
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bioeng. Biotechnol., 02 March 2023
Sec. Biosensors and Biomolecular Electronics
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1117871
This article is part of the Research TopicNanomaterials for Biosensing and BioassaysView all 5 articles
The recent pandemic has led to the fabrication of new nucleic acid sensors that can detect infinitesimal limits immediately and effectively. Therefore, various techniques have been demonstrated using low-dimensional materials that exhibit ultrahigh detection and accuracy. Numerous detection approaches have been reported, and new methods for impulse sensing are being explored. All ongoing research converges at one unique point, that is, an impetus: the enhanced limit of detection of sensors. There are several reviews on the detection of viruses and other proteins related to disease control point of care; however, to the best of our knowledge, none summarizes the various nucleotide sensors and describes their limits of detection and mechanisms. To understand the far-reaching impact of this discipline, we briefly discussed conventional and nanomaterial-based sensors, and then proposed the feature prospects of these devices. Two types of sensing mechanisms were further divided into their sub-branches: polymerase chain reaction and photospectrometric-based sensors. The nanomaterial-based sensor was further subdivided into optical and electrical sensors. The optical sensors included fluorescence (FL), surface plasmon resonance (SPR), colorimetric, and surface-enhanced Raman scattering (SERS), while electrical sensors included electrochemical luminescence (ECL), microfluidic chip, and field-effect transistor (FET). A synopsis of sensing materials, mechanisms, detection limits, and ranges has been provided. The sensing mechanism and materials used were discussed for each category in terms of length, collectively forming a fusing platform to highlight the ultrahigh detection technique of nucleotide sensors. We discussed potential trends in improving the fabrication of nucleotide nanosensors based on low-dimensional materials. In this area, particular aspects, including sensitivity, detection mechanism, stability, and challenges, were addressed. The optimization of the sensing performance and selection of the best sensor were concluded. Recent trends in the atomic-scale simulation of the development of Deoxyribonucleic acid (DNA) sensors using 2D materials were highlighted. A critical overview of the challenges and opportunities of deoxyribonucleic acid sensors was explored, and progress made in deoxyribonucleic acid detection over the past decade with a family of deoxyribonucleic acid sensors was described. Areas in which further research is needed were included in the future scope.
Deoxyribonucleic acid (DNA) carries the genetic information constituent of deoxyribose and nitrogenous bases known as nucleotides or fragments of DNA (Wilkins, 1956). These nucleotides contain genetic information that can encode life (Stegmann, 2005). As its size is comparable to the nanoscale, the double helix structure was reported in 1953 by Watson and Crick (1953). This well-known DNA double helix is formed from pairs of complementary single-stranded DNA (ssDNA). Double-stranded DNA (dsDNA) is a pair of bonded ssDNA (Watson and Crick, 1953).
Structural nanotechnology has made significant progress in terms of the rapid sensitivity of DNA sequences (Zhou et al., 2020). Each existing DNA is unique, indicating that DNA sequence is crucial for detecting, and it has been studied in the fight against sporadic pandemics. A small part of the DNA that holds genetic information is known as a gene. A complete DNA sequence of DNA is imperative for the fabrication of vaccines. Over the past two decades, many new methodologies have been developed for the detection of DNA, which has ultimately assisted in portable point-of-care diagnostics. Indigenous DNA is negatively charged (Zhang et al., 2007), which can be manipulated by several strategies, such as creating an electric field; thus, it behaves like electrons to attract positively charged particles (Liu and Hu, 2007). Therefore, the identification of specific DNA sequences is a crucial task achieved using modern nanotechnology, which consequently opens a new era of exploration ranging from detection to gene mutation. DNA sensors are capable of detecting changes in the form of electrical signals generated through the immune system in response to any perturbation. They convert biochemical reactions into a signal for further detection. Currently, integrated multiscale simulation and experimental techniques are used to study biosensing applications in a variety of interdisciplinary fields (Quan et al., 2018; Mohammadi et al., 2021; Babar et al., 2022).
Low-dimensional (LD) materials are a class of materials with extraordinary characteristics (Babar et al., 2022). They include graphene and carbon nanotubes (CNT), MXenes (Ti2C3), hexagonal boron nitride (h-BN), molybdenum disulfide (MoS2), and reduced graphene oxide (rGO), etc. (Mohammadi et al., 2021). They have displayed they are being used in a variety of applications. Currently, LD based sensors are replacing traditional sensors (Wu et al., 2022). LD materials are used for protective coating and biosensors due to their tunable, electrical, and optical, and excellent mechanical properties. They are also being used as substrate materials in electronic sensors due to their multilayered structures. Additionally, these materials modify their surface chemistry through associated functional groups due to which they respond to a specific analyte (Bolotsky et al., 2019). Other than normal 2D structures, very new types of nanostructured materials with improved optical properties have been reported (Kim and Benelmekki, 2019). They have diverse industrial applications, particularly suitable for biomolecular sensing. These are known as “smart nanosheets or nanoscrolls” (Kim et al., 2014; Kim and Benelmekki, 2018; Kim and Benelmekki, 2019; Benelmekki and Kim, 2023).
To achieve global market demand, the fabrication of DNA sensors should possess a low cost, low range of detection (LOD), simple and low processing time with high speed, and high selectivity. There are two main types of DNA detection techniques: conventional spectrophotometric and polymerase chain reactions (PCRs), and modern nanomaterial-based sensors, which are further subdivided into optical and electrical sensors. Optical sensors based on fluorescence (FL), surface plasmon resonance (SPR), colorimetric, and surface-enhanced Raman scattering (SERS) techniques, whereas electrical sensors based on electrochemical luminescence (ECL), chip-based, and field-effect transistors (FET) are described in Figure 1.
Detection methods based on electrical sensors have been gaining attention in the global market owing to their high multiplexing capability, high sensitivity, and wide dynamic range. The major contribution of biosensor involves a transduction mechanism for detection. The transducer transforms the electrical signal from the analyte and amplify it. Conventional sensors have low accuracy and expensive and complex instrumentation whereas nanomaterial-based sensors are more compatible and provide a proficient detection. However, the optical method is commonly used because it has numerous defects, such as it is difficult to reliably profile low-abundance genes, and it requires expensive fabrication of these optical devices and complex bioinformatics tools for fluorescence signal identification. FET-based nano electronic devices have attracted much attention owing to their small scale, simple design, and better performance compared with conventional devices.
Because of the discovery of DNA sensors, efforts have been made to demonstrate and optimize them for portable point-of-care applications. The continuous demand and advancement of nanotechnology has been pointed out by experts, and the DNA sensor market is predicted to reach up to 28 billion dollars. This set an annual growth rate of 8.4% in 2022, as illustrated in Figure 2. In particular, Figure 2 represents data rooted from Scopus using search title as “DNA/RNA/Nucleic acid sensor” and immunosensor/antibody sensor, and “Enzyme’’ sensor (Mujawar et al., 2020).
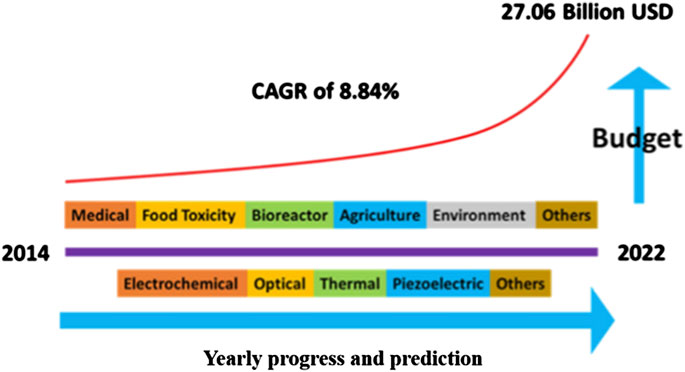
FIGURE 2. Representation of biosensors in the US industry and annual budget expenses (Mujawar et al., 2020).
A sensor is a combination of receptor-transducer, which converts the biochemical response to signals emitted from the source. Moreover, DNA sensors monitor involving biomolecular processes. A biosensor consists of an analyte, bioreceptor, transducer, electronics, and display setup (Naresh and Lee, 2021).
The interaction of light with matter, known as spectrometry, is an ancient technique that has been used in the fields of chemistry and biomedicine because of its low cost, simplicity, and convenience. This has led to the analytical detection of DNA derivatives, such as nucleic acids and nucleotides, owing to the presence of double bond systems as a consequence of these bonds responding to ultraviolet light in the spectrum. This type of specific absorbance will be helpful for the quantitative measurement of DNA by spectrophotometry in the form of micrograms/mL. The absorbance phenomenon corresponds to the transition of electrons from either the anti-bonding (π*) state to the non-bonding (n) state or from the anti-bonding (π*) state to the bonding state (π) associated with a specific energy (Mujawar et al., 2020). Infect nucleic acids have maximum absorbance of UV light at a wavelength of 260 nm; thus, a solution of DNA is exposed to quantify the exposed sample. The drawback of this methodology is that there is a considerably weak signal for single-stranded nucleic acids owing to the lack of bonding, anti-bonding, and non-bonding states. Furthermore, this technique cannot describe the detailed sequences of nucleic acids. As nucleic acids absorb UV light at 260 nm, the compound containing them absorbs UV at 280 nm. The purity of the sample can be calculated by the ratio of absorbance measurement following the criteria for being pure, i.e.,
Polymerase chain reaction (PCR) is an in vitro technique that can detect and amplify small DNA segments. Through this technique, millions of copies are developed in a fraction of the time. This method was proposed in early 1980 (Cantor, 1998), and has been used globally in various fields, such as food quality insurance, genetics, molecular biology, and forensic science. It is a well-established amplification technique during which millions of amplitudes are generated from a small segment of DNA in a short interval of time. Although it is a low-cost and universally used method, constraints, such as indirect melting temperature measurements and premier annealing, hinder their applications. Furthermore, specific amplification will only occur within a narrow range of reaction conditions that ultimately cause the failure of the PCR testing methodology.
For the sample preparation, errors in the volume of reagents significantly alter the result of the polymerase chain reactions cycle that may disrupt the melting temperature, premier annealing, and amplification. All of the building blocks of PCR hinder portability, as these types of measurements cannot be performed outside the laboratory in a well-controlled environment. Signal transduction mechanisms are another significant barrier that prevents PCR from becoming a popular portable device without the use of expensive instruments, such as PCR machines, and complicated processes, including electrophoresis. All these challenges limit the use of PCR as a portable device. Therefore, a simple modification was introduced using a colorimetric assay for the reserved transcription of DNA fragments known as real-time PCR (rt-PCR). RT-PCR was first described in 1990, and a schematic description is shown in Figure 3, which displays a comparison of the different PCR techniques. Conventional PCR, also known as end-point PCR for analysis of DNA amplification, was conducted at the end of fluorescent marking, whereas amplified DNA was analyzed during each cycle, also known as real-time PCR (Wang et al., 2018b). In general PCR, the fragmentation of DNA is pulled through the gel matrix using a centrifugal electric field that separates DNA segments. This process is called electrophoresis. The second PCR test known as qPCR is quantitative PCR, and it is a much more dynamic range of analysis than that of conventional PCR. It is also a modified form of PCR to qualitatively analyze the DNA by the introduction of florescent dyes during PCR cycles. The output of rPCR is typically displayed in the form of sinusoidal followed by a plateau. Digital PCR (dPCR) follows the random distribution of particles over numerous partitions. Each partition acts as an individual PCR through fluorescence detection. Poisson’s statistics were applied to the sample partitions to calculate the concentration of the target sequence from the proportion of the amplified positive concentration. The process constitutes independent partitioning and amplification followed by florescence detection.
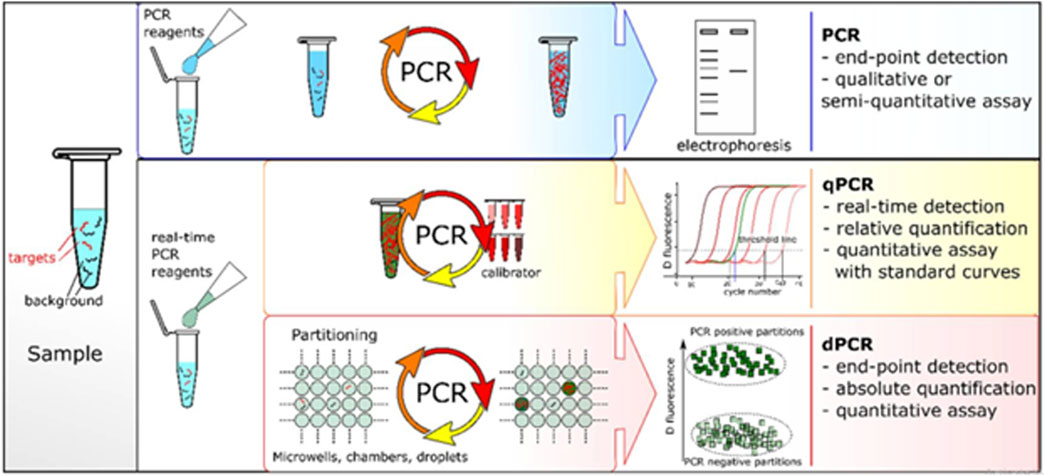
FIGURE 3. Comparison of different PCR-based techniques (Quan et al., 2018).
Nanomaterial-based sensors are considered efficient sensors with excellent signal absorption strength due to quantum effects. They have a high surface-to-volume ratio and higher optical and magnetic properties which make them reasonably good for sensing all types of analytes (Santhanam et al., 2020). Numerous NMBs have been used for DNA biocompatibility because of their higher signal detection capability and transduction technology which converts signals from an analyte during its biochemical reaction (Hlongwane et al., 2019). DNA sensing using NMBs has been included in the following sections.
These biosensors measure changes in optical properties, such as resonance, reflectance, absorbance, and luminescence from the sensor surface. This system measures the fluorescence from nanomaterials based on the detection of DNA hybridization (Vikrant et al., 2019). The optical sensors have been attracted due to their efficient detection level. The materials used in these detectors effectively amplify the detection signal. We summarized a few optical techniques used for developing nanomaterial-based DNA biosensors.
