucleus to the other (Torres et al., 2007). For example, Spencer et al. used a kar1 mutant as a vector to transfer starch (sta2) and melibiose (mel)-utilizing genes into industrial strains of S. cerevisiae by single-chromosomal transfer (Spencer et al., 1992). In another study, Guo et al. used the kar1 mutant approach; the four synthetic yeast chromosomes (synII, synV, synX, synXII) from the Synthetic Yeast Genome Project (Sc2.0) were transferred separately into wild-type yeast (Guo et al., 2022). In addition, Xu et al. used chromosome elimination via CRISPR-Cas9 to enable the chromosome transfer and demonstrated that chromosome XIV (chrXIV) is critical for the thermotolerance trait of the industrial strain Y12. In this study, the constructed heterozygous haploid, in which chrXIV from Y12 was transferred into BY4741 and the corresponding chrXIV of BY4741 was eliminated by CRISPR-Cas9, showed similar thermotolerance to the Y12 haploid. Through chromosome driving, the thermotolerance trait can be transferred into BY4741 (Xu et al., 2020).
Escherichia coli can maintain larger genomic fragments, which is also important for genome transfer. Here, we will mainly review E. coli as an assembly and transfer platform for genome or genomic fragments. There is a natural recombination system in E. coli, the RecA recombination system, which consists of the RecA and RecBCD proteins (Kowalczykowski, 2000). In practice, however, the RecA system has low recombination efficiency and requires a long homologous sequence (about 500 bp), which limits its application. Therefore, a more efficient in vivo recombination system has emerged in E. coli has emerged, λRed/ET, which relies on bacteriophage recombinases: either the Redα/Redβ recombinase from phage λ or the RecE/RecT recombinase from phage Rac (Zhang et al., 1998; Li et al., 2021). Redα and RecE are 5′-3′ ATP-independent nucleic acid exonucleases that can digest double-stranded DNA from the 5′end to the 3′end, exposing the 3′end of the DNA molecule, whereas Redβ and RecT are single-strand binding proteins with annealing and invasion functions. Redγ, another protein found in the λ phage, significantly enhances the recombination efficiency of Redα/Redβ. It was subsequently identified as an inhibitor of the RecB subunit of the RecBCD complex, preventing the degradation of linear DNA molecules by endogenous nucleases (Venkatesh and Radding, 1993; Paskvan et al., 2001; Murphy, 2007; Zhang et al., 2011). The λRed/ET technology can efficiently manipulate cloned genomes or genome-sized fragments (Yu et al., 2000). This technique was first used to construct a 43 kb gene cluster myxochromide S from Stigmatella aurantiaca in E. coli (Wenzel et al., 2005). Subsequently, biosynthetic gene clusters from other organisms have been constructed in E. coli, ranging in size from 11 kb to 106 kb (Wang et al., 2021). Previous recombination methods in E. coli relied on homologous recombination between linear and circular DNA molecules, which is less efficient (Zhang et al., 1998; Muyrers et al., 1999). However, the approaches using Redαβ or the truncated version of RecET are inefficient at mediating the homologous recombination between two linear DNA molecules. Therefore, Fu et al. used a full-length RecE/RecT, which significantly improved the recombination efficiency between two linear DNA molecules (Fu et al., 2012). In addition, genomic fragments can be cloned directly and transferred into E. coli by transformation. High-quality genomic sequences are obtained using low melting point agarose, ligated to vectors using enzymes, and then transformed to transfer genomic sequences into E. coli. Bacterial artificial chromosome (BAC) library construction technique is the traditional method for obtaining and transferring cross-species microbial genomic sequences into E. coli, but the method is time-consuming, labor-intensive, and the genomic sequences obtained are random. In recent years, many methods have emerged to obtain and transfer the targeted microbial genomic fragments into E. coli, such as CATCH, CAPTURE, ExoCET, TAPE, CAT-FISHING, etc. (Jiang et al., 2015; Wang et al., 2018; Enghiad et al., 2021; Cui et al., 2022; Liang et al., 2022). CATCH uses CRISPR/Cas9 technology to obtain the target genomic fragments and then uses Gibson assembly to clone the genomic fragments in vitro. Zhu et al. used this method to successfully transfer 150 kb of E. coli genomic sequence into E. coli. The CAPTURE method uses CRISPR/Cas12a to digest the genomic sequence and then uses a DNA assembly approach to obtain the genomic sequence. Zhao et al. obtained 113 kb of Actinomycetes genomic fragments in E. coli cells using this method. ExoCET combines nucleic acid exonuclease with the intracellular RecET protein of E. coli to synergistically obtain genomic DNA. Using this method, Zhang et al. acquired the 106 kb of genomic DNA from S. albus DSM41398. Li et al. developed the TAPE method, which uses the linear plasmid vector to target the genomic sequence, and they eventually transferred the 156 kb genomic sequence of E. coli into the E. coli. The CAT-FISHING technique used CRISPR/Cas12a to excise chromosome sequences and ligate them to the vector, Tan et al. captured 145 kb-long genomic DNA sequence from S. albus J1074 in E. coli by this method. However, it is difficult to obtain high quality genomic sequences by using low melting point agarose gels, and the length of the transferred genome is also limited.
