
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 18 May 2023
Sec. Bioprocess Engineering
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1181983
This article is part of the Research Topic Microbial Chain Elongation - Carbon Recovering Biorefineries for the Circular Economy View all 8 articles
Chain elongation is a relevant bioprocess in support of a circular economy as it can use a variety of organic feedstocks for production of valuable short and medium chain carboxylates, such as butyrate (C4), caproate (C6), and caprylate (C8). Alcohols, including the biofuel, butanol (C4), can also be generated in chain elongation but the bioreactor conditions that favor butanol production are mainly unknown. In this study we investigated production of butanol (and its precursor butyrate) during ethanol and acetate chain elongation. We used semi-batch bioreactors (0.16 L serum bottles) fed with a range of ethanol concentrations (100–800 mM C), a constant concentration of acetate (50 mM C), and an initial total gas pressure of ∼112 kPa. We showed that the butanol concentration was positively correlated with the ethanol concentration provided (up to 400 mM C ethanol) and to chain elongation activity, which produced H2 and further increased the total gas pressure. In bioreactors fed with 400 mM C ethanol and 50 mM C acetate, a concentration of 114.96 ± 9.26 mM C butanol (∼2.13 g L−1) was achieved after five semi-batch cycles at a total pressure of ∼170 kPa and H2 partial pressure of ∼67 kPa. Bioreactors with 400 mM C ethanol and 50 mM C acetate also yielded a butanol to butyrate molar ratio of 1:1. At the beginning of cycle 8, the total gas pressure was intentionally decreased to ∼112 kPa to test the dependency of butanol production on total pressure and H2 partial pressure. The reduction in total pressure decreased the molar ratio of butanol to butyrate to 1:2 and jolted H2 production out of an apparent stall. Clostridium kluyveri (previously shown to produce butyrate and butanol) and Alistipes (previously linked with butyrate production) were abundant amplicon sequence variants in the bioreactors during the experimental phases, suggesting the microbiome was resilient against changes in bioreactor conditions. The results from this study clearly demonstrate the potential of ethanol and acetate-based chain elongation to yield butanol as a major product. This study also supports the dependency of butanol production on limiting acetate and on high total gas and H2 partial pressures.
Microbial production of renewable and carbon-neutral chemicals is an important avenue to reduce carbon output and support a circular economy. The carboxylate platform, a microbial-based approach with global traction, can use diverse organic streams as feedstocks for production of butyrate (C4), caproate (C6), and caprylate (C8) (Steinbusch et al., 2010; Agler et al., 2011; Spirito et al., 2014; Roghair et al., 2018; De Groof et al., 2019; Han et al., 2019). These carboxylates are specialty chemicals for applications such as animal feed supplements, additives in chemical manufacturing, and biofuel precursors (Chen et al., 2017; Han et al., 2019; Wang and Yin, 2022). Chain elongation, the central metabolic process in the carboxylate platform, uses the reverse β-oxidation pathway to increase the carbon chain lengths by two carbons per completed cycle (Spirito et al., 2014; Angenent et al., 2016). In ethanol and acetate-based chain elongation, bacteria couple the oxidation of ethanol to the reductive elongation of the carboxylate. C. kluyveri, a strictly chain-elongating and model organism, is one of multiple key players in reactor microbiomes for chain elongation (Barker and Taha, 1942; Seedorf et al., 2008; Angenent et al., 2016; Candry and Ganigué, 2021).
Alcohols, such as butanol (C4) and hexanol (C6), can also be generated in ethanol and acetate-based chain elongation (Steinbusch et al., 2010; de Leeuw et al., 2021; Joshi et al., 2021; Robles et al., 2021; Huo et al., 2022), but their production is not always measured or reported. The value of an alcohol typically increases with carbon chain length (e.g., butanol vs. hexanol) because the higher carbon content yields the molecule a higher energy content and stability (Schiel-Bengelsdorf et al., 2013; Fernández-Naveira et al., 2017). Two pathways have been proposed for butanol production during chain elongation: (1) hydrogenotrophic carboxylate reduction (e.g., butyrate reduction to butanol) (Steinbusch et al., 2008), and (2) carboxyl-hydroxyl exchange, which couples excessive (hydrogenogenic) ethanol oxidation with hydrogenotrophic carboxylate reduction (de Leeuw et al., 2021). Butanol concentrations produced in chain elongation studies have ranged from trace concentrations up to 227 mM C (Richter et al., 2016). Several studies have reported concentrations on the range of 15–60 mM C butanol under a variety of chain elongation conditions in batch, semi-batch, or continuous bioreactors (Steinbusch et al., 2008; Ganigué et al., 2016; de Leeuw et al., 2021; Joshi et al., 2021; Robles et al., 2021; Fernández-Blanco et al., 2022; Huo et al., 2022; Vees et al., 2022; Bäumler et al., 2023). However, the conditions under which chain-elongating microbiomes can be directed to yield butanol as a major product during ethanol and acetate chain elongation are unknown.
Optimizing bioreactor conditions, identifying key microorganisms, and broadening the spectrum of possible end-products, including butanol and longer alcohols, are research endeavors required to increase the relevance of chain elongation as a bioprocess for a circular economy (de Leeuw et al., 2019; Han et al., 2019; Wang and Yin, 2022; Shrestha et al., 2023). Butanol can be used directly as a fuel or mixed with gasoline (Dekishima et al., 2011). It is also used as a sustainable solvent, a chemical intermediate, and it is used in the production of common-use items, such as cosmetics and pharmaceuticals (Ndaba et al., 2015). Butanol may also be a desirable H2-releasing substrate in subsurface bioremediation applications under anoxic conditions. For example, reductive dehalogenation of chlorinated ethenes (i.e., tetrachloroethene, trichloroethene, and vinyl chloride) by Dehalococcoides mccartyi sp. requires H2 as the obligate electron donor to convert the contaminants to non-toxic ethene (Löffler et al., 2013; Delgado et al., 2014; Mohana Rangan et al., 2020). Butanol has been shown to promote the complete reductive dehalogenation of tetrachloroethene and to enhance dehalogenation rates when tetrachloroethene was present as a dense non-aqueous phase liquid (Yu and Semprini, 2009). More recently, ethanol and acetate chain elongation was shown to support reductive dehalogenation of trichloroethene directly through production of H2 during elongation of acetate and indirectly through fermentation of the chain elongation products, butyrate and butanol (Robles et al., 2021). Understanding the conditions under which chain elongation may be steered to produce butanol and its precursor, butyrate, benefits applications for biochemical production and bioremediation.
The most studied pathway for bio-butanol production is acetone-butanol-ethanol (ABE) fermentation. In ABE fermentation, sugars and starches are converted to carboxylates (acetate and butyrate) and solvents (acetone, butanol, and ethanol) (Ndaba et al., 2015). Concentrations on the order of 539–1,000 mM C butanol (10–20 g L−1) (Xu et al., 2015; Wechgama et al., 2017) have been achieved via ABE fermentation with butanol yields ranging from ∼0.2 to 0.4 g g−1 glucose (Ndaba et al., 2015; Veza, Muhamad Said and Latiff, 2021). Butanol production via ABE is influenced by the medium composition (Al-Shorgani et al., 2018b), carbon source (Al-Shorgani et al., 2012), temperature (Ramió-Pujol et al., 2015), concentration of butyrate (Lee et al., 2008), and pH (Bahl et al., 1982), among other parameters. A limited number of chain elongation studies have reported that production of butanol by chain-elongating microbiomes can be encouraged by combining butyrate with a high H2 partial pressure (around 150 kPa) (Steinbusch et al., 2008) and by feeding syngas in bioreactors at an initial pH of around 6 (Ganigué et al., 2016). In anaerobic systems, including ABE fermentation, the total gas pressure in a bioreactor has also been shown to influence metabolic shifts from acidogenesis to solventogenesis (Doremus et al., 1985; Brosseau et al., 1986; Yan et al., 2017). Nonetheless, the total gas pressure is rarely directly considered in bioreactor design for chain elongation.
