
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 16 March 2023
Sec. Biosafety and Biosecurity
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1090814
 Yu-Guo Yuan1,2*†
Yu-Guo Yuan1,2*† Yi-Tian Xing1†
Yi-Tian Xing1† Song-Zi Liu1†
Song-Zi Liu1† Ling Li1,2
Ling Li1,2 Abu Musa Md Talimur Reza3
Abu Musa Md Talimur Reza3 He-Qing Cai1,2
He-Qing Cai1,2 Jia-Lin Wang1,2
Jia-Lin Wang1,2 Pengfei Wu1
Pengfei Wu1 Ping Zhong1*
Ping Zhong1* Il-Keun Kong4*
Il-Keun Kong4*The widespread use of graphene oxide-silver nanoparticle nanocomposites (GO-AgNPs) in biomedical sciences is increasing the chances of human and animal exposure to its chronic non-toxic doses. Exposure to AgNPs-related nanomaterials may result in the negative effect on the dam, fetus and offspring. However, there are only little available information for profound understanding of the epigenetic alteration in the cells and animals caused by low-dose chronic exposure of GO-AgNPs. The present study investigated the effect of 0.5 μg/mL GO-AgNPs for 10 weeks on the differential expression of circular RNAs (circRNAs) in caprine fetal fibroblast cells (CFFCs), and this dose of GO-AgNPs did not affect cell viability and ROS level. We predicted the functions of those differentially expressed (DE) circRNAs in CFFCs by bioinformatics analysis. Furthermore, we validated the expression of ten DE circRNAs using quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) to ensure the reliability of the sequencing data. Our results showed that the DE circRNAs may potentially regulate the GO-AgNPs-inducing epigenetic toxicity through a regulatory network consisted of circRNAs, miRNAs and messenger RNAs (mRNAs). Therefore, the epigenetics toxicity is essential to assess the biosafety level of GO-AgNPs.
The potential therapeutic and biomedical use of nanotechnology is not limited to the treatment of human diseases, it also has an enormous potential to be applied in veterinary biomedical sciences. Take an example, the recent integration of nanotechnology in different aspects of plant and animal production such as diagnosis of diseases, treatment, vaccination, reproduction, feeding, and hygienic management (Chariou et al., 2020; Kutawa et al., 2021). Researches have shown that graphene/silver nanocomposite (generated by coupling of silver nanoparticle nanocomposites (AgNPs) and graphene-based nanomaterials) has enhanced its antibacterial activity with the combination of therapeutic strategies (Tan et al., 2020; Porter et al., 2021). Besides, there are other issues that are being addressed by the practical applications of silver-nanoparticle-based materials, such as overcoming the challenges of antibiotic resistance and enhancing the overall veterinary practices (Yuan et al., 2017a; Estevez et al., 2021). This widespread use of nanotechnology, in particular, the use of graphene/silver nanocomposite has raised the concern of its potential effects on human health. Thus, a comprehensive analysis is required to know the possible long- and short-term effects of AgNP-related materials before its full-scale application in veterinary and animal sciences (Ema et al., 2017a; Yuan et al., 2021). A number of previous studies were conducted to evaluate possible mechanisms of graphene oxide or AgNPs-induced toxicity with various kinds of cells lines and animal models (Liao et al., 2019; Ghulam et al., 2022). Numerous studies have also shown the toxicological effects of graphene oxide-silver nanoparticle nanocomposites (GO-AgNPs) on human cells and animal (Ali et al., 2018; Courtois et al., 2019; Yuan et al., 2021), which even remains two generations post-maternal exposure (Ellis et al., 2020). These studies have identified distinctive cellular responses including long-term stress response, substantial modifications in the regulation of genes, and an upregulation of EGF signaling. However, it is not practical to correlate the findings of the acute in vitro nanoparticles exposure to the chronic in vitro/vivo model (Comfort et al., 2014). For example, chronic prenatal exposure to AgNPs may compromise the neonatal and postnatal development without showing any visible effect on the parents (Zhang et al., 2015; Becaro et al., 2021). Therefore, the potential reproductive and developmental toxicity in livestock is a valid concern related to the practical use of GO-AgNPs (Yuan et al., 2021), and indeed required to be investigated further (Ema et al., 2017b; Becaro et al., 2021; Wang et al., 2018; Dugershaw et al., 2020). As different cell types show varying sensitivity to nanoparticle, it is important to use varieties of cell types to get reliable observations. In our previous study, we have used caprine fetal fibroblast cells (CFFCs), which is considered as a suitable in vitro model for studying long term effect of the nanomaterial-mediated toxicity on the fetus (Yuan et al., 2021).
In the past, AgNPs or GO-AgNPs mediated toxicity has been studied focusing its impacts on the genetic and biological alterations, but recently nanoparticle mediated epigenetic toxicity becomes an attractive field of research (Gedda et al., 2019). The regulatory roles of epigenetic factors on cellular and biological processes are well evident, these factors include DNA methylation and hydroxymethylation, histone modification, chromatin remodeling, RNA methylation, and different non-coding RNAs such as microRNA (miRNA), transfer RNA (tRNA), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs). Recent research has identified that exposure of mammalian cells to AgNPs or GO-AgNPs has changed its DNA methylation, post-translational histone modifications, and microRNAs expression (Zhang et al., 2015; Zhang et al., 2020; Yuan et al., 2021). However, the expression pattern of circRNAs in AgNPs or GO-AgNPs-induced toxicity remains unexplored. CircRNA is a kind of ncRNA, whose structure comprises covalently closed loops without a 5′ end cap and 3′ end poly (A) tail. The majority of circRNAs are conserved across different species, however, it shows tissue or developmental-stage-specific expression (Memczak et al., 2013; Kristensen et al., 2019). The circRNAs are functionally different from miRNA, piRNA and lncRNA, and play an important role as microRNA sponges, regulating gene splicing and transcription, as well as RNA-binding protein sponges and protein/peptide translating (Patop et al., 2019). It also showed close involvement in a series of physiological processes that are regulating fetal growth, muscle formation, and lactation in mammals (Huang et al., 2020; He et al., 2021). CircRNAs are also found to be important in the regulation of post-transcriptional gene expression (Hansen et al., 2013).
Our previous study demonstrated that GO-AgNPs for 48 h can incite DNA hypomethylation, result in the production of reactive oxygen species (ROS) and apoptosis in caprine fetal fibroblast cells (Yuan et al., 2021). Since the synthesized GO-AgNPs is a potential antibacterial agent in veterinary medicine for the treatment of bovine and caprine mastitis during pregnancy, which take long time for treatment. Though developmental effects have been examined at non-maternal toxic doses to assess the direct effects on the embryo/fetus, the epigenetic alterations of which are not clear. The role of epigenetic modifications of circRNAs in GO-AgNPs’ toxicity remains unexplored. Thus, we suppose that circRNAs expression pattern may change in GO-AgNPs-treated cells. This study aims to evaluate the chronic in vitro effect of low concentration GO-AgNPs (0.5 μg/mL) on the circRNAs expression pattern in CFFCs. We have explored the expression pattern of both circRNAs and messenger RNAs (mRNAs) in CFFCs after 10-week of GO-AgNPs exposure and predicted a regulatory network (circRNAs-miRNA-mRNA) that potentially regulates the GO-AgNPs mediated epigenetic toxicity in CFFCs.
CFFCs that were isolated from caprine fetus and stored in our lab (Yuan et al., 2021). The goat owners were informed as to the purpose of this study and provided informed consent for their animals to be used in this study. CFFCs were recovered by dipping and continuous shaking the cryovial in 37°C water bath ∼1 min and seeded into 6-well plate containing 10% FBS-DMEM/F-12 (Thermo Fisher Scientifc, Waltham, MA, United States) supplemented with penicillin (100 units/mL), and streptomycin (100 μg/mL). The cells were cultured at 37°C in a humidified atmosphere containing 5% CO2 as described previously (Yuan et al., 2021). GO-AgNPs that was prepared and stored in our lab was sonicated for 20 min before use (Yuan and Gurunathan, 2017b). The viability and ROS level of CFFCs were not significantly reduced with the concentration of 1 μg/mL GO-AgNPs (Yuan et al., 2021). The practical applications of silver-nanoparticle-based materials to overcome antimicrobial resistance in goat and cow mastitis will be taken for more than some weeks. Therefore, the cells were exposed to continuous treatment with GO-AgNPs (0 and 0.5 μg/mL) for 10 weeks in 10% FBS-DMEM/F-12. During the treatment period, the cells were passaged every 2–3 days.
The cell viability of GO-AgNPs treated and control cells was measured every week using the cell counting kit-8 (CCK-8; Rockville, MD, United States) according to the manufacturer’s instruction as described previously (Yuan et al., 2021). In brief, at the end of treatment period, 10 μL CCK-8 was added into each well containing cells with 100 μL medium and incubated for 30 min at 37°C in dark. The absorbance at 450 nm was measured using a microplate reader (BioTek Synergy 2, United States).