The combination of genome engineering and genome transfer is a new approach for manipulating natural and synthetic genomes. Whole genomes and genomic segments in model organisms need to be transferred into recipient cells that are suitable for their expression. How to transfer the genome from the model organism into the final recipient cell is the difficult part of genome transfer technology. This requires the isolation and purification of large DNA in vitro or using cell fusion to transfer the donor genome into the recipient cell with the treatment of PEG. And due to genome size, phylogenetic distance, non-specific nucleases, restriction modification systems, and some unknown factors, the genome transfer is limited to a small set of mycoplasma species (Figure 3).
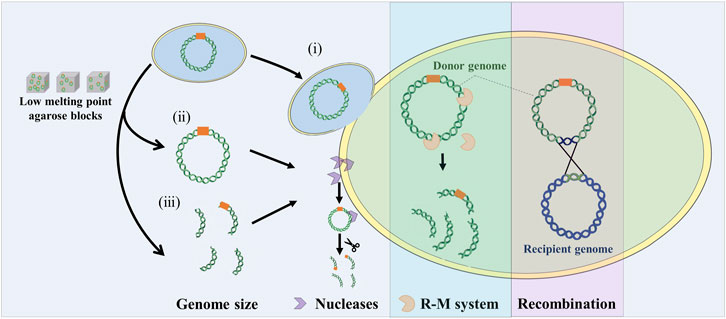
FIGURE 3. Influence factors on genome transfer. Whole genome transfer by cell fusion (i), or purified genomic DNA using low melt agarose block (ii). Genomic fragment transfer (iii). R-M system: restriction modification system. Recombination: two genomes can recombine to form a mosaic or hybrid genome.
Firstly, genome size is one of the important factors limiting genome transfer. Usually, the genome is extracted for transfer, but large DNA molecules are susceptible to breakage by shearing forces. Extracting high quality, large volume, intact genomes using low melting point agarose gels requires delicate technical manipulation. To circumvent this problem, Karas et al. directly transferred the bacterial genome into the yeast by PEG induction under conditions that promote cell fusion (Karas et al., 2013a). Currently, H. influenzae (1.8 Mb) is reported to be the largest genome transferred into yeast, but it is unclear whether the larger bacterial genome can be transferred into yeast. By improving cell fusion methods, this problem may be overcome. In addition, genome size is closely related to GC content. Genomes with relatively high GC content require the insertion of additional ARS to be stably maintained in yeast cells.
Secondly, the effect of the phylogenetic distance between donor and recipient on genome transfer needs to be better understood. When the donor genome enters the recipient cell, the recipient cell must be able to transcribe and translate the genes of the donor genome until the genome can replicate, transcribe, and translate on its own (Lartigue et al., 2007; Labroussaa et al., 2019). The molecular mechanisms of the recipient cell and the donor genome must be compatible. Labroussaa et al. investigated the effect of evolutionary distance between donor and recipient species on the efficiency of genome transfer (Labroussaa et al., 2016). The results showed that the closer the genomes of the donor and recipient cells were to each other, the higher the transfer efficiency.
Thirdly, the non-specific nucleases. These can be secreted into the environment or bound to membranes, and can even cleave the donor genome, so they need to be inactivated before genome transfer (Sharma et al., 2015). For example, the A. laylawii PG-8A genome mentioned above was successfully cloned into yeast after knocking out the gene encoding the extracellular nucleic acid endonuclease (Karas et al., 2012).
