
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 21 February 2023
Sec. Industrial Biotechnology
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1114946
This article is part of the Research Topic Proceedings of the 18th International Symposium on BioPolymers (ISBP2022) View all 12 articles
 Ramamoorthi M Sivashankari1
Ramamoorthi M Sivashankari1 Maierwufu Mierzati1
Maierwufu Mierzati1 Yuki Miyahara1
Yuki Miyahara1 Shoji Mizuno1
Shoji Mizuno1 Christopher T. Nomura2
Christopher T. Nomura2 Seiichi Taguchi3
Seiichi Taguchi3 Hideki Abe4
Hideki Abe4 Takeharu Tsuge1*
Takeharu Tsuge1*Polyhydroxyalkanoate (PHA) synthases (PhaCs) are key enzymes in PHA polymerization. PhaCs with broad substrate specificity are attractive for synthesizing structurally diverse PHAs. In the PHA family, 3-hydroxybutyrate (3HB)-based copolymers are industrially produced using Class I PhaCs and can be used as practical biodegradable thermoplastics. However, Class I PhaCs with broad substrate specificities are scarce, prompting our search for novel PhaCs. In this study, four new PhaCs from the bacteria Ferrimonas marina, Plesiomonas shigelloides, Shewanella pealeana, and Vibrio metschnikovii were selected via a homology search against the GenBank database, using the amino acid sequence of Aeromonas caviae PHA synthase (PhaCAc), a Class I enzyme with a wide range of substrate specificities, as a template. The four PhaCs were characterized in terms of their polymerization ability and substrate specificity, using Escherichia coli as a host for PHA production. All the new PhaCs were able to synthesize P(3HB) in E. coli with a high molecular weight, surpassing PhaCAc. The substrate specificity of PhaCs was evaluated by synthesizing 3HB-based copolymers with 3-hydroxyhexanoate, 3-hydroxy-4-methylvalerate, 3-hydroxy-2-methylbutyrate, and 3-hydroxypivalate monomers. Interestingly, PhaC from P. shigelloides (PhaCPs) exhibited relatively broad substrate specificity. PhaCPs was further engineered through site-directed mutagenesis, and the variant resulted in an enzyme with improved polymerization ability and substrate specificity.
The bacterial polyesters polyhydroxyalkanoates (PHAs) are considered excellent bio-based plastics and have been demonstrated to be biodegradable in various environments such as compost, soil, freshwater, and marine water (Suzuki et al., 2021). A myriad of microorganisms can synthesize PHA as an intracellular carbon and energy reserve under stressful conditions (Anderson and Dawes, 1990). Poly[(R)-3-hydroxybutyrate], P(3HB), is a major member of the PHA family and has been extensively studied since its discovery in 1926 (Lenz and Marchessault, 2005). Despite these merits, it is still challenging for PHA to compete with petroleum-based plastics because of the inherent flaws in P(3HB). The poor material properties of P(3HB) (Lehrle and Williams, 1994) such as its high crystallinity and narrow processing temperature window have greatly hampered the entry of this polymer into the commercial world. Fortunately, 3HB-based copolymers (Tsuge et al., 2005; Mizuno et al., 2010; Mierzati et al., 2020; Furutate et al., 2021) have been proven to overcome the material property limitations of P(3HB) to a certain extent, and have been used as a remedy for problems related to plastics (Sivashankari and Tsuge, 2021).
PHA synthases are key enzymes involved in PHA polymerization (Sudesh et al., 2000). Based on the substrate specificities and subunit compositions of PHA synthases, they are categorized into four classes (Rehm, 2003). Class I and II PHA synthases are homodimers of the PhaC subunits. Class I PHA synthases, represented by the Ralstonia eutropha enzyme, mainly polymerize short chain length (scl)-monomers (C3-C5), whereas Class II PHA synthases, represented by the Pseudomonas aeruginosa and Pseudomonas putida enzymes, polymerize medium chain length (mcl)-monomers (C6-C14). Class III PHA synthases such as Allochromatium vinosum and Synechocystis sp. PCC 6803 consists of two heterosubunits (PhaC and PhaE). Class IV PHA synthases, represented by Bacillus megaterium and Bacillus cereus, are similar to Class III PHA synthases and possess two subunits (PhaC and PhaR). Similar to Class I synthases, Class III and IV PHA synthases preferentially polymerize scl-monomers (C3-C5).
PhaCs with broad substrate specificities are attractive biocatalysts for PHA synthesis because they can naturally copolymerize different monomers to produce polymers with desirable physical properties. PhaC from Aeromonas caviae (PhaCAc) can naturally synthesize poly(3HB-co-3-hydroxyhexanoate) [P(3HB-co-3HHx)] from vegetable oils and fatty acids (Kobayashi et al., 1994; Shimamura et al., 1994; Doi et al., 1995; Tsuge et al., 2007a; Tsuge, 2016), distinguishing it from other Class I PhaCs because it exhibits polymerization activities toward 3HB monomers and mcl 3HHx monomers (Kobayashi et al., 1994). Therefore, PhaCAc is a marketable biocatalyst to produce P(3HB-co-3HHx) copolymers. The potential of PhaCAc has been fortified through evolutionary engineering with the development of the PhaCAcNSDG variant (Tsuge et al., 2007b). The PhaCAcNSDG variant has amino acid substitutions of asparagine 149 by serine (N149S) and aspartate 171 by glycine (D171G) and was shown to have the ability to synthesize the P(3HB-co-3HHx) copolymer with an enhanced 3HHx fraction compared to the wild-type enzyme, as well as recognize and incorporate other monomer units, such as 3-hydroxy-4-methylvalerate (3H4MV) (Tanadchangsaeng et al., 2009) and 3-hydroxy-2-methylbutyrate (3H2MB) (Watanabe et al., 2015). In addition, the molecular weight of P(3HB) synthesized by PhaCAcNSDG was higher than that of the wild-type enzyme (Tsuge et al., 2007b). These properties of PhaCAcNSDG variant are desirable for the development of PHA as an industrial biomaterial, making it a promising biocatalyst.
The partial crystal structures for several PhaCs have been solved (Wittenborn et al., 2016; Chek et al., 2017; Kim et al., 2017; Chek et al., 2020). The differences in the catalytic properties of these enzymes can be possibly due to their different structures (Chek et al., 2019). Although the crystal structure of PhaCAc has not yet been solved, a basic understanding of the enzymatic capability of PhaCAc could be elucidated using in silico homology modeling (Harada et al., 2021). Additionally, the use of structural information, namely the comparison of the amino acid residues that constitute the substrate-binding pocket of PhaCs, led to the generation of further engineered PhaCAcs (Harada et al., 2021).
