- 1Cancer Molecular Diagnostics Core, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Tianjin’s Clinical Research Center for Cancer, Tianjin, China
- 2Key Laboratory of Breast Cancer Prevention and Therapy, Tianjin Medical University, Ministry of Education, Tianjin, China
- 3Key Laboratory of Cancer Immunology and Biotherapy, Tianjin, China
- 4School of Computer Science and Technology, Harbin Institute of Technology, Harbin, China
Early myeloid-derived suppressor cells (eMDSCs) are a newly characterized subclass of MDSCs, which exhibit more potent immunosuppressive capacity than classical MDSCs. Previously, we found high eMDSCs infiltration was correlated with poor prognosis of breast cancer, though the regulatory mechanisms have not been fully understood. Here, we constructed a 21-gene signature to evaluate the status of eMDSCs infiltration within breast cancer tissues and found that highly infiltrated eMDSCs affected the prognosis of breast cancer patients, especially in luminal A subtype. We also found that eMDSCs promoted epithelial-mesenchymal transition (EMT) and accelerated cell migration and invasion in vitro. Meanwhile, eMDSCs significantly downregulated ARID1A expression in luminal A breast cancer, which was closely associated with EMT and was an important prognostic factor in breast cancer patients. Moreover, significant changes of EMT-related genes were detected in luminal A breast cancer cells after co-cultured with eMDSCs or ARID1A knock-down and overexpression of ARID1A significantly reversed this procedure. These results implied that eMDSCs might suppress the ARID1A expression to promote EMT in luminal A breast cancer cells, which might provide a new light on developing novel treatment regimens for relapsed luminal A breast cancer after conventional therapies.
Introduction
Breast cancer (BC) is one of the most common malignant tumors in women and can be divided into four subtypes: luminal A, luminal B, HER2+, and basal-like (Onitilo et al., 2009). Approximately 80% of breast cancer patients present as ER positive (luminal A and luminal B), and estrogen stimulates the development of ER+ breast cancers by increasing breast cancer cell survival and proliferation (Thomas and Gustafsson, 2011; Clarke et al., 2015; Thomas and Gustafsson, 2015). However, clinical investigations have shown that although surgery, chemotherapy, radiotherapy and anti-estrogen therapy are effective in reducing mortality in breast cancer patients, tumor recurrence and metastasis are the leading cause of mortality in these individuals (Carroll, 2016; Razavi et al., 2018). Accumulating evidence indicates that epithelial-mesenchymal transition (EMT), a process of initial step for tumor metastasis, gives cells the extra plasticity needed for invasion and metastasis to distant organs (Mittal, 2018; Dongre and Weinberg, 2019; Pastushenko and Blanpain, 2019). Furthermore, EMT endows tumor cells with cancer stem cell-like characteristics, making them resistant to therapies and prone to recurrence following therapy (Shibue and Weinberg, 2017; Erin et al., 2020). Therefore, a deeper knowledge of tumor growth and metastasis molecular pathways is essential to assist the creation of more accurate prognostic indicators as well as efficient treatment options.
Crosstalk between cancer cells and the surrounding microenvironment appears to play an essential role in the recurrence and metastasis of many malignancies, according to growing research (Wang et al., 2018; Hinshaw and Shevde, 2019; Lan et al., 2019; Zavros and Merchant, 2022). Myeloid-derived suppressor cells (MDSCs) are a prominent component of the tumor microenvironment, and their defining characteristic is their powerful immune suppressive action (Ma et al., 2011; Hegde et al., 2021). As a diverse group of immature myeloid cells, MDSCs have strong protumoral capacity by suppressing innate and adaptive immunity, promoting angiogenesis and stimulating tumor cell invasion and are further divided into three subsets: M-MDSCs (monocytic myeloid-derived suppressor cells), PMN-MDSCs (polymorphonuclear MDSCs) and eMDSCs (early-stage MDSCs) (Bronte et al., 2016). eMDSCs are a newly defined subset of MDSCs, whose phenotype in breast cancer were defined as Lin−HLA-DR-CD45+CD33+CD13+CD14−CD15− in humans and CD11b+Gr-1-F4/80-MHC-II- in mice (Zhou et al., 2010; Yu et al., 2013; Yu et al., 2014; Jiang et al., 2017; Zhang et al., 2018). Furthermore, eMDSCs are the most predominant subpopulation of MDSCs in breast cancer and exhibit enhanced immunosuppressive capacity in the tumor microenvironment compared to classical MDSCs (Yu et al., 2013; Yu et al., 2014; Jiang et al., 2017).
Although the concept of immunosuppressive MDSCs is well established, it could also contribute to tumor development through a variety of non-immunological pathways, such as promoting angiogenesis, enhancing tumor cell stemness, and maintaining pre-metastatic niche (Safarzadeh et al., 2018; Wang et al., 2019; Tang et al., 2021). Previously, we found high eMDSCs infiltration was correlated with poor prognosis of breast cancer (Jiang et al., 2017). However, whether eMDSCs promote tumor invasion and metastasis via EMT-related non-immunological pathways and then lead to tumor recurrence and poor prognosis of breast cancer is not fully understood.
Therefore, in this study we established a 21-gene signature to predict the infiltration of eMDSCs within breast cancer tissues which is a valuable predictive biomarker for luminal A breast cancer patients. Moreover, we found that eMDSCs suppress the ARID1A expression to promote EMT in luminal A breast cancer cells, which might provide a new light on developing novel treatment regimens for relapsed luminal A breast cancer after conventional therapies.