Finally, the restriction modification system is a defense mechanism against foreign DNA invasion, and the donor genome is recognized as exogenous DNA by the recipient cells. When transferring other bacterial genomes from yeast, it may be necessary to methylate the donor genome in vitro to protect it from restriction enzymes in recipient cells. Karas et al. found that removing the restriction modification system from the Mycoplasma mycoides JCVI-syn1.0 genome in yeast increased the efficiency of genome retransfer (Karas et al., 2013a; Karas et al., 2019).
In addition, there are some species-specific factors. For example, when a donor genome enters a recipient cell, the two genomes can recombine to form a mosaic or hybrid genome. The cytoskeleton and the nuclear membrane are also important influencing factors. Brown et al. proposed to synchronize recipient cells into the M (mitotic) phase, a period when the nuclear membrane and cytoskeleton are in a state of remodeling. Experimental results have shown that membrane fusion transfer using mammalian cells synchronized to mitosis can be almost 300 times more efficient (Brown et al., 2017).
Based on the above, the tasks required to adapt genome transfer technologies to other species should include: (1) Finding new recipient cells that are phylogenetically closer to the donor genome. (2) Modifying the recipient cells: one example is the introduction of genes associated with transcription, translation, and replication of the donor genome into recipient cells prior to transfer. (3) Improving protocols for the preparation of recipient cells: synchronizing the recipient cells to the M phase (mitosis) during cell fusion.
M-CZ and Y-ZC contributed equally to this work. All the authors together designed and wrote the manuscript.
This work was supported by the National Key Research and Development Program of China (2018YFA0900100); Tianjin Fund for Distinguished Young Scholars (19JCJQJC63300).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
B. subtilis, Bacillus subtilis; M. mycoides, Mycoplasma mycoides; M. capricolum, Mycoplasma capricolum; E. coli, Escherichia coli; S. cerevisiae, Saccharomyces cerevisiae; BGM, B. subtilis genome; LPS, Landing Pad Sequences; LPA, Landing Pad Sequences array; ARS, autonomously replicating sequence; CEN, centromere; M. genitalium, Mycoplasma. Genitalium; M. pneumoniae, Mycoplasma; M. mycoides subspecies capri, Mycoplasma subspecies capri; TAR, Transformation Associated Recombination; CRISPR/Cas9, Clustered Regularly Interspaced Short Palindromic Repeat; CReasPy cloning, a Method for Simultaneous Cloning and Engineering of Megabase-Sized Genomes in Yeast Using the CRISPR-Cas9 System; CasHRA, Cas9-facilitated Homologous Recombination Assembly; YAC, Yeast Artificial Chromosome; ATP, Adenosine-Triphosphate; BAC, Bacterial Artificial Chromosome; CATCH, Cas9-Assisted Targeting of Chromosome segments; CAPTURE, Cas12a-assisted Precise Targeted cloning Using in vivo Cre-lox Recombination; TAPE, the TelN/tos-assisted Precise Targeting of Chromosome segments; CAT-FISHING, CRISPR/Cas12a mediated fast direct biosynthetic gene cluster cloning; ExoCET, exonuclease in vitro assembly combined with RecET recombination; M phase, mitotic phase.
Baby, V., Labroussaa, F., Brodeur, J., Matteau, D., Gourgues, G., Lartigue, C., et al. (2018). Cloning and transplantation of the Mesoplasma florum genome. ACS Synth. Biol. 7 (1), 209–217. doi:10.1021/acssynbio.7b00279
Baby, V., Labroussaa, F., Lartigue, C., and Rodrigue, S. (2019). Synthetic chromosomes: Rewriting the code of life. Med. Sci. Paris. 35 (10), 753–760. doi:10.1051/medsci/2019153