PhaCAc and its PhaCAcNSDG variant are biocatalysts that produce PHA polymers with desirable material properties; however, the number of other naturally occurring PhaCs with broad substrate specificities are limited, hindering the development and commercial mass production of desirable PHAs. Thus, it is necessary to identify other novel PhaCs with broad substrate specificities to enable industrial-scale production of PHA copolymers to completely replace petroleum-based plastics. PhaCs, which can synthesize high-molecular-weight PHA, is essential to produce PHA as practical materials. The currently available PhaCAc is highly sensitive to ethanol (Hiroe et al., 2015), which is a metabolite of some bacteria, including Escherichia coli, and functions as a chain transfer agent to terminate polymerization reactions (Tsuge, 2016), resulting in the synthesis of relatively low-molecular-weight PHA when using E. coli as a production host. These low-molecular-weight PHA polymers have less desirable physical properties than their high-molecular-weight counterparts. Despite the unique ability of PhaCAc to polymerize various monomers, the relatively low molecular weight of PHA produced in recombinant E. coli using this enzyme has room for improvement.
In this study, to explore novel PhaCs with high polymerization ability and broad substrate specificity, four new PhaCs were identified by a bioinformatics approach using the PhaCAc amino acid sequence as a template for a basic local alignment search tool (BLAST) and included PhaCs from the bacteria Ferrimonas marina, Plesiomonas shigelloides, Shewanella pealeana, and Vibrio metschnikovii. PhaC proteins were individually expressed in E. coli LSBJ to synthesize P(3HB) and 3HB-based copolymers containing 3HHx, 3H4MV, 3H2MB, and 3-hydroxypivalate (3HPi) units. Furthermore, the effects of mutagenesis on polymerization activity and substrate specificity in the highest-performing PhaC enzyme were investigated.
A BLAST-protein (BLASTP) search was performed against the protein sub-sections of the National Center for Biotechnology Information (NCBI) and DNA Data Bank of Japan (DDBJ) databases using the PhaCAc amino sequence as a template (Accession No. BAA21815) (Altschul et al., 1990). PhaCs with more than 85% similarity index and an identity index of 50%–60% in the BLASTP search were targeted as potential PhaCs with broad substrate specificities. Among the various PhaCs from different organisms that satisfied the criteria in the BLASTP search, four PhaCs were selected based on the diversity of the N-terminal region for further evaluation. Phylogenetic analyses were performed using the maximum likelihood method in MEGA11 (Tamura et al., 2021) and the protein sequences were aligned using ClustalW. This analysis involved six amino acid sequences: PhaCAc, four newly selected PhaCs from BLASTP, and PhaC from Ralstonia eutropha (WP_011615085) as an outgroup.
Four PhaC amino acid sequences were chosen based on the BLASTP search results. These phaC genes were chemically synthesized with optimized codon usage in E. coli by Eurofins Genomics Co. Ltd. (Tokyo, Japan) for plasmid construction and evaluation. E. coli LSBJ, a fadB fadJ double-deletion strain of E. coli LS5218 [fadR601, atoC (Con)] (Tappel et al., 2012a), was used as the host strain for PHA biosynthesis. This strain is an ideal host for non-native PHA production because of its ability to take up a wide variety of substrates to be incorporated into PHA homo- and copolymers, and bench-level scale-up methodologies available for overall production (Tappel et al., 2012b; Levine et al., 2016; Pinto et al., 2016; Fadzil et al., 2018; Furutate et al., 2021; Scheel et al., 2021). A broad-host-range plasmid pBBR1MCS-2 (Kovach et al., 1995) harboring the genes encoding the PhaCs to be evaluated, the lac promoter region, the (R)-specific enoyl-CoA hydratase gene from A. caviae (phaJAc), the 3-ketothiolase gene (phaA) from Ralstonia eutropha H16, and the acetoacetyl-CoA reductase gene (phaB) from R. eutropha H16, termed pBBR1-phaCsABReJAc, was used for the expression of PhaCs (Supplementary Figure S1). For phaAB expression, the R. eutropha pha promoter and terminator regions were located upstream and downstream of their genes, respectively. To enhance the supply of 3HHx, 3H4MV, and 3H2MB monomers, the plasmid pTTQ-PCT (Furutate et al., 2017) containing the propionyl-CoA transferase (PCT) gene from Megasphaera elsdenii (pct) (Taguchi et al., 2008) was introduced into the E. coli LSBJ strain (Supplementary Figure S1).
Initially, recombinant E. coli LSBJ was incubated overnight at 37°C with reciprocal shaking (160 rpm) in a 50 mL baffle flask containing 20 mL of lysogeny broth (LB) medium as a seed culture. The LB medium contained 10 g/L Bacto-tryptone (Difco Laboratories, Detroit, MI, United States), 5 g/L Bacto-yeast extract (Difco Laboratories), and 10 g/L NaCl. For plasmid maintenance throughout the initial incubation period, 50 mg/L of kanamycin and 50 mg/L of carbenicillin were added.
Inoculations for PHA production were started with 5 mL of seed culture added to 500 mL shake flasks containing 95 mL of modified M9 medium (Furutate et al., 2021) (final volume:100 mL and 5% inoculum). The modified M9 medium comprised of 17.1 g/L Na2HPO4·12H2O, 3 g/L KH2PO4, 0.5 g/L NaCl, 2 mL of 1 M MgSO4·7H2O, 0.1 mL of 1 M CaCl2, and 2.5 g/L Bacto-yeast extract. For plasmid maintenance during PHA production, 50 mg/L of kanamycin and 50 mg/L of carbenicillin were added. Additionally, 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was used to induce phaJ and pct gene expression. The P(3HB) homopolymer was synthesized from 20 g/L glucose, which was added at the beginning of the culture at 30°C for 72 h. For the synthesis of 3HB-based copolymers, the total incubation time was set to 76 h, in which an initial step for 4 h at 30°C with reciprocal shaking (130 rpm) was performed before the addition of IPTG, precursors, and glucose, and further cultured for 72 h. Hexanoic acid, 4-methylvaleric acid, trans-2-methylbut-2-enoic acid (tiglic acid), and 2,2-dimethyl-3-hydroxypropionic acid (3-hydroxypivalic acid), which had previously been converted to their respective sodium salts, were used as precursors for the 3HHx, 3H4MV, 3H2MB, and 3HPi units, respectively (Füchtenbusch et al., 1998; Tanadchangsaeng et al., 2009; Watanabe et al., 2015). These precursors are known to inhibit cell growth, and a high concentration of glucose can repress phaJ and pct genes, otherwise induced by IPTG. Thus, lower concentrations of glucose and the precursors were added intermittently to the culture medium (at 4, 28, and 52 h). A total of 7.5 g/L glucose (2.5 g/L each time) and 0.6 g/L precursors (0.2 g/L each time) were added throughout the main incubation period. Finally, cells were harvested by centrifugation and lyophilized for further analysis. The relationship between the precursors used and biosynthesized polymers is shown in Figure 1.