Materials and methods
Clinical samples and healthy donors
We obtained 280 primary breast cancer tissue samples from two cohorts (Shanghai Outdo Biotech Co., Ltd.), whose tumor tissues were constructed into tissue arrays following surgical resection between January 2001 and December 2008. All patients were females, with a median age of 51 years (range: 29–83 years) in cohort 1 and 57 years (range: 37–88 years) in cohort 2. Infiltrating mammary-ductal cancer accounted for 93.6% (131/140) in cohort 1 and 97.8% (137/140) in cohort 2. The cohort 1 included 11 patients with clinical stage I, 80 patients with clinical stage II, and 47 patients with clinical stage III cancer, respectively. The cohort 2 included 3 patients with clinical stage I, 62 patients with clinical stage II, and 65 patients with clinical stage III cancer, respectively. The Cohort 1 included 79, 17, and 26 patients with luminal, HER2 overexpression, and TNBC cancer subtypes, respectively (Table 1). The Cohort 2 included 62, 43, and 35 patients with luminal, HER2 overexpression, and TNBC cancer subtypes, respectively (Table 1). Three individuals of the cohort 1 were dropped from the research due to non-cancer related deaths.
Fourteen fresh primary breast cancer tissue samples were acquired to validate the efficiency of the 21-gene signature in predicting eMDSCs infiltration in situ. Peripheral blood (PB) samples were taken from healthy volunteers in order to isolate CD33+ myeloid progenitors.
Cell line and cell culture
ATCC provided the breast cancer cell lines MCF-7, T47D, and EO771. MCF-7, and EO771 cell lines were grown in full DMEM (Gibco, Grand Island, NY, United States) with 10% FBS in a 5% CO2 incubator at 37°C. T47D cells were grown in full RPMI 1640 media (Gibco, Grand Island, NY, United States) with 10% fetal bovine serum in a 5% CO2 incubator at 37°C.
Animal experiments
The mice (Beijing Vital River Facility Animal Technology, Beijing, China) were kept at the Tianjin Medical University Cancer Institute and Hospital’s Specific pathogen free (SPF) animal laboratory. For constructing abundant tumor-infiltrating eMDSCs mice model, conditional targeting was performed as previously described (Yasukawa et al., 2003). An ES targeting technique was utilized to establish conditional SOCS3 knockout C57BL/6 mice. A targeting vector with recombination sites and selection markers was constructed. F1 SOCS3fl/+ mice were constructed by Cyagen Biosciences Inc (Guangzhou, China). F1 SOCS3fl/+ mice were intercrossed to generate SOCS3fl/fl mice. LysM-cre mice (cDNA for cre was inserted into the lysozyme M gene) were crossed with SOCS3fl/fl mice. These mice were further intercrossed to generate LysM-cre SOCS3fl/fl (SOCS3KO) mice. In addition, SOCS3fl/fl mice were used as control group.
For in vivo growth assay, EO771 cells (1×106 cells) were injected into the mammary fat pads of 6-week-old female SOCS3KO and SOCS3fl/fl mice. Every 3 days, the tumor volume and body weight were measured. After 15 days, all animals were euthanized under anesthesia by cervical dislocation, and the tumors were collected for future research.
Isolation of mice early myeloid-derived suppressor cells
The bone marrow or tumor tissues of SOCS3KO and SOCS3fl/fl mice were used to isolate murine CD11b+Gr-1- eMDSCs as described previously (Zhang et al., 2018). Bone marrow or tumor tissues were prepared into single-cell suspensions. CD11b+Gr-1− eMDSCs were isolated following erythrocytolysis using anti-mouse Gr-1 coupled with biotin and anti-biotin microbeads (Miltenyi Biotec, Germany). The BD FACSAria™ II cell sorter (BD Biosciences, San Jose, CA, United States) was used to isolate CD11b+Gr-1−F4/80−MHCII− eMDSCs. Flow cytometry and trypan blue staining were used to examine the vitality and purity of the retrieved cells.
Detection of primary early myeloid-derived suppressor cells in situ and induction of human early myeloid-derived suppressor cells in vitro
The extent of eMDSCs infiltration in the 14 fresh primary breast cancer tissue samples and 280 primary breast cancer tissue samples from two cohorts were examined using immunohistochemistry (IHC) assay and multispectral IF staining to detect the expression of CD33 protein as described previsouly (Yu et al., 2013, 2014; Jiang et al., 2017). The median number of positive staining was applied as the threshold of eMDSCs. Therefore, the samples with lower amount than the threshold were regarded as low infiltration of eMDSCs, while the samples with higher amount than the threshold were regarded as high infiltration of eMDSCs.
CD33+ cells were extracted from healthy PBMCs using human CD33 MicroBeads (130-045-501; Miltenyi Biotec, Germany) and co-cultured with MDA-MB-231 cells to create human eMDSCs, as previously described (Jiang et al., 2017).
Immunosuppressive capacity of early myeloid-derived suppressor cells
T cells were extracted from normal C57BL/6 mice using the Pan T Cell Isolation Kit (Miltenyi Biotec) and co-cultured in 24-well plates with either BM- or tumor-derived eMDSCs from tumor-bearing mice at a 1:3 ratio in RPMI-1640 media supplemented with 10% FBS. To excite T cells in vitro, anti-CD3/CD28 beads (20 μL/106 cells; Gibco) were used. T cell proliferation and apoptosis were investigated using the BrdU and Annexin V labeling assays described before (Yu et al., 2013).
