
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol., 16 September 2022
Sec. Biosensors and Biomolecular Electronics
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.986233
This article is part of the Research TopicCRISPR-Based BiosensorsView all 5 articles
 Kun Chen1
Kun Chen1 Ziyi Shen1
Ziyi Shen1 Guanzhen Wang1,2
Guanzhen Wang1,2 Wei Gu1
Wei Gu1 Shengchao Zhao1,2
Shengchao Zhao1,2 Zihan Lin1
Zihan Lin1 Wei Liu2
Wei Liu2 Yi Cai3
Yi Cai3 Gohar Mushtaq4
Gohar Mushtaq4 Jia Jia1*
Jia Jia1* Chunpeng (Craig) Wan5*
Chunpeng (Craig) Wan5* Tingdong Yan1*
Tingdong Yan1*CRISPR/Cas technology originated from the immune mechanism of archaea and bacteria and was awarded the Nobel Prize in Chemistry in 2020 for its success in gene editing. Molecular diagnostics is highly valued globally for its development as a new generation of diagnostic technology. An increasing number of studies have shown that CRISPR/Cas technology can be integrated with biosensors and bioassays for molecular diagnostics. CRISPR-based detection has attracted much attention as highly specific and sensitive sensors with easily programmable and device-independent capabilities. The nucleic acid-based detection approach is one of the most sensitive and specific diagnostic methods. With further research, it holds promise for detecting other biomarkers such as small molecules and proteins. Therefore, it is worthwhile to explore the prospects of CRISPR technology in biosensing and summarize its application strategies in molecular diagnostics. This review provides a synopsis of CRISPR biosensing strategies and recent advances from nucleic acids to other non-nucleic small molecules or analytes such as proteins and presents the challenges and perspectives of CRISPR biosensors and bioassays.
Accurate, rapidly available, and highly sensitive detection tools are essential to human health. In diagnosing diseases, early detection is significant for effective disease treatment. Early intervention can reduce the duration of untreated illness, slow down the disease progression and control the spread of infectious diseases (Parolo and Merkoci, 2013). One example is the ongoing worldwide epidemic of COVID-19, for which an increasing number of tests are being developed to detect nucleic acid markers of SARS-CoV-2 (Rahimi et al., 2021). In the field of food safety, fast detection of toxic substances in food allows for monitoring the quality and safety from raw material production to processing to packaging (Mao et al., 2022a). In addition, wearable devices with detection capabilities are being explored, with features such as easy sampling, high speed of detection, and easy reading of results (Nguyen et al., 2021), which provide a prototype for future out-of-lab testing.
Clustered, regularly interspaced short palindromic repeats (CRISPR) system was first discovered in 1987 (Ishino et al., 1987). CRISPR is a sequence of genes found in many bacteria and most archaea (Mojica et al., 2000). The CRISPR system is a defensive mechanism containing the CRISPR gene and the CRISPR-associated sequence proteins (Cas protein), which is an enzyme with the ability to cleave nucleic acids, relying on the guidance of RNA to target regions thereby defending against invasion by exogenous genes (Sander and Joung, 2014). These significant discoveries have provided new ideas and avenues for expanding the gene-editing tools (Cong et al., 2013).
In recent years, the classification of CRISPR-Cas proteins has gained increasing interest and attention as more Cas enzymes have been discovered. It has been reported that a multivariate classification system based on phylogenetic, comparative genomics, and structural analysis has been established (Makarova et al., 2011). Subsequently, Cas proteins have been divided into two class systems, Class I proteins, which have multiple subunits with CRISPR RNA (crRNA) complexes, and Class II, which rely on a single protein to function (Makarova et al., 2015). To date, the four Cas enzymes commonly used in nucleic acid detection all originate from the Class II system, which is capable of differential activity on single-stranded DNA (ssDNA), double-stranded DNA (dsDNA), and single-stranded RNA (ssRNA), respectively. Cas9 protein has a cis-cleavage function, recognizing the protospacer adjacent motif (PAM) sequence that binds specifically to its guide RNA, and efficient cleavage occurs. Cas12, Cas13, and Cas14 (Cas12f) all have a trans cleavage function in addition to cis cleavage, namely the ability to cleave non-selective collateral nucleic acids (Aman et al., 2020).
The CRISPR system-based assays are specific, programmable, and accessible. Further improvements have been reported for representative platforms targeting nucleic acid material, such as specific high sensitivity enzymatic reporter unlocking (SHERLOCK) and DNA endonuclease-targeted CRISPR trans-reporter (DETECTR), as well as more advanced platforms inspired by such platforms (Bonini et al., 2021). In previous reviews, the potential strengths of some platforms for nucleic acid detection and disease diagnostic applications have been summarized. However, some tools have recently been updated, with a growing number of analyses for non-nucleic acid material expanding the range of CRISPR-based tools, leading to significant advances in other areas of human health. Therefore, those newly evolved methods must be sorted out to illustrate the ongoing development and future trends. This review provides an overview of detection strategies ranging from nucleic acid to non-nucleic-acid materials based on improved platforms and an outlook on applying CRISPR system detection (Supplementary Material S1) in practical challenge scenarios such as disease diagnosis and food safety.
In archaea and bacteria, the CRISPR system functions by targeting nucleic acids to achieve immunity against foreign invasion (Barrangou et al., 2007). Although different CRISPR-Cas systems are found among them, crRNA is the core of their target molecules (Brouns et al., 2008). The editing of crRNA-employed genetic engineering allows the function of Cas proteins that hybridize with specific nucleic acid sequences (Cong et al., 2013). This exemplifies the programmability, simplicity, and high sensitivity of CRISPR, demonstrating the power of using CRISPR for detection.
Cas9 (part of Class II, Type II system) produces complementary trans-activating crRNA (tracrRNA) utilizing pre-crRNA repeat sequences for maturation (Jinek et al., 2012). In the construction of detection tools, tracrRNA is typically constructed in combination with crRNA as a small-guided-RNA (sgRNA), with its hybridized sequences limited to PAM or protospacer flanking sequences. Cas9 has only a cis-cleavage function and is capable of causing dsDNA cleavage (Chen and Doudna, 2017). Cleavage activity for ssDNA and ssRNA has also been reported in the presence of PAM-presenting oligonucleotides (PAMmers) sequence designed to complement the target sequence (O’Connell et al., 2014; Sternberg et al., 2014). This property has been successfully used in coupling with real-time fluorescence analysis modalities for the detection of the specific nucleic acid target (Zhou et al., 2018).
Cas12 belongs to the Class II, Type V system, with Cas12a being one of its widely used subtypes. Cas12 recognizes complementary ssDNA or dsDNA with a PAM sequence under the guidance of its crRNA, directing the cleavage of DNA through a single RuvC structural domain (Zetsche et al., 2015). Its family has another commendable category of nucleases, Cas12f (formerly Cas14) in the latest classification system (Makarova et al., 2020), which is less than half the size of Cas9, has promising ssDNA cleavage activity, and can be generated without the need for PAM sequences (Harrington et al., 2018; Karvelis et al., 2020). Cas12 has been reported to have efficient trans cleavage activity. It can induce robust and nonspecific ssDNA cleavage when it cleaves dsDNA in a sequence-specific manner, a property that has been widely used in assays since its discovery in 2018 (Li et al., 2018a; Chen et al., 2018).
Cas13, which belongs to the Class II, Type VI system, contains the HEPN structural domain that binds to RNA for cleavage and is guided by a single RNA (Abudayyeh et al., 2017). Cas13a (formerly C2c2) is one of its widely used subtypes. It has been found that the Cas13a-crRNA complex, once activated by binding to its target RNA, cleaves it as well as other non-specific RNAs. This trans cleavage capability expands the range of targets to be detected (Abudayyeh et al., 2016).
Colorimetric methods analyze the content of substances by comparing the shades of light in colored solutions and have unique readout systems in biosensing owing to their rapid, intuitive, and low-cost characteristics. Among the standard detection probes used in colorimetric systems, gold nanoparticles (AuNPs) are frequently used. They have particular optical properties such as high extinction coefficient, localized surface plasmon resonance (LSPR), and inherent photostability (Aldewachi et al., 2018). LSPR features lead to color changes in AuNP and are utilized to construct CRISPR-based colorimetric sensors (Figure 1A), as physiological processes and biomolecular interactions affect the dispersion and aggregation of AuNPs (Baptista et al., 2008).
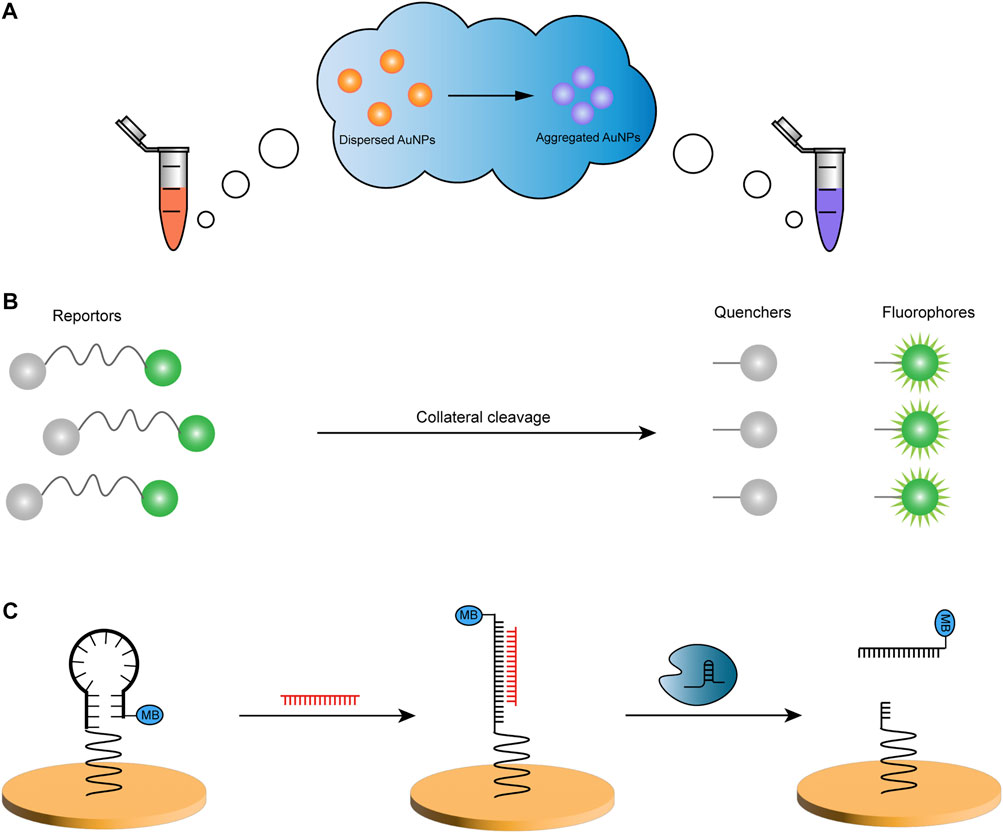
FIGURE 1. Examples of three common readout methods. (A) The color of the solution in the centrifuge tube changes due to the aggregation of AuNPs. (B) The reporter consists of a fluorophore and a quencher, linked by a nucleic acid molecule. After its collateral cleavage by the CRISPR system, the change in the distance leads to the generation of fluorescent energy. (C) The conformation of E-DNA changes from hairpin to linear dsDNA when the ssDNA of the gold electrode hairpin structure is complementary to the target ssDNA (red), and Cas12a cleaves the linear DNA to release the MB strand. MB, methylene blue.
However, colorimetric sensors also suffer from the same problems of false positives and negatives caused by the instability of the reporter molecule or due to the weak signal (Xie et al., 2021). In contrast to the other methods, the naked-eye readout system requires a stable optical signal, and the procedure does not rely on sophisticated equipment. Still, it can be a complex and time-consuming process. In addition, non-specific interference is a significant challenge for the colorimetric method, and refinements to AuNPs are continually required to improve the accuracy of their reporting signals (Xu et al., 2021).
In disease diagnosis, fluorescence technology is widely used in bioassays (Yue et al., 2018; Lu Y. et al., 2022). The fluorescence resonance energy transfer (FRET) sensor has the advantages of superb sensitivity, high specificity, and fast responsiveness (Zhang et al., 2019). FRET is a process in which non-radiative energy is transferred from donor fluorophore to acceptor chromophore, where the donor is electronically excited, with the acceptor both being photosensitive molecules (Zhang et al., 2019; Phan et al., 2022). In previous strategies, a pair of closely positioned donors and acceptors is usually designed to excite a reporter signal using specific recognition of the target molecule by the acceptor and conformational changes of the FRET system (Zhang et al., 2019). Quenchers are used with increasing frequency in acceptors due to their broad absorption spectra and high extinction coefficients and for the construction of detection systems containing pairs of fluorophore and quenchers of molecules linked by DNA or RNA probes (Figure 1B) (Phan et al., 2022). A CRISPR-based sensor using FRET technology would utilize the trans cleavage activity of the Cas enzyme to digest the probe by specific recognition followed by excitation of the fluorescent signal (Phan et al., 2022).