To construct mutated phaCPs, a substitution (N175G) was introduced into the gene by overlap extension PCR (Supplementary Figure S2) (Warrens et al., 1997). The primers for amino acid substitution were designed and chemically synthesized as follows:5′-GGCGGCCGCTCTAGAACTAGTGGATCCCGGGGCAA-3′ and 5′- CACTAAGTTTTGACCGCCGTTCTCCAAGGT-3′ for an amplification of the 1.4-kb fragment, 5′-GCGCTTGGAGGCCGGCACCG-3′ and 5′- GTGACCTTGGAGAACGGCGGTCAAAACTTA-3′ for an amplification of the 2.3-kb fragment. The underlined sequence in the primer indicates the codon used to replace Asn175 (AAT) with Gly (GGC). The resulting plasmid carrying the mutated gene was introduced into E. coli LSBJ along with pTTQ-PCT for PHA biosynthesis analysis.
The dry cell weight was gravimetrically measured after centrifuging the culture medium at 6,000 × g for 10 min at room temperature three times (once for collecting the cells, discarding the medium, and twice to wash away the remaining salts with water) and lyophilized for approximately 3 days.
PHA content, PHA yield, and 3HA monomer composition were determined by gas chromatography (GC) using a Shimadzu GC-2014s instrument (Shimadzu, Kyoto, Japan) with a flame ionization detector. Lyophilized cells were methanolyzed to convert PHA into 3HA-methyl ester constituents in the presence of 15% sulfuric acid for GC analysis. The methanolysis reaction was carried out at 100°C for 140 min, except for 3H2MB- and 3HPi-containing polymers, for which the reaction time was set to 8 h to increase the reaction yield. The methanolyzed samples were allowed to cool to room temperature, and 1 mL of deionized water was added to separate the polar components from the non-polar components. The non-polar fraction containing 3HA-methyl ester was filtered, and an equal volume of chloroform solution containing 0.1% (w/v) methyl-n-octanoate as an internal standard was added to prepare the final sample for GC analysis. The samples were injected through the GC capillary column InertCap 1 (30 m × 0.25 mm, GL Science, Tokyo, Japan). The column temperature was initially set at 90°C for 2 min, increased to 110°C at a rate of 5°C/min, and then increased to 280°C at a rate of 20°C/min. The signal peak areas obtained were calculated for the total PHA content and 3HA monomer composition.
The molecular weight of P(3HB) synthesized using various PhaC enzymes was determined by gel permeation chromatography (GPC) using a Shimadzu Nexera GPC system with an RI-504 refractive index detector (Shodex, Tokyo, Japan) equipped with two KF-406 LHQ joint-columns (at 40°C, Shodex, Tokyo, Japan). Chloroform was used as the mobile phase at a flow rate of 0.3 mL/min. The sample concentration and injection volume were set at 1 mg/mL and 10 μL, respectively. Polystyrene standards with low polydispersity were also analyzed as reference standards to construct a calibration curve.
A BLASTP search was performed against the protein sub-sections of the NCBI and DDBJ databases using the amino acid sequence of PhaCAc, the first enzyme characterized by the natural copolymerization of 3HB and 3HHx monomers to PHA copolymers. Four PhaCs from the bacteria Ferrimonas marina (Katsuta et al., 2005), Plesiomonas shigelloides (Ferguson and Henderson 1947; Janda et al., 2016), Shewanella pealeana (Leonardo et al., 1999), and Vibrio metschnikovii (Lee et al., 1978) were selected for further evaluation, because of the diversity of the N-terminal region such as positions 149 and 171 in PhaCAc. These PhaCs were identified as Class I PHA synthases, which have a high potential for synthesizing scl-mcl PHA copolymers in a manner similar to PhaCAc, based on their homology. Although these bacteria were discovered long ago, their ability to produce PHA has not yet been studied.
A comparison with the amino acid sequence of PhaCAc revealed that the four PhaC enzymes identified in this study shared 85%–91% similarity and approximately 55% identity with PhaCAc (Table 1). Multiple sequence alignment of PhaCs is shown in Figure 2. All new PhaCs have a PhaC box sequence at the active site, which is typically described as G-X-C-X-G-G (where X is an arbitrary amino acid), and cysteine (Cys319 in PhaCAc) is the active center (Nambu et al., 2020). In PhaCAc, the active sites Cys319, Asp475, and His503 have been proposed to form a catalytic triad (Tsuge et al., 2007a), which are all conserved in the newly identified PhaC enzymes. In contrast, PhaC from P. shigelloides has a primary sequence of approximately 30 amino acid residues greater than that of others and exhibits relatively low sequence homology in the C-terminal region. The phylogenetic tree shown in Figure 3 indicates that PhaC from F. marina is closely related to PhaCAc, whereas PhaC enzymes from S. pealeana and V. metchniskovii are evolutionarily distinct. PhaC from P. shigelloides is neither closely related nor evolutionarily distant from PhaCAc. To the best of our knowledge, no study has explored PhaC enzymes isolated from these bacteria for PHA production. Thus, genes encoding the four PhaC enzymes were chemically synthesized with optimized codon usage in E. coli. The DNA sequences are included in Supplementary Information.

FIGURE 2. Multiple sequence alignment of PHA synthases (PhaCs) from Aeromonas caviae, Ferrimonas marina, Plesiomonas shigelloides, Shewanella pealeana, and Vibrio metschnikovii. The active site residues of PhaCAc, cysteine (C319), aspartic acid (D475), and histidine (H503) are highlighted in blue. For the PhaCAcNSDG variant, the positions of two amino acid substitutions (N149 replaced with S and D171 replaced with G) are highlighted in orange.
