
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol., 27 February 2023
Sec. Synthetic Biology
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1090501
This article is part of the Research TopicFood to Medicine: Engineered Microbes for Human Health and PerformanceView all 3 articles
 Hua Ling1,2,3,4†‡
Hua Ling1,2,3,4†‡ Ruirui Liu1,2,3,4‡
Ruirui Liu1,2,3,4‡ Qi Hui Sam1,2,3
Qi Hui Sam1,2,3 Haosheng Shen1,2,3,4
Haosheng Shen1,2,3,4 Louis Yi Ann Chai1,2,5
Louis Yi Ann Chai1,2,5 Matthew Wook Chang1,2,3,4*
Matthew Wook Chang1,2,3,4*Candida albicans is an opportunistic pathogen, with its infection as one of the causes of morbidity or mortality. Notably, the probiotic yeast Saccharomyces cerevisiae var. boulardii has shown the potential to fight against Candida infections. In this study, we aimed to engineer a commercial boulardii strain to produce medium-chain fatty acids (MCFAs) with antagonistic effects against C. albicans. First, we identified and characterized a boulardii strain and created its auxotrophic strain Δura3. Next, we constructed and expressed a heterologous MCFA biosynthetic pathway under the control of inducible and constitutive promoters. Aside from examining MCFA production and secretion, we confirmed MCFAs’ effects on C. albicans’ anti-biofilm and anti-hyphal formations and the immunomodulatory effect of MCFA-containing supernatants on Caco-2 cells. We found that under constitutive promoters, the engineered boulardii strain constitutively produced and secreted a mixture of C6:0, C8:0, and C10:0. The secreted MCFAs then reduced biofilm and hyphal formations in C. albicans SC5314. We also confirmed that MCFAs upregulated the expression of virulence-related genes in SC5314. Furthermore, we found that the constitutively produced MCFAs in the supernatant induced the upregulation of immune response genes in Caco-2 cells co-cultured with SC5314, indicating MCFAs’ roles in immunomodulation. Overall, the engineered boulardii strain produced and secreted MCFAs, as well as demonstrated antagonistic effects against C. albicans SC5314 and immune-modulatory effects in Caco-2. To our knowledge, this represents the first study tackling the metabolic engineering of a commercial probiotic yeast strain to constitutively produce and secrete MCFAs showing anti-Candida effects. Our study forms the basis of the potential development of a live biotherapeutics probiotic yeast against Candida infections through metabolic engineering strategies.
Medium-chain fatty acids (MCFAs) are carboxylic acids with 6–12 carbons. MCFAs have demonstrated anti-pathogenic and immunomodulatory health effects, and are known to play a role in the regulation of body fat metabolism. Accordingly, MCFAs such as caprylic acid (C8:0), capric acid (C10:0), lauric acid (C12:0), or their combination with other compounds have been shown to inhibit pathogens like Candida albicans (CA) (Murzyn et al., 2010b; Lee et al., 2021), Clostridium difficile (Yang et al., 2017), Listeria monocytogenes (Hao et al., 2020), and Salmonella (Moschonas et al., 2012). In Candida spp, MCFAs may mimic the quorum sensing molecule farnesol that interferes with communications among fungal populations, leading to its potency against Candida (Lee et al., 2021). With regards to inflammation, C10:0 has been found to induce the upregulation of GPR84 and PPARγ and the downregulation of HIF-1α (Sam et al., 2021). In another study, C8:0 has been found to enhance inflammatory IL-8 secretion by >35% in human fetal intestinal epithelial cells (Andoh et al., 2000). Given MCFAs’ potency against pathogens and their roles in immunomodulation, MCFAs, their derivatives, and probiotics producing MCFAs have been widely used as anti-pathogenic agents and health supplements.
Saccharomyces cerevisiae var. boulardii (also named as Saccharomyces boulardii) is a probiotic yeast with versatile health effects. Not only are they anti-pathogenic and involved in immunomodulation, but they also promote the growth of beneficial gut microbiota—especially those associated with MCFAs produced and secreted by certain boulardii strains. For instance, Murzyn et al. found that a boulardii strain inhibits CA’s adhesion to intestinal cell lines and its extract reduces cytokine-induced inflammatory responses in Caco-2 cells as revealed by suppressed IL-8 expression (Murzyn et al., 2010a). Boulardii strains have also been found to diminish filamentation, biofilm formation, and CA translocation (Berg et al., 1993; Krasowska et al., 2009). Meanwhile, Tomicic and others reported how a boulardii strain significantly reduced the adherence ability of Candida glabrata ZIM2344 and ZIM2369 in a dose-dependent manner (Tomicic et al., 2016). The administration of the boulardii strains CNCM I-745, CNCM-I-1079, or CNCM-I-1079
C. albicans is a major opportunistic fungal pathogen. Clinically, current therapeutics for Candida infections mainly rely on the administration of anti-fungal drugs such as fluconazole, which has been implicated in multi-drug resistance (MDR). Notably, MDR associated with Candida has become a key clinical challenge in fighting fungal infections and a threat to public health (Berman and Krysan, 2020; Kunyeit et al., 2020). Consequently, alternative solutions are required to fight Candida infections more effectively. Given the proven anti-Candida effects of probiotic yeast and MCFAs, MCFA-producing boulardii strains could potentially offer an effective therapy against Candida infections. Nevertheless, MCFA production and secretion by wild type boulardii strains are strain- and condition-dependent, which could make it difficult to achieve desired anti-Candida effects. Therefore, a boulardii strain that stably produces and secretes MCFAs in a controllable manner would be ideal, and could be realized by expressing a heterologous MCFA biosynthetic pathway through synthetic biology-driven metabolic engineering strategies (Liu et al., 2016; Yu et al., 2016; Chen et al., 2018; Xia et al., 2019; Chen et al., 2020; Hossain et al., 2020).
In this study, we aimed to engineer a commercial probiotic yeast strain (S. cerevisiae var boulardii) to produce MCFAs with antagonistic effects against the opportunistic pathogen C. albicans. Briefly, we identified the ploidy of the boulardii strain, then constructed and expressed a heterologous MCFA biosynthetic pathway into Δura3. Next, we examined MCFA production and secretion, then confirmed MCFAs’ effects on anti-biofilm and anti-hyphal formations in C. albicans SC5314 cells and its immunomodulatory effects in the human epithelial cell line Caco-2. This represents the first study describing the metabolic engineering of a commercial probiotic yeast strain to produce MCFAs with anti-Candida effects. Our study provides useful insights into developing live biotherapeutics against Candida infections through metabolic pathway engineering.
