
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 13 April 2023
Sec. Cell and Gene Therapy
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1143866
This article is part of the Research Topic Application of Innovative Techniques in Genetic and Cellular Therapies View all 12 articles
Skin wound healing is a complex and multistage process, where any abnormalities at any stage can result in the accumulation of non-functional fibrotic tissue, leading to the formation of skin scars. Epigenetic modifications play a crucial role in regulating gene expression, inhibiting cell fate determination, and responding to environmental stimuli. m6A methylation is the most common post-transcriptional modification of eukaryotic mRNAs and long non-coding RNAs. However, it remains unclear how RNA methylation controls cell fate in different physiological environments. This review aims to discuss the current understanding of the regulatory pathways of RNA methylation in skin wound healing and their therapeutic implications with a focus on the specific mechanisms involved.
As the protective barrier of the body, the skin is often exposed to potential causes of injury that stimulate wound healing (Zeng et al., 2021). The healing of a normal tissue wound after injury is a complex multistage process that involves hemostasis, inflammation, proliferation, and remodeling (Amjadian et al., 2022; Yu et al., 2022). It is important to note that abnormalities in any of these stages can result in the accumulation of non-functional fibrotic tissue, leading to the formation of skin scars (Amini-Nik et al., 2018; Hu et al., 2018).
A pathological scar is formed in the process of wound healing and is a proliferative disease of skin connective tissue. Keloids are one of the most common pathologic scars that occur in people of color. Keloids typically present as a bulge extending beyond the original wound, nodular hyperplasia, and a hard, red, benign mass with itching, pain, and discomfort (Jones et al., 2017; Xie et al., 2022). Excessive skin scarring, hypertrophy, or keloids pose significant challenges for patients and physicians, as they can result in serious health problems, such as contracture, functional and aesthetic issues, and complications such as pain, thickness, and itching. These problems have a significant negative impact on the physical, mental health, and social quality of life of patients and lead to high medical costs (Goel and Shrivastava, 2010). Therefore, treatment for reducing skin scarring is necessary to improve the recovery of patients.
The pathogenesis of keloids is complex and involves wound tension, genetic factors, immune changes, and programmed cell death, among other factors (Wolfram et al., 2009; Hsu et al., 2017a; Limandjaja et al., 2020; Zhang et al., 2020; Macarak et al., 2021). Keloid formation is closely related to tumor-related genes (Ekstein et al., 2021), and keloids are considered benign fibrogenic skin tumors that possess many cancer-like features, such as uncontrolled proliferation, a lack of spontaneous recovery, and high recurrence rates (Atiyeh et al., 2005). Increasing evidence suggests that the interaction among various promoting or inhibiting factors in the tumor could explain the aggressive clinical behavior of keloids. The most similar genotypes and phenotypes between keloids and cancers are cell energy sources, epigenetic methylation signatures, and epithelial–mesenchymal transformation behavior (Tan et al., 2019).
The research into the wound healing process has a rich history and is rapidly evolving. In addition to traditional therapies, novel treatment options have emerged, such as growth factors, skin substitutes, cytokine stimulants, cytokine inhibitors, matrix metalloproteinase inhibitors, gene and stem therapies, extracellular matrix therapies, angiogenic stimulants, and nanopreparation therapies (Patel et al., 2019; Qian et al., 2020; Matoori et al., 2021).
Scarring and keloid formation can be prevented and treated with a variety of strategies, including pressure, silicone gels, corticosteroids, lasers, and surgery. Pressure suits restrict blood flow to the scar area reducing oxygen supply, increasing collagenase activity, and reducing adhesion between collagen fibers. Stress therapy also regulates the secretion of fibrocytokines and growth factors. Silicone gels mainly increase the temperature and hydration of the blocked scar. Corticosteroids inhibit inflammation, increase the vasoconstriction of the scar, reduce collagen and glycosaminoglycan production, and decrease fibroblast proliferation. Skin grafts are effective, however, they are limited by the availability of skin, graft tissue rejection, infection, and the prevention of wound overtension (Marshall et al., 2018).
As a result, traditional treatments lack specificity, and there is no evidence to support their absolute efficacy. Even with the best treatments, traumatic scarring is inevitable, and current treatments can only reduce scarring (Ekstein et al., 2021). Therefore, it is necessary to establish new methods to prevent or reduce dermal fibrosis by optimizing the wound healing process. This requires a better understanding of the mechanisms, key regulators, and risk factors associated with wound healing (Chen et al., 2021). Recently, epigenetic changes such as DNA methylation, histone modification, and non-coding RNAs (e.g., microRNAs and long non-coding RNAs [lncRNAs]), have been recognized as promising approaches for scar management. lncRNAs, in particular, offer new potential for targeted therapy to improve traditional and combination therapies.
