- 1Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education, School of Life Sciences and Health Engineering, Jiangnan University, Wuxi, China
- 2Yixing Institute of Food and Biotechnology Co., Ltd., Yixing, China
- 3Seragon Biosciences, Inc., Irvine, CA, United States
- 4National Engineering Research Center for Cereal Fermentation and Food Biomanufacturing, School of Biotechnology, Jiangnan University, Wuxi, China
Introduction: NADH pyrophosphatase, a hydrolase catalyzing the phosphate bond of NADH to reduced nicotinamide mononucleotide, has potential applications in the food, cosmetic and pharmaceutical industry.
Methods: Here, we investigated the effects of vector screening, promoter and RBS strategies on NADH pyrophosphatase expression and protein engineering on its enzymatic activity and thermal stability.
Results: In this study, we describe a NADH pyrophosphatase derived from Escherichia coli (EcNudc). Strategies focusing on expression regulation including screening vectors, optimizing promoters and ribosome binding sites were utilized to enhance the productivity of EcNudc (1.8 U/mL). Moreover, protein engineering was adopted to further improve the catalytic properties of EcNudc, achieving 3.3-fold higher activity and 3.6-fold greater thermostability at 50°C. Furthermore, fermentation for the combined mutant R148A-H149E (EcNudc-M) production in a 7 L fermenter was implemented and the enzyme activity of EcNudc-M reached 33.0 U/mL. Finally, the EcNudc-M was applied in the catalysis of NADH with the highest NMNH yield of 16.65 g/L.
Discussion: In conclusion, we constructed a commercially available genetically engineered strain with high activity and thermal stability of NADH pyrophosphatase, laying a broad foundation for the biocatalytic industrial production of NMNH and expand its application range.
1 Introduction
NAD+, also known as coenzyme I, is involved in multiple metabolic pathways in organisms, including the tricarboxylic acid cycle, glycolysis, and fatty acid oxidation (Hikosaka et al., 2019) along with regulation of cellular functions such as DNA repair and cellular senescence (Covarrubias et al., 2021). Studies have shown that NAD+ can activate mitochondrial function to mitigate neurodegenerative diseases such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) (Hikosaka et al., 2019), to enhance sirtuin activity in the treatment of heart and kidney diseases (Hershberger et al., 2017) and to stimulate Fndc5/irisin factor to relieve non-alcoholic fatty liver disease (Li et al., 2021).
NAD+ levels in organisms decrease with age. Supplementation with NAD+ precursors, such as nicotinic acid (NA), nicotinamide, nicotinamide riboside (NR), and nicotinamide mononucleotide (NMN), can be used as an effective way to increase NAD+ levels. However, NA activates the Gpr109A receptor leading to flushing and nicotinamide may inhibit sirtuin function and affect reverse cholesterol transport (Bogan and Brenner, 2008). Therefore, NR and NMN are considered to the leading products for efficacy. Due to the high-doses dependent limitation of NMN and NR, the reduced nicotinamide mononucleotide (NMNH) has recently attracted increasing attention as a stronger NAD+ enhancer (Liu et al., 2021; Zapata-Perez et al., 2021).
Through metabolomic analysis and in vitro experiments, Liu et al. demonstrated that NMNH inhibited glycolysis, the tricarboxylic acid cycle and cell growth (Liu et al., 2021). Zapata-Pérez et al. supplemented the same concentrations of NMN and NMNH in different mouse and human cell lines, respectively, and found that NMNH significantly enhanced the content of NAD+ and was more effective than NMN. In addition, by injecting 250 mg/kg NMN and NMNH into C57BL/6N mice, it was found that NMNH maintained NAD+ levels for a longer period of time than NMN. Furthermore, NMNH supplementation also protected renal tubular epithelial cells from hypoxia and reoxygenation injury (Zapata-Perez et al., 2021).
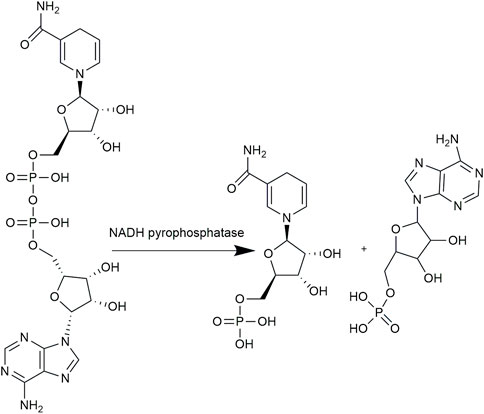
At present, there are two main methods for the synthesis of NMNH. The chemical reduction method reduces NMN into NMNH using reducing agent thiourea dioxide (TDO). Liu et al. (2021) reacted 340 mg NMN and 125 mg TDO in 10% ammonia solution at 40°C for 1 h. The product NMNH was then purified using preparative liquid phase and concentrated by vacuum drying. However, the reducing agent TDO is irritating to skin and mucous membranes, and may easily cause explosive decomposition under heat and impact conditions. The biological catalytic method depends on the catalysis of NADH pyrophosphatase (Nudc), which converts NADH into equal mole equivalent of NMNH and AMP (Figure 1). Compared with chemical method, the enzymatic synthesis reaction system is performed under mild reaction conditions with eco-friendly nature and shows high substrate specificity. NADH pyrophosphatase belongs to the Nudix hydrolase superfamily with the conserved sequence GX5EX7REVXEEXGU. In 1994, Frick and Bessman (1995) expressed Escherichia coli NADH pyrophosphatase in E. coli MG1655 strain. In catalyzing the hydrolysis of a series of dinucleotide pyrophosphates, the enzyme has a particular preference for NADH and the Vmax reached 7.6 µ/mg. In addition, other NADH pyrophosphatases have also been discovered and characterized in different organisms. For example, AbdelRaheim et al. (2001) found that the NADH diphosphatase encoded by Saccharomyces cerevisiae NPY1 Nudix hydrolase gene in peroxisome was successfully expressed and purified in E. coli BL21 (DE3) and the Vmax reached 2.0 μmol/min/mg. Wang et al. (2011) expressed the Nudc genes from M. tuberculosis H37Rv (NudCRv) and M. bovis BCG (NudCBCG) in E. coli BL21 (DE3). In the presence of Mg2+, the specific enzyme activities were 1.59 μ/mg and 0.02 μ/mg, respectively. Moreover, human NUDT12 and mouse NUDT13 were recombinantly expressed in insect cells by Abdelraheim et al. (2003; 2017).