Strains, growth conditions, plasmids, and chemicals. Strains and plasmids used are listed in Supplementary Table S1. S. cerevisiae S288C, S. cerevisiae var. boulardii, and C. albicans were grown in YPD (yeast extract 10 g/L, peptone 20 g/L, dextrose 20 g/L), YGD (yeast nitrogen base 6.7 g/L, dextrose 20 g/L, and yeast synthetic dropout medium supplements without uracil 1.92 g/L), or YRGD (yeast nitrogen base 6.7 g/L, D-raffinose 10 g/L, yeast synthetic dropout medium supplements without uracil 1.92 g/L, and D-(+)-galactose 20 g/L) at 30°C. Escherichia coli was grown in Luria–Bertani medium at 37°C. Hygromycin (200 mg/L) or ampicillin (100 mg/L) were added when applicable. DNA preparation kits were purchased from Qiagen (Germany), PCR reagents from Biorad (United States), restriction enzymes and DNA ligase from New England Biolabs (United Kingdom), and chemicals from Sigma-Aldrich (United States) unless specified.
Ploidy characterization by PCR. We isolated S. cerevisiae var. boulardii, designated as CLPY01, from Jarrow Formulas® yeast probiotics (Saccharomyces Boulardii MOS). To verify the boulardii strain, we performed PCR using genomic DNA as a template along with intron splice site primers EI1 and LA1 (Fietto et al., 2004). We then compared the amplicon’s profile with that of S. cerevisiae S288C, boulardii U28, and M2 (Hamedi et al., 2013). To elucidate the boulardii’s ploidy after strain verification, we performed PCR using genomic DNA as a template by primers that pair with the MATa locus (MatF) or MATα locus (MatαF) and the reverse primer MatR (Supplementary Table S1).
Gene deletion. We amplified the URA3 gene deletion cassette from pUG75, with this cassette containing a 45-bp upstream and downstream region flanking URA3’s coding sequence. The obtained gene deletion cassette was then transformed into CLPY01. To enable effective transformation, we used the following method for preparing competent cells: We prepared and diluted an overnight culture to OD600 0.3 in fresh YPD medium, after which we grew it at 30°C and 225 rpm until the culture reached OD600 1.6. Cells were collected and washed once with deionized H2O and then with an ice-chilled buffer (buffer 1: 1 M sorbitol
Gene cloning and overexpression. Genes including hSFP, mhFAS, and rTEII (sequences are shown in Supplementary Table S2) were synthesized by Life Technologies and cloned into pESC-URA standard protocols (Shao et al., 2009), resulting in the plasmid pESC-HR with hSFP under PGAL10 and mhFAS-rTEII under PGAL1. The recombinant plasmid pESC-HR was confirmed by sequencing and transformed into CLPY02 for MCFA production under D-galactose induction. Next, we replaced the promoters PGAL10 and PGAL1 to PTDH3 (for hSFP expression) and PTEF1 (for mhFAS-rTEII expression) in the opposite direction (sequences are shown in Supplementary Table S2), resulting in the plasmid pESC-gHR as confirmed by DNA sequencing. The used primers are listed in Supplementary Table S2.
Gene integration. We employed the DNA assembler method (Shao et al., 2009) to integrate the MCFA-biosynthetic pathway in pESC-gHR into CLPY02. Given the large size of the fragment, we divided the pathway into three fragments, including the terminator of CYC1 and ADH1 (HR1: 2965bp, HR2: 3016 bp, HR3: 4050 bp). HR3 was fused to a cassette expressing the hygromycin resistance gene (HygR) of pUG75 (HR3-HyrR) to facilitate colony screening with chromosomal integration. HR1, HR2, and HR3 contained 500-bp regions that overlapped with each other. HR1 and HR3-HyrR contained a 500-bp upstream or downstream region flanking the URA3 locus. The agar plate with both hygromycin and uracil added was used to select colonies integrated with the gene cassette, and marker rescue was performed prior to the second round of chromosomal integration. To integrate another copy of the MCFA biosynthetic pathway, HR3 was fused to a cassette with corresponding overlapping regions that express the URA3 of pUG72 (HR3-URA3) for chromosomal integration. HR1 and HR3-URA3 both contain a 500-bp upstream or downstream region flanking a URA3 locus. The agar plate excluding uracil was used to select colonies integrated with the gene cassette. The resulting strain carrying two copies of the MCFA biosynthetic pathway was named CLPY04 and used for subsequent experiments.
Fatty acid extraction and detection. To measure fatty acids produced in engineered strains, single colonies were pre-cultured in appropriate media overnight. Cells were then inoculated into a fresh medium and incubated at 30°C and 225 rpm. Five milliliters of cells were harvested by centrifugation, and cell pellets were washed using deionized water. We added an internal standard heptadecanoic acid and lysed cells by adding 1.5 mL hydrochloride (HCl) and incubating for 1 h at 70°C. To derivatize fatty acids, we added 1.5 mL 10% HCl–methanol (v/v) into the lysate, followed by vortexing and incubation for 3 h at 62°C. After cooling down, fatty acid methyl esters (FAMEs) were extracted by adding 2 mL hexane and vortexing. Lastly, we centrifuged the sample and collected the upper hexane layer for GC analysis. For the detection of extracellular fatty acids, we added the internal standard and 10% HCl–methanol (v/v) into the supernatant and proceeded with fatty acid derivatization. GC analysis was performed following a previously reported method using an HP 7890 B GC system with an Agilent 5977 A MSD equipped with a HP-5MS column (Yu et al., 2016; Ng et al., 2020). GC peaks were identified by comparing the retention times and mass spectra of FAME standards. Data analysis was performed using the Agilent Enhanced Data Analysis software.
Biofilm and hyphal formation assays. C. albicans SC5314 was inoculated for overnight cultivation. The overnight culture was diluted to OD600 0.1 in a microplate well containing supernatant (30%, v/v)
Co-culturing of Caco-2 and C. albicans SC5314. Caco-2 cells were seeded in a 6-well plate containing Gibco™ DMEM (DMEM2, with sodium bicarbonate and HEPES)
RNA extraction and real-time PCR (RT-PCR) analysis. The overnight culture of SC5314 was washed twice using customized DMEM and diluted in the appropriate medium to OD600 2 in a 6-well plate. Cells were collected after incubation at 75 rpm and 37°C at the appropriate timepoints. Total RNA was extracted using an RNeasy Kit (Qiagen) following the provided manual. Complementary DNA synthesis and RT-PCR were performed to analyze the transcription levels of BCR1, EFG1, HGC1, HWP1, and UME6 based on a reported method (Ling et al., 2015) using the 2−ΔΔCT method. Gene expression was normalized to ACT1.
The total RNA of the co-cultured Caco-2 cells was extracted using a Trizol reagent (Hwang et al., 2021). RT-PCR was then performed to analyze the transcription levels of IL-6, IL-8, LL-37, SA1009, CCL20, and hBD-3 using the 2−ΔΔCT method. Gene expression was normalized to the ACTB gene.
We firstly isolated the S. cerevisiae var. boulardii strain (named CLPY01 in this study) from a representative commercial yeast probiotic product (Jarrow Formulas) used to promote gut health.