In such sensors, a fluorescent nanomaterial, known as the transducer, is conjugated with a DNA molecule acting as a target molecule. A fluorescence spectrophotometer was used to measure the fluorescence emitted from the nanomaterial conjugated with the target DNA, which is known as fluorescence resonance energy transfer (FRET) (Ploetz et al., 2016). In this technique, excited electrons were transferred from DNA to nanomaterials at resonance separated by a nanoscale distance without emission of a photon; this process is called fluorescence quenching (FQ). In the presence of the probe and target DNA, both combine to form a hybrid that leads to a change in the fluorescence intensity of the nanomaterials, similar to the aptamer/DNA sensor. Figure 4 shows a scheme of target detection based on FRET with reduced graphene quantum dots (GQDs) combined with probe (complementary) DNA (single-stranded). A spectrophotometer was used to measure the resonance intensity at the graphene monolayer surface. The FRET signal emerging from the layer was analyzed for the DNA structures. Initially, inorganic GQDs reaction with NaBH4 (sodium borohydride) results in reduced GQDs called (rGQDs) followed by connecting DNA (cDNA). This reaction is further divided into three steps. In the first step single strand DNA (ssDNA) interact with GQDs via a condensation reaction. Later, this interaction is absorbed through electrostatic stacking making a base pair of ssDNA-rGQDs/GO. Lastly, the base pair interact with the target DNA (tDNA). This tDNA is replaced by a single-based mismatched DNA (mDNA) making a double-strand DNA (dsDNA)-rGQDs compound detached to produce florescence recovery (Qian et al., 2014).
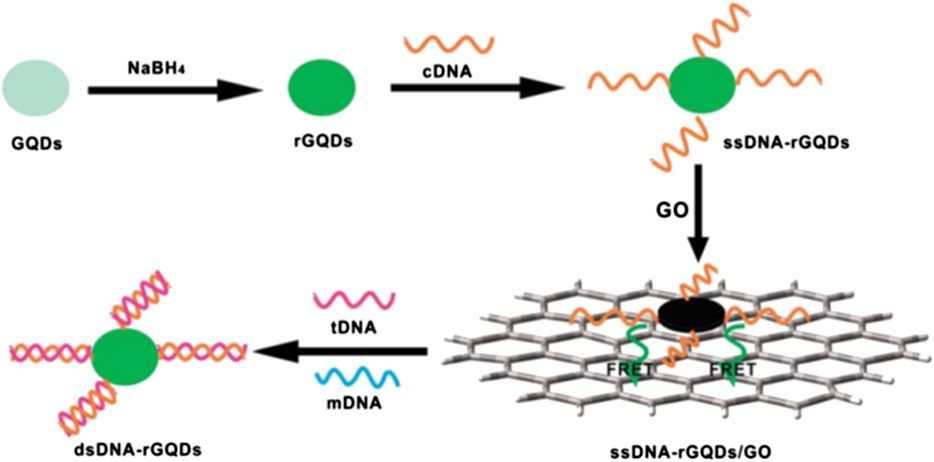
FIGURE 4. Schematic of a universal fiuorescence based sensor mechanism for the detection of DNA -FRET between GQDs and graphene oxide (GO) (Qian et al., 2014).
An overview of the variety of recently reported FRET-based DNA detection techniques is listed in Table 1. This table shows the complete information reading type of materials used, techniques and strategies of DNA detection, limit of detection, and range of analyte concentration. Herein, we can conclude that by using a tetrahedral DNA framework, we can achieve a DNA detection limit of 1 fM (Li et al., 2020b) while using Cu (I)-catalyzed alkyne-azide cycloaddition (CuAAC) as a fluorescent nanomaterial can achieve a detection range of 0.22 fM reported by Zheng et al. (2020). Furthermore, Zhang D et al. (2020) reported a 23.8 aM level detection limit using the composition of magnetic beads (MBs) that contain phosphate- Zr4+-carboxylate/Cu(II) Br/EDTA for lung cancer DNA. Another finding of magnetic nanoparticle-based on poly-enzyme nanobeads fluorescence assay show high detection of 1.6 aM, which is the highest ever reported by FRET techniques (Lapitan et al., 2019).
It is one of the most common optical sensing techniques which is based on surface plasmon which are electromagnetic waves originating from the metal interface. In this sensor, the incident light stimulates the resonance of the conduction electrons at the interface of the positive and negative permittivity materials. AuNPs with positive charge conjugate with the target DNA that is negatively charged; light stimulus resonance phenomena occur between them using conduction electrons, and there is a change in dielectric constants that generate surface plasmons. The schematic diagram shows the detection method by SPR, where the prism, transducer surface composed of nanomaterials, plane polarized light as a stimulus, and detector are the main components of the SPR system. During the conversion of the association phase to the disassociation phase, the refractive index changes, which ultimately deviate from the exiting light from the prism, are correlated with the concentration of the analyte, as shown in Figure 5. Specifically, Figure 5 represents the experimental illustration of SPR techniques (A) and a variation in the critical angle as a function of the intensity (B) and the response of the sensor during the experiment. This method offers label-free techniques; however, there is variation in the refractive index owing to changes in the transducer surface temperature and composition, which may significantly alter the detection results (Patching, 2014).
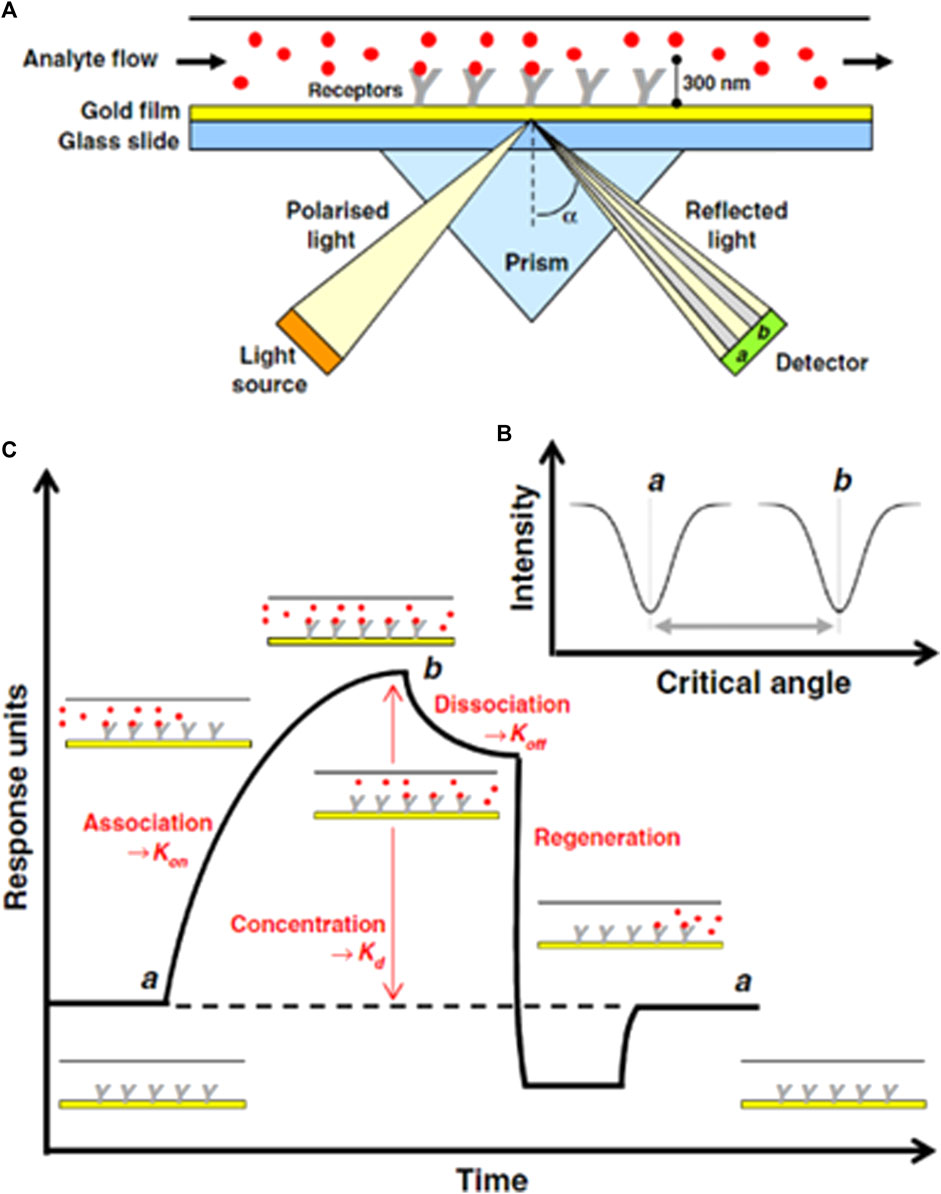
FIGURE 5. (A) Schematic of SPR-based DNA sensing technique for measuring the binding of an analyte molecule (B) varation in critical angle as fuction of intensity of incident light, (C) Evolution of sonogram's response during SPR experiment (Patching, 2014).
The SPR detection performs the analysis of biomolecular interaction ranging from organic compounds to proteins and nucleic acids and viruses based on real-time. The detection is non-invasive. i.e., it can analyze transparent or colored samples effectively. They are a label-free, specific, and sensitive method that is dependent on the changes in the refractive index of the material surface. A small change in the refractive index may induce a false signal during the detection of an analyte. Nanomaterials are the most ideal material for their fabrication and better signal detection and amplification. Most importantly they are available commercially (Deng et al., 2017; Das et al., 2021).
An overview of the various SPR-based DNA detection techniques reported recently is displayed in Table 2. We have listed all possible information, for example, the type of materials, techniques, and strategy of DNA detection used, limit of detection, and range of analyte concentration. Therefore, by employing a sandwich-like assay based on the selective capture of specific DNA targets, subsequent signal amplification can be obtained by a secondary DNA probe linked to Au nanostars with a detection limit of 3 fM to 6.9 aM (Mariani et al., 2015). Another study reported that a highly sensitive polarization control-modulated plasmonic biosensor based on monolayer graphene with Au film
In this technique, color tags or enzymes are used for the detection of DNA compatible with the substrate. Alternatively, nucleic acids functionalized with nanomaterial-based assays can be used for the detection and quantification of DNA (Krishnan and Syed, 2022). During colorimetric analysis, colloidal solution gold nanoparticles (GNPs) exhibit different colors based on their distance from red to blue. The detection is done through a change in the wavelength of the electrolyzed DNA. The color of GNPs is dependent on their dispersion and this help researchers to visually investigate assays that change owing to a decrease in the average distance between nanomaterials (Li and Rothberg, 2004). This method is straightforward, low-cost, and easy to perform for rapid onsite diagnostics. They have a poor limit of detection and prototype design. This DNA detection equipment is commercially available and more economical than other clinical diagnostics. Figure 6 displays schematic illustration of calorimeter based on the GNPs dispersion and aggregation (Liu et al., 2013). A series of DNA samples were electrolyzed and had different concentrations up to 6Nm. The color of the GNPs is gradually changing from pale to blue with the addition of concentration. The wavelength of absorption spectrum is in the range of 550–750 nm. This method is designed to detect a DNA sequence.
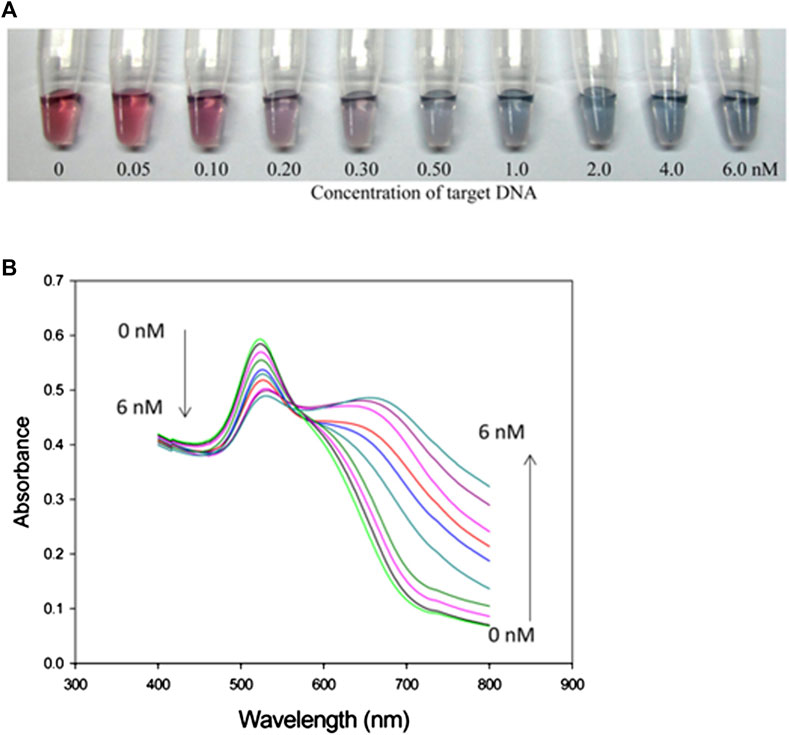
FIGURE 6. (A) Calorimetric response of concentration of DNA (0-6 nM), (B) absorption spectrum as a function of wavelength of electrolyzed DNA (Liu et al.,2013).
An overview of the various colorimetric DNA detection techniques recently reported is listed in Table 3. We summarized the types of materials used, techniques and strategies of DNA detection, limit of detection, and range of analyte concentration. Therefore, by employing electrophoretic streptavidin-coated MBs assisted with a magnetic field, we could introduce a new method, which uses active hybridization with a detection limit of 0.1 fM (Tian et al., 2019). Another report showed that Fe3O4 nanosheets in DNA/Fe3O4 networks display peroxidase-like catalytic activity, thereby enhancing detection to an extreme limit of 13 aM, which is the highest limit that has been reported for colorimetric techniques (Tang et al., 2019).