Tumor invasion induced by early myeloid-derived suppressor cells
The isolated human and mice eMDSCs were placed into a 0.4 μm co-incubation chamber and cocultured with T47D, MCF-7, or EO771 cell at 1:3 ratio. After 2 days, the cells were collected for Transwell assay and EMT markers detection.
Flow cytometry analysis
Human eMDSCs were labeled using CD45, CD13, CD33, CD14, and CD15 antibodies (BD Biosciences, United States). Mouse eMDSCs were detected using CD45, CD11b, Gr-1, F4/80, and MHC-II antibodies (Biolegend, United States). A BD FACS Canto™ II flow cytometer was used for the study (BD Biosciences, United States). Our prior paper detailed the gating technique (Jiang et al., 2017).
RNA-seq analysis and construction of the 21-genes signature prediction model
Novogene Co., Ltd. was tasked with library preparation and sequencing. To screen for potential candidates that might predict the infiltration of eMDSCs within breast cancer tissues, 424 differentially expressed genes were selected from the expression profiles of eMDSCsSOCS3KO, CD11b+GR-1+, and eMDSCsfl/fl cells by the Z-Score-like method as previously described (Li et al., 2010). Then, 50 genes that can separate the three groups well are selected and mapped to human genes to obtain 42 genes. We validated the 42 genes in human PBMC-derived eMDSCs using qRT-PCR. Finally, 21 genes with comparable transcriptional patterns in humans and mice were selected to construct the genetic prediction model.
siRNA interference and gene transfection
pcDNA3.1-ARID1A overexpression vector or pcDNA3.1 empty vector and/or siRNAs against ARID1A or negative control (Shanghai HanBio Co., Ltd.) were delivered using Lipofectamine® 2000 according to the manufacturer’s procedure (Invitrogen; Thermo Fisher Scientific, Inc.). After 6 h, the reduced-serum media was replaced with full medium. The cells were extracted after 48 h to be used in following studies.
Immunohistochemistry
Following surgical excision, fresh tissues were promptly fixed with formalin; tissues were then embedded in paraffin and cut into 4-µm serial sections. Deparaffinization and rehydration of breast cancer tissue samples followed. After inhibiting endogenous peroxidase with 3% H2O2, citrate-based antigen retrieval was carried out. Primary antibodies were incubated overnight at 4°C on the samples. Following an incubation with the secondary antibody, a DAB Substrate kit was used to stain the sections according to the manufacturer’s instructions. Five representative high-power fields (400 magnification) for each tissue section were selected for histology evaluation as previously described (Yu et al., 2013). The average amount of positive staining cells among 5 fields was calculated for each sample.
Multispectral IF staining
The breast cancer tissue samples were embedded in paraffin, deparaffinized and rehydrated, incubated with antibodies and imaged using the multiplex method as previously described (Ning et al., 2021). Five fields from each slide were imaged and scored. The average amount of positive staining cells among 5 fields was calculated for each sample.
Transwell assay
Before plating in the upper chamber, T47D, MCF-7, and EO771 cell lines were collected and resuspended in serum-free DMEM. 1640 or DMEM supplemented with 10% FBS was used to fill the lower chamber of the Transwell. The cell suspension was placed to the membrane, either with or without Mtrigel adhesive, and incubated for 48 h at 37°C. All of the trials were carried out three times.
qRT-PCR analysis
qRT-PCR was used to examine the mRNA expression of ARID1A, Vimentin and E-Cadherin in breast cancer cell lines. Table 2 displays the primers for ARID1A, Vimentin, and E-Cadherin. As an internal control, β-actin was employed. Each sample was calculated using the formula: 2−ΔCt (ΔCt = Ctgene–Ctβ-actin). Every test was carried out at least three times.
Western blot analysis
SDS-PAGE was used to separate cell lysates, which were then transferred to polyvinylidene difluoride membranes and incubated with rabbit antibody ARID1A (abcam, ab182560, United States), E-Cadherin (CST, 3195, United States), Vementin (CST, 5741, United States), and SNAIL (CST, 3879, United States) over night at 4°C. Following an incubation with the secondary antibody, the chemiluminescent substrate kit was used to visualize the protein bands. The relative densities of target protein were determined by comparing with GAPDH.
Immunofluorescence
Breast cancer cells were placed on cover glasses and allowed to cling to the surface. With 0.1 percent Triton X-100, the cells were fixed and permeabilized. They were then blocked with 5% BSA and treated at 4°C overnight with primary antibodies. Following an incubation with the secondary antibody for 1 h, the Iamger.Z2 (Zeiss, Oberkochen, Germany) was used to visualize the protein.
Statistical analysis
GraphPad Prism 8.0.2 software were used for statistical analyses. The Kaplan–Meier technique was employed to calculate the cumulative survival probability, and the log-rank test was utilized to compare the OS of each patient subgroup. All experiments were independently performed at least three times. The values are presented as mean ± standard deviation (SD). p < 0.05 was chosen as the level of statistical significance.
