- 1Medical School of Chinese People’s Liberation Army (PLA), Beijing, China
- 2Faculty of Hepato-Pancreato-Biliary Surgery, Chinese PLA General Hospital, Beijing, China
- 3Institute of Hepatobiliary Surgery of Chinese PLA, Beijing, China
- 4MOE Key Laboratory of Cell Proliferation and Differentiation, College of Life Sciences, Peking-Tsinghua Center for Life Sciences, Peking University, Beijing, China
- 5Peking University-Tsinghua University-National Institute of Biological Science Joint Graduate Program, College of Life Science, Peking University, Beijing, China
- 6Department of Rheumatology and Immunology, Peking University Third Hospital, Beijing, China
Objectives An acute injury is often accompanied by tissue regeneration. In this process, epithelial cells show a tendency of cell proliferation under the induction of injury stress, inflammatory factors, and other factors, accompanied by a temporary decline of cellular function. Regulating this regenerative process and avoiding chronic injury is a concern of regenerative medicine. The severe coronavirus disease 2019 (COVID-19) has posed a significant threat to people’s health caused by the coronavirus. Acute liver failure (ALF) is a clinical syndrome resulting from rapid liver dysfunction with a fatal outcome. We hope to analyze the two diseases together to find a way for acute failure treatment.
Methods COVID-19 dataset (GSE180226) and ALF dataset (GSE38941) were downloaded from the Gene Expression Omnibus (GEO) database, and the “Deseq2” package and “limma” package were used to identify differentially expressed genes (DEGs). Common DEGs were used for hub genes exploration, Protein-Protein Interaction (PPI) network construction, Gene Ontology (GO) functional enrichment, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. The real-time reverse transcriptase-polymerase chain reaction (RT-qPCR) was used to verify the role of hub genes in liver regeneration during in vitro expansion of liver cells and a CCl4-induced ALF mice model.
Results: The common gene analysis of the COVID-19 and ALF databases revealed 15 hub genes from 418 common DEGs. These hub genes, including CDC20, were related to cell proliferation and mitosis regulation, reflecting the consistent tissue regeneration change after the injury. Furthermore, hub genes were verified in vitro expansion of liver cells and in vivo ALF model. On this basis, the potential therapeutic small molecule of ALF was found by targeting the hub gene CDC20.
Conclusion We have identified hub genes for epithelial cell regeneration under acute injury conditions and explored a new small molecule Apcin for liver function maintenance and ALF treatment. These findings may provide new approaches and ideas for treating COVID-19 patients with ALF.
Introduction
COVID-19 is an acute respiratory infectious disease caused by Severe acute Breathing Syndrome Coronavirus 2 (SARS-CoV-2) (Sharma et al., 2021), which may cause damage to multiple systems, including the respiratory and digestive systems (Tregoning et al., 2021). While the pathogenesis of COVID-19 is well understood, the main clinical treatment is still symptomatic. Therefore, it is essential to investigate further the mechanism of epithelial cell regeneration under acute injury, find ways to maintain tissue and organ functions, and avoid chronic impairment (Li et al., 2020a; Delorey et al., 2021; Melms et al., 2021).
ALF is a severe clinical disease usually accompanied by multiple organ failures caused by acute liver damage. It can present as ALF or acute-on-chronic liver failure (ACLF) (Sarin et al., 2019). Overdose of acetaminophen (APAP) is a common cause in most Western countries. In China, acute liver failure caused by HBV infection also has a high incidence (Bunchorntavakul and Reddy, 2018; Dong et al., 2020). Treatment of liver transplantation is often limited by a shortage of liver sources (Olivo et al., 2018). Therefore, other interventions to mitigate liver damage are worth exploring. Despite some similar mechanisms, the prognosis of the ALF and ACLF is significantly different due to changes in liver regeneration properties. Continuous chronic injury destroys the microenvironment of liver cell regeneration, leading to continuous necrosis and apoptosis of the liver and eventually irreversible liver failure (Russo and Parola, 2011; Rastogi et al., 2014; Shubham et al., 2019).
A variety of epithelial cells, including renal tubule, respiratory epithelial, gastric epithelial cell, and liver cell, have certain regeneration capacities under acute injury (Sen et al., 2016; Wilkinson and Hardman, 2020). This regeneration process is the result of coordination among various cell types. Epithelial cells often undergo phenotypic changes, for example, cellular dedifferentiation and activation of regenerative pathways, including non-canonical nuclear factor-kappaB (NF-κB), Janus kinase-signal transducer and activator of transcription (JAK-STAT), phosphatidylinositol 3-kinase/protein kinase B (PI3K/Akt), Wnt, and AP1 (Sen et al., 2016; He and Barron, 2020; Michalopoulos and Bhushan, 2021; Giuliani et al., 2022). In this process, regenerated epithelium helps repair wounds and promote recovery. However, during the regeneration period, the epithelial cell may turn to progenitor-like cells, which affect normal cellular function, increasing the risk of damage in the short term (Yimlamai et al., 2014; Gárate-Rascón et al., 2021). Therefore, it is reasonable to balance motivating cellular regeneration with controlling inflammatory response and maintaining tissue function.
In this study, through bioinformatics analysis of the public databases, we studied the changes in gene expression in COVID-19 and ALF epithelial cells under acute injury and identified hub genes such as CDC20, CENPF, KIF4, KIF11, NUSAP1, TPX2, and PTTG1. The hub genes and their enrichment pathways helped to clarify the mechanisms of impairment in ALF and COVID-19. Further, the role of hub genes in liver regeneration or ALF was verified by in vitro and in vivo experiments. The therapeutic effect of an inhibitor for a hub gene CDC20 on acute liver failure was verified in the ALF mice model, which may provide a new method for treating ALF and COVID-19.
Methods
ALF and COVID-19 datasets collection
This article’s related datasets include COVID-19 and ALF from the Gene Expression Omnibus (GEO) database. The COVID-19 dataset (GSE180226) comprises 20 COVID-19 lung samples and three normal lung samples, and the ALF dataset (GSE38941) contains 17 HBV-infected ALF liver samples and ten healthy controls.
Identification of common DEGs between ALF and COVID-19
The “limma” package in RStudio software (version 4.2.2) was used to identify DEGs with standard | Log2Fold Change | > 1 and | adj. p-value | < 0.05 for the COVID-19 dataset (GSE180226). Meanwhile, DEGs with | Log2Fold Change | > 1 and | adj. p-value | < 0.05 for the ALF dataset (GSE38941) were also identified. We obtained COVID-19 and ALF common DEGs using the “Venn” package in R software.
Pathway enrichment analysis of common DEGs
The “clusterProfiler” package in R software (version 4.2.2) was used to perform GO and KEGG enrichment analysis. Other packages, including “dplyr,” “org.Hs.e.g.,.db,” “circlize,” “RColorBrewer,” “ggplot2,” “enrichplot,” “ggpubr,” and “ComplexHeatmap” were used for data annotation and visualization. p-value <0.05 was set as the cutoff criterion for common DEGs.
PPI network analysis based on common DEGs
PPI networks were established by Search Tool for the Retrieval of Interacting Genes (STRING) database based on the combined score >0.9 and visualized by Cytoscape 3.9.1 (version 4.2.2) to reveal the interactions among proteins of common DEGs.
Verification of hub genes
The expression of the common hub genes in GSE120652 for ALF and GSE139602 for ACLF was detected. GSE120652 contains three acetaminophen-induced ALF samples and three healthy samples, and GSE134431 contains eight ACLF samples and six healthy samples. Wilcoxon test was used to compare the two datasets.