In this technique, molecular vibrations which arise directly from analyte molecules were measured using Raman spectroscopy. Such types of resonance occur only when the target analyte is a nanostructure or a roughed metal surface. SERS sensing is based on the conjugation of nanomaterials and bioreceptor molecules (oligonucleotides) at the surface of a dye known as a Raman reporter or tag that enhances Raman signals during the detection of target DNA. SERS provides an enhanced Raman signal of 106–1014 order of magnitude owing to the electromagnetic interaction between the metal.
Nanostructures and the analyte. SERS-based biosensors have a comparatively low cost, high sensitivity, rapid results, and portability (Vikrant et al., 2019). Figure 7 shows the schematic setup for the detection of DNA based on AgNPs at the Si substrate as a sandwich-type sensing setup. Nanoparticles are functionalized with thiolated DNA as step 1, followed by conjugation of dye named Rhodamine as step 2. Therefore, a sandwich-like structure surrounded the target DNA by thiolated DNA and reported DNA that ultimately caused the formation of capture/target/reporter DNA ready for SERS detection as step 3, as shown in Figure 7. The optical sensors are summarized in Figure 8. Each optical sensor has been summarized in the flow chart.
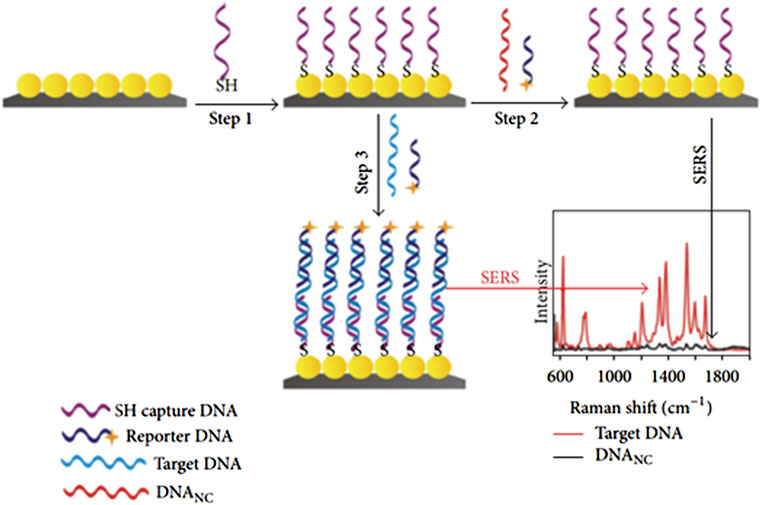
FIGURE 7. Schematic of the fabrication route for SERS sensor and variation of Raman shift as a function of intensity (Jiang et al., 2012).
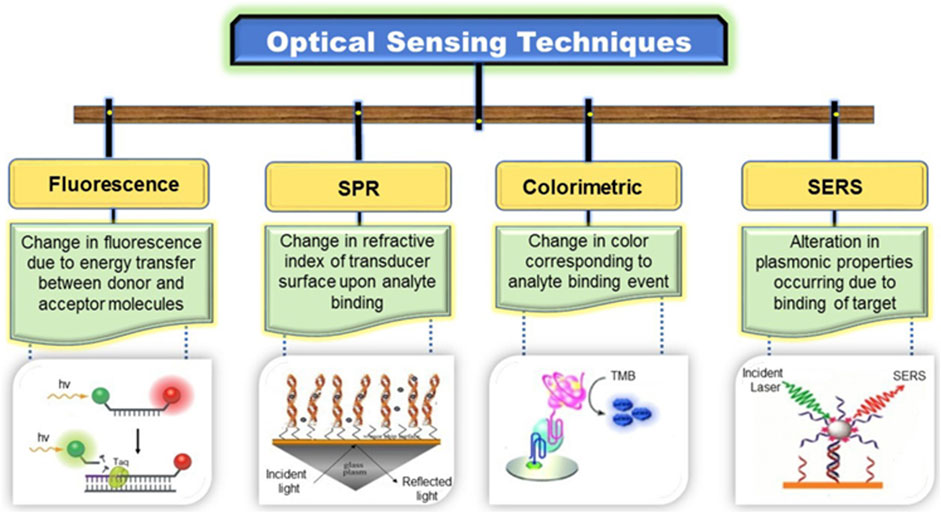
FIGURE 8. Division of optical sensors (Vikrant et al., 2019).
An overview of the various SERS-based DNA detection techniques reported recently is listed in Table 4. It contains the complete information type of materials used, techniques, a strategy of DNA detection, limit of detection, and range of analyte concentration. Thus, a detection limit of 10 uM–10 fM can be achieved by AuNPs on the surface of graphene oxide (GO) linking via hybridization (Khalil et al., 2019). He et al. (2019) reported a novel detection ratiometric sensor based on glucose oxidase (GOx) on Au and Si nanoflower substrates that enhances the detection to an extreme limit of 7.75 aM, which is the highest limit for colorimetric techniques. Table 4 lists earlier studies on SERS-based DNA sensors.
In this technique, chemical luminescence and electrochemical processes are combined and named as electro-chemiluminescence (ECL), which results in the emission of light. This photo emission occurs owing to the excitation and de-excitation of electrons between the ground and excited states stimulated by the electrochemical reaction in solution. Light is emitted because of the transfer of exergonic electrons at the electrode surface. This phenomenon is also known as electrogenerated chemical luminescence. The wavelength of the light emitted from the excited and relaxed states corresponds to the energy gap of the molecules. ssDNA is commonly used as a bioreceptor and printed screen, and pencil graphite glassy carbon, and gold are used as the working electrodes in the assembly of DNA sensors. Figure 9 shows the simple strategy and mechanism of detection. This design comprises a target DNA fragment that hybridizes with MBs, where streptadivine and biotin are used as linkers, and the ruthenium probe is conjugated at the other end (Figure 9A). This magnetic bead-assisted ECL reaction occurs in the presence of tripropylamine, where the ruthenium probe Ru (bpy)32+ amplifies the signal isothermally (Zhou et al., 2014).
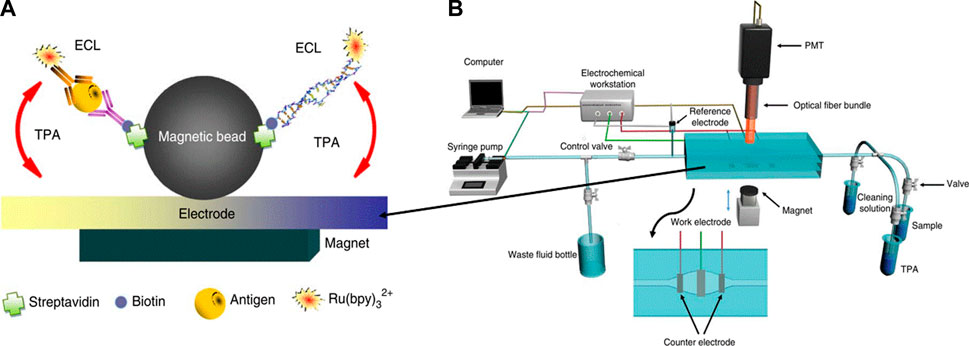
FIGURE 9. (A) Schematic illustration of magnetic based (MB) ECL for hepatitis B antigen model and nucleic acid target ,whereas antibody of nucleic acids are labeled with biotin and Ru(bpy), (B) MB based ECL measurement and detection system (Zhou et al. 2014).
Table 5 presents an overview of the various ECL-based DNA detection techniques reported in the last decade. Herein, we summarize the detection limit of 19.05 aM for biosensor based on the in situ generation of Cu nanoclusters as luminophores and TiO2 as a coreaction accelerator (Liao et al., 2018). Zhang et al. (2020e) constructed MXene (Ti3C2Tx)-based impedimetric aptasensing nanosheets and iron phthalocyanine quantum dots for an enhanced high detection sensitivity of up to 4.3 aM as compared to the individual component-based ECL technique.
Lab-on-a-chip (LOC) is a mini-integrated chip with small sensors in an array with an area of a few square centimeters. They are based on the micro-electrical-mechanical-based technology (MEMS) in the form of an integrated chip. These sensors are composed of a network of microfluidic channels in which the analyte can be manipulated at the microscale level. The compact design of microfluidic chips results in rapid heating and mixing; hence, providing ultrahigh sensitivity and portability for direct analysis of a crime scene (Temiz et al., 2015; Bruijns et al., 2016). This will also decrease the amount of analyte as well as reduces cross-contamination in the sealed environment. These systems are designed for single use, which will benefit from the chain of custody and contamination risk. A bandage-like flexible sensor that amplifies the DNA detection signal using microfluidic technology is shown in Figure 10. These sensors can be powered by body heat; thus, they are highly sensitive and portable for on-site detection (Bruijns et al., 2016). They are widely used for DNA amplification process and accelerate PCR process. They are small in dimension and instant detection and cost-effective instrument.
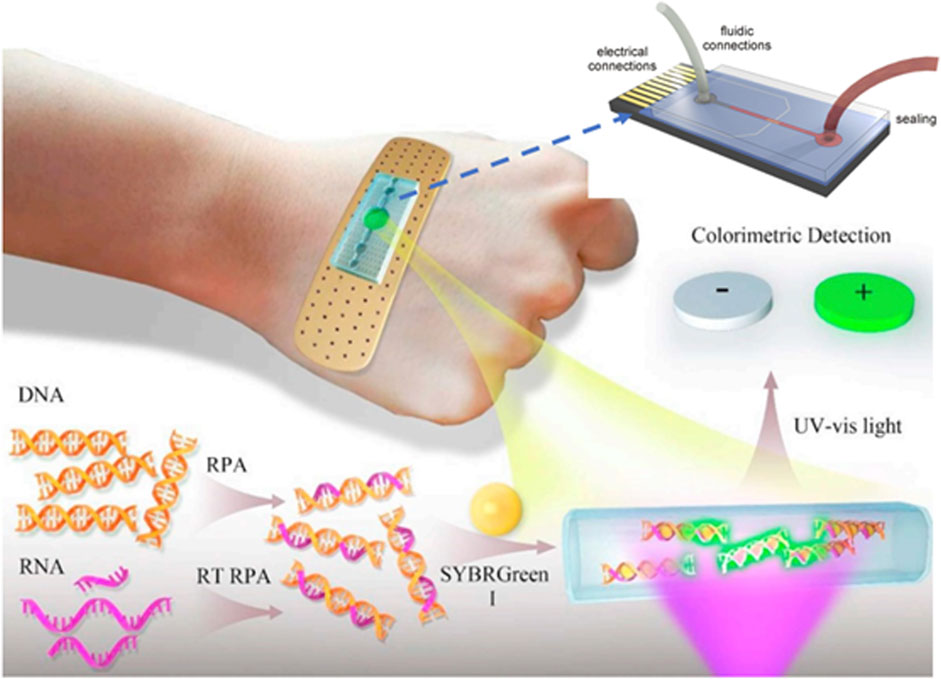
FIGURE 10. Schematic of the wearable microfluidic sensor for nucleic acids (Temiz et al., 2015; Yang et al., 2019).
An overview of the various microfluidic chip-based DNA detection techniques reported earlier is listed in Table 6. We can observe that biosensors based on microfluidic chips are designed as power-free chips. They follow the amplification of fluorescence signals after hybridization with laminar flow-assisted dendritic cells and have a detection limit of 0.045–0.45 pM (Kim et al., 2019). Microfluidic-based nucleic acid amplification tests as noise resistant quantitative PCR are used for rapid detection of ultralow-abundance DNA in real biofluids. They have the highest diagnostic limits of 0.05 aM to be reported for a microfluidic technique (Ye and De, 2017).
FET biosensors are highly sensitive detectors based on 2D materials. They have high carrier mobility owing to their nanoscale dimensions and high volume-to-charge ratio. MXene-based biosensors are a combination of metal carbides and nitrides that have attracted attention because of their unique characteristics (Babar et al., 2022; Guo et al., 2011). They are used for analyte detection and biosensing applications (Yadav et al., 2021). A combination of MXene-graphene-based FET has been used to detect influenza and 2019-Ncov (Li et al., 2021). A multiscale computer simulation method and experimental approach were employed to investigate the characteristics of flat and crumpled graphene-based biosensors by Hwang et al. (2020) ,who found that the detection limits of buffer and human samples are 600 aM and 20 aM, respectively. Furthermore, the atomic-scale simulation results revealed that the deformation mechanism resulted in electrical hotspots in the channel. This technique can be used to develop reliable fast-track biosensors for medical applications. Figure 11 displays a cross-sectional representation of flat and crumpled graphene FET sensors over a Debye length, represented by the blue curve (Figure 11A), whereas Figure 11B shows the fabrication route. Debye screening is weaker in crumpled graphene, which makes it more sensitive for detecting DNA (Hwang et al., 2020). The insets show the distribution of energy over the K-space.