Results
21-genes signature predicting highly early myeloid-derived suppressor cells infiltration in situ are significantly correlated with poor prognosis in breast cancer patients
Suppressor of cytokine signaling-3 (SOCS3), a known feedback inhibitor of the Janus kinase-signal transducer and activator of transcription 3 (JAK/STAT3) signaling pathway, is involved in the differentiation of eMDSCs (Jiang et al., 2017; Zhang et al., 2018). In this study, we successfully constructed a conditionally SOCS3 gene knockout mouse model in the myeloid lineage by using the murine Cre-loxP system (SOCS3KO) and detected a significant increase in the amount of CD11b+Gr-1− eMDSCs both in bone marrow (eMDSCsSOCS3KO) and tumor tissues (eMDSCsTumor) of tumor-bearing SOCS3KO mice. To analyze their immunosuppressive capacity, naïve T cells from wild-type C57BL/6 mice were co-cultured with eMDSCsSOCS3KO or eMDSCsTumor. We found that eMDSCsSOCS3KO and eMDSCsTumor significantly inhibited T cell proliferation (33.0 ± 3.4% vs. 18.1 ± 4.2% vs. 19.8 ± 3.6%, p = 0.0086 and 0.0097; Supplementary Figure S1A) and promoted T cell apoptosis (3.5 ± 0.9% vs. 17.3 ± 1.4% vs. 15.4 ± 1.5%, p = 0.0001 and 0.0003, Supplementary Figure S1B). This indicated that eMDSCs exerted exceptional T cell immunosuppressive ability in vitro and vivo.
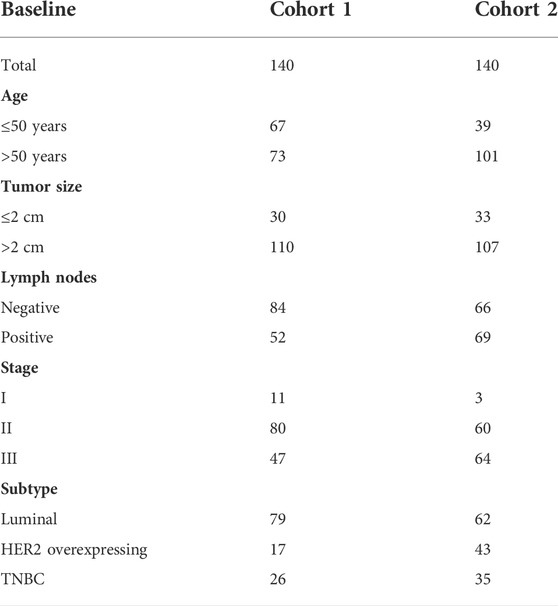
Using this mouse model, we obtained a sufficient number of eMDSCs to perform RNA sequencing and compared the genomic expression profile of eMDSCsSOCS3KO (CD11b+Gr-1-) cells with classical MDSCs (CD11b+Gr-1+) cells or with normal myeloid precursors eMDSCsfl/fl. The differentially expressed genes in eMDSCsSOCS3KO compared to those in CD11b+Gr-1+, and eMDSCsfl/fl were filtered to screen for potential candidates that might predict the infiltration of eMDSCs within breast cancer tissues as previously described (Li et al., 2010). 50 genes that can separate the three groups well are selected and validated them in human PBMC-derived eMDSCs using qRT-PCR (Figures 1A,B). Finally, 21-genes signature, including ABTB1, ACTN1, BTG1, BTG2, C5AR1, CCR1, CD300LF, CD33, CTSZ, DHRS7, FN1, FTH1, LDHA, PBXIP1, PLAC8, PYGL, PLK3, PLD4, S100A6, SORL1, and ST3GAL5 were selected to construct a genetic prediction model.
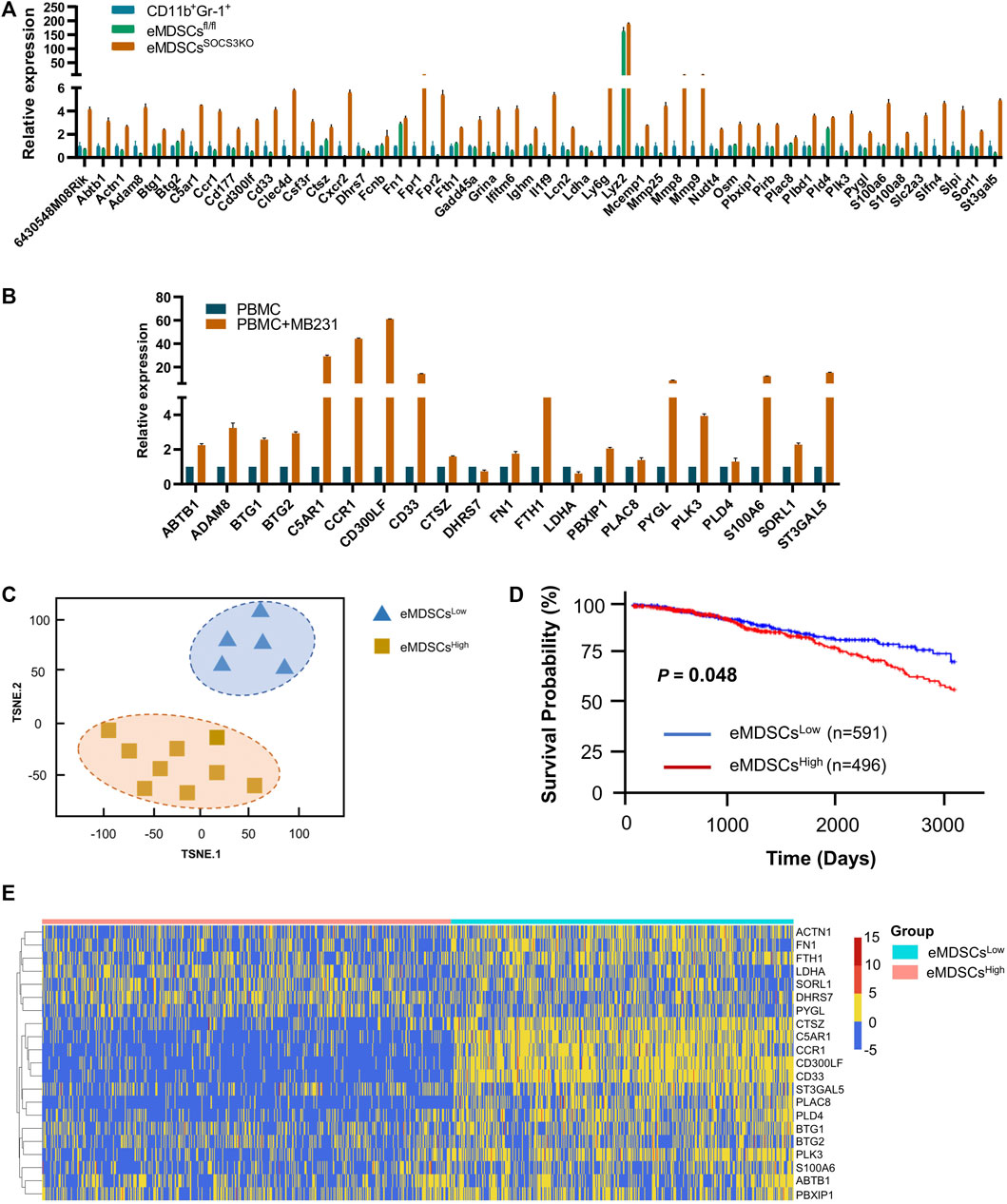
FIGURE 1. The 21-genes signature is able to predict the infiltration of eMDSCs in situ.(A) The relative expression of 50 genes that can separate the three groups of eMDSCsSOCS3KO, CD11b+Gr-1+ and eMDSCsfl/fl cells. (B) The relative expression of 21 genes in human PBMC-derived eMDSCs was confirmed by RT-qPCR. The transcriptional patterns of 21 genes are comparable both in humans and mice. CD33+ myeloid progenitors isolated from healthy PBMCs were co-cultured with MDA-MB-231 breast cancer cells to induce eMDSCs. (C) T-SNE analysis was used to visually display the classification results and the consistency between the 21-genes signature prediction and immunohistochemical observation was as high as 85.7% (12/14). (D) Kaplan-Meier survival analysis of differences between eMDSCshigh and eMDSCslow groups in TCGA breast cancer cohort showed that the OS was longer in the eMDSCslow group than the eMDSCshigh group. (E) Heatmap of the 21 genes expression profiles in the TCGA breast cancer cohort.