In this study, we investigated production of butanol (and its precursor butyrate) during ethanol and acetate-based chain elongation in semi-batch bioreactors (0.16 L serum bottles) fed with a range of ethanol concentrations (100–800 mM C), a constant and limiting acetate concentration (50 mM C), and an initial partial pressure of ∼112 kPa. Under conditions of high total gas pressure and high H2 partial pressure, we found that increasing the concentration of ethanol was positively correlated to chain elongation activity and ultimately yielded a butanol:butyrate molar ratio of 1:1. A butanol concentration of 114.96 ± 9.26 mM C was achieved in this study. The dependency of butanol production on high total gas pressure and high H2 partial pressure was experimentally verified by releasing gas from bioreactors. This study is the first to show butanol as a major end-product of ethanol and acetate chain elongation.
Anaerobic mineral medium was prepared as described in Robles et al. (2021). The medium contained 10 mL of a salt stock solution, 1 mL of a trace element A solution, 1 mL of a trace element B solution, and 1 mL of a vitamin solution per liter. The solutions were prepared as described in Löffler et al. (2005) with the modifications described in Robles et al. (2021). Additionally, the medium was amended with the reductants Na2S (0.2 mM) and L-cysteine (0.4 mM), vitamin B12 (0.5 mg L−1) and the buffer, potassium phosphate (10 mM). The initial pH of the medium was 7.5. The medium was bottled with UHP N2 in the headspace. Ethanol, 200 proof, molecular biology grade (Sigma Aldrich, St. Louis, MO, United States) and ReagentPlus sodium acetate trihydrate (Sigma Aldrich) were used as substrates for the bioreactors.
The inoculum for this study was an enrichment culture from a soil microcosm. The microcosm contained 10 g soil from Phoenix Goodyear Airport-North Superfund site. The soil microcosm was inoculated with 5 mL of a lactate-fermenting and trichloroethene-dehalogenating culture and 5 mL of an ethanol- and acetate-chain elongating culture in 90 mL anaerobic medium with 100 mM C ethanol, 100 mM C acetate, and 2.1 mmol L−1 trichloroethene (Robles et al., 2021). The enrichment culture from this soil microcosm converted ethanol and acetate to mainly butyrate with minimal methane production (≤1.7 mmol L−1 methane observed during incubation). The soil microcosm enrichment culture was maintained under these conditions prior to use in the experiments from this study. The microbial community composition of the enrichment culture primarily consisted of members of Clostridiales, Burkholderiales, and Eubacteriales. The most notable chain elongating microorganism in the culture was Clostridium kluyveri [100% similar to strain K1, ATCC 8527/DSM 555 using BLAST + consensus taxonomy classifier plugin (Camacho et al., 2009)].
Experiments in triplicate were setup in serum bottles sealed with rubber stoppers and aluminium crimps. The total volume of the bottles was 0.16 L (160 mL) with an initial liquid volume of 75 mL. For brevity, we hereafter refer to the experimental serum bottle reactors as bioreactors. The bioreactors were provided with the following initial concentration of substrates: 100 mM C ethanol + 50 mM C acetate, 200 mM C ethanol + 50 mM C acetate, 400 mM C ethanol + 50 mM C acetate, 800 mM C ethanol + 50 mM C acetate, and 800 mM C ethanol (Supplementary Table S1). Each bioreactor received 6 mL of inoculum culture at the start of the experiment. The bioreactors were operated in semi-batch cycles with draw and fill performed every 7 days. At the end of each 7-d cycle, one-third of the bioreactor’s liquid contents (25 mL) was removed and replaced with fresh medium (25 mL) containing the same initial substrate concentration as in Supplementary Table S1. The initial total gas pressure in the bioreactors was set at 110 ± 2 kPa (∼1.08 atm) by injecting UHP N2 gas. The initial pH in the bioreactors was set to ∼7.5. During the first seven semi-batch cycles, the bioreactors were allowed to accumulate H2 in the headspace and thus increasing total gas pressure. A pseudo steady-state was achieved by semi-batch cycle 4. The operating phase in semi-batch cycles 4 through 7 is referred to in the text and figures as “High H2 & total pressure.” At the beginning of semi-batch cycle 8, the bioreactors were intentionally perturbed by decreasing the total gas pressure to ∼112 kPa (similar to time 0 conditions). The operating phase for semi-batch cycles 8 through 11 is referred to in text and figures as “High H2 & low total pressure.” A total of 11 semi-batch cycles were completed in the study. The condition labelled “100 mM C EtOH + Acetate” was resupplied with 240 mM C ethanol at the beginning of cycle 8. An abiotic control with 200 mM C ethanol and 50 mM C acetate was setup and operated for two cycles. All bioreactors were incubated at 31°C on a platform shaker set to 150 rpm.
Ethanol, butanol, and hexanol, and acetate, butyrate, caproate, and caprylate were quantified at the beginning and end of each semi-batch cycle, except cycles 6 and 10 where samples were not preserved for analysis. The concentrations of carboxylates and alcohols were determined using a high-performance liquid chromatograph (HPLC) equipped with a refractive index detector, a photodiode-array detector, and an Aminex HPX-87H column (Bio-Rad Laboratories, Hercules, CA, United States). The HPLC method and sample preparation were completed as previously described (Joshi et al., 2021; Miranda et al., 2022). The method was run for a total of 120 min, with retention times ranging from 15 min for acetate to 79 min for hexanol. The detection limit of the analytes was 0.02–0.05 mM. Hexanol and caprylate were not detected in samples from this study. pH measurements were taken using a benchtop pH meter (Orion 2-star, Thermo Scientific, Waltham, MA, United States) equipped with an economy series pH electrode.
H2 concentration in the headspace of the bioreactors was quantified using a gas chromatograph with a thermal conductivity detector (GC-TCD) (Shimadzu GC-2010, Columbia, MD, United States) and a fused silica capillary column (Carboxen 1010 PLOT column, Supelco, Bellefonte, PA, United States). The sampling and GC-TCD method details were as previously published (Robles et al., 2021; Meinel et al., 2022). The calibration range for H2 was 0.013–10.22 mmol L−1 gas. The total gas volume in the bioreactors was measured with a frictionless syringe (Sigma-Aldrich) and was converted to a total gas pressure as described in the Supplementary Material.