Benders, G. A., Noskov, V. N., Denisova, E. A., Lartigue, C., Gibson, D. G., Assad-Garcia, N., et al. (2010). Cloning whole bacterial genomes in yeast. Nucleic Acids Res. 38 (8), 2558–2569. doi:10.1093/nar/gkq119
Bibi, A., and Ahmed, A. (2020). Synthetic biology: Approaches, opportunities, applications and challenges. Abasyn J. Life Sci., 25–40. doi:10.34091/ajls.3.2.3
Blount, Z. D. (2015). The unexhausted potential of E. coli. eLife 4, e05826. doi:10.7554/eLife.05826
Brown, D. M., Chan, Y. J. A., Desai, P. J., Grzesik, P., Oldfield, L. M., Vashee, S., et al. (2017). Efficient size-independent chromosome delivery from yeast to cultured cell lines. Nucleic Acids Res. 45 (7), e50. doi:10.1093/nar/gkw1252
Burgos-Morales, O., Gueye, M., Lacombe, L., Nowak, C., Schmachtenberg, R., Hörner, M., et al. (2021). Synthetic biology as driver for the biologization of materials sciences. Mat. Today Bio 11, 100115. doi:10.1016/j.mtbio.2021.100115
Cherry, J. M., Hong, E. L., Amundsen, C., Balakrishnan, R., Binkley, G., Chan, E. T., et al. (2012). Saccharomyces genome database: The genomics resource of budding yeast. Nucleic Acids Res. 40 (D1), D700–D705. doi:10.1093/nar/gkr1029
Clarke, L., and Kitney, R. (2020). Developing synthetic biology for industrial biotechnology applications. Biochem. Soc. Trans. 48 (1), 113–122. doi:10.1042/BST20190349
Cochrane, R. R., Brumwell, S. L., Soltysiak, M. P. M., Hamadache, S., Davis, J. G., Wang, J., et al. (2020). Rapid method for generating designer algal mitochondrial genomes. Algal Res. 50, 102014. doi:10.1016/j.algal.2020.102014
Coulson, A., Waterston, R., Kiff, J., Sulston, J., and Kohara, Y. (1988). Genome linking with yeast artificial chromosomes. Nature 335 (6186), 184–186. doi:10.1038/335184a0
Cui, Y. Z., Zhou, J. T., Li, B. Z., and Yuan, Y. J. (2022). The TelN/tos-assisted precise targeting of chromosome segments (TAPE). J. Adv. Res. 41, 169–177. doi:10.1016/j.jare.2022.01.017
Dutcher, S. K. (1981). Internuclear transfer of genetic information in kar1-1/KAR1 heterokaryons in Saccharomyces cerevisiae. Mol. Cell Biol. 1 (3), 245–253. doi:10.1128/mcb.1.3.245-253.1981
Enghiad, B., Huang, C. S., Guo, F., Jiang, G. D., Wang, B., Tabatabaei, S. K., et al. (2021). Cas12a-assisted precise targeted cloning using in vivo Cre-lox recombination. Nat. Commun. 12 (1), 1171. doi:10.1038/s41467-021-21275-4
Errington, J., and Aart, L. T. V. (2020). Microbe profile: Bacillus subtilis: Model organism for cellular development, and industrial workhorse. Microbiol. Read. 166 (5), 425–427. doi:10.1099/mic.0.000922
Fu, J., Bian, X., Hu, S., Wang, H., Huang, F., Seibert, P. M., et al. (2012). Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat. Biotechnol. 30 (5), 440–446. doi:10.1038/nbt.2183
Georgieva, B., and Rothstein, R. (2002). kar-mediated plasmid transfer between yeast strains: Alternative to traditional transformation methods. Methods Enzym. 350, 278–289. doi:10.1016/s0076-6879(02)50969-1
Gibson, D. G., Benders, G. A., Andrews-Pfannkoch, C., Denisova, E. A., Baden-Tillson, H., Zaveri, J., et al. (2008a). Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319 (5867), 1215–1220. doi:10.1126/science.1151721
Gibson, D. G., Benders, G. A., Axelrod, K. C., Zaveri, J., Algire, M. A., Moodie, M., et al. (2008b). One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc. Natl. Acad. Sci. U. S. A. 105 (51), 20404–20409. doi:10.1073/pnas.0811011106
Gibson, D. G., Glass, J. I., Lartigue, C., Noskov, V. N., Chuang, R.-Y., Algire, M. A., et al. (2010). Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329 (5987), 52–56. doi:10.1126/science.1190719