Human hepatocyte expansion
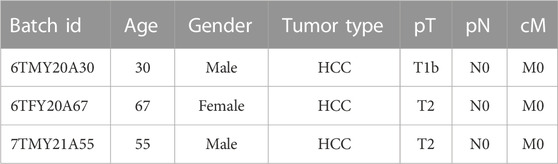
The cryopreserved human hepatocytes were plated on collagen type I-coated (Rat-tail collagen type I, Gibco) plate at a density of 1 × 105/cm2 in a modified liver expansion medium containing William’s E Medium supplemented with 2% B27 (without VA), 1% PS (All from Invitrogen), 50 ng/mL EGF, 20 ng/mL HGF (Both from Peprotech), 200 μM 2-phospho-L-ascorbic acid (pVc), 3 μM CHIR99021, 5 μM SB431542, 5 μM Lysophosphatidic acid (LPA), 0.5 μM Sphingosine-1-phosphate (S1P) (All from MCE), and 1% fetal bovine serum (FBS) (Gibco, US). After seeding for 7–10 days, expanded cells were passaged at a ratio of 1:2 after dissociation with Accutase (eBioscience). The expansion medium was changed every day. Hepatocyte donor information is shown in Table 1.
Real-time reverse transcriptase-polymerase chain reaction (RT-qPCR)
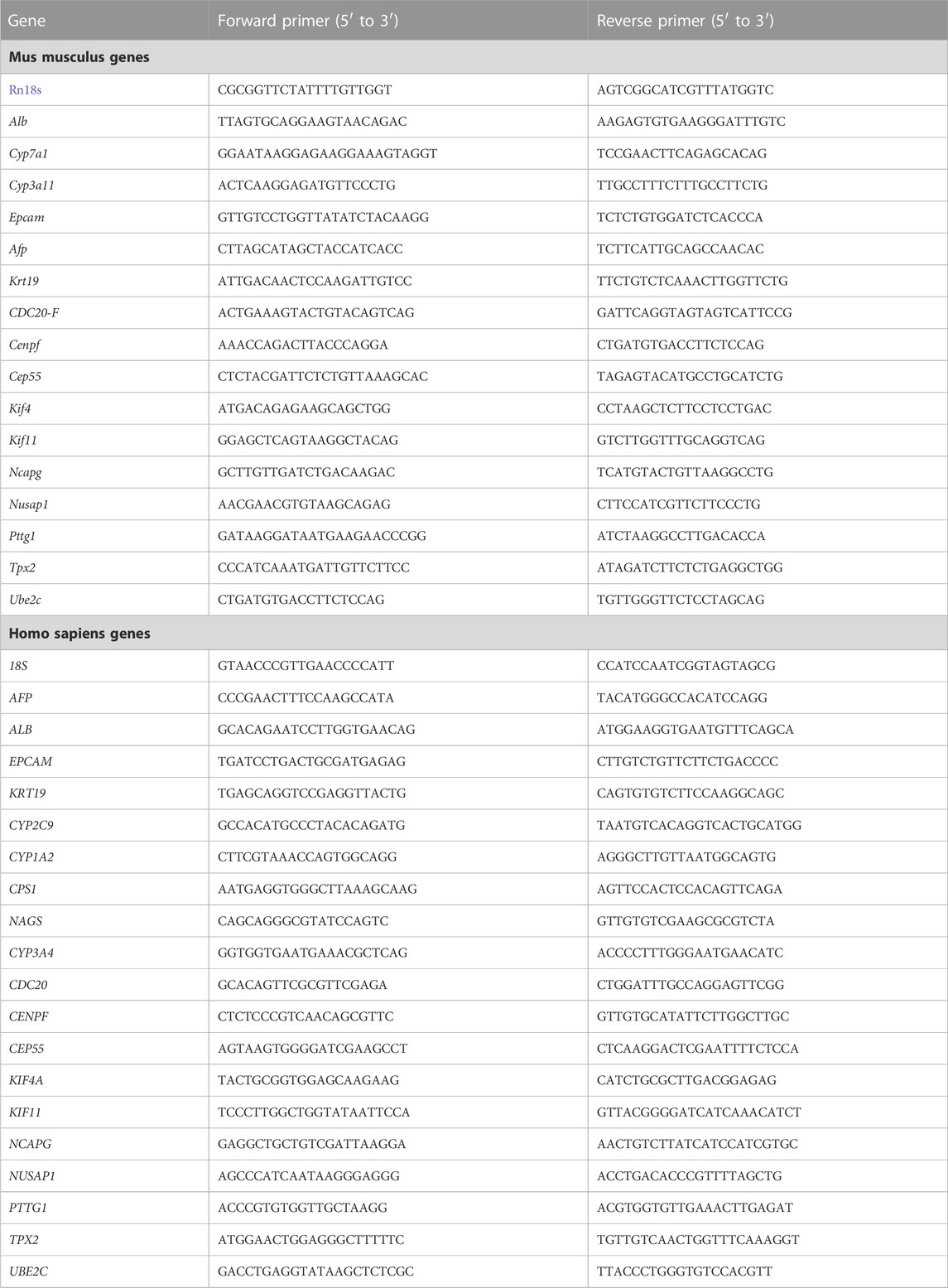
The Direct-zol RNA Kits (ZYMO Research, R2052) were used to isolate total RNA from liver tissues and hepatocytes, followed by reverse transcription using the TransScript First-Strand cDNA Synthesis SuperMix (TransGen, AT311). The KAPA SYBR® FAST qPCR Kit (KAPA Biosystems, KK4601) was used to perform relative gene expression on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). The housekeeping genes (RRN18S) were used as an internal reference to analyze RT-qPCR results using the 2^(−ΔΔCt) method. All the primer sequences for RT-qPCR are provided in Table 2.
The acute liver failure model
The animal experiments were conducted under a protocol (2019-X15-03) that the ethics committee approved by the Laboratory Animals Center of Chinese PLA General Hospital (Beijing). Twenty-one male C57BL/6J mice (weight 30 g, aged 8–10 weeks) were purchased from Sipeifu (Beijing, China) and randomly divided into three groups to obtain liver tissue for molecular and histological studies (n = 7/group): mice with corn oil only without CCl4 injury (NC), mice with corn oil and CCl4 injury (CCl4), and mice with Apcin and CCl4 injury (Apcin). Before the CCl4 injection, the Apcin mice group was intraperitoneally injected with Apcin (30 mg/kg) dissolved in corn oil. Eight hours later, the CCl4 and Apcin mice were given CCl4 2.9 mL/kg dissolved in corn oil (Solarbio, C7030) by intraperitoneal injection. The mice in the NC group were injected with corn oil as a control. After 24 h of CCl4 treatment, all mice were sacrificed to obtain serum and liver tissues. Another 21 mice were randomly divided into three groups (n = 7/groups) to obtain the survival rate of ALF, as described above. Seven days after injection, all mice were sacrificed, and the survival rate can be further analyzed.
Serum biochemical examination
0.8 mL serum was incubated for 30 min and then centrifuged at 3,000 rpm for 5 min to get the supernatant to collect the mouse blood sample. The automatic chemistry analyzer was used to measure the aspartate aminotransferase (AST) and alanine aminotransferase (ALT), with each biochemical parameter being repeated four times.