Although various Nudcs have been cloned and expressed, their properties and productivities still cannot meet the industrial applications, and there are few studies focusing on enzyme modification. In this study, we first expressed the Nudc in E. coli BL21 (DE3) (named EcNudc). In order to enhance the expression of EcNudc, a series of regulation strategies including optimization of the vector, promoter engineering and ribosome binding site (RBS) engineering were conducted. Furthermore, site-directed mutation was performed to enhance the catalytic activity and thermal stability of EcNudc. To assess the catalytic potential of NADH pyrophosphatase, fed-batch fermentation in a 7 L fermenter was performed. Finally, the NADH biocatalytic synthesis of NMNH reaction process was also evaluated to investigate industrial application potential. Overall, we constructed a commercially available genetically engineered strain with high activity and thermal stability of NADH pyrophosphatase, laying a broad foundation for biosynthetic NMNH application aspects.
2 Materials and methods
2.1 Materials
E. coli JM109 was used for gene cloning and E. coli BL21 (DE3) was used for recombinant protein expression, respectively. The expression plasmids including pET-28a (+), pET-3b, pRSFDuet-1, pCDFDuet-1, pACYCDuet-1 are all kept in the laboratory. The sequence of EcNudc was optimized and modified based on NADH pyrophosphatase from E. coli (GenBank: NEY29540.1).
The shake flask fermentation medium included 10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl. The seed medium included (g/L) (Gong et al., 2018): tryptone 20, yeast extract 5, KCl 0.19, NaCl 0.5, glucose 3.6, MgCl2 0.95, trace elements (FeCl3.6H2O 6, CuCl2.2H2O 0.2, CaCl2 0.2, EDTA 0.5, ZnSO4.7H2O 0.58, MnSO4·H2O 0.3) 2 mL/L. The on-tank fermentation medium included (g/L) (Gong et al., 2018): tryptone 20, yeast extract 5, KCl 0.19, NaCl 0.5, glucose 10, MgCl2 0.95, trace elements 2 mL/L. The feeding medium contained (g/L): glucose 700, MgSO4 14, trace elements 14 mL/L.
2.2 Engineering of promoters and RBSs for improving EcNudc expression
The promoters were kept in the laboratory. According to the translation rate of wild-type RBS on the plasmid calculated by the RBS calculator (Salis et al., 2009), RBS of different intensities (RBS5000-RBS50000) were designed to regulate the expression of the NADH pyrophosphatase. The RBS was replaced by inverse PCR technology. The PCR system consists of 50 μL total, including 20 μL ddH2O, 2 μL upstream primer, 2 μL downstream primer, 1 μL template, 25 μL 2 × Phanta Max buffer. The PCR amplification procedure was conducted as follows: 95°C for 3 min, then followed by 34 cycles (95°C for 15 s, 55°C for 15 s, 72°C for 6 min), and last extension 72°C for 5 min. The PCR products were digested with 1 μL DPNI enzyme for 2 h at 37°C (10 μL system includes 1 μL DPNI enzyme, 1 μL 10× Q.cut Buffer, 8 μL PCR product), and then transformed into E. coli hosts for cloning and expression. Correctly sequenced strains were subjected to fermentation. The primers are shown in Supplementary Table S1.
2.3 NADH pyrophosphatase assay
NADH pyrophosphatase activity was measured by determining the content of inorganic phosphate. The standard reaction mixture (50 μL) consisted of 5 mM MgCl2, 50 mM Tris-HCl, pH = 8.0; 25 mM NADH; 4 U alkaline phosphatase (calf intestine) and enzyme. After reaction with 37°C for 15 min, 250 μL of 4 mM EDTA was added to terminate the reaction. And then inorganic phosphate was measured by adding 700 μL of mixture (10% ascorbic acid and 0.42% ammonium molybdate tetrahydrate) at a wavelength of 820 nm (Ames and Dubin, 1960). One unit of NADH pyrophosphatase hydrolyzes 1 μmol NADH per minute under these conditions.
2.4 Site-directed mutagenesis of EcNudc
Through HotSpot Wizard web service, we selected eight residues (Sumbalova et al., 2018). The NADH pyrophosphatase site was mutated using inverse PCR. The method is the same as above.
2.5 Protein expression, purification of EcNudc and mutant, and kinetic parameter analysis
The recombinant strains and mutants were first cultivated at 37°C for 2 h. When OD600 reached 0.6–0.8, adding 0.5 mM IPTG to induce the expression of the strain at 25°C for 12 h. After fermentation, recombinant cells were harvested by centrifugation at 8,000 rpm, 4°C for 10 min. Each cell was washed and resuspended with Tris-HCl (PH = 8.0). After the resuspension was sonicated for 30 min, the supernatant was obtained by centrifugation at 8,000 rpm for 10 min at 4°C. Ni2+ column affinity chromatography was used for protein purification and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to assay protein purification. Protein concentration was determined by BCA Protein Assay Kit. Kinetic parameters were determined for EcNudc and mutant strains by using different substrate concentrations (0.25–10 mM).