To characterize the wild type strain CLPY01 (Genome Acc. No. JXBM00000000.1), we compared its genomic DNA amplicon profile and ploidy to other phylogenetically relative yeast strains. Figure 1A shows that strain CLPY01 has the same amplicon profiles as a known boulardii strain, namely Unique 28 (U28, Optibac Probiotics) and its mutant (M2) (Hamedi et al., 2013), both of which are distinct from S. cerevisiae S288C (SC). Next, we amplified the fragments located at the MATα and MATa sites in genome. Figure 1B shows that two fragments were amplified from CLPY01 along with U28 and M2, whereas only one fragment was amplified from SC. These results reveal that CLPY01 is a boulardii strain.
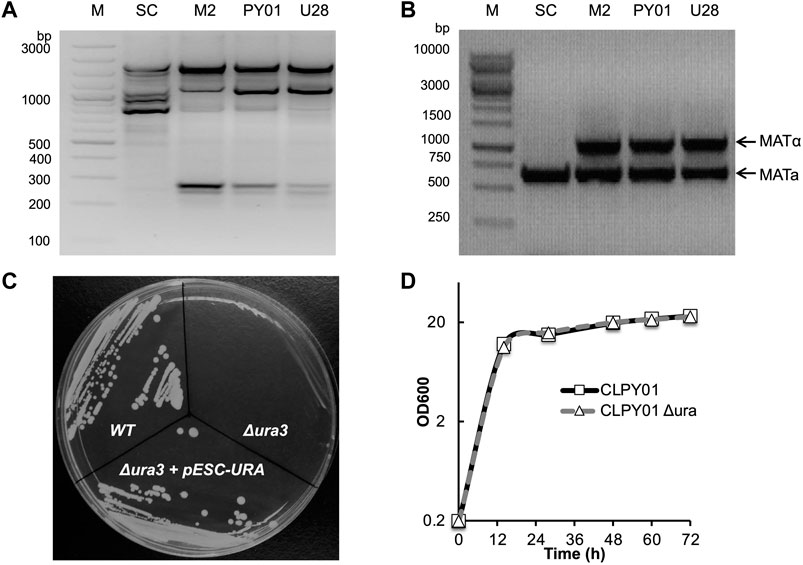
FIGURE 1. Identification of wild type and Δura3 boulardii strains. (A). PCR amplification of bands from genomic DNA. (B). Ploidy. (C). Phenotype identification of URA3 gene deletion. URA3 was deleted by using Cre-LoxP recombination. Cells were stroked onto an agar plate of minimal medium YGD without uracil. (D). Growth pattern comparison of S. boulardii wild type and Δura3 strains in YPD medium at 30°C. WT, CLPY01 wild type, Δura3, URA3 gene deletion mutant, Δura3+pESC-Ura, Δura3 harboring pESC-Ura.
To create an auxotrophic strain suitable for metabolic engineering, we designed a hygromycin-resistant gene cassette and deleted the URA3 gene by homologous recombination, resulting in the CLPY01 Δura3 that we named CLPY02. After obtaining CLPY02, we identified the auxotrophic phenotype of URA3 gene deletion by testing colony formation on uracil-free plates. Figure 1C shows that S. boulardii CLPY01 (WT) formed colonies on uracil-free plates, while CLPY02 did not form colonies, suggesting the deficiency of uracil biosynthesis in CLPY02 due to URA3’s absence. CLPY02 containing pESC-URA formed colonies, suggesting that URA3’s function is complemented by pESC-URA. This loss-and-gain function test confirms the successful deletion of URA3 gene in CLPY01. Furthermore, we compared CLPY02’s growth pattern to its wild type strain CLPY01 in YPD medium. Figure 1D shows that CLPY02 had the same growth pattern as CLPY01, suggesting that there was no growth defect due to URA3 deletion. Hence, the obtained auxotrophic boulardii strain CLPY02 was subsequently used for further experiments.
Despite several studies reporting the native production and secretion of MCFAs by S. cerevisiae var. boulardii strains, MCFA’s production level and composition remains subject to strain type and growing conditions (Murzyn et al., 2010b; Suchodolski et al., 2021). Therefore, we attempted to metabolically engineer CLPY02 to stably produce and secrete MCFAs by constructing and expressing a heterologous MCFA biosynthetic pathway.
In CLPY02, we cloned three genes for MCFA biosynthesis, including a mutant gene of Homo sapiens fatty acid synthase with its native thioesterase domain replaced by the Rattus norvegicus thioesterase II gene (mhFAS-rTEII) (Leber and Da Silva, 2014) and H. sapiens phosphopantetheinyl transferase gene (hSFP), resulting in pESC-HR (Table 1 and Supplementary Table S2). The expression of mhFAS-rTEII and hSFP are under the control of galactose-inducible promoters PGAL1 and PGAL10 (Figures 2A–C). After transforming pESC-HR into CLPY02, we measured the galactose-inducible production of MCFAs comprising of hexanoic acid (C6:0), octanoic acid (C8:0), decanoic acid (C10:0), and dodecanoic acid (C12:0). Figure 2D shows that the engineered strain CLPY03 produced about 2.4 mg/L MCFAs comprising of C8:0, C10:0, and C12:0 intracellularly. However, C6:0 could not be detected. Proportionally, C10:0 and C12:0 together accounted for over 60% of the produced MCFAs, whereas C8:0 had a lower abundance. Our time-course analysis shows the MCFA levels were the highest at 48 h and 72 h; Figure 2E shows that CLPY03 secreted a mixture of MCFAs into the extracellular environment, comprising of C6:0, C8:0, and C10:0 at up to 10.5 mg/L in total. Proportionally, C6:0 was slightly higher than C8:0, followed by C10:0 at 5% (at 48 h). C12:0 was not detectable. Time-course analysis shows that MCFA levels at 48 h was the highest. No MCFAs from CLPY02 carrying pESC-URA were detected in the supernatant. These results reveal that the engineered boulardii strain CLPY03 successfully produced and secreted the MCFA mixture (C6:0, C8:0 and C10:0) into the extracellular environment, up to 4.4-fold higher than the intracellular level.