Dichlorodihydrofluorescein diacetate (DCFH-DA) was used to detect intracellular ROS induced by 0 and 0.5 μg/mL of GO-AgNPs according to the manufacturer’s instructions as described previously (Yuan et al., 2021). In brief, CFFCs were incubated with 10 μM DCFH-DA for 30 min at 37°C, then were rinsed twice with PBS and harvested using 0.25% Trypsin-EDTA. The intracellular ROS accumulation was measured by flow cytometry (Beckman-Coulter, United States).
RNA samples were prepared from the cells using Trizol (TANGEN, Beijing, China) according to the manufacturer’s protocol and the optical density (OD260/280) value of the RNA concentration was measured with a NanoDrop ND-2000 instrument (Thermo Fisher Scientific, Waltham, MA, United States). Finally, the integrity of the isolated RNA was confirmed by agarose gel electrophoresis.
High-throughput transcriptome sequencing and bioinformatics analysis were performed using the commercial service provided by Cloud-Seq Biotech (Shanghai, China). In brief, the ribosomal RNA (rRNA) was depleted by treating the total RNA with a Ribo-Zero rRNA Removal kit (Illumina, San Diego, CA, United States). The purified RNA samples were used to generate the RNA library using the TruSeq Stranded Total RNA Library Prep kit (Illumina, San Diego, CA, United States) according to the manufacturer’s protocol. The quantification of the constructed libraries as well as their quality was measured using BioAnalyzer 2100 system. Finally, 10-pM libraries were denatured into single-stranded DNA molecules, captured on Illumina flow cells, amplified in situ, clustered, and 150 cycles of sequencing were performed on the Illumina HiSeq sequencer according to the manufacturer’s instructions.
The paired terminal readings were obtained from the Illumina HiSeq 4,000 sequencer, and Q30 was used to performed quality control. Cutadapt software (version 1.9.3) were used to primarily screened High-quality reads, and the low-quality reads were removed after performing 3′ adapter trimming. STAR software (version 2.5.1b) was used to map and align high-quality reads with the reference genome/transcriptome. After that, some nucleotide sequences from the reads were selected as anchor points and the results were inputted into the DCC software (version 0.4.4) to compare the connected and unconnected reads for identifying possible circRNAs. Finally, the data were normalized using the EdgeR software (version 3.16.5) and analyzed the identified DE circRNAs.
The DE circRNAs and mRNAs between the control and 0.5 μg/mL of GO-AgNPs groups was calculated using standardized readings. CircRNAs and mRNAs with a fold change of ≥ 2.0 and p < 0.05 were considered as DE as well as statistically significant.
GO (http://www.geneontology.org) and KEGG (http://www.genome.jp/kegg) were used to analyze genes that are related to the DE circRNAs and mRNA. GO analysis is divided into three aspects: molecular function (MF), biological process (BP), and cellular component (CC). The top 10 enriched GO terms were ranked according to the p-value. The possible biological functions of DE circRNA and mRNA were analyzed by KEGG pathway analysis.
In order to verify the data obtained from high-throughput RNA sequencing, the expression of five upregulated circRNAs and five downregulated circRNAs was verified by qRT-PCR while the expression of GAPDH was considered as the internal control. In brief, total RNA was reverse-transcribed into complementary DNA using the PrimeScript RT kit (Perfect Real Time; Takara, Osaka, Japan), and qRT-PCR was performed with SYBR Premix Ex Taq II (TaKaRa) using the Roche LightCycler 480II PCR instrument (Basel, Switzerland) for circRNA analyses according to the manufacturer’s protocols. The independent experiments were performed three times and the expression was determined using a threshold cycle while the relative expression level was calculated using the 2−ΔΔCT method. The primer sets that were used to amplify the selected circRNAs are enlisted in Table 1.
TargetScan (version 7.0) and MiRanda (version 3.3a) were used to predict the miRNA binding sites along with the target mRNAs. After that, Cytoscape software (version 3.1.0) was used to construct a network map of circRNA-miRNA-mRNA based on the results predicted before.
Analysis of the experimental data was performed using the GraphPad Prism 7.0 (GraphPad Software Inc., CA, United States). Student’s t-test and one-way ANOVA were used to compare differences between groups as appropriate. Data are presented as the mean ± standard deviation (SD), and a p-value < 0.05 was considered as statistically significant. The data are representative of at least three experiments.
To determining the chronic cytotoxic effect of the synthesized GO-AgNPs, the CFFCs were cultured in the medium containing 0.5 μg/mL GO-AgNPs for 10 weeks. The viability of the cells was measured every week using CCK-8 kit. As shown in Figure 1A, treatment with 0.5 μg/mL GO-AgNPs for 10 weeks did not cause significant reduction in the viability of CFFCs compared to the non-treated cells.
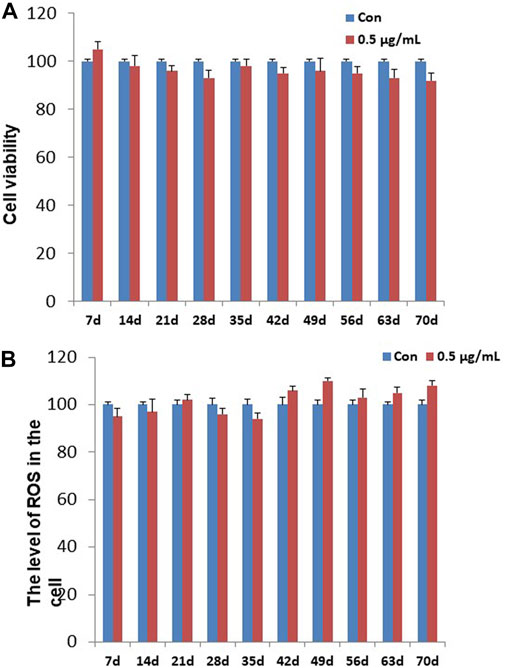
FIGURE 1. Effects of GO-AgNPs on the proliferation and ROS level of caprine fetal fibroblast cells (CFFCs). CFFCs were exposed to 0 and 0.5 μg/mL of GO-AgNPs for 70 days. (A) The percentage of CFFCs viability was then calculated relative to the control group (0 μg/mL). (B) Total reactive oxygen species (ROS) generation in GO-AgNP-treated cells. Caprine fetal fibro-blast cells (CFFCs) were checked for ROS every week. The percentage of ROS generation relative to the untreated control group (0 μg/mL). Values are presented as the mean ± SD of four independent experiments.
To detect the changes in ROS in the CFFCs that was exposed to the chronic dose of GO-AgNPs, intracellular ROS generation in both treated (0.5 μg/mL GO-AgNPs) and non-treated CFFCs was measured at every week. As shown in Figure 1B, treatment with 0.5 μg/mL GO-AgNPs for 10 weeks did not increase intracellular ROS production significantly.
To know the effect of chronic exposure to GO-AgNPs (0.5 μg/mL for 10 weeks) on circRNA expression, the RNA samples obtained from both treated and non-treated CFFCs were sequenced using Illumina high throughput sequencing platform. A total of 3,508 circRNAs were identified in CFFCs, which have not been reported before. As shown in Figure 2, most of the circRNAs were located on chromosomes 1–29 (Figure 2A) and the size of the detected circRNAs ranged between 62 and 90,000 nucleotides (Figure 2B). Although most of the circRNAs were of exonic origin (Figure 3C) but there were also circRNAs that were originated from non-exonic genome such as antisense, intergenetic and intronic (Figure 2D).
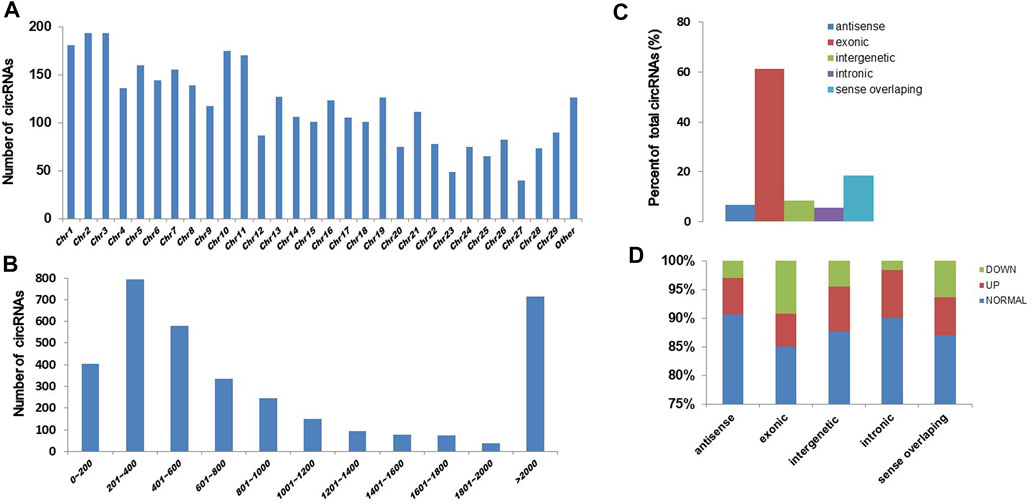
FIGURE 2. Expression pattern of circRNAs in the GO-AgNPs treated caprine fetal fibroblast cells (CFFCs). (A) Distribution of circRNAs on chromosomes. The x-axis represents the chromosome and the y-axis represents the number of circRNA. (B) The length distribution of total circRNAs. The x-axis represents the number of circRNAs and the y-axis represents the number of circRNA. (C) The genomic origin of the total circRNAs. (D) Percent of differentially expressed circRNAs.
