
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 22 June 2023
Sec. Bioprocess Engineering
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1159297
We investigated the optimum co-culture ratio with the highest biological nitrogen removal rate, revealing that chemical oxygen demand, total nitrogen (TN), and ammoniacal nitrogen (NH3-N) removal was increased in the Chlorella pyrenoidosa and Yarrowia lipolytica co-culture system at a 3:1 ratio. Compared with the control, TN and NH3-N content in the co-incubated system was decreased within 2–6 days. We investigated mRNA/microRNA (miRNA) expression in the C. pyrenoidosa and Y. lipolytica co-culture after 3 and 5 days, identifying 9885 and 3976 differentially expressed genes (DEGs), respectively. Sixty-five DEGs were associated with Y. lipolytica nitrogen, amino acid, photosynthetic, and carbon metabolism after 3 days. Eleven differentially expressed miRNAs were discovered after 3 days, of which two were differentially expressed and their target mRNA expressions negatively correlated with each other. One of these miRNAs regulates gene expression of cysteine dioxygenase, hypothetical protein, and histone-lysine N-methyltransferase SETD1, thereby reducing amino acid metabolic capacity; the other miRNA may promote upregulation of genes encoding the ATP-binding cassette, subfamily C (CFTR/MRP), member 10 (ABCC10), thereby promoting nitrogen and carbon transport in C. pyrenoidosa. These miRNAs may further contribute to the activation of target mRNAs. miRNA/mRNA expression profiles confirmed the synergistic effects of a co-culture system on pollutant disposal.
The nitrogen and phosphorus accumulation in agriculture and industrial wastewater leads to water eutrophication (Judd et al., 2015). It is more challenging to treat this kind of wastewater using traditional biological treatment methods. The wastewater treatment using algae has become a promising approach with a high effect on biological nitrogen removal (BNR) (Vyverman, 2016). Many types of filamentous algae have been used in wastewater treatment, including Oedogonium, Cladophora, Chlorella, Spirogyra, and Rhizoclonium (Liu et al., 2020). Nevertheless, treating wastewater using algae alone is more expensive and inefficient. Co-culture of microalgae and microorganisms is a cost-effective and efficient technology (Li et al., 2022). Microalgae are photosynthetic microorganisms (Patidar et al., 2015) that can recover and recycle nutrients from wastewater and accumulate large amounts of valuable biomass (Nwoba et al., 2019). It absorbs nitrogen from wastewater through photosynthesis and releases oxygen for microorganisms. Microorganisms can use the organic pollutants in the wastewater and release carbon dioxide through respiration to enhance the photosynthesis of microalgae (Lee et al., 2015; Singh et al., 2020). BNR is reached while forming a benign energy-materials cycle. Chlorella pyrenoidosa is one of the strains with a high BNR efficiency, which can be co-cultured with various microorganisms to provide resource recovery (Liu et al., 2019), nitrogen deep treatment, and generation of novel bioactive substances associated with sewage systems (Vaca-Garcia, 2014). In our previous study, we confirmed that the intracellular protein has structural differences in the co-culture system of C.pyrenoidosa and Yarrowia lipolytica (3:1), showing potent antioxidant activity (QinWang et al., 2019). Furthermore, trypsin-hydrolyzed peptides were obtained in our study through this specific co-culture system, which exerts remarkable antioxidant activity (Liu H, 2019).
The microalgae-yeast symbiosis system highlights the significance of wastewater treatment. It was reported that the algae-assisted sequencing batch biofilm reactor increased the TN removal efficiency from 38.5% to 65.8% (385–658 mg/L) (He, 2018). The mixture of algae, ammonia-rich oxidizing bacteria, and methanol can rapidly remove nitrogen in the reactor. (He, 2018). Recently, researchers have been paying more attention to the algae-yeast co-culture for the economical improvement of microalgae biomass. For example, Chlorella vulgaris (C. vulgaris) and Yarrowia lipolytica (Y. lipolytica) co-culture has a strong potential for the treatment of industrial wastewater (Dong, 2018; Asraful, 2019). It was reported that the co-culture of C.pyrenoidosa and yeast improves the nitrogen removal rate (Huankai Li, 2019). Our previously established novel method using a co-culture of C.pyrenoidosa and Rhodotorula glutinis showed removal efficiencies of 58.53 and 36.07 for ammoniacal nitrogen (NH3-N) and total nitrogen (TN) in piggery wastewater. This is due to the fact that inorganic nitrogen N) is a necessary nutrient for microalgae, as it is required for the synthesis of proteins, polysaccharides, and other biomolecules and usually exists in the form of nitrate, nitrite, and ammonium (Stefan Schmollinger et al., 2014). Ammonium is the preferred form of nitrogen. In general, algae prefer ammonium and consume nitrate only when ammonium is almost depleted (Cai et al., 2013a). Yeast converts organic nitrogen compounds into N, and microalgae preferentially take up NH3-N, which is the main component of TN (about 60%) and the most important pollutant in wastewater (Vlaeminck et al., 2009), and convert it to substances for growth through nitrogen metabolism. We consider that microalgae and yeast can exhibit a mutually beneficial relationship in the co-culture system and the co-culture system would affect microalgae nitrogen metabolism and gene expression, so as to enhance nitrogen removal in wastewater. MicroRNAs (miRNAs) are key regulators in gene expression that can control gene expression via messenger RNA (mRNA) cleavage or translational repression (Djami-Tchatchou et al., 2017; Lou et al., 2018). Meanwhile, miRNA-target genes are primarily involved in secondary metabolite biosynthesis (Azaman et al., 2020). Microalgae nitrogen metabolism can be influenced by both miRNA and mRNA, and the presence of yeast stimulates gene expression in microalgae. The mechanism of the interaction between the two species, however, is currently unclear.
Therefore, this study aimed the following: 1) Establishment of a co-culture system of C. pyrenoidosa and Y. lipolytica at different ratios (1:0, 1.5:1, 3:1, 6:1) for 6 days in stimulated wastewater, to find out the optimum ratio of the co-culture system with the highest BNR; 2) daily assessment of the BNR for the optimum co-culture ratio in stimulated wastewater to confirm the best day with the highest BNR; 3) identification of significant transcriptomic differences in metabolic pathways and systematical analysis of the significant differences in transcription pathways regulated by miRNAs, including nitrogen, amino acids, photosynthetic, and carbon metabolism; 4) quantitative PCR analysis of key genes for verification. This study provides some useful information on efficient BNR under the algal-organism symbiosis system.
Figure 1A shows the summarized experimental process. Chlorella pyrenoidosa was obtained from the Institute of Hydrobiology, Chinese Academy of Sciences, cultured in sterile Tris-acetate phosphate (TAP) medium. Y. lipolytica was obtained from the Laboratory of Guangdong Institute of Microbiology) and cultured in yeast mold medium (YM; Qingdao Hope Bio-Technology Co., Ltd.). They were placed in a light incubator and cultured in the light environment at 25°C, 36 μE/m2/s, shaking at 150 rpm.