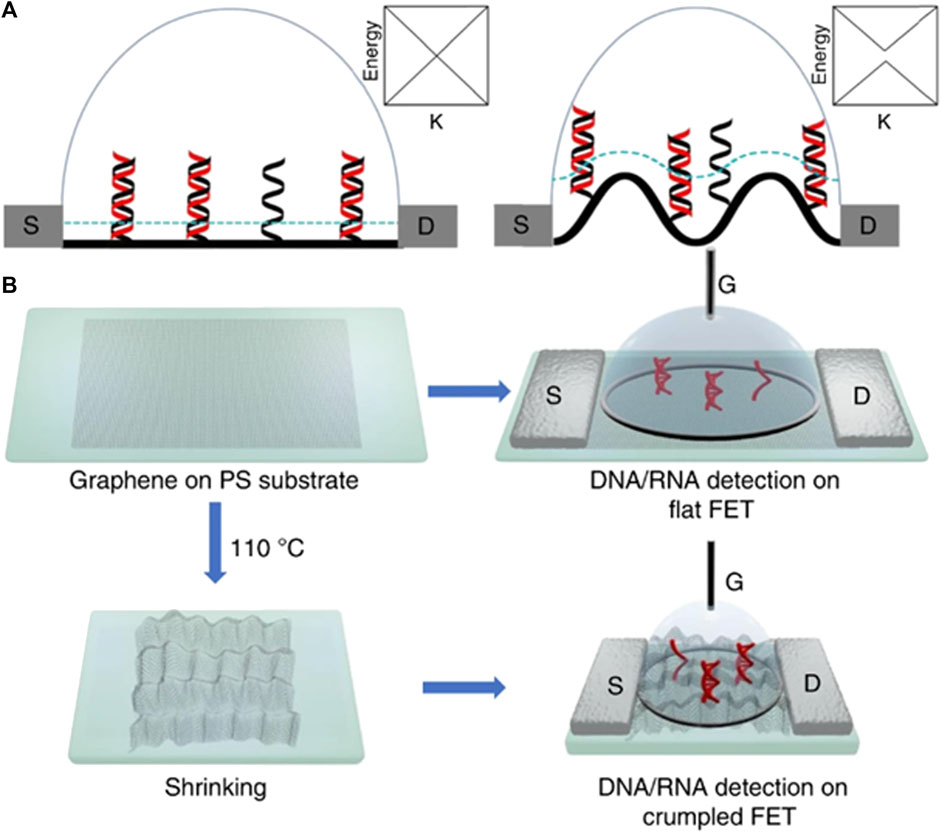
FIGURE 11. (A) Scheme of flat and crumpled graphene FET biosensor using single and double DNA strands over graphene surface. (B) Experimental route for fabrication and flow proces.
A computational study on single-layer MXene that has potential applications as a DNA detection material with high sensitivity and effectiveness was conducted (Yadav et al., 2021). Computer simulation methods have frequently been used to understand the mechanisms and interactions of materials at the atomic scale (Mustafa et al., 2017; Azeem et al., 2018a; Azeem et al., 2018b; Azeem et al., 2019b; Azeem et al., 2019a; Azeem et al., 2020; Azeem et al., 2021; Mustafa Azeem et al., 2018; Mustafa Azeem et al., 2019). An overview of the various FET-based DNA detection techniques reported earlier is listed in Table 7. It is important to point here that LD material has displayed clear advantage on other materials used for designing sensors for health application because of their tunable band structure and ultrathin nature. They also display an improved detection sensitivity. LD materials have been emerged as promising candidate for health industry. Figure 12 summarize LD materials and their application in sensing applications. The division is based on the synthesis, preparation methods, electrochemical and optical properties. The fabrication process of these materials involves process of exfoliation that depends on the chemical environment. Moreover, intercalation constitute chemical vapor deposition (CVD) and electrochemical exfoliation. The material based on LD sensors are used in theranostics image guide application and diagnostics (Bolotsky et al., 2019).
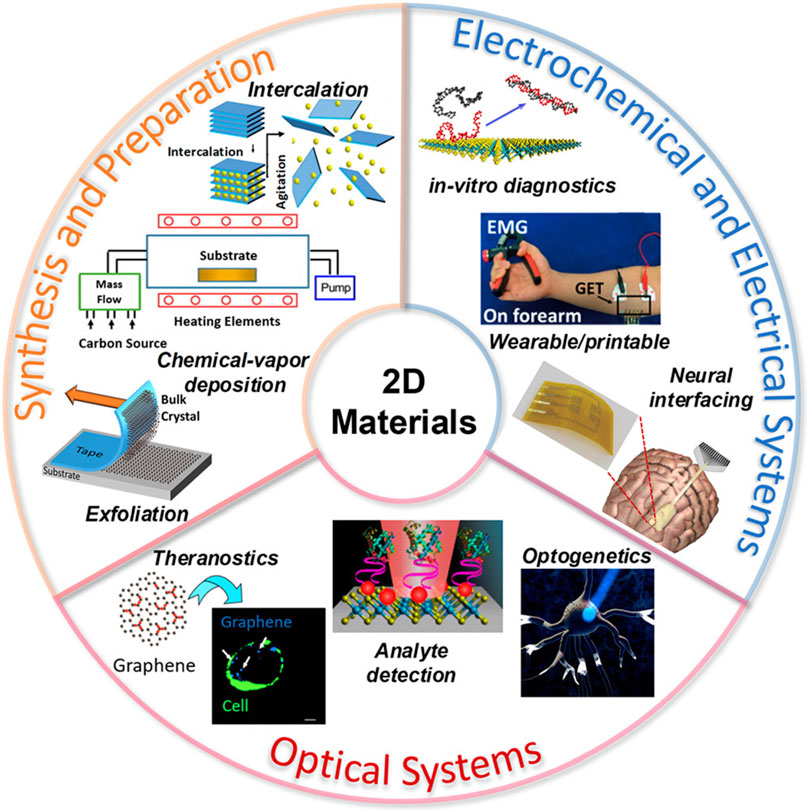
FIGURE 12. Preparation and synthesis of LD materials for optical and electrochemical applications (Bolotsky et al., 2019).
Sensors play an important role in our daily life specifically in the healthcare sector as DNA detection and its diagnostic involve curing diseases. The current trend and developing sensors are now hot topics. They have increasing demand after the nCov-19. A big challenge involving the development of DNA detection sensors requires components with on-site and rapid detection and detection limit. Before commercialization, it is very important to handle the stability and reproducibility of the samples. Another major challenge concerns the storage as handling the short lifetime of the samples and re-usage. The conventional detection methods are based on PCR techniques which are very expensive and time-consuming. It is one of the big challenges that involve the detection of ultra-low concentrations of any analyte. Another challenge involves the response time for detection. Developing accurate and economical DNA sensors is a present concern. Presently, DNA detection with nanopore technology has resolved this issue and these materials have already displayed unique characteristics. Compared to conventional methods 2D materials-based FET sensors involve the electric field to create a charge to interact with DNA and change in current results in the form of a signal to detect.
Infectious diseases have posed a challenge in the past few years. Biosensing and biotechnology are emerging fields. In particular, DNA biosensors have potential research applications because of their chemical properties as well as reliable and fast detection.
We have reviewed different types of biosensors and listed the fabrication techniques and materials used to develop them. Conventional methods for developing in vitro diagnostics are time-consuming and require multiple trials and centralized technologies. There are diverse techniques and strategies for developing sensors, including, but not limited to, collecting samples, and implementing integrated diagnostics for biological applications. LD materials are considered as alternate to traditional materials due to their nanosized thickness and compatible nature. Currently, understanding nature of these materials in biological environment is major challenge. Critical challenges exist in transforming the optimal clinical treatment of infectious diseases from trial to translational research.
MAz prepared the initial draft and collected the literature review, and MS collected most of the information. MZ, MAa, BS, and MWA reviewed and proofread the manuscript. All the authors have approved this manuscript.
This work is supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia, under the Reviewing Researcher Track (Grant No. 2389).
Authors would like to thank King Faisal University for supporting this research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdulwahab, M. H. M., Moser, N., Rodriguez-Manzano, J., and Georgiou, P. (2018). A CMOS bio-chip combining pH sensing, temperature regulation and electric field generation for DNA detection and manipulation. IEEE Int. Symp. Circuits Syst., 1–5.
Amini, B., Kamali, M., Salouti, M., and Yaghmaei, P. (2018). Spectrophotometric, colorimetric and visually detection of Pseudomonas aeruginosa ETA gene based gold nanoparticles DNA probe and endonuclease enzyme. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 199, 421–429. doi:10.1016/j.saa.2018.03.056
Amini, H., Ban, Ž., Ferger, M., Lorenzen, S., Rauch, F., Friedrich, A., et al. (2020). Tetracationic bis-triarylborane 1,3-butadiyne as a combined fluorimetric and Raman probe for simultaneous and selective sensing of various DNA, RNA, and proteins. Chemistry 26, 6017–6028. doi:10.1002/chem.201905328
Ayoib, A., Hashim, U., Gopinath, S. C. B., Thivina, V., and Arshad, M. K. M. (2020). Design and fabrication of PDMS microfluidics device for rapid and label-free DNA detection. Appl. Phys. A Mat. Sci. Process. 126, 193–198. doi:10.1007/s00339-020-3337-7
Azab, M. Y., Hameed, M. F. O., Nasr, A. M., and Obayya, S. S. A. (2018). Label free detection for DNA hybridization using surface plasmon photonic crystal fiber biosensor. Opt. Quantum Electron. 50, 68–13. doi:10.1007/s11082-017-1302-2
Azeem, M. M., Li, Z., Wang, Q., Amjad, Q. M. N., Zubair, M., and Ahmed, O. M. H. (2018a). “Classical molecular dynamics study for defect sink behavior in oxide dispersed strengthened alloys,” in Proceedings of 2018 15th International Bhurban Conference on Applied Sciences and Technology (Islamabad, Pakistan: IBCAST), 12–15. doi:10.1109/IBCAST.2018.8312177
Azeem, M. M., Li, Z., Wang, Q., and Hussian, A. (2018b). Molecular dynamics simulation study on the possible factors affecting stability of ODS steel. IOP Conf. Ser. Mater. Sci. Eng. 389, 012003. doi:10.1088/1757-899X/389/1/012003
Azeem, M. M., Wang, Q., Li, Z., and Zhang, Y. (2020). Dislocation-oxide interaction in Y2O3 embedded Fe: A molecular dynamics simulation study. Nucl. Eng. Technol. 52, 337–343. doi:10.1016/j.net.2019.07.011
Azeem, M. M., Wang, Q., Zhang, Y., Liu, S., and Zubair, M. (2019a). Effect of grain boundary on diffusion of P in alpha-Fe: A molecular dynamics study. Front. Phys. 7, 1–7. doi:10.3389/fphy.2019.00097
Azeem, M. M., Wang, Q., and Zubair, M. (2019b). Atomistic simulations of nanoindentation response of irradiation defects in iron. Sains Malays. 48, 2029–2039. doi:10.17576/jsm-2019-4809-24
Azeem, M. M., Yun, D., and Zubair, M. (2021). Atomic insights on interaction mechanism of dislocation with void/impurity/precipitates in bcc iron. Int. Conf. Nucl. Eng. Proc. ICONE 2, 1–7. doi:10.1115/ICONE28-65197
Babar, Z. U. D., Della Ventura, B., Velotta, R., and Iannotti, V. (2022). Advances and emerging challenges in MXenes and their nanocomposites for biosensing applications. RSC Adv. 12, 19590–19610. doi:10.1039/d2ra02985e
Bakshi, S. F., Guz, N., Zakharchenko, A., Deng, H., Tumanov, A. V., Woodworth, C. D., et al. (2017). Magnetic field-activated sensing of mRNA in living cells. J. Am. Chem. Soc. 139, 12117–12120. doi:10.1021/jacs.7b06022
Bakshi, S., Zakharchenko, A., Minko, S., Kolpashchikov, D. M., and Katz, E. (2019). Towards nanomaterials for cancer theranostics: A system of dna-modified magnetic nanoparticles for detection and suppression of rna marker in cancer cells. Magnetochemistry 5, 24. doi:10.3390/magnetochemistry5020024
Balaji, A., Yang, S., Wang, J., and Zhang, J. (2019). Graphene oxide-based nanostructured DNA sensor. Biosensors 9, 74. doi:10.3390/bios9020074
Banerjee, I., Aralaguppe, S. G., Lapins, N., Zhang, W., Kazemzadeh, A., Sönnerborg, A., et al. (2019). Microfluidic centrifugation assisted precipitation based DNA quantification. Lab. Chip 19, 1657–1664. doi:10.1039/c9lc00196d
Bechnak, L., El Kurdi, R., and Patra, D. (2020). Fluorescence sensing of nucleic acid by curcumin encapsulated poly(ethylene oxide)-block-poly(propylene oxide)-block-poly(ethylene oxide) based nanocapsules. J. Fluoresc. 30, 547–556. doi:10.1007/s10895-020-02528-9
Benelmekki, M., and Kim, J.-H. (2023). Stimulus-responsive ultrathin films for bioapplications: A concise review. Molecules 28, 1020. doi:10.3390/molecules28031020
Bi, Q., Gan, X., Yuan, R., and Xiang, Y. (2020). Copper-free click chemistry-mediated cyclic ligation amplification for highly sensitive and non-label electrochemical detection of gene mutation. J. Electrochem. Soc. 167, 027535. doi:10.1149/1945-7111/ab6a81
Bolotsky, A., Butler, D., Dong, C., Gerace, K., Glavin, N. R., Muratore, C., et al. (2019). Two-dimensional materials in biosensing and healthcare: From in vitro diagnostics to optogenetics and beyond. ACS Nano 13, 9781–9810. doi:10.1021/acsnano.9b03632
Bruijns, B., van Asten, A., Tiggelaar, R., and Gardeniers, H. (2016). Microfluidic devices for forensic DNA analysis: A review. Biosensors 6, 41–35. doi:10.3390/bios6030041
Campos, R., Borme, J., Guerreiro, J. R., Machado, G. J., Cerqueira, M. F., Petrovykh, D. Y., et al. (2019). Attomolar label-free detection of DNA hybridization with electrolyte-gated graphene field-effect transistors. ACS Sensors 4, 286–293. doi:10.1021/acssensors.8b00344
Cantor, D. (1998). Crafting science: A sociohistory of the quest for the genetics of cancer. Bull. Hist. Med. 72, 590–592. doi:10.1353/bhm.1998.0127
Cao, H., Zhou, X., and Zeng, Y. (2019). Microfluidic exponential rolling circle amplification for sensitive microRNA detection directly from biological samples. Sens. Actuators. B. Chem. 279, 447–457. doi:10.1016/j.snb.2018.09.121
Cao, X., Zhang, K., Yan, W., Xia, Z., He, S., Xu, X., et al. (2020). Calcium ion assisted fluorescence determination of microRNA-167 using carbon dots-labeled probe DNA and polydopamine-coated Fe(3)O(4) nanoparticles. Mikrochim. Acta 187, 212. doi:10.1007/s00604-020-4209-8
Chen, S., Sun, Y., Xia, Y., Lv, K., Man, B., and Yang, C. (2020). Donor effect dominated molybdenum disulfide/graphene nanostructure-based field-effect transistor for ultrasensitive DNA detection. Biosens. Bioelectron. 156, 1–7. doi:10.1016/j.bios.2020.112128
Chen, Y., Tai, T., Hsu, C., Sarangadharan, I., Kumar, A., Wang, H., et al. (2018). Sensors and Actuators B: Chemical Direct detection of DNA using electrical double layer gated high electron mobility transistor in high ionic strength solution with high sensitivity and speci fi city. Sensors Actuators B. Chem. 271, 110–117. doi:10.1016/j.snb.2018.05.119
Chen, Z., Peng, Y., Xie, X., Feng, Y., Li, T., Li, S., et al. (2019). Dendrimer-functionalized superparamagnetic nanobeacons for real-time detection and depletion of HSP90α mRNA and MR imaging. Theranostics 9, 5784–5796. doi:10.7150/thno.36545