DNA was extracted from bioreactor samples preserved in RNAprotect cell reagent (Qiagen, Germantown, MD, United States) at −80°C at the end of semi-batch cycles 5 (during “High H2 & total pressure” phase) when butanol concentrations were highest and 9 (during “High H2 & low total pressure” phase) when butanol concentrations were lowest. Pellets were pre-treated with an enzyme lysis buffer containing 20 mM Tris·HCl, 2 mM EDTA, 250 μg mL−1 achromopeptidase, and 20 mg mL−1 lysozyme (Mohana Rangan et al., 2023). After pre-treatment, genomic DNA was extracted using the Qiagen DNeasy Blood and Tissue kit (MO BIO Laboratories Inc., Carlsbad, CA, United States) following the protocol for Gram-positive bacteria.
Microbial community amplicon sequencing was performed on the Illumina platform with a Miseq instrument (San Diego, CA, United States) at the ASU Genomics Core Facility, Arizona State University, Tempe, AZ, United States. Sequencing used the universal primers 515F and 806R for the V4 hyper-variable region of the 16S rRNA gene of Bacteria and Archaea (Caporaso et al., 2012). Forward and reverse reads were processed using the Quantitative Insights into Microbial Ecology (QIIME 2.0 v. 2022.2) pipeline (Bolyen et al., 2019). Each sequence was truncated at 232 base pairs using DADA2 to maintain a quality score of 25 or better and produce amplicon sequence variants (ASVs). A pretrained Naïve Bayes classifier referencing the SILVA database (v.138) (Quast et al., 2012; Bokulich et al., 2018; Robeson et al., 2021) and the q2-feature-classifier plugin were used to assign taxonomy to amplicon sequence variants (DeSantis et al., 2006). For alpha diversity, Pielou’s evenness index was determined from sequences rarefied to a sampling depth of 16,224 counts. The raw sequences were submitted to the NCBI Sequence Read Archive and are available under the project number PRJNA913573 with accession numbers SRX18767348–SRX18767367.
The 16S rRNA gene of C. kluyveri was quantified in bioreactors at the end of semi-batch cycles 5 (“High H2 & total pressure”) and 9 (“High H2 & low total pressure”) via quantitative real-time PCR (qPCR) (Bio-Rad CFX96). The qPCR assays contained the following per 25 µL reaction: 2 µL DNA, 1.125 μL F′ primer, 1.125 μL R′ primer, 12.5 µL SYBR Green Master Mix (Bio-Rad) and 8.25 µL RNase-free water (MO Bio Laboratories Inc.). No-template controls were also included in the qPCR runs. A six-point calibration curve was created using a gBlock fragment (Integrated DNA Technologies, Inc., Coralville, IA, United States) as shown in the Supplementary Material. Triplicate reactions were setup for experimental samples, no-template controls, and the calibration. To infer concentrations of C. kluyveri in the bioreactors (in cells mL−1), copies per mL of the 16S rRNA gene were divided by 7, which is the number of 16S rRNA gene copies in the chromosomal DNA of C. kluyveri (Stoddard et al., 2014). Additional details about the qPCR analysis are presented in the Supplementary Material.
The concentrations of carbon (C)-containing chain elongation substrates and products were converted to mM carbon (C) by multiplying the concentration in mM by the corresponding C atom number in each compound: ethanol, 2; acetate, 2; butanol, 4; butyrate, 4; and caproate, 6. Total gas pressure was determined using the measured total gas volume and the ideal gas law. The average and maximum rates of butyrate and butanol production in units of mmol C L−1 d−1 were calculated using the data from the “High H2 & total pressure” phase (semi-batch cycles 4–7). Average ratios of butanol to butyrate (mol:mol) produced were calculated using data from semi-batch cycles 4–7 (for “High H2 & total pressure” phase) and 11 (three cycles after pressure was reduced to ∼112 kPa from “High H2 & low total pressure” phase). A Student’s t-test was used to evaluate statistical significance of chemical and microbiological data with a 95% confidence interval (p < 0.05 was considered statistically significant). Additional details on calculations are in the Supplementary Material.
Semi-batch bioreactors probing the potential of chain elongation for butanol production were fed with ethanol (100–800 mM C) and acetate (50 mM C) or ethanol only (800 mM C) at an initial total gas pressure of ∼112 kPa. Consumption of substrates (Supplementary Figure S1) coupled to the production of butyrate and butanol was observed in all bioreactors (Figures 1Ai–Av). In the absence of the inoculum culture, substrate consumption was not observed (Supplementary Figure S2). Increasing the ethanol concentration from 100 mM C to 400 mM C while keeping the acetate concentration constant enhanced chain elongation activity, including butanol production (Figures 1Ai–Aiii). It has been previously documented that a higher ethanol concentration and/or a higher molar ratio of ethanol to acetate can steer chain elongation from mainly butyrate to caproate and/or caprylate (Steinbusch et al., 2010; Coma et al., 2016; Liu et al., 2016; Lonkar et al., 2016; Spirito et al., 2018; Joshi et al., 2021). In our previous work, we showed that soil slurry semi-batch bioreactors fed with 200 mM C ethanol and 200 mM C acetate produced between 11.41 and 59.89 mM C butanol and 1.10 and 31.77 mM C hexanol (Joshi et al., 2021). However, butyrate, caproate, and caprylate were the dominant products (Joshi et al., 2021). In the present study, no caprylate or hexanol was detected and the caproate concentration remained low in all bioreactors throughout operation (0.37 ± 0.01–6.48 ± 0.87 mM C caproate, Figures 1Ai–Av). Two connected reasons likely explain the limited production of caproate. First, acetate, the electron acceptor in chain elongation, was limiting in our bioreactors relative to the concentration of ethanol, the electron donor. Partial ethanol oxidation to acetate and H2 is the ATP yielding reaction in chain elongation (Seedorf et al., 2008). Ethanol oxidation would provide a required carboxylate electron acceptor for chain elongation. However, the high H2 partial pressure in our bioreactors made this reaction unfavorable after four semi-batch cycles as evident by the plateau in H2 concentration (Figures 1Bii–Biv). Thus, butanol production became a major pathway through which electrons from H2 could be consumed in the system.
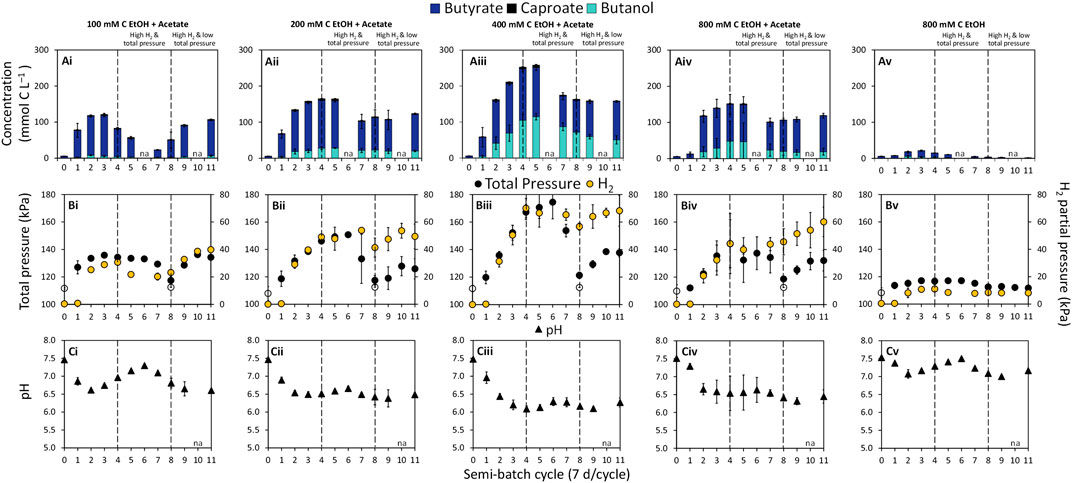
FIGURE 1. Concentrations of (A) butyrate, caproate, and butanol, (B) total gas pressure and H2 partial pressure (yellow circles on secondary y-axis), and (C) pH in semi-batch bioreactors. The concentration of acetate in the bioreactors was 50 mM C. The empty symbols for total pressure are measurements at time 0 and beginning of cycle 8 when total pressure was decreased to ∼112 kPa. The filled symbols are measurements at the end of a semi-batch cycle. The data are averages with standard deviations of triplicate bioreactors. The pH at the start of each cycle is reported in Supplementary Table S1. na = not analyzed.