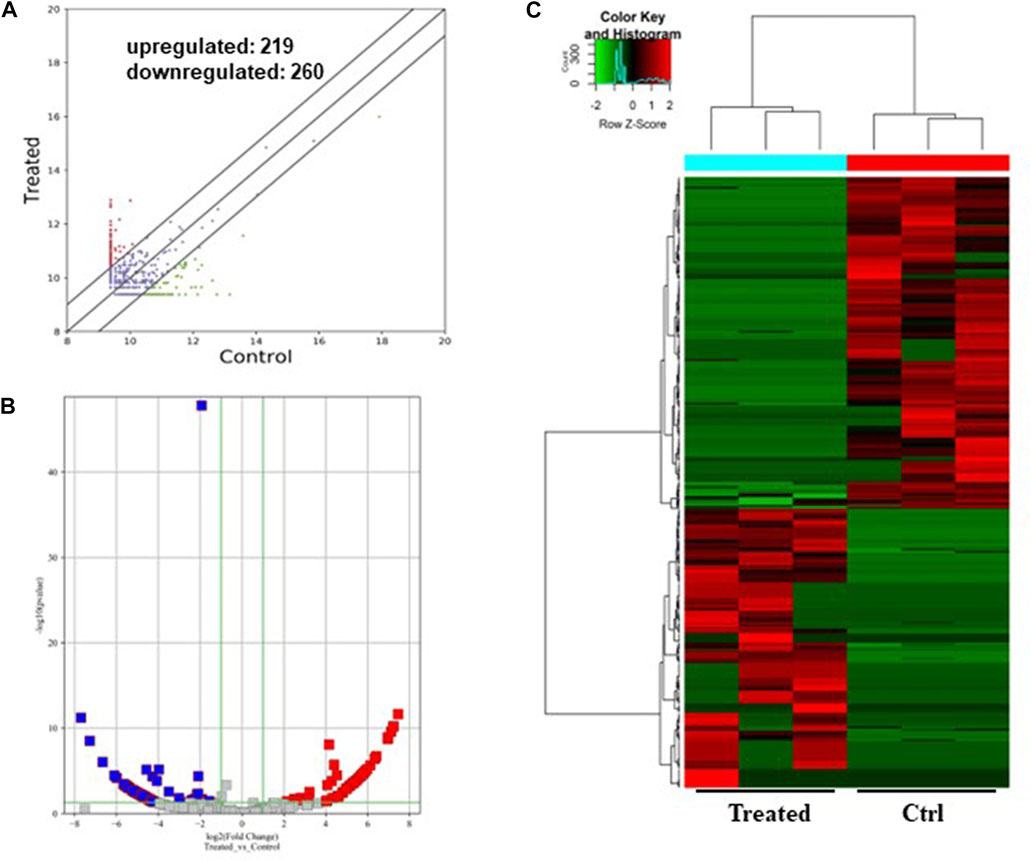
FIGURE 3. Differentially expressed circRNAs in the GO-AgNPs treated caprine fetal fibroblast cells. (A) Hierarchical cluster analysis of circRNAs in the cells between treated and control group. The values of the x and y-axes in the scatterplot are the averaged normalized signal values of groups of samples (log2 scaled). The oblique lines are fold change lines. The circRNAs with the red points (219) and blue points (260) indicate > 2.0 fold changes between control and treated cells. (B). Volcano plots were constructed using fold-change values and p-values. The red and blue points represent the differentially up- and downregulated circRNAs with statistical significance (fold-changes of > 2 and p values of < 0.05). (C) Clustered heatmap of differentially expressed circRNAs in the cells. The red and green represent upregulated and downregulated circRNAs, respectively.
As shown in Figure 3, a total of 479 circRNAs were expressed differentially between the control and treated groups: 219 upregulated and 260 downregulated (Figures 3A, B). Hierarchical clustering showed the circRNA expression pattern in the 0.5 μg/mL GO-AgNPs treated and non-treated CFFCs (Figure 3C).
GO and KEGG pathway enrichment analyses were performed to understand the biological roles of the DE circRNAs. GO analysis showed that the downregulated circRNAs are mainly involved in peptidyl-amino acid modification, intracellular, and GTPase regulator activity (Figures 4A–F) while the KEGG pathway enrichment indicated that the downregulated circRNAs are predominantly connected to the Ras signaling pathway (Figures 4G, H). The other KEGG pathways that are potentially connected to the downregulated circRNAs includes Axon guidance, FoxO signaling pathway, Hepatitis B, MAPK signaling pathway, Adherens junction, Focal adhesion, ErbB signaling pathway, TGF-beta signaling pathway, and Pancreatic cancer.
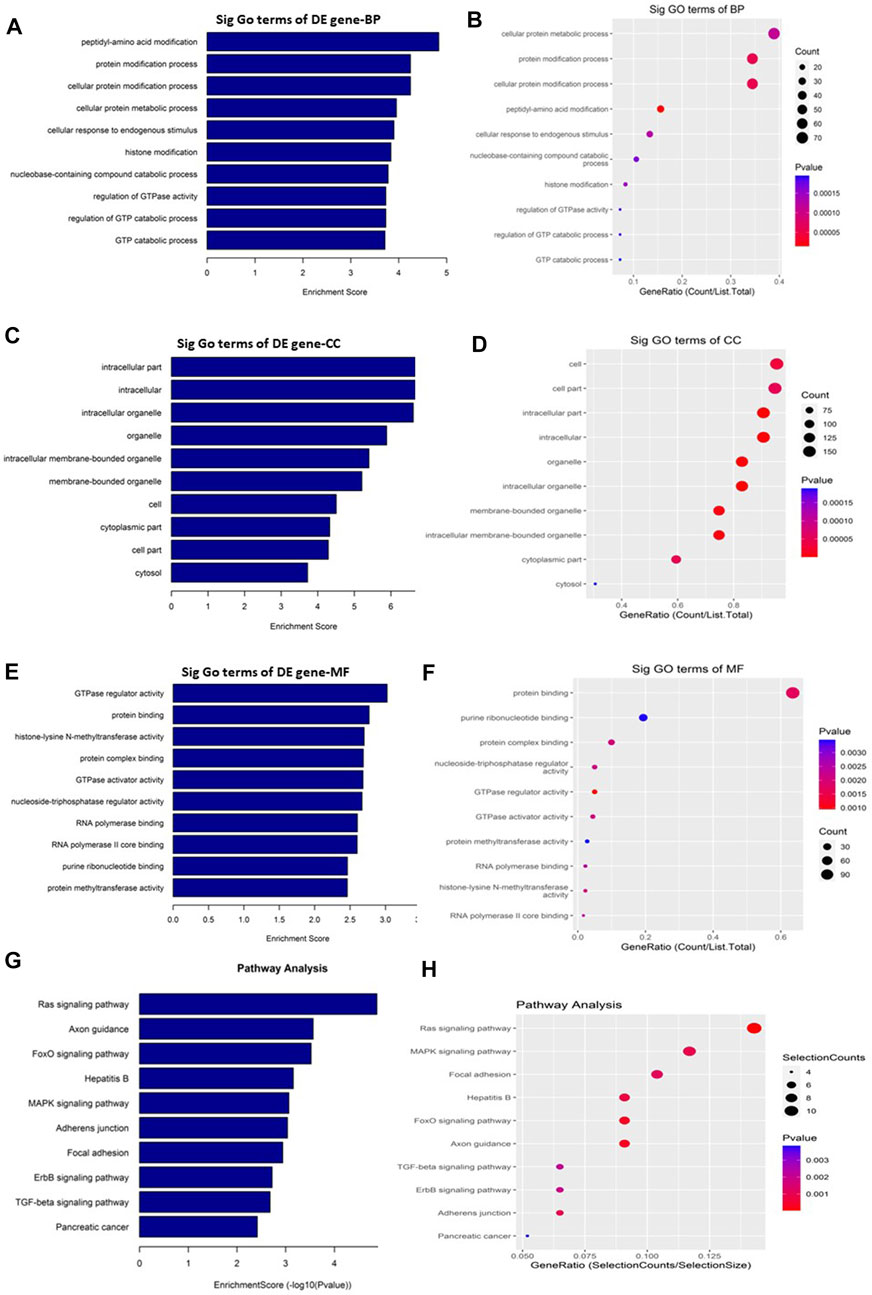
FIGURE 4. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of downregulated circRNAs in the GO-AgNPs treated caprine fetal fibroblast cells (CFFCs). GO analysis identified biological process (BP), cellular component (CC) and molecular function (MF) with the enrichment score [(A,C,E) respectively] and gene count (B,D,F). The top ten enriched KEGG pathways of significantly altered circRNA genes (G,H). The color intensity of the nodes shows the degree of enrichment of this analysis. The enrich-factor is defined as the ratio of the differential genes in the entire genome. The dot size represents the count of genes in a pathway.