Cheung, L. S., Wei, X., Martins, D., and Song, Y.-A. (2018). Rapid detection of exosomal microRNA biomarkers by electrokinetic concentration for liquid biopsy on chip. Biomicrofluidics 12, 014104. doi:10.1063/1.5009719
Chiticaru, E. A., Pilan, L., Damian, C. M., Vasile, E., Burns, J. S., and Ionita, M. (2019). Influence of graphene oxide concentration when fabricating an electrochemical biosensor for DNA detection. Biosensors 9, 113–118. doi:10.3390/bios9040113
Das, C. M., Guo, Y., Kang, L., Poenar, D. P., Xiong, J., Ramaswamy, Y., et al. (2021). Improving the sensitivity of SPR sensors with Au–Ag alloys and 2D materials — A simulation-based approach. Adv. Theory Simulations 4, 1–10. doi:10.1002/adts.202100292
Deng, H., Chai, Y., Yuan, R., and Yuan, Y. (2020). In situ formation of multifunctional DNA nanospheres for a sensitive and accurate dual-mode biosensor for photoelectrochemical and electrochemical assay. Anal. Chem. 92, 8364–8370. doi:10.1021/acs.analchem.0c00918
Deng, S., Wang, P., and Yu, X. (2017). Phase-sensitive surface plasmon resonance sensors: Recent progress and future prospects. Sensors Switz. 17, 2819. doi:10.3390/s17122819
Dinter, F., Burdukiewicz, M., Schierack, P., Lehmann, W., Nestler, J., Dame, G., et al. (2019). Simultaneous detection and quantification of DNA and protein biomarkers in spectrum of cardiovascular diseases in a microfluidic microbead chip. Anal. Bioanal. Chem. 411, 7725–7735. doi:10.1007/s00216-019-02199-x
Dong, X., Lau, C. M., Lohani, A., Mhaisalkar, S. G., Kasim, J., Shen, Z., et al. (2008). Electrical detection of femtomolar DNA via gold-nanoparticle enhancement in carbon-nanotube-network field-effect transistors. Adv. Mat. 20, 2389–2393. doi:10.1002/adma.200702798
Feng, E., Zheng, T., and Tian, Y. (2019). Dual-mode Au nanoprobe based on surface enhancement Raman scattering and colorimetry for sensitive determination of telomerase activity both in cell extracts and in the urine of patients. ACS Sensors 4, 211–217. doi:10.1021/acssensors.8b01244
Fuku, X., Baker, P., and Iwuoha, E. (2020). Influence of quantum dot surface on electrochemical DNA sensing mechanism. ChemElectroChem 7, 770–781. doi:10.1002/celc.201902079
Ganguli, A., Watanabe, Y., Hwang, M. T., Huang, J. C., and Bashir, R. (2018). Robust label-free microRNA detection using one million ISFET array. Biomed. Microdevices 20, 45. doi:10.1007/s10544-018-0290-8
Gao, J., Zhang, H., and Wang, Z. (2020a). A DNA tetrahedron nanoprobe-based fluorescence resonance energy transfer sensing platform for intracellular tumor-related miRNA detection. Analyst 145, 3535–3542. doi:10.1039/C9AN02610J
Gao, Y., Qiang, L., Chu, Y., Han, Y., Zhang, Y., and Han, L. (2020b). Microfluidic chip for multiple detection of miRNA biomarkers in breast cancer based on three-segment hybridization. AIP Adv. 10, 045022. doi:10.1063/1.5137784
Gao, Z., Xia, H., Zauberman, J., Tomaiuolo, M., Ping, J., Zhang, Q., et al. (2018). Detection of sub-fM DNA with target recycling and self-assembly ampli fi cation on graphene field-E ff ect biosensors, Nano Lett 18. 3509-3515. doi:10.1021/acs.nanolett.8b00572
García-Mendiola, T., Gutiérrez-Sánchez, C., Gibaja, C., Torres, I., Busó-Rogero, C., Pariente, F., et al. (2020). Functionalization of a few-layer antimonene with oligonucleotides for DNA sensing. ACS Appl. Nano Mat. 3, 3625–3633. doi:10.1021/acsanm.0c00335
Goel, A., Rewari, S., Verma, S., and Gupta, R. S. (2018). Dielectric modulated triple metal gate all around MOSFET (TMGAA) for DNA bio- molecule detection. 1, 24–25.
Guerrini, L., Morla-Folch, J., Gisbert-Quilis, P., Xie, H., and Alvarez-Puebla, R. A. (2016). Label-free direct surface-enhanced Raman scattering (SERS) of nucleic acids (Conference Presentation). SPIE Proceeding 9722, 23. doi:10.1117/12.2217653
Guo, J., Chen, Y., Jiang, Y., and Ju, H. (2017). Polyadenine-modulated DNA conformation monitored by surface-enhanced Raman scattering (SERS) on multibranched gold nanoparticles and its sensing application. Chemistry 23, 9332–9337. doi:10.1002/chem.201700883
Guo, Q., Yu, Y., Zhang, H., Cai, C., and Shen, Q. (2020). Electrochemical sensing of exosomal MicroRNA based on hybridization chain reaction signal amplification with reduced false-positive signals. Anal. Chem. 92, 5302–5310. doi:10.1021/acs.analchem.9b05849
Guo, S. R., Lin, J., Penchev, M., Yengel, E., Ghazinejad, M., Ozkan, C. S., et al. (2011). Label free DNA detection using large area graphene based field effect transistor biosensors. J. Nanosci. Nanotechnol. 11, 5258–5263. doi:10.1166/jnn.2011.3885
Hajian, R., Balderston, S., Tran, T., deBoer, T., Etienne, J., Sandhu, M., et al. (2019). Detection of unamplified target genes via CRISPR-Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 3, 427–437. doi:10.1038/s41551-019-0371-x
Hameed, M. F. O., Saadeldin, A. S., Elkaramany, E. M. A., and Obayya, S. S. A. (2017). Label-free highly sensitive hybrid plasmonic biosensor for the detection of DNA hybridization. J. Light. Technol. 35, 4851–4858. doi:10.1109/jlt.2017.2733720
Han, B., Zhang, Y., Zhu, L., Chen, X.-H., Ma, Z., Zhang, X.-L., et al. (2018a). Direct laser scribing of AgNPs@RGO biochip as a reusable SERS sensor for DNA detection. Sensors Actuators B Chem doi:10.1063/1.5137784
Han, S., Liu, W., Zheng, M., and Wang, R. (2020). Label-free and ultrasensitive electrochemical DNA biosensor based on urchinlike carbon nanotube-gold nanoparticle nanoclusters. Anal. Chem. 92, 4780–4787. doi:10.1021/acs.analchem.9b03520
Han, S., Locke, A. K., Oaks, L. A., Cheng, Y.-S. L., and Coté, G. L. (2018b). Development of a free-solution SERS-based assay for point-of-care oral cancer biomarker detection using DNA-conjugated gold nanoparticles. Proc. SPIE, 1050104. doi:10.1117/12.2290516
Han, S., Locke, A. K., Oaks, L. A., Cheng, Y. L., Coté, G. L., Han, S., et al. (2019). Nanoparticle-based assay for detection of S100P mRNA using surface-enhanced Raman spectroscopy. J. Biomed. Opt. 24, 1–9. doi:10.1117/1.jbo.24.5.055001
Hao, K., He, Y., Lu, H., Pu, S., Zhang, Y., Dong, H., et al. (2017). High-sensitive surface plasmon resonance microRNA biosensor based on streptavidin functionalized gold nanorods-assisted signal amplification. Anal. Chim. Acta 954, 114–120. doi:10.1016/j.aca.2016.12.006
He, Y., Yang, X., Yuan, R., and Chai, Y. (2019). A novel ratiometric SERS biosensor with one Raman probe for ultrasensitive microRNA detection based on DNA hydrogel amplification. J. Mat. Chem. B 7, 2643–2647. doi:10.1039/c8tb02894j
Hlongwane, G. N., Dodoo-Arhin, D., Wamwangi, D., Daramola, M. O., Moothi, K., and Iyuke, S. E. (2019). DNA hybridisation sensors for product authentication and tracing: State of the art and challenges. South Afr. J. Chem. Eng. 27, 16–34. doi:10.1016/j.sajce.2018.11.002
Hu, W. P., Tsai, C. C., Yang, Y. S., Chan, H. W. H., and Chen, W. Y. (2018). Synergetic improvements of sensitivity and specificity of nanowire field effect transistor gene chip by designing neutralized DNA as probe. Sci. Rep. 8, 12598–8. doi:10.1038/s41598-018-30996-4
Huang, Y., Tao, M., Luo, S., Zhang, Y., Situ, B., Ye, X., et al. (2020). A novel nest hybridization chain reaction based electrochemical assay for sensitive detection of circulating tumor DNA. Anal. Chim. Acta 1107, 40–47. doi:10.1016/j.aca.2020.02.006
Hügle, M., Behrmann, O., Raum, M., Hufert, F. T., Urban, G. A., and Dame, G. (2020). A lab-on-a-chip for free-flow electrophoretic preconcentration of viruses and gel electrophoretic DNA extraction. Analyst 145, 2554–2561. doi:10.1039/C9AN02333J
Hwang, M. T., Heiranian, M., Kim, Y., You, S., Leem, J., Taqieddin, A., et al. (2020). Ultrasensitive detection of nucleic acids using deformed graphene channel field effect biosensors. Nat. Commun. 11, 1543. doi:10.1038/s41467-020-15330-9
Hwang, M. T., Wang, Z., Ping, J., Ban, D. K., Shiah, Z. C., Antonschmidt, L., et al. (2018). DNA nanotweezers and graphene transistor enable label-free genotyping. Adv. Mat. 30, e1802440. doi:10.1002/adma.201802440
Jayadevimanoranjitham, J., and Narayanan, S. S. (2020). Fabrication of poly o-cresophthalein complexone film modified electrode and its application to simultaneous determination of purine bases in denatured DNA. Mat. Sci. Eng. C. Mat. Biol. Appl. 108, 110353. doi:10.1016/j.msec.2019.110353
Jia, L.-P., Feng, Z., Zhao, R.-N., Ma, R.-N., Zhang, W., Shang, L., et al. (2020). Enzyme-free and triple-amplified electrochemical sensing of 8-hydroxy-2′-deoxyguanosine by three kinds of short pDNA-driven catalyzed hairpin assemblies followed by a hybridization chain reaction. Analyst 145, 3605–3611. doi:10.1039/D0AN00233J
Jiang, J., Wu, H., Su, Y., Liang, Y., Shu, B., and Zhang, C. (2020a). Electrochemical cloth-based DNA sensors (ECDSs): A new class of electrochemical gene sensors. Anal. Chem. 92, 7708–7716. doi:10.1021/acs.analchem.0c00669
Jiang, P., Tan, M., Zhao, L., Wang, Y., Pan, T., Chomba, H., et al. (2020b). A novel electrochemical DNA detection method based on bio-barcode/gold label silver stain dual amplification. Mat. Express 10, 206–213. doi:10.1166/mex.2020.1633
Jiang, W., Yin, H., Zhou, Y., Duan, J., Li, H., Wang, M., et al. (2018). A novel electrochemiluminescence biosensor for the detection of 5-methylcytosine, TET 1 protein and β-glucosyltransferase activities based on gold nanoclusters-H2O2 system. Sensors Actuators B Chem. 274, 144–151. doi:10.1016/j.snb.2018.07.149
Jiang, Z. Y., Jiang, X. X., Su, S., Wei, X. P., Lee, S. T., and He, Y. (2012). Silicon-based reproducible and active surface-enhanced Raman scattering substrates for sensitive, specific, and multiplex DNA detection. Appl. Phys. Lett. 100, 203104. doi:10.1063/1.3701731
Jiao, Z., Zhao, L., Tang, C., Shi, H., Wang, F., and Hu, B. (2019). Droplet-based PCR in a 3D-printed microfluidic chip for miRNA-21 detection. Anal. Methods 11, 3286–3293. doi:10.1039/c9ay01108k
Kang, J., Lee, J., Kim, T. H., Park, J., Seong, M. J., and Hong, S. (2008). Large-scale assembly of carbon nanotube-based flexible circuits for DNA sensors. Nanotechnology 19, 135305. doi:10.1088/0957-4484/19/13/135305
Kannegulla, A., Liu, Y., Wu, B., and Cheng, L.-J. (2018a10728). Enhanced molecular beacon based DNA detection using plasmonic open-ring nanoarrays. Proc. SPIE. doi:10.1117/12.2321234
Kannegulla, A., Liu, Y., Wu, B., and Cheng, L. J. (2018b). Plasmonic open-ring nanoarrays for broadband fluorescence enhancement and ultrasensitive DNA detection. J. Phys. Chem. C 122, 770–776. doi:10.1021/acs.jpcc.7b09769
Khalil, I., Yehye, W. A., Julkapli, N. M., Rahmati, S., Sina, A. A. I., Basirun, W. J., et al. (2019). Graphene oxide and gold nanoparticle based dual platform with short DNA probe for the PCR free DNA biosensing using surface-enhanced Raman scattering. Biosens. Bioelectron. 131, 214–223. doi:10.1016/j.bios.2019.02.028
Khoder, R., and Korri-Youssoufi, H. (2020). E-DNA biosensors of M. tuberculosis based on nanostructured polypyrrole. Mat. Sci. Eng. C. Mat. Biol. Appl. 108, 110371. doi:10.1016/j.msec.2019.110371
Kim, B., Lee, J., Namgung, S., Kim, J., Park, J. Y., Lee, M. S., et al. (2012). DNA sensors based on CNT-FET with floating electrodes. Sensors Actuators, B Chem. 169, 182–187. doi:10.1016/j.snb.2012.04.063
Kim, H., Hyejeong, J., Moon, J., Byun, J., Jeong, J., Juyeon, J., et al. (2019). Metal–organic framework coating for the preservation of silver nanowire surface-enhanced Raman scattering platform. Adv. Mat. Interfaces 6, 1900427. doi:10.1002/admi.201900427
