
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol., 10 February 2023
Sec. Industrial Biotechnology
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1099999
 Anamika Singh1
Anamika Singh1 Virendra Kumar Yadav1
Virendra Kumar Yadav1 Rajendra Singh Chundawat1*
Rajendra Singh Chundawat1* Raya Soltane2
Raya Soltane2 Nasser S. Awwad3
Nasser S. Awwad3 Hala A. Ibrahium4,5
Hala A. Ibrahium4,5 Krishna Kumar Yadav6
Krishna Kumar Yadav6 Simona Ioana Vicas7*
Simona Ioana Vicas7*Plant Growth Promoting Rhizobacteria (PGPR) has gained immense importance in the last decade due to its in-depth study and the role of the rhizosphere as an ecological unit in the biosphere. A putative PGPR is considered PGPR only when it may have a positive impact on the plant after inoculation. From the various pieces of literature, it has been found that these bacteria improve the growth of plants and their products through their plant growth-promoting activities. A microbial consortium has a positive effect on plant growth-promoting (PGP) activities evident by the literature. In the natural ecosystem, rhizobacteria interact synergistically and antagonistically with each other in the form of a consortium, but in a natural consortium, there are various oscillating environmental conditions that affect the potential mechanism of the consortium. For the sustainable development of our ecological environment, it is our utmost necessity to maintain the stability of the rhizobacterial consortium in fluctuating environmental conditions. In the last decade, various studies have been conducted to design synthetic rhizobacterial consortium that helps to integrate cross-feeding over microbial strains and reveal their social interactions. In this review, the authors have emphasized covering all the studies on designing synthetic rhizobacterial consortiums, their strategies, mechanism, and their application in the field of environmental ecology and biotechnology.
Microorganisms are omnipresent in nature and are considered to be present with every living organism on the earth (Kirchman, 2018). From the vast literature, it has been found that microorganisms are present in all the parts of a plant for instance in the phylloplane areas, phyllosphere, or in the rhizosphere, the first two are mainly involved with the activities on the external surface of the plant and in severe cases may have a role in the plant diseases (Saeed et al., 2021). While the rhizospheric microbial species are of utmost importance due to their direct role in plant growth, health, and nutrition. In the last decade, rhizosphere-based microbial studies have gained huge attention among the scientific communities. The rhizosphere is an area where there are high biological and chemical activities occurs. In this zone, plant exudates are released which are used by the microbes as a source of energy (Prashar et al., 2013). The interaction of plants with microbes in the rhizospheric region helps in maintaining the soil fertility and health of the plant (Kumar et al., 2019a). One such group of bacteria which is present in the rhizosphere is planted growth-promoting bacteria (PGPR).
PGPR are free-living bacteria that have a direct role in the growth of the plant and rooting system among the plants (Hayat et al., 2010). It has also a role in nitrogen fixation, solubilization of insoluble phosphates, and secretion of several plant hormones. Due to the importance of PGPR in the ecosystem, it has drawn the attention of scientists in the last few years. Today PGPR is widely used in the field of agriculture, especially in biofertilizers, biogeochemical cycling of minerals, etc., (Shah et al., 2021a). From the pieces of literature, it has been found that only 2%–5% of rhizosphere bacteria are PGPR and they have been considered an important tool for sustainable agriculture (Antoun and Prevost (2005). From the various studies, it has been found that PGPR is adsorbed on the surface of the soil by the ion exchange process. Most plants are notable to use organic sources of elements so, these PGPR have a role in providing the plants with the inorganic form of elements. This maintains the fertility of the soil and is an important aspect of sustainable agriculture (Goswami et al., 2016). The PGPR bacteria have mainly members from the genera like Arthrobacter, Beijerinckia, Burkholderia, Derxia, Acetobacter, Acinetobacter, Klebsiella, Ochrobactrum, Bacillus, Enterobacter, Gluconacetobacter, Pantoae, Alcaligenes, Arthrobacter, Pseudomonas, Rhodococcus, Serratia, Azoarcus, Azospirillum, Azotobacter, Herbaspirillum, Stenotrophomonas, Lactobacillus, Paenobacillus, and Zoogloea (Vega-Celedón et al., 2021).
In the last few years, the trend toward PGPR studies has shown the role of microbial consortium in PGP activities. There are several reports where a vast variety of microorganisms are found in their hostile environment where they are interacting with other microorganisms intra and interspecifically. In the natural environment, 99% of microorganisms are found in the form of a microbial consortium (Dong et al., 2017). Till the 20th century, there are enormous studies that have shown that single microorganisms can play a beneficial role in plant growth, but in nature, it is noticeable that multiple species in microbial consortia can perform several beneficial functions for our ecosystem than a single microorganism. Interaction between PGPR and plants is synergistically driving benefits for the plant microbiome (Santos and Olivares 2021). Plants also promote PGPR growth through the production of various storage substances and also root exudates, which are used by PGPR for nutrition (Rahina et al., 2018). This tool is used to establish a symbiotic association between plants and PGPRs like mycorrhizal nitrogen-fixing root nodulation (Siddiqui et al., 2021) and this establishment can be endophytic or exophytic (Govindasamy et al., 2019). Once the association is established, PGPR shows benefits for plant growth, health, and traits through their direct and indirect mechanism, (Neshat et al., 2022). There are several reports which describe the role of monoculture in the case of PGPR but very few attempts were made to enhance the PGP activities by using microbial consortium. Some of the recent works done in this area are described below.
Fatima et al., 2022 reported the isolation of multifaced PGPR, from natural suppressive soils. Moreover, the investigators also studied the potential of such multifaced PGPR for bio-control activities and growth-promoting potential against the etiological agent of chickpea wilt (Fatima et al., 2022).
Recently a group of scientists reported the beneficial role of PGPR the in the remediation of nitrile pollutants from the cropland in order to enhance crop productivity. The investigators concluded that both nitrile and cyanide have a negative impact on plant health. Further investigators concluded that nitrile pollution can be managed by using nitrilase enzyme along with PGPR. The authors also suggested that the breakdown of nitrile by such activities will lead to the formation of nitrogen which could be utilized by the plant (Vaishnav et al., 2022).
Abbasi et al., 2022 developed a consortium of Streptomyces species and used them for the bell pepper fruits under controlled greenhouse conditions. The investigators reported that there was improved quality of bell pepper fruits, induced plant defencepriming, and moreover, the team led by Abbasi et al. (2022) reported that the Streptomyces consortium changed the microbial communities of the rhizosphere.
Samain et al., 2022 developed a PGPR microbial consortium using Paenibacillus sp. strain B2 and Arthrobacter sp. strain AA. The developed consortium was used for Zymoseptoria tritici and drought stress. The investigators reported that the developed consortium provided effective and durable systemic wheat-induced resistance (Samain et al., 2022).
By searching PGPR, bacterial consortium on science direct it was found that very less work is done in this area in comparison to other research activities. For instance, on searching PGPR, bacterial consortium, during the years 2017–2022, authors found that in 2017 about27 articles were published in 2018 about 34 articles, 65 articles in 2019, 95 articles in 2020, 126 in 2021, and 178 to date in 2022. The trend suggests that there is a gradual increase in PGPR-based research activities in the scientific community. In these last six years about a maximum were research articles i.e., 194, followed by book chapters i.e., 170, and review articles i.e., 108, the remaining were encyclopedias and other reports.
For the last six years researchers are working on designing rhizobacterial consortium for enhancing plant growth, their yield of production, soil health, as a biocontrol agent, and stabilizing the rhizobacterial interactions (Kamalnath et al., 2019; Swandi et al., 2019; Asyiah et al., 2020; Kumar et al., 2020; Shang and Liu 2020; Vaid et al., 2020; Redondo-Gómez et al., 2021; Karuppiah et al., 2022; Shabaan et al., 2022) In the current review, the authors have emphasized the role of rhizosphere microbes in PGP activities. Moreover, the authors have also highlighted the recent trends in PGPR-based research works. In addition to this, authors have emphasized the enhancement of the PGPR activities by using microbial consortium. The authors have emphasized the application of PGPR in environmental ecology and biotechnology. Such type of practices will lead to sustainable agriculture-led production of crops.
From the various pieces of literature, it has been found that there are basically two types of mechanisms associated with the beneficial effect of PGPR activities in the rhizosphere. For instance, it is the direct type and indirect type are shown below in Figure 1. Both the mechanism of PGPR activities is important and leads to sustainable agricultural crop production (Ahemad and Kibret 2014).
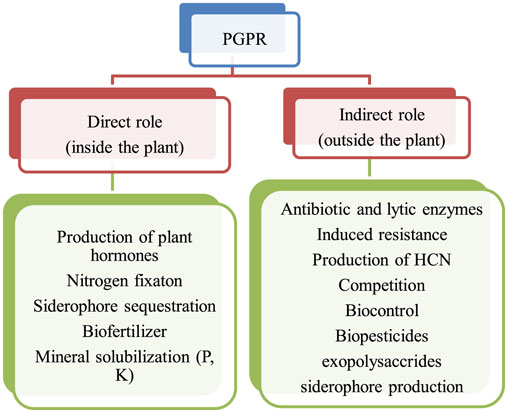
FIGURE 1. Types of PGPR activities in the rhizosphere (Kundan et al., 2015; Nath et al., 2018).
In direct mechanism, PGPR promotes plant growth by releasing various useful substances like phosphates, potassium, silicon, and zinc and the uptake of biologically fixed nitrogen, chelation of iron and other micronutrients and increase in available geospheric oxygen that stimulates aeriform biomass production, root development and also in stem elongation (Constantia and Ferniah 2020). PGPR also synthesizes phytohormones, such as gibberellins, auxins, indoleacetic acid, cytokinin, abscisic acid, and ethylene (Patel and Saraf 2017). The production of an enzyme 1-aminocyclopropane 1-carboxylate deaminase (ACC) reduces the level of ethylene in the roots of crops, therefore boosting the density and length of the roots (Singh and Jha 2017). Figure 2 shows the PGPR mechanism in the rhizosphere, while Figure 3 shows the movement of P in the soil. Figure 4 shows the various substances produced by phosphate-solubilizing bacteria (PSB), in the soil.
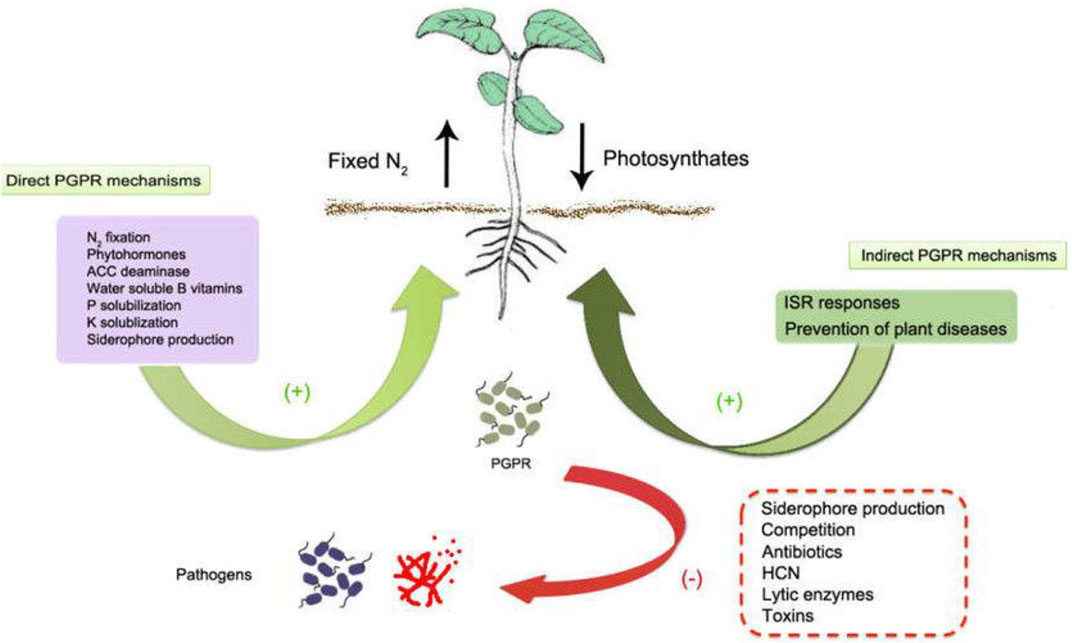
FIGURE 2. PGPR mechanism in the rhizosphere adopted from (Kamili et al., 2019; Nazir and Kamili, 2018).