In addition to FRET, researchers have similarly focused on luminescent resonance energy transfer (LRET) due to its more significant advantages by compensating for the shortcomings of FRET, for instance, micelles and liposomes scatter the light which complicates the FRET applications in membrane-protein research (Sohail et al., 2022). LRET uses luminescent lanthanide chelates as donors with millisecond lifetime, compared to FRET which uses a donor with millisecond lifetime. In addition to longer lifetime, LRET also has the advantage of using multiple acceptors with flexibility, which is better for detecting conformational changes and dynamics of proteins or other macromolecules (Dolino et al., 2014). In the application of integrated CRISPR sensing, Li established an LRET-based functional DNA (fDNA)-regulated CRISPR/Cas12a biosensor by combining holographic optical tweezers with energy-concentrating upconversion luminescence nanoparticle boosting enhancement of LRET (Li C.-Y. et al., 2021). This has a more significant advantage in detecting non-nucleic acid analytes since the approach improves sensitivity and imaging capability (Sohail et al., 2022).
Electrochemical biosensors can convert biochemical reactions into electrical signals that can be measured with simplicity, low cost, and high selectivity (Fritea et al., 2018). It has been combined with CRISPR to develop fast and less equipment-dependent detection tools through the use of membranes or microfluidics on working electrodes in association with different materials to eliminate interference and amplify signals (Bruch et al., 2019; Bruch et al., 2021; Phan et al., 2022), such as electrochemical DNA (E-DNA) sensors (Figure 1C) (Dai et al., 2019; Xu et al., 2020). In addition to the aforementioned fluorescence and luminescence techniques alone for bioanalysis, luminescence methods have also been exploited by integrating them with electrochemical techniques to create new electrochemiluminescent (ECL) biosensors. They work by generating substances on the electrode surface that change from stable precursors to unstable intermediates and consequently form luminescent excited states via electron transfer involving the conversion of electrical and radiative energy (Richter, 2004). ECL improves on conventional electrochemical sensors with high sensitivity, low background interference, electrochemical controllability, and excellent selectivity (He et al., 2020), which can build a more stable bioassay platform and play an essential role in the field of medical examinations (Qin et al., 2021; Suea-Ngam et al., 2021; Zhang et al., 2021).
In 2016, Collins et al. applied CRISPR/Cas technology to nucleic acid detection for the first time, successfully incorporating Cas9 protein to develop a paper-based sensor for the detection of Zika virus (ZIKV) (Pardee et al., 2016). In Collins’ study, they used an isothermal RNA amplification technique called NASBA (nucleic acid sequence-based amplification), where the first step of the process is the reverse transcription of the target RNA by a complementary reverse primer, thereby producing an RNA/DNA double-stranded body, followed by degradation of the RNA template in the presence of RNase H. Then, the T7 promoter containing the forward primer binds and initiates the extension of the complementary strand to produce a dsDNA product, which is finally transcribed into copies of the target RNA sequence. The reaction is carried out in an isothermal environment at 41°C after initial heating at 65°C. Once the nucleic acid amplification step is complete, the detection part is accomplished by a toehold switch, allowing the hairpin structure to release after the amplification product is added to the paper-based sensor. This would activate the synthesis of Lac Z, which reports by the colorimetric change of the yellow substrate into the red product. Due to the naturally occurring PAM sequence specificity of Cas9 protein, the NASBA-CRISPR cut (NASBACC) strategy (Figure 2A) was used for single-base recognition, enabling effective classification of different ZIKV strains while controlling the assay time to 3 h.
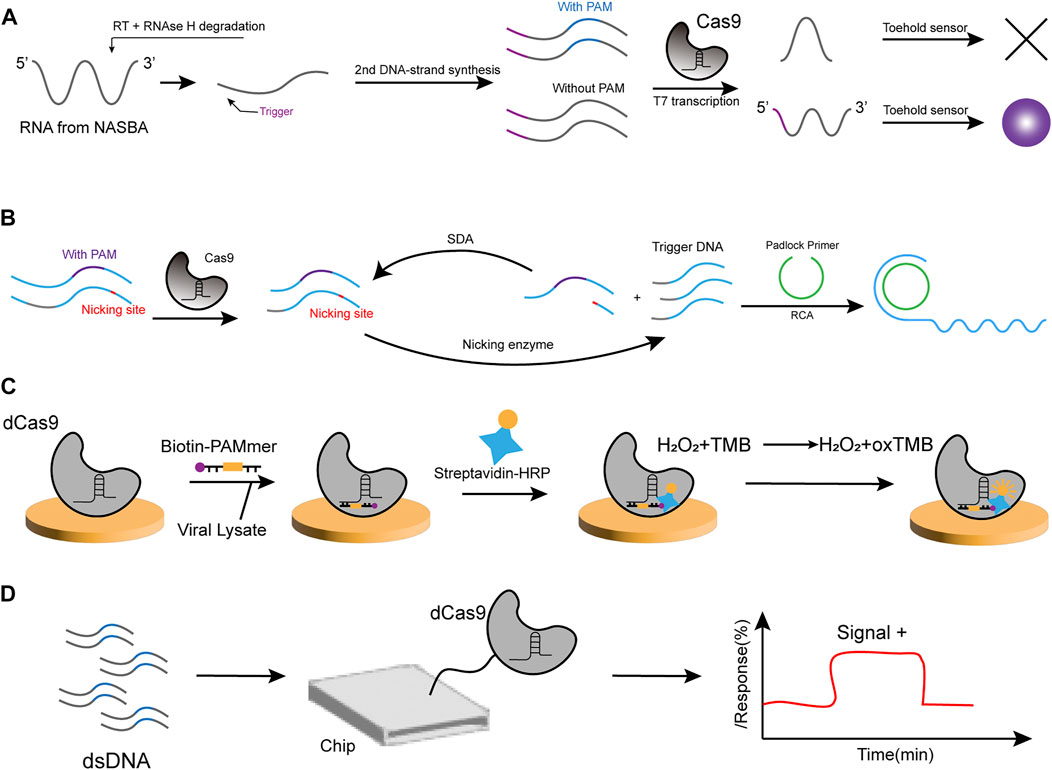
FIGURE 2. Strategies for Cas9-based nucleic acid sensor. (A) NASBA amplifies the RNA target, and the trigger sequence for the toehold switch is introduced. Subsequently, the RNA and DNA hybridization produces RNA that is degraded by RNAse H. The second DNA strand is synthesized, and it carries the T7 promoter sequence. The transcribed RNA can be used as starting material for NASBA and interact with the toehold switch. In the presence of PAM sequences, the RNA target site is shorter due to the cleaved template and cannot activate the sensor, and vice versa, producing a significant color change. (B) In the action of Cas9, broken dsDNA at the nicking site is further recognized by the nicking enzyme. Once the DNA polymerase reaches the nicking site, a new strand can be generated and the downstream strand is replaced. As a result, many short ssDNAs are generated to trigger rolling circle amplification (RCA). Subsequently, the RCA product can bind to oligonucleotide-spliced AuNPs, inducing the aggregation of AuNPs. SDA, Strand-displacement amplification. (C) Viral lysate and biotin-PAMmer are added to microplates immobilized with dCas9/gRNA complexes. Following, Streptavidin-HPR and TMB substrate solutions were added to the microplate. Finally, the yellow color is observed in the presence of the virus. (D) The CRISPR-chip is composed of a gFET structure with a complex of sgRNA and dCas9 formed on the graphene surface. When the target DNA is detected, dCas9 binds to the target DNA, which modulates the electrical properties of the gFET and leads to an electrical signal output. gEFT: graphene-based field-effect transistor.
Another material for the Cas9 detection strategy is AuNP, also by utilizing colorimetric methods. Liu et al. constructed an assay based on the CRISPR/Cas9 system that triggered signal amplification to identify the Phytophthora infestans genomic DNA (Chang et al., 2019). Oligonucleotide-functionalized AuNPs hybridize with Cas9/sgRNA digestion products that have been isothermally amplified, resulting in a color change from burgundy to purple after aggregation. Strand-displacement amplification (SDA) reactions and rolling circle amplification (RCA) reactions occur here. First, the cleaved dsDNA generates new strands in the presence of DNA polymerase, which further generates many short ssDNA to trigger RCA (Figure 2B). After the RCA reaction, the product is hybridized with oligonucleotide-functionalized AuNPs, thus causing the changes. The method has a visual detection limit of 2 pM DNA and shows an excellent linear relationship between the ratio of A 650 nm/A 525 nm and model DNA concentrations of 0.2 pM to 20 nM, with good sensitivity and high specificity.
Kang’s team designed a (CRISPR)/Cas9 endonuclease dead (dCas9)-based colorimetric sensor to detect viruses such as SARS-CoV-2. They attached biotin to a PAMmer and achieved color change by (HRP)/3,3′,5,5′-tetramethylbenzidine (TMB) reaction, thereby effectively reducing the assay time (Figure 2C) (Moon et al., 2020). The dCas9 is a catalytically inactive enzyme produced by mutations in the RuvC and HNH structural domains of wild-type SpCas9. Although it has lost its gene-editing function, the retained gRNA binding ability makes it a powerful tool in many fields, including disease diagnosis (Kanafi and Tavallaei, 2022). For example, the Paired dCas9 (PC) reporter system was designed (Zhang et al., 2017) and used the split luciferase system to construct two dCas9-based sensors that recognize two fragments of the same DNA sequence, each with half of the firefly luciferase enzyme attached to the N- and C-terminal, respectively. When specific recognition is achieved, the integral enzyme catalyzes the bioluminescent reaction and reportedly detects analytes at concentrations ranging from 5 × 10−5 to 6 × 10−3 nmol/ml.
Li et al. developed a CRISPR/Cas9-triggered hairpin probe-mediated biosensing method (CHP) that differentially hybridizes long-stranded RCA products with fluorophore quencher-labeled molecular beacons, enabling real-time fluorescent readout (Wang et al., 2021). The authors found that CHP is highly specific for detecting circulating tumor DNA (ctDNA) in human serum without requiring DNA extraction and purification. Additionally, to improve the detection efficiency, AuNPs have been used to develop lateral flow nucleic acid detection tools capable of accurately detecting viruses (Wang X. et al., 2020; Xiong et al., 2021). Zhang et al. developed a way to use engineered crRNAs introduced into DNA (as enzymes or probes) to bind fluorescent reporter groups for testing (Safdar et al., 2022).
Cas9 has also been used for electrochemical methods of the assay. Another assay using dCas9 is named CRISPR-Chip, which immobilizes the dCas9/CrRNA complex on a graphene-based field-effect transistor (gEFT) to construct a sensor that detects the variable currents caused by changes in graphene conductivity and does not require a reporter molecule for the deletion in the dystrophin genes of Duchenne muscular dystrophy patients (Figure 2D) (Hajian et al., 2019). Similarly, the team reported the CRISPR-SNP-Chip system in order to achieve typing of single-nucleotide polymorphisms (SNPs) to escape the dependence on advanced sequencing technologies (Balderston et al., 2021). Liu et al. explored a primer-exchange-reaction (PER)-based biochemical circuit cascade using a Cas9 enzyme with only single-stranded cleavage activity. Driving a pair with different sgRNAs of Cas9 generates dsDNA with sticky ends, ligated to a hairpin-structure substrate, which subsequently generates a large number of short fragments that report the electrical signal of the detected DNA (Dai et al., 2020). Huang’s team developed a dCas9-ECL probe for sensitive and single-base specific DNA detection with good resistance to nonspecific interference and a low false positive detection rate (Wu L. et al., 2022). This shows that electrochemical biosensors can modularize the circuitry with better programmability.
In 2018, Jennifer and associates took advantage of trans-cleavage activity by Cas12a (formerly Cpf1) to establish a method named DETECTR (Chen et al., 2018), which achieves attomolar sensitivity for DNA detection (Figure 3A). The authors built a platform for testing different strains of human papillomavirus (HPV) after discovering that Lachnospiraceae bacterium ND2006 Cas12a (LbCas12a) can cut ssDNA in a robust, nonspecific manner. Human samples are first processed for recombinase polymerase amplification (RPA) reactions and subsequently incubated in a fluorophore quencher (FQ) reporter substrate ligated by ssDNA and Cas12a-crRNA, in anticipation of a readout of the fluorescent reporter signal. Due to the importance of point-of-care testing (POCT) (Drain et al., 2014), a rapid and visual detection method named Cas12a-based Visual Detection (Cas12aVDet) was proposed to further reduce the time to approximately half an hour based on DETECTR. It is a one-pot reaction with bright fluorescence directly visible under blue light (Wang et al., 2019).