Kim, J.-H., and Benelmekki, M. (2018). “Bottom-up synthesis of hybrid carbon nanoscrolls,” in Emerging applications of nanoparticles and architecture nanostructures. Editors A. Barhoum,, and A. N. Makhlouf (Elsevier). doi:10.1016/B978-0-323-51254-1.00018-X
Kim, J.-H., and Benelmekki, M. (2019). “Two-dimensional nanostructures for biomedical applications,” in Nanostructured thin film: Fundamentals and applications. Editors M. Benelmekki,, and A. B. T.-F. of N. Erbe (Elsevier). doi:10.1016/B978-0-08-102572-7.00004-0
Kim, J. H., Bohra, M., Singh, V., Cassidy, C., and Sowwan, M. (2014). Smart composite nanosheets with adaptive optical properties. ACS Appl. Mat. Interfaces 6, 13339–13343. doi:10.1021/am5041708
Kim, J., Sahloul, S., Orozaliev, A., Do, V. Q., Pham, V. S., Martins, D., et al. (2020). Microfluidic electrokinetic preconcentration chips: Enhancing the detection of nucleic acids and exosomes. IEEE Nanotechnol. Mag. 14, 18–34. doi:10.1109/MNANO.2020.2966064
Kim, J. W., JanghaKu, Y. G. M., Kim, S., Lee, E., Cho, K., et al. (2018). Liquid coplanar-gate organic/graphene hybrid electronics for label-free detection of single and double-stranded DNA molecules. Org. Electron. 62, 163–167. doi:10.1016/j.orgel.2018.07.032
Kim, Y.-J., Hosokawa, K., and Maeda, M. (2019). Sensitivity enhancement of MicroRNA detection using a power-free microfluidic chip. Anal. Sci. Int. J. Jpn. Soc. Anal. Chem. 35, 1227–1236. doi:10.2116/analsci.19P211
Kong, M., Li, Z., Wu, J., Hu, J., Sheng, Y., Wu, D., et al. (2019). Talanta A wearable microfluidic device for rapid detection of HIV-1 DNA using recombinase polymerase amplification. Talanta 205, 1–8. doi:10.1016/j.talanta.2019.120155
Kowalczyk, A., Krajczewski, J., Kowalik, A., Weyher, J. L., Dzięcielewski, I., Chłopek, M., et al. (2019). New strategy for the gene mutation identification using surface enhanced Raman spectroscopy (SERS). Biosens. Bioelectron. 132, 326–332. doi:10.1016/j.bios.2019.03.019
Krishnan, S., and Syed, Z. Q. (2022). Colorimetric visual sensors for point-of-needs testing. Sensors Actuators Rep. 4, 100078. doi:10.1016/j.snr.2022.100078
Ku, G. M., Kim, J. W., Jang, Y. h., Kim, S., Lim, K., and Lee, W. H. (2018). Interfacial polymer brush layer for DNA sensors based on graphene transistors. Fibers Polym. 19, 2483–2488. doi:10.1007/s12221-018-8608-y
Kuo, W. C., Sarangadharan, I., Pulikkathodi, A. K., Chen, P. H., Wang, S. L., Wu, C. R., et al. (2019). Investigation of electrical stability and sensitivity of electric double layer gated field-effect transistors (FETs) for miRNA detection. Sensors Switz. 19, 1484. doi:10.3390/s19071484
Lapitan, L. D. S., Xu, Y., Guo, Y., and Zhou, D. (2019). Combining magnetic nanoparticle capture and poly-enzyme nanobead amplification for ultrasensitive detection and discrimination of DNA single nucleotide polymorphisms. Nanoscale 11, 1195–1204. doi:10.1039/c8nr07641c
Lednický, T., and Bonyár, A. (2020). Large scale fabrication of ordered gold nanoparticle-epoxy surface nanocomposites and their application as label-free plasmonic DNA biosensors. ACS Appl. Mat. Interfaces 12, 4804–4814. doi:10.1021/acsami.9b20907
Lee, S. H., Song, J., Cho, B., Hong, S. G., Hoxha, O., Kang, T., et al. (2019). Bubble-free rapid microfluidic PCR. Biosens. Bioelectron. 126, 725–733. doi:10.1016/j.bios.2018.10.005
Lee, Y., Trocchia, S. M., Warren, S. B., Young, E. F., Vernick, S., and Shepard, K. L. (2018). Electrically controllable single-point covalent functionalization of spin-cast carbon- nanotube field-effect transistor arrays. ACS Nano 12, 9922–9930. doi:10.1021/acsnano.8b03073
Lei, P., Zhou, Y., Zhu, R., Liu, Y., Dong, C., and Shuang, S. (2020). Novel strategy of electrochemical analysis of DNA bases with enhanced performance based on copper-nickel nanosphere decorated N,B-doped reduced graphene oxide. Biosens. Bioelectron. 147, 111735. doi:10.1016/j.bios.2019.111735
Li, C., Li, G., Liu, H., Xiao, Z., Jin, M., and Yuan, C. (2020a). A novel gold nanoparticles decorated magnetic microbead-based molecular beacon for DNA multiplexing detection by flow cytometry. Anal. Chim. Acta 1110, 19–25. doi:10.1016/j.aca.2020.02.063
Li, F., Li, F., Mao, X., Li, F., Li, M., Shen, J., et al. (2020b). Ultrafast DNA sensors with DNA framework-bridged hybridization reactions. J. Am. Chem. Soc. 142, 9975–9981. doi:10.1021/jacs.9b13737
Li, H., and Rothberg, L. (2004). Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 101, 14036–14039. doi:10.1073/pnas.0406115101
Li, J., Zhao, L., Wen, D., Li, X., Yang, H., Wang, D., et al. (2020c). Electrochemical CYFRA21-1 DNA sensor with PCR-like sensitivity based on AgNPs and cascade polymerization. Anal. Bioanal. Chem. 412, 4155–4163. doi:10.1007/s00216-020-02652-2
Li, L., Miao, B., Li, Z., Sun, Z., and Peng, N. (2019a). Sample-to-Answer hepatitis B virus DNA detection from whole blood on a centrifugal microfluidic platform with double rotation axes. ACS Sensors 4, 2738–2745. doi:10.1021/acssensors.9b01270
Li, M., Lin, H., Paidi, S. K., Mesyngier, N., Preheim, S., and Barman, I. (2020d). A fluorescence and surface-enhanced Raman spectroscopic dual-modal aptasensor for sensitive detection of cyanotoxins. ACS Sensors 5, 1419–1426. doi:10.1021/acssensors.0c00307
Li, S., Huang, K., Fan, Q., Yang, S., Shen, T., Mei, T., et al. (2019b). Highly sensitive solution-gated graphene transistors for label-free DNA detection. Biosens. Bioelectron. 136, 91–96. doi:10.1016/j.bios.2019.04.034
Li, X.-Y., Cui, Y.-X., Du, Y.-C., Tang, A.-N., and Kong, D.-M. (2020e). Isothermal cross-boosting extension-nicking reaction mediated exponential signal amplification for ultrasensitive detection of polynucleotide kinase. Analyst 145, 3742–3748. doi:10.1039/c9an02569c
Li, Y., Holl, N. J., Li, Y., Peng, Z., Holl, N. J., Hassan, R., et al. (2021). MXene–graphene field-effect transistor sensing of influenza virus and SARS-CoV-2. ACS Omega 6, 6643–6653. doi:10.1021/acsomega.0c05421
Liao, H., Zhou, Y., Chai, Y., and Yuan, R. (2018). An ultrasensitive electrochemiluminescence biosensor for detection of MicroRNA by in-situ electrochemically generated copper nanoclusters as luminophore and TiO(2) as coreaction accelerator. Biosens. Bioelectron. 114, 10–14. doi:10.1016/j.bios.2018.05.011
Liu, D., Zhang, X., Zhao, J., Chen, S., and Yuan, R. (2020a). An ultrasensitive sensing platform for microRNA-155 based on H(2)O(2) quenched hydroxide-dependent ECL emission of PFO Pdots. Biosens. Bioelectron. 150, 111872. doi:10.1016/j.bios.2019.111872
Liu, G., Ma, X., Tang, Y., and Miao, P. (2020b). Ratiometric fluorescence method for ctDNA analysis based on the construction of a DNA four-way junction. Analyst 145, 1174–1178. doi:10.1039/d0an00044b
Liu, P., Yang, X., Sun, S., Wang, Q., Wang, K., Huang, J., et al. (2013). Enzyme-free colorimetric detection of DNA by using gold nanoparticles and hybridization chain reaction amplification. Anal. Chem. 85, 7689–7695. doi:10.1021/ac4001157
Liu, Q., Sun, X., Liu, M., Jin, Y., and Li, B. (2020c). G-triplex molecular beacon‒based fluorescence biosensor for sensitive detection of small molecule-protein interaction via exonuclease III‒assisted recycling amplification. Sensors Actuators B-chemical 310, 127804. doi:10.1016/j.snb.2020.127804
Liu, S., Wei, M., Wang, X., Shi, C., Kuang, S., and Ma, C. (2020d). An ultrasensitive electrochemical DNA sensing strategy free from pre-immobilization via G-quadruplex based homogenous proximity hybridization. Talanta 210, 120628. doi:10.1016/j.talanta.2019.120628
Liu, Y., Chen, X., and Ma, Q. (2018). A novel amplified electrochemiluminescence biosensor based on Au NPs@PDA@CuInZnS QDs nanocomposites for ultrasensitive detection of p53 gene. Biosens. Bioelectron. 117, 240–245. doi:10.1016/j.bios.2018.06.023
Liu, Y., and Hu, N. (2007). Loading/release behavior of (chitosan/DNA)n layer-by-layer films toward negatively charged anthraquinone and its application in electrochemical detection of natural DNA damage. Biosens. Bioelectron. 23, 661–667. doi:10.1016/j.bios.2007.07.009
Liu, Y., Nie, Y., Wang, M., Zhang, Q., and Ma, Q. (2020e). Distance-dependent plasmon-enhanced electrochemiluminescence biosensor based on MoS(2) nanosheets. Biosens. Bioelectron. 148, 111823. doi:10.1016/j.bios.2019.111823
Liu, Y., Tu, Y., Wu, H., Zhang, H., Chen, H., Zhou, G., et al. (2020f). A renewable DNA biosensor for sensitive detection of DNA methyltransferase activity based on cascade signal amplification. Sensors Actuators B Chem. 313, 128029. doi:10.1016/j.snb.2020.128029
Liu, Z., Lei, S., Zou, L., Li, G., Xu, L., and Ye, B. (2020). Highly ordered 3D electrochemical DNA biosensor based on dual orientation controlled rolling motor and graftable tetrahedron DNA. Biosens. Bioelectron. 147, 111759. doi:10.1016/j.bios.2019.111759
Loan, P. T. K., Zhang, W., Lin, C. T., Wei, K. H., Li, L. J., and Chen, C. H. (2014). Graphene/MoS2 heterostructures for ultrasensitive detection of DNA hybridisation. Adv. Mat. 26, 4838–4844. doi:10.1002/adma.201401084
Lu, C., Liu, Y., Ying, Y., and Liu, J. (2017). Comparison of MoS2, WS2, and graphene oxide for DNA adsorption and sensing. Langmuir 33, 630–637. doi:10.1021/acs.langmuir.6b04502
Ma, D., Huang, C., Zheng, J., Tang, J., Li, J., Yang, J., et al. (2018). Quantitative detection of exosomal microRNA extracted from human blood based on surface-enhanced Raman scattering. Biosens. Bioelectron. 101, 167–173. doi:10.1016/j.bios.2017.08.062
Majd, S. M., Salimi, A., and Ghasemi, F. (2018). An ultrasensitive detection of miRNA-155 in breast cancer via direct hybridization assay using two-dimensional molybdenum disulfide field-effect transistor biosensor. Biosens. Bioelectron. 105, 6–13. doi:10.1016/j.bios.2018.01.009
Mariani, S., Scarano, S., Spadavecchia, J., and Minunni, M. (2015). A reusable optical biosensor for the ultrasensitive and selective detection of unamplified human genomic DNA with gold nanostars. Biosens. Bioelectron. 74, 981–988. doi:10.1016/j.bios.2015.07.071
Mei, J., Li, Y. T., Zhang, H., Xiao, M. M., Ning, Y., Zhang, Z. Y., et al. (2018). Molybdenum disulfide field-effect transistor biosensor for ultrasensitive detection of DNA by employing morpholino as probe. Biosens. Bioelectron. 110, 71–77. doi:10.1016/j.bios.2018.03.043
Min, S. K., Kim, W. Y., Cho, Y., and Kim, K. S. (2011). Fast DNA sequencing with a graphene-based nanochannel device. Nat. Nanotechnol. 6, 162–165. doi:10.1038/nnano.2010.283
Minero, G. A. S., Cangiano, V., Garbarino, F., Fock, J., and Hansen, M. F. (2019). Integration of microbead DNA handling with optomagnetic detection in rolling circle amplification assays. Mikrochim. Acta 186, 528. doi:10.1007/s00604-019-3636-x
Minero, G. A. S., Tefiku, E., Garbarino, F., Fock, J., and Hansen, M. F. (2020). On-chip DNA analysis of tuberculosis based on magnetic nanoparticle clustering induced by rolling circle amplification products. IEEE Magn. Lett. 11, 1–5. doi:10.1109/LMAG.2019.2959545
Miscourides, N., Yu, L.-S., Rodriguez-Manzano, J., and Georgiou, P. (2018). A 12.8 k current-mode velocity-saturation ISFET array for on-chip real-time DNA detection. IEEE Trans. Biomed. Circuits Syst. 12, 1202–1214. doi:10.1109/TBCAS.2018.2851448
Mohammadi, A. V., Rosen, J., and Gogotsi, Y. (2021). The world of two-dimensional carbides and nitrides (MXenes). Science 80-, eabf1581. doi:10.1126/science.abf1581