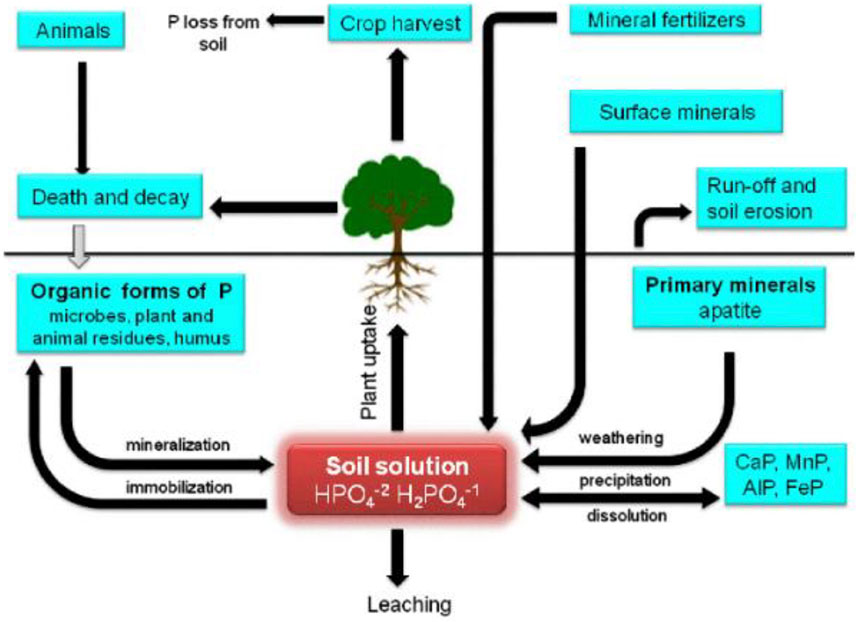
FIGURE 3. Movement of phosphorus in soil systems adopted from (Ahemad and Kibret 2014).
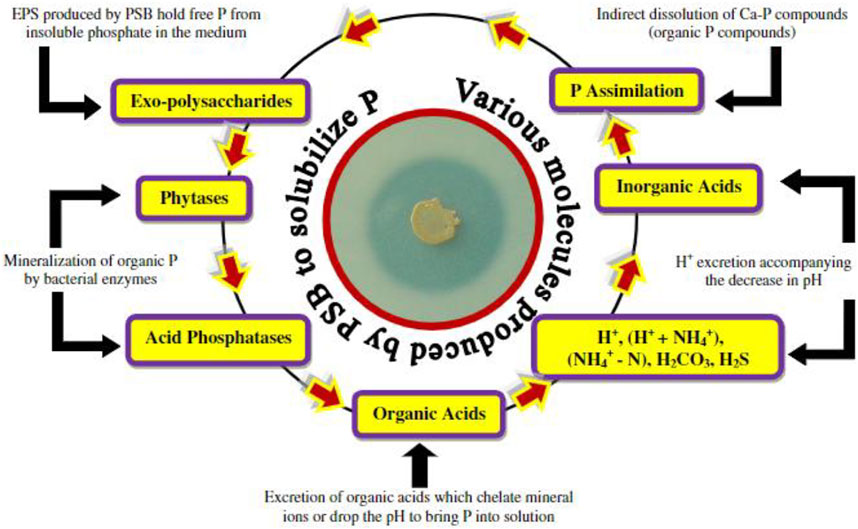
FIGURE 4. Different organic and inorganic substances produced by phosphate solubilizing bacteria for P, solubilization from soil adopted from (Ahemad and Kibret 2014).
In an indirect mechanism, PGPR is inducing systemic resistance and stimulates the innate resilience of the plant, and involves modification of the rhizosphere ecology (Adrees et al., 2019). PGPR release various substances that activate the protection mechanism of plants against various pathogenic microorganism and also promote the synthesis of physical and chemical barriers against abiotic stress, these substances are siderophore, pigments, antibiotics, organic acids, water-soluble vitamins, and different volatile organic compounds like monoterpene alcohols (Chandran et al., 2021). In return, plants allow PGPR to become more competitive in their niche colonization so that they can make interference with the quorum detection signal (Hartmann 2020) and also enables them to inhibit the formation of biofilms by pathogenic bacteria (Uruén et al., 2020). PGPR is also able to remediate contaminated soils (Vaishnav et al., 2022).
Because of their multifunctional benefits, PGPR is in leading demand in agroforestry management (Phale 2018). PGPR are key ecosystem service providers, they drive multifunctional processes and carry interaction with differential microbial communities (Lindemann 2019). This complex interconnected network of PGPRs responsible to maintain the transportation and circulation of energy and recirculation of resources to an entire ecosystem that affects soil microfauna and vegetational biome (Sylia et al., 2022).
Although both direct and indirect mechanism of PGPR equally provides benefits for plants’ health nitrogen fixation is the major mechanism of all PGPR (Mahmud et al., 2020), because all rhizobacteria have the capacity to fix atmospheric nitrogen, hence called rhizobacteria and there is the likelihood that each of the direct and indirect mechanism may present or absent in all rhizobacteria.
Most of the proteins, signaling molecules carries nitrogen as a key component. Nitrogen is also one of the crucial macronutrients for plant growth. Although nitrogen fertilizers are very costly in agricultural systems, thus affect the production of many crops. Exhaustive use of synthetic nitrogenous fertilizers in modern agriculture causes harmful effects on our natural environmental system. To preserve the health of the environment, PGPR is the resource of nitrogen and use as a natural biofertilizer to accomplish the requirement of nitrogen and other nutrients for plants without harming their productivity and the health of the environment (Monterio et al., 2021). Nitrogen is prominently present in the atmosphere in a gaseous form which is impassable to plants and animals. Assimilation of nitrogen by plants, atmospheric nitrogen is required to convert into ammonia, and this conversion is driven by nitrogenase enzymatic complex containing nitrogen-fixing microorganisms and the process called biological nitrogen fixation (Li et al., 2021). Rhizobia is the universal group of bacteria to fix nitrogen which is abundantly present in the rhizospheric area of soil (Shimonda et al., 2020). For sustainable production of crop production PGPR are the best source to provide nitrogen. Some examples of nitrogen-fixing microorganisms are, Actinobacteria, Micrococcales, Microbacteriaceae, Proteobacteria, Alphaproteobacteria, Rhodospirillales, Acetobacteraceae, Roseomonas (Zhang M. et al., 2020) Azotobacter vinelandii, Azospirillumzeae, Acetobacter diazotrophicus, Burkholderia tropica and Achromobacter insolitus etc., (Singh et al., 2020).
There are various examples where plant growth-promoting compounds are released by PGPR which are shown below in Table 1.
The PGPR activities could be influenced by various external factors for their enhanced activity. For instance, microbes could be used either in the form of monoculture or in the form of a consortium along with PGPR. This could enhance the activities performed by PGPR alone. There are several pieces of literary work thatshow the utilization of monoculture and microbial consortium for enhanced PGPR activities which of them are discussed below.
Microbial consortium (MC) carries flexibility to the oscillation of environmental factors such as temperature, pH, nutrient levels, and various toxic compounds (Pacheco et al., 2019). The diversity of metabolic pathways takes control of different members which can make it possible for the survival of consortia in fluctuating environmental conditions (Zhang and Zhang. 2022). The environmental fluctuations and metabolic diversity of the consortium are crucial to maintaining the desired functions of the PGP activities of rhizobacteria (Wang et al., 2022). For the improvement in plant growth, it is necessary to create a healthier soil environment, therefore evaluation of microbial consortium is the most important phase for their development (Kumar et al., 2019b).
In monoculture, a single strain of microorganism performs all the cell regulatory functions and a single cell carries all the burden of their metabolic pathways that leads to low efficiency and productivity of their beneficial activities (Backer et al., 2018). On the other hand, in consortia of two or more microorganisms, there is a division of labor of, therefore multiple cells in consortium exchange their levels of metabolism, energy, and function together like a cascade that results in high efficiency and productivity of their metabolic pathways (Roell et al., 2019). Monoculture suffers from the burden of the co-expression of heterologous proteins in a consortium and that results in achieving a maximum growth rate than monoculture (Liu et al., 2018). In the consortium, multiple cells are involved to perform the desired function and for that to achieve the maximum threshold of the prevailing performance of the consortium, it is required to optimize the ratio of the two cell types (Patowary et al., 2016).
For a better understanding of the benefits of the consortium over monoculture, in Figure 5 the degradation of lignocellulose is performed by a single microorganism and in Figures 6A, B by multiple microorganisms has been explained.
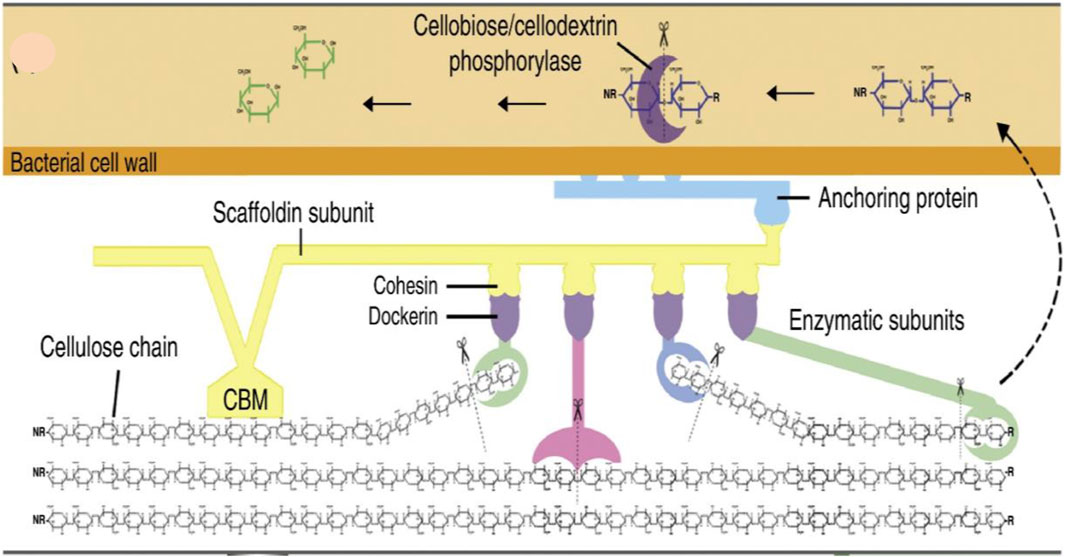
FIGURE 5. This diagram represents the cellular mechanism of lignocellulose degradation in the bacterial cell wall. Anaerobic degradation of lignocellulose is performed by a cellulosome complex which is attached to the cell wall of bacteria via an anchoring subunit. This complex consists of enzymes that are responsible for the hydrolysis of cellulose are and attached to a scaffolding subunit that anchors the enzymes and bacterial cells to the substrate by a carbohydrate-binding module (CBM) (Cragg et al., 2015).
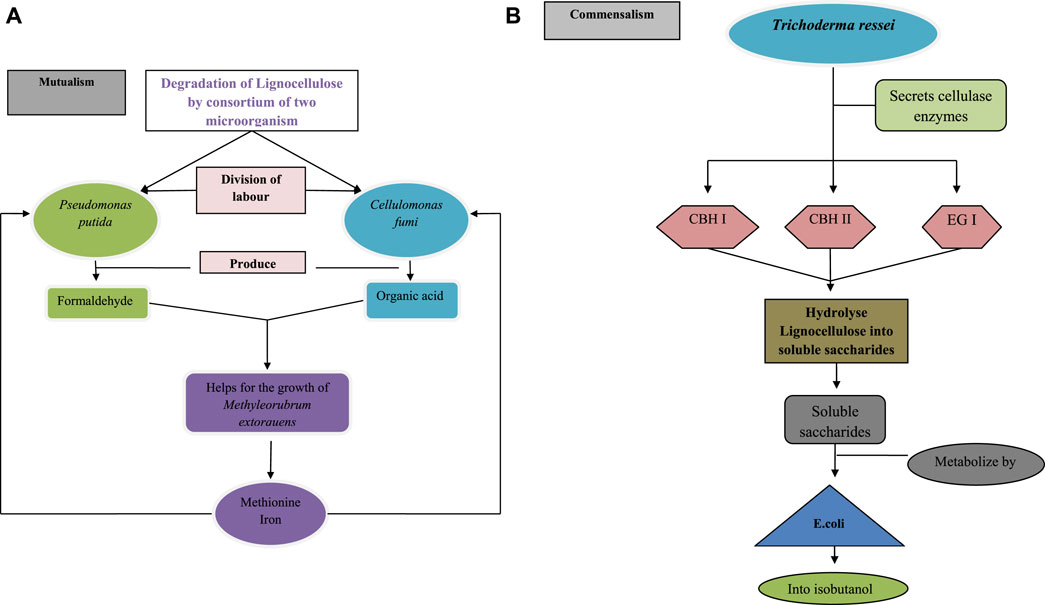
FIGURE 6. Lignocellulose degradation by (A) consortium of Pseudomonas putida and cellulomonas fungi by their mutualism interaction, division of labour between both the microorganism is present for the degradation of lignocellulose after the degradation Pseudomonas putida produce formaldehyde and Cellulomonas fumi produce organic acids, that helps in the growth of Methyl Rubrum extorquens which inturn secretes methionine and iron for the growth of Pseudomonas putida and Cellulomonas fumi (B) consortium of Trichoderma reesei and Escherichia coli by their commensalism interaction, in this interaction,Trichoderma ressei secretes cellobiohydrolaseI (CBH I), cellobiohydrolase II (CBH II) and endoglucanase (EG I) cellulase enzymes to hydrolyse lignocellulose into soluble saccharides and these soluble saccharides metabolize by E. coli into isobutanol.