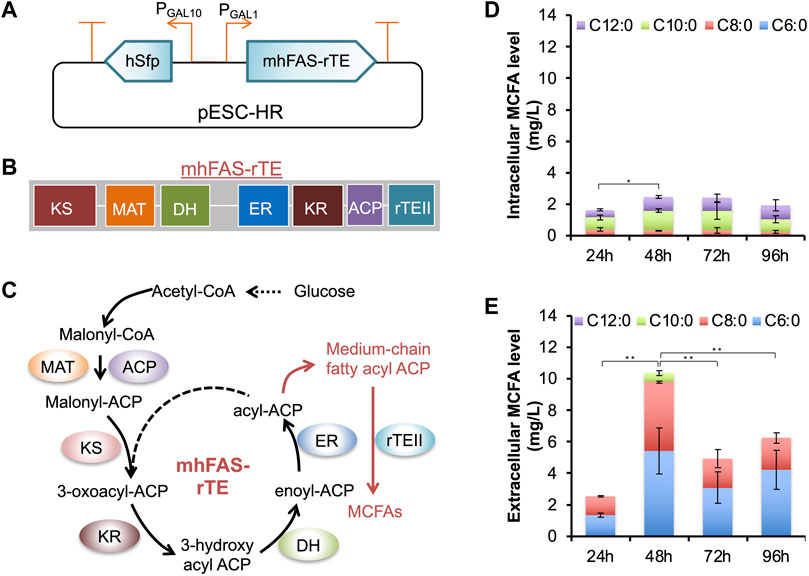
FIGURE 2. Plasmid-based inducible expression of an MCFA biosynthetic pathway and the analysis of MCFAs produced by the engineered strain CLPY03. (A). Schematic of the recombinant plasmid pESC-HR harboring the MCFA biosynthetic pathway under the control of galactose-inducible promoters (PGAL1 and PGAL10). (B). Organization of mhFAS-rTE domains. (C). Schematic of the MCFA biosynthetic pathway. (D). Measurement of the intracellular MCFAs. (E). The time-course production level of MCFAs secreted into the medium. The data was obtained from at least three independent experiments. *, p < 0.05, **, p < 0.01 (Student’s t-test). hSFP, human phosphopantetheinyl transferase gene (encoding NP_056238.2), mhFAS-rTEII, a human fatty acid synthase mutant gene (mhFAS, encoding aa1-2,218 of NP_004095.4) fused with a rat thioesterase gene (rTEII, encoding NP_073196.1). KS, β-ketoacyl synthase, MAT, Malonyl-CoA-/acetyl-CoA-ACP-transacylase, DH, Dehydratase, ER, β-enyl reductase, KR, β-ketoacyl reductase, ACP, Acyl carrier protein.
There have been few reports on engineering boulardii strains for producing MCFAs. However, a recent study described how S. cerevisiae co-expression of a human FAS mutant, rTEII, and Bacillus subtilis Sfp allowed MCFA production and secretion (C6:0, C8:0 and C10:0). In another study, the co-expression of yeast FAS with site mutations and a short chain thioesterase resulted in the production and secretion of C6∼C12 fatty acids in S. cerevisiae (Gajewski et al., 2017). We noted that in these strains, C8:0 is the major MCFA secreted into the medium (Leber and Da Silva, 2014). Meanwhile, in the engineered boulardii strain CLPY03, the proportion of C8:0 and C10:0 is comparable. The variations among MCFA profiles may be due to differences in the enzyme’s chain-length specificity, host strains, and growth conditions. Given the proven anti-fungal effects of C8:0 and C10:0 (Krasowska et al., 2009; Murzyn et al., 2010b; Khalandi et al., 2020; Suchodolski et al., 2021), the secreted MCFAs containing C8:0 and C10:0 (48 h) may show antagonistic effects against C. albicans.
To evaluate the anti-Candida effects of the secreted MCFAs by the engineered boulardii strain CLPY03, we analyzed the biofilm and hyphal formations of a representative C. albicans strain SC5314 in the presence of an MCFA-containing supernatant.
Figure 3A shows that adding 30% (v/v) of CLPY03’s supernatant at 48 h (pESC-HR) in YPD reduced biofilm formation by 4.3-fold over the control (CLPY02+pESC-URA). Under a microscope, we observed the formation of hyphae in the presence of the control supernatant. However, no obvious hypha formation was observed in the presence of CLPY03’s supernatant (HR) in a modified DMEM medium (Figure 3B). Given that biofilm formation and hyphal formation are related to Candida’s virulence, we were interested in exploring the expression profiles of virulence-relevant genes underlying HR-supernatant treatment in SC5314. Our real-time PCR analysis shows that EFG1 (encoding a filament-specific regulator), HGC1 (encoding a G1 cyclin-related protein), HWP1 (encoding hyphal wall protein 1), and UME6 (encoding a key filament-specific regulator) were downregulated by 2.0–4.6-fold (Figure 3C). These genes either positively regulate biofilm and hyphal formations or encode components of hyphae. In this study, their downregulation is in line with the reduced biofilm formation and hyphal formation in SC5314 with the HR-supernatant treatment. Notably, there was no change in the expression of BCR1, which encodes a biofilm formation regulator. These results suggest that the MCFA-containing supernatant of CLPY03 can reduce the expression of virulence-related genes, as well as biofilm and hyphal formations.
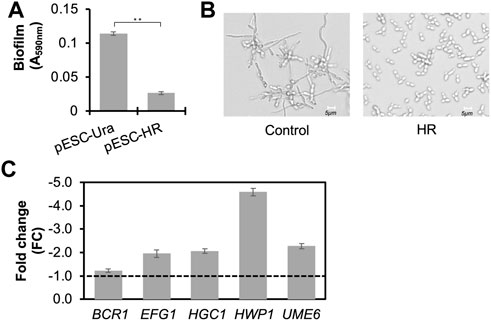
FIGURE 3. Functional analyses of the secreted MCFAs in the supernatant against biofilm and hyphal formations in C. albicans SC5314. (A). Effect of the MCFA-containing supernatant from the engineered boulardii producing MCFAs (HR) on biofilm formation. Control, the strain carrying pESC-URA. **, p < 0.01 (Student’s t-test). (B). Effect of MCFA-containing supernatant from the engineered boulardii producing MCFAs (HR) on hyphal formation at pH5.5. (C). Real-time PCR analysis of the gene profile in C. albicans co-cultured with MCFA-containing supernatant from the engineered boulardii strain producing and secreting MCFAs (HR). The expression of genes related to biofilm and hyphal formations at 12 h was analyzed. The supernatant from the strain carrying pESC-URA was used as a control. Reference gene, ACT1. A dotted line indicates FC1.0, representing the fold change of genes from the control. Minus indicates the genes’ downregulation.
The aforementioned anti-Candida effects may be due to the MCFAs, proteins, or other metabolites produced and secreted by CLPY03. To confirm the cause of biofilm and hyphal formations, we spiked the MCFA mixture (C6:0, C8:0, and C10:0) with the same amount as the MCFAs secreted by CLPY03 (48 h) into the induction medium. Next, we compared the biofilm amount and hyphal formations of SC5314 with the spiked MCFAs to the induction medium (control). Supplementary Figure S1 shows that an MCFA spike-in reduced biofilm amount by 3.3-fold and inhibited hyphal formation. This result confirms that the secreted MCFAs in CLPY03 supernatant can reduce biofilm and hyphal formations in SC5314. We also noted that SC5314 treated with the CLPY02 supernatant had less amounts of biofilm (∼15-fold) than the induction medium, suggesting that other compounds (proteins, metabolites, etc.) in the CLPY02+pESC-URA supernatant may reveal anti-Candida effects, with future efforts required to elucidate those compounds.