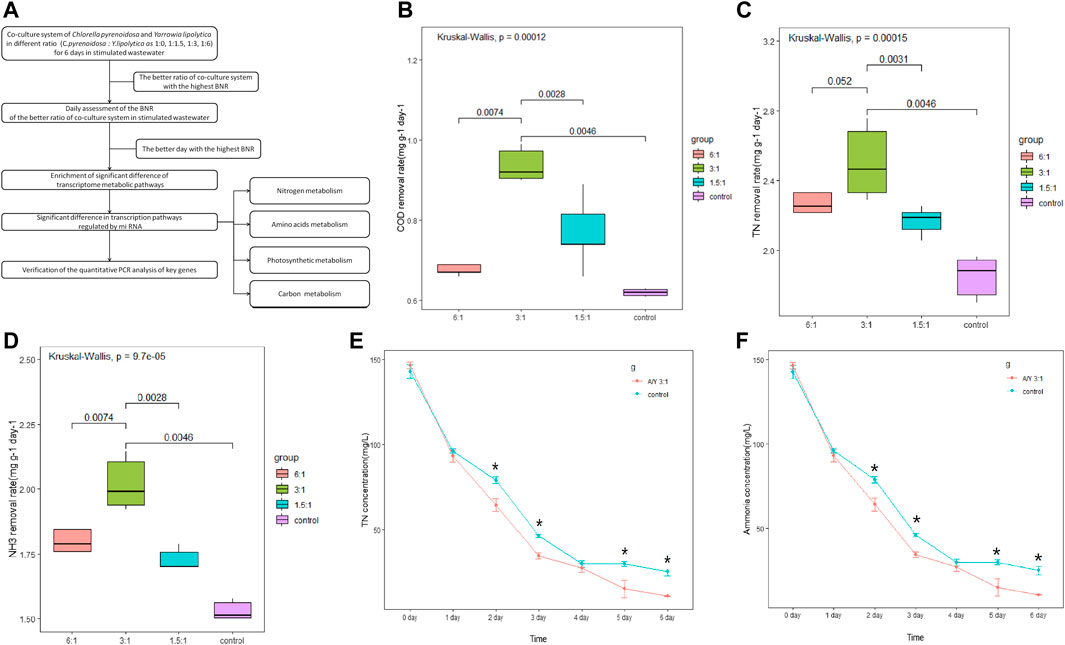
FIGURE 1. Cocultures of Y. lipolytica with Chlorella species. (A) The summarized experimental process, (B) COD removal rate, (C) TN removal rate, (D) NH3-N removal rate, (E) concentration of TN and (F) change in the amount of ammonia during 6 days of co-culture tests. Error bars represent the standard deviation of three cultures and the Kruskal test was performed.
The activated C. pyrenoidosa and Y. lipolytica were obtained after 3 and 2 days of incubation, respectively. The active C. pyrenoidosa and the active Y. lipolytica were mixed in 200 mL sterile stimulation wastewater at the ratio of 1: 0, 1.5: 1, 3: 1, and 6: 1 respectively for 6 days. The density of algae cells is maintained at 0.10–10.00×105 cells/mL (Cadena-Herrera et al., 2015). Refer to the article of Huankai Li (Li et al., 2019) to configure the nutrient concentration in the simulated wastewater. The content of chemical oxygen demand (COD), TN, and NH3-N were determined every day.
The growth curves were consistent with our previous studies, and the biomass yield of C. pyrenoidosa and Y. lipolytica peaked at an inoculation ratio of 3:1 (Huankai Li, 2019; Wang et al., 2019). After 3 and 5 days, active C. pyrenoidosa and active Y. lipolytica were mixed in sterile 200 mL Tris-acetate phosphate (TAP) medium (Wang et al., 2019) at the ratio of 1:0 and 3:1, respectively. The initial inoculation density was 5.00×105 cells/mL. The algal biomass was collected and transcriptome analysis was performed.
The concentration of the nutrients and other parameters of stimulated wastewater were measured, including COD, TN, and NH3-N (Liu et al., 2016). Where TN was measured on a TN analyzer (Jane Multi N/C 3000, Germany), COD and NH3-N were measured according to APHA standard methods (Gilcreas, 1966).
C. pyrenoidesa cells and the co-culture system on days 3 and 5 were collected via centrifugation. Total RNA was obtained from C. pyrenoidesa and the co-culture system using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s procedure, and sequencing libraries were generated using the NEBNext®Ultra™ RNA Library Prep Kit for Illumina® (NEB, United States). Briefly, the process includes the repair of fragmented DNA ends, the addition of A-tailing to the 3′ends, ligation adaptor, and PCR amplification. cDNA fragments were amplified using the AMPure XP system (Beckman Coulter, Beverly, United States). Finally, the Illumina HiSeq4000 (LC Sciences, United States) high throughput sequencing platform was used for sequencing.
Clean data in fastq format were obtained by removing reads containing the adapter, the ploy-N, and low-quality reads from the raw data through in-house Perl scripts. The reads were assembled using the Trinity software (Anders and Huber, 2010). The Unigene library was compared using bowtie software, and the transcriptional abundance was analyzed using RSEM software (Li and Dewey, 2011). Unigene expression was calculated and normalized to fragments per kilobase of transcript per million mapped reads (FPKM). The resulting p-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate (FDR) (Elowitz et al., 2002; Rajkumar et al., 2015). FDR<0.01 and fold change ≥2 were taken as filtering criteria for the differentially expressed genes (DEGs). The DEGs were compared using Nr, SwissProt, GO, KOG, EggNOG, and KEGG databases with blast software to obtain gene annotation information (Elowitz et al., 2002).
Total RNA was extracted from C. pyrenoidesa and the co-culture system using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) with the manufacturer’s protocol, and the miRNA sequencing library was generated using the Next Ultra-small RNA Sample Library Prep Kit for Illumina (NEB, United States). The library quality was determined using Agilent 2100 Bioanalyzer (Beckman Coulter, Beverly, United States) (Masotti and Preckel, 2006). Finally, the library was sequenced using the Illumina hiseq (LC Sciences, United States) high throughput sequencing platform.
Raw data in the fastq format were processed (Zhao et al., 2019) and clean data were obtained using the method described above for mRNAs. Clean data were obtained by removing the sequences smaller than 18 nucleotides (nt) or longer than 30 nt. The Q20, Q30, and GC content were evaluated the quality of sequencing. The mapped small RNA tags were then compared using the Bowtie tools (Langmead et al., 2009), and clean reads were aligned using the Silva, GtRNAdb, Rfam, and Repbase databases to remove certain RNA categories including rRNA, transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and other non-coding RNAs, as well as repeats. The known miRNA and novel miRNAs were obtained using the known miRNA database and mirdeep2 software, respectively. Then, the expression of miRNA was measured by the number of transcripts per million clean tags (TPM) (Mortazavi et al., 2008). |log2(FC)|≥0.58 was set as the threshold for a significantly differential expression. The target genes corresponding to miRNAs were predicted using Target Finder software.
The expression of several typical mRNAs and miRNAs was measured using qRT-PCR to verify the reliability of transcriptome sequencing data (Jozefczuk and Adjaye, 2011). The primers were designed using the Primer Premier 5.0 software (Ren et al., 2004), and mRNA and miRNA information for qRT-PCR are shown in Supplementary Table S1. The relative transcript abundance was calculated using the 2−ΔΔCT method (Jianhua et al., 2016). Each sample was analyzed in triplicates.
All experiments were conducted in triplicates. The mean values along with standard deviations were calculated. The significant difference was determined using Kruskal tests. Stars indicate significant differences between treatments (p < 0.05).
Heatmap, Volcano Plot, and bar plots were established using R packages pheatmap and ggplot2. The correlation tests were performed and verified using R packages psych and vegan. Other plots and analyses were conducted using R 3.52 and R packages.