In order to verify the efficacy of 21-genes signature to predict the infiltration of eMDSCs in situ, 14 primary breast cancer tissue samples were collected and eMDSCs in breast cancer tissues were detected by IHC staining. Breast cancer samples were separated into two groups based on the median number of CD33+ eMDSCs infiltrated locally: lowly infiltrated eMDSCs (eMDSCslow, n = 7) and highly infiltrated eMDSCs (eMDSCshigh, n = 7). The two groups of breast cancer tissue samples were subjected to RNA-seq analysis and were further analyzed using the 21-genes signature. As expected, the consistency between the 21-genes signature and IHC assay was as high as 85.7% (12/14), implying that the 21-genes signature accurately predicted the infiltration status of eMDSCs in situ (Figure 1C).
Therefore, we enrolled a total of 1087 cases of breast cancer samples in TCGA dataset and divided them into eMDSCshigh (n = 496) and eMDSCslow (n = 591) groups using the 21-genes signature (Figure 1D). The heatmap of the 21 genes expression profiles in TCGA breast cancer cohort is presented in Figure 1E. Of these 21 genes, high expression of ABTB1, ACTN1, BTG1, BTG2, C5AR1, CCR1, CD300LF, CD33, CTSZ, FN1, FTH1, PBXIP1, PLAC8, PYGL, PLK3, PLD4, S100A6, SORL1, and ST3GAL5 indicated to be associated with highly infiltrated eMDSCs, while high expression of DHRS7 and LDHA was associated with lowly infiltrated eMDSCs. The Kaplan-Meier survival analysis indicated that the median survival of eMDSCshigh breast cancer patients was significantly shorter than eMDSCslow breast cancer patients, consistent results were validated in 3 more breast cancer datasets, including GSE3143 (n = 158), GSE48408 (n = 164), and GSE9893 (n = 155) (Figure 1D; Supplementary Figure S2).
Multispectral IF staining was performed to detect the expression of CD33 protein in 280 primary breast cancer patients from two cohorts. According to the median number of CD33+ eMDSCs that infiltrated locally, breast cancer patients were categorized into eMDSCshigh and eMDSCslow groups (Figure 2A). We found that the OS of eMDSCshigh patients was significantly shorter than those of eMDSCslow patients in the cohort 1 (p = 0.033, Figure 2B), cohort 2(p = 0.0398, Figure 2C) and the combined cohorts 1 and 2 (p = 0.0004, Figure 2D). Furthermore, when we compared the OS among four different molecular subtypes of all 280 cases of breast cancer patients, highly infiltrated eMDSCs strongly correlated with poor patient outcome in luminal A subtype (p < 0.0001, Figure 2E), rather than in other subtypes such as luminal B, HER2 overexpression and Basal-like (p = 0.2465, p = 0.7607, p = 0.4780, Figures 2F–H).