Butanol production was observed at H2 partial pressures above ∼8.16 kPa, while significant butanol production occurred at H2 partial pressures above ∼50 kPa (p = 0.0241, Figures 1Bi–Bv). The highest concentration of butanol achieved in this study was 114.96 ± 9.26 mM C (∼2.13 g L−1) in condition “400 mM C EtOH + Acetate” at the end of semi-batch cycle 5 during the “High H2 & total pressure” phase (Figure 1Aiii). To the best of our knowledge, this is the highest reported butanol concentration in an ethanol and acetate-based chain elongation study. At the end of semi-batch cycle 5, the bioreactors fed with 400 mM C ethanol reached a H2 partial pressure of 66.58 ± 3.99 kPa and a total gas pressure of 170.63 ± 14.35 kPa (Figure 1Biii). Steinbusch et al. (2008) documented that bioreactors with a H2 headspace (150 kPa H2) and 200 mM C butyrate produced up to 14.64 mM C butanol. Additionally, total gas pressures between 156 and 184 kPa previously showed selection for alcohols over carboxylates in methanogenic bioreactors containing H2 and CO2 in the headspace (Yan et al., 2017). Findings from our study suggest that H2 partial pressure in combination with total gas pressure are powerful selection tools for butanol.
The highest butanol concentrations were achieved under conditions where H2 production stalled during the “High H2 & total pressure” phase (Figure 1, cycle 5). Enhanced production of butanol under this H2 stall phenomenon has been observed in ABE fermentation (Doremus et al., 1985; Brosseau et al., 1986; Yan et al., 2017). Stalls in H2 production can be attributed to the influence of dissolved H2 on NADH2 and H2 supersaturation in the medium, which can inhibit H2 production via the ferredoxin-linked hydrogenase (Doremus et al., 1985). Under this scenario, microorganisms can channel electrons through NADH:ferredoxin oxidoreductase, reducing butyrate to butanol (Doremus et al., 1985), supporting the high butanol production observed in our study.
Butanol (and butyrate) production rates in bioreactors increased with increasing concentration of ethanol up to 400 mM C (Supplementary Figure S3). The optimal range for ethanol-based chain elongation using mixed cultures has been reported to be between 216 and 434 mM C ethanol (5–10 g L−1 ethanol) (Lonkar et al., 2016). Improved production rates from our bioreactors with 200 and 400 mM C ethanol (+acetate) are in line with these previous findings. The bioreactors also showed a clear trend with respect to the molar ratio of butanol to butyrate produced. Specifically, the butanol:butyrate molar ratio increased from ∼1:6 at 100 mM C ethanol (+acetate) to ∼1:1 at 400 mM C ethanol (+acetate) during the “High H2 & total pressure” phase (Table 1).
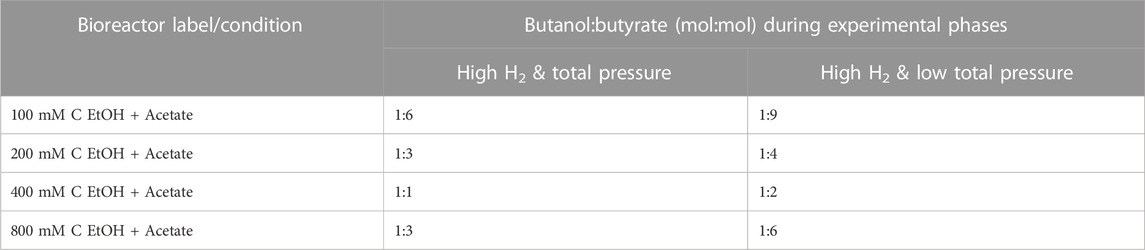
TABLE 1. Average butanol to butyrate molar ratio in bioreactors during semi-batch cycles 4–7 (“High H2 & total pressure” phase) and cycle 11 (“High H2 & low total pressure” phase).
Chain elongation activity in bioreactors significantly decreased when the concentration of ethanol was increased from 400 to 800 mM C (+50 mM C acetate) (p = 0.0002, Figure 1Aiii; Supplementary Figure S4). The decrease in chain elongation activity in bottles fed 800 mM C ethanol and 50 mM C acetate was likely a consequence of ethanol inhibition. Concentrations between ∼600 and 1,720 mM C ethanol (14 and 40 g L−1 ethanol) have been previously reported as inhibitory in chain elongation studies (Kucek et al., 2016; Lonkar et al., 2016). Butanol concentration of ∼50 mM C (1 g L−1) have been reported to inhibit growth of the butanol producer C. carboxidivorans (Fernández-Naveira et al., 2016). In our mixed culture bioreactors, ∼100 mM C butanol did not appear to have a prominent inhibitory effect as approximately the same percentage of substrates went to production of butyrate, butanol, caproate and H2 in bioreactors with 200 mM C ethanol and 400 mM C ethanol (Supplementary Figure S4). The absence of added electron acceptor, acetate, in the “800 mM C EtOH” condition further suppressed chain elongation activity, in agreement with previous studies (Spirito et al., 2018; de Leeuw et al., 2021; Joshi et al., 2021). Thus, the poor chain elongation extent at 800 mM C ethanol (Figure 1Av) is reflective of ethanol inhibition and acetate limitation.
A general decrease in pH was observed in all ethanol and acetate bioreactors from the start of incubation (time 0) and by the end of each semi-batch cycle (Figures 1Ci–Cv, pH at the start of cycles in Supplementary Table S2). The pH range across ethanol and acetate conditions was 6.09 ± 0.09–7.30 ± 0.04 at the end of semi-batch cycles (Figures 1Ci–Civ). The lowest pH was observed in “400 mM C EtOH + Acetate” bioreactors, where the highest butanol production occurred (pH ranged from 6.09 ± 0.09 to 6.29 ± 0.11, Figure 1Ciii). In ABE fermentation, butyrate in combination with mildly acidic pH (4.5–6.2) is a trigger for butanol production and increases solvent yields (Matta-El-Ammouri et al., 1987; Tashiro et al., 2004; Li et al., 2011; Al-Shorgani et al., 2018a). A pH range between 4.7 and 6.4 has also been found to support alcohol production in chain elongation systems with C. kluyveri (Ganigué et al., 2016; Richter et al., 2016). In this study, higher butanol production was observed at the pH range 6.1–6.3; however, the pH was not controlled during the experiment and thus a relationship between the extent of butanol production and pH could not be discerned based on the experimental design.