2.6 Molecular docking analysis
The protein structure model was built by SWISS-MODEL (Bienert et al., 2017; Waterhouse et al., 2018). Molecular docking was performed using AutoDock Vina (Trott and Olson, 2010; Eberhardt et al., 2021). The interactions between ligands and receptors were analyzed accordingly.
2.7 Fermentation experiments on tanks
The single colony of EcNudc-M from an agar plate (kanamycin) was selected into 10 mL of LB medium and cultivated 37°C and 220 rpm for 12 h. Then transferred to seed medium at 2% inoculum and cultivated at the same condition. Finally, the seed solution was then inoculated at a 10% inoculum into a 7 L fermenter with 4 L of medium, and when OD600 = 2, 0.1 mM IPTG was added and induced at 25°C. When the glucose concentration was below 2 g/L, the constant rate feeding was started at a rate of 0.1 mL/min. The speed and ventilation were regulated throughout the process to keep the dissolved oxygen above 10%, and the pH = 7.0 by adding ammonia.
2.8 Catalytic performance of the mutant strain and HPLC analysis
The catalytic performance of NADH pyrophosphatase for the bioconversion of NADH to NMNH was investigated. The supernatant of recombinant E. coli NADH pyrophosphatase was prepared and subjected to biotransformation reactions. The reaction solution was periodically sampled to detect the concentration of substrate NADH and product NMNH by HPLC. Using Diamonsil C18 column (5 μm, 250 × 4.6 mm) to perform HPLC detection at 340 nm. The mobile phase consists of two parts. Mobile phase A was methanol, water, acetic acid and tetrabutylammonium hydroxide (3/97/1/0.6), mobile phase B was methanol. The procedure was as follows: 0 min, 100% A; 5 min, 100% A; 25 min, 40% A; 30 min, 20% A; 40 min, 100% A.
3 Results and discussion
3.1 Screening of plasmids overexpressing Ec Nudc
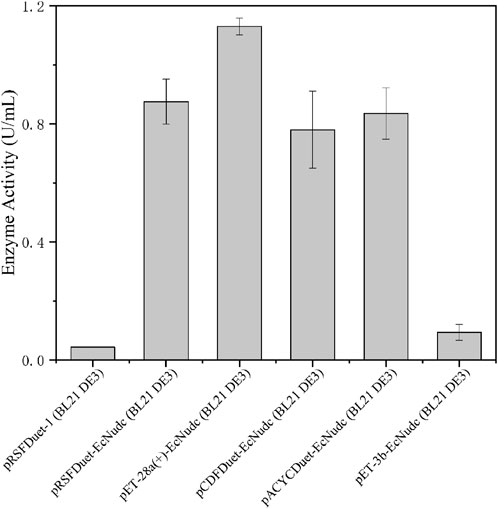
E. coli is widely used as a host for protein expression due to its simple genetic manipulation and rapid growth. Plasmids with different copy numbers are important factors affecting protein expression (Hayat et al., 2018). Therefore, we performed the initial expression of the NADH pyrophosphatase gene in E. coli and the enzyme activity of strain pRSFDuet-EcNudc/BL21 (DE3) was 0.88 U/mL, which was a high copy number plasmid (Shoji et al., 2021). SDS-PAGE analysis showed that the enzyme was successfully expressed in E. coli without inclusion body formation (Supplementary Figure S1). Then, plasmids with other copy numbers and expression patterns were also used to probe the effect on enzyme activity (Hayat et al., 2018). As shown in Figure 2, the pET-28a (+)-EcNudc/BL21 (DE3) strain had the highest enzyme activity, reaching 1.1 U/mL. In contrast, for the strain pRSFDuet-EcNudc carrying a high copy number plasmid, the enzyme activity was about the same as that of the strain pACYCDuet-EcNudc carrying a low copy number plasmid. This also indicates that the enzyme activity is not linearly correlated with the copy number (Shoji et al., 2021). Therefore, the pET-28a (+)-EcNudc/BL21 (DE3) strain was selected for further research.
3.2 Engineering of promoter and RBS sequences to enhance the enzyme activity of EcNudc
The promoters and ribosome binding sites (RBSs) are important elements for regulating gene transcription and the initiation rate of protein translation, respectively. Optimizing promoters and RBS sequences have become an effective strategy to rationally improve gene expression and increase productivity. Hou et al. (2013) increased the activity of D-amino acid oxidase (DAAO) by replacing the promoter and RBS, promoting its soluble expression. Duan et al. (2021) systematically studied the combined effect of promoter and RBS in C. glutamicum for the first time, and finally the production of arginine and citrulline in the 7 L reactor increased by 1.61 times and 2.35 times, respectively. Therefore, the promoter element was modified in the early stage. Among these promoters, adhA was the most effective in tandem with the T7 promoter and the enzyme activity reached 1.3 U/mL, increased by 18.2% (Figure 3A). On this basis, according to the RBS calculator, RBSs with different strengths were designed to further improve the catalytic ability of EcNudc and the constructed plasmids were transformed into E. coli BL21 (DE3) to compare their enzyme production levels. As shown in Figure 3B, there was no linear correlation between RBS intensity and expression, where the intensity region of 20,000–40,000 was suitable for EcNudc expression. The pyrophosphatase activity of RBS-20000 reached 1.8 U/mL, which was about 38.5% higher than that of the control strain. In addition, according to SDS-PAGE (Supplementary Figure S2), the expression level of RBS-20000 was increased compared with the control. Therefore, we successfully established an efficient expression system for producing NMNH in E. coli system.