Immunofluorescence staining
Hepatocytes on the glass bottom plate (Cellvis, P24-1.5H-N) were fixed with 4% paraformaldehyde in PBS at room temperature for 20 min. Then the cells permeabilized in 0.25% Triton X-100 and blocked with 2.5% normal donkey serum (Jackson ImmunoResearch Laboratories, 017-000-001) in PBS. Next, the hepatocytes were incubated with primary antibodies at room temperature for 1 h, including Anti-ALB (ab243894, Abcame, 1:200), Anti-CDC20 (sc-5296, Abcame, 1:200), and Anti-CK19(ab181557, Abcame, 1:250). Hepatocytes were incubated with the secondary antibodies, including DyLight® 650 Donkey anti-rabbit (ab96894, Abcame, 1:200) and DyLight® 650 Donkey anti-mouse (ab96878, Abcame, 1:200) for 1 h at room temperature. Finally, the samples were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen, D1306) for 5 min and observed and photographed under an inverted fluorescent microscope using Nikon Imaging System at the same exposure.
Immunohistochemistry and hematoxylin-eosin (H&E) staining
After paraffin-embedded, the liver tissue was cut into sections of 4 μm thickness. Then the slides immunohistochemistry for the TUNEL staining was performed using the TUNEL Assay Kit-HRP-DAB (Abcam, ab206386). The H&E staining was performed using a regular protocol. All the results were randomly taken at the same exposure.
Statistical analysis
Graph Pad 8.0 was used for statistical analysis. All the data were the means ± standard deviation (SD). Differences with p < 0.05 was considered statistically significant. Student’s t-test and Wilcox were used to analyze differences between two or multiple groups. Kaplan-Meier analysis was used for the overall survival.
Results
Acquirement of the common DEGs between ALF and COVID-19
To explore the common changes of epithelial cells under acute injury to find potential targets for injury recovery and tissue regeneration, we downloaded the transcriptome data of COVID-19 and ALF from the GEO database. We identified 6,592 and 2,191 DEGs by comparing them with healthy data (Figures 1A,B). Further, the volcano plots demonstrated DEGs of ALF and COVID-19, respectively (Figures 1C,D). One hundred ninety-two co-upregulated genes and 227 co-downregulated genes were shown in Venn diagrams between ALF and COVID-19 datasets (Figures 1E, F and Supplementary Material S1). These results indicated common mechanisms and genetic expression trends among different tissues under acute injury.
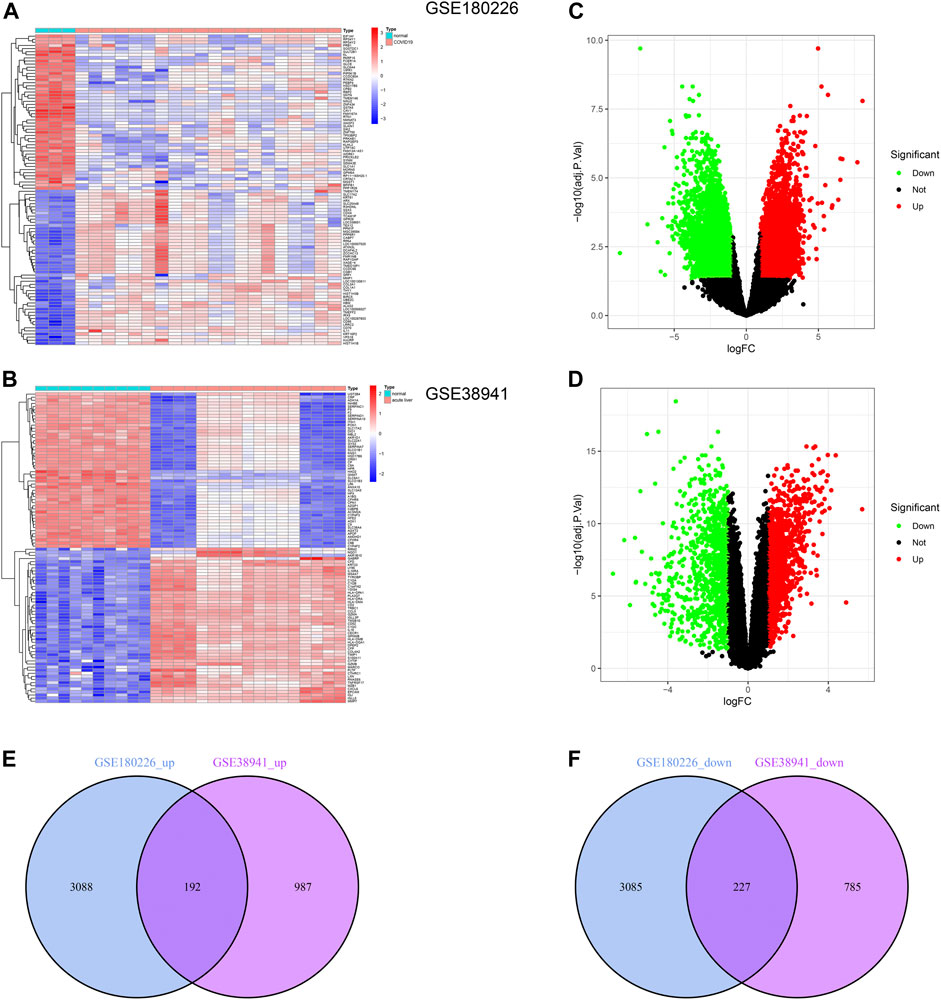
FIGURE 1. Identification of common DEGs between COVID-19 and ALF. (A) Heatmap of the DEGs in COVID-19 patients compared with healthy controls. (B) Heatmap of the DEGs in ALF patients compared with healthy controls. (C) Volcano plot of DEGs in COVID-19 patients, in which green represents downregulated DEGs, red represents upregulated DEGs, and black represents no difference. (D) Volcano plot of DEGs in ALF. And the colors represent the same sample as B. (E) Venn diagram showing the upregulated DEGs between COVID-19 and ALF patients. (F) Venn diagram showing the downregulated DEGs between COVID-19 and ALF patients.
GO and KEGG analysis of common DEGs
GO and KEGG enrichment analyses were used further to analyze the role and function of common DEGs. GO enrichment analysis included three categories: biological process (BP), cell composition (CC), and molecular function (MF). The common DEGs were mainly enriched in amino acid metabolic processes or metabolic processes of other substances (BP), the complex of collagen trimers, endoplasmic reticulum lumen, and fibrillar collagen trimer (CC), and extracellular matrix reconstruction and antioxidant stress response (MF) (Figure 2A). The KEGG enrichment revealed that the common DEGs mainly enriched in protein digestion and absorption, complement and coagulation cascades, and drug metabolism pathways (Figure 2B).
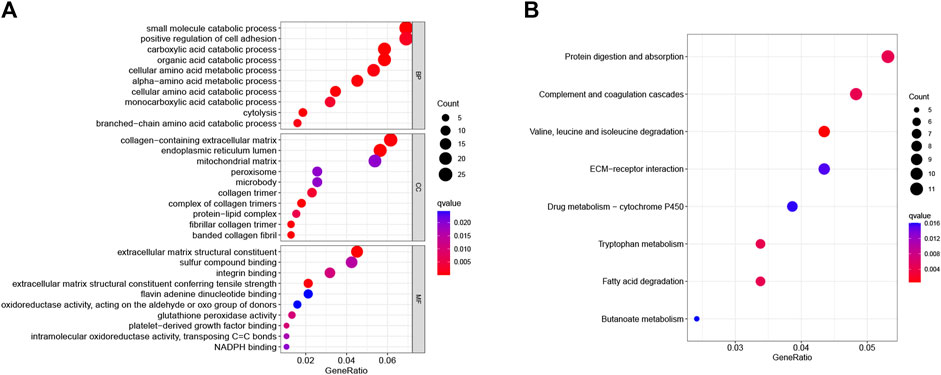
FIGURE 2. GO and KEGG pathway analyses of common DEGs. (A) The top ten GO functional enrichment analyses of the common DEGs in biological process (BP), cell composition (CC), and molecular function (MF) groups, respectively. The bubble size in the dotplot shows the first five terms, and the size represents p-adjusted values for these terms. (B) The top eight significant KEGG signal pathways of the common DEGs.