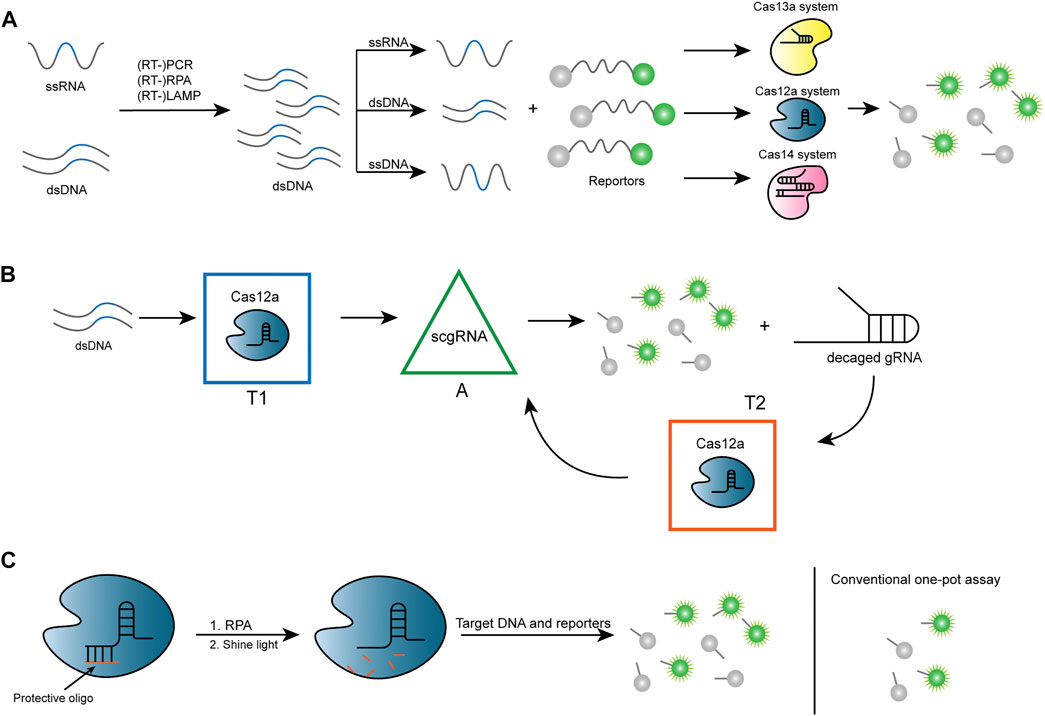
FIGURE 3. Strategies for Cas12 and Cas13-based nucleic acid sensor. (A) After amplification, ssRNA or dsDNA forms dsDNA and produces ssRNA, dsDNA, and ssRNA, depending on the different systems. After being mixed with fluorescent reporters, trans cleavage occurs through specific binding of different CRISPR/Cas systems, which subsequently generates a fluorescent signal. (B) In CONAN, there are three signal processors, namely transducer 1 (T1), transducer 2 (T2), and signal amplifier (A), where the parallel connection of A and T2 forms a positive feedback circuit and T1 is connected in series with the positive feedback circuit. Cas12a and gRNA for target dsDNA (gRNA-T) are pre-assembled as T1, which converts the signal input into active Cas12a protein (Cas12a/gRNA-T/DNA complex). After the action of a self-reporting scgRNA, an amplified fluorescent signal and multiple active (decaged) gRNA molecules are output. T2 contains Cas12a protein and a probe for decaged gRNA, which can transduce the resulting decaged gRNA into another active Cas12a protein to produce an amplified fluorescent signal. scgRNA: switchable-caged guide RNA. (C) The protective oligonucleotide is designed to partially pair with the crRNA, thereby altering the conformation of the crRNA so that it cannot bind to the target DNA and inhibit the cleavage activity of Cas12a. The protective oligonucleotide breaks when exposed to ultraviolet (UV) light at 365 nm isolated from crRNA, thereby restoring the function of CRISPR-Cas12a detection. This system ensures that the target DNA is not reduced by Cas12a during the amplification phase, resulting in a stronger fluorescent signal.
Another Cas12a-based assay is called HOLMES (one-hour low-cost multipurpose highly efficient system) (Li et al., 2018b). It enables the efficient detection of SNP in human genotypes. HOLMES uses LbCas12a and polymerase chain reaction (PCR) for amplification and is applied to detect DNA viruses and RNA viruses with detection limits as low as 1–10 aM. The detection system has also been updated into a second version (HOLMESv2) (Li et al., 2019), which employs Cas12b (previously known as C2c1). Cas12b is another type V-B CRISPR-Cas system subtype that has a more preferred trans cleavage activity for dsDNA than Cas12a (Chen et al., 2018). HOLMESv2 combines target detection and loop-mediated isothermal amplification (LAMP) reactions, accomplishing both in one system, thus significantly increasing the efficiency, along with the capability of single-molecule testing like DNA, RNA, and even the degree of methylation of DNA. LAMP has the characteristics of high sensitivity, short reaction time, and no need for expensive reagents and specialized apparatus for clinical use (Notomi et al., 2000). Improving on the CRISPR-Cas12-based assay DECTECTR (Broughton et al., 2020), a two-step assay was optimized for identifying SARS-CoV-2 with faster detection and reasonable accuracy (95% correct positives and 100% correct negatives) using reverse transcription LAMP (RT-LAMP) for pre-amplification. A Cas12b-based one-pot method termed STOPCovid (SHERLOCK testing in one pot) also uses RT-LAMP amplification with proper Cas proteins from Alicyclobacillus acidophilus (AspCas12b) that can adapt to the corresponding temperature for detection (Joung et al., 2020). Another system using Cas12b is called iSCAN; version 2 simplifies the previous two-pot system to a single-pot system using the reverse transcription RPA (RT-RPA) method for amplification, with optimization of the solution system as well as the steps to perform clinical assays at optimal temperatures (Aman et al., 2022). Shi et al. (2021) developed a CRISPR-Cas–only amplification network (termed CONAN) which was supported by a CRISPR-Cas–powered catalytic nucleic acid circuit for ultrasensitive nucleic acid diagnostics (Figure 3B). In this study, switchable-caged guide RNA (scgRNA) with self-reporting capability was designed for single-nucleotide mutation (SM) detection in combination with Cas12a, which has PAM sequence recognition and trans cleavage activity. Its one-step reaction has a greater advantage in detecting hepatitis B virus (HBV) infection and human bladder cancer–associated SM.
A new Cas12a-based assay platform was recently developed to detect rabies virus (RABV) using the ECL method (Liu S. et al., 2022). The target molecule is amplified in the presence of two DNA probes, generating a large amount of ssDNA to activate the trans cleavage activity of CRISPR/Cas12a. Subsequently, single-stranded trigger (ST) strands are non-specifically degraded and inactivated, resulting in the inability to close DNA nanotweezers (DTs), i.e., hemin cannot be captured at the electrode, thus generating a high concentration and triggering an ECL signal.
In 2021, Chen and Ji’s team reported a class 2 type V-F CRISPR-Cas genome-editing system from Acidibacillus sulfuroxidans (AsCas12f1, 422 amino acids) (Wu et al., 2021), capable of targeting dsDNA with 5′T-rich protospacer adjacent motifs. They subsequently developed a Cas12f-based assay with low detection limits and detection concentrations higher than 500 pM without amplification (Wu Z. et al., 2022). However, the trans cleavage activity of Cas12f is inferior to Cas12a and Cas12b (Harrington et al., 2018), and further optimization of the system is needed to bring the CRISPR/Cas12-based biosensor to the clinic.
Many have started with the reaction solution to improve sensitivity in order to achieve this goal. In previous studies, Mg2+ in the buffer solution was found to be essential for the cis cleavage ability of Cas12a (Fonfara et al., 2016). A study by Wang et al. found its potential to affect trans cleavage activity as well (Lv H. et al., 2021). Studies on the effect of different metal cations on the cis and trans cleavage activity of LbCas12a have been carried out (Ma et al., 2020; Nguyen et al., 2020; Yue et al., 2021), for example, in a recent study, Mn2+ was found to have the ability to promote both cis and trans cleavage activity (Xie et al., 2022). Other properties of the buffer solution also contribute, such as high pH (8.5–8.6) solutions being more favorable for Cas12a to exert trans cleavage activity. Several components, such as polyethylene glycol (PEG), also significantly enhance the cleavage signal (Lv H. et al., 2021). Bovine serum albumin (BSA) and L-proline markedly improved the detection efficiency of Cas12a (Li Z. et al., 2021). In addition to the buffer solution, the ratio of Cas12a, crRNA, and target dsDNA at 1:2:2 also exhibited the maximum kinetic efficiency (Lv H. et al., 2021). Besides, it was found that the base profile of the ssDNA linked to the FQ-reporter also affects the nonspecific cleavage of Cas12a, with the Poly-C reporter having the highest efficiency for being cleaved, while the Poly-G reporter, in contrast, can barely be cleaved. An increase in the length of this sequence up to 8 promotes an enhancement in fluorescence intensity. Still, the increase is not significant after the peak of 8 is reached, and the synthesis of excessively long ssDNA also leads to an additional cost of detection (Lv H. et al., 2021). In a recent study, researchers found enhanced trans cleavage activity and significantly improved signal intensity by linking the fluorophore to the quencher with hairpin DNA (Rossetti et al., 2022). These studies set the first step toward continuous improvement of the reaction system in the future. Researchers have also optimized the assay by crRNA engineering, introducing DNA, elongating one end of the crRNA, and chemical modification of the crRNA, all reported to have improved the assay sensitivity (Park et al., 2018; Nguyen et al., 2020; Ooi et al., 2021). Besides, Yin et al. found that most suboptimal (suboptimal) sequences (e.g., VTTV, TCTV, and TTVV) have better performance relative to standard PAM (TTTV) by comparing several PAM sequences used for crRNA and devised a method named sPAMC (for suboptimal PAM of Cas12a-based test with enhanced flexibility, speed, sensitivity, and reproducibility) to achieve fluorescence analysis of viruses in a one-pot reaction with less than 20 min (Lu S. et al., 2022). Hu et al. designed a light-controlled one-pot assay ingeniously by designing a protective oligonucleotide to bind to crRNA which reduces the cleavage of Cas12a at the RPA stage, after that, the protective oligonucleotide is cut off by UV irradiation at 365 nm and crRNA is released for the readout of the signal (Figure 3C) (Hu M. et al., 2022). The photocontrolled crRNA reaction provides a stronger detection signal than the conventional one-pot detection method. To change the current situation of the lateral flow test (LFT)-DETECTR system, where the test zone signal is prone to false positives, a system called DIRECT (DNA-Immunoglobulin Reporter Endonuclease Cleavage Test) was designed using a composite probe consisting of the DNA part (biotinylated dsDNA connected to ssDNA with fluorescein) (FAM) and the antibody part (mouse anti-FAM IgG), that was capable of improving the signal readout of the test zone2, (Bhatt et al., 2022). A method called CLEVER (CRISPR-Cas12 integrated RT-LAMP Easy, Visual and Extraction-free RNA) makes detection faster and more convenient by optimizing the detection of N and E genes of SARS-CoV-2 by removing the RNA extraction step in the process of protein digestion (Bhatt et al., 2022). Cheng’s team performed pre-amplification of Cas12a assay by LCR (ligase chain reaction) to achieve ultra-sensitive visualization of microRNA detection. However, a specific measure may be limited to certain Cas enzymes and reaction systems to be effective, and further identification of reaction mechanisms still requires in-depth research (Qiu M. et al., 2022).
In 2017, Zhang et al. initiated the groundbreaking use of Cas13a for highly sensitive detection (Figure 3A) by developing a SHERLOCK system targeting DNA or RNA (Gootenberg et al., 2017). The system first amplifies the target gene isothermally employing RPA or RT-RPA, a process that generates a large amount of DNA. Subsequently, T7 RNA polymerase comes into play and transcribes the DNA into RNA to serve as a substrate for Cas13a nuclease. The crRNA then complements this RNA. The technology was updated a year later with a second version (SHERLOCKv2) (Gootenberg et al., 2018). The new detection system achieved a 3.5-fold increase in signal sensitivity through the action of Csm6, a protein that activates collateral activity. In addition, SHERLOCKv2 also uses a lateral flow approach, thus providing rapid detection in areas lacking specialized equipment and promising to offer on-site diagnostic coverage in areas infected with viruses such as ZIKV and Gradenovirus (DENV).