Guo, Z., Yin, H., Ma, L., Li, J., Ma, J., Wu, Y., et al. (2022). Direct transfer and consolidation of synthetic yeast chromosomes by abortive mating and chromosome elimination. ACS Synth. Biol. 11 (10), 3264–3272. doi:10.1021/acssynbio.2c00174
Itaya, M., Fujita, K., Ikeuchi, M., Koizumi, M., and Tsuge, K. (2003). Stable positional cloning of long continuous DNA in the Bacillus subtilis genome vector. J. Biochem. 134 (4), 513–519. doi:10.1093/jb/mvg168
Itaya, M., Fujita, K., Kuroki, A., and Tsuge, K. (2008). Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat. Methods 5 (1), 41–43. doi:10.1038/NMETH1143
Itaya, M., Nagata, T., Shiroishi, T., Fujita, K., and Tsuge, K. (2000). Effcient cloning and engineering of giant DNAs in a novel Bacillus subtilis genome vector. J. Biochem. 128 (5), 869–875. doi:10.1093/oxfordjournals.jbchem.a022825
Itaya, M., Sato, M., Hasegawa, M., Kono, N., Tomita, M., and Kaneko, S. (2018). Far rapid synthesis of giant DNA in the Bacillus subtilis genome by a conjugation transfer system. Sci. Rep. 8 (1), 8792. doi:10.1038/s41598-018-26987-0
Itaya, M. (1995). Toward a bacterial genome technology: Integration of theEscherichia coli prophage lambda genome into the Bacillus subtilis 168 chromosome. Mol. General Genet. MGG 248 (1), 9–16. doi:10.1007/BF02456608
Itaya, M., Tsuge, K., Koizumi, M., and Fujita, K. (2005). Combining two genomes in one cell: Stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome. Proc. Natl. Acad. Sci. U. S. A. 102 (44), 15971–15976. doi:10.1073/pnas.0503868102
Jiang, W. J., Zhao, X. J., Gabrieli, T., Lou, C. B., Ebenstein, Y., and Zhu, T. F. (2015). Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat. Commun. 6, 8101. doi:10.1038/ncomms9101
Johnston, C., Martin, B., Fichant, G., Polard, P., and Claverys, J. P. (2014). Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12 (3), 181–196. doi:10.1038/nrmicro3199
Juhas, M., and Ajioka, J. W. (2016). Integrative bacterial artificial chromosomes for DNA integration into the Bacillus subtilis chromosome. J. Microbiol. Methods 125, 1–7. doi:10.1016/j.mimet.2016.03.017
Karas, B. J., Jablanovic, J., Irvine, E., Sun, L., Ma, L., Weyman, P. D., et al. (2014). Transferring whole genomes from bacteria to yeast spheroplasts using entire bacterial cells to reduce DNA shearing. Nat. Protoc. 9 (4), 743–750. doi:10.1038/nprot.2014.045
Karas, B. J., Jablanovic, J., Sun, L., Ma, L., Goldgof, G. M., Stam, J., et al. (2013a). Direct transfer of whole genomes from bacteria to yeast. Nat. Methods 10 (5), 410–412. doi:10.1038/nmeth.2433
Karas, B. J., Molparia, B., Jablanovic, J., Hermann, W. J., Lin, Y.-C., Dupont, C. L., et al. (2013b). Assembly of eukaryotic algal chromosomes in yeast. J. Biol. Eng. 7 (1), 30. doi:10.1186/1754-1611-7-30
Karas, B. J., Moreau, N. G., Deerinck, T. J., Gibson, D. G., Venter, J. C., Smith, H. O., et al. (2019). Direct transfer of a Mycoplasma mycoides genome to yeast is enhanced by removal of the mycoides glycerol uptake factor gene gIpF. ACS Synth. Biol. 8 (2), 239–244. doi:10.1021/acssynbio.8b00449
Karas, B. J., Suzuki, Y., and Weyman, P. D. (2015). Strategies for cloning and manipulating natural and synthetic chromosomes. Chromosome Res. 23 (1), 57–68. doi:10.1007/s10577-014-9455-3
Karas, B. J., Tagwerker, C., Yonemoto, I. T., Hutchison, C. A., and Smith, H. O. (2012). Cloning the Acholeplasma laidlawii PG-8A genome in Saccharomyces cerevisiae as a yeast centromeric plasmid. ACS Synth. Biol. 1 (1), 22–28. doi:10.1021/sb200013j
Kiga, D., and Yamamura, M. (2008). Synthetic biology. New gener. Comput. 26 (4), 347–364. doi:10.1007/s00354-008-0050-z