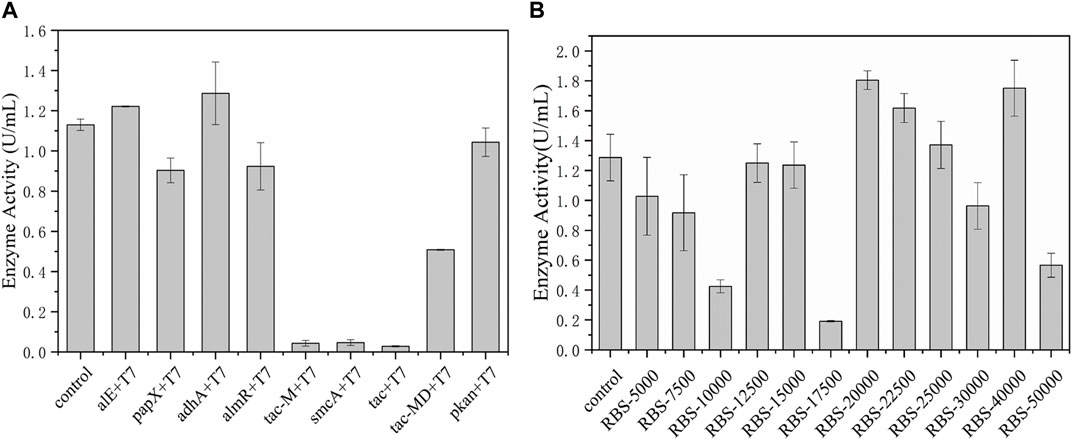
FIGURE 3. Adaptability modification of promoters and RBSs. (A) Adaptability modification of promoters. (B) Adaptability modification of RBSs with strengths.
3.3 Design and expression of recombinant EcNudc mutants through semi-rational strategies
It is important for industrial application to improve the catalytic capacity, thermal stability, and pH tolerance of enzymes. Thermally stable enzymes could greatly improve productivity and promote industrial applications (Lee et al., 2014; Zhu et al., 2021). HotSpot Wizard is a web server that allows semi-rational design of proteins based on bioinformatics analysis to enhance their catalytic activity, stability, etc., (Sumbalova et al., 2018). Therefore, in order to further improve the catalytic activity and thermostability of pyrophosphatase, based on the structural analysis and crucial residues prediction by HotSpot Wizard, we selected and mutated residue hotspots to the corresponding amino acids, resulting in eight mutation sites T100A, R122A, R148T, R150C, K218N, R120S, T147F, and H149M. As shown in Figure 4, the enzyme activities of R148T and H149M were significantly higher than the activity of the wild type by 91% and 64%, respectively. Therefore, two sites (R148, H149) with significantly elevated enzyme activity were subsequently subjected for site-directed saturation mutation.
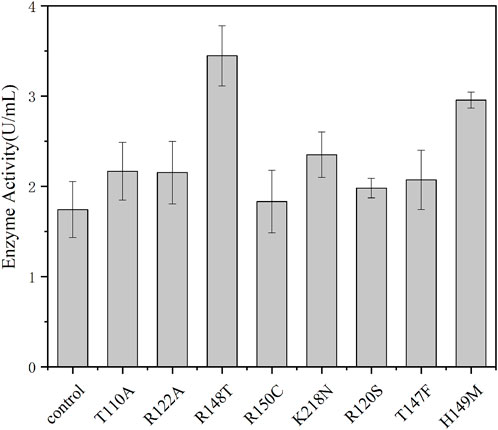
FIGURE 4. Enzyme activity of the EcNudc and its mutants under the standard reaction condition (37°C, pH 8.0).
3.4 Site-directed saturation mutagenesis of EcNudc
Saturation mutations were performed on the two selected residue sites. According to the results in Figure 5, the arginine at position 148 was mutated to the other 19 kinds of amino acids, and the mutant R148A achieved the most effective promotion with a 2.8-fold activity. For 149 site, the mutant H149E showed the highest activity of 5.1 U/mL. To further enhance the catalytic activity of NADH pyrophosphatase, a combined mutation of these two loci was constructed to obtain a double mutant strain R148A-H149E (named as EcNudc-M) with an enzyme activity of 6.0 U/mL. Multiple sequence alignment results indicate that the two sites are not fully conserved. This is the first report on these two mutant sites (Supplementary Figure S3).
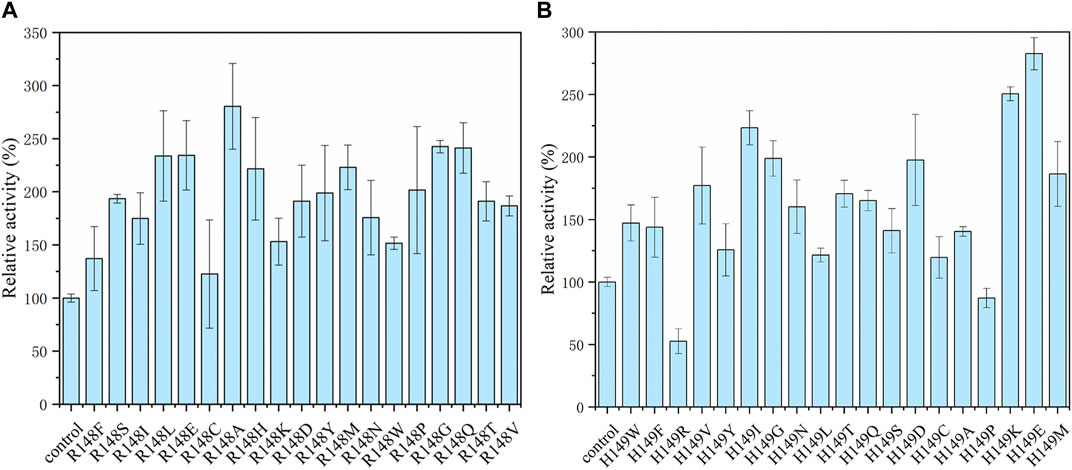
FIGURE 5. Site-directed saturation mutagenesis of EcNudc. (A) Relative activity with saturation mutations at the R148. (B) Relative activity with saturation mutations at the H149.