To detect ZIKV and DENV directly in the bodily fluids, the researchers developed HUDSON (heating unextracted diagnostic samples to obliterate nucleases) (Myhrvold et al., 2018), allowing colorimetric reporting of signal readout. HUDSON has expanded the assay to include samples that can be pre-amplified with RPA, such as blood, plasma, serum, urine, and saliva, by lysing the viral particles with heat and chemical reduction to inactivate the ribonuclease resulting in the release of nucleic acid molecules. The combination of HUDSON and SHERLOCK reduces the time to obtain results to 1–2 h with limited sample and equipment requirements. To improve HUDSON, a sensing method with sensitivity and specificity termed SHINE (Streamlined Highlighting of Infections to Navigate Epidemics) was developed (Arizti-Sanz et al., 2020). In addition to shortening the detection time, the authors also developed a companion smartphone application that allows straightforward interpretation of the fluorescence signal in the test tube. The method was tested in 30 SARS-CoV-2 positive patient samples and 20 negative patient samples with 100% specificity for negative samples and 90% sensitivity for positive samples (27/30). HUDSON reportedly accelerated the extraction of viruses from nasal swabs and saliva samples during this process. The method reduced the LOD (limit of detection) by adding RNase H and optimizing the concentration of various components of the buffer solution. Another Cas13a-based assay achieves quantitative measurement of RNA without pre-amplification of the genome (Fozouni et al., 2021). The authors used a Cas13a homolog from Leptotrichia buccalis (Lbu), which is considered to have robust trans cleavage activity and sensitivity, followed by designing multiple combinations of crRNAs for use towards separate target gene regions of the virus, capable of detecting ∼100 copies/µl of SARS-CoV-2 virus. Subsequently, a mobile phone-based fluorescence microscope and reaction chamber was used to quantify the fluorescence signal for quantitative measurements.
Another promising assay platform called CREST (Cas13-based, rugged, equitable, scalable testing) was developed by Rauch et al. to tackle the issue of reagents, equipment, and cost that prevented the use of Cas13 assay systems on a large scale (Rauch et al., 2021). Rauch et al. developed Bluetooth-enabled, field-ready thermocyclers in combination with Taq enzymes to overcome the previous restriction of PCR amplification only in specialized laboratories. Then, the authors produced a P51 cardboard fluorescence visualizer powered by a 9-V battery so that the activity of detecting Cas13 could be visualized, and the results of the FQ-reporter could be read out. Although CREST has further reduced the assay cost from several angles, RNA extraction continues to rely on commercial kits. Therefore, another method called PEARL (precipitation-enhanced analyte retrieval) was proposed to cut down the reliance on commercial kits (Ponce-Rojas et al., 2021).
Cas13d ribonuclease is derived from Ruminococcus flavefaciens (CasRx). To expand CRISPR-based diagnostic strategies, researchers have used Cas13d for the first time in molecular diagnostics by developing a system for detecting SARS-CoV-2 called SENSR (Sensitive Enzymatic Nucleic-acid Sequence Reporter) as a modification of SHERLOCK (Brogan et al., 2020). The first step of SENSR is isothermal amplification, an initial 45-minute isothermal pre-amplification reaction by RT-RPA that generates a short dsDNA amplicon containing the T7 promoter sequence that is processed into the final RNA target sequence. The reported signal is read out by fluorescence or lateral flow, with the LOD determined by fluorescence readout to be ∼100 copies/µl and 92% (11/12) concordance with SENSR fluorescence analysis on lateral flow analysis. These measurements indicate its potential to be used in rapid diagnostic tests.
Achieving rapid detection of multiple pathogens has become a major issue owing to many human infectious diseases and the significant threat they pose to humans. This led one group of researchers to develop a CARMEN detection system (Combinatorial Arrayed Reactions for Multiplexed Evaluation of Nucleic acids), a scalable multi-pathogen detection platform with considerable statistical robustness (Ackerman et al., 2020). Researchers have tested it against 169 human-associated viruses, and a minimum of 10 published genomic sequences found it to have excellent sensitivity along with low cost. The method uses PCR or RT-RPA for amplification. The assay reaction system is performed in a conventional microtitre plate, and the droplets are sorted by 1,050 color codes of the solution. The system is a breakthrough in reducing the cost of the SHERLOCK assay, achieving simplification of steps, and reducing the diagnosis time in epidemic situations.
Nucleic acid amplification is an essential part of assay reporting signal enhancement, but the process can be subject to problems such as sample contamination and loss. For this reason, Zhou’s team developed a confinement effect-inspired Cas13a assay called the ultra-localized Cas13a assay, which elevates molecular concentrations in picoliter-sized droplets by droplet microfluidics for the assay (Tian et al., 2021). The fluorescence signal of the highly concentrated droplet system was reported to be quite marked, achieving RNA quantification at the single-molecule level. The new method was used to accurately count the cell-free microRNA and bacterial 16S rRNA in clinical serum samples while also allowing the diagnosis of the N gene for SARS-CoV-2. The results showed that 40 nasopharyngeal swab samples reported results in complete agreement with RT-PCR, demonstrating its power as a tool for quantitative molecular biology and accurate clinical diagnosis.
The CASCADE (CRISPR/CAS-based Colorimetric Nucleic Acid Detection) method was recently reported for the rapid and specific diagnosis of SARS-CoV-2 under naked eye conditions (López-Valls et al., 2022). Since colorimetric methods using AuNPs are not commonly used in Cas13-based detection systems, the authors chose the Cas13a enzyme to extend the detection tool, thus increasing its versatility. Cas13a enzyme system has the advantage that it lacks dependence on PAM sequences. The detection process of this system involves activating the trans cleavage ability of Cas13a, which leads to the shortening of ssRNA on the surface of AuNPs, causing their aggregation to produce color changes. The authors emphasize that the destabilizing aggregation of ssRNA-AuNPs is stronger than the previously reported effects based on Cas12a and ssDNA-AuNPs, and the naked eye quickly observes the changes. In the detection of targeted SARS-CoV-2, the method shows sensitivity at the level of picomolar concentrations, and its detection limit can be reduced to femtomolar (3 fM), and even attomolar (40 aM) ranges when combined with optimization of isothermal amplification. Remarkably short time (15–30 min), and the inexpensive visual readout suggest its ability for point-of-care testing of infectious diseases and the feasibility of future large-scale use. Besides, Hu F. et al. (2022) designed a one-pot system named ECS-CRISPR (Easy to operate, Contamination-free, and Stable CRISPR). The system consists of a nested inner and outer tube, and the CRISPR Cas13a fluorescent detection reagent is pre-existing in the inner tube. After the amplification reaction is completed, the product is transferred to the outer tube by centrifugation through hydrophobic wells. Fluorescent visual detection stimulation of the target nucleic acid is achieved under blue light-emitting diodes (LED), enabling the detection of pathogenic nucleic acids within 25 min without the need for particular instruments, thus effectively reducing contamination.
Jung et al. combined Cas13a with an electrochemical sensor to achieve nucleic acid-free amplification detection of SARS-CoV-2 (Heo et al., 2022). In this study, a redox probe conjugated with ssRNA is immobilized on the electrode surface modified with a nanocomposite (NC) and gold nanoflower (AuNF), and Cas13a trans cleavage ability is activated after digestion of reRNA to generate current. The sensor achieves detection limits as low as 4.4 × 10−2 fg/ml for ORF and 8.1 × 10−2 fg/ml for S genes with a dynamic range of 1.0 × 10−1–1.0 × 105 fg/ml. Zhang et al. developed a new CRISPR Cas13a-gFET biosensor. In this system, the trans cleavage mechanism of CRISPR Cas13a is used to generate a current signal that can be combined with a graphene field-effect transistor (gFET) for amplification-free detection of nucleic acids with LODs down to the amolar range (Li et al., 2022d).
The above-mentioned methods for detecting nucleic acid analytes based on CRISPR are summarized in Table 1.
The most crucial step of CRISPR-based detection strategies when analyzing non-nucleic acid target molecules is to find ways to convert the non-nucleic acid signal into a nucleic acid signal. Therefore, some detection strategies have designed receptors containing DNA molecules that have in common the release of DNA upon the addition of a foreign target analyte to the system to act as an activator of the CRISPR system, thus triggering the trans cleavage ability to send a signal via a fluorescent method.
To address the lack of fluorescence intensity in complex samples, a novel luminescent material has been used in CRISPR detection, namely upconversion nanoparticle (UNCP) with high sensitivity and good resistance to environmental infections, and the trans cleavage ability of Cas12a allowing the departure of quenchers to trigger fluorescence (Li et al., 2020). Li et al. designed a polyacrylic acid-coated energy-confining UCNP with a sandwich structure (SUCNP), which was irradiated by a 980 nm portable laser to emit light, and the signal was able to be analyzed by a smartphone. The system cleverly utilizes fDNA, a DNA molecule complementary to crRNA, and a DNA aptamer that binds specifically to the target synthesized as a complex. When the target molecule is present, it competitively binds its aptamer, thus releasing DNA to activate Cas12a (Figure 4A). Similarly, Gao’s team constructed a UCNP-DNA-Fe3O4 probe to amplify fluorescent signals (Mao et al., 2022b). This biosensing approach can detect ochratoxin A, which appears in food and feed products. During the detection process, ochratoxin A binds to the aptamer and releases DNA to participate in subsequent reactions, achieving an overall detection time of approximately 1 h and a detection limit as low as 1.564 ng/ml in corn flour. In addition, Xiong et al. (2020) also designed a sensor using FQ-reporter by fDNA aptamer to detect ATP and Na+ rapidly. At ambient temperature (25°C), the method can complete the assay in two steps in less than 15 min and is suitable for on-site diagnosis or POCT. The other method for detecting ATP is different in that it is designed in such a way that crRNA is specifically recognized with the ATP aptamer so that in the presence of ATP, the trans cleavage ability of Cas12a is not activated due to the lack of the complementary strand of crRNA, and thus no fluorescent signal is generated (Peng et al., 2020).
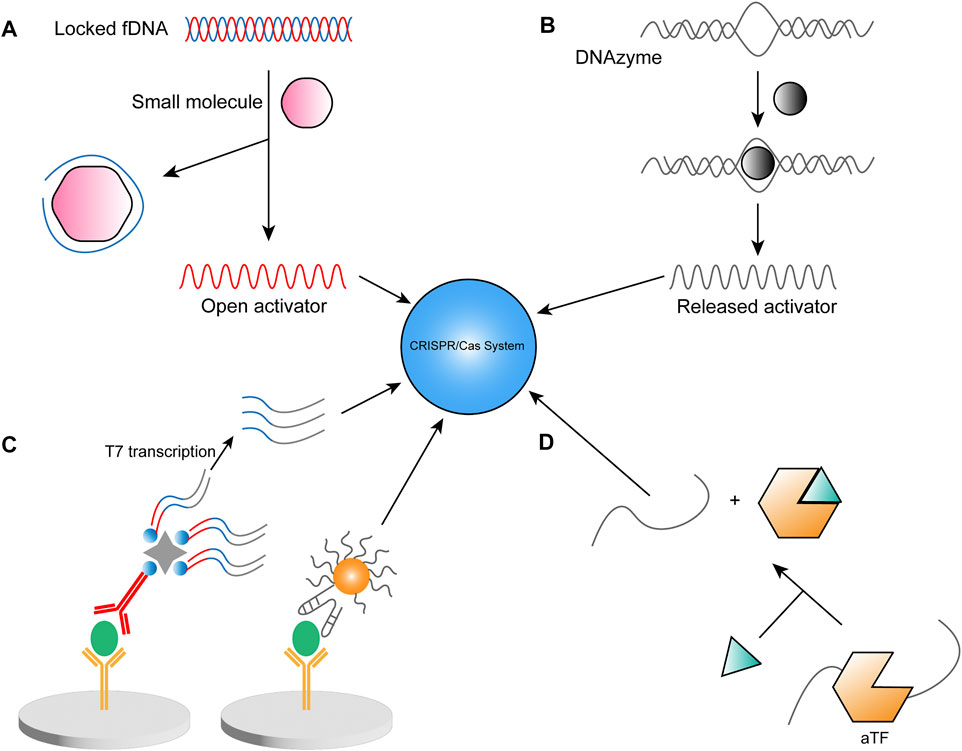
FIGURE 4. Strategies for CRISPR-based non-nucleic-acid detection. (A) Locked fDNA in the presence of a small molecule, one strand binds to it. It releases another complementary DNA strand to participate in the detection reaction (B) The DNAzyme starts in an inactive state and activates its cleavage ability with the involvement of specific ions, thus releasing short strands of DNA to participate in the detection system. (C) Left: Non-nucleic acid as antigen, linked to the CRISPR/Cas system and antibody (yellow)-antigen (green)-antibody (red) structure by a nucleic acid sequence containing the T7 promoter. Right: DNA-AuNP is used instead of antibody and is directly involved in the assay reaction. (D) Immobilized aTF-dsDNA complexes containing CRISPR target sequences change the conformation in the presence of aTF target small molecules, resulting in the release of dsDNA.