PPI network construction and hub genes extraction
STRING was used to construct the PPI network to explore gene interactions further and establish a hub gene network, in which 81 nodes and 250 edges were included (Figure 3A). CytoHubba was used to identify the top 15 genes in Cytoscape based on the degree method scores in the PPI network, including BUB1B, CDC20, ASPM, NUSAP1, CEP55, UBE2C, KIF11, RRM2, PBK, PTTG1, NCAPG, TPX2, KIF4A, NUF2, and CENPF (Figures 3B,C). We believe these targets may reflect the change of epithelial cells in the state of acute injury. Validation was performed for ALF and ACLF from GEO, and the results showed that these hub genes were upregulated in ALF compared to normal tissues, although without significant differences (Figures 4A, B). However, the trend is not evident in ACLF (Figures 4A, B). These results indicate that chronic injury weakens the regenerative ability of epithelial cells, consistent with current theoretical studies, suggesting the role of hub genes in acute injury from the side.
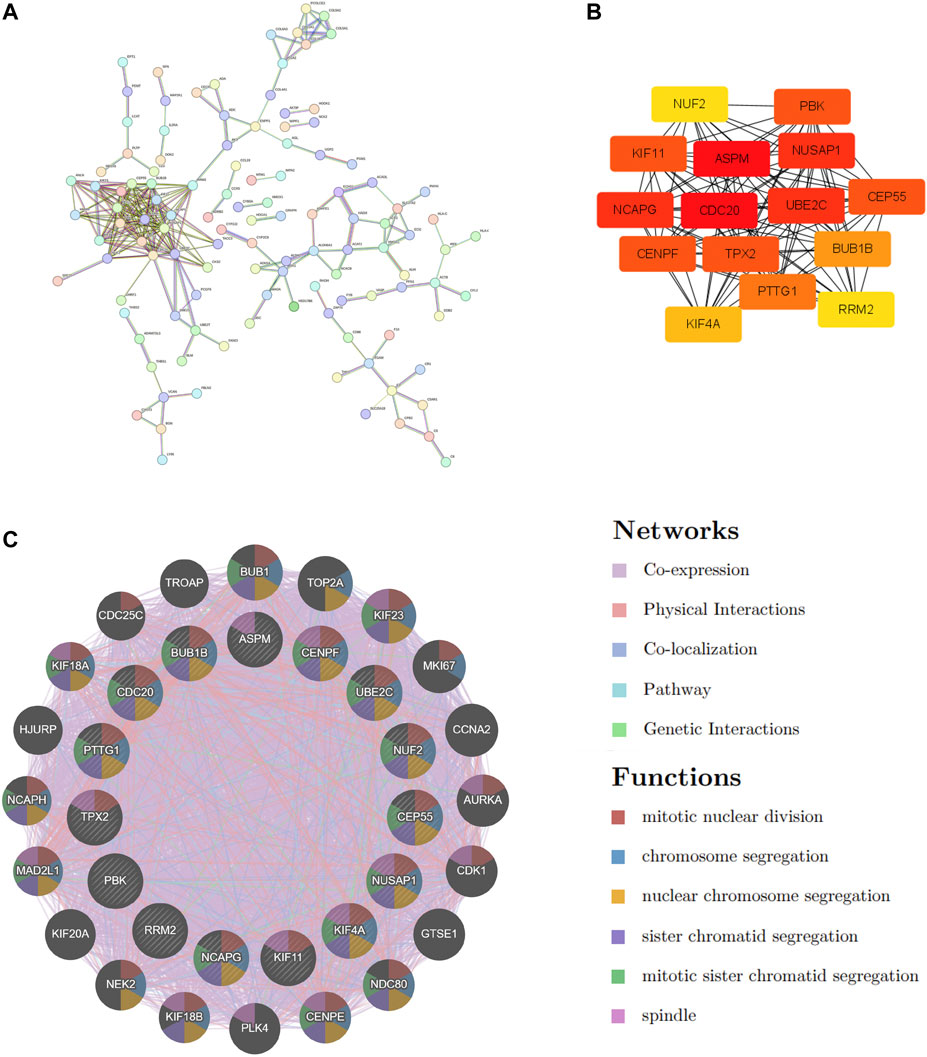
FIGURE 3. PPI Network Construction and hub genes extraction. (A) COVID-19 and ALF common DEGs in the PPI network constructed by STRING. (B) The top 15 hub genes and their interactions with each other. (C) The gene–gene interaction network was visualized by the GeneMANIA database. The top 20 mostly related neighboring genes are shown in the outer circle of hub genes.
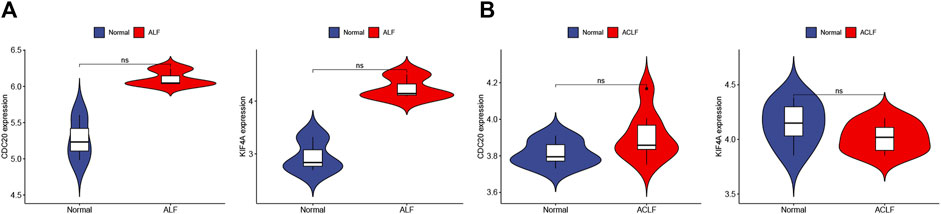
FIGURE 4. Validation of hub genes in ALF and ACLF by Violin plots. (A) The expression of representative hub genes between livers of ALF patients with normal livers was compared. (B) The expression of representative hub genes in ACLF and normal livers was compared. Eight ACLF and six control healthy livers were obtained from GSE139602. Data were statistically analyzed using the Wilcoxon test (ns indicates no significant difference).
Functional enrichment analysis of hub genes
To further explore hub genes’ biological effects on tissue injury, GO and KEGG analyses were conducted again on the 15 hub genes. Interestingly, the enrichment status of hub genes was apparent. In GO enrichment, the BP category, CC category, and MF category all showed functional enrichment related to cell proliferation, mitosis, and spindle formation. KEGG enrichment analysis demonstrated pathways associated with cell proliferation and viral infection (e.g., Cell cycle, Human T−cell leukemia virus one infection, and Oocyte meiosis) (Figures 5A, B). These results indicate that tissue regeneration is the most apparent phenomenon under the stress of acute failure.