Epigenetic modifications refer to changes in the gene expression that occur without altering the underlying DNA sequence. These modifications can be passed down from one generation to another and can be influenced by environmental factors such as diet, stress, and exposure to toxins. Epigenetic modifications play a crucial role in development, differentiation, and disease. Dysregulation of epigenetic modifications can lead to a range of disorders, including cancer, neurological disorders, and cardiovascular disease. Understanding epigenetic modifications can provide insight into disease mechanisms and potential targets for therapeutic intervention.
Epigenetic modifications including DNA deposition and histone modifications are well-established mechanisms that regulate gene expression to suppress cell fate determination and the response to environmental stimuli. DNA methylation involves the addition of a methyl group to a cytosine residue in DNA, which can result in the suppression of gene expression. Histone modification involves the addition or removal of chemical groups to histone proteins, which can affect the way that DNA is packaged and therefore impact gene expression. However, the role of transcriptional modifications (RNA) in gene expression regulation is only beginning to be revealed (Hwang et al., 2017; Livneh et al., 2020). mRNA is not just an intermediate molecule, but also an important regulator of gene expression. The process of post-transcriptional regulation of RNA involves a variety of mechanisms, both cis- and trans-acting, that are essential for controlling gene expression programs that determine cell function and fate. These mechanisms include alternative splicing, RNA editing, mRNA stability, and translation initiation, among others. Importantly, these mechanisms can be rapidly and dynamically regulated in response to changes in the cellular environment, allowing for precise control of gene expression in the face of changing conditions (Vu et al., 2019).
m6A methylation, a prevalent post-transcriptional modification of eukaryotic mRNAs and lncRNAs, has garnered increasing attention in recent years (Wang et al., 2018). RNA methylation of transcripts was first discovered in the 1970s when a poly(A) sequence was found at the 3′end of mRNA (Desrosiers et al., 1974). However, the methylation site of m6A methylation remained in an early stage of characterization until the recent discovery of fat mass and obesity-associated protein (FTO) (Jia et al., 2011) and AlkB homolog 5 (ALKBH5) as erasers of m6A methylation (Zheng et al., 2013). These findings revealed that this modification could directly control gene expression. With the development of m6A-specific antibodies and several next-generation sequencing techniques, such as m6A sequencing (methylated RNA immunoprecipitation sequencing, meRIP-seq (Dominissini et al., 2012)), and single-nucleotide mapping of m6A methylation (miCLIP-seq (Linder et al., 2015)), the m6A modification of specific mRNAs can now be analyzed in the entire transcriptome.
m6A methylation is a widely distributed and dynamically regulated modification in the transcriptome, whose understanding is primarily focused on its “readers” and “writers.” m6A writers are methyltransferases that catalyze the transfer of methyl groups from S-adenosine methionine (SAM) to adenosine at N-6 a). The m6A “writer” complex mainly includes methyltransferase-like 3 (METTL3), METTL14, Wilms’ tumor 1-associated protein (WTAP), RNA-binding motif protein 15 (RBM15), Vir-like m6A methyltransferase associated (VIRMA)/KIAA1429, and zinc finger CCCH domain-containing protein 13 (ZC3H13) (Uddin et al., 2021), Among them, METTL3 and its homolog METTL14 form stable heterodimers and exhibit methyltransferase activity. Whereas WTAP was later identified as another component of the mammalian methyltransferase complex (Liu et al., 2021b; Oerum et al., 2021; Guo et al., 2022; Huang et al., 2022). RBM15 and its paramotifs, including RBM15B, KIAA1429, HAKAI, and ZC3H13, have also been reported in recent years, with different important functions such as recruitment and catalysis (Patil et al., 2016; Yue et al., 2018).
“Erasers” of m6A methylation refers to enzymes that can remove or reverse the addition of a methyl group to an adenosine base in RNA molecules. These enzymes are also known as demethylases (Edupuganti et al., 2017). There are several known erasers of m6A methylation, including FTO (Fat Mass and Obesity-associated protein) and ALKBH5 (AlkB Homolog 5) (Zheng et al., 2013). These enzymes belong to the family of alpha-ketoglutarate-dependent dioxygenases and use molecular oxygen to oxidize the methyl group of m6A, which is then removed as formaldehyde. FTO is a member of the AlkB family of non-heme iron and 2-oxoglutarate-dependent dioxygenases. It was the first identified m6A demethylase, and has been shown to be involved in regulating a variety of physiological processes, including adipogenesis, energy metabolism, and circadian rhythm. ALKBH5 is another m6A demethylase that has been shown to regulate RNA metabolism, including RNA splicing, translation, and decay. ALKBH5 is also involved in the regulation of various cellular processes, including cell differentiation and proliferation. (Wei et al., 2018). Future research could lead to the discovery of novel m6A demethylases and alternative mechanisms that mediate m6A modification removal and m6A-labeled transcriptional clearance.