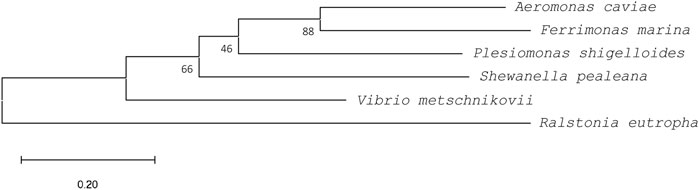
FIGURE 3. A phylogenetic tree of PhaCs rooted by outgroup (PhaC from Ralstonia eutrophus, WP_011615085). Sequences were aligned using ClustalW, and the phylogenetic tree was generated using MEGA11 software. PhaCs from Aeromonas caviae (BAA21815), Ferrimonas marina (WP_067661665), Plesiomonas shigelloides (WP_116546999), Shewanella pealeana (WP_012154995), and Vibrio metschnikovii (WP_154168902) were used. Bootstrap values (expressed as percentages of 1,000 replications) are shown at the branch points. Scale bar = 0.2 substitution per amino acid position.
The biosynthesis of P(3HB) from 20 g/L glucose using one of the four PhaC enzymes is summarized in Table 2. P(3HB) accumulation ranging from 38.4 wt% to 54.2 wt% was achieved using the new PhaC enzymes, which was comparable to PhaCAc and its variant PhaCAcNSDG. Thus, all newly identified PhaC enzymes showed great potential as biocatalysts for P(3HB) production. However, PhaCs from P. shigelloides (PhaCPs) showed the highest P(3HB) accumulation among the wild-type PhaCs tested.
The molecular weight is a crucial aspect in determining the suitability of a material for various commercial uses (Sudesh et al., 2000). The weight-average molecular weight (Mw) is more closely related to material properties than the number-average molecular weight (Mn). For PHA, ultrahigh molecular weight polymers can form strong fibers (Tsuge, 2016), thus meeting the requirements for practical use. In addition, a low polydispersity index (PDI) (Tsuge, 2016) also plays a significant role in determining the suitability of PHA for specific applications. However, not all PhaC enzymes can synthesize PHAs with high Mw and low PDI. In this study, PhaCPs synthesized P(3HB) with an ultrahigh Mw, which exceeded 3 × 106, with a relatively low PDI below 1.5 (Table 2). Moreover, the other two identified PhaC enzymes from F. marina and S. pealeana could also synthesize P(3HB) with Mw of approximately 2 × 106 with PDIs ranging from 1.3 to 1.5. PhaCVm from V. metschnikovii proved to be an exception, with PDI >2.5. The currently available PhaCAc is highly sensitive to ethanol (Hiroe et al., 2015), which is a metabolite of some bacteria, including E. coli, and functions as a chain transfer agent to terminate polymerization reactions (Tsuge, 2016), resulting in the synthesis of relatively low-molecular-weight PHA. The new PhaCs reported in this study may be less sensitive toward ethanol, thereby producing PHA with high Mw and low PDI. These new PhaC enzymes, especially PhaCPs, exhibited superior Mw and PDI values compared with PhaCAc and its NSDG variant, which could benefit PHA processing and material properties.
The new PhaC enzymes were evaluated for their substrate specificities alongside PhaCAc and its NSDG variant for incorporating 3HHx, 3H4MV, 3H2MB, and 3HPi monomers. Biosynthesis was performed using four precursors (hexanoic acid, 4-methylvaleric acid, tiglic acid, and 3-hydroxypivalic acid) in the presence of glucose (Figure 1). These precursors are toxic to cells, thus inhibiting cell growth and subsequently lowering PHA accumulation in bacteria. As PHA production is associated with cell growth (Sudesh et al., 2000), it is imperative to eliminate or reduce the risk of toxicity induced by such precursors. Therefore, the precursors were introduced into the culture medium after 4 h, once substantial cell growth was achieved, mainly for better tolerance (Furutate et al., 2021). Meanwhile, a high glucose concentration can cause catabolic repression of phaJ and pct genes induced by IPTG (Furutate et al., 2021); thus, the glucose concentration was maintained at a minimum to promote cell growth only. Glucose and its precursors were intermittently added to allow for better uptake of the second monomer, with no or fewer unanticipated effects on the cells. The details of the biosynthesis results are summarized in Tables 3–6.
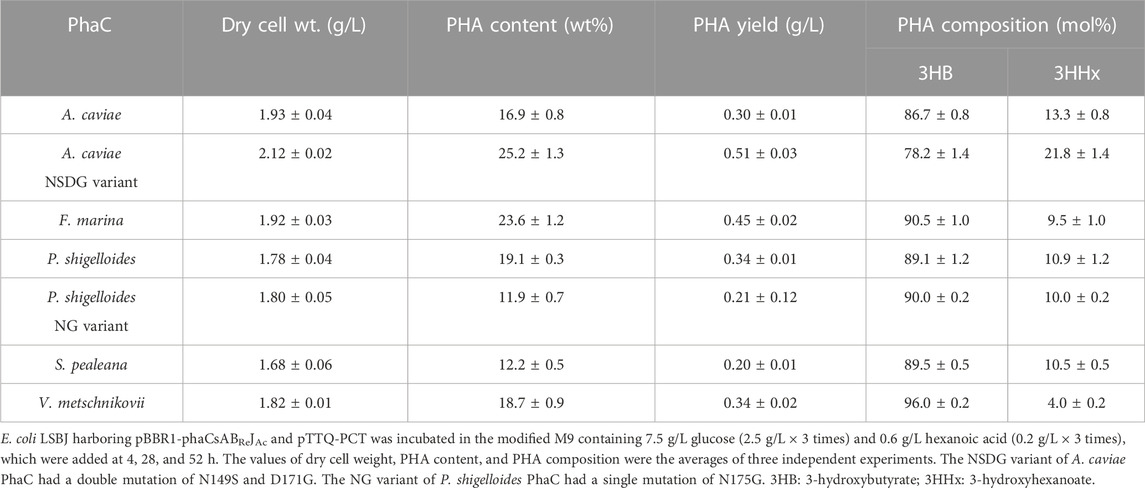
TABLE 3. Biosynthesis of P (3HB-co-3HHx) by E. coli LSBJ expressing various PhaCs from glucose and hexanoic acid.