Examples of lignocelluloses degradation mechanisms performed by single bacteria and in the consortium (Figures 5, 6).
Various attempts have been made to find out potential consortium groups of rhizobacteria to intensify their PGP activities. Various combinations of PGPR are found in soil that is living symbiotically and performs varieties of efficient, robust activities to maintain the soil and plant health (Alves et al., 2022). The Rhizobacterial consortium also provides protection from biotic and abiotic stress to plants (Karuppiah et al., 2022). A single microorganism cannot provide protection against vast varieties of plant pathogens, that’s why there is an increasing demand for efficient consortium groups to target multiple pathogens that work as dynamic biocontrol agent (Yaduwanshi et al., 2021). Stability and compatibility between rhizobacteria are very important for their survival in nature that’s why in a rhizobacterial consortium, every single microorganism should be compatible with each other for their non-competitive establishment in the rhizosphere (El-Tarabily et al., 2021). The stability and functionality of consortia is depending upon their cooperative relationship and interactions (Che and Men. 2019). The fundamental mode of interaction between microorganisms with combinations of positive, negative, and neutral effects between two species are mutualism, commensalism, parasitism or predation, competition, amensalism, and neutralism (Yuan et al., 2021). In the consortium, rhizobacteria communicate by the exchange of metabolites and signals to coordinate their activities and to provide benefits to each other by division of labour, which permit consortia to perform a complex function and sequential processes for resource employment (Atkinson et al., 2022). In rhizobacterial consortia, the diversity of biochemical reactions enhances the utilization of resources and brings down the formation of byproducts (Zhang L. N. et al., 2019). Besides, in natural consortia, many members may metabolize harmful or toxic byproducts of primary substances like acetic acid which if not supplementary consumed will certainly waste the energy and carbon and also inhibit the production of biomass because of anion accumulation and acidification (Sun et al., 2021).
Belimov et al., 2020 developed a microbial consortium of PGPR comprising rhizobia and mycorrhizal fungi. The developed consortium applied to mutant pea species SGECdtwhich was comparable to Indian mustard in Cd tolerance and accumulation (Belimov et al., 2020).
Engineering consortiums may be an essential way to maintain the long-term stability and functionality of the rhizobacterial consortium (Han et al., 2019). The metabolic interactions have an enormous effect on the application of the rhizobacterial consortium (Woo and Pepe 2018). The metabolic compounds secreted by specific species can enhance or defeat the growth of other species and make alterations between them and also affect the function of the whole community (Berg et al., 2020). Synthetic rhizobacterial consortia are one that is created artificially by co-culturing two or more species on the basis of their compatibility (Kapoore et al., 2021). These synthetic consortia are different from natural microbial consortia in their feasible redesign of metabolic pathways to obtain convenient functions (Frioux et al., 2018). Their novel intuition will unveil the common and unique attributes of microorganisms.
Different parameters of tomato growth have been studied by the four rhizobacterial species Bacillus subtilis, Pseudomonas, and the consortium, on-screen house result from the biological activity of these PGPR (Shukla et al., 2022). The phosphate solubilization ability of Bacillus subtilis and Pseudomonas aeruginosa are similar to the potent performance of Bacillus strain BPR7, but the consortium was magnificent in all the growth parameters. 33 isolates studied and evaluated the effect of a rhizobacterial consortium of Bacillus spp. on two micro-propagated bananas and concluded that bacterial consortium is the best-anticipated method to improve vegetative health and survival rates in commercial nurseries. Four isolates, Bacillus subtilis, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Citrobacter youngae were the most effective and consistent PGPR identified (Tzirita et al., 2018; Oloyede et al., 2021).
Apart from these advantages, there are many provocations yet remain, while engineering consortium between inter and intraspecies, like their incompatibility and stabilization of their interaction to control their growth, optimize their metabolic pathways, and also understand their role in population dynamics.
There are variousdrawbacks to natural consortiums because of their difficulty in culture, long operational cycle, poor stability and controllability that make a hurdle in their practical application in various fields of biotechnology (Mccully et al., 2017). The construction of a synthetic microbial consortium would be able to overcome these difficulties of the natural consortium by stabilizing their interactions and making the consortium more bearable and sustainable towards oscillating environmental conditions (Lu et al., 2020; Xu and Yu. 2021).
Designing a synthetic consortium of single species by cell-cell communication is quorum sensing (QS). In QS signalling some molecules are autoinducers, they diffuse from intracellular to extracellular (Tan et al., 2020). Autoinducers trigger or coordinate the expression of certain genes when they cross a certain threshold (Almutairi 2017). OS signalling is a communication mechanism between bacteria that allow for the formation of biofilms, the production of secondary metabolites, and stress adaptation including their secretion system (Isah 2019). The secretion system plays a crucial role in cell-cell communication (Pena et al., 2019). The population density of rhizobacteria is coupled by a few special modules with the help of fluorescent protein insaptio-a temporal form of QS signalling (Mostek et al., 2020). In E. coli the coupling of Las/RLasL as a cell density module control with their motility control, cell density is inversely proportional to the motility (Bi and Sourjik. 2018). There is also two-way interaction by which consortium is constructed with their LasR/LasI and RhIR/RhII QS system, in this system gene expression will only respond if the population is present over a threshold cell density (Ding et al., 2018).
Omics (Transcriptomics, trans proteomics, and metabolomics) tool that illuminates the understanding of the interaction between microorganisms in the consortium and provides precious information about functional diversity of consortium in their gene expression levels and also their metabolites profiling that helps to design and decipher the interaction of natural microbial consortium (Jia et al., 2016; Manzoni et al., 2016). Profiling of proteins and metabolites helps to artificially assemble and understand the interaction between Ketogulonicigenium vulgare and Bacillus megaterium that produce 2-keto-gluconic acid, in this interaction B. megaterium helps K. vulgare to resist reactive oxygen stress, and B. megaterium also provide necessary nutrients, like purine for the growth of K. vulgare for the production of 2-keto-gluconic acid (Wushensky et al., 2018; Yang et al., 2020).
For the successful construction of a synthetic rhizobacterial consortium, it is necessary to select members who are highly dependable on each other (Tabacchioni et al., 2021) to form stable relationships and embrace cross-feeding, detoxification, and biofilm formation to obtain desired functions (Castro et al., 2019; Tang et al., 2020; Vieira et al., 2021).
For the emergence of top-down and bottom-up strategies (San León and Nogales. 2022) a model consumer-resource model (Marsland et al., 2020) has been studied to understand the occurrence of coalescence of rhizobacteria at their community level, according to their model, collective invasions can be looked forward to producing ecological co-selection as a result of cross-feeding (Diaz-Colunga et al., 2022). There are three strategies for the selection of consortium members i.e., top-down strategy, bottom-up strategy and enrichment strategy which are shown in Figure 7.
This strategy occurs in natural microbial interdependence. Top-down engineering involves modifying environmental factors (pH, redox conditions, carbon source, and salt content) for assembling a natural microbial community (San León and Nogales, 2022). For the degradation of toxic chlorinated contaminants, the addition of electron donors helps in the growth of microbial consortium and also in the microbial bioremediation process (Lovley, 2022). Although, the identification of microbial culture is an important precondition for the consortium designing strategy. In spite of the fact there are various techniques, like in situ culture, resuscitation stimulation, and cell sorting, have encouraged research on uncultured microorganisms (Zhang X. H. et al., 2020).
A study showed that incubation time, oxygen availability, exchange in nutrient content, and spatial colocalization between members of bacterial consortium might be responsible factors to operate population dynamics in the rhizobacterial consortium (Leal et al., 2018; Santoyo et al., 2021).
An innovative top-down strategy is developed by co-cultivation of LAB (Lactic acid bacteria) and LB (lignocellulolytic bacteria) populations by using a sequential and rational modification of MRS broth by overall dilution to stimulation approach (Díaz-García et al., 2021a). In this strategy, it is very important to manipulate the whole microbial consortium to obtain the desired function.
This top-down strategy provides benefits to designing more stable and potential synthetic microbial consortia by utilizing environmental microorganisms.
For the construction of synthetic microbial consortia, this strategy is based on microbial metabolism and their interaction (Lin L. 2022). It is quite challenging to construct microbial consortia synthetically because of their complex interlinking metabolic pathways but the development of the multi-omics tool (Mallick et al., 2017; Raju and Bidlan 2017) helps to understand the genetic and molecular factors of these complex metabolic pathways, therefore it is easy to design consortium by following a bottom-up strategy and also for various quantitative models that used to uncover the dynamics of rhizobacterial consortium (Wyrwicka et al., 2019). But still, some incomplete data on genes and proteins of the metabolic network is a backbreaking challenge to engineering microbial consortia.
By using this bottom-up strategy of microbial consortium a study has been developed for the degradation of chitin (Vortmann et al., 2021). According to this study, Step-wise development of a mutualistic and non-competitive consortium in which lysine-auxotrophic Escherichia coli substrate converter cleaves the chitin monomer N-acetylglucosamine (GlcNAc) into glucosamine (GlcN) and acetate, but uses only acetate while leaving GlcN for growth of the lysine-secreting Corynebacterium glutamicum producer strain. They engineered the substrate converter strain for growth on GlcN and not for acetate. Two strains were co-cultured in the presence of a mixture of GlcN and acetate, in this mixture the growth is stabilized through the lysine cross-feeding. By using this strategy PGP activities and functions of the rhizobacterial consortium can be stabilized and optimized and make the consortium more vigorous than its natural consortium.
In this strategy, there is an increase in the population of desired microorganisms by stimulating their growth and that leads to the shifting of microbial communities on the basis of their PGP activities from least potent to highly robust species (Raju and Bidlan. 2017).
Usually, a modified single microbial population with genetic circuits carries all the designated tasks and components in their monoculture environment, therefore single microorganisms bearing the burden of their whole metabolic pathway and the end product will form along with their ‘unprocessed’ intermediary metabolic components (Atkinson et al., 2022). Thus, there is a requirement to divide and combine the metabolic burden among members of the consortium, for the formation of the end product with their completely processed intermediary components (Czajka et al., 2017). For instance, the degradation of pyrene by a synthetic rhizobacterial consortium of three rhizobacteria (Mycobacterium, Novosphingobium, and Ochrobactrum) has been studied, initial degradation step of pyrene is performed by the Mycobacterium strain and the degradation of pyrene intermediates is efficiently performed by Novosphingobium and Ochrobactrum strains, however, Mycobacterium strain own the complete set of genes for pyrene degradation but these genes did not metabolize pyrene, therefore Ochrobactrum and Novosphingobium degrade intermediates (phthalate or protocatechuate) more efficiently then Mycobacterium strain (Wanapaisan et al., 2018; De Lorenzo, 2019; Singha and Pandey. 2020).
Therefore, engineering microbial consortia enable the division of labour between their members where each cell population performs one function. The use of plasmid, genome integration and spatial separation of the population is a common method to engineer these consortiums (Jia et al., 2016).
The different populations within the community may have accidental interactions and their consequences can influence the dynamics of the consortia (Gupta et al., 2020; Dos Santos et al., 2022). If the stabilization mechanism is absent among consortium members, therefore a fast-growing population can navigate the extinction of a slow-growing population (Duncker et al., 2021).
For the successful design of SRC, each step must be precise and flawless in terms of environmental sustainability. The various steps involved in designing SRC are as follows; Isolation of core microbes from the rhizosphere of plant roots, development of strategies for the selection of compatible rhizobacterial microorganisms, next-generation sequencing and network analysis, maintaining suitable culturable conditions by selecting suitable media by using the KOMODO tool (Chung et al., 2017). This is followed by synergy testing of core microbes’ population dynamics and their plant growth-promoting activities, a comparative study of cellular functions between monoculture and their consortium groups, a multi-omics tool to understand the genetic and molecular levels of metabolic pathways of consortium members, stabilization of the interaction between members of the consortium in a fluctuating environment; optimization of the metabolic pathway of interlinking consortium members. This is followed by testing of the long-term effects of the consortium, testing of SMC in consideration of the environmental impacts and finally ecological risk evaluation of SMC.
In a natural ecosystem, microorganism survives in microbial communities, which organized of many interacting species, in these communities’ microbes exchange their metabolites, cross-feed and plays important role in the global cycling of gases and these natural consortia uses for decades for various purpose (Ben Said and Or, 2017).