The biological function of m6A modification is primarily mediated through the specific recognition and binding of readers, which are responsible for the diverse effects of m6A methylation on gene expression (Huang et al., 2020; Zhang et al., 2021b). These effects include the regulation of RNA splicing, output, decay, stabilization, and translation (Wang et al., 2014; Alarcon et al., 2015; Spitale et al., 2015; Xiao et al., 2016; Roundtree et al., 2017b; Slobodin et al., 2017). A major reader of m6A methylation is the protein family containing the YTH domain, including YTHDF1, YTHDF2, YTHD3, YTHDC1, and YTHDC2 (Luo and Tong, 2014). For instance, YTHDF1 has been found to enhance translation by binding to translation primers and ribosomes (Wang et al., 2014). In addition, the insulin-like growth factor 2 mRNA-binding protein (IGF2BP) protein family includes IGF2BP1/2/3, comprising reported readers of m6A methylation (Huang et al., 2018). Moreover, heterogeneous nuclear ribonucleoproteins C (HNRNPC) and G (HNRNPG) are considered “indirect” readers of m6A methylation because they preferentially bind to the RNA structural “switches” induced by m6A modification (Hsu et al., 2017b; Zong et al., 2021; Chen et al., 2022). For example, HNRNPC is an important physiological modulator of 3′-untranslated region processing and miRNA maturation and acts as a reader protein during m6A modification. It recognizes m6A modification groups and mediates the selective splicing of mRNA precursors (Gruber et al., 2018). Thus, RNA methylation is dynamically controlled by writers, readers, erasers, and other proteins that might affect these regulators (Figure 1). Although RNA methylation is recognized to play a crucial role in gene expression regulation and many cellular processes (Roundtree et al., 2017a; Meyer and Jaffrey, 2017; Huang et al., 2020; Livneh et al., 2020), how it controls cell fate in different physiological environments remains unclear. In skin regeneration, epigenetic modifications are essential for the proper activation and maintenance of stem cells responsible for tissue regeneration. Epigenetic changes also play a role in the differentiation of keratinocyte and fibroblast. This review aims to discuss the current understanding of the regulatory pathways of RNA methylation in skin wound healing and their therapeutic implications.
During the wound healing process, various cellular and molecular events are activated, including the regulation of gene expression through RNA modifications such as m6A methylation. The relationship between wound healing and epigenetic inheritance is complicated and unclear (Mi et al., 2020; Zhi et al., 2021). Several studies have demonstrated that m6A methylation can affect wound healing by regulating the expression of genes involved in various cellular processes, such as proliferation, differentiation, and migration of cells (Table 1). Zhou et al. identified adipose-derived stem cells (ADSCs) via flow cytometry and tested their pluripotency in terms of differentiation into adipocytes and bone. Based on this, ADSCs were found to accelerate LEC proliferation, migration, and lymphangiogenesis through the METTL3 pathway and regulate vascular endothelial growth factor C (VEGF-C) expression and VEGF-C-mediated lymphatic angiogenesis through the METTL3/IGF2BP2-m6A pathway, thus promoting the repair of diabetic foot ulcer (DFU) wounds. The modification of ADSCs by METTL3-mediated VEGF-C m6A methylation could be a promising therapeutic strategy for promoting DFU wound healing (Zhou et al., 2021).
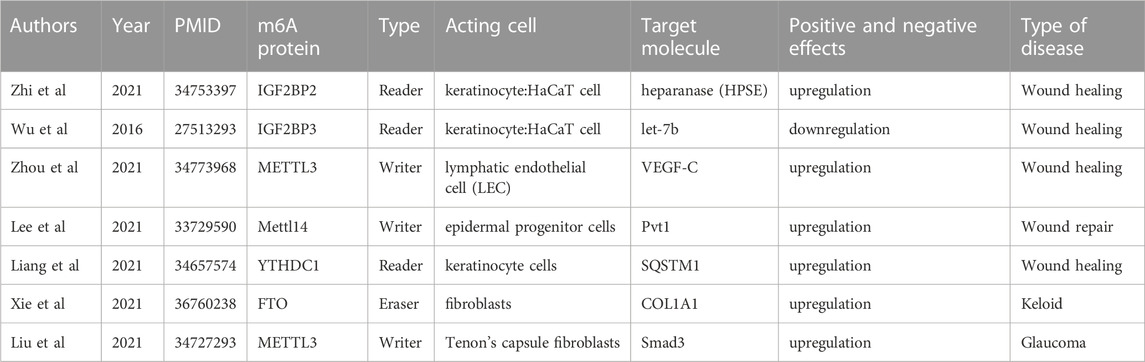
TABLE 1. Current research progresses of m6A methylation in wound healing, hypertrophic scars, and keloids.