On the other hand, the upregulated circRNAs showed involvement mainly in organelle organization, intracellular part and ras GTPase binding (Figures 5A–F). They are also potentially involved in the epigenetic regulation through histone H4K8 and H4K5 acetyla-tion. In addition, the upregulated circRNAs showed high enrichment for KEGG apoptosis pathway (Figures 5G, H). Among the other KEGG pathways that showed enrichment for circRNAs includes focal adhesion, propanoate metabolism, chronic myeloid leukemia, osteoclast differentiation, bacterial invasion of epithelial cells, tight junction, T cell recep-tor signaling pathway, lysine degradation, and VEGF signaling pathway.
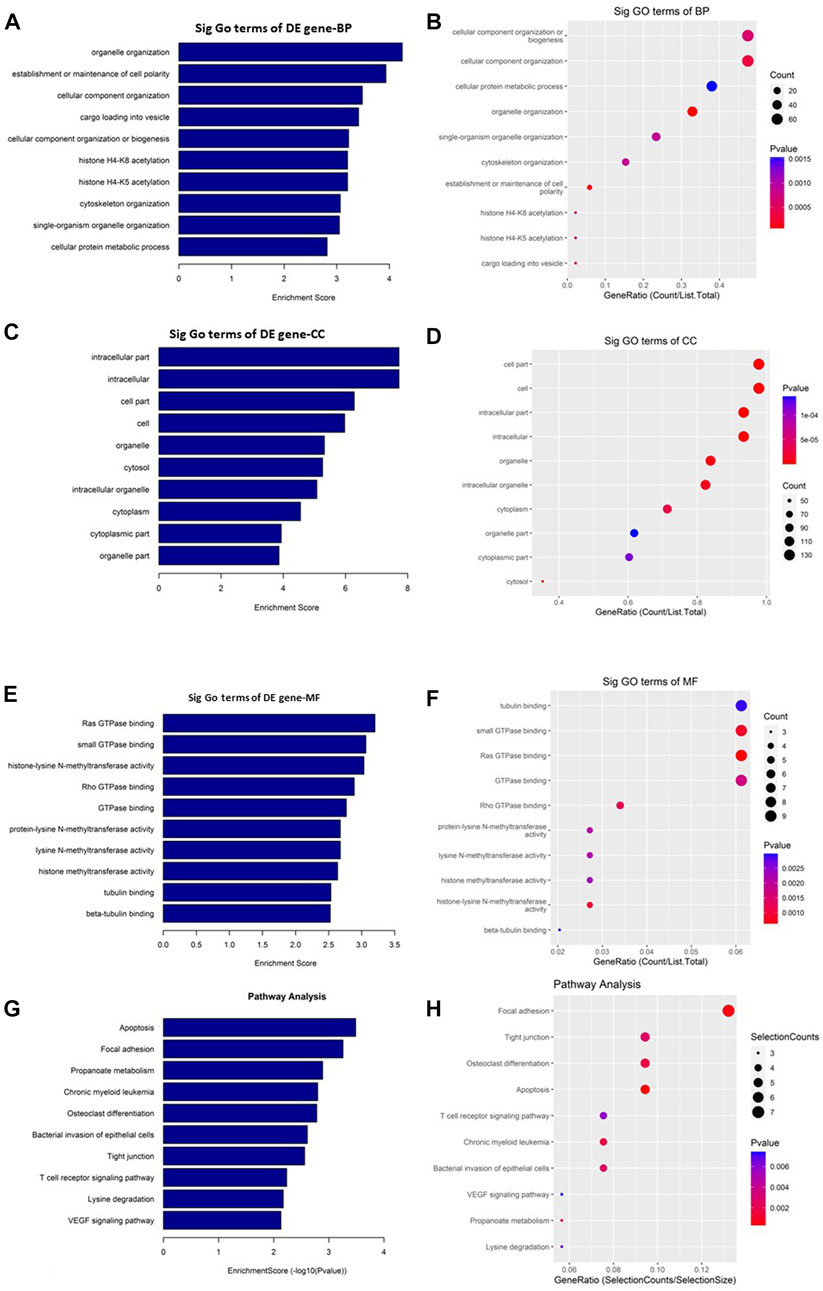
FIGURE 5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of upregulated circRNAs in the GO-AgNPs treated caprine fetal fibroblast cells (CFFCs). GO analysis identified biological process (BP), cellular compo-nent (CC) and molecular function (MF) with the enrichment score [(A,C,E) respectively] and gene count (B,D,F). The top ten enriched KEGG pathways of significantly altered circRNA genes (G,H). The color intensity of the nodes shows the degree of enrichment of this analysis. The enrich-factor is defined as the ratio of the differential genes in the entire genome. The dot size represents the count of genes in a pathway.
As shown in Figure 6, a total of 11374 messenger RNAs (mRNAs) were differentially expressed in the CFFCs where 5,159 mRNAs upregulated, and 6,215 mRNAs downregulated (Figures 6A, B). The hierarchical cluster showed the differential expression pattern of mRNAs between 0.5 μg/mL GO-AgNPs treated and non-treated CFFCs (Figure 6C).
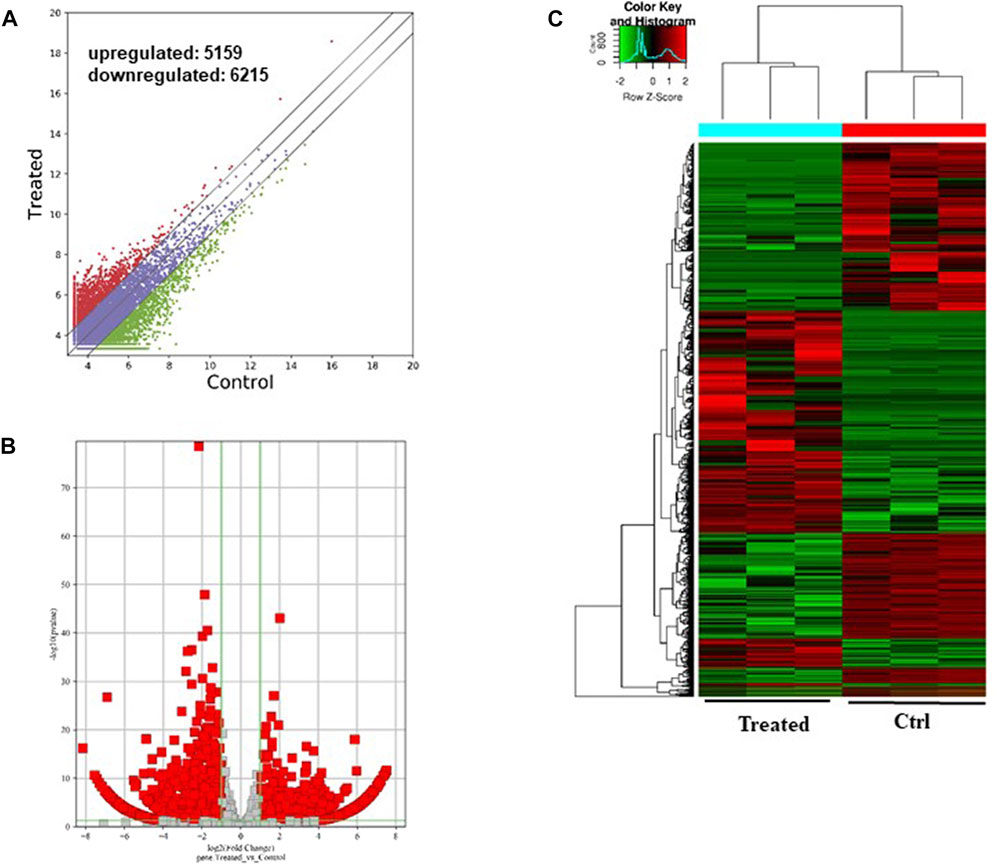
FIGURE 6. Differentially expressed mRNAs in the GO-AgNPs treated caprine fetal fibroblast cells (CFFCs). (A) Hierarchical cluster analysis of mRNAs in the cells. The mRNAs with the red points and blue points indicate > 2.0 fold changes between control and treated cells. (B) Volcano plots were constructed using fold-change values and p-values. The red and blue points represent the differentially up- and downregulated mRNAs with statistical significance (fold-changes of > 2 and p-values of < 0.05). (C) Clustered heatmap of differentially expressed mRNAs in the cells.