The protein structure model of EcNudc was built by SWISS-MODEL (Supplementary Figure S4) (Bienert et al., 2017; Waterhouse et al., 2018). The results of molecular docking showed that the R148 and H149 sites and catalytic residues (Garcia-Saura et al., 2019) (E174, E177, and E178) were on both sides of the substrate NADH. The positive charge of R148 formed an electrostatic interaction with the negative charge of the phosphate bond on the substrate NADH, hindering the contact of the phosphate bond with the catalytic residue and affecting the catalytic efficiency. Therefore, when R148 was mutated to other amino acids, the enzyme activities were all enhanced. Especially, R148A has the best performance of all variants (Figures 6A, B). The mutations in adjacent residues generally display synergistic effects (Ellis et al., 2022). When H149 was mutated to glutamic acid, it formed electrostatic repulsion with the O− of the phosphate bond on NADH (Figure 6C). Therefore, the enzyme activity was enhanced. Furthermore, the enzyme activity of the combined mutant strain EcNudc-M reached 3.3-fold of the wild type. This is due to the loss of electrostatic interaction as well as electrostatic repulsion, which promoted the contact of the substrate NADH with the catalytic residue (Figure 6D).
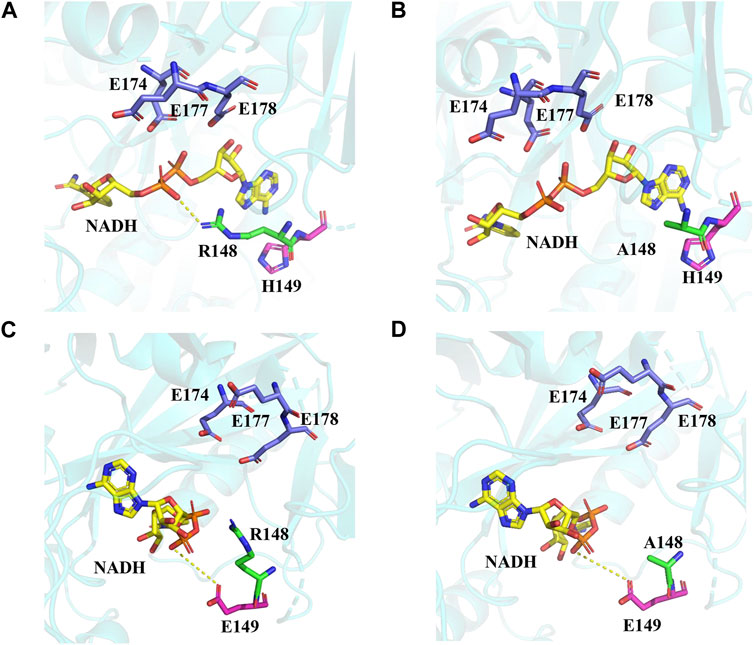
FIGURE 6. Structural analysis of the activity-related residues. (A) WT. (B) Mutant (R148A). (C) Mutant (H149E). (D) Mutant (EcNudc-M).
3.5 Purification and kinetic parameter of WT and mutants
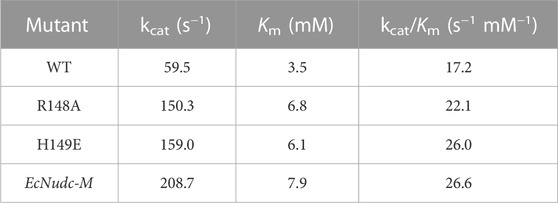
In this study, a single protein band of approximately 30 KDa appeared on SDS-PAGE after purification of WT and mutants by a nickel column (Supplementary Figure S5). Then, the kinetic parameters were measured separately. As shown in Table 1, the Km value was increased from 3.5 (WT) to 7.9 (EcNudc-M), indicating a decreased affinity of the mutant toward NADH. However, compared with the wild type, all mutants exhibited higher catalytic efficiency. In particular, the catalytic efficiency of the mutant EcNudc-M was increased by 1.5-fold.
3.6 Thermal stability assessment of mutants
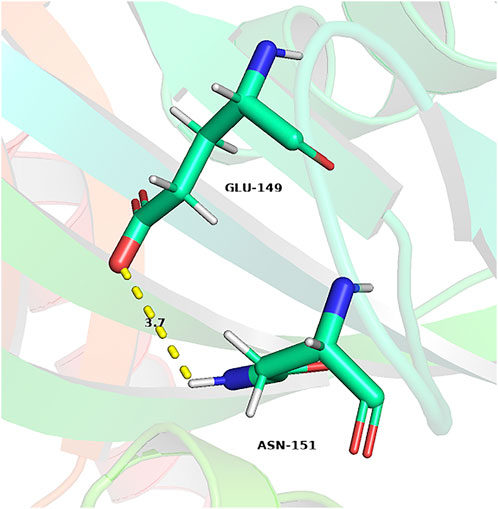
In addition to the enhanced enzyme activity, the stability of the mutant strains was also improved. As shown in Figure 7, the half-life of WT, R148A, H149E and EcNudc-M were 8 h, 10.9 h, 54 h, and 53.2 h at 40°C, respectively; and 4.8 min, 6.8 min, 14.1 min, and 17.5 min at 50°C, respectively. This indicated that the thermostability of EcNudc-M were significantly improved at 40°C and 50°C by 6.65 and 3.6 folds. The mutation of EcNudc-M was refinement by Rosetta relax and the enhanced stability is mainly due to the formation of hydrogen bonding between E149 and N151 (Figure 8).