The fDNA probes can also be combined with electrochemical approaches to achieve more accurate and sensitive detection. Lu et al. designed an inductively coupled plasma mass spectrometry (ICPMS)-based sensor. They selected Thulium (Tm) as the material for the reporter molecule because of the extremely low concentration and interference in the organism and the environment. After the trans cleavage event, Tm was captured in the supernatant by a streptavidin-coated magnetic bead (SA-MB) and subsequently monitored by ICPMS. The assay platform was used to analyze kanamycin residues, reducing the workflow to less than 30 min and obtaining detection limits as low as 4.06 pM (Hu et al., 2021). In response to the SARS-CoV-2 pandemic, an electrochemical sensor based on the viral nucleocapsid protein was designed (Han et al., 2022). A gold electrode surface was used as an electrochemical sensing interface with methylene blue labeled poly adenines DNA sequence immobilized on its surface as a signal reporter molecule. Np aptamers were hybridized with the activation strand as an arched probe to release the activator in the presence of a target molecule. The cleavage of the reporter molecule caused a reduction in the current of differential pulse voltammetry, lowering the LOD to 16.5 pg/ml within 30 min. The authors tested the detection potential in different complex samples such as tap water, milk, and serum, to explore the future industrialization of the sensor.
A small molecule modification strategy for fDNA has been reported in recent studies (Kim et al., 2021). Small molecules can serve as key components for the recognition of target proteins, and the presence or absence of complexes composed of target proteins and small molecules affects the binding efficiency of crRNA during downstream signaling. The eventual reduction in fluorescent signal due to target protein binding can help quantify the detection.
Based on the selectivity of DNAzyme for metal ions, Wu’s team has developed a DNAzyme-based lead ion detection system for signal amplification via CRISPR (Chen et al., 2022). The method uses an enzyme consisting of two chains, the enzyme chain, and the substrate chain. When lead ions are present, the enzyme strand is energized to cut the substrate strand to release short ssDNA, activating Cas12a or Cas14a (Figure 4B). The authors verified the specificity of DNAzyme for the lead among various ions and purchased bottled water samples from the local area for testing, confirming its ability to detect actual water samples. The combination of CRISPR-Cas12a with DNAzyme achieved a detection limit as low as 0.48 nM for lead ions.
Although organophosphorus pesticides (OPs) have great potential to protect crops, their improper handling can lead to environmental pollution. A team of researchers has designed a dual enzyme assay combined with Cas12a to detect Ops (Fu et al., 2022). Briefly, the presence of OPs will affect the ability of acetylcholinesterase (AChE) to hydrolyze acetylthiocholine to thiocholine (TCh). In response, TCh can induce the degradation of MnO2 nanosheets and generate enough Mn2+ to stimulate the Mn2+-dependent DNAzyme activity. The method has been verified to possess strong resistance to interference in food matrices.
Enzyme-linked immunosorbent assay (ELISA) is a widely used assay technique in routine diagnostics. Zhou’s team developed a sensor called CLISA (the CRISPR/Cas13a signal amplification linked immunosorbent assay) to improve ELISA sensitivity using CRISPR/Cas13a for signal amplification (Chen et al., 2020). In this study, a primary antibody-target (antigen)-secondary antibody sandwich structure was established in which the T7 promoter, with the help of biotin and streptavidin, was recognized by T7 polymerase and initiated transcription, producing many copies of single-stranded RNA (Figure 4C). These RNAs were identified by Cas13a, which cleaved the reporter to produce fluorescence. The method has been validated for detecting an inflammatory factor, human interleukin-6 (human IL-6), and a tumor marker, human vascular endothelial growth factor (human VEGF), with LODs of 45.81 and 32.27 fg/ml, respectively, thereby achieving a hundredfold improvement in LOD. Subsequently, Song’s team designed Nano-CLISA by employing DNA-AuNPs as secondary antibodies to activate the trans cleavage ability of Cas12a (Figure 4C). The platform detects carcinoembryonic antigen (CEA) and prostate-specific antigen (PSA) biomarkers and achieves a 1000-fold higher sensitivity relative to ELISA, enabling quantitative analysis of proteins at the attomolar levels (Zhao et al., 2021). Similarly, an assay called CAFI (CRISPR/Cas12a assisted on-fiber immunosensor) was designed to examine small proteins in complex biological samples. The authors tested IFN-γ in human serum, sweat, saliva, and whole blood samples with detection limits as low as 58.8 aM (Deng et al., 2022).
In 2019, Zhang’s team used the recognition ability of allosteric transcription factors (aTF) to design a small molecule detection platform called CaT-SMelor (CRISPR-Cas12a- and aTF-mediated small molecule detector) (Liang et al., 2019). In this study, aTF has two structural domains, the DNA-binding structural domain, and the effector-binding structural domain. When the presence of small molecules initiates the metastable regulation of aTF, the binding ability of aTF to dsDNA is diminished, resulting in the release of the activation strand (Figure 4D). The aTF fused to the cellulose-binding domain (CBD-aTF) is immobilized on microcrystalline cellulose to analyze uric acid, a biomarker of chronic gout, in human blood samples. The fact that only 1 μl of blood is sufficient for analysis and that no sample preparation is required offer significant advantages for future detection of small molecules. A similar approach, SPRINT (SHERLOCK-based profiling of in vitro transcription), was devised to detect small molecules or enzymatic reaction products (Iwasaki and Batey, 2020). In these studies, aTF was used as a switch for transcription by RNA polymerase, just like a riboswitch, which activates the in vitro RNA polymerase transcription when a target molecule is present, and produces a large amount of RNA to provide an activator for Cas13a. SPRINT has been reported to have the ability to detect a wide range of small molecule compounds since aTF and riboswitch can have their specificities modified to target different target molecules. Chen et al. introduced Exo III, a nuclease that removes nucleotides, in their protocol to achieve the detection of TF (Li B. et al., 2021). When a TF called NF-κB p50 binds to dsDNA, its large spatial site block prevents the cleavage of Exo III from preserving dsDNA that can be used as an activator.
In addition to aTF, specific allosteric probes (APs) can also be used as a link in the signal transition. Usually, single-stranded DNA is used as the AP, containing a target recognition structural domain, i.e., a hairpin structure whose activity can be altered. In a detection system called APC-Cas (allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction), the AP contains the aptamer domain of the pathogen, the primer binding domain, and the T7 promoter domain. The existence of the pathogen causes the AP hairpin structure to unfold to facilitate amplification of ssDNA into dsDNA, which subsequently initiates transcription to produce large amounts of ssRNA (Shen et al., 2020). Similarly, in the study by Li D. et al. (2022), AP was used for the recognition of tobramycin and contained an aptamer domain, a DNA amplification template domain, and an enzymatic cleavage site domain. Binding of tobramycin results in a conformational change of the AP to facilitate polymerase amplification and subsequent cleavage to provide substrate for the SDA reaction, which can perform a considerable amount of amplification to enable recognition of Cas12a.
PNKP (polynucleotide kinase/phosphatase) catalyzes the dephosphorylation of DNA 3′- and phosphorylation of DNA 5′- and plays a vital role in DNA repair in cells (Wang D.-X. et al., 2020). To achieve the detection of PNKP, a dual amplification sensing strategy was designed. The substrate DNA also includes a hairpin structural domain, a DNA amplification structural domain, and a nicking endonuclease (Nb.BbvCI) site structural domain. PNKP hydrolyzes the 3′-phosphate group to a hydroxyl group, amplified by DNA polymerase to initiate the SDA reaction.
Circulating tumor cells (CTCs) are tumor markers of great prognostic significance. In a recent study to detect CTCs, Yang’s team designed a CRISPR/Cas12a-based sensor (Lv Z. et al., 2021). In this study, a duplexed aptamer (DA) binds to a protein receptor on the cell surface upon encounter with CCRF-CEM cells, causing conformational reorganization and the release of an aptamer-complementary DNA strand (ACD) to the aptamer. Long ssDNA molecules with hundreds of repetitive aptamer units were constructed on MB by RCA reaction to regulate MDANs (Multivalent Duplexed-aptamer Networks). The CTC assay workflow is simple (two-step reaction) and fast (less than 60 min).
Since its discovery, CRISPR has been widely used in gene editing and biosensing and has excellent potential for clinical applications, especially in POCT. Currently, among all Cas enzyme types, the three most applied are Cas9, Cas12, Cas13, and their respective subtypes. Both the cis-cleavage ability of Cas9 and the trans-cleavage ability of Cas12 and Cas13 serve as the key to reporter signal amplification, with the hope that future researchers can discover or modify more Cas proteins with advantages. For example, in a recent study, Cas3 proteins from type I were also used to develop detection systems. Cas3-Operated Nucleic Acid detection (CONAN) was designed for targeted detection of nucleic acid molecules, based on the authors’ finding that EcoCas3 also has non-specific single-stranded DNA cleavage activity (Yoshimi et al., 2022). In one study, Cas12a and Cas13a were used in combination to improve orthogonal cleavage activity and multiplex detection. Interestingly, dual-gene amplified products from the multiplex RPA were simultaneously detected in a single tube (Tian et al., 2022). Future investigations on different Cas proteins and their mechanisms of action will help select the most appropriate protocols for other specific target molecules. In conclusion, the CRISPR/Cas system is an excellent tool for signal amplification, and its unique specificity advantage can play a significant role in testing.
Epidemic infectious diseases have always been a significant problem plaguing human progress. In particular, the COVID-19 pandemic is a massive challenge for many poorer countries lacking specialized equipment and laboratories. To address this crisis, developing biosensors with rapid, easy, and non-device-dependent detection is paramount. The CRISPR/Cas system has been integrated with biosensors, and many CRISPR-based detection platforms have emerged in the medical field. For example, the Omicron variant of SARS-CoV-2 was typed by designing different crRNAs (Liang et al., 2022), or some methods could test for various pathogens (Li C. et al., 2022; Lu P. et al., 2022; Liu X. P. et al., 2022; Qiu X. T. et al., 2022; Li F. A. et al., 2022; Huang et al., 2022; Shi et al., 2022; Wei et al., 2022; Zhang et al., 2022). Whether colorimetric, fluorescent, or electrochemical and electrochemiluminescence methods, CRISPR-based sensors are moving towards short and efficient detection, and even companion applications or smartphones have emerged, allowing diagnosis to be made at the comfort of one’s home. In addition to some of the classical detection strategies, systems have also arisen by engineering crRNA and combining it with PCR. Recently, a method called LEOPARD (leveraging engineered tracrRNAs and on-target DNAs for parallel RNA detection) was reported (Jiao et al., 2021), which describes its principle of detecting different RNA sequences with single nucleotide specificity by reprogramming tracrRNAs to bind desired cellular transcripts and finally forming Cas-crRNA complexes. This complex with intracellular RNA can target its complementary DNA and undergo cleavage, followed by a reporter signal by analyzing the size of the DNA. Wang and coworkers established a method named CRISPR/Cas9-typing PCR version 4.0 (ctPCR4.0), which simplifies the cumbersome steps of the previous version with ingredients by simply including the sample, Cas9-sgRNA, insert oligos, universal primer, and other PCR constituents. It is a one-pot assay that can be performed on a PCR only once and can be used to detect HPV DNA in cervical cancer cells with reasonable specificity and operability (Gao et al., 2021).
Detection of non-nucleic acid targets is an upcoming area where CRISPR and biosensing are combined. CRISPR is known to be a nucleic acid-sensitive detection system with specificity. Researchers must explore how to interpret the signals of small molecules, proteins, and other substances into nucleic acid signals. We still expect to see a detection system as good as SHERLOCK for large-scale commercial use in the future. Nevertheless, current research on non-nucleic acid targets is also of considerable interest. Many detection systems have developed, especially in the food and environmental fields, to detect pathogenic small molecules or parasites (Lei et al., 2022; You et al., 2022), or pathogens in agriculture (Jiao et al., 2022; Zhu et al., 2022).
Since the emergence of CRISPR technology in the Nobel Prize arena, increasing research and discoveries have been generated in this field. However, many issues remain in the application of CRISPR-based assays. For example, amplification of nucleic acid-like target molecules is still required for initial signal enhancement before Cas proteins cleave FQ-reporters to release fluorescence for signal amplification. Nevertheless, for some usage scenarios where rapid detection is needed, there is more potential to develop amplification-free approaches, and these detection systems should focus on improving test sensitivity. Next, for the wide range of biomarkers, from nucleic acid molecules to non-nucleic acid analytes there is still a need for more sensitive and specific detection systems to fill some of the gaps in the detection of target substances. More advanced and reliable assay strategies are being explored, from nucleic acid analytes to non-nucleic acid detectors. Future research will continue to improve detection strategies for both the amplification (-free) method and the CRISPR system. Newer Cas enzymes, more powerful readout systems, and more flexible ways of signal conversion and amplification will represent the possible directions of improvement. The prospect is enormous for artificial intelligence, novel materials, and engineering developments to empower CRISPR research.