As shown in Figure 7, the downregulated transcripts have potential regulatory roles in cellular macromolecule catabolic process, lysosome formation, and catalytic activity (Figures 7A–F). In addition, these downregulated transcripts might have important role in the signaling pathways related to cancer (Figures 7G, H). On the other hand, the upregu-lated mRNAs are potentially involved in the biogenesis and organization of cellular components such as actomyosin (Figures 8A–F) as well as signaling pathways related to focal adhesion, ECM-receptor interaction, and biosynthesis of amino acids (Figures 8G, H).
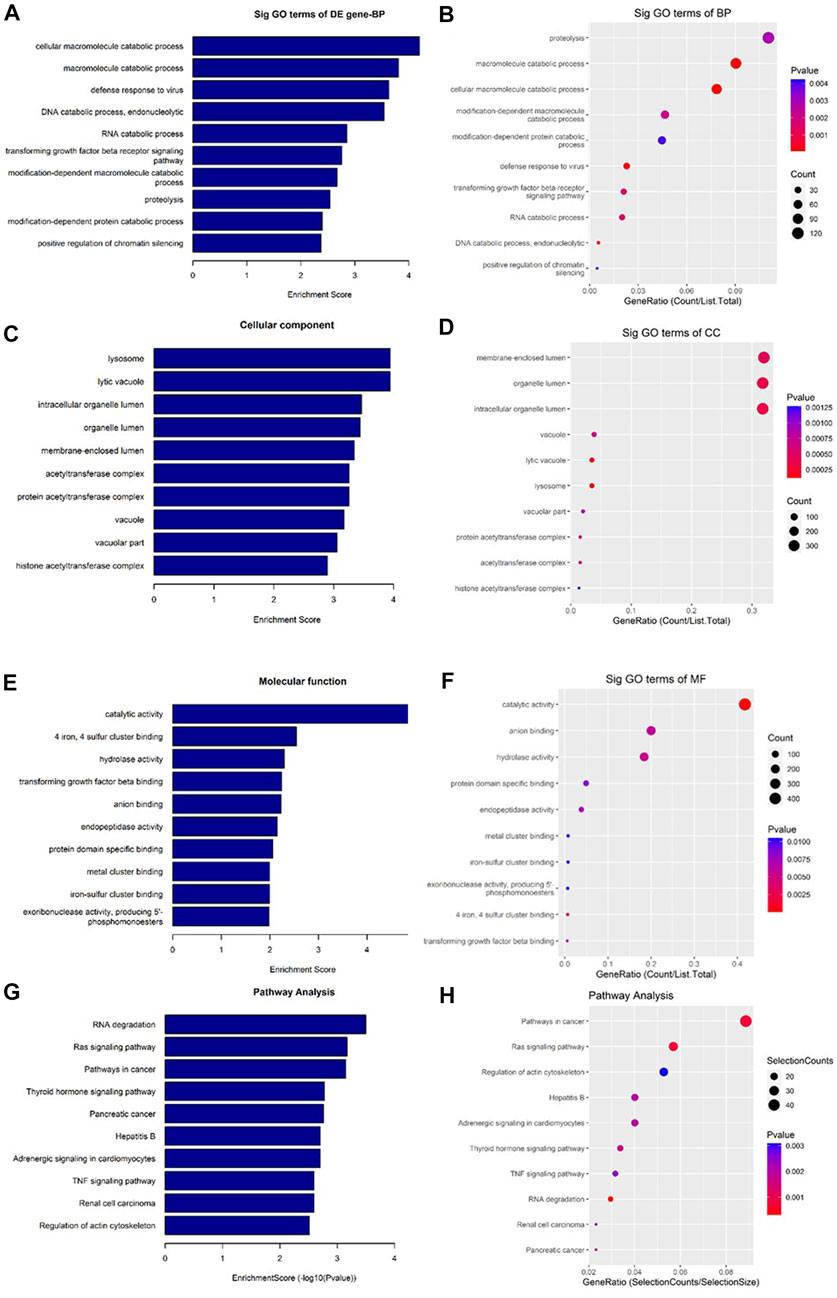
FIGURE 7. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of downregulated mRNAs in the GO-AgNPs treated caprine fetal fibroblast cells (CFFCs). GO analysis identified biological process (BP), cellular component (CC) and molecular function (MF) with the enrichment score [(A,C,E) respectively] and gene count (B,D,F), The top ten enriched KEGG pathways of significantly altered mRNA genes (G,H). The color intensity of the nodes shows the degree of enrichment of this analysis. The enrich-factor is defined as the ratio of the differential genes in the entire genome. The dot size represents the count of genes in a pathway.
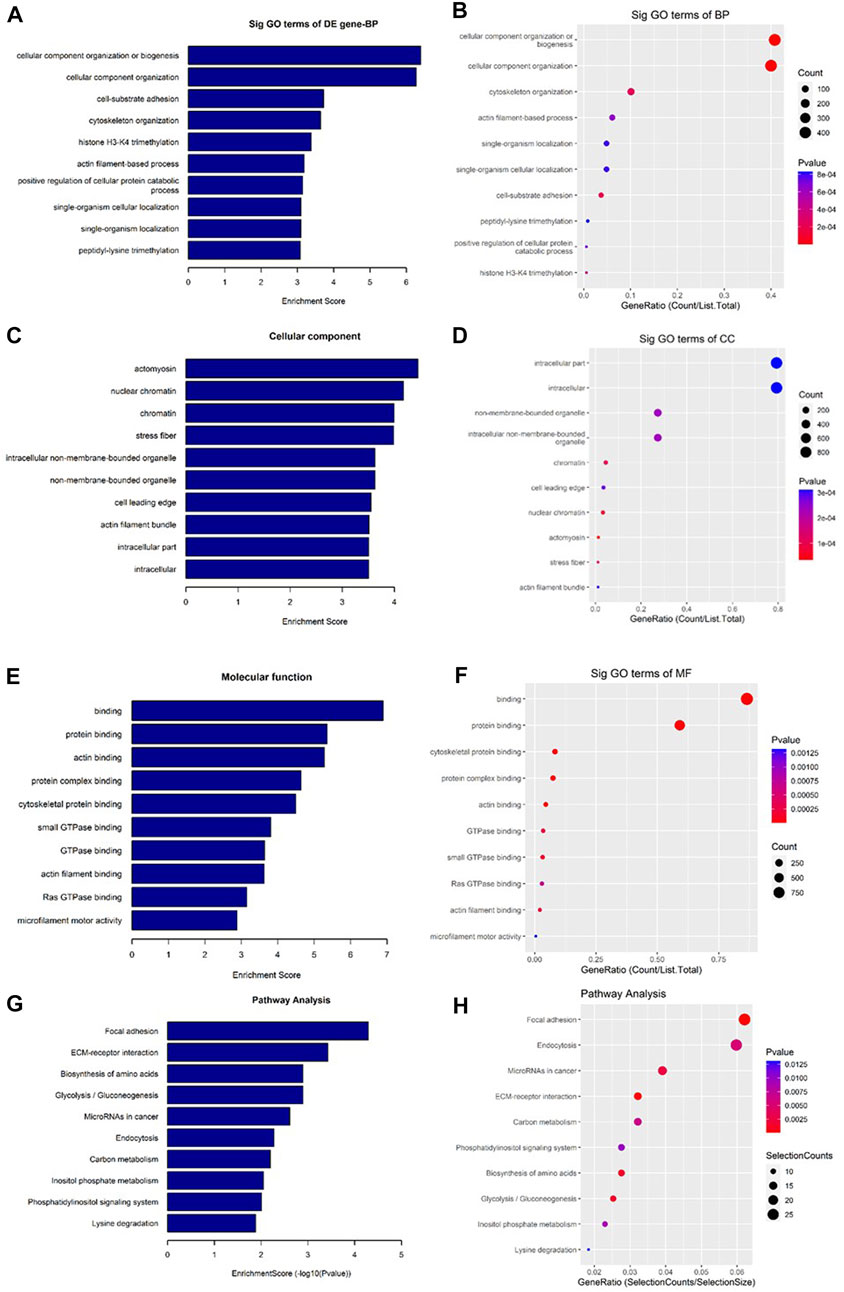
FIGURE 8. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of upregulated mRNAs in the GO-AgNPs treated caprine fetal fibroblast cells (CFFCs). GO analysis identified biological process (BP), cellular component (CC) and molecular function (MF) with the enrichment score [(A,C,E) respectively] and gene count (B,D,F). The top ten enriched KEGG pathways of significantly altered mRNA genes (G,H). The color intensity of the nodes shows the degree of enrichment of this analysis. The enrich-factor is defined as the ratio of the differential genes in the entire genome. The dot size represents the count of genes in a pathway.
To confirm the reliability of the sequencing results of top differentially expressed circRNAs (Tables 2, 3), the expression level of 10 DE circRNAs (five upregulated and five downregulated) were tested by qRT-PCR. The results showed similar pattern of circRNAs expression as it was initially detected by high throughput sequencing (Figure 9).