Koster, C. C., Postma, E. D., Knibbe, E., Cleij, C., and Daran-Lapujade, P. (2022). Synthetic genomics from a yeast perspective. Front. Bioeng. Biotechnol. 10, 869486. doi:10.3389/fbioe.2022.869486
Kouprina, N., and Larionov, V. (2003). Exploiting the yeast Saccharomyces cerevisiae for the study of the organization and evolution of complex genomes. FEMS Microbiol. Rev. 27 (5), 629–649. doi:10.1016/S0168-6445(03)00070-6
Kouprina, N., and Larionov, V. (2016). Transformation-associated recombination (TAR) cloning for genomics studies and synthetic biology. Chromosoma 125 (4), 621–632. doi:10.1007/s00412-016-0588-3
Kowalczykowski, S. C. (2000). Initiation of genetic recombination and recombination-dependent replication. Trends biochem. Sci. 25 (4), 156–165. doi:10.1016/S0968-0004(00)01569-3
Labroussaa, F., Baby, V., Rodrigue, S., and Lartigue, C. (2019). Whole genome transplantation: Bringing natural or synthetic bacterial genomes back to life. Med. Sci. Paris. 35 (10), 761–770. doi:10.1051/medsci/2019154
Labroussaa, F., Lebaudy, A., Baby, V., Gourgues, G., Matteau, D., Vashee, S., et al. (2016). Impact of donor-recipient phylogenetic distance on bacterial genome transplantation. Nucleic Acids Res. 44 (17), 8501–8511. doi:10.1093/nar/gkw688
Larin, Z., Monaco, A. P., and Lehrach, H. (1991). Yeast artificial chromosome libraries containing large inserts from mouse and human DNA. Proc. Natl. Acad. Sci. U. S. A. 88 (10), 4123–4127. doi:10.1073/pnas.88.10.4123
Lartigue, C., Glass, J. I., Alperovich, N., Pieper, R., Parmar, P. P., Hutchison, C. A., et al. (2007). Genome transplantation in bacteria: Changing one species to another. Science 317 (5838), 632–638. doi:10.1126/science.1144622
Lartigue, C., Vashee, S., Algire, M. A., Chuang, R.-Y., Benders, G. A., Ma, L., et al. (2009). Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science 325 (5948), 1693–1696. doi:10.1126/science.1173759
Lee, N. C. O., Larionov, V., and Kouprina, N. (2015). Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic Acids Res. 43 (8), e55. doi:10.1093/nar/gkv112
Li, R., Zhao, X., Yang, R., Liu, Y., Yan, F., Wang, H., et al. (2021). Recombineering mediated by bacteriophage recombinases. Microbiol. China 48 (9), 3230–3248.
Liang, M. D., Liu, L. S., Xu, F., Zeng, X. Q., Wang, R. J., Yang, J. L., et al. (2022). Activating cryptic biosynthetic gene cluster through a CRISPR-Cas12a-mediated direct cloning approach. Nucleic Acids Res. 50 (6), 3581–3592. doi:10.1093/nar/gkac181
Liu, X., Wang, H., Xiong, Y., Zhao, G., and Wang, J. (2019). Progress in gene synthesis and genome editing. Chin. J. Cell Biol. 41 (11), 2072–2082.
Lu, J., Luo, Z., Jiang, S., Shen, Y., Wu, Y., Yang, H., et al. (2018). Technologies for DNA synthesis, assembly, and transplantation. Bull. Chin. Acad. Sci. 33 (11), 1174–1183.
Malci, K., Watts, E., Roberts, T. M., Auxillos, J. Y., Nowrouzi, B., Boll, H. O., et al. (2022). Standardization of synthetic biology tools and assembly methods for Saccharomyces cerevisiae and emerging yeast species. ACS Synth. Biol. 11 (8), 2527–2547. doi:10.1021/acssynbio.1c00442
Mukai, T., Yoneji, T., Yamada, K., Fujita, H., Nara, S., and Su’etsugu, M. (2020). Overcoming the challenges of megabase-sized plasmid construction in Escherichia coli. ACS Synth. Biol. 9 (6), 1315–1327. doi:10.1021/acssynbio.0c00008
Murphy, K. C. (2007). The λ gam protein inhibits RecBCD binding to dsDNA ends. J. Mol. Biol. 371 (1), 19–24. doi:10.1016/j.jmb.2007.05.085
Muyrers, J. P. P., Zhang, Y. M., Testa, G., and Stewart, A. F. (1999). Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27 (6), 1555–1557. doi:10.1093/nar/27.6.1555