An example of a natural consortium is two coexisting bioleaching bacteria called Ferroplasma acidiphilum and Leptopirillum ferrriphilum (Merino et al., 2016), their symbiotic association helps in oxidizing sulfur and iron consisting minerals. The natural reservoir of microbial and fungal consortia is herbivore guts, these consortia work synergistically to release a manifold range of cellulolytic enzymes for the degradation of plant biomass (Xu and Yu 2021).
The natural consortium is not stable in fluctuating environmental conditions because of their incompatibility, metabolic burden, and less productivity of biomolecules and products. Microbial synthetic biology helps to engineer rhizobacterial consortium, to accomplish all the drawbacks of natural consortium and enhance productivity.
Designing synthetic rhizobacterial consortium in concern with an environmental ecology, that promotes the beneficial activities of PGPR for a long time and stabilizes their interactions, leads to revealing microbial ecosystem, and for those various studies have been conducted in the last decade to obtain benefits from the vigorous multifunctional rhizobacterial consortium. Here we are compiling all the applications of formulated rhizobacterial consortium (Table 2).
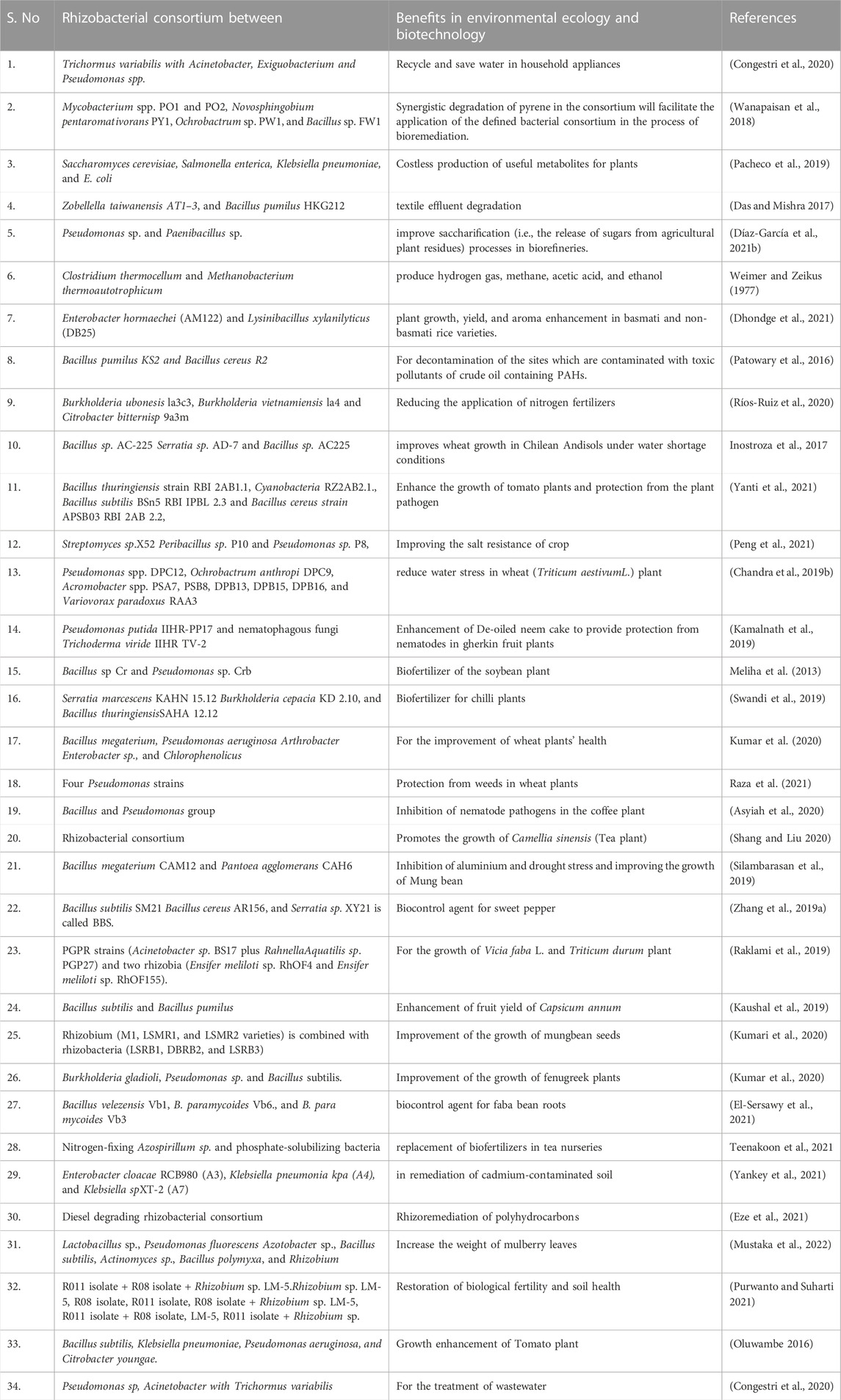
TABLE 2. Engineered rhizobacterial consortium and their applications in environmental ecology and biotechnology.
Biological control of plant disease is defeating plant pathogens by using varieties of biological agents such as plant metabolites, anti-pathogenic microorganisms, root exudates, synthetic fertilizers etc. Similarly, the rhizobacterial consortium is used as a biological control agent for plants and improves plant productivity.
The study shows rhizobacterial consortia from arid ecosystems improve wheat growth in Chilean Andisols under water shortage conditions and reduce water stress by ACC deaminase-producing rhizobacteria consortium (Inostroza et al., 2017; Chandra et al., 2019a). Many studies enlighten that rhizobacterial consortium is a biocontrol agent and provide anti pathogenicity for tomato and wheat plants four Pseudomonas strains were selected for the formulation of consortium on the basis of their efficient PGP activities and their combination with sorghum allelopathic water extract was found to be more effective to control Phalaris minor Retz. and Avenafatu L. weed of wheat plants for sustainable production (Raza et al., 2021), Consortium of Pseudomonas putida IIHR-PP17 and nematophagous fungi Trichoderma viride IIHR TV-2 were used to enrich the de-oiled neem cake for the protection from Meloidogyne incognita and Fusarium oxysporum f sp. in gherkin (Cucumis anguria L.) fruit plant (Kamalnath et al., 2019). The consortium of Bacillus thuringiensis, Bacillus cereus, Bacillus subtilis strains, and cyanobacteria have the best synergistic ability that helps to increase the growth of tomato plants and provide protection from plant pathogens (Yanti et al., 2021). Tetra combination of consortium used for the wheat growth (Kumar et al., 2020) and also the consortium of Bacillus and Pseudomonas group that efficiently inhibit the growth of Pratylenchus coffee and enhance the growth of Robusta coffee (Asyiah et al., 2020). Consortium against Fusarium oxysporum helps to provide protection from wilt disease to faba bean plant roots (El-Sersawy et al., 2021). Three PGPR strains Bacillus cereus AR156, Bacillus subtilis SM21 and Serratia sp. XY21 called BBS treatment in the soil leads to the shifting of the microbial community that suppressed soil-borne disease and improves the soil’s chemical properties (Zhang L. et al., 2019).
Plants acclimatize their physiology and morphology to thrive in high-stress environmental conditions. The different rhizobacterial consortiums are designed to bring about more plant tolerance against biotic and abiotic stresses. Figure 8 shows PGPR and the interaction of plants in the rhizosphere.
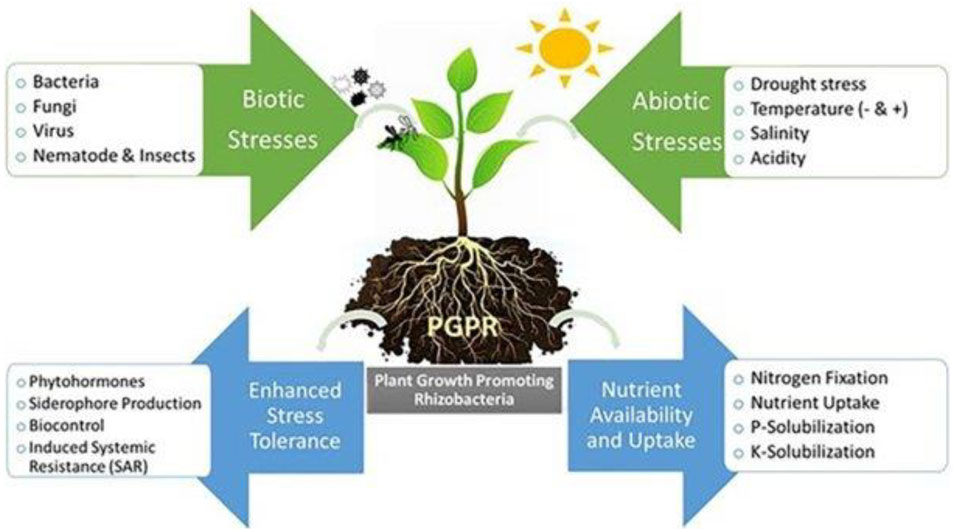
FIGURE 8. PGPR and the interaction of plants in the rhizosphere (Shah et al., 2021b).
PGPR consortium improves salt tolerance in maize plants by using the consortium of sp. P8, Paenibacillus sp. P10 and streptomyces sp. X52 (Cavalu and Damian, 2003; Peng et al., 2021). The consortium of Bacillus sp Cr and Pseudomonas sp. Crb shows a significant increase in soybean plant growth and mineral uptake (Meliha et al., 2013) and treatment of Zn malnutrition in staple rice grains by the formulation of the rhizobacterial consortium (Vaid et al., 2020). Bacillus megaterium CAM12 and Pantoea agglomerans CAH6 were selected on the basis of their ability to tolerate high Al (8 mM) and drought stress. By using this consortium Vigna radiata plants can tolerate more abiotic stresses and efficiently grow in high levels of Al soil (Silambarasan et al., 2019). Two studies show the enhancement of yield and aroma of basmati and non-basmati rice by using Enterobacter hormaechei (AM122) and Lysinibacillus xylanilyticus (DB25) (Dhondge et al., 2021). Saccharomyces cerevisiae, Salmonella enterica, Klebsiella pneumoniae, and E. coli, members of the consortium frequently release and exchange beneficial metabolites that are used by plants (Pacheco et al., 2019).
Two studies show the engineering of rhizobacterial consortium for the treatment of contaminated soil and wastewater (Yankey et al., 2021) (Congestri et al., 2020) a study is conducted for the metagenomic analysis of PGPR consortium members to uncover the varieties of diesel degrading rhizobacterial consortium and their genetic variation (Eze et al., 2021). The degradation of pyrene by the synergistically rhizobacterial consortium has been studied (Cavalu and Damian, 2003; Wanapaisan et al., 2018).
Varieties of synthetic fertilizers are used simultaneously to maintain the nutrient balance for plants but these synthetic fertilizers also cause harmful effects on our soil microbiome, therefore engineered rhizobacterial consortium can also be used as a biofertilizer tool. Many studies have been conducted to provide natural fertilizers, various phosphate solubilizing rhizobacteria and Bradyrhizobium japonicum as a biofertilizer in a soybean plant (Meliha et al., 2013), reducing the application of nitrogen fertilizers by using rhizobacterial consortium (Ríos-Ruiz et al., 2020).
A study shows the growth enhancement of tomato plants by the cooperative PGP activities in the consortium (Oluwambe 2016; Ríos-Ruiz et al., 2020) and the restoration of biological fertility and soil health (Purwanto and Suharti 2021). The consortium of Lactobacillus sp., Bacillus subtilis, Actinomyces sp., Azotobacter sp., Bacillus polymyxa, Pseudomonas fluorescens, and Rhizobium Has been studied on the basis of their PGP activities that help to increase the weight of mulberry leaves (Morus indica) than single rhizobacteria (Mustaka et al., 2022). Isolation of nitrogen-fixing Azospirillum sp. and phosphate solubilizing bacteria from the three-soil series of Sri Lanka was used as a consortium to fix the nutrient uptake in tea plants and as biofertilizers (Tennakoon et al., 2021). Rhizobacterial consortiums are used as biofertilizers to increase the growth of chilli plants (Swandi et al., 2019). Microbial consortium TCM was selected in a study for field trial and showed that TCM consortium improves the effectiveness of rhizospheric nutrition and promotes the growth of Camellia sinensis (Shang and Liu 2020). Fenugreek plant and its yield is significantly increased in consortium than their single inoculation (Patowary et al., 2016; Kumar et al., 2020). In a study, mung bean seeds (SML668 and SML832) were inoculated with single rhizobium and in combination with rhizobacteria. The combination of rhizobacteria and rhizobium improves mungbean overall growth as compared to their single inoculation with rhizobium (Kumari et al., 2020). A consortium of two PGPR isolates (Bacillus subtilis and Bacillus pumilus) was used by the consortium to increase the yield of the bell pepper plant and disease resistance under field conditions (Kaushal et al., 2019). Two PGPR strains (Acinetobacter sp. BS17 and Rahnella Aquatilis sp. PGP27) and two rhizobia (Ensifer Meliloti sp. RhOF4 and Ensifer Meliloti sp. RhOF155) were used to improve the growth parameters of plants. The experimental results show that rhizobacterial and mycorrhizal consortiums appear to adapt to the soil’s native microflora when applied in the field for the growth of Vici faba L. and Triticum durum (Raklami et al., 2019).