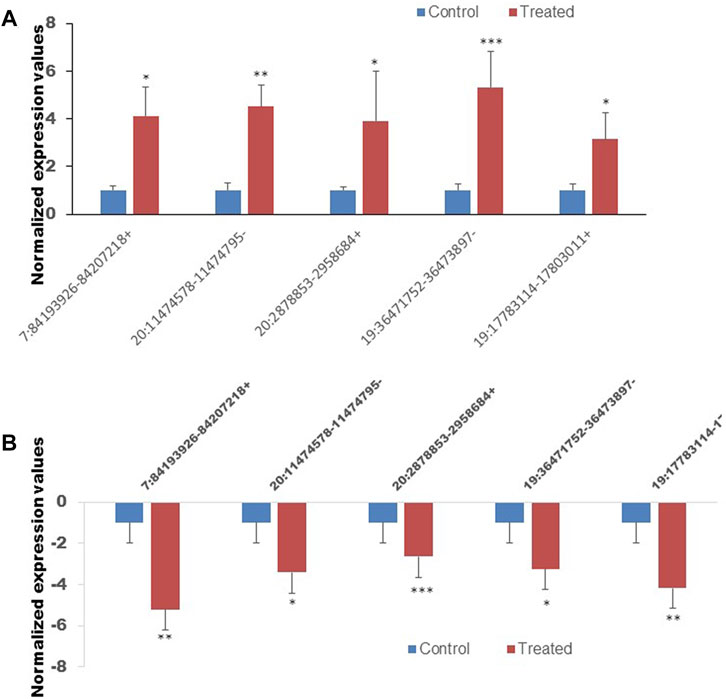
FIGURE 9. The expression of ten circRNAs confirmed by the quan-titative real-time reverse transcription-polymerase chain reaction (qRT-PCR). (A) The expression of five selected upregulated circRNAs. (B) The expression of five selected downregulated circRNAs. *p < 0.05, **p < 0.01, ***p < 0.001. The primer sets that were used to amplify the selected circRNAs are enlisted in Table 1.
Five confirmed circRNAs were considered to predict their regulatory network using bioinformatics analysis. As shown in Figure 10, all five circRNAs are connected to the miRNAs that play regulatory roles on the differentially expressed mRNAs. This indicates that the circRNA-miRNA-mRNA creates an intimate regulatory loop.
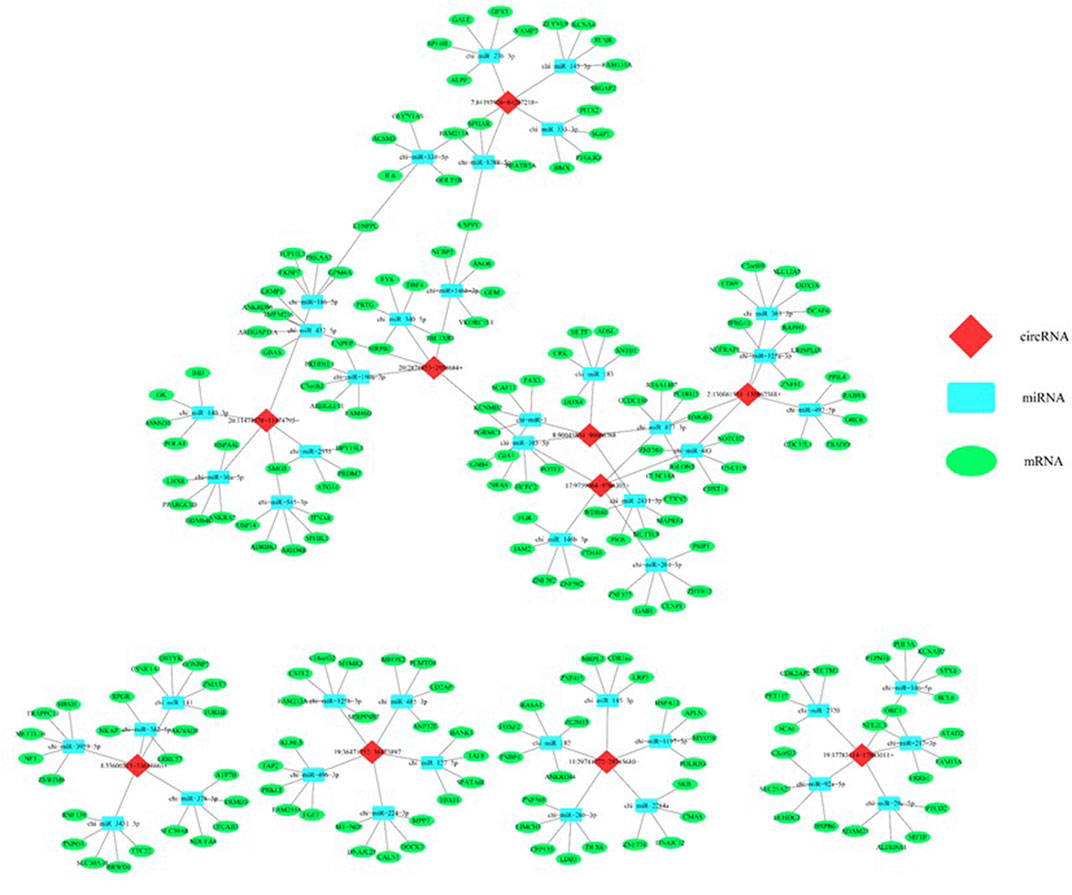
FIGURE 10. In silico analysis predicted a regulatory network consisted of circRNAs, miRNAs, and mRNAs that potentially regulates the epigenetic toxicity in caprine fetal fibroblast cells (CFFCs) induced by the chronic exposure to the GO-AgNPs. Each of the top five circRNAs related to five miRNAs and mRNAs were selected to construct the network.
CircRNAs is a large class of non-coding RNA which has regulatory roles on different molecules including mRNAs, non-coding RNAs, DNAs and proteins (Chen, 2020) as well as on various biological processes (Huang et al., 2020; Yang et al., 2021). To understand how nanotoxicity interferes the ex-pression pattern and functions of circRNAs, we have induced GO-AgNPs mediated chronic toxicity in CFFCs. This does not cause oxidative stress and cell death but resulted in differential expression of mRNAs and circRNAs. Further, the in silico analysis predicted that these differentially expressed circRNAs and mRNAs along with the miRNAs creates a regulatory loop to induce the GO-AgNPs mediated epigenetic toxicity in CFFCs.
The unaltered cell viability and ROS level upon treatment with 0.5 μg/mL GO- AgNPs for 10 weeks has ensured that the experimental does was indeed non-cytotoxic for CFFCs. Previous experiment showed that human dermal fibroblast remains viable up to 20 weeks under the exposure of gold nanoparticles (Falagan-Lotsch et al., 2016) while HaCaTs have insignificant change in cell viability after 14 weeks’ exposure of silver nanoparticles (Comfort et al., 2014). Long-term exposure to AgNPs also induces cell transformation (Choo et al., 2016; Vila et al., 2017). Therefore, our results along with the previous findings suggest that a chronic exposure to different nanoparticles could be visibly non-cytotoxic and non-apoptotic. Of course, an acute dose of nanoparticle is always cytotoxic, as we have shown that only 24 h exposure to 4–8 μg/mL GO-AgNPs is detrimental to the CFFCs (Yuan et al., 2021). Although, the cytotoxicity varies among the type of cells (Yuan and Gurunathan, 2017b; Yuan et al., 2017c), the graphene originated nanomaterials mediated toxicity is well proven in both animal and human cells (Ema et al., 2017a; Wu et al., 2021), and showed much more activity against cancerous cells than normal ones by increasing the expression level of the intracellular ROS and Bax/Bcl-2 genes (Gurunathan et al., 2019).
Accumulated evidence demonstrates that epigenetic modification have been used to check the toxicity inducing by engineered nanomaterials and nanoparticles and, more importantly, to predict their toxicity in preclinical assessments. Several types of nanoparticles have been proved to induce epigenetic modification in DNA methylation patterns, histone modifications and miRNA expressions (Wong et al., 2017). The expression of miRNAs was significantly altered by treatment with graphene oxide or AgNPs, which involved in various apoptosis-related biological pathways (Brzóska et al., 2019; Gurunathan et al., 2019). However, how the chronic exposure to na-noparticles changes epigenetic signature of the cells is not well understood. In the present research, we have found that even a visibly non-cytotoxic and non-apoptotic dose of GO-AgNPs can significantly alter the epigenetic status of CFFCs. The sequencing data showed that 479 circRNAs were differentially expressed in CFFCs after exposed to 0.5 μg/mL GO-AgNPs, of which 219 genes were upregulated and 260 genes were downregulated. 5,159 mRNAs were upregulated and 6,215 mRNAs were downregulated. Interestingly, the altered circRNAs are connected to a group of miRNAs (such as miR-186-5p, miR432-5p, miR-190a-3p) that potentially regulate the expression of the deregulated mRNAs. Further, our in silico prediction showed that several signaling pathways and biological processes are associated with the differentially expressed circRNAs. This means that the circRNAs, miRNAs and mRNAs created a regulatory loop to response the chronic toxicity induced by GO-AgNPs. It is known that different nanoparticles can alter the expression of genes (Ganjouzadeh et al., 2022), and miRNAs (Eom et al., 2014; Oh et al., 2016). But the alteration of circRNAs expression in response to the chronic exposure to the GO-AgNPs has not been reported yet. Hence, our findings revealed the GO-AgNPs induced alteration of circRNAs expression in CFFCs and its potential relevance to the biological processes.