Nielsen, J. (2019). Yeast systems biology: Model organism and cell factory. Biotechnol. J. 14 (9), e1800421. doi:10.1002/biot.201800421
O'Neill, B. M., Mikkelson, K. L., Gutierrez, N. M., Cunningham, J. L., Wolff, K. L., Szyjka, S. J., et al. (2012). An exogenous chloroplast genome for complex sequence manipulation in algae. Nucleic Acids Res. 40 (6), 2782–2792. doi:10.1093/nar/gkr1008
Ogawa, T., Iwata, T., Kaneko, S., Itaya, M., and Hirota, J. (2015). An inducible recA expression Bacillus subtilis genome vector for stable manipulation of large DNA fragments. BMC Genomics 16 (1), 209. doi:10.1186/s12864-015-1425-4
Paskvan, I., Salaj-Smic, E., Ivancic-Bace, I., Zahradka, K., Trgovcevic, Z., and Brcic-Kostic, K. (2001). The genetic dependence of RecBCD-Gam mediated double strand end repair in Escherichia coli. FEMS Microbiol. Lett. 205 (2), 299–303. doi:10.1111/j.1574-6968.2001.tb10964.x
Rideau, F., Le Roy, C., Descamps, E. C. T., Renaudin, H., Lartigue, C., and Bébéar, C. (2017). Cloning, stability, and modification of Mycoplasma hominis genome in yeast. ACS Synth. Biol. 6 (5), 891–901. doi:10.1021/acssynbio.6b00379
Ruiz, E., Talenton, V., Dubrana, M.-P., Guesdon, G., Lluch-Senar, M., Salin, F., et al. (2019). CReasPy-cloning: A method for simultaneous cloning and engineering of megabase-sized genomes in yeast using the CRISPR-cas9 system. ACS Synth. Biol. 8 (11), 2547–2557. doi:10.1021/acssynbio.9b00224
Ruiz, N., and Silhavy Thomas, J. (2022). How Escherichia coli became the flagship bacterium of molecular biology. J. Bacteriol. 204 (9), e00230–22. doi:10.1128/jb.00230-22
Sharma, S., Tivendale, K. A., Markham, P. F., and Browning, G. F. (2015). Disruption of the membrane nuclease gene (MBOVPG45_0215) of Mycoplasma bovis greatly reduces cellular nuclease activity. J. Bacteriol. 197 (9), 1549–1558. doi:10.1128/JB.00034-15
Spencer, F., Hugerat, Y., Simchen, G., Hurko, O., Connelly, C., and Hieter, P. (1994). Yeast kar1 mutants provide an effective method for YAC transfer to new hosts. Genomics 22 (1), 118–126. doi:10.1006/geno.1994.1352
Spencer, J. F. T., Spencer, D. M., de Figueroa, L., Nougues, J. M., and Heluane, H. (1992). Transfer of genes for utilization of starch (sta2) and melibiose (mel) to industrial strains of Saccharomyces cerevisiae by single-chromosome transfer, using a kar1 mutant as vector. Appl. Microbiol. Biotechnol. 37 (2), 230–234. doi:10.1007/BF00178176
Tagwerker, C., Dupont, C. L., Karas, B. J., Ma, L., Chuang, R.-Y., Benders, G. A., et al. (2012). Sequence analysis of a complete 1.66 Mb Prochlorococcus marinus MED4 genome cloned in yeast. Nucleic Acids Res. 40 (20), 10375–10383. doi:10.1093/nar/gks823
Tan, X. Y., Wu, X. L., Han, M. Z., Wang, L., Xu, L., Li, B. Z., et al. (2021). Yeast autonomously replicating sequence (ARS): Identification, function, and modification. Eng. Life Sci. 21 (7), 464–474. doi:10.1002/elsc.202000085
Torres, E. M., Sokolsky, T., Tucker, C. M., Chan, L. Y., Boselli, M., Dunham, M. J., et al. (2007). Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317 (5840), 916–924. doi:10.1126/science.1142210
Vashee, S., Arfi, Y., and Lartigue, C. (2020). Budding yeast as a factory to engineer partial and complete microbial genomes. Curr. Opin. Syst. Biol. 24, 1–8. doi:10.1016/j.coisb.2020.09.003
Venetz, J. E., Del Medico, L., Wölfle, A., Schächle, P., Bucher, Y., Appert, D., et al. (2019). Chemical synthesis rewriting of a bacterial genome to achieve design flexibility and biological functionality. Proc. Natl. Acad. Sci. U. S. A. 116 (16), 8070–8079. doi:10.1073/pnas.1818259116