All the studies of engineering rhizobacterial consortium are based on their PGP activities that help to improve plants’ growth, productivity, their anti-pathogenicity, makes them tolerant against biotic and abiotic stresses, and helps in the degradation of various Polycyclic Aromatic Hydrocarbons (PAHs) compounds. Synthesis of the rhizobacterial consortium is a multistep process from the selection of the members to their implications in the field, that’s why in all the studies we found a common method of designing experiments which is a comparison of PGP activities between single rhizobacteria and with their consortium and that establish the functional dominance of rhizobacterial consortium over the single rhizobacteria.
All the field experimental studies show, that the rhizobacterial activities in the consortium are more robust and dynamic than single rhizobacterial activities but it is necessary to perceive the “long-term consortium effect and stability” while performing in field trials and for that, existing studies must be practised more in the field and diversified studies should be conducted to promote understanding of consortium mechanisms. Therefore, the studies suggested that there is an urgent requirement to design and establish the rhizobacterial consortium in vitro to find out their communication mechanism and confirmations are required that consortium groups can be applied in the field for plant growth promotion, soil health, and our environmental ecology. A lot of research-based experiments are required in this field involving microbial consortiums. Moreover, we need to test their findings more in field trials for many years to establish the very long-term effect of consortium in all environmental conditions.
The rhizosphere region is a highly active zone which harbours various important microbes. The release of root and plant exudates which are rich in nutrients is the major factor in the microbial activities in the rhizosphere region. PGPR has become important for almost all activities today including sustainable agriculture, biofertilizers etc. The utilization of microbial consortium in PGPR has shown better adaptation of consortium groups in the natural environment. The microbial consortium-based PGPR showed significant activities that refine and upgrade the health and productivity of plants. Experimental evidence proved that rhizobacteria perform more beneficial activities in their consortium than single rhizobacteria. Thereare still some areas to be revealed for the development of the consortium. The most important challenge to designing consortiums is to understand the complexity of microbial interactions in the natural environment, therefore while designing consortia it will become uncomplicated to understand the metabolic pathways, compatibility between microorganisms and their limitations. Another challenge while designing a consortium is the exposure of mutants. Minimizing the mutants is the utmost priority to be fulfilled. Due to the breathtaking robust applications of the rhizobacterial consortium, in the future, furthermore, studies must be conducted to design and discover more beneficial rhizobacterial consortium groups and also to make awareness among farmers to utilize its applications for sustainable environment and human wellbeing.
AS: Original writing, review editing, project administration, data curation and interpretation. RSC, VKY, RS, and SIV: Supervision, writing analysis, validation, software, review editing and critical revision. HAI, NSA, and KKY: formal analysis, critical revision, software, editing and data approval.
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for supporting this work through research group's program undergrant number RGP2/164/43. This research was also funded by the University of Oradea. For the completion of this work, we would like to thank our department, School of Liberal Arts and Sciences of Mody University of Science and Technology, Laskshmangarh, Sikar for providing infrastructural facilities.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
PGPR, plant growth promoting rhizobacteria; MC, microbial consortium; RC, rhizobacterial consortium; SRC, synthetic rhizobacterial consortium; ERC, engineered rhizobacterial consortium; ACC, 1-aminocyclopropane 1-carboxylate deaminase; IAA, indole acetic acid; PSB, phosphate solubilizing bacteria.
Abbasi, S., AlipourKafi, S., Karimi, E., and Sadeghi, A. (2022). Streptomyces consortium improved quality attributes of bell pepper fruits, induced plant defense priming, and changed microbial communities of Rhizosphere under commercial greenhouse conditions. Rhizosphere 23, 100570. doi:10.1016/j.rhisph.2022.100570
Adrees, H., Haider, M., Anjum, T., and Akram, W. (2019). Inducing systemic resistance in cotton plants against charcoal root rot pathogen using indigenous rhizospheric bacterial strains and chemical elicitors. Crop Prot. 115, 75–83. doi:10.1016/j.cropro.2018.09.011
Ahemad, M., and Kibret, M. (2014). Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ. - Sci. 26, 1–20. doi:10.1016/j.jksus.2013.05.001
Almutairi, Z. (2017). Expression profiling of certain MADS-box genes in Arabidopsis thaliana plant treated with silver nanoparticles. Czech J. Genet. Plant Breed. 53, 30–36. doi:10.17221/130/2015-cjgpb
Alves, A., Yin, Q., Oliveira, R., Silva, E., and Novo, L. (2022). Plant growth-promoting bacteria in phytoremediation of metal-polluted soils: Current knowledge and future directions. Sci. Total Environ. 838, 156435. doi:10.1016/j.scitotenv.2022.156435
Antoun, H., and Prevost, D. (2005). “Ecology of plant growth promoting rhizobacteria,” in Pgpr: Biocontrol and biofertilization. Editor Z. A. Siddiqui (Netherlands: Springer), 2.
Asyiah, I., Mudakir, I., Hoesain, M., Pradana, A., Djunaidy, A., and Sari, R. (2020). Consortium of endophytic bacteria and rhizobacteria effectively suppresses the population of Pratylenchuscoffeae and promotes the growth of Robusta coffee. Biodiversitas J. Biol. Divers. 21. doi:10.13057/biodiv/d211032
Atkinson, E., Tuza, Z., Perrino, G., Stan, G., and Ledesma-Amaro, R. (2022). Resource-aware whole-cell model of division of labour in a microbial consortium for complex-substrate degradation. Microb. Cell Factories 21, 115. doi:10.1186/s12934-022-01842-0
Baber, M., Fatima, M., Abbas, R., Qaisrani, M. M., Naz, S., Hanif, M. K., et al. (2018). Weed rhizosphere: A source of novel plant growth promoting rhizobacteria (PGPR). Int. J. Biosci. (IJB) 13, 224–234. doi:10.12692/ijb/13.1.224-234
Backer, R., Rokem, J., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9, 1473. doi:10.3389/fpls.2018.01473
Belimov, A. A., Shaposhnikov, A. I., Azarova, T. S., Makarova, N. M., Safronova, V. I., Litvinskiy, V. A., et al. (2020). Microbial Consortium of PGPR, rhizobia and arbuscular mycorrhizal fungus makes pea mutant sgecdt comparable with Indian mustard in cadmium tolerance and accumulation. Plants 9, 975. doi:10.3390/plants9080975
Ben Said, S., and Or, D. (2017). Synthetic microbial ecology: Engineering habitats for modular consortia. Front. Microbiol. 8, 1125. doi:10.3389/fmicb.2017.01125
Berg, G., Rybakova, D., Fischer, D., Cernava, T., Vergès, M., Charles, T., et al. (2020). Microbiome definition re-visited: Old concepts and new challenges. Microbiome 8, 103. doi:10.1186/s40168-020-00875-0
Bhattacharyya, P. N., and Jha, D. K. (2011). Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. doi:10.1007/s11274-011-0979-9
Bi, S., and Sourjik, V. (2018). Stimulus sensing and signal processing in bacterial chemotaxis. Curr. Opin. Microbiol. 45, 22–29. doi:10.1016/j.mib.2018.02.002
Castro, L., Blázquez, M., González, F., Muñoz, J., and Ballester, A. (2019). Anaerobic bioreduction of jarosites and biofilm formation by a natural microbial consortium. Minerals 9, 81. doi:10.3390/min9020081
Cavalu, S., and Damian, G. (2003). Rotational correlation times of 3-carbamoyl-2,2,5,5-tetramethyl-3-pyrrolin-1-yloxy spin label with respect to heme and nonheme proteins. Biomacromolecules 4 (6), 1630–1635. doi:10.1021/bm034093z
Chandra, D., Srivastava, R., Gupta, V., Franco, C., and Sharma, A. (2019a). Evaluation of ACC-deaminase-producing rhizobacteria to alleviate water-stress impacts in wheat (Triticum aestivum L.) plants. Can. J. Microbiol. 65, 387–403. doi:10.1139/cjm-2018-0636
Chandra, D., Srivastava, R., Gupta, V. V. S. R., Franco, C. M. M., and Sharma, A. K. (2019b). Evaluation of ACC-deaminase-producing rhizobacteria to alleviate water-stress impacts in wheat (triticumaestivumL.) plants. Can. J. Microbiol. 65, 387–403. doi:10.1139/cjm-2018-0636
Chandran, H., Meena, M., and Swapnil, P. (2021). Plant growth-promoting rhizobacteria as a green alternative for sustainable agriculture. Sustainability 13, 10986. doi:10.3390/su131910986
Che, S., and Men, Y. (2019). Synthetic microbial consortia for biosynthesis and biodegradation: Promises and challenges. J. Industrial Microbiol. Biotechnol. 46, 1343–1358. doi:10.1007/s10295-019-02211-4
Choudhary, D. K., Prakash, A., and Johri, B. N. (2007). Induced systemic resistance (ISR) in plants: Mechanism of action. Indian J. Microbiol. 47, 289–297. doi:10.1007/s12088-007-0054-2
Chung, E., Dean, S., Propst, C., Bishop, B., and van Hoek, M. (2017). Komodo dragon-inspired synthetic peptide DRGN-1 promotes wound-healing of a mixed-biofilm infected wound. npj Biofilms Microbiomes 3, 9. doi:10.1038/s41522-017-0017-2
Congestri, R., Savio, S., Farrotti, S., Amati, A., Krasojevic, K., Perini, N., et al. (2020). Developing a microbial consortium for removing nutrients in dishwasher wastewater: Towards a biofilter for its up-cycling. Water Sci. Technol. 82, 1142–1154. doi:10.2166/wst.2020.325
Constantia, J., and Ferniah, R. (2020). Vegetative growth of rainbow chili (capsicum annuum l.) in the treatment of pgpr (plant growth promoting rhizobacteria), PGPR-NPK fertilizer, and PGPR-compost combination. Agric 32, 95–104. doi:10.24246/agric.2020.v32.i2.p95-104
Cragg, S., Beckham, G., Bruce, N., Bugg, T., Distel, D., Dupree, P., et al. (2015). Lignocellulose degradation mechanisms across the tree of life. Curr. Opin. Chem. Biol. 29, 108–119. doi:10.1016/j.cbpa.2015.10.018
Czajka, J., Wang, Q., Wang, Y., and Tang, Y. (2017). Synthetic biology for manufacturing chemicals: Constraints drive the use of non-conventional microbial platforms. Appl. Microbiol. Biotechnol. 101, 7427–7434. doi:10.1007/s00253-017-8489-9
Das, A., and Mishra, S. (2017). Removal of textile dye reactive green-19 using bacterial consortium: Process optimization using response surface methodology and kinetics study. J. Environ. Chem. Eng. 5, 612–627. doi:10.1016/j.jece.2016.10.005
De Lorenzo, V. (2019). “Biodegradation and bioremediation: An introduction,” in Consequences of microbial interactions with Hydrocarbons, oils, and lipids: Biodegradation and bioremediation. Handbook of hydrocarbon and lipid microbiology. Editor R. Steffan (Cham: Springer), 1–20. doi:10.1007/978-3-319-50433-9_1
Dhondge, H. V., Pable, A. A., Barvkar, V. T., Dastager, S. G., and Nadaf, A. B. (2021). Rhizobacterial consortium mediated aroma and yield enhancement in Basmati and non-basmati rice (Oryza sativa L.). J. Biotechnol. 328, 47–58. doi:10.1016/j.jbiotec.2021.01.012
Diaz-Colunga, J., Lu, N., Sanchez-Gorostiaga, A., Chang, C. Y., Cai, H. S., Goldford, J. E., et al. (2022). Top-down and bottom-up cohesiveness in microbial community coalescence. Proc. Natl. Acad. Sci. 119, e2111261119. doi:10.1073/pnas.2111261119
Díaz-García, L., Chaparro, D., Jiménez, H., Gómez-Ramírez, L. F., Bernal, A. J., Burbano-Erazo, E., et al. (2021a). Top-down enrichment strategy to co-cultivate lactic acid and lignocellulolytic bacteria from the megathyrsus Maximus Phyllosphere. Front. Microbiol. 12, 744075. doi:10.3389/fmicb.2021.744075
Díaz-García, L., Huang, S., Spröer, C., Sierra-Ramírez, R., Bunk, B., Overmann, J., et al. (2021b). Dilution-to-stimulation/extinction method: A combination enrichment strategy to develop a minimal and versatile lignocellulolytic bacterial consortium. Appl. Environ. Microbiol. 87, 024277–e2520. doi:10.1128/aem.02427-20