In CLYP03, the inducible plasmid-based expression of the MCFA biosynthetic pathway resulted in the secretion of MCFAs with anti-Candida effects. However, MCFA production by CLPY03 is subject to the availability of galactose as an inducer, which is generally absent in vivo conditions and subject to the stability of the gene expression level and plasmid. To overcome this potential limitation, we expressed the MCFA biosynthetic pathway under constitutive promoters for the stable production and secretion of MCFAs.
To modify MCFA biosynthetic pathway expression, we replaced the inducible promoters (PGAL1 and PGAL10) with constitutive promoters (PTEF1 for mhFAS-rTEII, and PTDH3 for hSFP), and integrated each copy of the modified pathway with a URA3 cassette and an HygR cassette (hygromycin resistance gene) respectively into the chromosome of CLPY02 at the original URA3 locus (Figure 4A). We named the resulting strain CLPY04, which carries two copies of the MCFA biosynthetic pathway in the chromosome. Next, we measured the MCFAs secreted by CLPY04 expressing the MCFA biosynthetic pathway driven by constitutive promoters. Figure 4B shows that in the medium YGD, CLPY04 produced and secreted all three MCFAs (C6:0, C8:0 and C10:0) simultaneously over 72 h. The secreted MCFA amount by CLPY04 at 24 h (3.4 mg/L) was the highest, followed by 48 h (3.0 mg/L) and 72 h (2.9 mg/L). Proportionally, C8:0 was the highest MCFA secreted (∼52%), followed by C6:0 (∼37%), and C10:0 (∼11%). Overall, the composition and amount of the secreted MCFAs by CLPY04 remained almost the same, indicating that their stable production and secretion does not require a chemical inducer like galactose.
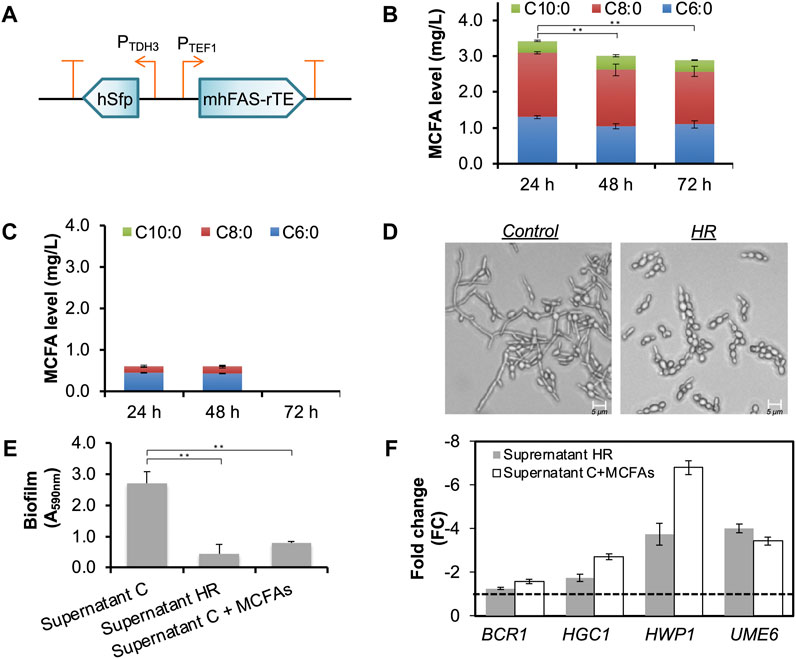
FIGURE 4. Chromosomal constitutive expression of an MCFA biosynthetic pathway and the functional analysis of the secreted MCFAs. (A). Schematic of the recombinant plasmid pESC-HR harboring the MCFA biosynthetic pathway under the control of constitutive promoters (PTEF1 and PTDH3). (B). The time-course production level of MCFAs secreted into the medium. The data was obtained from at least three independent experiments. (C). MCFAs secreted by CLPY02 in YGD medium. (D). Effect of the MCFA-containing supernatant from CLPY04 (HR) on hyphal formation at pH5.5. Control, the supernatant of CLPY01 which does not contain MCFAs. (E). Effect of Supernatant HR on biofilm formations in the customized DMEM. Supernatant C, the supernatant of CLPY01. Supernatant C
Interestingly, we detected a small amount of C6:0 and C8:0 (0.6 mg/L in total) in the supernatant of CLPY02+pESC-URA (Supernatant C), while no C10:0 was detected at 24 h and 48 h (Figure 4C). In contrast, CLPY02 under the induction medium (YRGD) did not secrete C6:0 nor C8:0 into the supernatant. Such differences in MCFA secretion by the same strain could be attributed to different growth conditions, consistent with the aforementioned case-dependence of MCFA production by the boulardii strains.
Upon the confirmation of MCFA production by CLPY04 under constitutive promoters, we next evaluated the anti-Candida effects of CLPY04’s MCFA-containing supernatant (24 h), including effects such as biofilm and hyphal formations.
To facilitate subsequent functional assays involving cell culture, we selected a commercial cell culture medium, Dulbecco’s Modified Eagle Medium (DMEM), to evaluate the anti-Candida effects of CLPY04’s supernatant (Supernatant HR). Surprisingly, we did not observe reduced biofilm and hyphal formations in the SC5314 in Supernatant HR (30% v/v) in DMEM. We noted that commercial DMEM maintains a high pH (pH7.5) due to a sodium bicarbonate buffer system. We hypothesized that the high pH maintained in commercial DMEM may have reduced or abolished the MCFAs’ anti-Candida effects. To confirm this possibility, we modified the pH of commercial DMEM by excluding sodium bicarbonate. Next, we adjusted CLPY01’s (Supernatant C) and Supernatant HR’s pH from 3.4 to 6.0. After adding them (30% v/v) to the customized DMEM, we observed hyphal formation, specifically finding that SC5314 formed hyphae when Supernatant C was at pH
Figure 4E shows that adding 30% (v/v) of CLPY04’s supernatant (Supernatant HR, pH5.5) at 24 h into the modified DMEM reduced biofilm formation by 6.0-fold than Supernatant C. The spike-in of MCFAs into Supernatant C (Supernatant C + MCFAs) at an equal amount as Supernatant HR also reduced biofilm formation (3.5-fold) compared to Supernatant C. This result suggests that MCFAs secreted by CLPY04 reduce SC5314 biofilm formation. To further confirm biofilm and hyphal formations, we analyzed the expression of genes responsible for hyphal formation. Our real-time PCR analysis shows that EFG1, HGC1, HWP1, and UME6 were downregulated by
In the present study, the use of Supernatant HR comprising C6:0 (1.3 mg/L), C8:0 (1.8 mg/L) and C10:0 (0.3 mg/L) reduced both biofilm and hyphal formations in SC5314. Lee et al. reported that C8:0 and C10:0 at 1.0 mg/L respectively reduced biofilm formation in C. albicans DAY185 (Lee et al., 2021). In contrast, C6:0 at 37.8 mg/L or C8:0 at 17.9 mg/L does not affect the biofilm formation of SC5314 (Murzyn et al., 2010b). Given the relatively low concentration of the secreted MCFAs, the reduction of biofilm and hyphal formations by Supernatant HR might be attributed to the synergistic effects of C6:0, C8:0 and C10:0.