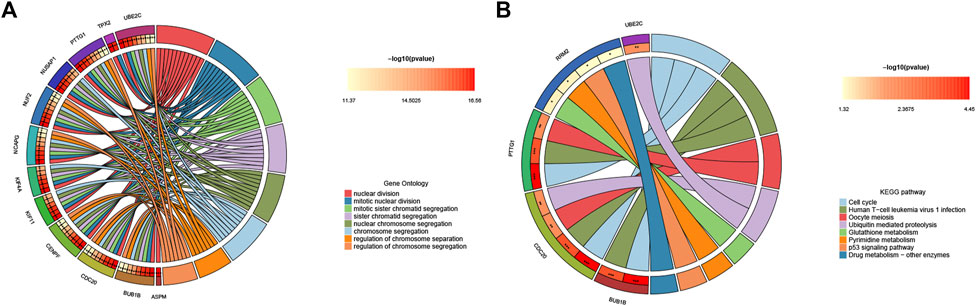
FIGURE 5. Functional enrichment analysis of hub genes. (A) Circle diagram visualization of GO terms, and (B) top eight significant KEGG pathways. For each graph, the right semicircle represents significant pathways. Different color modules represent different pathways. And the left semicircle represents the genes enriched in the same color pathways.
In vitro and in vivo models of liver regeneration
In the in vitro hepatocyte regeneration model, serum and cytokines simulating the injury environment were used to induce hepatocyte proliferation (Figure 6A). Hepatocytes entered the progenitor cell state with the upregulation of hub genes such as CDC20, CENPF, CEP55, KIF4A, KIF11, NCAPG, NUSAP1, PTTG1, TPX2, and UBE2C (Figure 6B, Supplementary Material S2A). Compared to the primary hepatocytes, progenitor markers such as EPCAM, AFP, and CK19 expressed much higher in liver progenitors while liver function genes including cytochrome P450 (CYP) enzymes and urea metabolism genes downregulated (Figures 6C, D, Supplementary Material S2A). Immunofluorescence staining confirmed the changes in cellular identity and function (Figures 6E, F). These results demonstrated the relationship between hub genes and liver regeneration. The in vivo experiment schematic is represented in Figure 7A. Similarly, in the CCL4-induced ALF model to monitor liver injury and regeneration, we also observed significantly upregulated hub genes in the ALF liver than healthy control (Figure 7C, Supplementary Material S2B). In addition, progenitor genes were upregulated, while CYP enzymes and other functional genes were down (Figures 7D, E, Supplementary Material S2B). These results reflect the consistent changes in response to stress stimulation and following regeneration patterns in liver cells.
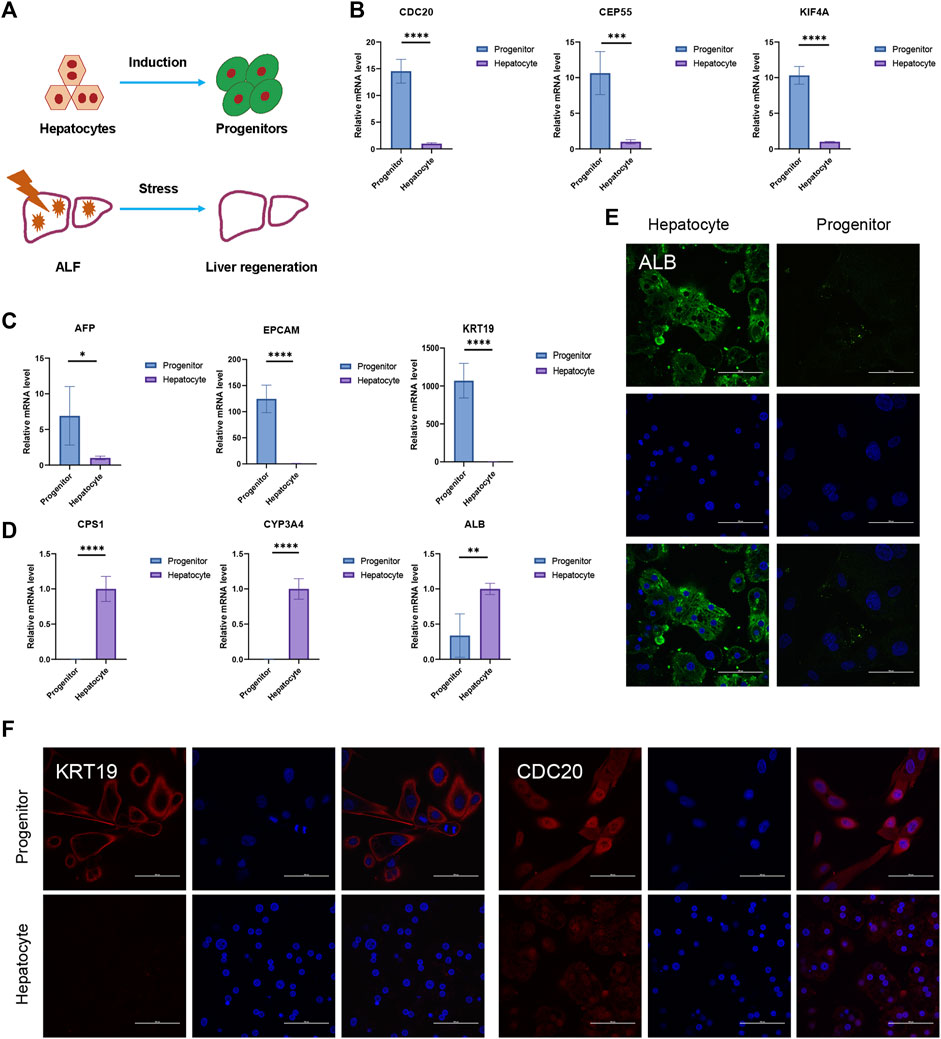
FIGURE 6. In vitro liver regeneration under physiological state. (A) Damage and regeneration pattern diagram. The process of hepatocytes turning into progenitors represents in vitro regeneration model. Liver regeneration occurs during ALF and represents in vivo regeneration model. (B–D) The relative expression of hub genes (B), progenitor genes (C), and liver functional genes (D) between hepatocytes and progenitors. N = 4. Data were statistically analyzed using Student’s t-test, and values were presented as mean ± SD. *indicates significant difference at p < 0.05, ** indicates significant difference at p < 0.01, *** indicates significant difference at p < 0.001, **** indicates significant difference at p < 0.0001. (E–F) Representative immunofluorescence staining images of ALB (E), KRT19, and CDC20 (F) (Scale bars = 100 µm).