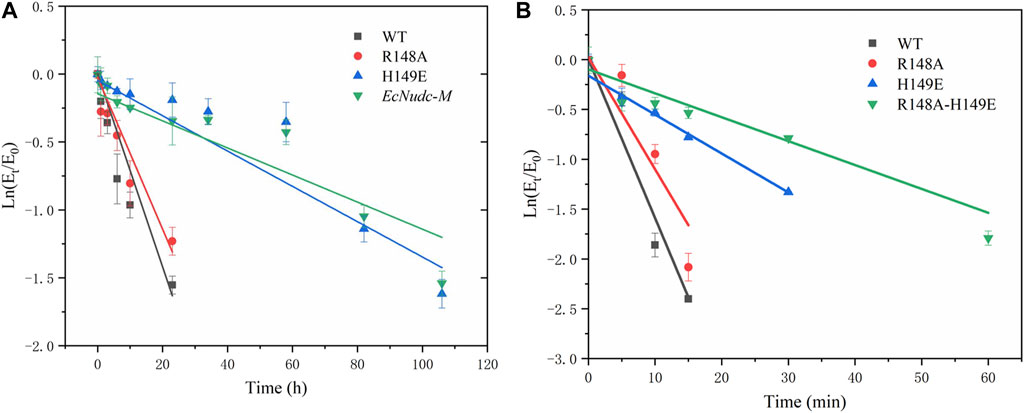
FIGURE 7. Thermal stability of wild type and mutants. (A) Thermal stability of wild type and mutants at 40°C. (B) Thermal stability of wild type and mutants at 50°C.
3.7 Scale-up production of NADH pyrophosphatase in 7 L fermenter
To further evaluate the production capacity of NADH pyrophosphatase, the fementation of the mutant EcNudc-M was expanded on a 7 L fermenter based on a constant rate replenishment strategy. In the early stage of fermentation, glucose was consumed at a rapid rate. The supplemented medium was added when the glucose concentration was below 2 g/L (12 h) and the growth was in an upward trend, but no significant increase was observed for the enzyme activity. It was thought that the supplemented glucose was mainly used for the growth of the strain. With the replenishment found that the overall rate of glucose consumption was on a downward trend, the growth of the strain also tended to level off, and the enzyme activity also decreased. The highest NADH pyrophosphatase activity reached 33 U/mL at 33 h, which was 3.5-fold higher than that of the flask level (after induction optimization) (Figure 9A). SDS-PAGE analysis indicated that the expression level of EcNudc-M was further enhanced (Figure 9B).
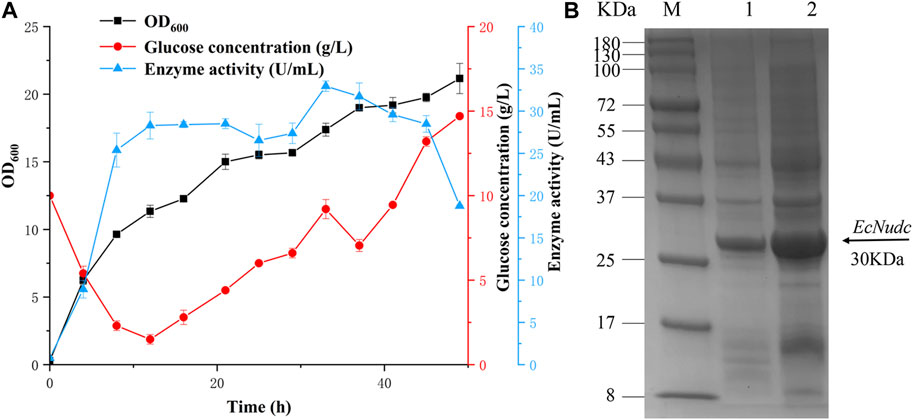
FIGURE 9. Fermentation curve of EcNudc-M and SDS-PAGE analysis of recombinant EcNudc expression. (A) Fermentation curve of EcNudc-M under constant rate replenishment strategy. (B) SDS-PAGE analysis of recombinant EcNudc expression in shake flask and fermenter level. M: protein standard marker; Lane 1: The supernatant of EcNudc-M in shake flask level; Lane 2: The supernatant of EcNudc-M in the fermenter level.
3.8 Biosynthesis of NMNH by mutant EcNudc-M
In order to evaluate the application of NADH pyrophosphatase for catalytic production of NMNH, the crude enzyme of EcNudc-M was used as a catalyst for biocatalytic reactions. However, in order to fully understand the catalytic properties of NADH pyrophosphatase recombinant bacteria on the substrate NADH, the optimal conditions for the catalytic reaction were first explored. The results are shown in Figure 10A, with the increase of reaction temperature, the NMNH yield also gradually increased, and the highest yield was reached at 40°C. pH is also an important factor affecting the catalytic reaction, as shown in Figure 10B, the conversion reaction had higher yield under alkaline conditions, in which, the highest yield was reached at pH = 9. Therefore, the best biotransformation experiment was performed at 40°C in pH = 9 buffer.
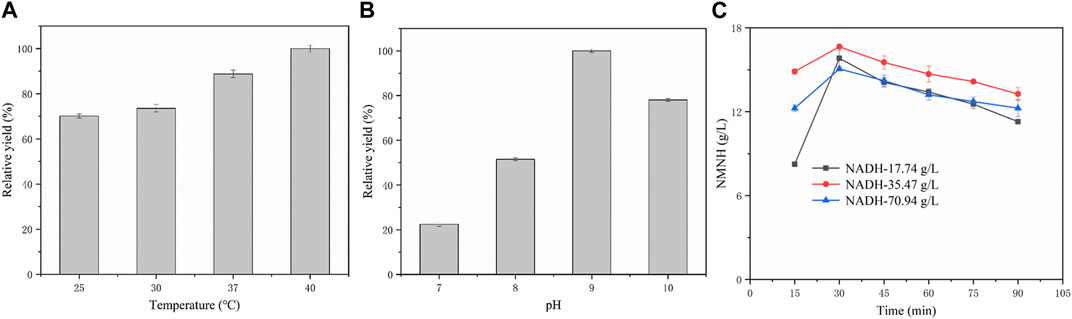
FIGURE 10. Exploration of catalytic process and biosynthesis of NMNH. (A) Relative yields of NMNH at different reaction temperatures. (B) Relative yields of NMNH at different reaction pH. (C) Biotransformation curve of NMNH at different substrate concentrations.