To understand the role of the MCFA-containing supernatant of CLPY04 (Supernatant HR) in immunomodulation, we examined transcriptional levels of immunomodulation genes in a model human epithelial cell line Caco-2.
Firstly, in a trans-well, we co-cultured Caco-2 cells with live SC5314 cells and Supernatant HR (30%, v/v, pH5.5) in the modified DMEM lacking the pH buffering capacity. Next, we analyzed the transcriptional level of immunomodulatory genes via real-time PCR. Figure 5A shows that the supplement of Supernatant HR upregulated the expression of IL-6 (FC4.1), IL-8 (FC2.1), CCL20 (FC1.8), and hBD-3 (FC3.2) in Caco-2 cells in the presence of C. albicans SC5314 cells. IL-6 gene encodes Interleukin-6 (IL-6), while IL-8 gene encodes Interleukin-8 (IL-8), both of which are pro-inflammatory cytokines produced in response to pathogenic infections. The two cytokines contribute to host defense by stimulating an immune response (Tanaka et al., 2014; D’enfert et al., 2021; Sam et al., 2021). The hBD-3 gene encodes an anti-microbial peptide β-Defensin 3 (hBD-3) that shows a wide range of immunomodulatory functions and protects the gut during Candida infections (Fusco et al., 2021). CCL20 gene encodes the chemokine ligand-20, which plays a role in attracting immune cells to infectious sites during the early stages of infection (Ma’ayeh et al., 2018). The upregulation of these genes indicates the immunomodulatory effects of the Supernatant HR in Caco-2 cells in response to the Candida infections.
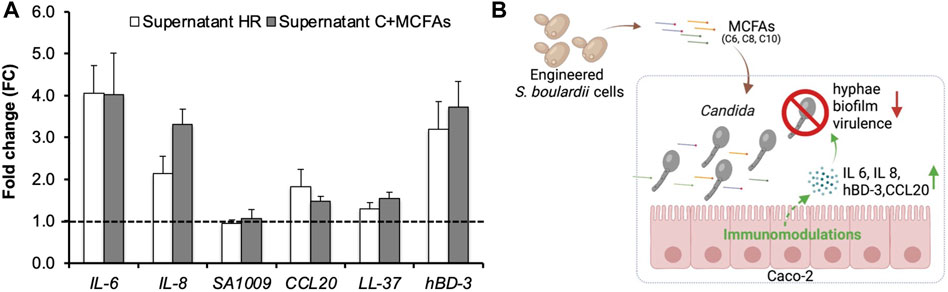
FIGURE 5. RT-PCR analysis of genes involved in immunomodulation in Caco-2 cells and the proposed mechanism of the MCFAs’ anti-Candida effects. Caco-2 cells were co-cultured with C. albicans and the MCFA-containing supernatant of CLPY04. (A). Real-time PCR analysis. Expression of genes related to immunomodulation at 3.5 h was analyzed. The supernatant from the strain CLPY01 (Supernatant C) was used as a control. MCFAs were spiked into Supernatant C (Supernatant C + MCFAs) and used as a positive control. Reference gene, ACT1. A dotted line indicates FC1.0, representing the fold change of genes from the control. The pH of Supernatant C and Supernatant HR was adjusted to 5.5 prior to co-culturing. (B). The proposed mechanism of anti-Candida effect by MCFAs. Reduction of Candida’s hyphae and biofilm formations, and virulence-related genes as well as the upregulation of IL-6, IL-8, human beta-defensin 3 (hBD-3), and CCL20 genes are indicated by arrows. The box in blue indicates the co-culturing of C. albicans cells, MCFA-containing supernatant, and Caco-2 cells.
To confirm the MCFAs’ immunomodulatory effects, we spiked commercial MCFAs into CLPY02’s supernatant with an equal amount of Supernatant HR and compared the gene profiles in Caco-2 cells co-cultured with SC5314. Figure 5A shows that MCFA spike-in (Supernatant C + MCFAs) resulted in the upregulation of IL-6 (FC4.0), IL-8 (FC3.3), CCL20 (FC1.5) and hBD-3 (FC3.7), sharing a similar induction trend to Supernatant HR. This result confirms that MCFAs present in the Supernatant HR can upregulate these genes. The different expression between Supernatant C + MCFAs and Supernatant HR might be due to other compounds in the supernatants. Hence, the MCFAs secreted by the engineered boulardii strain CLPY04 can alter immunomodulation in Caco-2, and likely trigger host defenses against C. albicans infections. Given the relatively low concentration of MCFAs in the supernatant, the upregulation of these genes by Supernatant HR might be attributed to the synergistic effects of C6:0, C8:0 and C10:0. The hyphal development of C. albicans might also impact cytokine gene expression. These results demonstrate the potential of applying CLPY04 as a therapeutic agent against Candida infections.
In this study, we characterized and metabolically engineered a commercial probiotic yeast S. cerevisiae var. boulardii strain for antagonistic effects against the opportunistic pathogen C. albicans. The engineered boulardii strain was shown to stably produce and secrete an MCFA mixture comprising C6:0, C8:0, and C10:0. We found that the secreted MCFAs showed antagonistic effects to the opportunistic pathogen C. albicans, as demonstrated by reduced biofilm and hyphal formations. Through RT-PCR analysis, we confirmed that the secreted MCFAs downregulated the expression of virulence-related genes in C. albicans. Furthermore, we found that MCFAs induced the upregulation of immune response genes in Caco-2 cells co-cultured with C. albicans, indicating MCFAs’ roles in immunomodulation (Figure 5B). To our knowledge, our study represents the first study tackling the metabolic engineering of a commercial probiotic yeast for anti-Candida applications. Overall, our findings provide the basis for potentially developing a live biotherapeutics probiotic yeast against fungal infections such as Candida strains.