The growth rate of Y.lipolytica in the symbiotic system is faster than that of C. pyrenoidosa, and it holds a dominant position in 6 days (Wang et al., 2019). The biomass yield reached the peak when the inoculation ratio of Y.lipolytica to C. pyrenoidosa was 3: 1, and reached 5.35 g/L after 6 days (Huankai Li, 2019). To analyze the effect of Y.lipolytica on the degradation of different organic compounds in single and mixed culture systems, we measured the COD, TN, and NH3-N removal rates on day 6 of incubation at 1.5:1, 3:1, and 6:1 ratios and the results are shown in Figures 1B–F. Results suggested that the COD removal rate was significantly increased in the co-incubation system, especially with the co-culture of C. pyrenoidosa to Y. lipolytica at a 3:1 ratio (Figure 1B), and the removal rate reached 56.25% after 6 days of culture, from 1,200 mg/L to 525 mg/L. Some studies have shown that 50%–90% (500–900 mg/L) COD can be removed by C. pyrenoidosa (Gupta et al., 2017; Li et al., 2019; Bai et al., 2022). However, it seems that COD cannot be completely removed only by C. pyrenoidosa, which is probably due to the COD content of the refractory organic compounds as a result of the anaerobic respiration in the C. pyrenoidosa culture (Xia and Murphy, 2016). Compared with a single culture system, the co-culture system had significant advantages in wastewater pollutant removal, which may be because Y. lipolytica degraded these compounds so that they can be utilized by C. pyrenoidosa. As for nitrogen, the co-incubated system also increased the TN and NH3-N removal rate, especially at a 3:1 ratio (Figures 1C,D). Compared with the control, the TN and NH3-N content in the co-incubation system at a 3:1 ratio was significantly decreased within 2–6 days (Figures 1E, F). The removal of TN and NH3-N from the wastewater after 6 days of co-cultivation was 147.82 mg/L and 98.54 mg/L, respectively. Both removal efficiencies were above 90.00%, and the changing trend of NH3-N was similar to that of TN. Nitrogen is an essential element for the synthesis of various biological substances, such as proteins, nucleic acids, and phospholipids (Xu et al., 2015). NH3-N is the major component of TN in the wastewater and it is a major source of nitrogen in wastewater, which was considered the utilizable form of nitrogen for C. pyrenoidosa and Y. lipolytica growth (Cai et al., 2013a). Some studies have shown that the difference in ammonia stripping and assimilative capacity may be the reason why the removal efficiency of the co-culture system was higher than that of the single culture (Qin et al., 2018).
RNA-seq of six libraries from C. pyrenoidosa cultured for 5 days with and without Y. lipolytica resulted in 186.48 Gb of clean data, with >93.82% of the data exhibiting a quality score of Q30 (Supplementary Table S2). Using Trinity, 258,147 high-quality clean reads were assembled de novo into 34,010 unigenes with a mean length of 934.53 base pairs (bp) and an N50 length of 1704 bp. The length of all unigenes was >200 bp and the length of 29.53% (10,044) of the unigenes was >1,000 bp (Supplementary Table S3), which indicated that the obtained transcriptome data were acceptable and reliable with good quality.
The expression levels of 12 libraries from C.pyrenoidosa with or without Y.lipolytica after 3 or 5 days, which were calculated based on the FPKM values, showed that the incubation time and Y. lipolytica had a certain impact on the transcriptome of C. pyrenoidosa (Figure 2A). Based on the expression levels, we identified DEGs. There were 9885 DEGs, including 2575 downregulated and 7310 upregulated genes for C. pyrenoidosa after 3 days (Figure 2B). Exactly 3976 DEGs were detected, including 1,149 downregulated and 2827 upregulated genes for C. pyrenoidosa after 5 days (Figure 2C). The functional annotation of C. pyrenoidosa unigenes with or without Y. lipolytica co-culture in each database is listed in Supplementary Table S4. Therefore, Y.lipolytica had a great effect on gene expression in C.pyrenoidosa, especially after 3 days of culture.
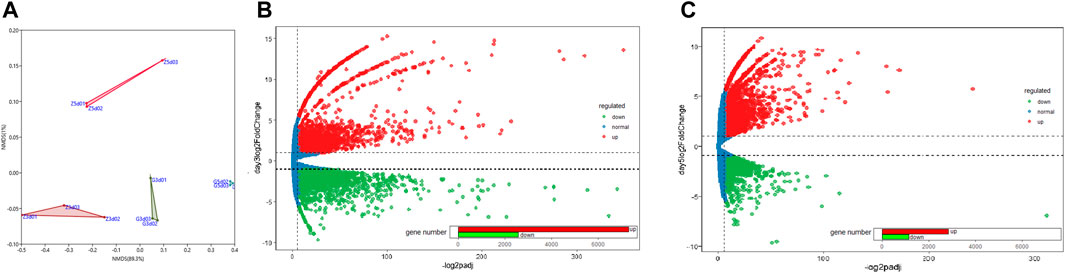
FIGURE 2. (A) NMDS plots of differentially expressed genes. MA plots of differentially expressed genes after (B) 3 and (C) 5 days of culture.
Carbon and nitrogen metabolisms were identified as the center metabolic pathways, as well as photosynthesis, in C. pyrenoidosa. Exactly 4 DEGs were involved in the nitrogen metabolism, which are displayed in Figure 3 and Table 1, such as genes encoding ammonia transporter, nitrate/nitrite transporter, and fungal nitric oxide reductase. According to the biomass analysis, Y. lipolytica could significantly improve TN and ammonia removal rates (Figures 1C–F). Compared with a single culture system, ammonia transporter-related genes were upregulated, nitrate/nitrite transporter-related genes were mix-regulated, and fungal nitric oxide reductase was downregulated in the co-culture system (Figure 3).
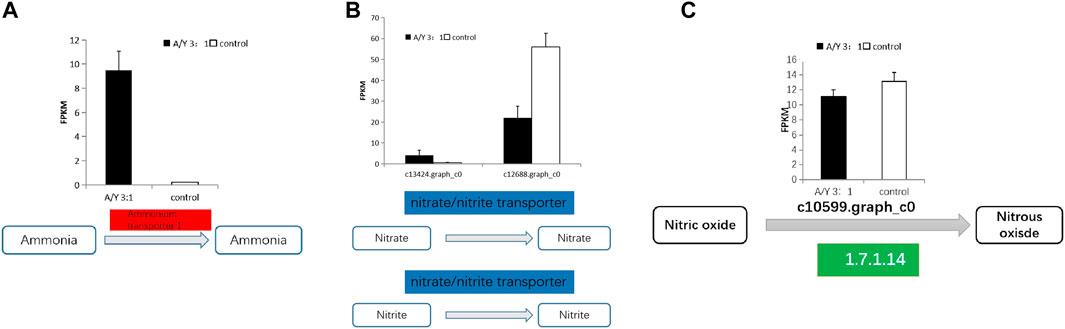
FIGURE 3. The representative differentially expressed genes associated with (A–C) the nitrogen metabolism of C. pyrenoidesa in response to Y. lipolytica.
Ammonia and nitrate transport were identified as the main pathway of nitrogen assimilation and nitrogen metabolism in C. pyrenoidosa. Because ammonia was the only nitrogen source in our batch tests, the ammonia transporter was related to varying nitrogen sources and levels (Klahn et al., 2015). Nitrate/nitrite is transported into the cell by a nitrate transporter (Mtolera, 2003; Sanz-Luque et al., 2013). Nitrate reductase activity was related to the content of nitrate, so it can control the nitrate assimilation rates (Scherholz and Curtis, 2013). Nitric oxide (NO) is reduced to nitrous oxide (N2O) by fungal nitric oxide reductase in the NO pathway (Zhang and Shoun, 2008). The ammonia transporter and nitrate transporter were upregulated by Y. lipolytica. We inferred that Y. lipolytica fixed nitrogen and might provide nitrogen for C. pyrenoidosa, leading to the change of nitrogen form and concentration in the co-culture system in the co-culture system. A similar observation has been reported between Chlorella variabilis and Rhizobium (CongFei et al., 2019).