Moser, N., Rodriguez-Manzano, J., Lande, T. S., and Georgiou, P. (2018). A scalable ISFET sensing and memory array with sensor auto-calibration for on-chip real-time DNA detection. IEEE Trans. Biomed. Circuits Syst. 12, 390–401. doi:10.1109/TBCAS.2017.2789161
Movilli, J., Kolkman, R. W., Rozzi, A., Corradini, R., Segerink, L. I., and Huskens, J. (2020). Increasing the sensitivity of electrochemical DNA detection by a micropillar-structured biosensing surface. Langmuir 36, 4272–4279. doi:10.1021/acs.langmuir.0c00144
Mujawar, M. A., Gohel, H., Bhardwaj, S. K., Srinivasan, S., Hickman, N., and Kaushik, A. (2020). Nano-enabled biosensing systems for intelligent healthcare: Towards COVID-19 management. Mat. Today Chem. 17, 100306. doi:10.1016/j.mtchem.2020.100306
Mustafa, A. M., Li, Z., and Shao, L. (2017). “Molecular dynamics simulations of damage cascades creation in oxide-particle-embedded Fe,” in 25th International Conference on Nuclear Engineering,ASME, 1–5. doi:10.1115/ICONE25-67356
Mustafa Azeem, M., Abd El Gawad, K., Zubair, M., Ibrahim, S. A., Ado, M., and Mehdi, G. (2019). “Radiation damage effects in oxide dispersion strengthened steel alloys,” in Proceedings, ICONE International Conference on Nuclear Engineering. doi:10.1299/jsmeicone.2019.27.2086
Mustafa Azeem, M., Li, Z., Wang, Q., and Zubair, M. (2018). Molecular dynamics studies and irradiation effects in ODSS alloys. Int. J. Nucl. Energy Sci. Technol. 12, 381–399. doi:10.1504/IJNEST.2018.097200
Na, H.-K., Wi, J.-S., Son, H. Y., Ok, J. G., Huh, Y.-M., and Lee, T. G. (2018). Discrimination of single nucleotide mismatches using a scalable, flexible, and transparent three-dimensional nanostructure-based plasmonic miRNA sensor with high sensitivity. Biosens. Bioelectron. 113, 39–45. doi:10.1016/j.bios.2018.04.033
Napoli, C., Lai, S., Giannetti, A., Tombelli, S., Baldini, F., Barbaro, M., et al. (2018). Electronic detection of DNA hybridization by coupling organic field-effect transistor-based sensors and hairpin-shaped probes. Sensors Switz. 18, 990. doi:10.3390/s18040990
Naresh, V., and Lee, N. (2021). A review on biosensors and recent development of nanostructured materials-enabled biosensors. Sensors Switz. 21, 1109–1135. doi:10.3390/s21041109
Ng, E., Yao, C., Shultz, T. O., Ross-Howe, S., and Wang, S. X. (2019). Magneto-nanosensor smartphone platform for the detection of HIV and leukocytosis at point-of-care. Nanomedicine 16, 10–19. doi:10.1016/j.nano.2018.11.007
Nguyen, T. T. T., Legallais, M., Morisot, F., Cazimajou, T., Stambouli, V., Mouis, M., et al. (2019). First evidence of superiority of Si nanonet field effect transistors over multi-parallel Si nanowire ones in view of electrical DNA hybridization detection. Mat. Res. Express 6, 016301. doi:10.1088/2053-1591/aae0d5
Novara, C., Chiadò, A., Paccotti, N., Catuogno, S., Esposito, C. L., Condorelli, G., et al. (2017). SERS-active metal-dielectric nanostructures integrated in microfluidic devices for label-free quantitative detection of miRNA. Faraday Discuss. 205, 271–289. doi:10.1039/C7FD00140A
Ouyang, L., Hu, Y., Zhu, L., Cheng, G. J., and Irudayaraj, J. (2017). A reusable laser wrapped graphene-Ag array based SERS sensor for trace detection of genomic DNA methylation. Biosens. Bioelectron. 92, 755–762. doi:10.1016/j.bios.2016.09.072
Pal, S., Chakraborty, M., and Sarkar, N. (2020). Graphene oxide functionalized with 5-aminophenanthroline for selective detection of adenine through fluorescence “turn-off–on” response. ACS Appl. Nano Mat. 3, 3532–3539. doi:10.1021/acsanm.0c00212
Patching, S. G. (2014). Surface plasmon resonance spectroscopy for characterisation of membrane protein-ligand interactions and its potential for drug discovery. Biochim. Biophys. Acta - Biomembr. 1838, 43–55. doi:10.1016/j.bbamem.2013.04.028
Perez-Toralla, K., Pereiro, I., Garrigou, S., Di Federico, F., Proudhon, C., Bidard, F. C., et al. (2019). Microfluidic extraction and digital quantification of circulating cell-free DNA from serum. Sensors Actuators, B Chem. 286, 533–539. doi:10.1016/j.snb.2019.01.159
Ploetz, E., Lerner, E., Husada, F., Roelfs, M., Chung, S., Hohlbein, J., et al. (2016). Förster resonance energy transfer and protein-induced fluorescence enhancement as synergetic multi-scale molecular rulers. Sci. Rep. 6, 1–18. doi:10.1038/srep33257
Prada, J., Cordes, C., Harms, C., and Lang, W. (2019). Design and manufacturing of a disposable, cyclo-olefin copolymer, microfluidic device for a biosensor. Sensors Switz. 19, 1178. doi:10.3390/s19051178
Pramanik, S., Nandy, A., Chakraborty, S., Pramanik, U., Nandi, S., and Mukherjee, S. (2020). Preferential binding of thioflavin T to AT-rich DNA: White light emission through intramolecular förster resonance energy transfer. J. Phys. Chem. Lett. 11, 2436–2442. doi:10.1021/acs.jpclett.0c00237
Qi, X., and Bi, J. (2019). Plasmonic sensors relying on nanoparticle arrays created by a template-directed dewetting process. Opt. Commun. 453, 124328. doi:10.1016/j.optcom.2019.124328
Qian, Z. S., Shan, X. Y., Chai, L. J., Ma, J. J., Chen, J. R., and Feng, H. (2014). A universal fluorescence sensing strategy based on biocompatible graphene quantum dots and graphene oxide for the detection of DNA. Nanoscale 6, 5671–5674. doi:10.1039/c3nr06583a
Quan, P. L., Sauzade, M., and Brouzes, E. (2018). Dpcr: A technology review. Sensors Switz. 18, 1271. doi:10.3390/s18041271
Rahman, M., Cui, D., Zhou, S., Zhang, A., and Chen, D. (2020). A graphene oxide coated gold nanostar based sensing platform for ultrasensitive electrochemical detection of circulating tumor DNA. Anal. Methods 12, 440–447. doi:10.1039/C9AY01620A
Ramshani, Z., Zhang, C., Richards, K., Chen, L., Xu, G., Stiles, B. L., et al. (2019). Extracellular vesicle microRNA quantification from plasma using an integrated microfluidic device. Commun. Biol. 2, 189–9. doi:10.1038/s42003-019-0435-1
Ren, R., Bi, Q., Yuan, R., and Xiang, Y. (2020). An efficient, label-free and sensitive electrochemical microRNA sensor based on target-initiated catalytic hairpin assembly of trivalent DNAzyme junctions. Sensors Actuators, B Chem. 304, 127068. doi:10.1016/j.snb.2019.127068
Roether, J., Chu, K.-Y., Willenbacher, N., Shen, A. Q., and Bhalla, N. (2019). Real-time monitoring of DNA immobilization and detection of DNA polymerase activity by a microfluidic nanoplasmonic platform. Biosens. Bioelectron. 142, 111528. doi:10.1016/j.bios.2019.111528
Saad, S. M., Abdullah, J., Abd Rashid, S., Fen, Y. W., Salam, F., and Yih, L. H. (2020). A carbon dots based fluorescence sensing for the determination of Escherichia coli O157:H7. Measurement 160, 107845. doi:10.1016/j.measurement.2020.107845
Sánchez-Paniagua, M., Palenzuela-Batista, S., Manzanares-Palenzuela, C. L., and López-Ruiz, B. (2020). Electrochemical genosensor for Klotho detection based on aliphatic and aromatic thiols self-assembled monolayers. Talanta 212, 120735. doi:10.1016/j.talanta.2020.120735
Santhanam, M., Algov, I., and Alfonta, L. (2020). DNA/RNA electrochemical biosensing devices a future replacement of PCR methods for a fast epidemic containment. Sensors Switz. 20, 1–15. doi:10.3390/s20164648
Seo, G., Lee, G., Kim, M. J., Baek, S. H., Choi, M., Ku, K. B., et al. (2020). Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 14, 5135–5142. doi:10.1021/acsnano.0c02823
Sforzi, J., Ferrauto, G., Aime, S., and Geninatti Crich, S. (2020). A simple and fast assay based on carboxyfluorescein-loaded liposome for quantitative DNA detection. ACS Omega 5, 1764–1772. doi:10.1021/acsomega.9b01457
Shen, C.-H. (2019). “Detection and analysis of nucleic acids,” in Diagnostic molecular biology. Editor C.-H. B. T.-D. M. B. Shen (Academic Press), 167–185. doi:10.1016/B978-0-12-802823-0.00007-9
Shen, X., Liu, D., Zhu, C., Li, Y., Liu, Y., and You, T. (2020). Photoelectrochemical and electrochemical ratiometric aptasensing: A case study of streptomycin. Electrochem. Commun. 110, 106637. doi:10.1016/j.elecom.2019.106637
Shen, Z., He, L., Wang, W., Tan, L., and Gan, N. (2020). Highly sensitive and simultaneous detection of microRNAs in serum using stir-bar assisted magnetic DNA nanospheres-encoded probes. Biosens. Bioelectron. 148, 111831. doi:10.1016/j.bios.2019.111831
Shlyapnikov, Y. M., Malakhova, E. A., and Shlyapnikova, E. A. (2019). Rapid amplification-free microarray-based ultrasensitive detection of DNA. Anal. Chem. 91, 11209–11214. doi:10.1021/acs.analchem.9b02149
Si, Y., Xu, L., Wang, N., Zheng, J., Yang, R., and Li, J. (2020). Target MicroRNA-responsive DNA hydrogel-based surface-enhanced Raman scattering sensor arrays for MicroRNA-marked cancer screening. Anal. Chem. 92, 2649–2655. doi:10.1021/acs.analchem.9b04606
Song, G., Zhang, R., Jiang, X., Liu, F., and Liu, X. (2020a). Nitrogen-doped reduced graphene oxide as a sensing platform for detection of guanine and application in cell necrosis. Chem. Pap. 74, 89–98. doi:10.1007/s11696-019-00856-0
Song, R., Tian, M., Li, Y., Liu, J., and Liu, G. (2020b). Detection of MicroRNA based on three-dimensional graphene field-E ® ect transistor biosensor. Nano 15, 1–8. doi:10.1142/S1793292020500393
Špaček, J., Eksin, E., Havran, L., Erdem, A., and Fojta, M. (2020). Fast enzyme-linked electrochemical sensing of DNA hybridization at pencil graphite electrodes. Application to detect gene deletion in a human cell culture. J. Electroanal. Chem. 862, 113951. doi:10.1016/j.jelechem.2020.113951
Stegmann, U. E. (2005). Genetic information as instructional content. Philos. Sci. 72, 425–443. doi:10.1086/498472
Sun, J., Xie, X., Xie, K., Xu, S., Jiang, S., Ren, J., et al. (2019a). Magnetic graphene field-effect transistor biosensor for single-strand DNA detection. Nanoscale Res. Lett. 14, 248–8. doi:10.1186/s11671-019-3048-1
Sun, Y., Cai, H., Qiao, X., and Wang, X. (2019b). High-performance polarization control modulated surface plasmon resonance sensor based on monolayer graphene/Au-NPs architecture for detection of DNA hybridization. Meas. Sci. Technol. 30, 125701. doi:10.1088/1361-6501/ab383a
Sun, Y., Peng, Z., Li, H., Wang, Z., Mu, Y., Zhang, G., et al. (2019c). Suspended CNT-Based FET sensor for ultrasensitive and label-free detection of DNA hybridization. Biosens. Bioelectron. 137, 255–262. doi:10.1016/j.bios.2019.04.054
Tadimety, A., Zhang, Y., Kready, K. M., Palinski, T. J., Tsongalis, G. J., and Zhang, J. X. J. (2019). Design of peptide nucleic acid probes on plasmonic gold nanorods for detection of circulating tumor DNA point mutations. Biosens. Bioelectron. 130, 236–244. doi:10.1016/j.bios.2019.01.045
Tang, S., Li, Y., Zhu, A., Yao, Y., Sun, J., Zheng, F., et al. (2019). A triple-amplification strategy based on the formation of peroxidase-like two-dimensional DNA/Fe3O4 networks initiated by the hybridization chain reaction for highly sensitive detection of microRNA. Chem. Commun. 55, 8386–8389. doi:10.1039/C9CC03194D
Temiz, Y., Lovchik, R. D., Kaigala, G. V., and Delamarche, E. (2015). Lab-on-a-chip devices: How to close and plug the lab? Microelectron. Eng. 132, 156–175. doi:10.1016/j.mee.2014.10.013
Tian, B., Han, Y., Fock, J., Strömberg, M., Leifer, K., and Hansen, M. F. (2019). Self-Assembled magnetic nanoparticle-graphene oxide nanotag for optomagnetic detection of DNA. ACS Appl. Nano Mat. 2, 1683–1690. doi:10.1021/acsanm.9b00127
Tian, M. (2018). RNA detection based on graphene field-effect transistor biosensor. Adv. Condens. Matter Phys., 27–32.