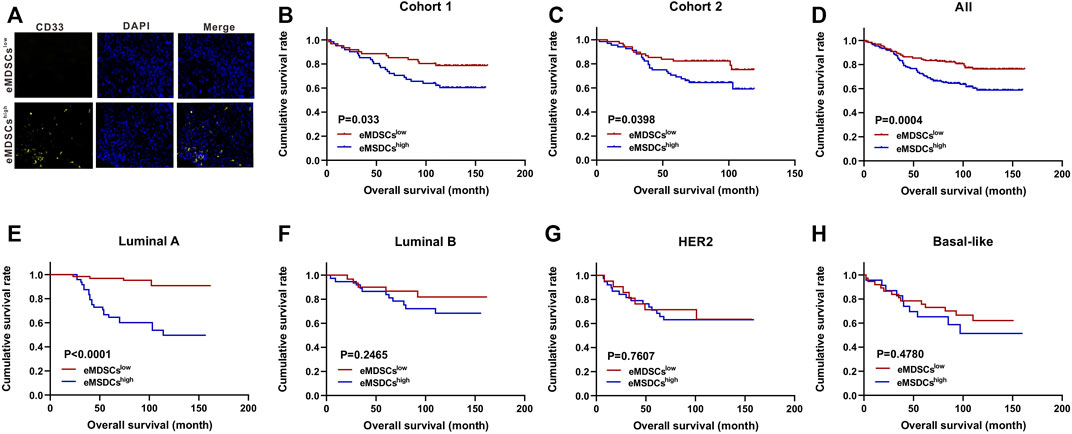
FIGURE 2. Highly-infiltrated eMDSCs are significantly correlated with poor prognosis in breast cancer patients (A) Multispectral IF staining analysis showed the infiltration of CD33+ cells in tumor sections from 280 cases of breast cancer patients. (B–H) Kaplan-Meier survival analysis of overall survival between eMDSCshigh and eMDSCslow groups among 4 different subtypes of 280 cases of breast cancer patients.
Early myeloid-derived suppressor cells promote migration and invasion of luminal A breast cancer cells via stimulating epithelial-mesenchymal transition
We performed gene expression and pathway enrichment analysis between eMDSCshigh and eMDSCslow groups in TCGA database using the 21-genes signature, and found that the cell adhesion molecules, focal adhesion, gap junction and adherens junction were significantly enriched as suggested by KEGG analysis (Figure 3A). It has been observed that cells communicate with one another through subapical tight junctions, adherens junctions and desmosomes at lateral surfaces, and scattered gap junctions at lateral surfaces, all of which are required for epithelial integrity (Huang et al., 2012). When EMT begins, these junctions are dismantled, and the junction proteins are relocalized and/or destroyed. These data suggested that eMDSCs may promote breast cancer metastasis via accelerating EMT.
FIGURE 3. eMDSCs promote migration and invasion of luminal A breast cancer cells via stimulating EMT (A) KEGG enrichment analysis of the differential expressed genes between eMDSCshigh and eMDSCslow groups divided by the 21-genes signature in TCGA breast cancer cohort. (B) Distribution of FPKM values for TCGA breast cancer samples between eMDSCshigh and eMDSCslow groups for the three EMT-related genes. (C) Immunoblotting was used to assess the proteins of E-Cadherin, Vimentin, and SNAIL in the indicated breast cancer cells. (D) Immunofluorescence was used to examine the levels of E-Cadherin in EO771, T47D, and MCF7 cells cocultured with eMDSCs. (E) Transwell assays revealed that eMDSCs dramatically improved the migratory and invasion capacities of EO771, T47D, and MCF7 cells.
The loss of E-Cadherin protein was thought to be the start of EMT, whereas the gain of Vimentin and SNAIL gave tumor cells increased migratory ability (Lamouille et al., 2014). TCGA database analysis showed that E-Cadherin was reduced and Vimentin and SNAIL were elevated in eMDSCshigh breast cancer patients relative to eMDSCslow group (Figure 3B). To verify the above results, we co-cultured eMDSCs with 3 luminal A breast cancer cell lines (mouse mammary tumor cell line EO771, human mammary tumor cell line MCF7 and T47D) in vitro. Western blot and immunofluorescence analyses were also done to identify the expression levels of E-Cadherin, Vimentin and SNAIL in luminal A breast cancer cell lines after being co-cultured with eMDSCs. In line with the results shown above, eMDSCs significantly suppressed the expression of E-Cadherin and enhanced the expression of Vimentin and SNAIL (Figures 3C,D). Moreover, the migration and invasion of EO771, MCF7, and T47D cells were greatly accelerated following co-culture with eMDSCs (Figure 3E). Taken together, these results suggest that eMDSCs promoted migration and invasion of luminal A breast cancer cells via stimulating EMT.
Early myeloid-derived suppressor cells downregulate the expression of ARID1A in luminal A breast cancer cells
We performed a proteomics analysis on mice mammary breast cancer cell line EO771 xenografts collected from SOCS3KO mice (eMDSCshigh group) or SOCS3fl/fl mice (eMDSCslow group). Bioinformatic analysis of GO based on the strongest differentially expressed proteins between eMDSCshigh and eMDSCslow groups was further employed to conduct functional annotation. Interestingly, the ARID1A-containing BAF chromatin remodeling complexes (mammalian SWI/SNF) ranked among the top 30 of GO enrichment (Figure 4A).