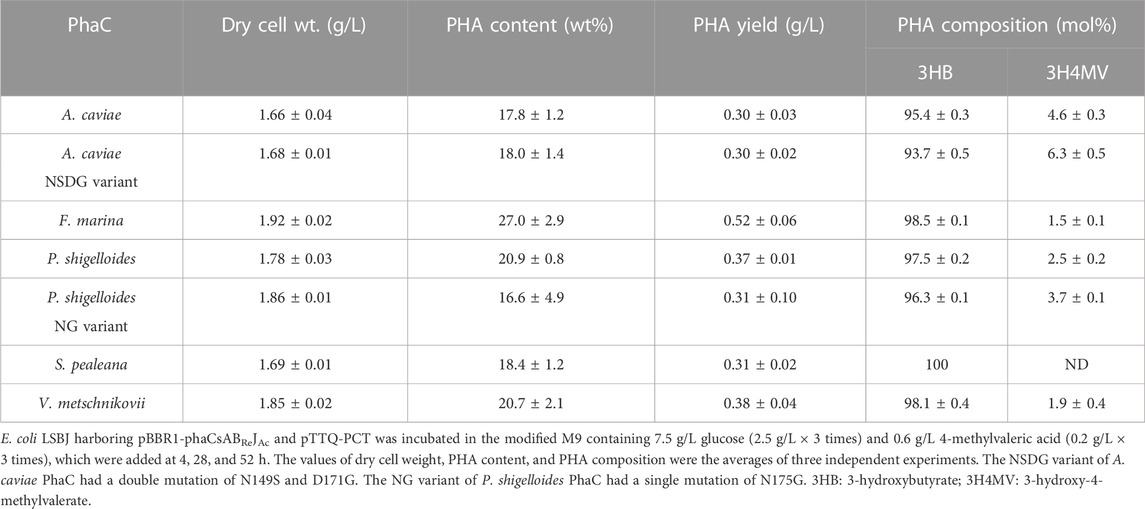
TABLE 4. Biosynthesis of P (3HB-co-3H4MV) by E. coli LSBJ expressing various PhaCs from glucose and 4-methylvaleric acid.
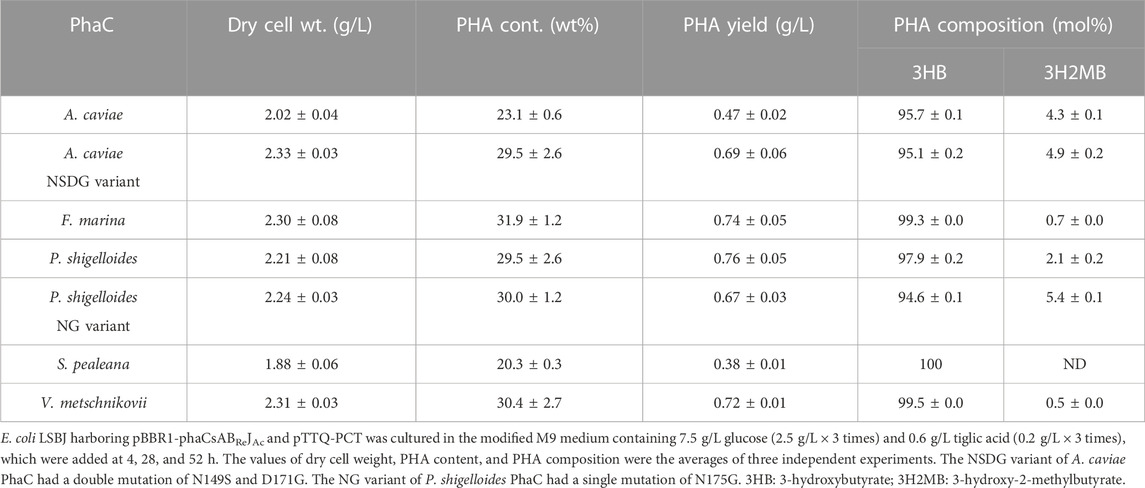
TABLE 5. Biosynthesis of P (3HB-co-3H2MB) by E. coli LSBJ expressing various PhaCs from glucose and tiglic acid.
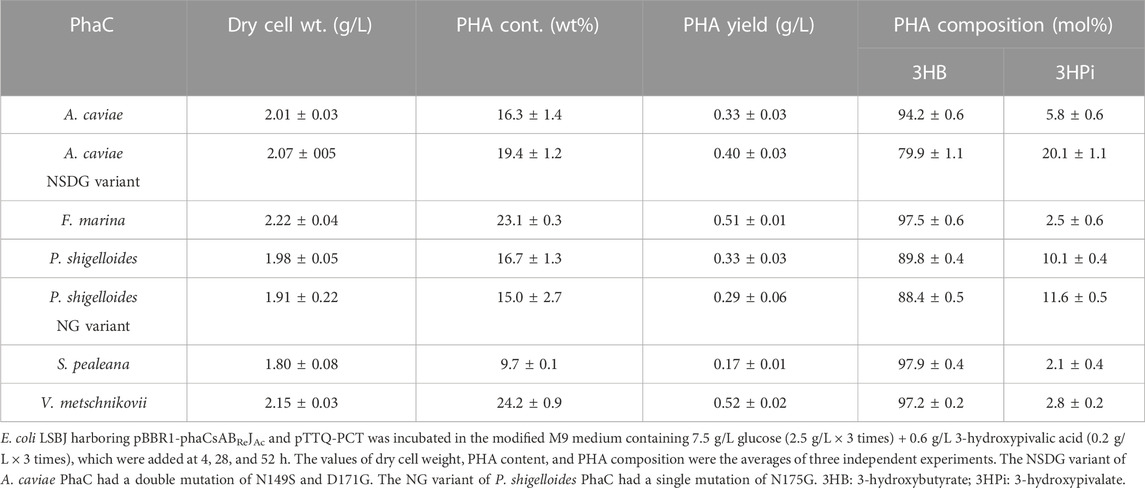
TABLE 6. Biosynthesis of P (3HB-co-3HPi) by E. coli LSBJ expressing various PhaCs from glucose and 3-hydoxypivalic acid.