Tian, Y., Lu, Q., Guo, X., Wang, S., Gao, Y., and Wang, L. (2020). Au nanoparticles deposited on ultrathin two-dimensional covalent organic framework nanosheets for in vitro and intracellular sensing. Nanoscale 12, 7776–7781. doi:10.1039/C9NR08220D
Tripathy, S., Joseph, J., Pothuneedi, S., Das, D., Vanjari, S. R. K., Rao, A. V. S. S. N., et al. (2019). A miniaturized electrochemical platform with an integrated PDMS reservoir for label-free DNA hybridization detection using nanostructured Au electrodes. Analyst 144, 6953–6961. doi:10.1039/C9AN01076A
Trotter, M., Juric, D., Bagherian, Z., Borst, N., Glaser, K., Meissner, T., et al. (2020). Inkjet-printing of nanoparticle gold and silver ink on cyclic olefin copolymer for DNA-sensing applications. Sensors (Basel) 20, 1–15. doi:10.3390/s20051333
Verschueren, D. V., Pud, S., Shi, X., De Angelis, L., Kuipers, L., and Dekker, C. (2019). Label-free optical detection of DNA translocations through plasmonic nanopores. ACS Nano 13, 61–70. doi:10.1021/acsnano.8b06758
Vikrant, K., Bhardwaj, N., Bhardwaj, S. K., Kim, K. H., and Deep, A. (2019). Nanomaterials as efficient platforms for sensing DNA. Biomaterials 214, 119215. doi:10.1016/j.biomaterials.2019.05.026
Wang, H. N., Register, J. K., Fales, A. M., Gandra, N., Cho, E. H., Boico, A., et al. (2018a). Surface-enhanced Raman scattering nanosensors for in vivo detection of nucleic acid targets in a large animal model. Nano Res. 11, 4005–4016. doi:10.1007/s12274-018-1982-3
Wang, H., Tang, H., Yang, C., and Li, Y. (2019a). Selective single molecule nanopore sensing of microRNA using PNA functionalized magnetic core–shell Fe3O4–Au nanoparticles. Anal. Chem. 91, 7965–7970. doi:10.1021/acs.analchem.9b02025
Wang, J., Zhao, W.-W., Zhou, H., Xu, J.-J., and Chen, H.-Y. (2013). Amplified electrochemiluminescence detection of DNA-binding protein based on the synergy effect of electron and energy transfer between CdS nanocrystals and gold nanoparticles. Biosens. Bioelectron. 41, 615–620. doi:10.1016/j.bios.2012.09.041
Wang, Q., Zhang, B., Xu, X., Long, F., and Wang, J. (2018b). CRISPR-typing PCR (ctPCR), a new Cas9-based DNA detection method. Sci. Rep. 8, 1–13. doi:10.1038/s41598-018-32329-x
Wang, Q., Zou, L., Yang, X., Liu, X., Nie, W., Zheng, Y., et al. (2019b). Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens. Bioelectron. 135, 129–136. doi:10.1016/j.bios.2019.04.013
Wang, X., Zhang, J., Wei, Y., Xing, T., Cao, T., Wu, S., et al. (2020). A copper-based metal–organic framework/graphene nanocomposite for the sensitive and stable electrochemical detection of DNA bases. Analyst 145, 1933–1942. doi:10.1039/C9AN02398D
Wang, Z., and Jia, Y. (2018). Graphene solution-gated field effect transistor DNA sensor fabricated by liquid exfoliation and double glutaraldehyde cross-linking. Carbon N. Y. 130, 758–767. doi:10.1016/j.carbon.2018.01.078
Watson, J. D., and Crick, F. H. C. (1953). Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 171, 737–738. doi:10.1038/171737a0
Wilkins, M. H. (1956). Physical studies of the molecular structure of deoxyribose nucleic acid and nucleoprotein. Cold Spring Harb. Symp. Quant. Biol. 21, 75–90. doi:10.1101/sqb.1956.021.01.007
Wu, C., Manga, Y. B., Yang, M., Chien, Z., and Lee, K. (2018). Label-free detection of BRAF V599E gene mutation using side-gated nanowire field effect transistors. J. Electrochem. Soc. 165, B576–B581. doi:10.1149/2.0641813jes
Wu, C., Zhang, X., Wang, R., Chen, L. J., Nie, M., Zhang, Z., et al. (2022). Low-dimensional material based wearable sensors. Nanotechnology 33, 072001. doi:10.1088/1361-6528/ac33d1
Wu, T., Li, X., Fu, Y., Ding, X., Li, Z., Zhu, G., et al. (2020a). A highly sensitive and selective fluorescence biosensor for hepatitis C virus DNA detection based on δ-FeOOH and exonuclease III-assisted signal amplification. Talanta 209, 120550. doi:10.1016/j.talanta.2019.120550
Wu, Y., Choi, N., Chen, H., Dang, H., Chen, L., and Choo, J. (2020b). Performance evaluation of surface-enhanced Raman scattering-polymerase chain reaction sensors for future use in sensitive genetic assays. Anal. Chem. 92, 2628–2634. doi:10.1021/acs.analchem.9b04522
Xiao, G., Chen, B., He, M., Li, X., and Hu, B. (2019). A highly sensitive assay of DNA based on inductively coupled plasma mass spectrometry detection with gold nanoparticle amplification and isothermal circular strand-displacement polymerization reaction. Talanta 202, 207–213. doi:10.1016/j.talanta.2019.05.018
Xu, M., Fu, P., Xing, S., Zhao, Y., and Zhao, C. (2020a). A PNA-DNA(2) triple-helix molecular switch-based colorimetric sensor for sensitive and specific detection of microRNAs from cancer cells. Chembiochem 21, 2667–2675. doi:10.1002/cbic.202000155
Xu, S., Chang, Y., Wu, Z., Li, Y., Yuan, R., and Chai, Y. (2020b). One DNA circle capture probe with multiple target recognition domains for simultaneous electrochemical detection of miRNA-21 and miRNA-155. Biosens. Bioelectron. 149, 111848. doi:10.1016/j.bios.2019.111848
Yadav, P., Cao, Z., and Barati Farimani, A. (2021). DNA detection with single-layer Ti3C2MXene nanopore. ACS Nano 15, 4861–4869. doi:10.1021/acsnano.0c09595
Yang, B., Kong, J., and Fang, X. (2019). Bandage-like wearable flexible microfluidic recombinase polymerase amplification sensor for the rapid visual detection of nucleic acids. Talanta 204, 685–692. doi:10.1016/j.talanta.2019.06.031
Ye, H., and De, S. (2017). Thermal injury of skin and subcutaneous tissues: A review of experimental approaches and numerical models. J. Int. Soc. Burn Inj. 43, 909–932. doi:10.1016/j.burns.2016.11.014
Yi, J. C., Kim, Y. J., Kim, Y. Y., Lee, S., Woo, D. H., and Kim, C. (2019). “Detection of single-stranded DNA using the Dirac voltage change of graphene-based FETs,” in IEEE Region 10 Annual International Conference, Proceedings/TENCON, 2051–2053. doi:10.1109/TENCON.2019.8929535
Yin, Z., Cui, H., Shu, Q., Jin, C., Lin, Y., Su, J., et al. (2020). Multi-signal amplification electrochemical DNA biosensor based on exonuclease III and tetraferrocene. J. Mat. Chem. B 8, 4143–4150. doi:10.1039/D0TB00204F
You, L., Li, R., Dong, X., Wang, F., Guo, J., and Wang, C. (2017). Micron-sized surface enhanced Raman scattering reporter/fluorescence probe encoded colloidal microspheres for sensitive DNA detection. J. Colloid Interface Sci. 488, 109–117. doi:10.1016/j.jcis.2016.10.086
Zhang, D., Yan, Y., Que, H., Yang, T., Cheng, X., Ding, S., et al. (2020). CRISPR/Cas12a-Mediated interfacial cleaving of hairpin DNA reporter for electrochemical nucleic acid sensing. ACS sensors 5, 557–562. doi:10.1021/acssensors.9b02461
Zhang, H., Fu, C., Wu, S., Shen, Y., Zhou, C., Neng, J., et al. (2019a). Magnetic-capture-based SERS detection of multiple serum microRNA biomarkers for cancer diagnosis. Anal. Methods 11, 783–793. doi:10.1039/C8AY02423E
Zhang, H. W., Zhang, L., Sun, X., and Zhang, Z. R. (2007). Successful transfection of hepatoma cells after encapsulation of plasmid DNA into negatively charged liposomes. Biotechnol. Bioeng. 96, 118–124. doi:10.1002/bit.21146
Zhang, J., Ba, Y., Liu, Q., Zhao, L., Wang, D., Yang, H., et al. (2020). CuBr(2)/EDTA-mediated ATRP for ultrasensitive fluorescence detection of lung cancer DNA. J. Adv. Res. 22, 77–84. doi:10.1016/j.jare.2019.11.006
Zhang, J., Hu, S., Wang, Z., and Jia, Y. (2019b). Detection of methylated cytosine on single strand DNA based on immune-functionalized and glutaraldehyde modified liquid exfoliated graphene field effect transistor. Proc. SPIE, 113841D. doi:10.1117/12.2559509
Zhang, J., Song, C., Zhou, H., Jia, J., Dai, Y., Cui, D., et al. (2020d). A dual signal amplification strategy for the highly sensitive fluorescence detection of nucleic acids. Analyst 145, 1219–1226. doi:10.1039/c9an02183c
Zhang Y, Y., Hao, L., Zhao, Z., Yang, X., Wang, L., and Liu, S. (2020). Immuno-DNA binding directed template-free DNA extension and enzyme catalysis for sensitive electrochemical DNA methyltransferase activity assay and inhibitor screening. Analyst 145, 3064–3072. doi:10.1039/D0AN00008F
Zhang, Z., Pettinari, R., Zhou, L., Song, Y., Wang, M., He, L., et al. (2020e). Sensors and Actuators B: Chemical Construction of the 0D/2D heterojunction of Ti 3 C 2 T x MXene nanosheets and iron phthalocyanine quantum dots for the impedimetric aptasensing of. Sensors Actuators B. Chem. 310, 127844. doi:10.1016/j.snb.2020.127844
Zhao, L., Kong, D., Wu, Z., Liu, G., Gao, Y., Yan, X., et al. (2020). Interface interaction of MoS2 nanosheets with DNA based aptameric biosensor for carbohydrate antigen 15–3 detection. Microchem. J. 155, 104675. doi:10.1016/j.microc.2020.104675
Zheng, X., Zhao, L., Wen, D., Wang, X., Yang, H., Feng, W., et al. (2020). Ultrasensitive fluorescent detection of HTLV-II DNA based on magnetic nanoparticles and atom transfer radical polymerization signal amplification. Talanta 207, 120290. doi:10.1016/j.talanta.2019.120290
Zhou, W., Han, Y., Beliveau, B. J., and Gao, X. (2020). Combining qdot nanotechnology and DNA nanotechnology for sensitive single-cell imaging. Adv. Mat. 32, 1908410. doi:10.1002/adma.201908410
Zhou, X., Zhu, D., Liao, Y., Liu, W., Liu, H., Ma, Z., et al. (2014). Synthesis, labeling and bioanalytical applications of a tris(2,2’-bipyridyl)ruthenium(II)-based electrochemiluminescence probe. Nat. Protoc. 9, 1146–1159. doi:10.1038/nprot.2014.060
Zhou, Y., Jiang, W., Wu, H., Liu, F., Yin, H., Lu, N., et al. (2019). Amplified electrochemical immunoassay for 5-methylcytosine using a nanocomposite prepared from graphene oxide, magnetite nanoparticles and β-cyclodextrin. Mikrochim. Acta 186, 488. doi:10.1007/s00604-019-3575-6
Ziółkowski, R., Oszwałdowski, S., Kopyra, K. K., Zacharczuk, K., Zasada, A. A., and Malinowska, E. (2020). Toward Fluorimetric-Paired-Emitter-Detector-Diode test for Bacillus anthracis DNA based on graphene oxide. Microchem. J. 154, 104592. doi:10.1016/j.microc.2019.104592
Keywords: detection, deoxyribonucleic acid, sensors, polymerase chain reaction, surface plasmon resonance, field effect transistor, nucleotide sensors
Citation: Azeem MM, Shafa M, Aamir M, Zubair M, Souayeh B and Alam MW (2023) Nucleotide detection mechanism and comparison based on low-dimensional materials: A review. Front. Bioeng. Biotechnol. 11:1117871. doi: 10.3389/fbioe.2023.1117871
Received: 07 December 2022; Accepted: 13 February 2023;
Published: 02 March 2023.
Edited by:
Jeong-Hwan Kim, Yokohama City University, JapanReviewed by:
Weiling Song, Qingdao University of Science and Technology, ChinaCopyright © 2023 Azeem, Shafa, Aamir, Zubair, Souayeh and Alam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Mustafa Azeem, bXVzdGFmYUBtc3QuZWR1; Muhammad Aamir, bXNhZGlxQGtmdS5lZHUuc2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.