To evaluate the dependency of butanol production on gas pressure and composition, the total gas pressure was adjusted at the start of semi-batch cycle 8 by releasing gas from the bioreactors and resetting the total gas pressure to ∼112 kPa (Figure 1, “High H2 & low total pressure” phase). The decreases in total gas pressure immediately decreased butanol production at the end of cycle 8 (Figures 1Aii–Aiv) but butanol production continued throughout this experimental phase. The H2 stall observed during the first phase was overcome during the “High H2 & low total pressure” phase. During the second phase, H2 partial pressures reached 7.75 ± 1.33–68.19 ± 1.33 kPa and total gas pressure ranged from 111.71 ± 1.33 to 138.49 ± 1.33 kPa (Figures 1Bi–Bv). The butanol:butyrate molar ratio was consistently lower in all conditions during the “High H2 & low total pressure” phase after the total gas pressure was decreased (Figures 1Ai–Aiv; Table 1). The lower total gas pressure observed in “High H2 & low total pressure” phase and lower butanol:butyrate molar ratios supports previous findings where total gas pressures between 101 and 124 kPa previously selected for production of carboxylate and H2 over alcohols (Yan et al., 2017).
The microbial community composition was determined to identify any potential linkage between observed activity, particularly butanol and butyrate production, during the “High H2 & total pressure” phase and the “High H2 & low total pressure” phase. Regardless of the experimental phase, the most abundant or second most abundant phylum in the microbial community was Firmicutes (recently renamed Bacillota) (Supplementary Figure S5). Firmicutes contains the majority of the identified chain elongating species (Elsden et al., 1956; Wallace et al., 2004; Seedorf et al., 2008; Angenent et al., 2016; Zhu et al., 2017; Han et al., 2018; Candry and Ganigué, 2021; Joshi et al., 2021). During the “High H2 & total pressure” phase, Firmicutes ASVs accounted for 76.2% of the microbial community in bioreactors fed 400 mM C ethanol and acetate, which also produced the highest concentration of butanol and butyrate (Figure 1Aiii; Supplementary Figure S5). ASVs for strict carboxydotrophic microbes known to produce butanol were not identified in these bioreactors. Furthermore, methanogenic ASVs were either not detected in samples or were at ≤ 0.7% of total sequences, consistent with the characteristics of the inoculum where methane production was absent or minimal even at a pH range of 6–7 (Joshi et al., 2021; Robles et al., 2021).
In our study, C. kluyveri ASVs, which classified in Clostridium sensu stricto 12, were most abundant in bioreactors fed 200 and 400 mM C ethanol (+acetate) in semi-batch cycle 5, accounting for ∼56% of the total sequences (Figure 2). In our study, the C. kluyveri ASV showed 100% sequence match to strain K1 (Barker and Taha, 1942) in a BLAST consensus sequence search. C. kluyveri produces carboxylates as the major metabolites. Butanol (Thauer et al., 1968; Kenealy and Waselefsky, 1985) and propanol (Candry et al., 2020) have also been reported in pure culture studies with C. kluyveri. It is possible that C. kluyveri was a major player in butanol production, although other microorganisms could have contributed to production of this metabolite. Quantification of C. kluyveri through qPCR supported its prominent role in chain elongation (and potentially butanol production). Specifically, the highest concentration of C. kluyveri cells (up to 6.71×108 ± 3.61×107 cells mL−1) was observed during the “High H2 & total pressure” phase at 200 mM C ethanol (Figure 2, cycle 5). A lower concentrations of C. kluyveri was quantified at 800 mM C ethanol than at 200 mM C ethanol (+acetate), even though the cumulative concentration of butyrate and butanol were similar in these conditions (Figures 1Aii, Aiv).
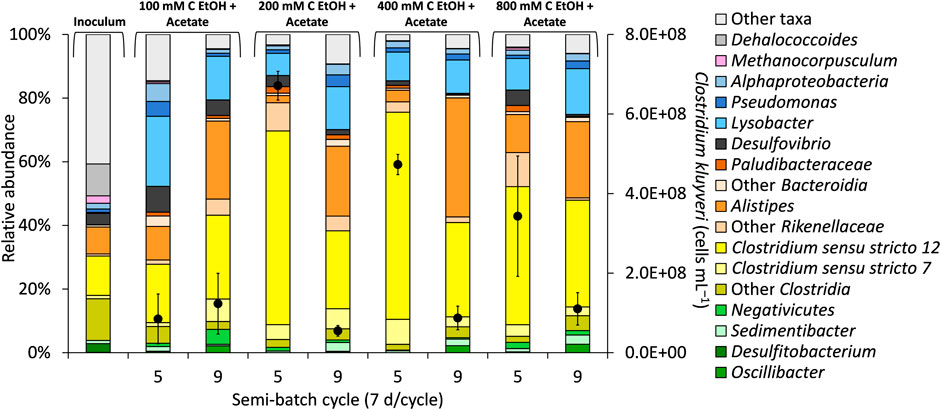
FIGURE 2. Selected taxa in the bioreactors and quantification of Clostridium kluyveri (black circles, secondary y-axis) during “High H2 & total pressure” phase (end of semi-batch cycle 5) and during “High H2 & low total pressure” phase (end of semi-batch cycle 9). The concentration of acetate in the bioreactors was 50 mM C. The total pressure in the bioreactors was decreased at the end of semi-batch cycle 7. The data are averages from triplicate bioreactors.
Microbial diversity and richness were not affected by increasing concentration of ethanol or by the experimental phase (Supplementary Figure S6, Pielou’s evenness range 0.49–0.67, p = 0.4199), highlighting the overall stability of the microbiome. A noteworthy trend was observed during “High H2 & low total pressure” where Clostridium sensu stricto 12 ASV decreased in relative abundance while Alistipes ASV (up to 37% of the total sequences, Figure 2) and other Rikenellaceae ASVs from Bacteroidota increased in relative abundance (Figure 2). Alistipes has been recently linked to butyrate production in anaerobic digestion of cellulose using anaerobic sludge as inoculum (Rico et al., 2021). Alistipes ASVs possibly also contributed to butyrate production in our bioreactors. Sedimentibacter, Oscillibacter, and Pseudomonas are ASVs commonly reported to also enrich in chain elongation bioreactors (Rühl et al., 2009; Candry and Ganigué, 2021; Joshi et al., 2021). In our study, these ASVs were more abundant during the “High H2 & low total pressure” phase, although to a much lower extent than Clostridium sensu stricto 12 and Alistipes (Figure 2).
In this study, we show that selective conditions in bioreactor can be imposed for a chain-elongating microbiome to yield butanol as a major product in ethanol and acetate-based chain elongation. The highest chain elongation activity was observed in bioreactors fed 400 mM C ethanol during the “High H2 & total pressure” phase where up to 114.95 ± 9.26 mM C butanol (∼2.13 g L−1) was produced. We showed that bioreactors operated under high total pressure and high H2 partial pressure with limited acetate relative to ethanol produce butanol instead of the longer carboxylate, caproate. The individual effect of H2 partial pressure, total gas pressure, and acetate concentration were not isolated in our work. However, results from this work clearly support that these parameters can be applied for selective production of butanol in chain elongation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA913573: SRX18767348–SRX18767367.
SS, AR, and SM completed the experimental work and analyzed the data. AR generated the first draft of the manuscript. AD conceived and directed the study and contributed to the draft of the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was funded by US National Science Foundation (NSF) award CBET-2221805 and a Non-Academic Research Internships for Graduate Students (INTERN) supplement to award NSF EEC-1449501. Any opinions, findings and conclusions or recommendations expressed in this article are those of the authors and do not necessarily reflect those of the NSF.