The engineered boulardii strain was proven to produce and secrete MCFAs showing antagonistic effects against C. albicans. We confirmed the antagonistic effects through in vitro assays by supplying the MCFA-containing supernatant from the engineered boulardii strain into the co-culturing system. In the future, we could further investigate the attribution of individual MCFAs, as well as specific combinations of different MCFAs at varying doses to the downregulation of virulence-related genes in Candida strains. Future efforts could also consider further optimizing the engineered strains and validating their antagonistic effects. For instance, native fatty acid synthesis could be tuned and blocked, while other competing pathways could be removed to increase the metabolic fluxes towards MCFA biosynthesis. We observed a slower growth and lower cell density of CLPY04 compared to the control CLPY02 (Supplementary Figure S3), suggesting a need to further improve cell growth. Moreover, it would be interesting to determine CLPY04’s genetic stability. Given the boulardii strains’ relatively low engineering efficiency, strain engineering could be facilitated by using synthetic biology toolkits such as CRISPR-based genome editing (Liu et al., 2016) and an optimal method with higher transformation efficiency. The engineered MCFA-producing strain could be directly co-cultured to validate its antagonistic effects against Candida. In addition, the efficacy of the engineered boulardii strain could be validated using an in vivo model.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
HL, RL, QS, and HS performed the experiments and analyzed experimental data. HL, MC, and LC oversaw the project and provided guidance. HL, RL, and QS wrote, reviewed, and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
The NUS Medicine Synthetic Biology Translational Research Program (NUHSRO/2020/077/MSC/02/SB), the Summit Research Program of the National University Health System (NUHSRO/2016/053/SRP/05), the Synthetic Biology Initiative of the National University of Singapore (DPRT/943/09/14), the Synthetic Biology R&D Program (SBP-P2, SBP-P7, SBP-P9), National Research Foundation Investigatorship (NRF-NRFI05-2019-0004), the U.S. Army (ARO/2019/74459LS), and the Ministry of Education, Singapore (MOE/2014/T2/2/128, NUHSRO/2020/046/T1/3).
We thank Wei Suong Teo and Yuxuan Lee for their assistance in gene cloning and pathway construction. We thank Kamila Isabelle Alabado Navarro for the comments made to the manuscript. Figure 5B was created with BioRender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1090501/full#supplementary-material
Andoh, A., Takaya, H., Araki, Y., Tsujikawa, T., Fujiyama, Y., and Bamba, T. (2000). Medium- and long-chain fatty acids differentially modulate interleukin-8 secretion in human fetal intestinal epithelial cells. J. Nutr. 130, 2636–2640. doi:10.1093/jn/130.11.2636
Berg, R., Bernasconi, P., Fowler, D., and Gautreaux, M. (1993). Inhibition of Candida albicans translocation from the gastrointestinal tract of mice by oral administration of Saccharomyces boulardii. J. Infect. Dis. 168, 1314–1318. doi:10.1093/infdis/168.5.1314
Berman, J., and Krysan, D. J. (2020). Drug resistance and tolerance in fungi. Nat. Rev. Microbiol. 18, 319–331. doi:10.1038/s41579-019-0322-2
Chen, B., Foo, J. L., Ling, H., and Chang, M. W. (2020). Mechanism-driven metabolic engineering for bio-based production of free r-lipoic acid in Saccharomyces cerevisiae mitochondria. Front. Bioeng. Biotechnol. 8, 965. doi:10.3389/fbioe.2020.00965
Chen, B., Lee, H. L., Heng, Y. C., Chua, N., Teo, W. S., Choi, W. J., et al. (2018). Synthetic biology toolkits and applications in Saccharomyces cerevisiae. Biotechnol. Adv. 36, 1870–1881. doi:10.1016/j.biotechadv.2018.07.005
D'enfert, C., Kaune, A. K., Alaban, L. R., Chakraborty, S., Cole, N., Delavy, M., et al. (2021). The impact of the fungus-host-microbiota interplay upon Candida albicans infections: Current knowledge and new perspectives. FEMS Microbiol. Rev. 45 (3), fuaa060. doi:10.1093/femsre/fuaa060
Duysburgh, C., Van Den Abbeele, P., Morera, M., and Marzorati, M. (2021). Lacticaseibacillus rhamnosus GG and Saccharomyces cerevisiae boulardii supplementation exert protective effects on human gut microbiome following antibiotic administration in vitro. Benef. Microbes 12 (4), 365–379. doi:10.3920/bm2020.0180
Fietto, J. L., Araujo, R. S., Valadao, F. N., Fietto, L. G., Brandao, R. L., Neves, M. J., et al. (2004). Molecular and physiological comparisons between Saccharomyces cerevisiae and Saccharomyces boulardii. Can. J. Microbiol. 50, 615–621. doi:10.1139/w04-050
Fusco, A., Savio, V., Donniacuo, M., Perfetto, B., and Donnarumma, G. (2021). Antimicrobial peptides human beta-defensin-2 and -3 protect the gut during Candida albicans infections enhancing the intestinal barrier integrity: In vitro study. Front. Cell Infect. Microbiol. 11, 666900. doi:10.3389/fcimb.2021.666900
Gajewski, J., Pavlovic, R., Fischer, M., Boles, E., and Grininger, M. (2017). Engineering fungal de novo fatty acid synthesis for short chain fatty acid production. Nat. Commun. 8, 14650. doi:10.1038/ncomms14650
Hamedi, H., Misaghi, A., Modarressi, M. H., Salehi, T. Z., Khorasanizadeh, D., and Khalaj, V. (2013). Generation of a uracil auxotroph strain of the probiotic yeast Saccharomyces boulardii as a host for the recombinant protein production. Avicenna J. Med. Biotechnol. 5, 29–34.