All PhaCs, except PhaC from S. pealeana (PhaCSp), were able to incorporate all targeted monomers (3HHx, 3H4MV, 3H2MB, and 3HPi). PhaCSp copolymerized 3HB with 3HHx or 3HPi, but not 3H4MV or 3H2MB. The number of PhaC enzymes with broad substrate specificity is scarce; thus, the new PhaC enzymes reported in this study are highly intriguing for future studies. Furthermore, PHAs containing α-carbon methylated units are potentially attractive bio-based materials (Füchtenbusch et al., 1998; Furutate et al., 2021); thus, PhaCs with the ability to polymerize 3H2MB and 3HPi are of great interest. PhaCPs demonstrated superior performance in the polymerization of 3HPi to PhaCAc. Additionally, all PhaCPs-expressing strains showed higher PHA content than PhaCAc-expressing strains. Therefore, the potential of PhaCPs was further explored using site-directed mutagenesis.
In vitro evolution of PhaC is a powerful approach for enhancing the productivity and quality of PHA (Kichise et al., 2002; Taguchi and Doi, 2004). For instance, PhaCAcNSDG, a variant of PhaCAc, exhibits enhanced performance (such as production yield and substrate specificity) compared to that of the wild-type enzyme (Tsuge et al., 2007b). In addition, various studies have proven the efficacy of PhaC engineering in PHA production towards the formation of super biocatalysts for tailor-made PHAs (Taguchi and Doi, 2004; Nomura and Taguchi, 2007). Therefore, PhaCPs, which exhibited the best performance among the new PhaCs, were selected for site-directed mutagenesis to study their potential positive effects on PHA production. Considering that the double mutation of PhaCAcNSDG, amino acid substitutions of N149S and D171G drastically enhanced the performance of the enzyme (Tsuge et al., 2007b; Harada et al., 2021), similar efforts were adopted to generate a PhaCPs variant. According to the alignment (Figure 2), PhaCPs naturally contain a serine residue at the corresponding position of 149 in PhaCAcNSDG. Thus, a single amino acid substitution was performed in PhaCPs in which asparagine 175 was changed to glycine (N175G). The resultant variant was termed PhaCPsNG, and its PHA production ability was examined.
Interestingly, PhaCPsNG showed enhanced P(3HB) synthesis, while maintaining a high molecular weight (Table 2). PhaCPsNG exhibited enhanced activity for the incorporation of the α-methylated monomer 3H2MB compared with the parent enzyme and PhaCAcNSDG (Table 5). This indicates the potential of PhaCPsNG to surpass the currently best-performing enzyme (PhaCAcNSDG) for the incorporation of the 3H2MB monomer. Moreover, PhaCPsNG was shown to have a better ability to incorporate 3H4MV and 3HPi than the parent enzyme but less so than PhaCAcNSDG (Table 4 and Table 6). Finally, PhaCPsNG exhibited an almost similar level of 3HHx incorporation as the parent enzyme, which was inferior to PhaCAcNSDG (Table 3).
In conclusion, four Class I PhaC enzymes from different bacteria were identified using BLASTP and were characterized for PHA production. To the best of our knowledge, this is the first report to characterize PhaC enzymes from F. marina, P. shigelloides, S. pealeana, and V. metschnikovii. These PhaCs exhibited a relatively high potential for polymerizing P(3HB) in recombinant E. coli. PhaC enzymes identified in this study, with the exception of PhaCSp from S. pealeana, were able to incorporate all the targeted monomers, namely 3HHx, 3H4MV, α-carbon methylated 3H2MB, and α-carbon dimethylated 3HPi. Among the four new PhaCs, PhaCPs from P. shigelloides displayed the best performance; thus, we attempted to further improve their attributes through protein engineering. The resultant variant PhaCPsNG exhibited superior capability in polymerizing the 3H2MB monomer compared to PhaCAc and its NSDG variant. Furthermore, PhaCPsNG showed the enhanced synthesis of P(3HB) with ultrahigh molecular weight and low PDI. Finally, these newly identified PhaC enzymes show great versatility, suggesting their potential as workhorse enzymes for the industrial-scale production of 3HB-based copolymers.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
RS, MM, YM, SM, CN, ST, HA, and TT jointly conceived the study. RS, MM, YM, and SM performed the experiments. RS wrote the manuscript in consultation with CN, ST, HA, and TT. All authors read and approved the final manuscript.
JSPS KAKENHI Grant Number 21H03640.
The authors thank S. Ozawa (Teijin Co., Tokyo, Japan) and T. Yamamoto (Teijin Co., Tokyo, Japan) for helpful discussions. The authors also thank the Biomaterials Analysis Division of the Tokyo Institute of Technology for the DNA sequencing analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1114946/full#supplementary-material
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi:10.1016/S0022-2836(05)80360-2
Anderson, A. J., and Dawes, E. (1990). Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54, 450–472. doi:10.1128/mr.54.4.450-472.1990
Chek, M. F., Hiroe, A., Hakoshima, T., Sudesh, K., and Taguchi, S. (2019). PHA synthase (PhaC): Interpreting the functions of bioplastic-producing enzyme from a structural perspective. Appl. Microbiol. Biotechnol. 103, 1131–1141. doi:10.1007/s00253-018-9538-8
Chek, M. F., Kim, S. Y., Mori, T., Arsad, H., Samian, M. R., Sudesh, K., et al. (2017). Structure of polyhydroxyalkanoate (PHA) synthase PhaC from Chromobacterium sp. USM2, producing biodegradable plastics. Sci. Rep. 7, 5312. doi:10.1038/s41598-017-05509-4