The biotransformation reactions were studied at 25 mM, 50 mM, and 100 mM substrate concentrations, respectively. The time courses of the bioconversion of NADH to NMNH were shown in Figure 10C. The highest yield of 16.65 g/L was achieved at 30 min at a substrate concentration of 50 mM and the conversion rate was 99.1%. However, after a second batch of substrate was put in at 30 min, the NMNH yield decreased, which is due to the instability of NMNH (Liu et al., 2021). When the substrate concentration was 25 mM, the highest yield reached 15.82 g/L and the conversion rate was 94.2% at 30 min (the second batch of substrate was put in at 15 min). However, the yield of NMNH also decreased after the third batch of substrate was put in at 30 min. When the substrate concentration was 100 mM, the yield reached a maximum of 15.06 g/L and the conversion rate was 44.8%. Therefore, the maximum yield of EcNudc-M in this study could be achieved at a substrate concentration of 50 mM, which is the highest level reported so far and further lays the foundation for industrial application.
4 Conclusion
NMNH has received substantial attention due to stimulating a rapid increase in NAD+ levels and its potential for applications such as the treatment of acute kidney injury. However, there are few studies on the enzymatic catalytic production of NMNH. Herein we successfully constructed an E. coli system for the efficient expression of NADH pyrophosphatase. The enzyme activity of NADH pyrophosphatase was significantly improved from 1.1 U/mL to 6.0 U/mL via expression regulation strategy and site-directed saturation mutagenesis. Furthermore, the thermal stability of EcNudc-M was also improved at 50°C by 3.6-folds. The molecular docking results showed that the weakened electrostatic interaction and the formation of hydrogen bonds were responsible for the enhancement of enzyme activity and thermal stability, respectively. The highest activity reached 33 U/mL by a continuous replenishment strategy in an expanded culture within a 7 L fermenter. Finally, the highest yield of NMNH was 16.65 g/L under the catalysis of EcNudc-M, which is the highest level reported so far. Overall, the mutant strain in this work improved the production efficiency of NMNH, which laid the foundation for its further industrial application.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
YL: Conceptualization, investigation, writing–original draft, visualization. J-SG: Investigation, writing–review and editing, supervision, project administration, funding acquisition. GM and CS: Writing–review and editing. MH, HL, G-QX, J-SS, and Z-HX: Investigation, supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Key R&D Program of China (No. 2021YFC2100900), the National Natural Science Foundation of China (No. 32171261), and the Fundamental Research Funds for the Central Universities (No. JUSRP22047).
Conflict of interest
Authors YL, GM, MH, J-SS, and Z-HX are employed by Yixing Institute of Food and Biotechnology Co., Ltd. and Seragon Biosciences.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1159965/full#supplementary-material
References
AbdelRaheim, S. R., Cartwright, J. L., Gasmi, L., and McLennan, A. G. (2001). The NADH diphosphatase encoded by the Saccharomyces cerevisiae NPY1 nudix hydrolase gene is located in peroxisomes. Arch. Biochem. Biophys. 388 (1), 18–24. doi:10.1006/abbi.2000.2268
Abdelraheim, S. R., Spiller, D. G., and McLennan, A. G. (2003). Mammalian NADH diphosphatases of the nudix family: Cloning and characterization of the human peroxisomal NUDT12 protein. Biochem. J. 374 (2), 329–335. doi:10.1042/bj20030441
Abdelraheim, S. R., Spiller, D. G., and McLennan, A. G. (2017). Mouse Nudt13 is a mitochondrial nudix hydrolase with NAD(P)H pyrophosphohydrolase activity. Protein J. 36 (5), 425–432. doi:10.1007/s10930-017-9734-x
Ames, B. N., and Dubin, D. T. (1960). The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J. Biol. Chem. 235, 769–775. doi:10.1016/S0021-9258(19)67936-6
Bienert, S., Waterhouse, A., de Beer, T. A., Tauriello, G., Studer, G., Bordoli, L., et al. (2017). The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 45 (D1), D313–d319. doi:10.1093/nar/gkw1132
Bogan, K. L., and Brenner, C. (2008). Nicotinic acid, nicotinamide, and nicotinamide riboside: A molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 28, 115–130. doi:10.1146/annurev.nutr.28.061807.155443
Covarrubias, A. J., Perrone, R., Grozio, A., and Verdin, E. (2021). NAD(+) metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 22 (2), 119–141. doi:10.1038/s41580-020-00313-x
Duan, Y., Zhai, W., Liu, W., Zhang, X., Shi, J. S., Zhang, X., et al. (2021). Fine-tuning multi-gene clusters via well-characterized gene expression regulatory elements: Case study of the arginine synthesis pathway in C. glutamicum. ACS Synth. Biol. 10 (1), 38–48. doi:10.1021/acssynbio.0c00405