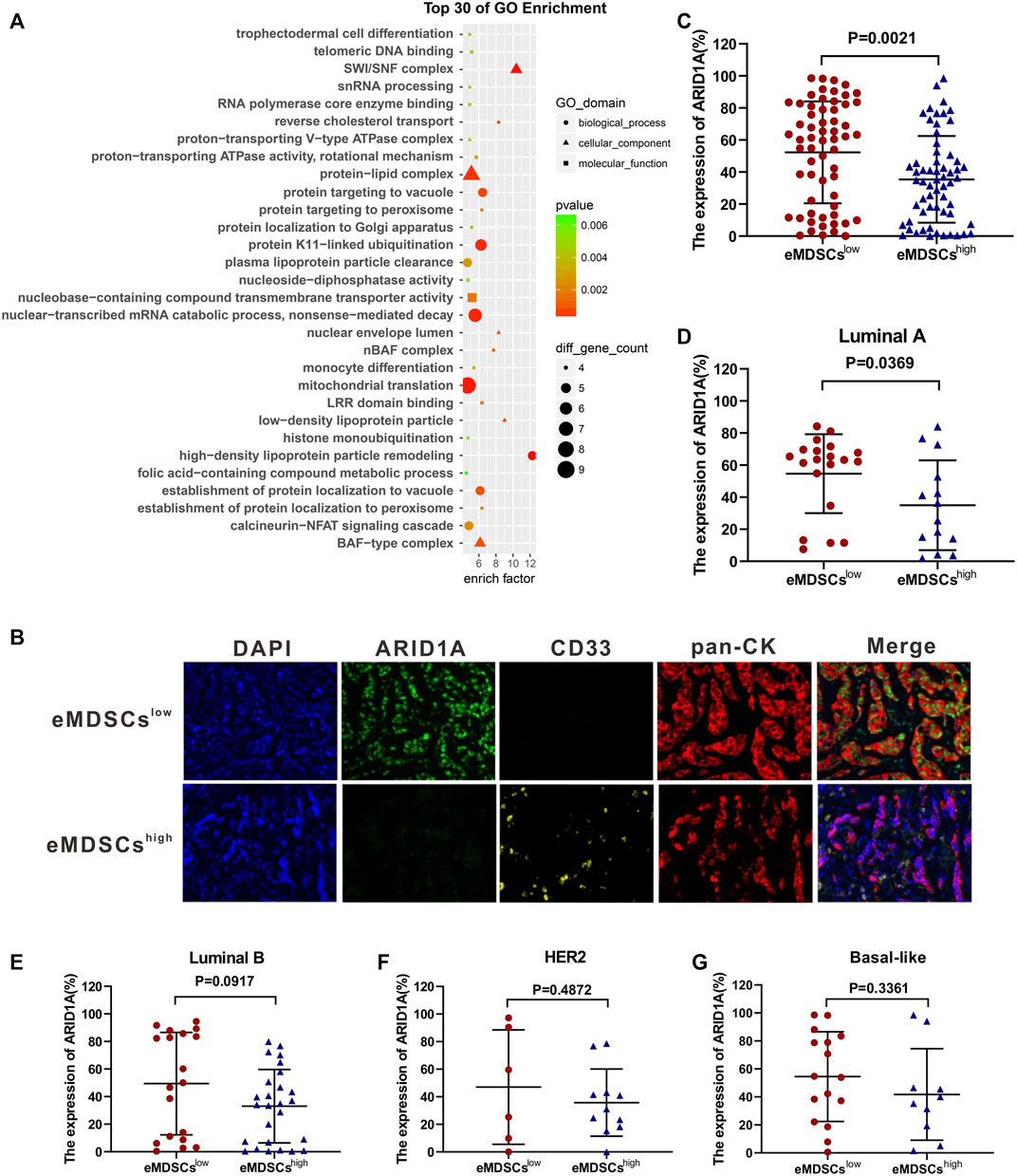
FIGURE 4. eMDSCs downregulate the expression of ARID1A in luminal A breast cancer cells (A) Gene ontology enrichment analysis of the strongest differential expressed proteins between eMDSCshigh and eMDSCslow groups. (B) A multispectral IF staining assay showed that the changes of ARID1A expression between eMDSCshigh and eMDSCslow groups in 140 primary breast cancer samples from cohort 1. (C–G) The ARID1A expression between different groups in (B) was analyzed.
It’s reported that ARID1A depletion accelerates EMT, increases cancer stemness and promotes migration, invasion and angiogenesis in various cancers by reducing the chromatin accessibility of target genes or influencing the post-transcriptional modification process (Wilson et al., 2019; Luo et al., 2020; Wang et al., 2020; Tomihara et al., 2021). We compared the correlation between the number of infiltrated eMDSCs and the expression level of ARID1A protein in 140 primary breast cancer tissues from cohort 1. Consistently, the expression level of ARID1A protein dramatically decreased in eMDSCshigh group compared to that in eMDSCslow group using multispectral immunofluorescence IF staining assay (p = 0.0021, Figures 4B,C). Further analysis of breast cancer patients with different molecular subtypes revealed that ARID1A expression was significantly lower in eMDSCshigh luminal A breast cancer (p = 0.0369, Figure 4D), rather than in the luminal B, HER2 and Basal-like subtype (p = 0.0917, p = 0.4872, p = 0.3361, Figures 4E–G). These data are highly consistent with our previous observation that eMDSCs affected the prognosis of patients with luminal A breast cancer. Furthermore, Kaplan-Meier analysis in 140 primary breast cancer tissues from cohort 1 demonstrated that patients with lower ARID1A expression showed a worse outcome (Supplementary Figure S3A), which was reconfirmed by online Kaplan–Meier-Plotter database-analyzed overall survival plot of breast cancer (Supplementary Figure S3B) (Gyorffy et al., 2010). These data suggest that eMDSCs suppress the ARID1A expression to cause poor prognosis in breast cancer patients.