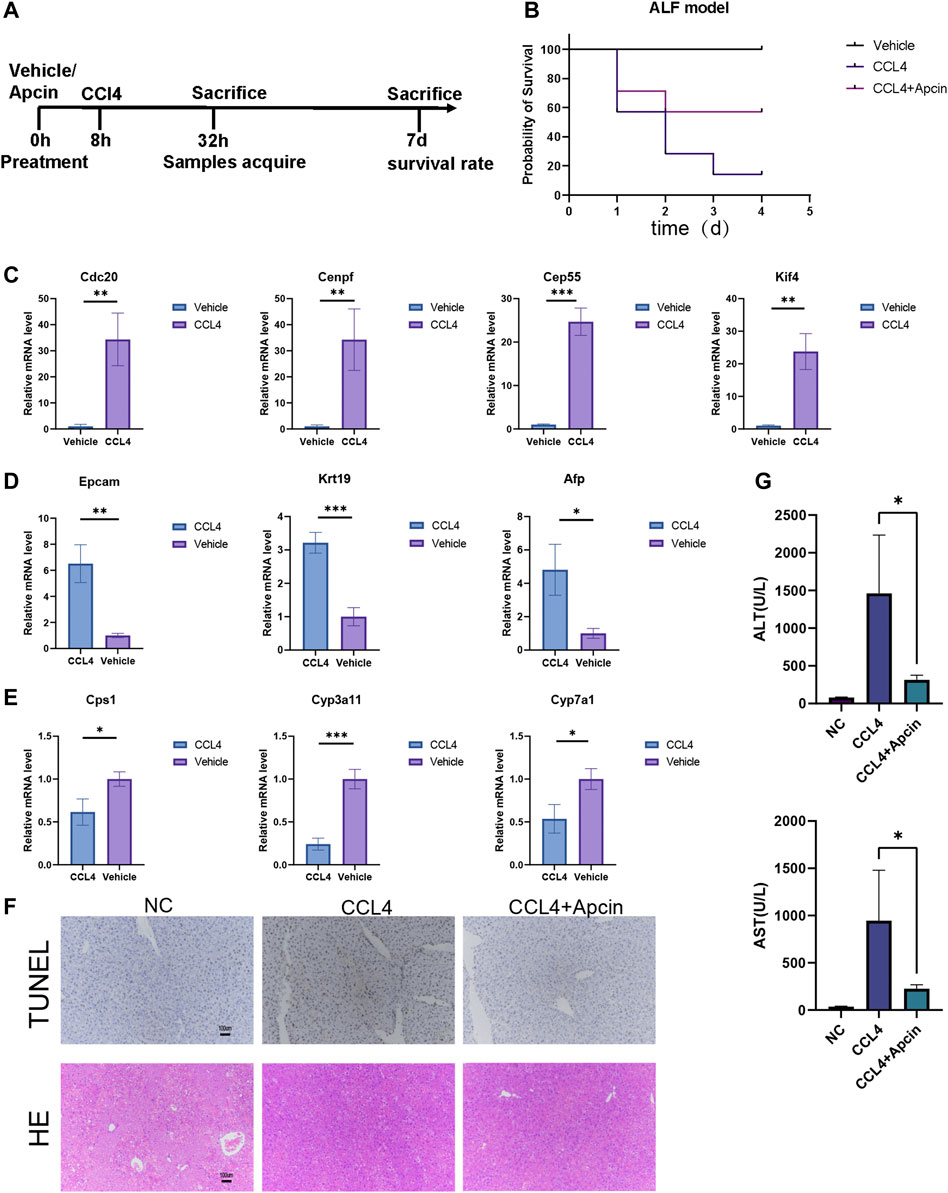
FIGURE 7. CDC20 inhibitor Apcin alleviates ALF. (A) The flow diagram demonstrates the ALF model construction and treatment. (B) The survival rate of ALF mice with or without Apcin pretreatment. N = 7. (C–E) The relative expression of hub genes (C), progenitor genes (D), and functional genes (E) between ALF and vehicle mice. N = 3. (F) Representative TUNEL (upper panels) and H&E (lower panels) staining of liver tissues harvested at 24 h post CCL4 induction with or without Apcin pretreatment. N = 3 (Scale bars = 100 µm). (G) The AST and ALT levels in NC, CCl4 with or without Apcin mice groups. N = 4. Data were statistically analyzed using Student’s t-test, and values were presented as mean ± SD. *indicates significant difference at p < 0.05, ** indicates significant difference at p < 0.01, *** indicates significant difference at p < 0.001.
The inhibitor Apcin for bub gene CDC20 alleviates ALF
During ALF, extensive hepatocytes undergo dedifferentiation, and proliferation may influence liver function. Therefore, we wonder if maintaining the identity of hepatocytes and moderate inhibition of proliferation will contribute to the survival of ALF. CDC20 inhibitor Apcin has been reported to inhibit cancer cell proliferation by competitively inhibiting anaphase-promoting complex/Cyclosome (APC/C)-dependent ubiquitination. It was found that Apcin pretreatment can attenuate liver failure and improve the survival rate in a mouse model (Figure 7B). Liver damage and inflammatory response were relieved by Tunnel and H&E staining (Figure 7F). Serum of mice was collected for biochemical detection, including alanine aminotransferase (ALT) and aspartate transaminase AST (Figure 7G). These data illustrate the positive effect of Apcin on ALF, which may also help with the treatment of COVID-19.
In conclusion, these results indicate that regardless of COVID-19, ALF, or other acute failures, the epithelium entered the cell proliferation stage induced by injury, and this regeneration contributed to repairing and recovery in the long run. However, short-term loss of function may aggravate the general condition. Therefore, proper regulation of the balance between cell proliferation and function maintenance may be beneficial for patients to recover from acute failure. The hub genes identified in this paper and their corresponding targets, such as CDC20 inhibitor-Apcin, may be a new possible approach for ALF and COVID-19 treatment.
Discussion
In the study of tissue regeneration after acute injury, we analyzed the database of ALF and COVID-19 by bioinformatics method and found common hub genes, including CDC20, CENPF, KIF4, KIF11, NUSAP1, TPX2, and PTTG1, which were related to mitosis and cell cycle regulation. They were highly expressed in patients with ALF and COVID-19, reflecting epithelial tissue’s regeneration and repair response to acute injury. We confirmed that these hub genes showed significantly enhanced expression during in vitro hepatocyte regeneration and mouse models with ALF. Therefore, we conclude that these genes may play an essential role in epithelial damage and regeneration. Further in vivo studies confirmed that targeting CDC20 to maintain hepatocyte identity and liver function is beneficial for mice to survive ALF. Perhaps it can be further extended to the treatment of COVID-19.
The lungs are the primary site of immunopathology induced by SARS-CoV-2, and angiotensin-converting enzyme 2 (ACE2) are receptors for the virus to enter host cells (Mulay et al., 2021). Complex lung regeneration and repair programs exist to replenish damaged cell populations. In addition to clinical treatments being applied, it is necessary to promote lung regeneration for COVID-19 patients (Zhao et al., 2022).
For treating ALF, the current research mainly focuses on two aspects. The liver is the organ with the strongest regenerative capacity in the human body, and after a temporary injury, the liver can still regenerate and recover. Therefore, inhibiting inflammatory response, eliminating inhibition signal, and promoting hepatocyte regeneration are effective means for treating ALF (Rutherford and Chung, 2008; Larsen, 2019). However, studies have found that liver cells cannot maintain their properties under ALF conditions and undergo dedifferentiation and proliferation, which may lead to impaired liver function, increased blood ammonia and bilirubin, and aggravated inflammation (Ko et al., 2020). Therefore, maintaining the function of the liver is also an essential means of ALF treatment.
In conclusion, it is of great significance to find the hub genes in the state of acute injury for treating COVID-19 infection and ALF.
CDC20, part of the anaphase-promoting complex/cyclosome (APC/C), has been recognized as necessary in cell cycle regulation. APC/CCDC20 promotes the ubiquitination and degradation of critical proteins to influence various cellular processes (Wang et al., 2015). CDC20 and KIF11 were listed as hub genes for COVID-19 treatment and novel therapeutic targets against SARS-CoV-2 infections (Auwul et al., 2021). Moreover, they were identified as biomarkers of HIV-infected COVID-19 patients (Yan et al., 2022). Based on the study of myocardial injury and myocardial regeneration differences among different ages, CDC20 was identified as the critical factor of myocardial regeneration (Locatelli et al., 2020). In addition, the promoting effect of CDC20 was found in muscle regeneration, corneal regeneration, and liver regeneration studies (Kuwata et al., 2011; Song et al., 2022). CDC20 has been reported as a promoter in human tumorigenesis. Overexpression of CDC20 is associated with clinicopathologic features of various human tumors. These tumors include hepatocellular carcinoma (HCC), non-small cell lung cancer (NSCLC), pancreatic tumor, and colorectal cancer (Li et al., 2017; Li et al., 2020b; Dai et al., 2021; Gong et al., 2021). All these demonstrated the role of CDC20 in normal tissue regeneration and tumorigenesis. However, whether CDC20 works in acute liver failure remains unclear. This study innovatively adopted the CDC20 inhibitor Apcin for treating ALF, and a significant therapeutic effect was achieved in the mouse model of ALF (Ramavath et al., 2021).