Eberhardt, J., Santos-Martins, D., Tillack, A. F., and Forli, S. (2021). AutoDock Vina 1.2.0: New docking methods, expanded force field, and Python bindings. J. Chem. Inf. Model. 61 (8), 3891–3898. doi:10.1021/acs.jcim.1c00203
Ellis, J. M., Campbell, M. E., Kumar, P., Geunes, E. P., Bingman, C. A., and Buller, A. R. (2022). Biocatalytic synthesis of non-standard amino acids by a decarboxylative aldol reaction. Nat. Catal. 5 (2), 136–143. doi:10.1038/s41929-022-00743-0
Frick, D. N., and Bessman, M. J. (1995). Cloning, purification, and properties of a novel NADH pyrophosphatase. J. Biol. Chem. 270 (4), 1529–1534. doi:10.1074/jbc.270.4.1529
Garcia-Saura, A. G., Zapata-Perez, R., Martinez-Monino, A. B., Hidalgo, J. F., Morte, A., Perez-Gilabert, M., et al. (2019). The first comprehensive phylogenetic and biochemical analysis of NADH diphosphatases reveals that the enzyme from Tuber melanosporum is highly active towards NAD+. Sci. Rep. 9 (1), 16753. doi:10.1038/s41598-019-53138-w
Gong, J. S., Zhang, Q., Gu, B. C., Dong, T. T., Li, H., Li, H., et al. (2018). Efficient biocatalytic synthesis of nicotinic acid by recombinant nitrilase via high density culture. Bioresour. Technol. 260, 427–431. doi:10.1016/j.biortech.2018.03.109
Hayat, S. M. G., Farahani, N., Golichenari, B., and Sahebkar, A. (2018). Recombinant protein expression in Escherichia coli (E.coli): What we need to know. Curr. Pharm. Des. 24 (6), 718–725. doi:10.2174/1381612824666180131121940
Hershberger, K. A., Martin, A. S., and Hirschey, M. D. (2017). Role of NAD(+) and mitochondrial sirtuins in cardiac and renal diseases. Nat. Rev. Nephrol. 13 (4), 213–225. doi:10.1038/nrneph.2017.5
Hikosaka, K., Yaku, K., Okabe, K., and Nakagawa, T. (2019). Implications of NAD metabolism in pathophysiology and therapeutics for neurodegenerative diseases. Nutr. Neurosci. 24 (5), 371–383. doi:10.1080/1028415x.2019.1637504
Hou, J., Liu, Y., Li, Q., and Yang, J. (2013). High activity expression of D-amino acid oxidase in Escherichia coli by the protein expression rate optimization. Protein Expr. Purif. 88 (1), 120–126. doi:10.1016/j.pep.2012.11.020
Lee, C. W., Wang, H. J., Hwang, J. K., and Tseng, C. P. (2014). Protein thermal stability enhancement by designing salt bridges: A combined computational and experimental study. PLoS One 9 (11), e112751. doi:10.1371/journal.pone.0112751
Li, D. J., Sun, S. J., Fu, J. T., Ouyang, S. X., Zhao, Q. J., Su, L., et al. (2021). NAD(+)-boosting therapy alleviates nonalcoholic fatty liver disease via stimulating a novel exerkine Fndc5/irisin. Theranostics 11 (9), 4381–4402. doi:10.7150/thno.53652
Liu, Y., Luo, C., Li, T., Zhang, W., Zong, Z., Liu, X., et al. (2021). Reduced nicotinamide mononucleotide (NMNH) potently enhances NAD(+) and suppresses glycolysis, the TCA cycle, and cell growth. J. Proteome Res. 20 (5), 2596–2606. doi:10.1021/acs.jproteome.0c01037
Salis, H. M., Mirsky, E. A., and Voigt, C. A. (2009). Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 27 (10), 946–950. doi:10.1038/nbt.1568
Shoji, S., Yamaji, T., Makino, H., Ishii, J., and Kondo, A. (2021). Metabolic design for selective production of nicotinamide mononucleotide from glucose and nicotinamide. Metab. Eng. 65, 167–177. doi:10.1016/j.ymben.2020.11.008
Sumbalova, L., Stourac, J., Martinek, T., Bednar, D., and Damborsky, J. (2018). HotSpot wizard 3.0: Web server for automated design of mutations and smart libraries based on sequence input information. Nucleic Acids Res. 46 (W1), W356–w362. doi:10.1093/nar/gky417
Trott, O., and Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31 (2), 455–461. doi:10.1002/jcc.21334
Wang, X. D., Gu, J., Wang, T., Bi, L. J., Zhang, Z. P., Cui, Z. Q., et al. (2011). Comparative analysis of mycobacterial NADH pyrophosphatase isoforms reveals a novel mechanism for isoniazid and ethionamide inactivation. Mol. Microbiol. 82 (6), 1375–1391. doi:10.1111/j.1365-2958.2011.07892.x
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–w303. doi:10.1093/nar/gky427
Zapata-Perez, R., Tammaro, A., Schomakers, B. V., Scantlebery, A. M. L., Denis, S., Elfrink, H. L., et al. (2021). Reduced nicotinamide mononucleotide is a new and potent NAD(+) precursor in mammalian cells and mice. FASEB J. 35 (4), e21456. doi:10.1096/fj.202001826R
Keywords: reduced nicotinamide mononucleotide, NADH pyrophosphatase, expression, semi-rational engineering, protein engineering
Citation: Liu Y, Gong J-S, Marshall G, Su C, Hall M, Li H, Xu G-Q, Shi J-S and Xu Z-H (2023) Protein engineering of NADH pyrophosphatase for efficient biocatalytic production of reduced nicotinamide mononucleotide. Front. Bioeng. Biotechnol. 11:1159965. doi: 10.3389/fbioe.2023.1159965
Received: 06 February 2023; Accepted: 24 March 2023;
Published: 04 April 2023.
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaCopyright © 2023 Liu, Gong, Marshall, Su, Hall, Li, Xu, Shi and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Song Gong, amluc29uZ2dvbmcuYmlvQGhvdG1haWwuY29t
 Ye Liu1,2
Ye Liu1,2 Jin-Song Gong
Jin-Song Gong Guo-Qiang Xu
Guo-Qiang Xu Jin-Song Shi
Jin-Song Shi Zheng-Hong Xu
Zheng-Hong Xu