We thank Prof. Kerry Hamilton and Ms. Sayalee Joshi from Biodesign Center for Environmental Health Engineering, Arizona State University, for instrument access and expert advice on qPCR methods. We also thank our undergraduate researchers, Alex Gaura, for assistance with DNA extractions and Anh Quan Truong, for help in preparing the qPCR assays.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1181983/full#supplementary-material
Agler, M. T., Wrenn, B. A., Zinder, S. H., and Angenent, L. T. (2011). Waste to bioproduct conversion with undefined mixed cultures: The carboxylate platform. Trends Biotechnol. 29 (2), 70–78. doi:10.1016/j.tibtech.2010.11.006
Al-Shorgani, N. K. N., Ali, E., Kalil, M. S., and Yusoff, W. M. W. (2012). Bioconversion of butyric acid to butanol by Clostridium saccharoperbutylacetonicum N1-4 (ATCC 13564) in a limited nutrient medium. BioEnergy Res. 5 (2), 287–293. doi:10.1007/s12155-011-9126-6
Al-Shorgani, N. K. N., Kalil, M. S., Yusoff, W. M. W., and Hamid, A. A. (2018a). Impact of pH and butyric acid on butanol production during batch fermentation using a new local isolate of Clostridium acetobutylicum YM1. Saudi J. Biol. Sci. 25 (2), 339–348. doi:10.1016/j.sjbs.2017.03.020
Al-Shorgani, N. K. N., Shukor, H., Abdeshahian, P., Kalil, M. S., Yusoff, W. M. W., and Hamid, A. A. (2018b). Enhanced butanol production by optimization of medium parameters using Clostridium acetobutylicum YM1. Saudi J. Biol. Sci. 25 (7), 1308–1321. doi:10.1016/j.sjbs.2016.02.017
Angenent, L. T., Richter, H., Buckel, W., Spirito, C. M., Steinbusch, K. J. J., Plugge, C. M., et al. (2016). Chain elongation with reactor microbiomes: Open-culture biotechnology to produce biochemicals. Environ. Sci. Technol. 50, 2796–2810. doi:10.1021/acs.est.5b04847
Bahl, H., Andersch, W., Braun, K., and Gottschalk, G. (1982). Effect of pH and butyrate concentration on the production of acetone and butanol by Clostridium acetobutylicum grown in continuous culture. Eur. J. Appl. Microbiol. Biotechnol. 14 (1), 17–20. doi:10.1007/bf00507998
Barker, H. A., and Taha, S. M. (1942). Clostridium kluyverii, an organism concerned in the formation of caproic acid from ethyl alcohol. J. Bacteriol. 43 (3), 347–363. doi:10.1128/jb.43.3.347-363.1942
Bäumler, M., Burgmaier, V., Herrmann, F., Mentges, J., Schneider, M., Ehrenreich, A., et al. (2023). Continuous production of ethanol, 1-butanol and 1-hexanol from CO with a synthetic co-culture of Clostridia applying a cascade of stirred-tank bioreactors. Microorganisms 11 (4), 1003. doi:10.3390/microorganisms11041003
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6 (1), 90. doi:10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi:10.1038/s41587-019-0209-9
Brosseau, J. D., Yan, J.-Y., and Lo, K. V. (1986). The relationship between hydrogen gas and butanol production by Clostridium saccharoperbutylacetonicum. Biotechnol. Bioeng. 28 (3), 305–310. doi:10.1002/bit.260280302
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: Architecture and applications. BMC Bioinforma. 10 (1), 421. doi:10.1186/1471-2105-10-421
Candry, P., and Ganigué, R. (2021). Chain elongators, friends, and foes. Curr. Opin. Biotechnol. 67, 99–110. doi:10.1016/j.copbio.2021.01.005
Candry, P., Ulcar, B., Petrognani, C., Rabaey, K., and Ganigué, R. (2020). Ethanol:propionate ratio drives product selectivity in odd-chain elongation with Clostridium kluyveri and mixed communities. Bioresour. Technol. 313, 123651. doi:10.1016/j.biortech.2020.123651
Caporaso, G. J., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6 (8), 1621–1624. doi:10.1038/ismej.2012.8
Chen, W. S., Strik, D. P. B. T. B., Buisman, C. J. N., and Kroeze, C. (2017). Production of caproic acid from mixed organic waste: An environmental life cycle perspective. Environ. Sci. Technol. 51 (12), 7159–7168. doi:10.1021/acs.est.6b06220
Coma, M., Vilchez-Vargas, R., Roume, H., Jauregui, R., Pieper, D. H., and Rabaey, K. (2016). Product diversity linked to substrate usage in chain elongation by mixed-culture fermentation. Environ. Sci. Technol. 50 (12), 6467–6476. doi:10.1021/acs.est.5b06021
De Groof, V., Coma, M., Arnot, T., Leak, D. J., and Lanham, A. B. (2019). Medium chain carboxylic acids from complex organic feedstocks by mixed culture fermentation. Molecules 24 (3), 398. doi:10.3390/molecules24030398
de Leeuw, K. D., Buisman, C. J. N., and Strik, D. P. B. T. B. (2019). Branched medium chain fatty acids: Iso-caproate formation from iso-butyrate broadens the product spectrum for microbial chain elongation. Environ. Sci. Technol. 53 (13), 7704–7713. doi:10.1021/acs.est.8b07256
de Leeuw, K. D., Ahrens, T., Buisman, C. J. N., and Strik, D. P. B. T. (2021). Open culture ethanol-based chain elongation to form medium chain branched carboxylates and alcohols. Front. Bioeng. Biotechnol. 9, 697439. doi:10.3389/fbioe.2021.697439
Dekishima, Y., Lan, E. I., Shen, C. R., Cho, K. M., and Liao, J. C. (2011). Extending carbon chain length of 1-butanol pathway for 1-hexanol synthesis from glucose by engineered Escherichia coli. J. Am. Chem. Soc. 133 (30), 11399–11401. doi:10.1021/ja203814d
Delgado, A. ., G., Kang, D.-W., Nelson, K. G., Fajardo-Williams, D., Miceli, J. F., Done, H. Y., et al. (2014). Selective enrichment yields robust ethene-producing dechlorinating cultures from microcosms stalled at cis-dichloroethene. PLoS ONE 9 (6), e100654. doi:10.1371/journal.pone.0100654
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72 (7), 5069–5072. doi:10.1128/aem.03006-05
Doremus, M. G., Linden, J. C., and Moreira, A. R. (1985). Agitation and pressure effects on acetone-butanol fermentation. Biotechnol. Bioeng. 27 (6), 852–860. doi:10.1002/bit.260270615
Elsden, S. R., Volcani, B. E., Gilchrist, F. M. C., and Lewis, D. (1956). Properties of fatty acid forming organism isolated from the rumen of sheep. J. Bacteriol. 72 (5), 681–689. doi:10.1128/jb.72.5.681-689.1956
Fernández-Blanco, C., Veiga, M. C., and Kennes, C. (2022). Efficient production of n-caproate from syngas by a co-culture of Clostridium aceticum and Clostridium kluyveri. J. Environ. Manag. 302, 113992. doi:10.1016/j.jenvman.2021.113992