Impaired physiological functions of keratinocytes induced by a high-glucose environment lead to delayed healing of diabetic wounds (Hu and Lan, 2016). In a study of diabetic skin wound healing by Liang et al., m6A RNA modification and the regulation of autophagy were found to play a key role in diabetic skin wound healing. The m6A reader protein YTHDC1 (YTH domain 1), which interacts with autophagy receptor SQSTM1 mRNA, is downregulated in keratinocytes in both acute and chronic hyperglycemia. The knockdown of YTHDC1 affects keratinocyte biological functions, including an increased apoptosis rate and impaired wound-healing ability. Further studies found that YTHDC1 regulates autophagy in diabetic keratinocytes by regulating the stability of SQSTM1 nuclear mRNA, ultimately affecting diabetic skin wound healing functions (Liang et al., 2022). Overall, the evidence suggests that m6A methylation plays an important role in regulating gene expression and cellular processes involved in wound healing, making it a promising target for future therapeutic interventions.
Hypertrophic scars (HSs) frequently arise following burns and trauma, posing a formidable challenge in the field of wound repair and plastic surgery. Conventional methods, such as surgery, radiotherapy, and hormone therapy, have proven insufficient in achieving complete HS remission, particularly in regions susceptible to HS recurrence (Jaloux et al., 2017), Current studies generally support the pathological features of HSs, including abnormal inflammation, excessive proliferation and differentiation of fibroblasts, increased angiogenesis, and excessive deposition of extracellular matrix (ECM); however, the causative factors and molecular mechanisms underlying the unremitting collagen synthesis in HSs remain indeterminate (Keane et al., 2018; Roderfeld, 2018; Zhang et al., 2021a). Whether m6A methylation, as a mechanism of targeting abnormal epigenetic modification, can serve as a new target and mechanism for fibrotic diseases and provide a new therapeutic strategy for the treatment of skin fibrosis remains to be determined.
Liu et al. (2021a) conducted a study to investigate the mechanism of m6A modification in HSs and normal skin tissues. The study employed m6A methylation and RNA sequencing, followed by bioinformatics analysis, immunoprecipitation of m6A-related RNA, real-time quantitative polymerase chain reaction verification, and other methods. The presence of 14,791 new m6A methylation peaks in HS samples was accompanied by the disappearance of 7,835 peaks. Unique m6A-related genes in HS were associated with fibrosis-related pathways, and differentially expressed mRNA transcripts were identified in HS samples with hypermethylated or hypomethylated m6A methylation peaks. The m6A transcriptome map of human HS helped to elucidate the possible mechanism underlying m6A-mediated gene expression regulation (Liu et al., 2021b).
Furthermore, Liu et al. (2021b) reported the m6A methylation mechanism of scar formation caused by human Tenon’s fibroblasts (HTFs). First, they isolated and identified primary HTFs and found that transforming growth factor beta 1 (TGF-β1) enhanced cell viability and proliferation and ECM deposition in HTFs. Subsequently, TGF-β1 was found to increase the number of m6A methylation events and promote the expression of an m6A “reader.” The study further revealed that the downregulation of METTL3 inhibited the TGF-β1-induced promotion of cell viability and proliferation and ECM deposition in HTFs. The results indicated that METTL3 can indeed regulate the expression of Smad3, playing a critical role in the regulation of TGF-β1-induced HTFs and providing new theoretical strategies for regulating scar formation based on METTL3 (Liu et al., 2021b).
The emergence of a public database of collaborative studies has made a vast amount of gene expression data from RNA sequencing available not only for tumor tissues, but also for keloid-associated tissues. This provides great convenience for exploring and identifying gene expression differences in keloid tissue analysis. For example, tumor suppressor genes, such as p53, p16, Fas, and p27, were found to lose their inhibitory effect on fibroblast proliferation after mutations (Tan et al., 2019). Moreover, the overexpression of c-MYC and c-FOS promotes fibroblast proliferation and inhibits apoptosis (He et al., 2017).
Recently, Xie et al. (2022) collected and integrated keloid-related sequencing results from public datasets (GSE44270 and GSE145725) to establish a new keloid risk diagnosis model, extracted m6A-related genes from the literature, and compared their expression matrices between high-risk and low-risk groups to explore differences in m6A methylation. It was found that ALKBH5, FTO, and HNRNPA2B1 were highly expressed, while YTHDF2 was lowly expressed in the high-risk keloid group (Xie et al., 2022). This study provides an idea for treatment planning based on clinical risk grouping.