After ammonia was transported into the cell, we also identified 1 DEG related to carbon metabolism and 11 DEGs related to photosystem metabolism as shown in Figures 4A, B and Table 2. Photosystem metabolism was mainly related to F-type H + -transporting ATPase subunit b, photosystem II oxygen-evolving enhancer protein 1, cytochrome b6-f complex iron-sulfur subunit, ferredoxin--NADP + reductase, F-type H + -transporting ATPase subunit gamma, ferredoxin, cytochrome b6-f complex subunit 8, and carbonic anhydrase activity. Assimilation carbon pathways were identified as other associate pathways related to the nitrogen pathway. Combined with the biomass analysis, Y. lipolytica could significantly increase the removal rate of COD (Figure 1C). Y. lipolytica could significantly enhance the expression level of carbonic anhydrase (EC:4.2.1.1), indicating that the conversion of carbon dioxide to HCO3− strengthened under the effect of CO2 assimilation in the co-culture system (Figure 4A). It has been reported that C. pyrenoidosa and other microalgae can utilize CO2 by yeast respiration (Lei et al., 2018). Yeast oxidizes and decomposes carbonaceous organic matter to CO2. Some of the CO2 escapes into the atmosphere, while some is absorbed and utilized by microalgae cells. O2 generated by microalgae photosynthesis enhances yeast’s oxidative decomposition of carbonaceous organic matter, allowing inorganic carbon to be removed from wastewater. As a result of algal photosynthesis consuming a large amount of acidic CO2, the pH of the wastewater rises and becomes alkaline, and some N is removed by overflowing as NH3 (Vadiveloo et al., 2020). Besides, a series of genes of C. pyrenoidosa were downregulated in the photosynthesis pathway (Figure 4B), which could limit the synthesis of NADPH and ATP required for dark reactions (Markou et al., 2016). Therefore, we inferred that the presence of Y. lipolytica in the mixed culture reduced the available light for C. pyrenoidosa, limited its photosynthesis, and further limited the fixation of CO2 by C. pyrenoidosa. Y. lipolytica enhanced the exchange between oxygen and CO2 (Zhang et al., 2018a). Therefore, CO2 and organic matter from Y. lipolytica and COD in wastewater may be used as a carbon source for C. pyrenoidosa growth.
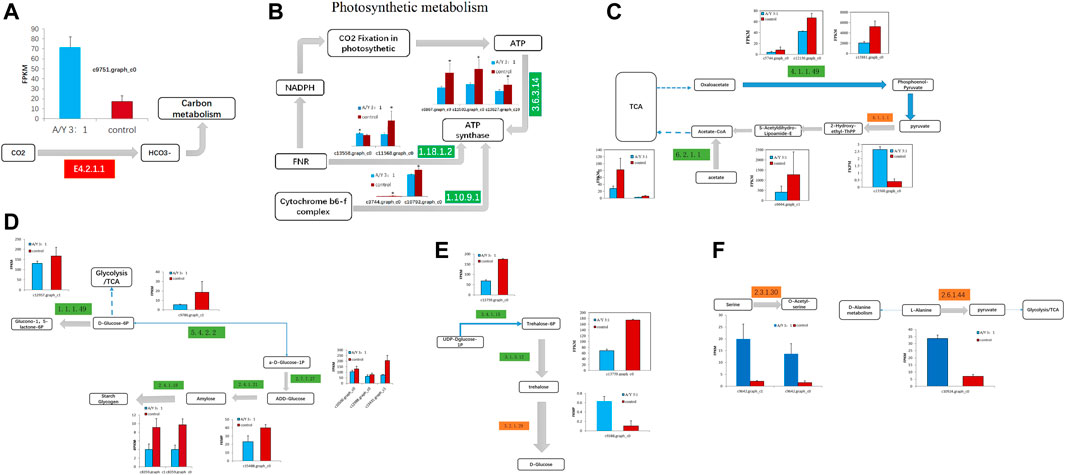
FIGURE 4. The representative differentially expressed genes associated with (A–B) the photosynthesis metabolism, (C) TCA cycle, (D–E) glycometabolism, and (F) amino acid metabolism of C. pyrenoidesa in response to Y. lipolytica.
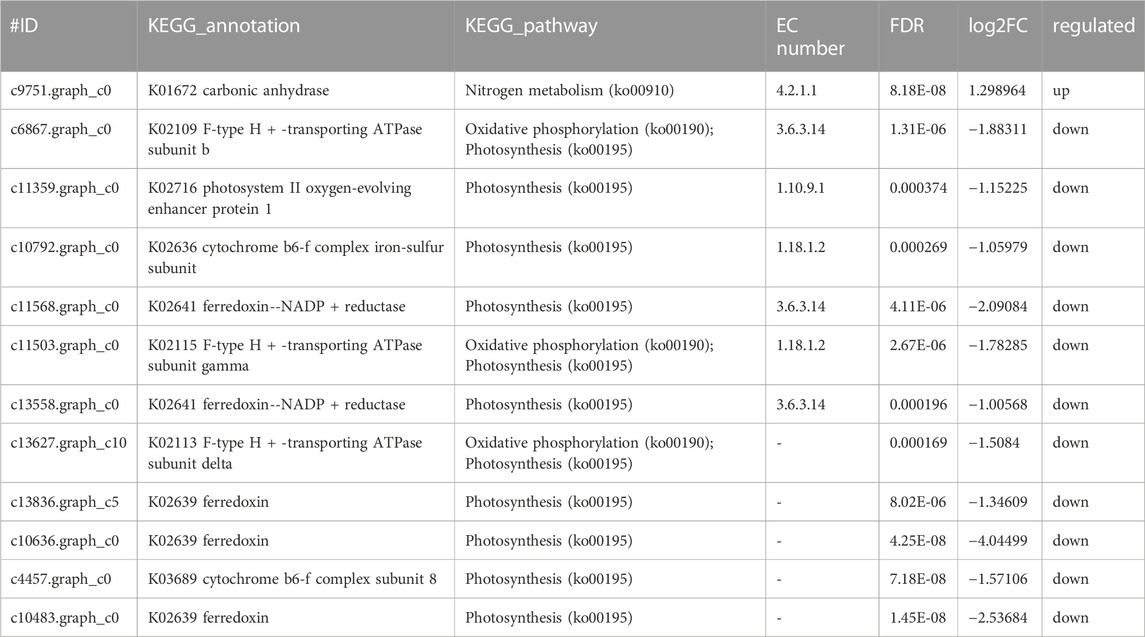
TABLE 2. Genes involved in Photosynthetic metabolism by co-culture system of C.pyrenoidosa and Y.lipolytica.
Glucose was a key material of the stress response, as well as carbon and nitrogen metabolisms, which was related to the phosphorylation of glucose by regulating gene expression in signal transduction pathways and a variety of plant hormone signal pathways (Moore and Rolland, 2003). Meanwhile, glycolysis/gluconeogenesis, pentose phosphate, starch, and sucrose pathway-related genes encoding two potentially rate-limiting enzymes, phosphoglucomutase (EC:5.4.2.2) and glucose-6-phosphate 1-dehydrogenase (EC: 1.1.1.49), were downregulated after Y. lipolytica treatment. Due to several downregulated genes, D-glucose-6P might not be converted to glucono-1, 5-lactone-6P, and D-glucose-1P, which is related to the starch/glycogen synthesis (Figure 4C; Table 3). D-glucose-6P is related to the tricarboxylic acid cycle (TCA) and synthesis. Moreover, glucose 6 phosphate dehydrogenase (EC: 1.1.1.49) was the first rate limiting enzyme in the well-known pentose phosphate oxidation pathway, catalyzing glucono-1,5-lactone-6P and gluconate synthesis (Huntzinger and Elisa, 2011). These reactions produce moieties of reducing equivalents, such as NADPH, which is utilized for nitrogen assimilation in algae (Zhang et al., 2018b). The upregulation of the glycolysis reactions of fructose and mannose and trehalose metabolism with the genes of carbon metabolism, may suggest an ammonia transport that increased the applicability of carbon and further promoted the synthesis of amino acids.
Meanwhile, the results of carbon metabolic analysis showed that the mixed culture with Y. lipolytica weakened the photosystem of C. pyrenoidosa and strengthened glycolysis/gluconeogenesis and starch pathways. The CO2 fixing pathway was downregulated and CO2 assimilation was enhanced in C. pyrenoidosa, which would deeply affect other carbon pathways, such as glycolysis/gluconeogenesis and starch synthesis. In starch synthesis, alpha and alpha-trehalase [EC:3.2.1.28] were upregulated after Y. lipolytica treatment. Trehalose conversion to D-glucose was enhanced, which suggested that glucose came from the trehalose storage, while the trehalose synthesis pathway was weakened in the mixed culture (Figure 4D; Table 4).