Fernández-Naveira, Á., Abubackar, H. N., Veiga, M. C., and Kennes, C. (2016). Carbon monoxide bioconversion to butanol-ethanol by Clostridium carboxidivorans: Kinetics and toxicity of alcohols. Appl. Microbiol. Biotechnol. 100 (9), 4231–4240. doi:10.1007/s00253-016-7389-8
Fernández-Naveira, Á., Veiga, M. C., and Kennes, C. (2017). H-B-E (hexanol-butanol-ethanol) fermentation for the production of higher alcohols from syngas/waste gas. J. Chem. Technol. Biotechnol. 92 (4), 712–731. doi:10.1002/jctb.5194
Ganigué, R., Sánchez-Paredes, P., Bañeras, L., and Colprim, J. (2016). Low fermentation pH is a trigger to alcohol production, but a killer to chain elongation. Front. Microbiol. 7, 702. doi:10.3389/fmicb.2016.00702
Han, W., He, P., Shao, L., and Lü, F. (2018). Metabolic interactions of a chain elongation microbiome. Appl. Environ. Microbiol. 84 (22), e01614–e01618. doi:10.1128/aem.01614-18
Han, W., He, P., Shao, L., and Lü, F. (2019). Road to full bioconversion of biowaste to biochemicals centering on chain elongation: A mini review. J. Environ. Sci. 86, 50–64. doi:10.1016/j.jes.2019.05.018
Huo, W., Fu, X., Bao, M., Ye, R., Shao, Y., Liu, Y., et al. (2022). Strategy of electron acceptors for ethanol-driven chain elongation from kitchen waste. Sci. Total Environ. 846, 157492. doi:10.1016/j.scitotenv.2022.157492
Joshi, S., Robles, A., Aguiar, S., and Delgado, A. G. (2021). The occurrence and ecology of microbial chain elongation of carboxylates in soils. ISME J. 15, 1907–1918. doi:10.1038/s41396-021-00893-2
Kenealy, W. R., and Waselefsky, D. M. (1985). Studies on the substrate range of Clostridium kluyveri; the use of propanol and succinate. Archives Microbiol. 141, 187–194. doi:10.1007/bf00408056
Kucek, L. A., Spirito, C. M., and Angenent, L. T. (2016). High n-caprylate productivities and specificities from dilute ethanol and acetate: Chain elongation with microbiomes to upgrade products from syngas fermentation. Energy Environ. Sci. 9 (11), 3482–3494. doi:10.1039/c6ee01487a
Lee, S.-M., Cho, M. O., Park, C. H., Chung, Y.-C., Kim, J. H., Sang, B.-I., et al. (2008). Continuous butanol production using suspended and immobilized Clostridium beijerinckii NCIMB 8052 with supplementary butyrate. Energy & Fuels 22 (5), 3459–3464. doi:10.1021/ef800076j
Li, S.-Y., Srivastava, R., Suib, S. L., Li, Y., and Parnas, R. S. (2011). Performance of batch, fed-batch, and continuous A–B–E fermentation with pH-control. Bioresour. Technol. 102 (5), 4241–4250. doi:10.1016/j.biortech.2010.12.078
Liu, Y., Lü, F., Shao, L., and He, P. (2016). Alcohol-to-acid ratio and substrate concentration affect product structure in chain elongation reactions initiated by unacclimatized inoculum. Bioresour. Technol. 218, 1140–1150. doi:10.1016/j.biortech.2016.07.067
Löffler, F., Sanford, R., and Ritalahti, K. (2005). Enrichment, cultivation, and detection of reductively dechlorinating bacteria. Methods Enzym. 397, 77–111. doi:10.1016/S0076-6879(05)97005-5
Löffler, F. E., Yan, J., Ritalahti, K. M., Adrian, L., Edwards, E. A., Konstantinidis, K. T., et al. (2013). Dehalococcoides mccartyi gen. nov., sp nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 63, 625–635. doi:10.1099/ijs.0.034926-0
Lonkar, S., Fu, Z., and Holtzapple, M. (2016). Optimum alcohol concentration for chain elongation in mixed-culture fermentation of cellulosic substrate. Biotechnol. Bioeng. 113 (12), 2597–2604. doi:10.1002/bit.26024
Matta-El-Ammouri, G., Janati-Idrissi, R., Junelles, A.-M., Petitdemange, H., and Gay, R. (1987). Effects of butyric and acetic acids on acetone-butanol formation by Clostridium acetobutylicum. Biochimie 69 (2), 109–115. doi:10.1016/0300-9084(87)90242-2
Meinel, M., Delgado, A. G., Ilhan, Z. E., Aguero, M. L., Aguiar, S., Krajmalnik-Brown, R., et al. (2022). Organic carbon metabolism is a main determinant of hydrogen demand and dynamics in anaerobic soils. Chemosphere 303, 134877. doi:10.1016/j.chemosphere.2022.134877
Miranda, E. M., Severson, C., Reep, J. K., Hood, D., Hansen, S., Santisteban, L., et al. (2022). Continuous-mode acclimation and operation of lignocellulosic sulfate-reducing bioreactors for enhanced metal immobilization from acidic mining-influenced water. J. Hazard. Mater. 425, 128054. doi:10.1016/j.jhazmat.2021.128054
Mohana Rangan, S., Mouti, A., LaPat-Polasko, L., Lowry, G. V., Krajmalnik-Brown, R., and Delgado, A. G. (2020). Synergistic zerovalent iron (Fe0) and microbiological trichloroethene and perchlorate reductions are determined by the concentration and speciation of Fe. Environ. Sci. Technol. 54 (22), 14422–14431. doi:10.1021/acs.est.0c05052
Mohana Rangan, S., Rao, S., Robles, A., Mouti, A., LaPat-Polasko, L., Lowry, G. V., et al. (2023). Decoupling Fe0 application and bioaugmentation in space and time enables microbial reductive dechlorination of trichloroethene to ethene: Evidence from soil columns. Environ. Sci. Technol. 57, 4167. doi:10.1021/acs.est.2c06433
Ndaba, B., Chiyanzu, I., and Marx, S. (2015). n-Butanol derived from biochemical and chemical routes: A review. Biotechnol. Rep. 8, 1–9. doi:10.1016/j.btre.2015.08.001
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41 (D1), D590–D596. doi:10.1093/nar/gks1219
Ramió-Pujol, S., Ganigué, R., Bañeras, L., and Colprim, J. (2015). Incubation at 25°C prevents acid crash and enhances alcohol production in Clostridium carboxidivorans P7. Bioresour. Technol. 192, 296–303. doi:10.1016/j.biortech.2015.05.077
Richter, H., Molitor, B., Diender, M., Sousa, D. Z., and Angenent, L. T. (2016). A narrow pH range supports butanol, hexanol, and octanol production from syngas in a continuous co-culture of Clostridium ljungdahlii and Clostridium kluyveri with in-line product extraction. Front. Microbiol. 7, 1773. doi:10.3389/fmicb.2016.01773
Rico, J. L., Reardon, K. F., and De Long, S. K. (2021). Inoculum microbiome composition impacts fatty acid product profile from cellulosic feedstock. Bioresour. Technol. 323, 124532. doi:10.1016/j.biortech.2020.124532