Hao, K., Meng, R., Bu, X., Liu, Z., Yan, H., Zhang, Y., et al. (2020). Antibacterial effect of caprylic acid and potassium sorbate in combination against Listeria monocytogenes ATCC 7644. J. Food Prot. 83, 920–927. doi:10.4315/0362-028x.jfp-19-458
Hossain, G. S., Saini, M., Miyake, R., Ling, H., and Chang, M. W. (2020). Genetic biosensor design for natural product biosynthesis in microorganisms. Trends Biotechnol. 38, 797–810. doi:10.1016/j.tibtech.2020.03.013
Hwang, I. Y., Kim, H. R., De Sotto, R., and Chang, M. W. (2021). Engineered probiotics modulate the endocannabinoid system. Biotechnol. Notes 2, 33–38. doi:10.1016/j.biotno.2021.08.001
Jung, H., Ling, H., Tan, Y. Q., Chua, N. H., Yew, W. S., and Chang, M. W. (2021). Heterologous expression of cyanobacterial gas vesicle proteins in Saccharomyces cerevisiae. Biotechnol. J. 16, e2100059. doi:10.1002/biot.202100059
Khalandi, H., Masoori, L., Farahyar, S., Delbandi, A. A., Raiesi, O., Farzanegan, A., et al. (2020). Antifungal activity of capric acid, nystatin, and fluconazole and their in vitro interactions against Candida isolates from neonatal oral thrush. Assay. Drug Dev. Technol. 18, 195–201. doi:10.1089/adt.2020.971
Krasowska, A., Murzyn, A., Dyjankiewicz, A., Lukaszewicz, M., and Dziadkowiec, D. (2009). The antagonistic effect of Saccharomyces boulardii on Candida albicans filamentation, adhesion and biofilm formation. FEMS Yeast Res. 9, 1312–1321. doi:10.1111/j.1567-1364.2009.00559.x
Kunyeit, L., K, A. A., and Rao, R. P. (2020). Application of probiotic yeasts on Candida species associated infection. J. Fungi (Basel) 6 (4), 189. doi:10.3390/jof6040189
Leber, C., and Da Silva, N. A. (2014). Engineering of Saccharomyces cerevisiae for the synthesis of short chain fatty acids. Biotechnol. Bioeng. 111, 347–358. doi:10.1002/bit.25021
Lee, J. H., Kim, Y. G., Khadke, S. K., and Lee, J. (2021). Antibiofilm and antifungal activities of medium-chain fatty acids against Candida albicans via mimicking of the quorum-sensing molecule farnesol. Microb. Biotechnol. 14, 1353–1366. doi:10.1111/1751-7915.13710
Ling, H., Pratomo Juwono, N. K., Teo, W. S., Liu, R., Leong, S. S., and Chang, M. W. (2015). Engineering transcription factors to improve tolerance against alkane biofuels in Saccharomyces cerevisiae. Biotechnol. Biofuels 8, 231. doi:10.1186/s13068-015-0411-z
Liu, J. J., Kong, I., Zhang, G. C., Jayakody, L. N., Kim, H., Xia, P. F., et al. (2016). Metabolic engineering of probiotic Saccharomyces boulardii. Appl. Environ. Microbiol. 82, 2280–2287. doi:10.1128/aem.00057-16
Ma'ayeh, S. Y., Knorr, L., Skold, K., Garnham, A., Ansell, B. R. E., Jex, A. R., et al. (2018). Responses of the differentiated intestinal epithelial cell line Caco-2 to infection with the Giardia intestinalis GS Isolate. Front. Cell Infect. Microbiol. 8, 244. doi:10.3389/fcimb.2018.00244
More, M. I., and Swidsinski, A. (2015). Saccharomyces boulardii CNCM I-745 supports regeneration of the intestinal microbiota after diarrheic dysbiosis - a review. Clin. Exp. Gastroenterol. 8, 237–255. doi:10.2147/CEG.S85574
Moschonas, G., Geornaras, I., Stopforth, J. D., Wach, D., Woerner, D. R., Belk, K. E., et al. (2012). Activity of caprylic acid, carvacrol, epsilon-polylysine and their combinations against Salmonella in not-ready-to-eat surface-browned, frozen, breaded chicken products. J. Food Sci. 77, M405–M411. doi:10.1111/j.1750-3841.2012.02757.x
Murzyn, A., Krasowska, A., Augustyniak, D., Majkowska-Skrobek, G., Lukaszewicz, M., and Dziadkowiec, D. (2010a). The effect of Saccharomyces boulardii on Candida albicans-infected human intestinal cell lines Caco-2 and Intestin 407. FEMS Microbiol. Lett. 310, 17–23. doi:10.1111/j.1574-6968.2010.02037.x
Murzyn, A., Krasowska, A., Stefanowicz, P., Dziadkowiec, D., and Lukaszewicz, M. (2010b). Capric acid secreted by S. boulardii inhibits C. albicans filamentous growth, adhesion and biofilm formation. PLoS One 5, e12050. doi:10.1371/journal.pone.0012050
Ng, T. K., Yu, A. Q., Ling, H., Pratomo Juwono, N. K., Choi, W. J., Leong, S. S. J., et al. (2020). Engineering Yarrowia lipolytica towards food waste bioremediation: Production of fatty acid ethyl esters from vegetable cooking oil. J. Biosci. Bioeng. 129, 31–40. doi:10.1016/j.jbiosc.2019.06.009
Sam, Q. H., Ling, H., Yew, W. S., Tan, Z., Ravikumar, S., Chang, M. W., et al. (2021). The divergent immunomodulatory effects of short-chain fatty acids and medium-chain fatty acids. Int. J. Mol. Sci. 22 (12), 6453. doi:10.3390/ijms22126453
Shao, Z., Zhao, H., and Zhao, H. (2009). DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 37, e16. doi:10.1093/nar/gkn991
Suchodolski, J., Derkacz, D., Bernat, P., and Krasowska, A. (2021). Capric acid secreted by Saccharomyces boulardii influences the susceptibility of Candida albicans to fluconazole and amphotericin B. Sci. Rep. 11, 6519. doi:10.1038/s41598-021-86012-9
Tanaka, T., Narazaki, M., and Kishimoto, T. (2014). IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 6, a016295. doi:10.1101/cshperspect.a016295
Tomicic, Z., Zupan, J., Matos, T., and Raspor, P. (2016). Probiotic yeast Saccharomyces boulardii (nom. nud.) modulates adhesive properties of Candida glabrata. Med. Mycol. 54, 835–845. doi:10.1093/mmy/myw026
Xia, P. F., Ling, H., Foo, J. L., and Chang, M. W. (2019). Synthetic genetic circuits for programmable biological functionalities. Biotechnol. Adv. 37, 107393. doi:10.1016/j.biotechadv.2019.04.015
Yang, H. T., Chen, J. W., Rathod, J., Jiang, Y. Z., Tsai, P. J., Hung, Y. P., et al. (2017). Lauric acid is an inhibitor of Clostridium difficile growth in vitro and reduces inflammation in a mouse infection model. Front. Microbiol. 8, 2635. doi:10.3389/fmicb.2017.02635
Yu, A. Q., Pratomo Juwono, N. K., Foo, J. L., Leong, S. S. J., and Chang, M. W. (2016). Metabolic engineering of Saccharomyces cerevisiae for the overproduction of short branched-chain fatty acids. Metab. Eng. 34, 36–43. doi:10.1016/j.ymben.2015.12.005
Zhang, W., Bao, C., Wang, J., Zang, J., and Cao, Y. (2020). Administration of Saccharomyces boulardii mafic-1701 improves feed conversion ratio, promotes antioxidant capacity, alleviates intestinal inflammation and modulates gut microbiota in weaned piglets. J. Anim. Sci. Biotechnol. 11, 112. doi:10.1186/s40104-020-00516-4
Keywords: probiotic yeast, medium-chain fatty acids, Candida albicans, virulence, immunomodulation
Citation: Ling H, Liu R, Sam QH, Shen H, Chai LYA and Chang MW (2023) Engineering of a probiotic yeast for the production and secretion of medium-chain fatty acids antagonistic to an opportunistic pathogen Candida albicans. Front. Bioeng. Biotechnol. 11:1090501. doi: 10.3389/fbioe.2023.1090501
Received: 05 November 2022; Accepted: 01 February 2023;
Published: 27 February 2023.
Edited by:
Aiqun Yu, Tianjin University of Science and Technology, ChinaReviewed by:
Ning Xu, Tianjin Institute of Industrial Biotechnology (CAS), ChinaCopyright © 2023 Ling, Liu, Sam, Shen, Chai and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew Wook Chang, YmNoY213QG51cy5lZHUuc2c=
†Present Address: Hua Ling, Wilmar Innovation Centre, Wilmar International Limited, Singapore, Singapore
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.