Furthermore, Lin et al. (2022) investigated the overall m6A-modified RNA pattern and the possible mechanism of keloid pathogenesis (Lin et al., 2022). They collected 14 pairs of normal skin and keloid tissues and found that WTAP and METTL3 protein expression was higher in keloid tissues than in normal tissues. MeRIP and RNA sequencing and bioenrichment analysis revealed 21,020 unique m6A methylation peaks and 6,573 unique m6A-related gene transcripts in keloid samples. In normal tissues, 4,028 unique m6A methylation peaks were found, including 779 m6A-related modified genes. Subsequent functional verification also showed that the m6A-methylated genes and the differentially upregulated genes between the two tissues were mainly related to the Wnt signaling pathway. These findings directly confirmed that keloid fibroblasts were in a state of m6A methylation activation and that the high expression of the Wnt/β-catenin pathway in skin fibroblasts, modified and activated via m6A methylation, might promote the occurrence of keloid.
Wound healing is a complex biological process involving a series of molecular events to promote skin regeneration. However, abnormal wound healing can lead to the formation of thick, painful, and itchy scars, which can cause aesthetic and functional complications. Therefore, it is crucial to develop more effective and targeted treatments to prevent the deposition of excess fibrous tissue. This can be achieved through a correct understanding of the regulatory mechanisms of wound healing and scar formation.
Given the significant role of genetic and epigenetic differences in physiological or pathological wound repair, we emphasize the critical role of m6A epigenetic changes in regulating the switch during wound healing. Regulatory epigenetic molecules are considered promising therapeutic tools for scar management. However, their exact roles need to be further explored in terms of unexpected genetic disorders.
In conclusion, recent studies have suggested that RNA methylation is a major pathway regulating skin wound healing. This finding highlights the therapeutic potential of genetic and epigenetic approaches for optimizing wound healing and scar management. However, the precise function of m6A methylation in regulating skin wound healing at the cellular level and its potential mechanism in regulating gene expression after transcription require further investigation. Whereas it is exciting to explore RNA methylation as a promising avenue from a therapeutic perspective, translation of these new findings to the clinic remains an urgent challenge. Successful application of these emerging strategies will require advances in existing tools and technologies for the discovery, delivery, and control of genetic and epigenetic regulation, which can help overcome associated challenges and provide a new approach to wound healing management.
All authors contributed to the design of the study and writing of the manuscript. XL, SZ, and JL performed the research, XL, HW, YW, and ZN. wrote the main manuscript text and prepared the figures. LW and ZL. revised the article critically for important intellectual content and provided final approval of the version to be submitted. All authors reviewed the manuscript.
This study was supported by financial support from the National Natural Science Foundation of China (grant number: 82202461) and Hubei Provincial Natural Science Foundation of China (2022CFB233).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YX declared a shared affiliation with the authors to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
lncRNAs, long non-coding RNAs; ALKBH5, AlkB homolog 5; meRIP-seq, methylated RNA immunoprecipitation sequencing; miCLIP-seq; single nucleotide mapping of m6A; SAM, S-adenosine methionine; METTL3, methyltransferase-like 3; METTL14, methyltransferase-like 14; WTAP, Wilms’ tumor 1-associated protein; RBM15, RNA-binding motif protein 15; VIRMA, Vir-like m6A methyltransferase associated; ZC3H13, Zinc finger CCCH domain-containing protein 13; HNRNPC, heterogeneous nuclear ribonucleoproteins C; VEGF-C, vascular endothelial growth factor C; ECM, extracellular matrix; TGF-β1, transforming growth factor beta 1.