Robeson, M. S., II, O’Rourke, D. R., Kaehler, B. D., Ziemski, M., Dillon, M. R., Foster, J. T., et al. (2021). RESCRIPt: Reproducible sequence taxonomy reference database management. PLOS Comput. Biol. 17 (11), e1009581. doi:10.1371/journal.pcbi.1009581
Robles, A., Yellowman, T. L., Joshi, S., Mohana Rangan, S., and Delgado, A. G. (2021). Microbial chain elongation and subsequent fermentation of elongated carboxylates as H2-producing processes for sustained reductive dechlorination of chlorinated ethenes. Environ. Sci. Technol. 55 (15), 10398–10410. doi:10.1021/acs.est.1c01319
Roghair, M., Liu, Y., Strik, D. P. B. T., Weusthuis, R. A., Bruins, M. E., and Buisman, C. J. N. (2018). Development of an effective chain elongation process from acidified food waste and ethanol into n-caproate. Front. Bioeng. Biotechnol. 6, 50. doi:10.3389/fbioe.2018.00050
Rühl, J., Schmid, A., and Blank, L. M. (2009). Selected Pseudomonas putida strains able to grow in the presence of high butanol concentrations. Appl. Environ. Microbiol. 75 (13), 4653–4656. doi:10.1128/aem.00225-09
Schiel-Bengelsdorf, B., Montoya, J., Linder, S., and Dürre, P. (2013). Butanol fermentation. Environ. Technol. 34 (13-14), 1691–1710. doi:10.1080/09593330.2013.827746
Seedorf, H., Fricke, W. F., Veith, B., Brüggemann, H., Liesegang, H., Strittmatter, A., et al. (2008). The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc. Natl. Acad. Sci. 105 (6), 2128–2133. doi:10.1073/pnas.0711093105
Shrestha, S., Xue, S., and Raskin, L. (2023). Competitive reactions during ethanol chain elongation were temporarily suppressed by increasing hydrogen partial pressure through methanogenesis inhibition. Environ. Sci. Technol. 57 (8), 3369–3379. doi:10.1021/acs.est.2c09014
Spirito, C. M., Richter, H., Rabaey, K., Stams, A. J., and Angenent, L. T. (2014). Chain elongation in anaerobic reactor microbiomes to recover resources from waste. Curr. Opin. Biotechnol. 27, 115–122. doi:10.1016/j.copbio.2014.01.003
Spirito, C. M., Marzilli, A. M., and Angenent, L. T. (2018). Higher substrate ratios of ethanol to acetate steered chain elongation toward n-caprylate in a bioreactor with product extraction. Environ. Sci. Technol. 52 (22), 13438–13447. doi:10.1021/acs.est.8b03856
Steinbusch, K. J. J., Hamelers, H. V. M., and Buisman, C. J. N. (2008). Alcohol production through volatile fatty acids reduction with hydrogen as electron donor by mixed cultures. Water Res. Oxf. 42 (15), 4059–4066. doi:10.1016/j.watres.2008.05.032
Steinbusch, K. J. J., Hamelers, H. V. M., Plugge, C. M., and Buisman, C. J. N. (2010). Biological formation of caproate and caprylate from acetate: Fuel and chemical production from low grade biomass. Energy & Environ. Sci. 4 (1), 216–224. doi:10.1039/c0ee00282h
Stoddard, S. F., Smith, B. J., Hein, R., Roller, B. R. K., and Schmidt, T. M. (2014). rrnDB: Improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 43 (D1), D593–D598. doi:10.1093/nar/gku1201
Tashiro, Y., Takeda, K., Kobayashi, G., Sonomoto, K., Ishizaki, A., and Yoshino, S. (2004). High butanol production by Clostridium saccharoperbutylacetonicum N1-4 in fed-batch culture with pH-Stat continuous butyric acid and glucose feeding method. J. Biosci. Bioeng. 98 (4), 263–268. doi:10.1016/s1389-1723(04)00279-8
Thauer, R. K., Jungermann, K., Henninger, H., Wenning, J., and Decker, K. (1968). The energy metabolism of Clostridium kluyveri. Eur. J. Biochem. 4 (2), 173–180. doi:10.1111/j.1432-1033.1968.tb00189.x
Vees, C. A., Herwig, C., and Pflügl, S. (2022). Mixotrophic co-utilization of glucose and carbon monoxide boosts ethanol and butanol productivity of continuous Clostridium carboxidivorans cultures. Bioresour. Technol. 353, 127138. doi:10.1016/j.biortech.2022.127138
Veza, I., Muhamad Said, M. F., and Latiff, Z. A. (2021). Recent advances in butanol production by acetone-butanol-ethanol (ABE) fermentation. Biomass Bioenergy 144, 105919. doi:10.1016/j.biombioe.2020.105919
Wallace, R. J., Chaudhary, L. C., Miyagawa, E., McKain, N., and Walker, N. D. (2004). Metabolic properties of Eubacterium pyruvativorans, a ruminal ‘hyper-ammonia-producing’ anaerobe with metabolic properties analogous to those of Clostridium kluyveri. Microbiology 150 (9), 2921–2930. doi:10.1099/mic.0.27190-0
Wang, J., and Yin, Y. (2022). Biological production of medium-chain carboxylates through chain elongation: An overview. Biotechnol. Adv. 55, 107882. doi:10.1016/j.biotechadv.2021.107882
Wechgama, K., Laopaiboon, L., and Laopaiboon, P. (2017). Enhancement of batch butanol production from sugarcane molasses using nitrogen supplementation integrated with gas stripping for product recovery. Industrial Crops Prod. 95, 216–226. doi:10.1016/j.indcrop.2016.10.012
Xu, M., Zhao, J., Yu, L., Tang, I. C., Xue, C., and Yang, S.-T. (2015). Engineering Clostridium acetobutylicum with a histidine kinase knockout for enhanced n-butanol tolerance and production. Appl. Microbiol. Biotechnol. 99 (2), 1011–1022. doi:10.1007/s00253-014-6249-7
Yan, B. H., Selvam, A., and Wong, J. W. C. (2017). Influence of acidogenic headspace pressure on methane production under schematic of diversion of acidogenic off-gas to methanogenic reactor. Bioresour. Technol. 245, 1000–1007. doi:10.1016/j.biortech.2017.08.173
Yu, S., and Semprini, L. (2009). Enhanced reductive dechlorination of PCE DNAPL with TBOS as a slow-release electron donor. J. Hazard. Mater. 167 (1), 97–104. doi:10.1016/j.jhazmat.2008.12.087
Keywords: butanol, butyrate, hydrogen partial pressure, carboxylate reduction, Clostridium kluyveri, microbial chain elongation, total gas pressure
Citation: Robles A, Sundar SV, Mohana Rangan S and Delgado AG (2023) Butanol as a major product during ethanol and acetate chain elongation. Front. Bioeng. Biotechnol. 11:1181983. doi: 10.3389/fbioe.2023.1181983
Received: 08 March 2023; Accepted: 08 May 2023;
Published: 18 May 2023.
Edited by:
David Strik, Wageningen University and Research, NetherlandsReviewed by:
Christian Kennes, University of A Coruña, SpainCopyright © 2023 Robles, Sundar, Mohana Rangan and Delgado. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aide Robles, YXJvYmxlczlAYXN1LmVkdQ==; Anca G. Delgado, YW5jYS5kZWxnYWRvQGFzdS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.