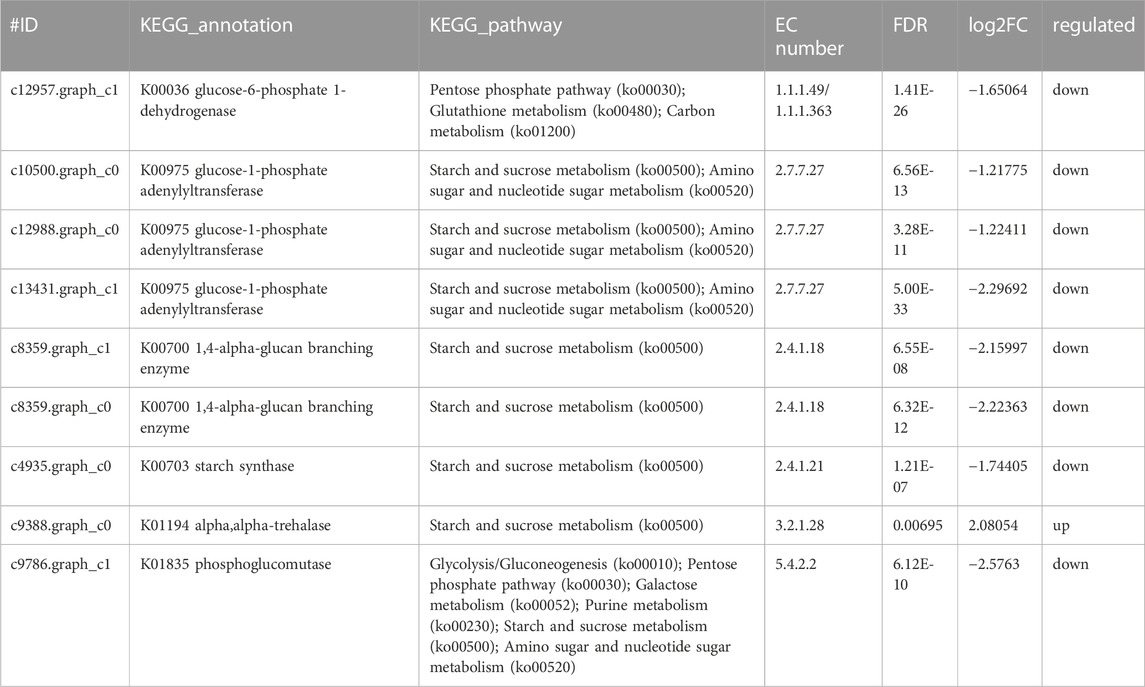
TABLE 4. Genes involved in carbon metabolism by co-culture system of C.pyrenoidosa and Y.lipolytica.
Amino acids are one of the organic nitrogen sources for microalgae (Näsholm et al., 2009), which contain a large number of amino acid transporters (Rentsch et al., 2007). Amino acids are degraded to provide ammonium, which is then taken up by microalgae and contributes to the denitrification of wastewater (Serna and Martin, 2006). Co-culture systems can promote amino acid synthesis in microalgae (Zhang et al., 2011). Exactly six DEGs were involved in amino acid biosynthesis, which are displayed in Figure 4F and Table 4, such as genes encoding serine O-acetyltransferase, alanine-glyoxylate transaminase/(R)-3-amino-2-methylpropionate-pyruvate transaminase, glutamine synthetase, and cysteine dioxygenase. In the co-culture, alanine-glyoxylate transaminase/(R)-3-amino-2-methylpropionate-pyruvate transaminase expression was upregulated 1.35 times, which is related to alanine aspartate and glutamate metabolism. It controls the conversion from L-alanine to pyruvate, which is related to the D-alanine metabolism and TCA cycle. Glutamine synthetase, a key enzyme in amino acid metabolism, is regulated in multiple ways in response to varying nitrogen sources and levels. Some studies have shown that the absorption of NH3-N is an important process of arginine synthesis in C. pyrenoidosa (Llacer et al., 2007). We found two kinds of glutamine synthetases in the co-culture, which were downregulated. The results showed that Y. lipolytica and C.pyrenoidosa could choose different ammonia sources for the glutamate metabolism. Both Y. lipolytica and C.pyrenoidosa could achieve assimilation of ammonia, which contributed to the synthesis of different amino acids.
Several miRNAs from C.pyrenoidosa in the control and co-culture were identified. The low quality and contaminated reads, adaptor sequences, poly-A-containing sequences, and sequences shorter than 14 nt were removed, and more than 10 million clean reads were obtained in each sample, with Q30 ≥ 85%. Moreover, the length distribution of miRNAs in this study was mainly n 18–24 nt, and 18, 22, and 24 nt miRNAs showed a higher abundance, which accounted for 43.8, 22.8, and 15.5%, respectively (Figure 5A). miRNAs mainly play a role in regulating gene expression in many eukaryotes (Goossens et al., 2003).
In the present study, we identified 11 differentially expressed C. pyrenoidesa miRNAs after 3 days, including three upregulated and eight downregulated miRNAs. According to the expression correlation between miRNAs and their targets, some of them had the same change in expression, which may involve other regulatory mechanisms (Huntzinger and Elisa, 2011). However, two miRNA/mRNA pairs had the opposite trend (Table 5), which mainly focused on amino acid biosynthesis and the ABC transport pathway (Zhang et al., 2018b). Y. lipolytica activated the expression of unconservative_c13405.graph_c0_13924, which plays a role in regulating the expressions of its predicted target mRNA, including cysteine dioxygenase, hypothetical protein, and histone-lysine N-methyltransferase SETD1. The catabolism of cysteine was catalyzed by cysteine dioxygenase (CDO), which was the first major step in cysteine catabolism (Chai et al., 2006). Histone-lysine N-methyltransferase SETD1 mediates lysine methylation or demethylation (Florian Maumus et al., 2011). Therefore, it may reduce the metabolic capacity of amino acids by regulating the expression of related genes via unconservative_c13405.graph_c0_13924. Furthermore, the unconservative_c11633.graph_c0_4666 may further contribute to the upregulation of genes encoding the ATP-binding cassette, subfamily C (CFTR/MRP), member 10 (ABCC10), which play important roles in ABC transport (Duan et al., 2019). Peptides, carbohydrates, lipids, heavy metal chelates, inorganic acids, steroids, and xenobiotics are transported by ABC transporter proteins (Alain et al., 2003). Therefore, Y. lipolytica may promote nitrogen and carbon transport in C. pyrenoidesa.
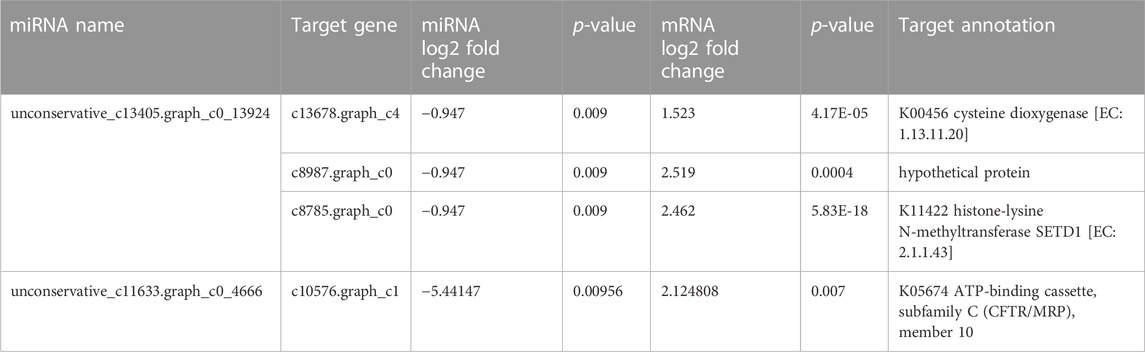
TABLE 5. Correlations analysis between miRNAs and mRNAs by co-culture system of C.pyrenoidosa and Y.lipolytica.
miRNAs can regulate gene expression through destabilization of the mRNA and translational repression (Chung et al., 2017). Most miRNAs are involved in regulating the biosynthesis of secondary metabolites, followed by protein metabolism (Azaman et al., 2020). From the above results, it can be seen that the transcription level of genes related to amino acid synthesis in C. pyrenoidesa is regulated by miRNAs, and miRNAs have a certain influence on nitrogen entering C. pyrenoidesa to synthesize amino acids. To validate the sequencing results, 26 mRNAs and two miRNAs were selected for qRT-PCR validation. Expressions of 20 mRNAs and two miRNAs were determined and compared with the RNA-seq data, which was similar to our sequencing data including seven upregulated and 12 downregulated mRNAs (Figure 6). Furthermore, the expression of unconservative_c13405.graph_c0_13924 and the unconservative_c11633.graph_c0_4666 was downregulated (Figure 6), which closely matched with the sequencing data. The results showed that miRNA and mRNA data were credible.