Venkatesh, T. V., and Radding, C. M. (1993). Ribosomal protein S1 and NusA protein complexed to recombination protein beta of phage lambda. J. Bacteriol. 175 (6), 1844–1846. doi:10.1128/jb.175.6.1844-1846.1993
Venter, J. C., Glass, J. I., Hutchison, C. A., and Vashee, S. (2022). Synthetic chromosomes, genomes, viruses, and cells. Cell 185 (15), 2708–2724. doi:10.1016/j.cell.2022.06.046
Wang, H. L., Li, Z., Jia, R. N., Yin, J., Li, A. Y., Xia, L. Q., et al. (2018). ExoCET: Exonuclease in vitro assembly combined with RecET recombination for highly efficient direct DNA cloning from complex genomes. Nucleic Acids Res. 46 (5), e28. doi:10.1093/nar/gkx1249
Wang, W. F., Zheng, G. S., and Lu, Y. H. (2021). Recent advances in strategies for the cloning of natural product biosynthetic gene clusters. Front. Bioeng. Biotechnol. 9, 692797. doi:10.3389/fbioe.2021.692797
Wenzel, S. C., Gross, F., Zhang, Y., Fu, J., Stewart, A. F., and Müller, R. (2005). Heterologous expression of a myxobacterial natural products assembly line in pseudomonads via red/ET recombineering. Chem. Biol. 12 (3), 349–356. doi:10.1016/j.chembiol.2004.12.012
Xu, H., Han, M., Zhou, S., Li, B.-Z., Wu, Y., and Yuan, Y.-J. (2020). Chromosome drives via CRISPR-Cas9 in yeast. Nat. Commun. 11 (1), 4344. doi:10.1038/s41467-020-18222-0
Yadav, T., Carrasco, B., Myers, A. R., George, N. P., Keck, J. L., and Alonso, J. C. (2012). Genetic recombination in Bacillus subtilis: A division of labor between two single-strand DNA-binding proteins. Nucleic Acids Res. 40 (12), 5546–5559. doi:10.1093/nar/gks173
Yadav, T., Carrasco, B., Serrano, E., and Alonso, J. C. (2014). Roles of Bacillus subtilis DprA and SsbA in RecA-mediated genetic recombination. J. Biol. Chem. 289 (40), 27640–27652. doi:10.1074/jbc.M114.577924
Yang, Z. Y., and Kuang, D. R. (1996). Study on the transferring of YACs to new host by means of KAR cross. Shiyan Shengwu Xuebao 29 (3), 247–253.
Yu, D., Ellis, H. M., Lee, E. C., Jenkins, N. A., Copeland, N. G., and Court, D. L. (2000). An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97 (11), 5978–5983. doi:10.1073/pnas.100127597
Zhang, J., McCabe, K. A., and Bell, C. E. (2011). Crystal structures of λ exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc. Natl. Acad. Sci. U. S. A. 108 (29), 11872–11877. doi:10.1073/pnas.1103467108
Zhang, W. M., Mitchell, L. A., Bader, J. S., and Boeke, J. D. (2020). “Synthetic genomes,” in Annual review of biochemistry. Editor R. D. Kornberg, Vol. 89, 77–101.
Zhang, X.-E., Liu, C., Dai, J., Yuan, Y., Gao, C., Feng, Y., et al. (2023). Enabling technology and core theory of synthetic biology. Sci. China Life Sci., 1–44. doi:10.1007/s11427-022-2214-2
Zhang, Y., Buchholz, F., Muyrers, J. P. P., and Stewart, A. F. (1998). A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20 (2), 123–128. doi:10.1038/2417
Zhou, F., Li, Q., and Gao, S.-J. (2009). A sequence-independent in vitro transposon-based strategy for efficient cloning of genomes of large DNA viruses as bacterial artificial chromosomes. Nucleic Acids Res. 37 (1), e2. doi:10.1093/nar/gkn890
Keywords: synthetic genomics, microorganisms, genome transfer, cross-species, host platform
Citation: Zhu M-C, Cui Y-Z, Wang J-Y, Xu H, Li B-Z and Yuan Y-J (2023) Cross-species microbial genome transfer: a Review. Front. Bioeng. Biotechnol. 11:1183354. doi: 10.3389/fbioe.2023.1183354
Received: 10 March 2023; Accepted: 24 April 2023;
Published: 04 May 2023.
Edited by:
Sanjay Vashee, J. Craig Venter Institute (La Jolla), United StatesCopyright © 2023 Zhu, Cui, Wang, Xu, Li and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing-Zhi Li, YnpsaUB0anUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.