Ding, F., Oinuma, K., Smalley, N., Schaefer, A., Hamwy, O., Greenberg, E., et al. (2018). The pseudomonas aeruginosa orphan quorum sensing signal receptor QscR regulates global quorum sensing gene expression by activating a single linked operon. mBio 9, 012744–e1318. doi:10.1128/mbio.01274-18
Dong, W., Liu, K., Wang, F., Xin, F., Zhang, W., Zhang, M., et al. (2017). The metabolic pathway of metamifop degradation by consortium ME-1 and its bacterial community structure. Biodegradation 28, 181–194. doi:10.1007/s10532-017-9787-8
Dos Santos, A. R., Di Martino, R., Testa, S. E., and Mitri, S. (2022). Classifying interactions in a synthetic bacterial community is hindered by inhibitory growth medium. mSystems 7, e0023922. doi:10.1128/msystems.00239-22
Duncker, K. E., Holmes, Z. A., and You, L. (2021). Engineered microbial consortia: Strategies and applications. Microb. Cell Factories 20, 211. doi:10.1186/s12934-021-01699-9
El-Sersawy, M., Hassan, S., El-Ghamry, A., El-Gwad, A., and Fouda, A. (2021). Implication of plant growth-promoting rhizobacteria of Bacillus spp. as biocontrol agents against wilt disease caused by Fusarium oxysporum Schlecht. in Vicia faba L. Vicia faba L. Biomol. Concepts 12, 197–214. doi:10.1515/bmc-2021-0020
El-Tarabily, K. A., Sham, A., Elbadawi, A. A., Hassan, A. H., Alhosani, B. K., El-Esawi, M. A., et al. (2021). A consortium of Rhizosphere-competent Actinobacteria exhibiting multiple plant growth-promoting traits improves the growth of Avicennia marina in the United Arab Emirates. Front. Mar. Sci. 8. doi:10.3389/fmars.2021.715123
Eze, M. O., Hose, G. C., George, S. C., and Daniel, R. (2021). Diversity and metagenome analysis of a hydrocarbon-degrading bacterial consortium from Asphalt Lakes located in Wietze, Germany. Amb. Express 11, 89. doi:10.1101/2021.03.25.436929
Fatima, I., Hakim, S., Imran, A., Ahmad, N., Imtiaz, M., Ali, H., et al. (2022). Exploring biocontrol and growth-promoting potential of multifaceted PGPR isolated from natural suppressive soil against the causal agent of chickpea wilt. Microbiol. Res. 260, 127015. doi:10.1016/j.micres.2022.127015
Fazeli-Nasab, B., and Sayyed, R. Z. (2019). “Plant growth-promoting rhizobacteria and salinity stress: A journey into the soil,” in Plant growth promoting rhizobacteria for sustainable stress management. Microorganisms for sustainability. Editors R. Sayyed, N. Arora, and M. Reddy (Singapore: Springer), Vol. 12, 21–34. doi:10.1007/978-981-13-6536-2_2
Frioux, C., Fremy, E., Trottier, C., and Siegel, A. (2018). Scalable and exhaustive screening of metabolic functions carried out by microbial consortia. Bioinformatics 34, i934–i943. doi:10.1093/bioinformatics/bty588
Goswami, D., Thakker, J. N., and Dhandhukia, P. C. (2016). Portraying mechanics of plant growth promoting rhizobacteria (PGPR): A review. Cogent Food & Agric. 2. doi:10.1080/23311932.2015.1127500
Govindasamy, V., George, P., Kumar, M., Aher, L., Raina, S., Rane, J., et al. (2019). Multi-trait PGP rhizobacterial endophytes alleviate drought stress in a senescent genotype of sorghum [Sorghum bicolor (L.) Moench]. 3 Biotech. 10, 13. doi:10.1007/s13205-019-2001-4
Govindasamy, V., Senthilkumar, M., Magheshwaran, V., Kumar, U., Bose, P., Sharma, V., et al. (2010). Bacillus and paenibacillus spp.: Potential PGPR for sustainable agriculture. Plant Growth Health Promot. Bact. 18, 333–364. doi:10.1007/978-3-642-13612-2_15
Gowtham, H. G., Brijesh, S. S., Murali, M., Shilpa, N., Prasad, M., Aiyaz, M., et al. (2020). Induction of drought tolerance in tomato upon the application of ACC deaminase producing plant growth promoting Rhizobacterium Bacillus subtilis Rhizo SF 48. Microbiol. Res. 234, 126422. doi:10.1016/j.micres.2020.126422
Gupta, S., Ross, T. D., Gomez, M. M., Grant, J. L., Romero, P. A., and Venturelli, O. S. (2020). Investigating the dynamics of microbial consortia in spatially structured environments. bioRxiv. doi:10.1101/2020.02.17.953240
Han, W., He, P., Lin, Y., Shao, L., and Lü, F. (2019). A methanogenic consortium was active and exhibited long-term survival in an extremely acidified thermophilic bioreactor. Front. Microbiol. 10, 2757. doi:10.3389/fmicb.2019.02757
Hartmann, A. (2020). Quorum sensing N-acyl-homoserine lactone signal molecules of plant beneficial Gram-negative rhizobacteria support plant growth and resistance to pathogens. Rhizosphere 16, 100258. doi:10.1016/j.rhisph.2020.100258
Hayat, R., Ali, S., Amara, U., Khalid, R., and Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 60, 579–598. doi:10.1007/s13213-010-0117-1
Hussin, N. A., and Ab Majid, A. H. (2020). Termiticidal activity of chitinase enzyme of bacillus licheniformis, a symbiont isolated from the gut of GlobitermesSulphureusWorker. Biocatal. Agric. Biotechnol. 24, 101548. doi:10.1016/j.bcab.2020.101548
Inostroza, N., Barra, P., Wick, L., Mora, M., and Jorquera, M. (2017). Effect of rhizobacterial consortia from undisturbed arid- and agro-ecosystems on wheat growth under different conditions. Lett. Appl. Microbiol. 64, 158–163. doi:10.1111/lam.12697
Isah, T. (2019). Stress and defense responses in plant secondary metabolites production. Biol. Res. 52, 39. doi:10.1186/s40659-019-0246-3
Jaleel, R. A., Ahsan, K. P. J., Bhattacharjee, S., and Priya, S. (2021). Isolation, molecular identification and application of exogenous indole acetic acid (IAA) producing plant growth promoting rhizobacteria (PGPR) from soil. IARJSET 8. doi:10.17148/iarjset.2021.81007
Jayaprakashvel, M., and Mathivanan, N. (2011). Management of plant diseases by microbial metabolites. Bact. Agrobiology Plant Nutrient Manag. 2011, 237–265. doi:10.1007/978-3-642-21061-7_10
Jia, X., Liu, C., Song, H., Ding, M., Du, J., Ma, Q., et al. (2016). Design, analysis and application of synthetic microbial consortia. Synthetic Syst. Biotechnol. 1, 109–117. doi:10.1016/j.synbio.2016.02.001
Kamalnath, M., Rao, M., and Umamaheswari, R. (2019). Rhizophere engineering with beneficial microbes for growth enhancement and nematode disease complex management in gherkin (Cucumis anguriaL.). Sci. Hortic. 257, 108681. doi:10.1016/j.scienta.2019.108681
Kapoore, R., Padmaperuma, G., Maneein, S., and Vaidyanathan, S. (2021). Co-Culturing microbial consortia: Approaches for applications in biomanufacturing and bioprocessing. Crit. Rev. Biotechnol. 42, 46–72. doi:10.1080/07388551.2021.1921691
Karuppiah, V., Natarajan, S., Gangatharan, M., Aldayel, M., Alsowayeh, N., and Thangavel, K. (2022). Development of siderophore-based rhizobacterial consortium for the mitigation of biotic and abiotic environmental stresses in tomatoes: An in vitro and in planta approach. J. Appl. Microbiol. 133, 3276–3287. doi:10.1111/jam.15625
Kaushal, M., Mandyal, P., and Kaushal, R. (2019). Field based assessment of capsicum annuum performance with inoculation of Rhizobacterial Consortia. Microorganisms 7, 89. doi:10.3390/microorganisms7030089
Kirchman, D. L. (2018). The physical-chemical environment of microbes. Oxford Scholarship Online. doi:10.1093/oso/9780198789406.003.0003
Kumar, A., Kumari, M., Swarupa, P., and Shireen, S. (2019a). Characterization of pH dependent growth response of agriculturally important microbes for development of plant growth promoting bacterial consortium. J. Pure Appl. Microbiol. 13, 1053–1061. doi:10.22207/jpam.13.2.43
Kumar, A., Patel, J. S., Meena, V. S., and Ramteke, P. W. (2019b). Plant growth-promoting rhizobacteria: Strategies to improve abiotic stresses under sustainable agriculture. J. Plant Nutr. 42, 1402–1415. doi:10.1080/01904167.2019.1616757
Kumar, P., Aeron, A., Shaw, N., Singh, A., Bajpai, V., Pant, S., et al. (2020). Seed bio-priming with tri-species consortia of phosphate solubilizing rhizobacteria (PSR) and its effect on plant growth promotion. Heliyon 6, e05701. doi:10.1016/j.heliyon.2020.e05701
Kumari, P., Sharma, P., and Sharma, S. (2020). Synergism of rhizobium and rhizobacteria on growth, symbiotic parameters, soil quality and grain yield in summer mungbean (Vigna radiata L. Wilczek). Int. J. Curr. Microbiol. Appl. Sci. 9, 136–151. doi:10.20546/ijcmas.2020.903.017
Kundan, R., Pant, G., Jadon, N., and Agrawal, P. K. (2015). Plant growth promoting rhizobacteria: Mechanism and current prospective. J. Fertilizers Pesticides 06. doi:10.4172/2471-2728.1000155
Leal, P. L., Pinheiro Dadalto, S., de Fernandes, R., Queiroz, M. E., and Tótola, M. R. (2018). <b>Enrichment of population density of a bacterial consortium during bioremediation of a soil under successive contaminations with diesel oil. Acta Sci. Biol. Sci. 40, 36904. doi:10.4025/actascibiolsci.v40i1.36904
Li, W., Li, F., Zeng, H., Ma, L., Qi, L., Wang, X., et al. (2021). Diversity and variation of asymbiotic nitrogen-fixing microorganisms in alpine grasslands on the Tibetan Plateau. Front. Ecol. Evol. 9. doi:10.3389/fevo.2021.702848
Lin, L. (2022). Bottom-up synthetic ecology study of microbial consortia to enhance lignocellulose bioconversion. Biotechnol. Biofuels Bioprod. 15, 14. doi:10.1186/s13068-022-02113-1
Lindemann, S. (2019). Microbial Ecology: Functional ‘modules’ drive assembly of polysaccharide-degrading marine microbial communities. Curr. Biol. 29, R330–R332. doi:10.1016/j.cub.2019.03.056
Linu, M., Asok, A., Thampi, M., Sreekumar, J., and Jisha, M. (2019). Plant growth promoting traits of indigenous phosphate solubilizing Pseudomonas aeruginosa isolates from chilli (Capsicumannuum L.) Rhizosphere. Commun. Soil Sci. Plant Analysis 50, 444–457. doi:10.1080/00103624.2019.1566469
Liu, Y., Tu, X., Xu, Q., Bai, C., Kong, C., Liu, Q., et al. (2018). Engineered monoculture and co-culture of methylotrophic yeast for de novo production of Monacolin J and Lovastatin from methanol. Metab. Eng. 45, 189–199. doi:10.1016/j.ymben.2017.12.009
Lovley, D. (2022). Electrotrophy: Other microbial species, iron, and electrodes as electron donors for microbial respirations. Bioresour. Technol. 345, 126553. doi:10.1016/j.biortech.2021.126553
Lu, J., Chen, C., Huang, C., and Lee, D. (2020). Glucose fermentation with biochar-amended consortium: Microbial consortium shift. Bioengineered 11, 272–280. doi:10.1080/21655979.2020.1735668
Mabood, F., Zhou, X., and Smith, D. L. (2014). Microbial signaling and plant growth promotion. Can. J. Plant Sci. 94, 1051–1063. doi:10.4141/cjps2013-148
Mahmud, K., Makaju, S., Ibrahim, R., and Missaoui, A. (2020). Current progress in nitrogen fixing plants and microbiome research. Plants 9, 97. doi:10.3390/plants9010097
Mallick, H., Ma, S., Franzosa, E., Vatanen, T., Morgan, X., and Huttenhower, C. (2017). Experimental design and quantitative analysis of microbial community multiomics. Genome Biol. 18, 228. doi:10.1186/s13059-017-1359-z
Manzoni, C., Kia, D. A., Vandrovcova, J., Hardy, J., Wood, N. W., Lewis, P. A., et al. (2016). Genome, transcriptome and proteome: The rise of OMICS data and their integration in Biomedical Sciences. Briefings Bioinforma. 19, 286–302. doi:10.1093/bib/bbw114
Marsland, R., Cui, W., Goldford, J., and Mehta, P. (2020). The community simulator: A Python package for microbial ecology. Plos One 15, e0230430. doi:10.1371/journal.pone.0230430
McCully, A., LaSarre, B., and McKinlay, J. (2017). Growth-independent cross-feeding modifies boundaries for coexistence in a bacterial mutualism. Environ. Microbiol. 19, 3538–3550. doi:10.1111/1462-2920.13847
Meena, M., and Swapnil, P. (2019). Regulation ofwrkygenes in plant defence with beneficial fungustrichoderma: Current perspectives and future prospects. Archives Phytopathology Plant Prot. 52, 1–17. doi:10.1080/03235408.2019.1606490
Meliah, S., Wahyudi, A. T., and Nawangsih, A. A. (2013). The formulation of consortium of phosphate solubilizing rhizobacteria with bradyrhizobium japonicum as biofertilizer and their application to soyabean plant. Scientific repository IPB University. Available at: http://repository.ipb.ac.id/handle/123456789/63130.