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015). N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485. doi:10.1038/nature14281
Amini-Nik, S., Yousuf, Y., and Jeschke, M. G. (2018). Scar management in burn injuries using drug delivery and molecular signaling: Current treatments and future directions. Adv. Drug Deliv. Rev. 123, 135–154. doi:10.1016/j.addr.2017.07.017
Amjadian, S., Moradi, S., and Mohammadi, P. (2022). The emerging therapeutic targets for scar management: Genetic and epigenetic landscapes. Skin. Pharmacol. Physiol. 35, 247–265. doi:10.1159/000524990
Atiyeh, B. S., Costagliola, M., and Hayek, S. N. (2005). Keloid or hypertrophic scar: The controversy: Review of the literature. Ann. Plast. Surg. 54, 676–680. doi:10.1097/01.sap.0000164538.72375.93
Chen, D., Cheung, H., Lau, H. C., Yu, J., and Wong, C. C. (2022). N(6)-Methyladenosine RNA-binding protein YTHDF1 in gastrointestinal cancers: Function, molecular mechanism and clinical implication. Cancers (Basel) 14, 3489. doi:10.3390/cancers14143489
Chen, H., Hou, K., Wu, Y., and Liu, Z. (2021). Use of adipose stem cells against hypertrophic scarring or keloid. Front. Cell Dev. Biol. 9, 823694. doi:10.3389/fcell.2021.823694
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71, 3971–3975. doi:10.1073/pnas.71.10.3971
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi:10.1038/nature11112
Edupuganti, R. R., Geiger, S., Lindeboom, R. G. H., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878. doi:10.1038/nsmb.3462
Ekstein, S. F., Wyles, S. P., Moran, S. L., and Meves, A. (2021). Keloids: A review of therapeutic management. Int. J. Dermatol 60, 661–671. doi:10.1111/ijd.15159
Goel, A., and Shrivastava, P. (2010). Post-burn scars and scar contractures. Indian J. Plast. Surg. 43, S63–S71. doi:10.4103/0970-0358.70724
Gruber, A. J., Schmidt, R., Ghosh, S., Martin, G., Gruber, A. R., Van Nimwegen, E., et al. (2018). Discovery of physiological and cancer-related regulators of 3' UTR processing with KAPAC. Genome Biol. 19, 44. doi:10.1186/s13059-018-1415-3
Guo, S., Zhao, C., Fang, L., Xu, W., Dalia, S., Glass, J., et al. (2022). The m(6)A methyltransferase WTAP plays a key role in the development of diffuse large B-cell lymphoma via regulating the m(6)A modification of catenin beta 1. Ann. Transl. Med. 10, 779. doi:10.21037/atm-22-3027
He, Y., Deng, Z., Alghamdi, M., Lu, L., Fear, M. W., and He, L. (2017). From genetics to epigenetics: New insights into keloid scarring. Cell Prolif. 50, e12326. doi:10.1111/cpr.12326
Hsu, K. C., Luan, C. W., and Tsai, Y. W. (2017a). Review of silicone gel sheeting and silicone gel for the prevention of hypertrophic scars and keloids. Wounds. 29, 154–158.
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017b). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127. doi:10.1038/cr.2017.99
Hu, M. S., Borrelli, M. R., Hong, W. X., Malhotra, S., Cheung, A. T. M., Ransom, R. C., et al. (2018). Embryonic skin development and repair. Organogenesis 14, 46–63. doi:10.1080/15476278.2017.1421882
Hu, S. C., and Lan, C. E. (2016). High-glucose environment disturbs the physiologic functions of keratinocytes: Focusing on diabetic wound healing. J. Dermatol Sci. 84, 121–127. doi:10.1016/j.jdermsci.2016.07.008
Huang, H., Weng, H., and Chen, J. (2020). The biogenesis and precise control of RNA m(6)A methylation. Trends Genet. 36, 44–52. doi:10.1016/j.tig.2019.10.011
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. doi:10.1038/s41556-018-0045-z
Huang, Q., Mo, J., Liao, Z., Chen, X., and Zhang, B. (2022). The RNA m(6)A writer WTAP in diseases: Structure, roles, and mechanisms. Cell Death Dis. 13, 852. doi:10.1038/s41419-022-05268-9
Hwang, J. Y., Aromolaran, K. A., and Zukin, R. S. (2017). The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat. Rev. Neurosci. 18, 347–361. doi:10.1038/nrn.2017.46
Jaloux, C., Bertrand, B., Degardin, N., Casanova, D., Kerfant, N., and Philandrianos, C. (2017). Keloid scars (part II): Treatment and prevention. Ann. Chir. Plast. Esthet. 62, 87–96. doi:10.1016/j.anplas.2016.04.006
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi:10.1038/nchembio.687
Jones, M. E., Mclane, J., Adenegan, R., Lee, J., and Ganzer, C. A. (2017). Advancing keloid treatment: A novel multimodal approach to ear keloids. Dermatol Surg. 43, 1164–1169. doi:10.1097/dss.0000000000001145