PTTG1, CENPF, KIF4, KIF11, NUSAP1, and TPX2 have also been implicated in many biological functions, such as DNA repair, organ development, and regulation of mitosis (Försti et al., 2016). CENPF was viewed as a prognostic indicator of lung cancer patients with COVID-19 infection (Cury et al., 2023). TPX2 and KIF11 were upregulated in peripheral blood mononuclear cells and contributed to diagnosing and managing COVID-19 patients (Hasan et al., 2022). It is worth noting that KIF4 and KIF11, molecular motor proteins, have been identified as powerful targets contributing to the regeneration of injured axons. CENPF, NUSAP1, UBE2C, and CDC20 are identified as markers of transit-amplifying cells (TACs) by scRNA-seq and are related to proliferation stages. They are also considered independent prognostic markers for various tumors and play an essential role in tumor progression, metastasis, tumor stem cell formation, and epithelial-mesenchymal transition (Parte et al., 2019; Guo et al., 2020).
In conclusion, these hub genes are essential in tumorigenesis, acute injury, and tissue regeneration and may become potential targets for treating these diseases.
The study had several limitations. First, we have emphasized the role of hub genes in tissue injury and regeneration but lack further exploration of the mechanisms. Second, the therapeutic effect of Apcin for COVID-19 is mainly based on theoretical speculation and lacks some experimental verification. Third but not negligible, in current study, we compared sequencing data between liver and lung tissues, which may have biased the experimental results. Therefore, acquiring and analyzing liver tissue samples of ALF induced by different etiologies including COVID-19 infection may be essential for the next study.
Conclusion
In this paper, we have identified hub genes that worked in different epithelial tissue regeneration after injury. The results were verified by hepatocyte expansion and ALF model under physiological conditions. In addition, the treatment of ALF by Apcin, a CDC20 inhibitor, was reported for the first time. This discovery could help develop new means or drugs to treat COVID-19 and ALF.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE180226, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE38941, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE120652, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE139602.
Ethics Statement
The studies involving human participants were reviewed and approved by Chinese People’s Liberation Army General Hospital Medical Ethics Committee. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by The Laboratory Animals Center of Chinese PLA General Hospital (Beijing). Written informed consent was obtained from the individual (s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
JS, GL, and XY conceptualized and designed the study. JS, GL, XY, YW, and MG performed the experiments and analyzed the data. JS, GL, CL, and XG contributed significantly to the interpretation of the data. JS, GL, and XY wrote the manuscript. SL designed, supervised the research, and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1135997/full#supplementary-material
References
Auwul, M. R., Rahman, M. R., Gov, E., Shahjaman, M., and Moni, M. A. (2021). Bioinformatics and machine learning approach identifies potential drug targets and pathways in COVID-19. Brief. Bioinform 22 (5), bbab120. doi:10.1093/bib/bbab120
Bunchorntavakul, C., and Reddy, K. R. (2018). Acetaminophen (APAP or N-Acetyl-p-Aminophenol) and acute liver failure. Clin. Liver Dis. 22 (2), 325–346. doi:10.1016/j.cld.2018.01.007
Cury, S. S., Oliveira, J. S., Biagi-Júnior, C. A. O., Silva, W. A., Reis, P. P., Cabral-Marques, O., et al. (2023). Transcriptional profiles and common genes link lung cancer with the development and severity of COVID-19. Gene 852, 147047. doi:10.1016/j.gene.2022.147047
Dai, L., Song, Z-X., Wei, D-P., Zhang, J-D., Liang, J. Q., Wang, B. B., et al. (2021). CDC20 and PTTG1 are important biomarkers and potential therapeutic targets for metastatic prostate cancer. Adv. Ther. 38 (6), 2973–2989. doi:10.1007/s12325-021-01729-3
Delorey, T. M., Ziegler, C. G. K., Heimberg, G., Normand, R., Yang, Y., Segerstolpe, Å., et al. (2021). COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 595 (7865), 107–113. doi:10.1038/s41586-021-03570-8
Dong, V., Nanchal, R., and Karvellas, C. J. (2020). Pathophysiology of acute liver failure. Nutr. Clin. Pract. 35 (1), 24–29. doi:10.1002/ncp.10459
Försti, A., Frank, C., Smolkova, B., Kazimirova, A., Barancokova, M., Vymetalkova, V., et al. (2016). Genetic variation in the major mitotic checkpoint genes associated with chromosomal aberrations in healthy humans. Cancer Lett. 380 (2), 442–446. doi:10.1016/j.canlet.2016.07.011
Gárate-Rascón, M., Recalde, M., Jimenez, M., Elizalde, M., Azkona, M., Uriarte, I., et al. (2021). Splicing factor SLU7 prevents oxidative stress-mediated hepatocyte nuclear factor 4α degradation, preserving hepatic differentiation and protecting from liver damage. Hepatology 74 (5), 2791–2807. doi:10.1002/hep.32029
Giuliani, G., Rosina, M., and Reggio, A. (2022). Signaling pathways regulating the fate of fibro/adipogenic progenitors (FAPs) in skeletal muscle regeneration and disease. FEBS J. 289 (21), 6484–6517. doi:10.1111/febs.16080
Gong, K., Zhou, H., Liu, H., Xie, T., Luo, Y., Guo, H., et al. (2021). Identification and integrate analysis of key biomarkers for diagnosis and prognosis of non-small cell lung cancer based on bioinformatics analysis. Technol. Cancer Res. Treat. 20, 153303382110602. doi:10.1177/15330338211060202
Guo, H., Zou, J., Zhou, L., Zhong, M., He, Y., Huang, S., et al. (2020). NUSAP1 promotes gastric cancer tumorigenesis and progression by stabilizing the YAP1 protein. Front. Oncol. 10, 591698. doi:10.3389/fonc.2020.591698
Hasan, M. I., Rahman, M. H., Islam, M. B., Islam, M. Z., Hossain, M. A., and Moni, M. A. (2022). Systems Biology and Bioinformatics approach to Identify blood based signatures molecules and drug targets of patient with COVID-19. Inf. Med. Unlocked 28, 100840. doi:10.1016/j.imu.2021.100840
He, J-Q., and Barron, C. (2020). Signaling pathways in modulation of tissue and organ regeneration in vertebrates. Semin. Cell. Dev. Biol. 100, 1–2. doi:10.1016/j.semcdb.2019.12.001
Ko, S., Russell, J. O., Molina, L. M., and Monga, S. P. (2020). Liver progenitors and adult cell plasticity in hepatic injury and repair: Knowns and unknowns. Annu. Rev. Pathol. 15, 23–50. doi:10.1146/annurev-pathmechdis-012419-032824