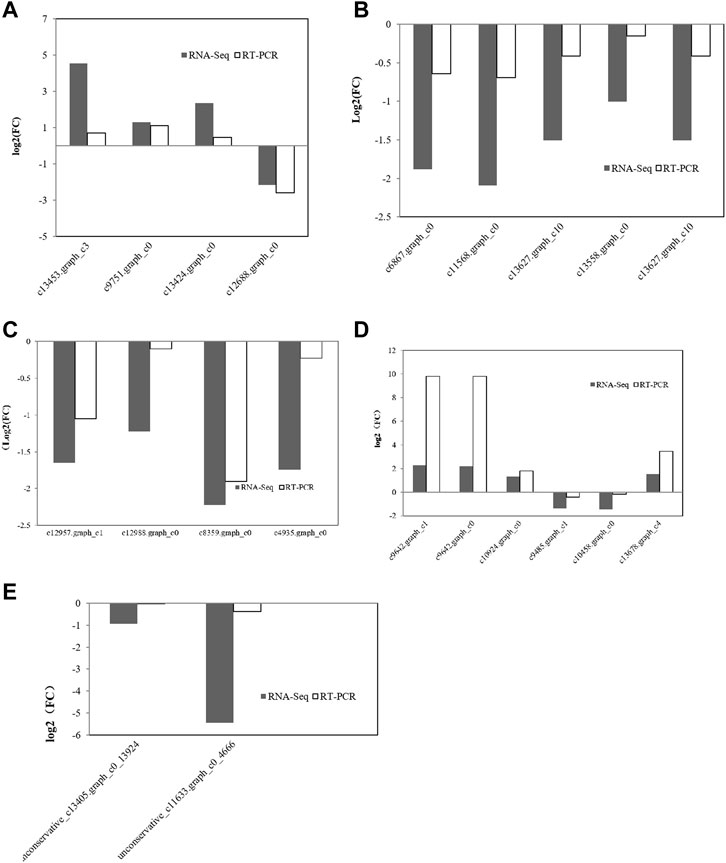
FIGURE 6. Validation of mRNA sequencing data for (A) nitrogen metabolism, (B) photosynthesis metabolism, (C) glycometabolism, and (D) amino acid metabolism via qRT-PCR. (E) Validation of miRNA sequencing data via qRT-PCR.
In this study, the peak biomass yield was reached at a 3:1 inoculation ratio of Y. lipolytica to C. pyrenoidosa. The co-culture system promoted the removal of COD, TN, and NH3-N, particularly at 3:1 ratio, the removal of COD reached 56.25% (675 mg/L) and the removal of both TN and NH3-N could reach 90.27% (147.82 mg/L and 98.54 mg/L, respectively) after 6 days of co-culture. Y. lipolytica had an effect on the transcriptome of C. pyrenoidosa, especially with the co-culture of C. pyrenoidosa and Y. lipolytica at a 3:1 ratio after 3 days. Therefore, we focused on Y. lipolytica-responsive mRNAs and miRNAs by performing transcriptome sequencing and RNA-seq of C. pyrenoidosa after 3 days. Our sequencing results revealed a large number of Y. lipolytica-responsive known miRNAs or novel miRNAs in C. pyrenoidosa, which regulate the target gene expression. The transcript levels of most of the genes related to nitrogen metabolism, carbon metabolism, amino acid synthesis, and other transporter proteins were upregulated in the co-culture system of C. pyrenoidosa. In addition, two differentially expressed miRNAs were negatively correlated with their target mRNA expressions. One of these miRNAs (unconservative_c13405.graph_c0_13924) regulates gene expression of cysteine dioxygenase, hypothetical protein, and histone-lysine N-methyltransferase SETD1, thereby reducing amino acid metabolic capacity; the other miRNA (unconservative_c11633.graph_c0_4666) may promote upregulation of genes encoding the ATP-binding cassette, subfamily C (CFTR/MRP), member 10 (ABCC10), thereby promoting nitrogen and carbon transport in C. pyrenoidosa. Based on the present results, we propose a pollutant disposal model for the mode of action of Y. lipolytica in C. pyrenoidosa. It seems that Y. lipolytica may function as a stress inducer to promote the removal of nitrogen and carbon via nitrogen, photosynthesis, and amino acid metabolism, as well as glycometabolism, which further prompts the activation of a signaling cascade of miRNAs. Our results may provide a valuable resource for further investigation of the regulation mechanisms of Y. lipolytica on the removal of COD, TN, and NH3-N.
The data presented in the study are deposited in the GEO repository, accession number GSE231399.
Conceptualization, YZ and HFL; methodology and software, YZ; validation, YZ, DL, and SL; investigation, YZ; resources, HL; data curation, DL; writing-original draft preparation, HFL and YZ; writing-review, and editing, DL; visualization, LM; project administration, HFL and YZ; funding acquisition, QW and HL. All authors contributed to the article and approved the submitted version.
The authors gratefully acknowledge the Natural Science Foundation of the Guangzhou Science and Technology Project (Nos. 201907010005, 201807010056) Guangdong Province (2018A030313425) for partially funding this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1159297/full#supplementary-material
Alain, G., Suvi, Häkkinen, Into, Laakso, Oksman-Caldentey, K. M., and Inzé, D. (2003). Secretion of secondary metabolites by ATP-binding cassette transporters in plant cell suspension cultures. Plant Physiol. 131, 1161–1164. doi:10.1104/pp.102.016329
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. doi:10.1186/gb-2010-11-10-r106
Asraful, A. M., Wei, D., Wang, Z., and Alam, M. A. (2019). Advantage assessment of mixed culture of Chlorella vulgaris and Yarrowia lipolytica for treatment of liquid digestate of yeast industry and cogeneration of biofuel feedstock. Appl. Biochem. Biotechnol. 187 (3), 856–869. doi:10.1007/s12010-018-2854-8
Azaman, S. N. A., Satharasinghe, D. A., Tan, S. W., Nagao, N., Yusoff, F. M., and Yeap, S. K. (2020). Identification and analysis of microRNAs in Chlorella sorokiniana using high-throughput sequencing. Genes 11 (10), 1131. doi:10.3390/genes11101131
Bai, Y., Li, Y., Tang, Y., Zhou, R., and Fan, Y. (2022). Rhizopus oryzae fermentation wastewater nutrient removal coupling protein fodder production by employing Chlorella pyrenoidosa. Bioresour. Technol. 362, 127858. doi:10.1016/j.biortech.2022.127858
Cadena-Herrera, D., Esparza-De Lara, J. E., Ramírez-Ibañez, N. D., López-Morales, C. A., Pérez, N. O., Flores-Ortiz, L. F., et al. (2015). Validation of three viable-cell counting methods: Manual, semi-automated, and automated. Biotechnol. Rep. Amst. Neth. 7, 9–16. doi:10.1016/j.btre.2015.04.004
Cai, T., Park, S. Y., and Li, Y. (2013a). Nutrient recovery from wastewater streams by microalgae: Status and prospects. Renew. Sustain. Energy Rev. 19, 360–369. doi:10.1016/j.rser.2012.11.030
Chai, S. C., Bruyere, J. R., and Maroney, M. J. (2006). Probes of the catalytic site of cysteine dioxygenase. J. Biol. Chem. 281, 15774–15779. doi:10.1074/jbc.m601269200
Chung, B. Y., Deery, M. J., Groen, A. J., Howard, J., and Baulcombe, D. C. (2017). Endogenous miRNA in the green alga Chlamydomonas regulates gene expression through CDS-targeting. Nat. Plants 3 (10), 787–794. doi:10.1038/s41477-017-0024-6
CongFei, T., AbeselomWoldemicael, MeilinHe, ShanmeiZou, ChanghaiWang, He, M., Zou, S., and Wang, C. (2019). Nitrogen supplemented by symbiotic Rhizobium stimulates fatty-acid oxidation in Chlorella variabilis. Algal Res. 44, 101692. doi:10.1016/j.algal.2019.101692
Djami-Tchatchou, A. T., Sanan-Mishra, N., Ntushelo, K., and Dubery, I. A. (2017). Functional roles of microRNAs in agronomically important plants—potential as targets for crop improvement and protection. Front. Plant Sci. 8, 378. doi:10.3389/fpls.2017.00378
Dong, W., Chen, J., Lim, P. E., and Wei, D., Dual-species cultivation of microalgae and yeast for enhanced biomass and microbial lipid production. J. Appl. Phycol. 2018; 30: 2997. doi:10.1007/s10811-018-1526-y
Duan, L., Chen, Q., and Duan, S. (2019). Transcriptional analysis of Chlorella pyrenoidosa exposed to bisphenol A. Int. J. Environ. Res. Public Health 16, 1374. doi:10.3390/ijerph16081374
Elowitz, M. B., Levine, A. J., Siggia, E. D., and Swain, P. S. (2002). Stochastic gene expression in a single cell. Science 297, 1183–1186. doi:10.1126/science.1070919
Florian Maumus, P. R., Bowler, C., and Rivarola, M. (2011). Stemming epigenetics in marine stramenopiles. Curr. Genomics 12, 357–370. doi:10.2174/138920211796429727
Gilcreas, F. W. (1966). Standard methods for the examination of water and waste water. Am. J. Public Health Nation Health 56 (3), 387–388. doi:10.2105/ajph.56.3.387
Goossens, A., Hakkinen, S. T., Laakso, I., Oksman-Caldentey, K. M., and Inze, D. (2003). Secretion of secondary metabolites by ATP-binding cassette transporters in plant cell suspension cultures. Plant Physiol. 131, 1161–1164. doi:10.1104/pp.102.016329