Moreover, previous researches have shown that circRNAs regulate epigenetic status by mediating histone modification in response to both in vivo and in vitro exposure of nanomaterials (Pogribna and Hammons, 2021). Treatment of mouse erythroleukemia cells or A549 cells with 8 or 10 μg/mL AgNPs modified level of acetylation of histone H3 (Qian et al., 2015; Blanco et al., 2017). Our GO results showed that the differentially expressed circRNAs are potentially connected to a group of genes that are related to epigenetic modifications such as histone modification, histone-lysine N-methyltransferase activity, histone H4K5/8 acetylation, lysine N-methyltransferase activity and histone methyltransferase activity while the genes that are connected to the DE circRNAs includes KAT7, SMYD3, KMT2C/D and TET3, which are related to histone modification. CircMRPS35 can recruit KAT7 to the promoters in FOXO1 and FOXO3a genes, which elicits acetylation of H4K5 in their promoters (Jie et al., 2020). SMYD3 and KMT2C/D is histone H3 lysine 4 dimethyltransferase and trimethyltransferase (Hamamoto et al., 2004; Allis et al., 2007), resulting in a significant reduction of global methylation for gene activation (Qian et al., 2015). Previous studies demonstrated that the reduced level of global DNA methylation by nanoparticle were accompanied by a decrease in total activity of DNA methyltransferase such as DNMT, TET (Patil et al., 2016; Choudhury et al., 2017). Overall, our data may highlight the cross-talk between histone modification and toxicity induced by nanoparticle.
The interaction between the top five up- and five downregulated circRNAs and their respected 5 miRNA targets were predicted by conserved seed-matching sequencing using TargetScan and miRanda database, and Cytoscape software was used to construct a network. The circRNA-miRNA network indicated that all of the DE circRNAs have their respective miR response elements, which may provide direction for further studies.
In the present study, 0.5 μg/mL GO-AgNPs did not increase intracellular ROS production in CFFCs after tested for 10 weeks but induced epigenetic alterations. Enriched GO terms and KEGG pathways for differentially circRNAs showed involvement mainly in organelle organization, intracellular part, ras GTPase binding and the Ras signaling pathway. In conclusion, chronic exposure of GO-AgNPs causes epigenetic alterations in CFFCs that may be potentially controlled by a network consisted of circRNAs, miRNAs, and mRNAs. However, further studies are required to understand whether alteration of circRNAs expression is the ultimate cause of epigenetic changes or it is just another component of the regulatory loop. Future studies may focus on understanding of the mechanisms and processes in epigenetic alterations induced by the nano-materials and the role of epigenetic alterations as biomarkers of the nanomaterials’ toxicity. The role of special circRNA in governing histone modification also need to be further identified in the GO-AgNPs treated CFFCs.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE199594.
The animal study was reviewed and approved by the Animal Ethics Committee of Yangzhou University (No. 2018.1109). Written informed consent was obtained from the owners for the participation of their animals in this study.
Y-GY: conceptualization, investigation, supervision, review and editing. Y-TX and LL: investigation, visualization, methodology. S-ZL: investigation, methodology. AR: review and editing. H-QC: software, data curation, formal analysis. J-LW: conceptualization, methodology. PW: data cura-tion. PZ: supervision. I-KK: review.
This project was partly funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), development of a new precise cytosine base editor (JBGS[2021]025), the 111 Project D18007, Yangzhou city and Yangzhou University corporation (YZ2021161) and the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (Grant No. NRF 2020R1A2C2006614/2020-IT-RD-0193-01-101, RS-2023-00208894).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ali, D., Alarifi, S., Alkahtani, S., and Almeer, R. S. (2018). Silver-doped graphene oxide nanocomposite triggers cytotoxicity and apoptosis in human hepatic normal and carcinoma cells. Int. J. Nanomedicine 13, 5685–5699.
Allis, C. D., Berger, S. L., Cote, J., Dent, S., Jenuwien, T., Kouzarides, T., et al. (2007). New nomenclature for chromatin-modifying enzymes. Cell 131, 633–636. doi:10.1016/j.cell.2007.10.039
Becaro, A. A., de Oliveira, L. P., de Castro, V. L. S., Siqueira, M. C., Brandão, H. M., Correa, D. S., et al. (2021). Effects of silver nanoparticles prenatal exposure on rat offspring development. Environ. Toxicol. Pharmacol. 81, 103546. doi:10.1016/j.etap.2020.103546
Blanco, J., Lafuente, D., Gómez, M., García, T., Domingo, J. L., and Sánchez, D. J. (2017). Polyvinyl pyrrolidone-coated silver nanoparticles in a human lung cancer cells: Time- and dose-dependent influence over p53 and caspase-3 protein expression and epigenetic effects. Arch. Toxicol. 91, 651–666. doi:10.1007/s00204-016-1773-0
Brzóska, K., Grądzka, I., and Kruszewski, M. (2019). Silver, gold, and iron oxide nanoparticles alter miRNA expression but do not affect DNA methylation in HepG2 cells. Mater. (Basel) 12, 1038. doi:10.3390/ma12071038
Chariou, P. L., Ortega-Rivera, O. A., and Steinmetz, N. F. (2020). Nanocarriers for the delivery of medical, veterinary, and agricultural active ingredients. Acs. Nano. 14, 2678–2701. doi:10.1021/acsnano.0c00173
Chen, L. L. (2020). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell. Biol. 21, 475–490. doi:10.1038/s41580-020-0243-y
Choo, W. H., Park, C. H., Jung, S. E., Moon, B., Ahn, H., Ryu, J. S., et al. (2016). Long-term exposures to low doses of silver nanoparticles enhanced in vitro malignant cell transformation in non-tumorigenic BEAS-2B cells. Toxicol. Vitro. 37, 41–49. doi:10.1016/j.tiv.2016.09.003
Choudhury, S. R., Ordaz, J., Lo, C. L., Damayanti, N. P., Zhou, F., and Irudayaraj, J. (2017). From the cover: Zinc oxide nanoparticles-induced reactive oxygen species promotes multimodal cyto- and epigenetic toxicity. Toxicol. Sci. 156, 261–274. doi:10.1093/toxsci/kfw252
Comfort, K. K., Braydich-Stolle, L. K., Maurer, E. I., and Hussain, S. M. (2014). Less is more: Long-term in vitro exposure to low levels of silver nanoparticles provides new insights for nanomaterial evaluation. Acs. Nano. 8, 3260–3271. doi:10.1021/nn5009116
Courtois, P., Rorat, A., Lemiere, S., Guyoneaud, R., Attard, E., Levard, C., et al. (2019). Ecotoxicology of silver nanoparticles and their derivatives introduced in soil with or without sewage sludge: A review of effects on microorganisms, plants and animals. Environ. Pollut. 253, 578–598. doi:10.1016/j.envpol.2019.07.053
Dugershaw, B. B., Aengenheister, L., Hansen, S. S. K., Hougaard, K. S., and Buerki-Thurnherr, T. (2020). Recent insights on indirect mechanisms in developmental toxicity of nanomaterials. Part. Fibre. Toxicol. 17, 31. doi:10.1186/s12989-020-00359-x
Ellis, L. A., Kissane, S., Hoffman, E., Brown, J. B., Valsami-Jones, E., Colbourne, J., et al. (2020). Multigenerational exposures of Daphnia magna to pristine and aged silver nanoparticles: Epigenetic changes and phenotypical ageing related effects. Small 16, e2000301. doi:10.1002/smll.202000301
Ema, M., Gamo, M., and Honda, K. (2017b). A review of toxicity studies on graphene-based nanomaterials in laboratory animals. Regul. Toxicol. Pharmacol. 85, 7–24. doi:10.1016/j.yrtph.2017.01.011
Ema, M., Okuda, H., Gamo, M., and Honda, K. (2017a). A review of reproductive and developmental toxicity of silver nanoparticles in laboratory animals. Reprod. Toxicol. 67, 149–164. doi:10.1016/j.reprotox.2017.01.005