KC: Investigation, data analysis, and writing-original draft. ZS, GW, WG, SZ, and ZL: Resources and investigation. WL: Investigation. YC and JJ: Funding acquisition, resources, and supervision. GM: Writing-review and editing. CW and TY: Funding acquisition, conceptualization, supervision, resources, and writing-review and editing. All authors have read and agreed to the final manuscript draft.
This work was supported by grants from the National Natural Science Foundation of China (No. 31972899), by the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning, and Shanghai Overseas Talents Introduction Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer HL declared a shared parent affiliation with the author YC at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.986233/full#supplementary-material
Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., et al. (2017). RNA targeting with CRISPR–Cas13. Nature 550 (7675), 280–284. doi:10.1038/nature24049
Abudayyeh, O. O., Gootenberg, J. S., Konermann, S., Joung, J., Slaymaker, I. M., Cox, D. B. T., et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Sci. (New York, N.Y.) 353 (6299), aaf5573. doi:10.1126/science.aaf5573
Ackerman, C. M., Myhrvold, C., Thakku, S. G., Freije, C. A., Metsky, H. C., Yang, D. K., et al. (2020). Massively multiplexed nucleic acid detection with Cas13. Nature 582 (7811), 277–282. doi:10.1038/s41586-020-2279-8
Aldewachi, H., Chalati, T., Woodroofe, M. N., Bricklebank, N., Sharrack, B., and Gardiner, P. (2018). Gold nanoparticle-based colorimetric biosensors. Nanoscale 10 (1), 18–33. doi:10.1039/c7nr06367a
Aman, R., Mahas, A., and Mahfouz, M. (2020). Nucleic acid detection using CRISPR/Cas biosensing technologies. ACS Synth. Biol. 9 (6), 1226–1233. doi:10.1021/acssynbio.9b00507
Aman, R., Marsic, T., Sivakrishna Rao, G., Mahas, A., Ali, Z., Alsanea, M., et al. (2022). iSCAN-V2: A one-pot RT-RPA–CRISPR/Cas12b assay for point-of-care SARS-CoV-2 detection. Front. Bioeng. Biotechnol. 9, 800104. doi:10.3389/fbioe.2021.800104
Arizti-Sanz, J., Freije, C. A., Stanton, A. C., Petros, B. A., Boehm, C. K., Siddiqui, S., et al. (2020). Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 11 (1), 5921. doi:10.1038/s41467-020-19097-x
Balderston, S., Taulbee, J. J., Celaya, E., Fung, K., Jiao, A., Smith, K., et al. (2021). Discrimination of single-point mutations in unamplified genomic DNA via Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 5 (7), 713–725. doi:10.1038/s41551-021-00706-z
Baptista, P., Pereira, E., Eaton, P., Doria, G., Miranda, A., Gomes, I., et al. (2008). Gold nanoparticles for the development of clinical diagnosis methods. Anal. Bioanal. Chem. 391 (3), 943–950. doi:10.1007/s00216-007-1768-z
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 (5819), 1709–1712. doi:10.1126/science.1138140
Bhatt, A., Fatima, Z., Ruwali, M., Misra, C. S., Rangu, S. S., Rath, D., et al. (2022). CLEVER assay: A visual and rapid RNA extraction-free detection of SARS-CoV-2 based on CRISPR-Cas integrated RT-LAMP technology. J. Appl. Microbiol. 133, 410–421. doi:10.1111/jam.15571
Bonini, A., Poma, N., Vivaldi, F., Kirchhain, A., Salvo, P., Bottai, D., et al. (2021). Advances in biosensing: The CRISPR/Cas system as a new powerful tool for the detection of nucleic acids. J. Pharm. Biomed. Analysis 192, 113645. doi:10.1016/j.jpba.2020.113645
Brogan, D. J., Chaverra-Rodriguez, D., Lin, C. P., Smidler, A. L., Yang, T., Alcantara, L. M., et al. (2020). A sensitive, rapid, and portable CasRx-based diagnostic assay for SARS-CoV-2. medRxiv. doi:10.1101/2020.10.14.20212795
Broughton, J. P., Deng, X., Yu, G., Fasching, C. L., Servellita, V., Singh, J., et al. (2020). CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 38 (7), 870–874. doi:10.1038/s41587-020-0513-4
Brouns, S. J. J., Jore, M. M., Lundgren, M., Westra, E. R., Slijkhuis, R. J. H., Snijders, A. P. L., et al. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321 (5891), 960–964. doi:10.1126/science.1159689
Bruch, R., Baaske, J., Chatelle, C., Meirich, M., Madlener, S., Weber, W., et al. (2019). CRISPR/Cas13a-Powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv. Mater. 31 (51), 1905311. doi:10.1002/adma.201905311
Bruch, R., Johnston, M., Kling, A., Mattmüller, T., Baaske, J., Partel, S., et al. (2021). CRISPR-powered electrochemical microfluidic multiplexed biosensor for target amplification-free miRNA diagnostics. Biosens. Bioelectron. 177, 112887. doi:10.1016/j.bios.2020.112887
Chang, W., Liu, W., Liu, Y., Zhan, F., Chen, H., Lei, H., et al. (2019). Colorimetric detection of nucleic acid sequences in plant pathogens based on CRISPR/Cas9 triggered signal amplification. Microchim. Acta 186 (4), 243. doi:10.1007/s00604-019-3348-2
Chen, J. S., and Doudna, J. A. (2017). The chemistry of Cas9 and its CRISPR colleagues. Nat. Rev. Chem. 1 (10), 0078. doi:10.1038/s41570-017-0078
Chen, J. S., Ma, E., Harrington, L. B., Da Costa, M., Tian, X., Palefsky, J. M., et al. (2018). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Sci. (New York, N.Y.) 360 (6387), 436–439. doi:10.1126/science.aar6245
Chen, Q., Tian, T., Xiong, E., Wang, P., and Zhou, X. (2020). CRISPR/Cas13a signal amplification linked immunosorbent assay for femtomolar protein detection. Anal. Chem. 92 (1), 573–577. doi:10.1021/acs.analchem.9b04403
Chen, Y., Wu, H., Qian, S., Yu, X., Chen, H., and Wu, J. (2022). Applying CRISPR/Cas system as a signal enhancer for DNAzyme-based lead ion detection. Anal. Chim. Acta 1192, 339356. doi:10.1016/j.aca.2021.339356
Cong, L., Ran, F. A., Cox, D., Lin, S. L., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339 (6121), 819–823. doi:10.1126/science.1231143
Dai, Y. F., Somoza, R. A., Wang, L., Welter, J. F., Li, Y., Caplan, A. I., et al. (2019). Exploring the trans-cleavage activity of CRISPR-Cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew. Chem. Int. Ed. 58 (48), 17399–17405. doi:10.1002/anie.201910772
Dai, Y., Xu, W., Somoza, R. A., Welter, J. F., Caplan, A. I., and Liu, C. C. (2020). An integrated multi-function Heterogeneous biochemical circuit for high-Resolution Electrochemistry-based genetic analysis. Angew. Chem. Int. Ed. 59 (46), 20545–20551. doi:10.1002/anie.202010648
Deng, F., Li, Y., Qiao, L., and Goldys, E. (2022). A CRISPR/Cas12a-assisted on-fibre immunosensor for ultrasensitive small protein detection in complex biological samples. Anal. Chim. Acta 1192, 339351. doi:10.1016/j.aca.2021.339351
Dolino, D. M., Ramaswamy, S. S., and Jayaraman, V. (2014). Luminescence resonance energy transfer to study conformational changes in membrane proteins expressed in Mammalian cells. J. Vis. Exp. 2014(91), 51895. doi:10.3791/51895
Drain, P. K., Hyle, E. P., Noubary, F., Freedberg, K. A., Wilson, D., Bishai, W. R., et al. (2014). Diagnostic point-of-care tests in resource-limited settings. Lancet Infect. Dis. 14 (3), 239–249. doi:10.1016/s1473-3099(13)70250-0
Fonfara, I., Richter, H., Bratovič, M., Le Rhun, A., and Charpentier, E. (2016). The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532 (7600), 517–521. doi:10.1038/nature17945
Fozouni, P., Son, S., Díaz de León Derby, M., Knott, G. J., Gray, C. N., D’Ambrosio, M. V., et al. (2021). Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 184 (2), 323–333.e9. doi:10.1016/j.cell.2020.12.001
Fritea, L., Tertis, M., Sandulescu, R., and Cristea, C. (2018). Enzyme–graphene platforms for electrochemical biosensor design with Biomedical applications. Methods Enzymol. 609, 293–333. doi:10.1016/bs.mie.2018.05.010
Fu, R., Wang, Y., Liu, Y., Liu, H., Zhao, Q., Zhang, Y., et al. (2022). CRISPR-Cas12a based fluorescence assay for organophosphorus pesticides in agricultural products. Food Chem. 387, 132919. doi:10.1016/j.foodchem.2022.132919
Gao, J., Wu, L., Yang, D., Gong, W., and Wang, J. (2021). A one-pot CRISPR/Cas9-Typing PCR for DNA detection and genotyping. J. Mol. Diagnostics 23 (1), 46–60. doi:10.1016/j.jmoldx.2020.10.004
Gootenberg, J. S., Abudayyeh, O. O., Kellner, M. J., Joung, J., Collins, J. J., and Zhang, F. (2018). Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360 (6387), 439–444. doi:10.1126/science.aaq0179
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., et al. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356(6336), 438-442. doi:10.1126/science.aam9321
Hajian, R., Balderston, S., Tran, T., deBoer, T., Etienne, J., Sandhu, M., et al. (2019). Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 3 (6), 427–437. doi:10.1038/s41551-019-0371-x
Han, C., Li, W., Li, Q., Xing, W., Luo, H., Ji, H., et al. (2022). CRISPR/Cas12a-Derived electrochemical aptasensor for ultrasensitive detection of COVID-19 nucleocapsid protein. Biosens. Bioelectron. 200, 113922. doi:10.1016/j.bios.2021.113922
Harrington, L. B., Burstein, D., Chen, J. S., Paez-Espino, D., Ma, E., Witte, I. P., et al. (2018). Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362 (6416), 839–842. doi:10.1126/science.aav4294
He, Y., Hu, X., Gong, Z., Chen, S., and Yuan, R. (2020). A novel electrochemiluminescence biosensor based on the self-ECL emission of conjugated polymer dots for lead ion detection. Microchim. Acta 187 (4), 237. doi:10.1007/s00604-020-4212-0
Heo, W., Lee, K., Park, S., Hyun, K.-A., and Jung, H.-I. (2022). Electrochemical biosensor for nucleic acid amplification-free and sensitive detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA via CRISPR/Cas13a trans-cleavage reaction. Biosens. Bioelectron. 201, 113960. doi:10.1016/j.bios.2021.113960
Hu, F., Liu, Y., Zhao, S., Zhang, Z., Li, X., Peng, N., et al. (2022a). A one-pot CRISPR/Cas13a-based contamination-free biosensor for low-cost and rapid nucleic acid diagnostics. Biosens. Bioelectron. 202, 113994. doi:10.1016/j.bios.2022.113994
Hu, J., Song, H., Zhou, J., Liu, R., and Lv, Y. (2021). Metal-tagged CRISPR/Cas12a bioassay enables ultrasensitive and highly selective evaluation of kanamycin Bioaccumulation in fish samples. Anal. Chem. 93 (42), 14214–14222. doi:10.1021/acs.analchem.1c03094
Hu, M., Qiu, Z., Bi, Z., Tian, T., Jiang, Y., and Zhou, X. (2022b). Photocontrolled crRNA activation enables robust CRISPR-Cas12a diagnostics. Proc. Natl. Acad. Sci. U. S. A. 119 (26), e2202034119. doi:10.1073/pnas.2202034119
Huang, M. Q., Liu, S. H., Xu, Y. A., Li, A. Q., Wu, W., Liang, M. F., et al. (2022). CRISPR/Cas12a technology combined with RPA for rapid and portable SFTSV detection. Front. Microbiol. 13, 754995. doi:10.3389/fmicb.2022.754995
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169 (12), 5429–5433. doi:10.1128/jb.169.12.5429-5433.1987