Gupta, S., Pandey, R. A., and Pawar, S. B. (2017). Bioremediation of synthetic high–chemical oxygen demand wastewater using microalgal species Chlorella pyrenoidosa. Bioremediation J. 21, 38–51. doi:10.1080/10889868.2017.1282936
He, Z. W., Tian, Y., Liang, H., Zuo, W., Wang, Z. W., Zhang, J., et al. (2018). Enhanced nitrogen and phosphorus removal from domestic wastewater via algae-assisted sequencing batch biofilm reactor. Bioresour. Technol. 250, 185–190. doi:10.1016/j.biortech.2017.11.028
Huntzinger, E. I., and Elisa, P. (2011). Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12, 99–110. doi:10.1038/nrg2936
Jianhua, F., Hui, Xu, and Yuanguang, Li (2016). Transcriptome-based global analysis of gene expression in response to carbon dioxide deprivation in the green algae Chlorella pyrenoidosa. Algal Res. 16, 12–19. doi:10.1016/j.algal.2016.02.032
Jozefczuk, J., and Adjaye, J. (2011). Quantitative real-time PCR-based analysis of gene expression. Met. Enzymol. 500, 99–109. doi:10.1016/B978-0-12-385118-5.00006-2
Judd, S., van den Broeke, L. J., Shurair, M., Kuti, Y., and Znad, H. (2015). Algal remediation of CO2 and nutrient discharges: A review. Water Res. 87, 356–366. doi:10.1016/j.watres.2015.08.021
Klahn, S., Schaal, C., Georg, J., Baumgartner, D., Knippen, G., Hagemann, M., et al. (2015). The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc. Natl. Acad. Sci. U. S. A. 112, 6243–6252. doi:10.1073/pnas.1508412112
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Gen. Biol. 10 (3), R25. doi:10.1186/gb-2009-10-3-r25
Lee, C. S., Lee, S. A., Ko, S. R., Oh, H. M., and Ahn, C. Y. (2015). Effects of photoperiod on nutrient removal, biomass production, and algal-bacterial population dynamics in lab-scale photobioreactors treating municipal wastewater. Water Res. 68, 680–691. doi:10.1016/j.watres.2014.10.029
Lei, Q., Dong, W., Zhongming, W., and Asraful, A. M. (2018). Advantage assessment of mixed culture of Chlorella vulgaris and Yarrowia lipolytica for treatment of liquid digestate of yeast industry and cogeneration of biofuel feedstock. Appl. Biochem. Biotechnol. 187, 856–869. doi:10.1007/s12010-018-2854-8
Li, B., and Dewey, C. N. (2011). Rsem: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinforma. 12, 323. doi:10.1186/1471-2105-12-323
Li, C., Ping, W., Ge, J., and Lin, Y. (2022). [Advances in the co-culture of microalgae with other microorganisms and applications]. Sheng wu gong cheng xue bao = Chi. J. Biotechnol. 38 (2), 518–530. doi:10.13345/j.cjb.210337
Li, H., Zhong, Y., Lu, Q., Zhang, X., Wang, Q., Liu, H., et al. (2019). Co-cultivation of Rhodotorula glutinis and Chlorella pyrenoidosa to improve nutrient removal and protein content by their synergistic relationship. RSC Adv. 9 (25), 14331–14342. doi:10.1039/c9ra01884k
Liu, H. L. S., Zhong, Y., Liu, J., Liu, H., Cheng, J., Ma, L., et al. (2019). Quantitative structure-activity relationship model to predict antioxidant effects of the peptide fraction extracted from a Co-culture system of Chlorella pyrenoidosa and Yarrowia lipolytica. Mar. Drugs 17, 633. doi:10.3390/md17110633
Liu, H., Wang, Q., Sun, Y., Zhou, K., Liu, W., Lu, Q., et al. (2016). Isolation of a non-fermentative bacterium, Pseudomonas aeruginosa, using intracellular carbon for denitrification and phosphorus-accumulation and relevant metabolic mechanisms. Bioresour. Technol. 211, 6–15. doi:10.1016/j.biortech.2016.03.051
Liu, J., Pemberton, B., Lewis, J., Scales, P. J., and Martin, G. J. O. (2020). Wastewater treatment using filamentous algae – a review. Bioresour. Technol., 298, 122556. doi:10.1016/j.biortech.2019.122556
Llacer, J. L., Contreras, A., Forchhammer, K., Marco-Marin, C., Gil-Ortiz, F., Maldonado, R., et al. (2007). The crystal structure of the complex of P II and acetylglutamate kinase reveals how P II controls the storage of nitrogen as arginine. Proc. Natl. Acad. Sci. U. S. A. 104, 17644–17649. doi:10.1073/pnas.0705987104
Lou, S., Sun, T., Li, H., and Hu, Z. 2018, Mechanisms of microRNA-mediated gene regulation in unicellular model alga Chlamydomonas reinhardtii. Biotechnol. Biofuels 11, 244. doi:10.1186/s13068-018-1249-y
Markou, G., Depraetere, O., and Muylaert, K. (2016). Effect of ammonia on the photosynthetic activity of arthrospira and Chlorella: A study on chlorophyll fluorescence and electron transport. Algal Res. 16, 449–457. doi:10.1016/j.algal.2016.03.039
Masotti, A., and Preckel, T. (2006). Analysis of small RNAs with the agilent 2100 bioanalyzer. Nat. Methods 3, 658. doi:10.1038/nmeth908
Moore, B. Z. L., Rolland, F., Hall, Q., Cheng, W. H., Liu, Y. X., et al. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336. doi:10.1126/science.1080585
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5 (7), 621–628. doi:10.1038/nmeth.1226
Mtolera, M. (2004). Some properties of glutamate dehydrogenase from the marine red alga gracilaria sordida (harv) W nelson. West. Indian Ocean J. Mar. Sci. 2 (2), 179–186. doi:10.4314/wiojms.v2i2.28438
Näsholm, T., Kielland, K., and Ganeteg, U. (2009). Uptake of organic nitrogen by plants. New Phytol. 182, 31–48. doi:10.1111/j.1469-8137.2008.02751.x
Nwoba, E. G., Mickan, B. S., and Moheimani, N. R. (2019). Chlorella sp. growth under batch and fed-batch conditions with effluent recycling when treating the effluent of food waste anaerobic digestate. J. Appl. Phycol. 31, 3545–3556. doi:10.1007/s10811-019-01878-7
Patidar, S. K., Mishra, S. K., Bhattacharya, S., Ghosh, T., Paliwal, C., Goel, S., et al. (2015). Naturally floating microalgal mat for in situ bioremediation and potential for biofuel production. Algal Res. 9, 275–282. doi:10.1016/j.algal.2015.03.019
Qin, L., Liu, L., Wang, Z., Chen, W., and Wei, D. (2018). Efficient resource recycling from liquid digestate by microalgae-yeast mixed culture and the assessment of key gene transcription related to nitrogen assimilation in microalgae. Bioresour. Technol. 264, 90–97. doi:10.1016/j.biortech.2018.05.061
Qin Wang, S., Yuming, Z., Huifan, Liu, Jianliang, L., Hui, L., Jian, C., et al. (2019). Protein extracted from symbiotic culture of Chlorella pyrenoidosa and Yarrowia lipolytica shows structure-related detoxifying effects against 2, 2’-azobis (2-methyl-propanimidamidine) dihydrochloride induced oxidative stress. Algal Res. 44, 101701. doi:10.1016/j.algal.2019.101701
Rajkumar, A. P., Qvist, P., Lazarus, R., Lescai, F., Ju, J., Nyegaard, M., et al. (2015). Experimental validation of methods for differential gene expression analysis and sample pooling in RNA-seq. BMC Genomics 16, 548. doi:10.1186/s12864-015-1767-y
Ren, L., Zhu, B. Q., Zhang, Y. B., Wang, H. Y., and Ba, C. F. (2004). The research of applying primer premier 5.0 to design PCR primer.