Eom, H. J., Chatterjee, N., Lee, J., and Choi, J. (2014). Integrated mRNA and microRNA profiling reveals epigenetic mechanism of differential sensitivity of Jurkat T cells to AgNPs and Ag ions. Toxicol. Lett. 229, 311–318. doi:10.1016/j.toxlet.2014.05.019
Estevez, M. B., Casaux, M. L., Fraga, M., Faccio, R., and Alborés, S. (2021). Biogenic silver nanoparticles as a strategy in the fight against multi-resistant Salmonella enterica isolated from dairy calves. Front. Bioeng. Biotechnol. 9, 644014. doi:10.3389/fbioe.2021.644014
Falagan-Lotsch, P., Grzincic, E. M., and Murphy, C. J. (2016). One low-dose exposure of gold nanoparticles induces long-term changes in human cells. Proc. Natl. Acad. Sci. 113, 13318–13323. doi:10.1073/pnas.1616400113
Ganjouzadeh, F., Khorrami, S., and Gharbi, S. (2022). Controlled cytotoxicity of Ag-GO nanocomposite biosynthesized using black peel pomegranate extract against MCF-7 cell line. J. Drug. Deliv. Sci. Tec. 71, 103340. doi:10.1016/j.jddst.2022.103340
Gedda, M. R., Babele, P. K., Zahra, K., and Madhukar, P. (2019). Epigenetic aspects of engineered nanomaterials: Is the collateral damage inevitable? Front. Bioeng. Biotechnol. 7, 228. doi:10.3389/fbioe.2019.00228
Ghulam, A. N., Dos Santos, O. A. L., Hazeem, L., Pizzorno, B. B., Bououdina, M., and Bellucci, S. (2022). Graphene oxide (GO) materials-applications and toxicity on living organisms and environment. J. Funct. Biomater. 13, 77. doi:10.3390/jfb13020077
Gurunathan, S., ArsalanIqbal, M., Qasim, M., Park, C. H., Yoo, H., Hwang, J. H., et al. (2019). Evaluation of graphene oxide induced cellular toxicity and transcriptome analysis in human embryonic kidney cells. Nanomater. (Basel) 9, 969. doi:10.3390/nano9070969
Hamamoto, R., Furukawa, Y., Morita, M., Iimura, Y., Silva, F. P., Li, M., et al. (2004). SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell. Biol. 6, 731–740. doi:10.1038/ncb1151
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi:10.1038/nature11993
He, T., Chen, Q., Tian, K., Xia, Y., Dong, G., and Yang, Z. (2021). Functional role of circRNAs in the regulation of fetal development, muscle development, and lactation in livestock. Biomed. Res. Int. 2021, 5383210. doi:10.1155/2021/5383210
Huang, Y., Wang, Y., Zhang, C., and Sun, X. (2020). Biological functions of circRNAs and their progress in livestock and poultry. Reprod. Domest. Anim. 55, 1667–1677. doi:10.1111/rda.13816
Jie, M., Wu, Y., Gao, M., Li, X., Liu, C., Ouyang, Q., et al. (2020). CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol. Cancer. 19, 56. doi:10.1186/s12943-020-01160-2
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. doi:10.1038/s41576-019-0158-7
Kutawa, A. B., Ahmad, K., Ali, A., Hussein, M. Z., Abdul Wahab, M. A., Adamu, A., et al. (2021). Trends in nanotechnology and its potentialities to control plant pathogenic fungi: A review. Biology 10, 881. doi:10.3390/biology10090881
Liao, C., Li, Y., and Tjong, S. C. (2019). Bactericidal and cytotoxic properties of silver nanoparticles. Int. J. Mol. Sci. 20, 449. doi:10.3390/ijms20020449
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi:10.1038/nature11928
Oh, J. H., Son, M. Y., Choi, M. S., Kim, S., Choi, A. Y., Lee, H. A., et al. (2016). Integrative analysis of genes and miRNA alterations in human embryonic stem cells-derived neural cells after exposure to silver nanoparticles. Toxicol. Appl. Pharmacol. 299, 8–23. doi:10.1016/j.taap.2015.11.004
Patil, N. A., Gade, W. N., and Deobagkar, D. D. (2016). Epigenetic modulation upon exposure of lung fibroblasts to TiO2 and ZnO nanoparticles: Alterations in DNA methylation. Int. J. Nanomedicine. 11, 4509–4519. doi:10.2147/ijn.s110390
Patop, I. L., Wüst, S., and Kadener, S. (2019). Past, present, and future of circRNAs. EMBO. J. 38, e100836. doi:10.15252/embj.2018100836
Pogribna, M., and Hammons, G. (2021). Epigenetic effects of nanomaterials and nanoparticles. J. Nanobiotechnology. 19, 2. doi:10.1186/s12951-020-00740-0
Porter, G. C., Duncan, W. J., Jude, A., Abdelmoneim, D., Easingwood, R. A., and Coates, D. E. (2021). Endocytosed silver nanoparticles degrade in lysosomes to form secondary nanoparticle structures during expression of autophagy genes in osteogenic cells. Nanomedicine 33, 102355. doi:10.1016/j.nano.2020.102355
Qian, Y., Zhang, J., Hu, Q., Xu, M., Chen, Y., Hu, G., et al. (2015). Silver nanoparticle-induced hemoglobin decrease involves alteration of histone 3 methylation status. Biomaterials 70, 12–22. doi:10.1016/j.biomaterials.2015.08.015
Tan, S., Wu, X., Xing, Y., Lilak, S., Wu, M., and Zhao, J. X. (2020). Enhanced synergetic antibacterial activity by a reduce graphene oxide/Ag nanocomposite through the photothermal effect. Colloids. Surf. B. Biointerfaces. 185, 110616. doi:10.1016/j.colsurfb.2019.110616
Vila, L., Marcos, R., and Hernandez, A. (2017). Long-term effects of silver nanoparticles in caco-2 cells. Nanotoxicology 11, 771–780. doi:10.1080/17435390.2017.1355997
Wang, R., Song, B., Wu, J., Zhang, Y., Chen, A., and Shao, L. (2018). Potential adverse effects of nanoparticles on the reproductive system. Int. J. Nanomedicine. 13, 8487–8506. doi:10.2147/ijn.s170723
Wong, B. S. E., Hu, Q., and Baeg, G. H. (2017). Epigenetic modulations in nanoparticle-mediated toxicity. Food. Chem. Toxicol. 109, 746–752. doi:10.1016/j.fct.2017.07.006
Wu, K., Zhou, Q., and Ouyang, S. (2021). Direct and indirect genotoxicity of graphene family nanomaterials on DNA-A review. Nanomater. (Basel). 11, 2889. doi:10.3390/nano11112889
Yang, Z., He, T., and Chen, Q. (2021). The roles of CircRNAs in regulating muscle development of livestock animals. Front. Cell. Dev. Biol. 9, 619329. doi:10.3389/fcell.2021.619329
Yuan, Y. G., Cai, H. Q., Wang, J. L., Mesalam, A., Talimur, M. D., Reza, A. M., et al. (2021). Graphene oxide-silver nanoparticle nanocomposites induce oxidative stress and aberrant methylation in caprine fetal fibroblast cells. Cells 10, 682. doi:10.3390/cells10030682
Yuan, Y. G., and Gurunathan, S. (2017b). Combination of graphene oxide&ndash;silver nanoparticle nanocomposites and cisplatin enhances apoptosis and autophagy in human cervical cancer cells. Int. J. Nanomedicine. 12, 6537–6558. doi:10.2147/ijn.s125281
Yuan, Y. G., Peng, Q. L., and Gurunathan, S. (2017a). Effects of silver nanoparticles on multiple drug-resistant strains of Staphylococcus aureus and Pseudomonas aeruginosa from mastitis-infected goats: An alternative approach for antimicrobial therapy. Int. J. Mol. Sci. 18, 569. doi:10.3390/ijms18030569
Yuan, Y. G., Wang, Y. H., Xing, H. H., and Gurunathan, S. (2017c). Quercetin-mediated synthesis of graphene oxide&ndash;silver nanoparticle nanocomposites: A suitable alternative nanotherapy for neuroblastoma. Int. J. Nanomedicine. 12, 5819–5839. doi:10.2147/ijn.s140605
Zhang, W., Liu, S., Han, D., and He, Z. (2020). Engineered nanoparticle-induced epigenetic changes: An important consideration in nanomedicine. Acta. Biomater. 117, 93–107. doi:10.1016/j.actbio.2020.09.034
Keywords: nanotoxicity, circRNA, reduced graphene oxide, silver nanoparticles, caprine fetal fibroblast cells
Citation: Yuan Y-G, Xing Y-T, Liu S-Z, Li L, Reza AMMT, Cai H-Q, Wang J-L, Wu P, Zhong P and Kong I-K (2023) Identification of circular RNAs expression pattern in caprine fetal fibroblast cells exposed to a chronic non-cytotoxic dose of graphene oxide-silver nanoparticle nanocomposites. Front. Bioeng. Biotechnol. 11:1090814. doi: 10.3389/fbioe.2023.1090814
Received: 06 November 2022; Accepted: 24 February 2023;
Published: 16 March 2023.
Edited by:
Giada Graziana Genchi, Italian Institute of Technology (IIT), ItalyReviewed by:
Gianvito Lanave, University of Bari Aldo Moro, ItalyCopyright © 2023 Yuan, Xing, Liu, Li, Reza, Cai, Wang, Wu, Zhong and Kong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Guo Yuan, eXlnOTc3NjQzMEAxNjMuY29t; Ping Zhong, emhvbmdwaW5nNzgxMDEyQDE2My5jb20=; Il-Keun Kong, aWtvbmc3OTAwQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.