Iwasaki, R. S., and Batey, R. T. (2020). Sprint: A Cas13a-based platform for detection of small molecules. Nucleic Acids Res. 48 (17), e101. doi:10.1093/nar/gkaa673
Jiao, C. L., Sharma, S., Dugar, G., Peeck, N. L., Bischler, T., Wimmer, F., et al. (2021). Noncanonical crRNAs derived from host transcripts enable multiplexable RNA detection by Cas9. Science 372 (6545), 941–948. doi:10.1126/science.abe7106
Jiao, J., Yang, M. J., Zhang, T. F., Zhang, Y. L., Yang, M. L., Li, M., et al. (2022). A sensitive visual method for onsite detection of quarantine pathogenic bacteria from horticultural crops using an LbCas12a variant system. J. Hazard. Mater. 426, 128038. doi:10.1016/j.jhazmat.2021.128038
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 (6096), 816–821. doi:10.1126/science.1225829
Joung, J., Ladha, A., Saito, M., Kim, N.-G., Woolley, A. E., Segel, M., et al. (2020). Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. Overseas. Ed. 383 (15), 1492–1494. doi:10.1056/NEJMc2026172
Kanafi, M. M., and Tavallaei, M. (2022). Overview of advances in CRISPR/deadCas9 technology and its applications in human diseases. Gene 830, 146518. doi:10.1016/j.gene.2022.146518
Karvelis, T., Bigelyte, G., Young, J. K., Hou, Z. L., Zedaveinyte, R., Budre, K., et al. (2020). PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 48 (9), 5016–5023. doi:10.1093/nar/gkaa208
Kim, H., Lee, S., Yoon, J., Song, J., and Park, H. G. (2021). CRISPR/Cas12a collateral cleavage activity for simple and rapid detection of protein/small molecule interaction. Biosens. Bioelectron. 194, 113587. doi:10.1016/j.bios.2021.113587
Lei, R., Li, L., Wu, P., Fei, X., Zhang, Y., Wang, J., et al. (2022). RPA/CRISPR/Cas12a-Based on-site and rapid nucleic acid detection of Toxoplasma gondii in the environment. ACS Synth. Biol. 11, 1772–1781. doi:10.1021/acssynbio.1c00620
Li, B., Shao, Z., and Chen, Y. (2021a). An exonuclease protection and CRISPR/Cas12a integrated biosensor for the turn-on detection of transcription factors in cancer cells. Anal. Chim. Acta 1165, 338478. doi:10.1016/j.aca.2021.338478
Li, C.-Y., Zheng, B., Li, J.-T., Gao, J.-l., Liu, Y.-H., Pang, D.-W., et al. (2021b). Holographic optical tweezers and boosting upconversion luminescent resonance energy transfer combined Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas12a biosensors. ACS Nano 15 (5), 8142–8154. doi:10.1021/acsnano.0c09986
Li, C.-Y., Zheng, B., Liu, Y.-H., Gao, J.-L., Zheng, M.-Q., Pang, D.-W., et al. (2020). A boosting upconversion luminescent resonance energy transfer and biomimetic periodic chip integrated CRISPR/Cas12a biosensor for functional DNA regulated transduction of non-nucleic acid targets. Biosens. Bioelectron. 169, 112650. doi:10.1016/j.bios.2020.112650
Li, C., Chen, X., Wen, R. Q., Ma, P., Gu, K., Li, C., et al. (2022a). Immunocapture magnetic beads enhanced the LAMP-CRISPR/Cas12a method for the sensitive, specific, and visual detection of Campylobacter jejuni. Biosens. (Basel). 12 (3), 154. doi:10.3390/bios12030154
Li, D., Ling, S., Wu, H., Yang, Z., and Lv, B. (2022b). CRISPR/Cas12a-based biosensors for ultrasensitive tobramycin detection with single- and double-stranded DNA activators. Sensors Actuators B Chem. 355, 131329. doi:10.1016/j.snb.2021.131329
Li, F. A., Xiao, J., Yang, H. M., Yao, Y., Li, J. Q., Zheng, H. W., et al. (2022c). Development of a rapid and efficient RPA-CRISPR/Cas12a assay for Mycoplasma pneumoniae detection. Front. Microbiol. 13, 858806. doi:10.3389/fmicb.2022.858806
Li, H. J., Yang, J., Wu, G. F., Weng, Z. Y., Song, Y., Zhang, Y. X., et al. (2022d). Amplification-free detection of SARS-CoV-2 and respiratory Syncytial virus using CRISPR Cas13a and graphene field-effect transistors. Angew. Chem. Int. Ed. Engl. 61, e202203826. doi:10.1002/anie.202203826
Li, L., Li, S., Wu, N., Wu, J., Wang, G., Zhao, G., et al. (2019). HOLMESv2: A CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 8 (10), 2228–2237. doi:10.1021/acssynbio.9b00209
Li, S.-Y., Cheng, Q.-X., Liu, J.-K., Nie, X.-Q., Zhao, G.-P., and Wang, J. (2018a). CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 28 (4), 491–493. doi:10.1038/s41422-018-0022-x
Li, S.-Y., Cheng, Q.-X., Wang, J.-M., Li, X.-Y., Zhang, Z.-L., Gao, S., et al. (2018b). CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 4 (1), 20. doi:10.1038/s41421-018-0028-z
Li, Z., Zhao, W., Ma, S., Li, Z., Yao, Y., and Fei, T. (2021c). A chemical-enhanced system for CRISPR-Based nucleic acid detection. Biosens. Bioelectron. X. 192, 113493. doi:10.1016/j.bios.2021.113493
Liang, M., Li, Z., Wang, W., Liu, J., Liu, L., Zhu, G., et al. (2019). A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules. Nat. Commun. 10 (1), 3672. doi:10.1038/s41467-019-11648-1
Liang, Y., Lin, H., Zou, L., Deng, X., and Tang, S. (2022). Rapid detection and tracking of Omicron variant of SARS-CoV-2 using CRISPR-Cas12a-based assay. Biosens. Bioelectron. X. 205, 114098. doi:10.1016/j.bios.2022.114098
Liu, S., Wang, C., Wang, Z., Xiang, K., Zhang, Y., Fan, G.-C., et al. (2022a). Binding induced isothermal amplification reaction to activate CRISPR/Cas12a for amplified electrochemiluminescence detection of rabies viral RNA via DNA nanotweezer structure switching. Biosens. Bioelectron. 204, 114078. doi:10.1016/j.bios.2022.114078
Liu, X. P., Qiu, X. T., Xu, S., Che, Y. L., Han, L. C., Kang, Y. T., et al. (2022b). A CRISPR-Cas12a-assisted fluorescence platform for rapid and accurate detection of Nocardia cyriacigeorgica. Front. Cell. Infect. Microbiol. 12, 835213. doi:10.3389/fcimb.2022.835213
López-Valls, M., Escalona-Noguero, C., Rodríguez-Díaz, C., Pardo, D., Castellanos, M., Milán-Rois, P., et al. (2022). Cascade: Naked eye-detection of SARS-CoV-2 using Cas13a and gold nanoparticles. Anal. Chim. Acta 1205, 339749. doi:10.1016/j.aca.2022.339749
Lu, P., Chen, J. L., Li, Z. P., Li, Z., Zhang, J. Y., Kan, B., et al. (2022a). Visual identification and Serotyping of Toxigenic Vibrio cholerae Serogroups O1 and O139 with CARID. Front. Cell. Infect. Microbiol. 12, 863435. doi:10.3389/fcimb.2022.863435
Lu, S., Tong, X., Han, Y., Zhang, K., Zhang, Y., Chen, Q., et al. (2022b). Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a. Nat. Biomed. Eng. 6, 286–297. doi:10.1038/s41551-022-00861-x
Lu, Y., Xu, J., Jia, Z., Kong, S., Qiao, Y., Li, L., et al. (2022c). A near-infrared multifunctional fluorescent probe for hypoxia monitoring and tumor-targeted therapy. Chin. Chem. Lett. 33 (3), 1589–1594. doi:10.1016/j.cclet.2021.09.013
Lv, H., Wang, J., Zhang, J., Chen, Y., Yin, L., Jin, D., et al. (2021a). Definition of CRISPR Cas12a T rans-cleavage Units to facilitate CRISPR diagnostics. Front. Microbiol. 12, 766464. doi:10.3389/fmicb.2021.766464
Lv, Z., Wang, Q., and Yang, M. (2021b). Multivalent duplexed-aptamer networks regulated a CRISPR-Cas12a system for circulating tumor cell detection. Anal. Chem. 93 (38), 12921–12929. doi:10.1021/acs.analchem.1c02228
Ma, P., Meng, Q., Sun, B., Zhao, B., Dang, L., Zhong, M., et al. (2020). MeCas12a, a highly sensitive and specific system for COVID-19 detection. Adv. Sci. (Weinh). 7 (20), 2001300. doi:10.1002/advs.202001300
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J. J., Charpentier, E., Horvath, P., et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9 (6), 467–477. doi:10.1038/nrmicro2577
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 13 (11), 722–736. doi:10.1038/nrmicro3569
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2020). Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 18 (2), 67–83. doi:10.1038/s41579-019-0299-x
Mao, Z., Chen, R., Wang, X., Zhou, Z., Peng, Y., Li, S., et al. (2022a). CRISPR/Cas12a-based technology: A powerful tool for biosensing in food safety. Trends food Sci. Technol. 122, 211–222. doi:10.1016/j.tifs.2022.02.030
Mao, Z., Wang, X., Chen, R., Zhou, Z., Ren, S., Liang, J., et al. (2022b). Upconversion-mediated CRISPR-Cas12a biosensing for sensitive detection of ochratoxin A. Talanta 242, 123232. doi:10.1016/j.talanta.2022.123232
Mojica, F. J. M., Diez-Villasenor, C., Soria, E., and Juez, G. (2000). Biological significance of a family of regularly spaced repeats in the genomes of Archaea, bacteria and mitochondria. Mol. Microbiol. 36 (1), 244–246. doi:10.1046/j.1365-2958.2000.01838.x
Moon, J., Kwon, H.-J., Yong, D., Lee, I.-C., Kim, H., Kang, H., et al. (2020). Colorimetric detection of SARS-CoV-2 and drug-resistant pH1N1 using CRISPR/dCas9. ACS Sens. 5 (12), 4017–4026. doi:10.1021/acssensors.0c01929
Myhrvold, C., Freije, C. A., Gootenberg, J. S., Abudayyeh, O. O., Metsky, H. C., Durbin, A. F., et al. (2018). Field-deployable viral diagnostics using CRISPR-Cas13. Science 360 (6387), 444–448. doi:10.1126/science.aas8836
Nguyen, L. T., Smith, B. M., and Jain, P. K. (2020). Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 11 (1), 4906. doi:10.1038/s41467-020-18615-1
Nguyen, P. Q., Soenksen, L. R., Donghia, N. M., Angenent-Mari, N. M., de Puig, H., Huang, A., et al. (2021). Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nat. Biotechnol. 39 (11), 1366–1374. doi:10.1038/s41587-021-00950-3
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28 (12), 63e. doi:10.1093/nar/28.12.e63
O’Connell, M. R., Oakes, B. L., Sternberg, S. H., East-Seletsky, A., Kaplan, M., and Doudna, J. A. (2014). Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516 (7530), 263–266. doi:10.1038/nature13769
Ooi, K. H., Liu, M. M., Tay, J. W. D., Teo, S. Y., Kaewsapsak, P., Jin, S., et al. (2021). An engineered CRISPR-Cas12a variant and DNA-RNA hybrid guides enable robust and rapid COVID-19 testing. Nat. Commun. 12 (1), 1739. doi:10.1038/s41467-021-21996-6
Pardee, K., Green, A. A., Takahashi, M. K., Braff, D., Lambert, G., Lee, J. W., et al. (2016). Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165 (5), 1255–1266. doi:10.1016/j.cell.2016.04.059
Park, H. M., Liu, H., Wu, J., Chong, A., Mackley, V., Fellmann, C., et al. (2018). Extension of the crRNA enhances Cpf1 gene editing in vitro and in vivo. Nat. Commun. 9 (1), 3313. doi:10.1038/s41467-018-05641-3
Parolo, C., and Merkoci, A. (2013). Paper-based nanobiosensors for diagnostics. Chem. Soc. Rev. 42 (2), 450–457. doi:10.1039/c2cs35255a
Peng, L., Zhou, J., Liu, G. Z., Yin, L. J., Ren, S. Y., Man, S. L., et al. (2020). CRISPR-Cas12a based aptasensor for sensitive and selective ATP detection. Sensors Actuators B Chem. 320, 128164. doi:10.1016/j.snb.2020.128164