Rentsch, D., Schmidt, S., and Tegeder, M. (2007). Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett. 581, 2281–2289. doi:10.1016/j.febslet.2007.04.013
Sanz-Luque, E., Ocaña-Calahorro, F., Llamas, A., Galvan, A., and Fernandez, E. (2013). Nitric oxide controls nitrate and ammonium assimilation in Chlamydomonas reinhardtii. J. Exp. Bot. 64 (11), 3373–3383. doi:10.1093/jxb/ert175
Scherholz, M. L., and Curtis, W. R. (2013). Achieving pH control in microalgal cultures through fed-batch addition of stoichiometrically-balanced growth media. BMC Biotechnol. 13, 39. doi:10.1186/1472-6750-13-39
Serna, L., and Martin, C. (2006). Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 11 (6), 274–280. doi:10.1016/j.tplants.2006.04.008
Singh, R., Paliwal, C., Nesamma, A. A., Narula, A., and Jutur, P. P. (2020). Nutrient deprivation mobilizes the production of unique tocopherols as a stress-promoting response in a new indigenous isolate monoraphidium sp. Front. Mar. Sci. 7, 1. doi:10.3389/fmars.2020.575817
Stefan Schmollinger, T. M., NanetteBoyle, Ra, Blaby, Ian K., Casero, a. David, Mettler, Tabea, JeffreyMoseley, L. J. K., et al. (2014). Nitrogen-sparing mechanisms in chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26, 1410–1435. doi:10.1105/tpc.113.122523
Vaca-Garcia, C., Zebib, B., Merah, O., and Pontalier, P. Y. (2014). Morphology, composition, production, processing and applications of Chlorella vulgaris: A review. Renew. Sustain. Energy Rev. 35, 265–278. doi:10.1016/j.rser.2014.04.007
Vadiveloo, A., Matos, A. P., Chaudry, S., Bahri, P. A., and Moheimani, N. R. (2020). Effect of CO2 addition on treating anaerobically digested abattoir effluent (ADAE) using Chlorella sp. (Trebouxiophyceae). J. CO2 Util. 38, 273–281. doi:10.1016/j.jcou.2020.02.006
Vlaeminck, S. E., Terada, A., Smets, B. F., Linden, D. Vd, Boon, N., Verstraete, W., et al. (2009). Nitrogen removal from digested black water by one-stage partial nitritation and anammox. Environ. Sci. Technol. 43, 5035–5041. doi:10.1021/es803284y
Vyverman, W., Danneels, B., and Vanormelingen, P. (2016). Nutrient removal from horticultural wastewater by benthic filamentous algae Klebsormidium sp., Stigeoclonium spp. and their communities: From laboratory flask to outdoor Algal Turf Scrubber (ATS). Water Res. 92, 61–68. doi:10.1016/j.watres.2016.01.049
Wang, Q., Li, S., Zhong, Y., Liu, H., Liu, J., Liu, H., et al. (2019). Protein extracted from symbiotic culture of Chlorella pyrenoidosa and Yarrowia lipolytica shows structure-related detoxifying effects against 2, 2’-azobis (2-methyl-propanimidamidine) dihydrochloride induced oxidative stress. Algal Res. 44, 101701. doi:10.1016/j.algal.2019.101701
Xia, A., and Murphy, J. D. (2016). Microalgal cultivation in treating liquid digestate from biogas systems. Trends Biotechnol. 34, S016777991500270X. doi:10.1016/j.tibtech.2015.12.010
Xu, J., Zhao, G., Zhang, H., and Zhao, Y. (2015). Nutrient removal and biogas upgrading by integrating freshwater algae cultivation with piggery anaerobic digestate liquid treatment. Appl. Microbiol. Biotechnol. 7598, 6493–6501. doi:10.1007/s00253-015-6537-x
Zhang, L., and Shoun, H. (2008). Purification and functional analysis of fungal nitric oxide reductase cytochrome P450nor. Methods Enzymol. 437, 117–133. doi:10.1016/S0076-6879(07)37007-9
Zhang, R., Wang, Y., Wang, C., Wei, Z., Xia, D., Wang, Y., et al. (2011). Time-course analysis of levels of indole-3-acetic acid and expression of auxin-responsive GH3 genes in betula platyphylla. Plant Mol. Biol. Report. 29, 898–905. doi:10.1007/s11105-011-0306-5
Zhang, X., Li, K., Xing, R., Liu, S., Chen, X., Yang, H., et al. (2018b). miRNA and mRNA expression profiles reveal insight into chitosan-mediated regulation of plant growth. J. Agric. Food Chem. 66, 3810–3822. doi:10.1021/acs.jafc.7b06081
Zhang, Xiaoqian, Li, Kecheng, Xing, Ronge, Liu, S., Chen, X., Yang, H., et al. (2018a). miRNA and mRNA expression profiles reveal insight into chitosan-mediated regulation of plant growth. J. Agric. Food Chem. 66, 3810–3822. doi:10.1021/acs.jafc.7b06081
Keywords: Chlorella Pyrenoidosa, co-cluture, Yarrowia Lipolytica, transcriptomics, nitrogen metabolism pathway
Citation: Zhong Y, Lin D, Li S, Wang Q, Liu H, Ma L and Liu H (2023) Enhanced nitrogen removal via Yarrowia lipolytica-mediated nitrogen and related metabolism of Chlorella pyrenoidosa from wastewater. Front. Bioeng. Biotechnol. 11:1159297. doi: 10.3389/fbioe.2023.1159297
Received: 05 February 2023; Accepted: 24 May 2023;
Published: 22 June 2023.
Edited by:
Eleonora Sforza, University of Padua, ItalyReviewed by:
Peter J. Lammers, Arizona State University, United StatesCopyright © 2023 Zhong, Lin, Li, Wang, Liu, Ma and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huifan Liu, bG1femtuZ0AxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.