Merino, M. P., Andrews, B. A., Parada, P., and Asenjo, J. A. (2016). Characterization ofFerroplasmaAcidiphilumgrowing in pure and mixed culture withleptospirillumferriphilum. Biotechnol. Prog. 32, 1390–1396. doi:10.1002/btpr.2340
Mohanram, S., and Kumar, P. (2019). Rhizosphere microbiome: Revisiting the synergy of plant-microbe interactions. Ann. Microbiol. 69, 307–320. doi:10.1007/s13213-019-01448-9
Monteiro, G., Nogueira, G., Neto, C., Nascimento, V., and Freitas, J. (2021). “Promotion of nitrogen assimilation by Plant Growth-promoting rhizobacteria,” in Nitrogen in agriculture - physiological, agricultural and ecological aspects [working title]. doi:10.5772/intechopen.96634
Mostek, A., Janta, A., Majewska, A., and Ciereszko, A. (2020). Fluorescent protein labeling coupled with thiol-staining reveals protein candidates involved in redox signaling accompanying bull sperm capacitation. Animal Reproduction Sci. 220, 106405. doi:10.1016/j.anireprosci.2020.106405
Mustaka, Z., Patandjengi, B., Alam, G., and Melina, M. (2022). Potential of rhizobacterial consortium in increasing area and weight of mulberry leaves (Morus indica). Biodiversitas J. Biol. Divers. 23. doi:10.13057/biodiv/d230322
Nath, M., Bhatt, D., Bhatt, M. D., Prasad, R., and Tuteja, N. (2018). Microbe-mediated enhancement of nitrogen and phosphorus content for crop improvement. Crop Improv. Through Microb. Biotechnol. 2018, 293–304. doi:10.1016/b978-0-444-63987-5.00014-1
Nazir, N., and Kamili, A. N. (2018). Mechanism of plant growth promoting rhizobacteria (PGPR) in enhancing plant growth – a review. Durdana Shah Int. J. Manag. Technol. Eng. 8 (7). JULY/2018 ISSN NO : 2249-7455.
Neshat, M., Abbasi, A., Hosseinzadeh, A., Sarikhani, M., Dadashi Chavan, D., and Rasoulnia, A. (2022). Plant growth promoting bacteria (PGPR) induce antioxidant tolerance against salinity stress through biochemical and physiological mechanisms. Physiology Mol. Biol. Plants 28, 347–361. doi:10.1007/s12298-022-01128-0
Nguyen, N. H., Trotel-Aziz, P., Villaume, S., Rabenoelina, F., Schwarzenberg, A., Nguema-Ona, E., et al. (2020). Bacillus subtilis and pseudomonas fluorescens trigger common and distinct systemic immune responses in Arabidopsis thaliana depending on the pathogen lifestyle. Vaccines 8, 503. doi:10.3390/vaccines8030503
Oloyede, A., Ogbuagor, C., Afolabi, C., and Akintokun, A. (2021). Biological control of bacterial canker of tomato (Lycopersicon esculentum Mill.) by use of non-native strains of plant growth-promoting rhizobacteria. Archives Phytopathology Plant Prot. 54, 1182–1203. doi:10.1080/03235408.2021.1895477
Oluwambe, T. (2016). Comparison of single culture and the consortium of growth-promoting rhizobacteria from three tomato (Lycopersicon esculentum Mill) varieties. Adv. Plants Agric. Res. 5. doi:10.15406/apar.2016.05.00167
Oteino, N., Lally, R. D., Kiwanuka, S., Lloyd, A., Ryan, D., Germaine, K. J., et al. (2015). Plant growth promotion induced by phosphate solubilizing endophytic pseudomonas isolates. Front. Microbiol. 6, 745. doi:10.3389/fmicb.2015.00745
Pacheco, A., Moel, M., and Segrè, D. (2019). Costless metabolic secretions as drivers of interspecies interactions in microbial ecosystems. Nat. Commun. 10, 103. doi:10.1038/s41467-018-07946-9
Patel, T., and Saraf, M. (2017). Biosynthesis of phytohormones from novel rhizobacterial isolates and their in vitro plant growth-promoting efficacy. J. Plant Interact. 12, 480–487. doi:10.1080/17429145.2017.1392625
Patowary, K., Patowary, R., Kalita, M. C., and Deka, S. (2016). Development of an efficient bacterial consortium for the potential remediation of hydrocarbons from contaminated sites. Front. Microbiol. 7, 1092. doi:10.3389/fmicb.2016.01092
Pena, R. T., Blasco, L., Ambroa, A., González-Pedrajo, B., Fernández-García, L., López, M., et al. (2019). Relationship between quorum sensing and secretion systems. Front. Microbiol. 10, 1100. doi:10.3389/fmicb.2019.01100
Peng, J., Ma, J., Wei, X., Zhang, C., Jia, N., Wang, X., et al. (2021). Accumulation of beneficial bacteria in the rhizosphere of maize (Zea mays L.) grown in a saline soil in responding to a consortium of plant growth promoting rhizobacteria. Ann. Microbiol. 71, 40. doi:10.1186/s13213-021-01650-8
Phale, S. (2018). Isolation and screening of multifunctional plant growth promoting rhizobacteria (PGPR) from onion rhizosphere (Allium cepa). J. Bioprocess. Biotech. 08. doi:10.4172/2155-9821.1000335
Podile, A. R., and Kishore, G. K. (2006). in Plant growth-promoting rhizobacteria. Plant-Associated Bacteria. Editor S. S. Gnanamanickam (Dordrecht, Netherlands: Springer), 195–230. doi:10.1007/978-1-4020-4538-7_6
Prashar, P., Kapoor, N., and Sachdeva, S. (2013). Rhizosphere: Its structure, bacterial diversity and significance. Rev. Environ. Sci. Bio/Technology 13, 63–77. doi:10.1007/s11157-013-9317-z
Purwanto, P., and Suharti, W. (2021). Nutrient uptake, chlorophyll content, and yield of rice (Oryza sativa L.) under the application of PGPR consortium. Biosaintifika J. Biol. Biol. Educ. 13, 336–344. doi:10.15294/biosaintifika.v13i3.31990
Rahina, K., Muhammad, I., Muhammad, I., Muhammad, R., Obaid, U., Ayesha, M., et al. (2018). Response of groundnut (Arachis hypogaea L.) to plant growth promoting rhizobacteria in degraded soils. Afr. J. Agric. Res. 13, 904–910. doi:10.5897/ajar2018.13081
Raju, S., and Bidlan, R. (2017). Isolation, enrichment and metagenomic characterization of simultaneous DDT and lindane degrading microbial consortium. J. Pure Appl. Microbiol. 11, 1947–1952. doi:10.22207/jpam.11.4.36
Raklami, A., Bechtaoui, N., Tahiri, A., Anli, M., Meddich, A., and Oufdou, K. (2019). Use of rhizobacteria and mycorrhizae consortium in the open field as a strategy for improving crop nutrition, productivity and soil fertility. Front. Microbiol. 10, 1106. doi:10.3389/fmicb.2019.01106
Raza, T., Yahya Khan, M., Mahmood Nadeem, S., Imran, S., Nazir Qureshi, K., Naeem Mushtaq, M., et al. (2021). Biological management of selected weeds of wheat through co-application of allelopathic rhizobacteria and sorghum extract. Biol. Control 164, 104775. doi:10.1016/j.biocontrol.2021.104775
Redondo-Gómez, S., Mesa-Marín, J., Pérez-Romero, J. A., López-Jurado, J., García-López, J. V., Mariscal, V., et al. (2021). Consortia of plant-growth-promoting rhizobacteria isolated from halophytes improve response of eight crops to soil salinization and climate change conditions. Agronomy 11, 1609. doi:10.3390/agronomy11081609
Ríos-Ruiz, W., Torres-Chávez, E., Torres-Delgado, J., Rojas-García, J., Bedmar, E., and Valdez-Nuñez, R. (2020). Inoculation of bacterial consortium increases rice yield (Oryza sativa L.) reducing applications of nitrogen fertilizer in San Martin region, Peru. Rhizosphere 14, 100200. doi:10.1016/j.rhisph.2020.100200
Roell, G., Zha, J., Carr, R., Koffas, M., Fong, S., and Tang, Y. (2019). Engineering microbial consortia by division of labor. Microb. Cell Factories 18, 35. doi:10.1186/s12934-019-1083-3
Saeed, Q., Xiukang, W., Haider, F. U., Kučerik, J., Mumtaz, M. Z., Holatko, J., et al. (2021). Rhizosphere bacteria in plant growth promotion, biocontrol, and bioremediation of contaminated sites: A comprehensive review of effects and mechanisms. Int. J. Mol. Sci. 22, 10529. doi:10.3390/ijms221910529
Sagar, A., Sayyed, R. Z., Ramteke, P. W., Sharma, S., Marraiki, N., Elgorban, A. M., et al. (2020). ACC deaminase and antioxidant enzymes producing halophilic Enterobacter sp. PR14 promotes the growth of rice and millets under salinity stress. Physiology Mol. Biol. Plants 26, 1847–1854. doi:10.1007/s12298-020-00852-9
Samain, E., Ernenwein, C., Aussenac, T., and Selim, S. (2022). Effective and durable systemic wheat-induced resistance by a plant-growth-promoting rhizobacteria consortium of paenibacillus sp. strain B2 and Arthrobacter spp. strain AA against zymoseptoriatritici and drought stress. Physiological Mol. Plant Pathology 119, 101830. doi:10.1016/j.pmpp.2022.101830
San León, D., and Nogales, J. (2022). Toward merging bottom–UP and top–down model-based designing of synthetic microbial communities. Curr. Opin. Microbiol. 69, 102169. doi:10.1016/j.mib.2022.102169
Santos, L., and Olivares, F. (2021). Plant microbiome structure and benefits for sustainable agriculture. Curr. Plant Biol. 26, 100198. doi:10.1016/j.cpb.2021.100198
Santoyo, G., Urtis-Flores, C. A., Loeza-Lara, P. D., Orozco-Mosqueda, M. del, and Glick, B. R. (2021). Rhizosphere colonization determinants by Plant Growth-promoting rhizobacteria (PGPR). Biology 10, 475. doi:10.3390/biology10060475
Saraf, M., Pandya, U., and Thakkar, A. (2014). Role of allelochemicals in plant growth promoting rhizobacteria for biocontrol of phytopathogens. Microbiol. Res. 169, 18–29. doi:10.1016/j.micres.2013.08.009
Shabaan, M., Asghar, H., Zahir, Z., Sardar, M., Parveen, R., Faiza, , et al. (2022). Halotolerant rhizobacterial consortium confers salt tolerance to maize under naturally salt-affected soil. Soil Sci. Soc. Am. J. 86, 1264–1279. doi:10.1002/saj2.20438
Shah, A., Nazari, M., Antar, M., Msimbira, L., Naamala, J., Lyu, D., et al. (2021a). PGPR in agriculture: A sustainable approach to increasing climate change resilience. Front. Sustain. Food Syst. 5. doi:10.3389/fsufs.2021.667546
Shah, A., Nazari, M., Antar, M., Msimbira, L. A., Naamala, J., Lyu, D., et al. (2021b). PGPR in agriculture: A sustainable approach to increasing climate change resilience. Front. Sustain. Food Syst. 5. doi:10.3389/fsufs.2021.667546
Shang, J., and Liu, B. (2020). Application of a microbial consortium improves the growth of Camellia sinensis and influences the indigenous rhizosphere bacterial communities. J. Appl. Microbiol. 130, 2029–2040. doi:10.1111/jam.14927
Sharifi, R., Lee, S. M., and Ryu, C. M. (2017). Microbe-induced plant volatiles. New Phytol. 220, 684–691. doi:10.1111/nph.14955
Sharma, V., Singh, A., Sharma, D., Sharma, A., Phogat, S., Chakraborty, N., et al. (2022). Stress mitigation strategies of plant growth-promoting rhizobacteria: Plant growth-promoting rhizobacteria mechanisms. Plant Sci. Today 8, 25–32. doi:10.14719/pst.1543
Shimoda, Y., Nishigaya, Y., Yamaya-Ito, H., Inagaki, N., Umehara, Y., Hirakawa, H., et al. (2020). The Rhizobial Autotransporter determines the symbiotic nitrogen fixation activity of lotus japonicus in a host-specific manner. Proc. Natl. Acad. Sci. 117, 1806–1815. doi:10.1073/pnas.1913349117