Chek, M. F., Kim, S. Y., Mori, T., Tan, H. T., Sudesh, K., and Hakoshima, T. (2020). Asymmetric open-closed dimer mechanism of polyhydroxyalkanoate synthase PhaC. iScience 23, 101084. doi:10.1016/j.isci.2020.101084
Doi, Y., Kitamura, S., and Abe, H. (1995). Microbial synthesis and characterization of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate). Macromolecules 28, 4822–4828. doi:10.1021/ma00118a007
Fadzil, F. I. M., Mizuno, S., Hiroe, A., Nomura, C. T., and Tsuge, T. (2018). Low carbon concentration feeding improves medium-chain-length polyhydroxyalkanoate production in Escherichia coli strains with defective β-oxidation. Front. Bioeng. Biotechnol. 6, 178. doi:10.3389/fbioe.2018.00178
Ferguson, W. W., and Henderson, N. D. (1947). Description of strain C27: A motile organism with the major antigen of Shigella sonnei phase I. J. Bacteriol. Res. 54, 179–181. doi:10.1128/jb.54.2.179-181.1947
Füchtenbusch, B., Fabritius, D., Wältermann, M., and Steinbüchel, A. (1998). Biosynthesis of novel copolyesters containing 3-hydroxypivalic acid by Rhodococcus ruber NCIMB 40126 and related bacteria. FEMS Microbiol. Lett. 159, 85–92. doi:10.1111/j.1574-6968.1998.tb12845.x
Furutate, S., Kamoi, J., Nomura, C. T., Taguchi, S., Abe, H., and Tsuge, T. (2021). Superior thermal stability and fast crystallization behavior of a novel, biodegradable α-methylated bacterial polyester. NPG Asia Mater 13, 31. doi:10.1038/s41427-021-00296-x
Furutate, S., Nakazaki, H., Maejima, K., Hiroe, A., Abe, H., and Tsuge, T. (2017). Biosynthesis and characterization of novel polyhydroxyalkanoate copolymers consisting of 3-hydroxy-2-methylbutyrate and 3-hydroxyhexanoate. J. Polym. Res. 24, 221. doi:10.1007/s10965-017-1392-3
Harada, K., Kobayashi, S., Oshima, K., Yoshida, S., Tsuge, T., and Sato, S. (2021). Engineering of Aeromonas caviae polyhydroxyalkanoate synthase through site-directed mutagenesis for enhanced polymerization of the 3-hydroxyhexanoate unit. Front. Bioeng. Biotechnol. 9, 627082. doi:10.3389/fbioe.2021.627082
Hiroe, A., Shiraishi, M., Mizuno, K., and Tsuge, T. (2015). Behavior of different polyhydroxyalkanoate synthases in response to the ethanol level in Escherichia coli cultures. Polym. J. 47, 767–770. doi:10.1038/pj.2015.53
Janda, J. M., Abbott, S. L., and McIver, C. J. (2016). Plesiomonas shigelloides revisited. Clin. Microbiol. Rev. 29, 349–374. doi:10.1128/CMR.00103-15
Katsuta, A., Adachi, K., Matsuda, S., Shizuri, Y., and Kasai, H. (2005). Ferrimonas marina sp. nov. Int. J. Syst. Evol. Microbiol. 55, 1851–1855. doi:10.1099/ijs.0.63689-0
Kichise, T., Taguchi, S., and Doi, Y. (2002). Enhanced accumulation and changed monomer composition in polyhydroxyalkanoate (PHA) copolyester by in vitro evolution of Aeromonas caviae PHA synthase. Appl. Environ. Microbiol. 68, 2411–2419. doi:10.1128/AEM.68.5.2411-2419.2002
Kim, J., Kim, Y. J., Choi, S. Y., Lee, S. Y., and Kim, K. J. (2017). Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms. Biotechnol. J. 12, 1600648. doi:10.1002/biot.201600648
Kobayashi, G., Shiotani, T., Shima, Y., and Doi, Y. (1994). “Biosynthesis and characterization of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from oils and fats by Aeromonas sp. OL-338 and Aeromonas sp. FA-440,” in Biodegradable plastics and polymers. Editors Y. Doi, and K. Fukuda (Amsterdam: Elsevier), 410–416. doi:10.1016/b978-0-444-81708-2.50044-0
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi:10.1016/0378-1119(95)00584-1
Lee, J. V., Donovan, T. J., and Furniss, A. L. (1978). Characterization, taxonomy, and emended description of Vibrio metschnikovii. Vibrio metschnikovii Int. J. Syst. Evol. Microbiol. 28, 99–111. doi:10.1099/00207713-28-1-99
Lehrle, R., and Williams, R. J. (1994). Thermal degradation of bacterial poly(hydroxybutyric acid): Mechanisms from the dependence of pyrolysis yields on sample thickness. Macromolecules 27, 3782–3789. doi:10.1021/ma00092a017
Lenz, R. W., and Marchessault, R. H. (2005). Bacterial polyesters: Biosynthesis, biodegradable plastics and biotechnology. Biomacromolecules 6, 1–8. doi:10.1021/bm049700c
Leonardo, M. R., Moser, D. P., Barbieri, E., Brantner, C. A., MacGregor, B. J., Paster, B. J., et al. (1999). Shewanella pealeana sp. nov., a member of the microbial community associated with the accessory nidamental gland of the squid Loligo pealei. Int. J. Syst. Evol. Microbiol. 49, 1341–1351. doi:10.1099/00207713-49-4-1341
Levine, A. C., Heberlig, G. W., and Nomura, C. T. (2016). Use of thiol-ene click chemistry to modify mechanical and thermal properties of polyhydroxyalkanoates (PHAs). Int. J. Biol. Macromol. 83, 358–365. doi:10.1016/j.ijbiomac.2015.11.048
Mierzati, M., Mizuno, S., and Tsuge, T. (2020). Biosynthesis and characterization of poly(3-hydroxybutyrate-co-2-hydroxyalkanoate) with different comonomer fractions. Polym. Degrad. Stabil. 178, 109193. doi:10.1016/j.polymdegradstab.2020.109193
Mizuno, K., Ohta, A., Hyakutake, M., Ichinomiya, Y., and Tsuge, T. (2010). Isolation of polyhydroxyalkanoate-producing bacteria from a polluted soil and characterization of the isolated strain Bacillus cereus YB-4. Polym. Degrad. Stabil. 95, 1335–1339. doi:10.1016/j.polymdegradstab.2010.01.033
Nambu, Y., Ishii-Hyakutake, M., Harada, K., Mizuno, S., and Tsuge, T. (2020). Expanded amino acid sequence of the PhaC box in the active center of polyhydroxyalkanoate synthases. FEBS Lett. 594, 710–716. doi:10.1002/1873-3468.13651
Nomura, C. T., and Taguchi, S. (2007). PHA synthase engineering toward superbiocatalysts for custom-made biopolymers. Appl. Microbiol. Biotechnol. 73, 969–979. doi:10.1007/s00253-006-0566-4