Kuwata, K., Shibutani, M., Hayashi, H., Shimamoto, K., Hayashi, S-M., Suzuki, K., et al. (2011). Concomitant apoptosis and regeneration of liver cells as a mechanism of liver-tumor promotion by β-naphthoflavone involving TNFα-signaling due to oxidative cellular stress in rats. Toxicology 283 (1), 8–17. doi:10.1016/j.tox.2011.01.020
Larsen, F. S. (2019). Artificial liver support in acute and acute-on-chronic liver failure. Curr. Opin. Crit. Care 25 (2), 187–191. doi:10.1097/mcc.0000000000000584
Li, J., Wang, Y., Wang, X., and Yang, Q. (2020). CDK1 and CDC20 overexpression in patients with colorectal cancer are associated with poor prognosis: Evidence from integrated bioinformatics analysis. World J. Surg. Oncol. 18 (1), 50. doi:10.1186/s12957-020-01817-8
Li, L., Lei, Q., Zhang, S., Kong, L., and Qin, B. (2017). Screening and identification of key biomarkers in hepatocellular carcinoma: Evidence from bioinformatic analysis. Oncol. Rep. 38 (5), 2607–2618. doi:10.3892/or.2017.5946
Li, Z., Niu, S., Guo, B., Gao, T., Wang, L., Wang, Y., et al. (2020). Stem cell therapy for COVID-19, ARDS and pulmonary fibrosis. Cell. Prolif. 53 (12), e12939. doi:10.1111/cpr.12939
Locatelli, P., Belaich, M. N., López, A. E., Olea, F. D., Uranga Vega, M., Giménez, C. S., et al. (2020). Novel insights into cardiac regeneration based on differential fetal and adult ovine heart transcriptomic analysis. Am. J. Physiol. Heart Circ. Physiol. 318 (4), H994–H1007. doi:10.1152/ajpheart.00610.2019
Melms, J. C., Biermann, J., Huang, H., Wang, Y., Nair, A., Tagore, S., et al. (2021). A molecular single-cell lung atlas of lethal COVID-19. Nature 595 (7865), 114–119. doi:10.1038/s41586-021-03569-1
Michalopoulos, G. K., and Bhushan, B. (2021). Liver regeneration: Biological and pathological mechanisms and implications. Nat. Rev. Gastroenterol. Hepatol. 18 (1), 40–55. doi:10.1038/s41575-020-0342-4
Mulay, A., Konda, B., Garcia, G., Yao, C., Beil, S., Villalba, J. M., et al. (2021). SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell. Rep. 35 (5), 109055. doi:10.1016/j.celrep.2021.109055
Olivo, R., Guarrera, J. V., and Pyrsopoulos, N. T. (2018). Liver transplantation for acute liver failure. Clin. Liver Dis. 22 (2), 409–417. doi:10.1016/j.cld.2018.01.014
Parte, S., Virant-Klun, I., Patankar, M., Batra, S. K., Straughn, A., and Kakar, S. S. (2019). PTTG1: A unique regulator of stem/cancer stem cells in the ovary and ovarian cancer. Stem Cell. Rev. Rep. 15 (6), 866–879. doi:10.1007/s12015-019-09911-5
Ramavath, N. N., Gadipudi, L. L., Provera, A., Gigliotti, L. C., Boggio, E., Bozzola, C., et al. (2021). Inducible T-cell costimulator mediates lymphocyte/macrophage interactions during liver repair. Front. Immunol. 12, 786680. doi:10.3389/fimmu.2021.786680
Rastogi, A., Maiwall, R., Bihari, C., Trehanpati, N., Pamecha, V., and Sarin, S. K. (2014). Two-tier regenerative response in liver failure in humans. Virchows Arch. 464 (5), 565–573. doi:10.1007/s00428-014-1547-0
Russo, F. P., and Parola, M. (2011). Stem and progenitor cells in liver regeneration and repair. Cytotherapy 13 (2), 135–144. doi:10.3109/14653249.2010.545386
Rutherford, A., and Chung, R. T. (2008). Acute liver failure: Mechanisms of hepatocyte injury and regeneration. Semin. Liver Dis. 28 (2), 167–174. doi:10.1055/s-2008-1073116
Sarin, S. K., Choudhury, A., Sharma, M. K., Maiwall, R., Al Mahtab, M., Rahman, S., et al. (2019). Acute-on-chronic liver failure: Consensus recommendations of the asian pacific association for the study of the liver (APASL): An update. Hepatol. Int. 13 (4), 353–390. doi:10.1007/s12072-019-09946-3
Sen, C. K., Ghatak, S., Gnyawali, S. C., Roy, S., and Gordillo, G. M. (2016). Cutaneous imaging technologies in acute burn and chronic wound care. Plast. Reconstr. Surg. 138 (3), 119S–28S. doi:10.1097/prs.0000000000002654
Sharma, A., Ahmad Farouk, I., and Lal, S. K. (2021). COVID-19: A review on the novel coronavirus disease evolution, transmission, detection, control and prevention. Viruses 13 (2), 202. doi:10.3390/v13020202
Shubham, S., Kumar, D., Rooge, S., Maras, J. S., Maheshwari, D., Nautiyal, N., et al. (2019). Cellular and functional loss of liver endothelial cells correlates with poor hepatocyte regeneration in acute-on-chronic liver failure. Hepatol. Int. 13 (6), 777–787. doi:10.1007/s12072-019-09983-y
Song, Y., Li, M., Lei, S., Hao, L., Lv, Q., Liu, M., et al. (2022). Silk sericin patches delivering miRNA-29-enriched extracellular vesicles-decorated myoblasts (SPEED) enhances regeneration and functional repair after severe skeletal muscle injury. Biomaterials 287, 121630. doi:10.1016/j.biomaterials.2022.121630
Tregoning, J. S., Flight, K. E., Higham, S. L., Wang, Z., and Pierce, B. F. (2021). Progress of the COVID-19 vaccine effort: Viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 21 (10), 626–636. doi:10.1038/s41577-021-00592-1
Wang, L., Zhang, J., Wan, L., Zhou, X., Wang, Z., and Wei, W. (2015). Targeting Cdc20 as a novel cancer therapeutic strategy. Pharmacol. Ther. 151, 141–151. doi:10.1016/j.pharmthera.2015.04.002
Wilkinson, H. N., and Hardman, M. J. (2020). Wound healing: Cellular mechanisms and pathological outcomes. Open Biol. 10 (9), 200223. doi:10.1098/rsob.200223
Yan, C., Niu, Y., and Wang, X. (2022). Blood transcriptome analysis revealed the crosstalk between COVID-19 and HIV. Front. Immunol. 13, 1008653. doi:10.3389/fimmu.2022.1008653
Yimlamai, D., Christodoulou, C., Galli, G. G., Yanger, K., Pepe-Mooney, B., Gurung, B., et al. (2014). Hippo pathway activity influences liver cell fate. Cell. 157 (6), 1324–1338. doi:10.1016/j.cell.2014.03.060
Keywords: COVID-19, ALF, liver regeneration, cdc20, Apcin
Citation: Shi J, Li G, Yuan X, Wang Y, Gong M, Li C, Ge X and Lu S (2023) Exploration and verification of COVID-19-related hub genes in liver physiological and pathological regeneration. Front. Bioeng. Biotechnol. 11:1135997. doi: 10.3389/fbioe.2023.1135997
Received: 02 January 2023; Accepted: 13 February 2023;
Published: 23 February 2023.
Edited by:
Yawei Du, Shanghai Jiao Tong University, ChinaReviewed by:
Yuling Han, Cornell University, United StatesTuo Yang, University of California, San Diego, United States
Copyright © 2023 Shi, Li, Yuan, Wang, Gong, Li, Ge and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shichun Lu, bHVzY18zMDFAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Jihang Shi
Jihang Shi Guangya Li4,5†
Guangya Li4,5† Yafei Wang
Yafei Wang Shichun Lu
Shichun Lu