Keane, T. J., Horejs, C. M., and Stevens, M. M. (2018). Scarring vs. functional healing: Matrix-based strategies to regulate tissue repair. Adv. Drug Deliv. Rev. 129, 407–419. doi:10.1016/j.addr.2018.02.002
Liang, D., Lin, W. J., Ren, M., Qiu, J., Yang, C., Wang, X., et al. (2022). m(6)A reader YTHDC1 modulates autophagy by targeting SQSTM1 in diabetic skin. Autophagy 18, 1318–1337. doi:10.1080/15548627.2021.1974175
Limandjaja, G. C., Niessen, F. B., Scheper, R. J., and Gibbs, S. (2020). The keloid disorder: Heterogeneity, histopathology, mechanisms and models. Front. Cell Dev. Biol. 8, 360. doi:10.3389/fcell.2020.00360
Lin, C. X., Chen, Z. J., Peng, Q. L., Xiang, K. R., Xiao, D. Q., Chen, R. X., et al. (2022). The m(6)A-methylated mRNA pattern and the activation of the Wnt signaling pathway under the hyper-m(6)A-modifying condition in the keloid. Front. Cell Dev. Biol. 10, 947337. doi:10.3389/fcell.2022.947337
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772. doi:10.1038/nmeth.3453
Liu, S. Y., Wu, J. J., Chen, Z. H., Zou, M. L., Teng, Y. Y., Zhang, K. W., et al. (2021a). The m(6)A RNA modification modulates gene expression and fibrosis-related pathways in hypertrophic scar. Front. Cell Dev. Biol. 9, 748703. doi:10.3389/fcell.2021.748703
Liu, Y., Gu, C., Li, X., Wang, T., and Yu, L. (2021b). Involvement of METTL3/m6Adenosine and TGFβ/Smad3 signaling on Tenon’s fibroblasts and in a rabbit model of glaucoma surgery. J. Mol. Histol. 52, 1129–1144. doi:10.1007/s10735-021-10028-8
Livneh, I., Moshitch-Moshkovitz, S., Amariglio, N., Rechavi, G., and Dominissini, D. (2020). The m(6)A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 21, 36–51. doi:10.1038/s41583-019-0244-z
Luo, S., and Tong, L. (2014). Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. U. S. A. 111, 13834–13839. doi:10.1073/pnas.1412742111
Macarak, E. J., Wermuth, P. J., Rosenbloom, J., and Uitto, J. (2021). Keloid disorder: Fibroblast differentiation and gene expression profile in fibrotic skin diseases. Exp. Dermatol 30, 132–145. doi:10.1111/exd.14243
Marshall, C. D., Hu, M. S., Leavitt, T., Barnes, L. A., Lorenz, H. P., and Longaker, M. T. (2018). Cutaneous scarring: Basic science, current treatments, and future directions. Adv. Wound Care (New Rochelle) 7, 29–45. doi:10.1089/wound.2016.0696
Matoori, S., Veves, A., and Mooney, D. J. (2021). Advanced bandages for diabetic wound healing. Sci. Transl. Med. 13, eabe4839. doi:10.1126/scitranslmed.abe4839
Meyer, K. D., and Jaffrey, S. R. (2017). Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342. doi:10.1146/annurev-cellbio-100616-060758
Mi, B., Xiong, Y., Yan, C., Chen, L., Xue, H., Panayi, A. C., et al. (2020). Methyltransferase-like 3-mediated N6-methyladenosine modification of miR-7212-5p drives osteoblast differentiation and fracture healing. J. Cell Mol. Med. 24, 6385–6396. doi:10.1111/jcmm.15284
Oerum, S., Meynier, V., Catala, M., and Tisne, C. (2021). A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 49, 7239–7255. doi:10.1093/nar/gkab378
Patel, S., Srivastava, S., Singh, M. R., and Singh, D. (2019). Mechanistic insight into diabetic wounds: Pathogenesis, molecular targets and treatment strategies to pace wound healing. Biomed. Pharmacother. 112, 108615. doi:10.1016/j.biopha.2019.108615
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. doi:10.1038/nature19342
Qian, Z., Wang, H., Bai, Y., Wang, Y., Tao, L., Wei, Y., et al. (2020). Improving chronic diabetic wound healing through an injectable and self-healing hydrogel with platelet-rich plasma release. ACS Appl. Mater Interfaces 12, 55659–55674. doi:10.1021/acsami.0c17142
Roderfeld, M. (2018). Matrix metalloproteinase functions in hepatic injury and fibrosis. Matrix Biol. 68-69, 452–462. doi:10.1016/j.matbio.2017.11.011
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017a). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi:10.1016/j.cell.2017.05.045
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017b). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6, e31311. doi:10.7554/elife.31311
Slobodin, B., Han, R., Calderone, V., Vrielink, J., Loayza-Puch, F., Elkon, R., et al. (2017). Transcription impacts the efficiency of mRNA translation via Co-transcriptional N6-adenosine methylation. Cell 169, 326–337.e12. doi:10.1016/j.cell.2017.03.031