Shukla, V., Kumar, S., Tripathi, Y. N., and Upadhyay, R. S. (2022). Bacillus subtilis- and pseudomonas fluorescens-mediated systemic resistance in tomato against sclerotium rolfsii and study of physio-chemical alterations. Front. Fungal Biol. 3. doi:10.3389/ffunb.2022.851002
Siddiqui, Z., Ali, F., and Uddin, Z. (2021). Sustainable effect of a symbiotic nitrogen-fixing bacterium Sinorhizobiummeliloti on nodulation and photosynthetic traits of four leguminous plants under low moisture stress environment. Lett. Appl. Microbiol. 72, 714–724. doi:10.1111/lam.13463
Silambarasan, S., Logeswari, P., Cornejo, P., and Kannan, V. (2019). Role of plant growth–promoting rhizobacterial consortium in improving the Vigna radiata growth and alleviation of aluminum and drought stresses. Environ. Sci. Pollut. Res. 26, 27647–27659. doi:10.1007/s11356-019-05939-9
Singh, R., and Jha, P. (2017). The PGPR Stenotrophomonas maltophilia SBP-9 augments resistance against biotic and abiotic stress in wheat plants. Front. Microbiol. 8, 1945. doi:10.3389/fmicb.2017.01945
Singh, R. K., Singh, P., Li, H.-B., Song, Q.-Q., Guo, D.-J., Solanki, M. K., et al. (2020). Diversity of nitrogen-fixing rhizobacteria associated with sugarcane: A comprehensive study of plant-microbe interactions for growth enhancement in saccharum spp. BMC Plant Biol. 20, 220. doi:10.1186/s12870-020-02400-9
Singha, L., and Pandey, P. (2020). Rhizobacterial community of Jatropha curcas associated with pyrene biodegradation by consortium of PAH-degrading bacteria. Appl. Soil Ecol. 155, 103685. doi:10.1016/j.apsoil.2020.103685
Sun, Y., Li, X., Wu, L., Li, Y., Li, F., Xiu, Z., et al. (2021). The Advanced Performance of Microbial Consortium for simultaneous utilization of glucose and xylose to produce lactic acid directly from dilute sulfuric acid pretreated corn stover. doi:10.21203/rs.3.rs-778235/v1
Swandi, M. K., Mubarik, N. R., and Tjahjoleksono, A. (2019). Rhizobacterial Inoculants: The formulation as biofertilizer and its application on chili plants (Capsicum annum L.). Malays. J. Microbiol. 15, 44. doi:10.21161/mjm.112517
Sylia, A., Corrêa, A., Cruz, C., Yadav, A., and Nabti, E. (2022). Plant growth promoting microbes as biofertilizers: Promising solutions for sustainable agriculture under climate change associated abiotic stresses. Plant Sci. Today 8, 60–76. doi:10.14719/pst.1608
Tabacchioni, S., Passato, S., Ambrosino, P., Huang, L., Caldara, M., Cantale, C., et al. (2021). Identification of beneficial microbial consortia and bioactive compounds with potential as plant biostimulants for a sustainable agriculture. Microorganisms 9, 426. doi:10.3390/microorganisms9020426
Tan, W., Law, J., Law, L., Letchumanan, V., and Chan, K. (2020). Insights into quorum sensing (QS): QS-regulated biofilm and inhibitors. Prog. Microbes amp; Mol. Biol. 3. doi:10.36877/pmmb.a0000141
Tang, S., Song, X., Wang, Q., and Wang, S. (2020). Effects of two surfactants on microbial diversity of a PCE-degrading microbial consortium. Chemosphere 261, 127685. doi:10.1016/j.chemosphere.2020.127685
Tennakoon, P., Rajapaksha, R., and Hettiarachchi, L. (2021). Indigenous plant growth-promoting rhizobacterial consortia greatly reduces fertilizer need for tea nurseries: Characterization and evaluation. Sri Lanka J. Food Agric. 7, 41. doi:10.4038/sljfa.v7i1.93
Tzirita, M., Papanikolaou, S., Chatzifragkou, A., and Quilty, B. (2018). Waste fat biodegradation and biomodification by Yarrowialipolytica and a bacterial consortium composed of Bacillus spp. and Pseudomonas putida. Eng. Life Sci. 18, 932–942. doi:10.1002/elsc.201800067
Uruén, C., Chopo-Escuin, G., Tommassen, J., Mainar-Jaime, R. C., and Arenas, J. (2020). Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics 10, 3. doi:10.3390/antibiotics10010003
Vaid, S., Srivastava, P., Pachauri, S., Sharma, A., Rawat, D., Shankhadhar, S., et al. (2020). Effective zinc mobilization to rice grains using rhizobacterial consortium. Israel J. Plant Sci. 67, 145–157. doi:10.1163/22238980-20201085
Vaishnav, A., Kumar, R., Singh, H., and Sarma, B. (2022). Extending the benefits of PGPR to bioremediation of nitrile pollution in crop lands for enhancing crop productivity. Sci. Total Environ. 826, 154170. doi:10.1016/j.scitotenv.2022.154170
Vega-Celedón, P., Bravo, G., Velásquez, A., Cid, F. P., Valenzuela, M., Ramírez, I., et al. (2021). Microbial diversity of psychrotolerant bacteria isolated from Wild Flora of Andes Mountains and Patagonia of Chile towards the selection of plant growth-promoting bacterial consortia to alleviate cold stress in plants. Microorganisms 9, 538. doi:10.3390/microorganisms9030538
Vieira, G., Cabral, L., Otero, I., Ferro, M., Faria, A., Oliveira, V., et al. (2021). Marine associated microbial consortium applied to RBBR textile dye detoxification and decolorization: Combined approach and metatranscriptomic analysis. Chemosphere 267, 129190. doi:10.1016/j.chemosphere.2020.129190
Vortmann, M., Stumpf, A. K., Sgobba, E., Dirks-Hofmeister, M. E., Krehenbrink, M., Wendisch, V. F., et al. (2021). A bottom-up approach towards a bacterial consortium for the biotechnological conversion of chitin to L-lysine. Appl. Microbiol. Biotechnol. 105, 1547–1561. doi:10.1007/s00253-021-11112-5
Wanapaisan, P., Laothamteep, N., Vejarano, F., Chakraborty, J., Shintani, M., Muangchinda, C., et al. (2018). Synergistic degradation of pyrene by five culturable bacteria in a mangrove sediment-derived bacterial consortium. J. Hazard. Mater. 342, 561–570. doi:10.1016/j.jhazmat.2017.08.062
Wang, J., He, Y., Li, T., Li, C., Xu, X., Xiang, H., et al. (2022). Complex biochemical synergistic interactions between two rhizobacteria grown in consortium, bacillus subtilis SL-44 and enterobacterhormaechei Wu-15. Rhizosphere 24, 100587. doi:10.1016/j.rhisph.2022.100587
Weimer, P. J., and Zeikus, J. G. (1977). Fermentation of cellulose and cellobiose by clostridium thermocellum in the absence of MethanobacteriumThermoautotrophicum. Appl. Environ. Microbiol. 33, 289–297. doi:10.1128/aem.33.2.289-297.1977
Woo, S., and Pepe, O. (2018). Microbial Consortia: Promising probiotics as plant biostimulants for sustainable agriculture. Front. Plant Sci. 9, 1801. doi:10.3389/fpls.2018.01801
Wushensky, J., Youngster, T., Mendonca, C., and Aristilde, L. (2018). Flux connections between gluconate pathway, glycolysis, and pentose–phosphate pathway during carbohydrate metabolism in Bacillus megaterium QM B1551. Front. Microbiol. 9, 2789. doi:10.3389/fmicb.2018.02789
Wyrwicka, A., Urbaniak, M., Siebielec, G., Siebielec, S., Chojak-Koźniewska, J., Przybylski, M., et al. (2019). The influence of bottom sediments and inoculation with rhizobacterial inoculants on the physiological state of plants used in urban plantings. Water 11, 1792. doi:10.3390/w11091792
Xu, C., and Yu, H. (2021). Insights into constructing a stable and efficient microbial consortium. Chin. J. Chem. Eng. 30, 112–120. doi:10.1016/j.cjche.2020.12.012
Yaduwanshi, B., Sahu, R., Mitra, N., and Dwivedi, B. (2021). Impact of microbial consortia on microbial population and available nutrients in soil under soybean crop. J. Indian Soc. Soil Sci. 69, 187–194. doi:10.5958/0974-0228.2021.00029.3
Yang, W., Sun, H., Dong, D., Ma, S., Mandlaa, W. Z., Xu, H., et al. (2020). Enhanced 2-keto-l-gulonic acid production by a mixed culture of Ketogulonicigenium vulgare and Bacillus megaterium using three-stage temperature control strategy. Braz. J. Microbiol. 52, 257–265. doi:10.1007/s42770-020-00396-w
Yankey, R., Karajna, J., Okal, E., Omoor, I., Lin, H., Bodjremou, D., et al. (2021). A consortium of plant growth promoting rhizobacteria strains synergistically assists jujuncao (Pennisetum giganteum) to remediate cadmium contaminated soils. Appl. Ecol. Environ. Res. 19, 2425–2442. doi:10.15666/aeer/1903_24252442
Yanti, Y., Hamid, H., and Reflin, (2021). Development of the PGPR and cyanobacteria consortium for growth promotion and control ralstoniasyzigii subsp. indonesiensis of tomato. IOP Conf. Ser. Earth Environ. Sci. 709, 012085. doi:10.1088/1755-1315/709/1/012085
Yuan, M., Guo, X., Wu, L., Zhang, Y., Xiao, N., Ning, D., et al. (2021). Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 11, 343–348. doi:10.1038/s41558-021-00989-9
Zhang, L., Wang, D., Hu, Q., Dai, X., Xie, Y., Li, Q., et al. (2019a). Consortium of plant growth-promoting rhizobacteria strains suppresses sweet pepper disease by altering the rhizosphere microbiota. Front. Microbiol. 10, 1668. doi:10.3389/fmicb.2019.01668
Zhang, L. N., Wang, D.-C., Hu, Q., Dai, X.-Q., Xie, Y.-S., Li, Q., et al. (2019b). Consortium of Plant Growth-promoting rhizobacteria strains suppresses sweet pepper disease by altering the rhizosphere microbiota. Front. Microbiol. 10, 1668. doi:10.3389/fmicb.2019.01668
Zhang, M., Yang, L., Hao, R., Bai, X., Wang, Y., and Yu, X. (2020). Drought-tolerant plant growth-promoting rhizobacteria isolated from jujube (ziziphus jujuba) and their potential to enhance drought tolerance. Plant Soil 452, 423–440. doi:10.1007/s11104-020-04582-5
Zhang, T., and Zhang, H. (2022). Microbial consortia are needed to degrade soil pollutants. Microorganisms 10, 261. doi:10.3390/microorganisms10020261
Zhang, X. H., Ahmad, W., Zhu, X. Y., Chen, J., and Austin, B. (2020). Viable but nonculturable bacteria and their resuscitation: Implications for cultivating uncultured marine microorganisms. Mar. Life Sci. Technol. 3, 189–203. doi:10.1007/s42995-020-00041-3
Keywords: rhizobacteria, rhizosphere, synergistic relation, consortium, sustainable development, cross-feeding
Citation: Singh A, Yadav VK, Chundawat RS, Soltane R, Awwad NS, Ibrahium HA, Yadav KK and Vicas SI (2023) Enhancing plant growth promoting rhizobacterial activities through consortium exposure: A review. Front. Bioeng. Biotechnol. 11:1099999. doi: 10.3389/fbioe.2023.1099999
Received: 16 November 2022; Accepted: 16 January 2023;
Published: 10 February 2023.
Edited by:
Panagiotis Madesis, University of Thessaly, GreeceReviewed by:
Kalpana Bhatt, Purdue University, United StatesCopyright © 2023 Singh, Yadav, Chundawat, Soltane, Awwad, Ibrahium, Yadav and Vicas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rajendra Singh Chundawat, cnNjaHVuZGF3YXQuc2xhc0Btb2R5dW5pdmVyc2l0eS5hYy5pbg==; Simona Ioana Vicas, c2ltX3ZpY2FzQHlhaG9vLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.