Pinto, A., Ciesla, J., Palucci, A., Sutliff, B., and Nomura, C. T. (2016). Chemically intractable no more: In vivo incorporation of “click”-ready fatty acids into poly-[(R)-3-hydroxyalkanoates in Escherichia coli. ACS Macro. Lett. 5, 215–219. doi:10.1021/acsmacrolett.5b00823
Rehm, B. H. (2003). Polyester synthases: Natural catalysts for plastics. Biochem. J. 376, 15–33. doi:10.1042/bj20031254
Scheel, R. A., Ho, T., Kageyama, Y., Masisak, J., McKenney, S., Lundgren, B. R., et al. (2021). Optimizing a fed-batch high-density fermentation process for medium-chain-length poly(3-hydroxyalkanoates) in Escherichia coli. 9, 134. Front. Bioeng. Biotechnol. 9, 618259. doi:10.3389/fbioe.2021.618259
Shimamura, E., Kasuya, K., Kobayashi, G., Shiotani, T., Shima, Y., and Doi, Y. (1994). Physical properties and biodegradability of microbial poly(3-hydroxybutyrate-co-3-hydroxyhexanoate). Macromolecules 27, 878–880. doi:10.1021/ma00081a041
Sivashankari, R., and Tsuge, T. (2021). “Development of polyhydroxyalkanoate (PHA) and its copolymers as a possible “cure” for the plastic pollution,” in Environmental and Microbial Biotechnology book series. Editor R. Prasad (Singapore: Springer), 59–79. doi:10.1007/978-981-15-5499-5_3
Sudesh, K., Abe, H., and Doi, Y. (2000). Synthesis, structure and properties of polyhydroxyalkanoates: Biological polyesters. Prog. Polym. Sci. 25, 1503–1555. doi:10.1016/S0079-6700(00)00035-6
Suzuki, M., Tachibana, Y., and Kasuya, K. I. (2021). Biodegradability of poly(3-hydroxyalkanoate) and poly(ε-caprolactone) via biological carbon cycles in marine environments. Polym. J. 53, 47–66. doi:10.1038/s41428-020-00396-5
Taguchi, S., and Doi, Y. (2004). Evolution of polyhydroxyalkanoate (PHA) production system by "enzyme evolution": Successful case studies of directed evolution. Macromol. Biosci. 4, 145–156. doi:10.1002/mabi.200300111
Taguchi, S., Yamada, M., Matsumoto, K. I., Tajima, K., Satoh, Y., Munekataet, M., et al. (2008). A microbial factory for lactate-based polyesters using a lactate-polymerizing enzyme. Proc. Natl. Acad. Sci. U.S.A. 105, 17323–17327. doi:10.1073/pnas.0805653105
Tamura, K., Stecher, G., and Kumar, S. (2021). Mega 11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi:10.1093/molbev/msab120
Tanadchangsaeng, N., Kitagawa, A., Yamamoto, T., Abe, H., and Tsuge, T. (2009). Identification, biosynthesis, and characterization of polyhydroxyalkanoate copolymer consisting of 3-hydroxybutyrate and 3-hydroxy-4-methylvalerate. Biomacromolecules10 2866-10, 2866–2874. doi:10.1021/bm900696c
Tappel, R. C., Kucharski, J. M., Mastroianni, J. M., Stipanovic, A. J., and Nomura, C. T. (2012b). Biosynthesis of poly[(R)-3-hydroxyalkanoate] copolymers with controlled repeating unit compositions and physical properties. Biomacromolecules 13, 2964–2972. doi:10.1021/bm301043t
Tappel, R. C., Wang, Q., and Nomura, C. T. (2012a). Precise control of repeating unit composition in biodegradable poly(3-hydroxyalkanoate) polymers synthesized by Escherichia coli. Escherichia coli J. Biosci. Bioeng. 113, 480–486. doi:10.1016/j.jbiosc.2011.12.004
Tsuge, T. (2016). Fundamental factors determining the molecular weight of polyhydroxyalkanoate during biosynthesis. Polym. J. 48, 1051–1057. doi:10.1038/pj.2016.78
Tsuge, T., Watanabe, S., Sato, S., Hiraishi, T., Abe, H., Doi, Y., et al. (2007a). Variation in copolymer composition and molecular weight of polyhydroxyalkanoate generated by saturation mutagenesis of Aeromonas caviae PHA synthase. Macromol. Biosci. 7, 846–854. doi:10.1002/mabi.200700023
Tsuge, T., Watanabe, S., Shimada, D., Abe, H., Doi, Y., and Taguchi, S. (2007b). Combination of N149S and D171G mutations in Aeromonas caviae polyhydroxyalkanoate synthase and impact on polyhydroxyalkanoate biosynthesis. FEMS Microbiol. Lett. 277, 217–222. doi:10.1111/j.1574-6968.2007.00958.x
Tsuge, T., Yano, K., Imazu, S. I., Numata, K., Kikkawa, Y., Abe, H., et al. (2005). Biosynthesis of polyhydroxyalkanoate (PHA) copolymer from fructose using wild-type and laboratory evolved PHA synthases. Macromol. Biosci. 5, 112–117. doi:10.1002/mabi.200400152
Warrens, A. N., Jones, M., and Lechler, R. I. (1997). Splicing by overlap extension by PCR using asymmetric amplification: An improved technique for the generation of hybrid proteins of immunological interest. Gene 186, 29–35. doi:10.1016/S0378-1119(96)00674-9
Watanabe, Y., Ishizuka, K., Furutate, S., Abe, H., and Tsuge, T. (2015). Biosynthesis and characterization of novel poly(3-hydroxybutyrate-co-3-hydroxy-2-methylbutyrate): Thermal behavior associated with α-carbon methylation. RSC Adv. 5, 58679–58685. doi:10.1039/C5RA08003G
Keywords: PHA synthases, broad substrate specificities, molecular weight, blast, copolymer
Citation: Sivashankari RM, Mierzati M, Miyahara Y, Mizuno S, Nomura CT, Taguchi S, Abe H and Tsuge T (2023) Exploring Class I polyhydroxyalkanoate synthases with broad substrate specificity for polymerization of structurally diverse monomer units. Front. Bioeng. Biotechnol. 11:1114946. doi: 10.3389/fbioe.2023.1114946
Received: 03 December 2022; Accepted: 06 February 2023;
Published: 21 February 2023.
Edited by:
Kumar Sudesh, University of Science Malaysia (USM), MalaysiaReviewed by:
Maciej Guzik, Jerzy Haber Institute of Catalysis and Surface Chemistry, Polish Academy of Sciences, PolandCopyright © 2023 Sivashankari, Mierzati, Miyahara, Mizuno, Nomura, Taguchi, Abe and Tsuge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takeharu Tsuge, dHN1Z2UudC5hYUBtLnRpdGVjaC5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.