Spitale, R. C., Flynn, R. A., Zhang, Q. C., Crisalli, P., Lee, B., Jung, J. W., et al. (2015). Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490. doi:10.1038/nature14263
Tan, S., Khumalo, N., and Bayat, A. (2019). Understanding keloid pathobiology from a quasi-neoplastic perspective: Less of a scar and more of a chronic inflammatory disease with cancer-like tendencies. Front. Immunol. 10, 1810. doi:10.3389/fimmu.2019.01810
Uddin, M. B., Wang, Z., and Yang, C. (2021). The m(6)A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol. Cancer 20, 61. doi:10.1186/s12943-021-01356-0
Vu, L. P., Cheng, Y., and Kharas, M. G. (2019). The biology of m(6)A RNA methylation in normal and malignant hematopoiesis. Cancer Discov. 9, 25–33. doi:10.1158/2159-8290.cd-18-0959
Wang, S., Chai, P., Jia, R., and Jia, R. (2018). Novel insights on m(6)A RNA methylation in tumorigenesis: A double-edged sword. Mol. Cancer 17, 101. doi:10.1186/s12943-018-0847-4
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi:10.1038/nature12730
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71, 973–985.e5. doi:10.1016/j.molcel.2018.08.011
Wolfram, D., Tzankov, A., Pulzl, P., and Piza-Katzer, H. (2009). Hypertrophic scars and keloids--a review of their pathophysiology, risk factors, and therapeutic management. Dermatol Surg. 35, 171–181. doi:10.1111/j.1524-4725.2008.34406.x
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 925–519. doi:10.1016/j.molcel.2016.03.004
Xie, J., Zhang, X., Zhang, K., Wu, C., Yao, G., Shi, J., et al. (2022). Construction and validation of the diagnostic model of keloid based on weighted gene co-expression network analysis (WGCNA) and differential expression analysis. J. Plast. Surg. Hand Surg. 57, 163–171. doi:10.1080/2000656x.2021.2024557
Yu, H., Wang, Y., Wang, D., Yi, Y., Liu, Z., Wu, M., et al. (2022). Landscape of the epigenetic regulation in wound healing. Front. Physiol. 13, 949498. doi:10.3389/fphys.2022.949498
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10. doi:10.1038/s41421-018-0019-0
Zeng, N., Chen, H., Wu, Y., and Liu, Z. (2021). Adipose stem cell-based treatments for wound healing. Front. Cell Dev. Biol. 9, 821652. doi:10.3389/fcell.2021.821652
Zhang, J., Zheng, Y., Lee, J., Hua, J., Li, S., Panchamukhi, A., et al. (2021a). A pulsatile release platform based on photo-induced imine-crosslinking hydrogel promotes scarless wound healing. Nat. Commun. 12, 1670. doi:10.1038/s41467-021-21964-0
Zhang, T., Wang, X. F., Wang, Z. C., Lou, D., Fang, Q. Q., Hu, Y. Y., et al. (2020). Current potential therapeutic strategies targeting the TGF-β/Smad signaling pathway to attenuate keloid and hypertrophic scar formation. Biomed. Pharmacother. 129, 110287. doi:10.1016/j.biopha.2020.110287
Zhang, Y., Chen, W., Zheng, X., Guo, Y., Cao, J., Zhang, Y., et al. (2021b). Regulatory role and mechanism of m(6)A RNA modification in human metabolic diseases. Mol. Ther. Oncolytics 22, 52–63. doi:10.1016/j.omto.2021.05.003
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. doi:10.1016/j.molcel.2012.10.015
Zhi, S., Li, J., Kong, X., Xie, X., Zhang, Q., and Fang, G. (2021). Insulin-like growth factor 2 mRNA binding protein 2 regulates proliferation, migration, and angiogenesis of keratinocytes by modulating heparanase stability. Bioengineered 12, 11267–11276. doi:10.1080/21655979.2021.2002495
Zhou, J., Wei, T., and He, Z. (2021). ADSCs enhance VEGFR3-mediated lymphangiogenesis via METTL3-mediated VEGF-C m(6)A modification to improve wound healing of diabetic foot ulcers. Mol. Med. 27, 146. doi:10.1186/s10020-021-00406-z
Keywords: m6A methylation, skin regeneration, wound healing, scars, keloids
Citation: Luo X, Zhu S, Li J, Zeng N, Wang H, Wu Y, Wang L and Liu Z (2023) Potential genetic therapies based on m6A methylation for skin regeneration: Wound healing and scars/keloids. Front. Bioeng. Biotechnol. 11:1143866. doi: 10.3389/fbioe.2023.1143866
Received: 13 January 2023; Accepted: 06 April 2023;
Published: 13 April 2023.
Edited by:
Yori Endo, Brigham and Women’s Hospital and Harvard Medical School, United StatesReviewed by:
Kui Zhang, The University of Chicago, United StatesCopyright © 2023 Luo, Zhu, Li, Zeng, Wang, Wu, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Le Wang, bHdhbmcxMjMyMDIyQDE2My5jb20=; Zeming Liu, Nm15dEAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.