Phan, Q. A., Truong, L. B., Medina-Cruz, D., Dincer, C., and Mostafavi, E. (2022). CRISPR/Cas-powered nanobiosensors for diagnostics. Biosens. Bioelectron. 197, 113732. doi:10.1016/j.bios.2021.113732
Ponce-Rojas, J. C., Costello, M. S., Proctor, D. A., Kosik, K. S., Wilson, M. Z., Arias, C., et al. (2021). A fast and Accessible method for the isolation of RNA, DNA, and protein to facilitate the detection of SARS-CoV-2. J. Clin. Microbiol. 59 (4), e02403-20. doi:10.1128/jcm.02403-20
Qin, N., Deng, L., Wang, M., and Hun, X. (2021). Gold nanoparticles/Mo2C/MoO2-modified electrodes for nucleic acid detection through CRISPR/Cas12a Photoelectrochemical assay. ACS Appl. Nano Mater. 4 (10), 10701–10707. doi:10.1021/acsanm.1c02164
Qiu, M., Zhou, X.-M., and Liu, L. (2022a). Improved strategies for CRISPR-Cas12-based nucleic acids detection. J. Anal. Test. 6 (1), 44–52. doi:10.1007/s41664-022-00212-4
Qiu, X. T., Xu, S., Liu, X. P., Han, L., Zhao, B., Che, Y. L., et al. (2022b). A CRISPR-based nucleic acid detection platform (CRISPR-CPA): Application for detection of Nocardia farcinica. J. Appl. Microbiol. 132 (5), 3685–3693. doi:10.1111/jam.15424
Rahimi, H., Salehiabar, M., Barsbay, M., Ghaffarlou, M., Kavetskyy, T., Sharafi, A., et al. (2021). CRISPR systems for COVID-19 diagnosis. ACS Sens. 6 (4), 1430–1445. doi:10.1021/acssensors.0c02312
Rauch, J. N., Valois, E., Solley, S. C., Braig, F., Lach, R. S., Audouard, M., et al. (2021). A scalable, easy-to-Deploy protocol for Cas13-based detection of SARS-CoV-2 genetic material. J. Clin. Microbiol. 59 (4), e02402-20. doi:10.1128/jcm.02402-20
Richter, M. M. (2004). Electrochemiluminescence (ECL). Chem. Rev. 104 (6), 3003–3036. doi:10.1021/cr020373d
Rossetti, M., Merlo, R., Bagheri, N., Moscone, D., Valenti, A., Saha, A., et al. (2022). Enhancement of CRISPR/Cas12a trans-cleavage activity using hairpin DNA reporters. Nucleic Acids Res. 2022, gkac578. doi:10.1093/nar/gkac578
Safdar, S., Driesen, S., Leirs, K., De Sutter, D., Eyckerman, S., Lammertyn, J., et al. (2022). Engineered tracrRNA for enabling versatile CRISPR-dCas9-based biosensing concepts. Biosens. Bioelectron. 206, 114140. doi:10.1016/j.bios.2022.114140
Sander, J. D., and Joung, J. K. (2014). CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32 (4), 347–355. doi:10.1038/nbt.2842
Shen, J., Zhou, X., Shan, Y., Yue, H., Huang, R., Hu, J., et al. (2020). Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat. Commun. 11 (1), 267. doi:10.1038/s41467-019-14135-9
Shi, K., Xie, S. Y., Tian, R. Y., Wang, S., Lu, Q., Gao, D. H., et al. (2021). A CRISPR-Cas autocatalysis-driven feedback amplification network for supersensitive DNA diagnostics. Sci. Adv. 7 (5), eabc7802. doi:10.1126/sciadv.abc7802
Shi, Y. Q., Kang, L., Mu, R. R., Xu, M., Duan, X. Q., Li, Y. J., et al. (2022). CRISPR/Cas12a-Enhanced loop-mediated isothermal amplification for the visual detection of Shigella flexneri. Front. Bioeng. Biotechnol. 10, 845688. doi:10.3389/fbioe.2022.845688
Sohail, M., Xie, S., Zhang, X., and Li, B. (2022). Methodologies in visualizing the activation of CRISPR/Cas: The last mile in developing CRISPR-Based diagnostics and biosensing – a review. Anal. Chim. Acta 1205, 339541. doi:10.1016/j.aca.2022.339541
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C., and Doudna, J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507 (7490), 62–67. doi:10.1038/nature13011
Suea-Ngam, A., Howes, P. D., and DeMello, A. J. (2021). An amplification-free ultra-sensitive electrochemical CRISPR/Cas biosensor for drug-resistant bacteria detection. Chem. Sci. 12 (38), 12733–12743. doi:10.1039/d1sc02197d
Tian, T., Qiu, Z. Q., Jiang, Y. Z., Zhu, D. B., and Zhou, X. M. (2022). Exploiting the orthogonal CRISPR-Cas12a/Cas13a trans-cleavage for dual-gene virus detection using a handheld device. Biosens. Bioelectron. X. 196, 113701. doi:10.1016/j.bios.2021.113701
Tian, T., Shu, B., Jiang, Y., Ye, M., Liu, L., Guo, Z., et al. (2021). An Ultralocalized Cas13a assay enables universal and nucleic acid amplification-free single-molecule RNA diagnostics. ACS Nano 15 (1), 1167–1178. doi:10.1021/acsnano.0c08165
Wang, B., Wang, R., Wang, D., Wu, J., Li, J., Wang, J., et al. (2019). Cas12aVDet: A CRISPR/Cas12a-based platform for rapid and visual nucleic acid detection. Anal. Chem. 91 (19), 12156–12161. doi:10.1021/acs.analchem.9b01526
Wang, D.-X., Wang, J., Du, Y.-C., Ma, J.-Y., Wang, S.-Y., Tang, A.-N., et al. (2020a). CRISPR/Cas12a-based dual amplified biosensing system for sensitive and rapid detection of polynucleotide kinase/phosphatase. Biosens. Bioelectron. 168, 112556. doi:10.1016/j.bios.2020.112556
Wang, M., Han, D., Zhang, J., Zhang, R., and Li, J. (2021). High-fidelity detection of DNA combining the CRISPR/Cas9 system and hairpin probe. Biosens. Bioelectron. 184, 113212. doi:10.1016/j.bios.2021.113212
Wang, X., Xiong, E., Tian, T., Cheng, M., Lin, W., Wang, H., et al. (2020b). Clustered regularly interspaced short palindromic repeats/Cas9-mediated lateral flow nucleic acid assay. ACS Nano 14 (2), 2497–2508. doi:10.1021/acsnano.0c00022
Wei, N., Zheng, B. H., Niu, J. J., Chen, T., Ye, J., Si, Y. H., et al. (2022). Rapid detection of genotype II African swine Fever virus using CRISPR Cas13a-based lateral flow Strip. Viruses 14 (2), 179. doi:10.3390/v14020179
Wu, L., Zhou, T., and Huang, R. (2022a). A universal CRISPR/Cas9-based electrochemiluminescence probe for sensitive and single-base-specific DNA detection. Sensors Actuators B Chem. 357, 131411. doi:10.1016/j.snb.2022.131411
Wu, Z., Tang, N., and Ji, Q. (2022b). Detection system of CRISPR/Cas12f, comprises Cas12f nuclease, sgRNA, fluorescent probe and the sequence to be tested, and the described sequence to be tested includes a target sequence that can base complementary pairing.
Wu, Z., Zhang, Y., Yu, H., Pan, D., Wang, Y., Wang, Y., et al. (2021). Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat. Chem. Biol. 17 (11), 1132–1138. doi:10.1038/s41589-021-00868-6
Xie, S., Ji, Z., Suo, T., Li, B., and Zhang, X. (2021). Advancing sensing technology with CRISPR: From the detection of nucleic acids to a broad range of analytes – a review. Anal. Chim. Acta 1185, 338848. doi:10.1016/j.aca.2021.338848
Xie, S., Xu, B., Tang, R., Chen, S., Lei, C., and Nie, Z. (2022). Kinetics accelerated CRISPR-Cas12a enabling Live-cell monitoring of Mn2+ Homeostasis. Anal. Chem. 94, 10159–10167. doi:10.1021/acs.analchem.2c01461
Xiong, E., Jiang, L., Tian, T. A., Hu, M. L., Yue, H. H., Huang, M. Q., et al. (2021). Simultaneous dual-gene diagnosis of SARS-CoV-2 based on CRISPR/Cas9-Mediated lateral flow assay. Angew. Chem. Int. Ed. 60 (10), 5307–5315. doi:10.1002/anie.202014506
Xiong, Y., Zhang, J., Yang, Z., Mou, Q., Ma, Y., Xiong, Y., et al. (2020). Functional DNA regulated CRISPR-Cas12a sensors for point-of-care diagnostics of non-nucleic-acid targets. J. Am. Chem. Soc. 142 (1), 207–213. doi:10.1021/jacs.9b09211
Xu, N., Jin, S., and Wang, L. (2021). Metal nanoparticles-based nanoplatforms for colorimetric sensing: A review. Rev. Anal. Chem. 40 (1), 1–11. doi:10.1515/revac-2021-0122
Xu, W., Jin, T., Dai, Y., and Liu, C. C. (2020). Surpassing the detection limit and accuracy of the electrochemical DNA sensor through the application of CRISPR Cas systems. Biosens. Bioelectron. 155, 112100. doi:10.1016/j.bios.2020.112100
Yoshimi, K., Takeshita, K., Yamayoshi, S., Shibumura, S., Yamauchi, Y., Yamamoto, M., et al. (2022). CRISPR-Cas3-based diagnostics for SARS-CoV-2 and influenza virus. iScience 25 (2), 103830. doi:10.1016/j.isci.2022.103830
You, H., Gordon, C. A., MacGregor, S. R., Cai, P. F., and McManus, D. P. (2022). Potential of the CRISPR‐Cas system for improved parasite diagnosis. Bioessays 44 (4), 2100286. doi:10.1002/bies.202100286
Yue, D., Wang, M., Deng, F., Yin, W., Zhao, H., Zhao, X., et al. (2018). Biomarker-targeted fluorescent probes for breast cancer imaging. Chin. Chem. Lett. 29 (5), 648–656. doi:10.1016/j.cclet.2018.01.046
Yue, H., Shu, B., Tian, T., Xiong, E., Huang, M., Zhu, D., et al. (2021). Droplet Cas12a assay enables DNA quantification from unamplified samples at the single-molecule level. Nano Lett. 21 (11), 4643–4653. doi:10.1021/acs.nanolett.1c00715
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163 (3), 759–771. doi:10.1016/j.cell.2015.09.038
Zhang, K., Fan, Z., Yao, B., Ding, Y., Zhao, J., Xie, M., et al. (2021). Exploring the trans-cleavage activity of CRISPR-Cas12a for the development of a Mxene based electrochemiluminescence biosensor for the detection of Siglec-5. Biosens. Bioelectron. 178, 113019. doi:10.1016/j.bios.2021.113019
Zhang, X., Hu, Y., Yang, X., Tang, Y., Han, S., Kang, A., et al. (2019). FÖrster resonance energy transfer (FRET)-based biosensors for biological applications. Biosens. Bioelectron. 138, 111314. doi:10.1016/j.bios.2019.05.019
Zhang, X. Y., Tian, Y., Xu, L., Fan, Z. H., Cao, Y. L., Ma, Y. M., et al. (2022). CRISPR/Cas13-assisted hepatitis B virus covalently closed circular DNA detection. Hepatol. Int. 16 (2), 306–315. doi:10.1007/s12072-022-10311-0
Zhang, Y., Qian, L., Wei, W., Wang, Y., Wang, B., Lin, P., et al. (2017). Paired design of dCas9 as a Systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synth. Biol. 6 (2), 211–216. doi:10.1021/acssynbio.6b00215
Zhao, Q., Pan, Y., Luan, X., Gao, Y., Zhao, X., Liu, Y., et al. (2021). Nano-immunosorbent assay based on Cas12a/crRNA for ultra-sensitive protein detection. Biosens. Bioelectron. 190, 113450. doi:10.1016/j.bios.2021.113450
Zhou, W., Hu, L., Ying, L., Zhao, Z., Chu, P. K., and Yu, X.-F. (2018). A CRISPR–Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 9 (1), 5012. doi:10.1038/s41467-018-07324-5
Keywords: molecular diagnostics, CRISPR/Cas, biosensor, non-nucleic-acid analytes, nucleic acid detection
Citation: Chen K, Shen Z, Wang G, Gu W, Zhao S, Lin Z, Liu W, Cai Y, Mushtaq G, Jia J, Wan C and Yan T (2022) Research progress of CRISPR-based biosensors and bioassays for molecular diagnosis. Front. Bioeng. Biotechnol. 10:986233. doi: 10.3389/fbioe.2022.986233
Received: 04 July 2022; Accepted: 10 August 2022;
Published: 16 September 2022.
Edited by:
Xiaoming Zhou, South China Normal University, ChinaReviewed by:
Chunyang Lei, Hunan University, ChinaCopyright © 2022 Chen, Shen, Wang, Gu, Zhao, Lin, Liu, Cai, Mushtaq, Jia, Wan and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia Jia, amlhamlhMTI2MTI2QHNodS5lZHUuY24=; Chunpeng (Craig) Wan, Y2h1bnBlbmd3YW5AanhhdS5lZHUuY24=; Tingdong Yan, eWFudGRudHUyMDE4QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.