Early myeloid-derived suppressor cells accelerate epithelial-mesenchymal transition in luminal A breast cancer cells by downregulating ARID1A
After co-cultured with eMDSCs, ARID1A showed significant downregulated in both mRNA and protein expression in EO771, MCF-7, and T47D cells, which is consistent with our previous observations in clinical samples (Figures 5A,B). To further explore if downregulation of ARID1A expression is crucial for eMDSCs to promote EMT of luminal A breast cancer cells, we knocked down ARID1A by RNA interference and compared the expression of multiple functional genes by RT-PCR (Figure 5C). As expected, the expression of EMT-related genes was significantly regulated, in which the expression of Vimentin increased but the expression of E-Cadherin decreased (Figure 5C). Subsequently, we established stable EO771, MCF-7, and T47D clones overexpressing ARID1A respectively for further confirming that the expression level of ARID1A is crucial for eMDSCs to promote EMT of luminal A breast cancer cells (Figure 5D). The breast cancer cells transfected with the pCNDA3.1-ARID1A or pCNDA3.1 plasmids were independently cocultured with eMDSCs. Compared to negative control, the E-Cadherin gene increased significantly and Vimentin decreased after ARID1A overexpression (Figure 5D).
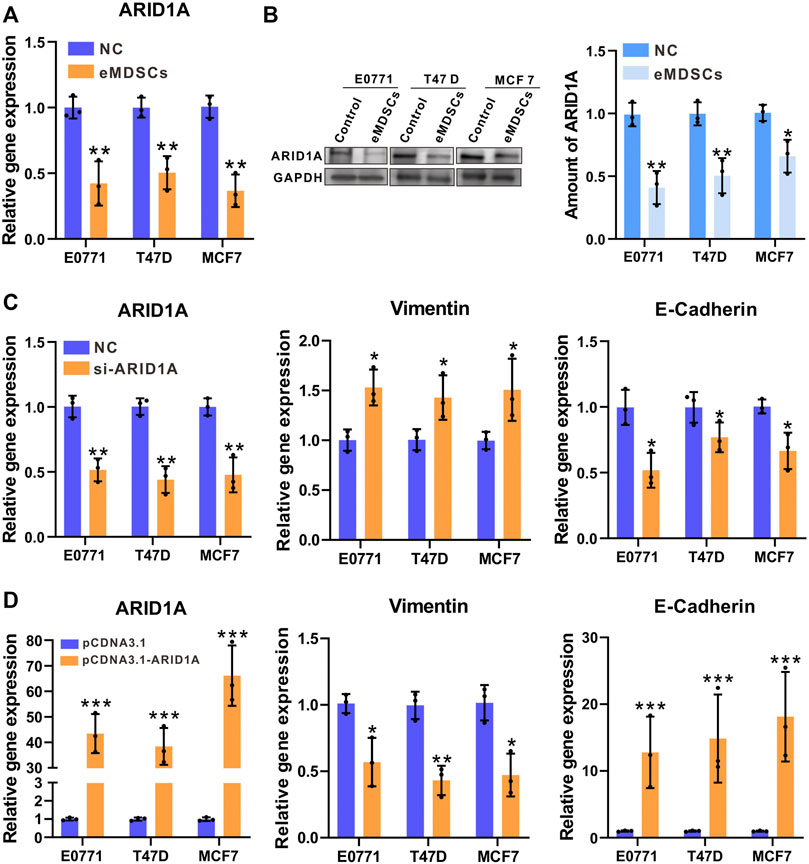
FIGURE 5. eMDSCs accelerate epithelial-mesenchymal transition in luminal A breast cancer cells by downregulating ARID1A (A,B) The mRNA and protein level of ARID1A were determined by RT-PCR and immunoblotting in EO771, T47D and MCF7 cells after cocultured with eMDSCs. (C) Quantitative RT-PCR showing expression changes of ARID1A and EMT-related genes in EO771, T47D and MCF7 cells upon ARID1A knocked-down with the transfection of siRNA or vehicle. (D) Quantitative RT-PCR showing expression changes of ARID1A and EMT-related genes in EO771, T47D, and MCF7 cells cocultured with eMDSCs upon ARID1A overexpression plasmids or vehicle. *p < 0.05, **p < 0.01, ***p < 0.001.
To assess the impact of eMDSCs on luminal A breast cancer cells in vivo, EO771 cells were used to establish subcutaneous xenograft model in SOCS3fl/fl and SOCS3KO mice for further experiments. We found that conditional SOCS3 knockout in myeloid linage strikingly promoted tumor growth (Figure 6A). After 15 days, the weight and volume of tumor in SOCS3KO group were much more than that in control group (Figures 6B,C), whereas there was no discernible change in body weight between the two groups (Figure 6D). Flow cytometry analysis revealed that the number of eMDSCs increased in the tumor tissues of tumor-bearing SOCS3KO mice compared to SOCS3fl/fl mice (Figure 6E; Supplementary Figure S4). Meanwhile, IHC staining showed attenuated levels of ARID1A and E-Cadherin and enhanced expression of Vimentin in highly eMDSCs-infiltrated xenografts (Figure 6F), which suggested that eMDSCs accelerated EMT process and may further promote the initiation of metastasis. Collectively, the above results suggested that eMDSCs accelerated epithelial-mesenchymal transition by downregulating ARID1A in luminal A breast cancer cells.
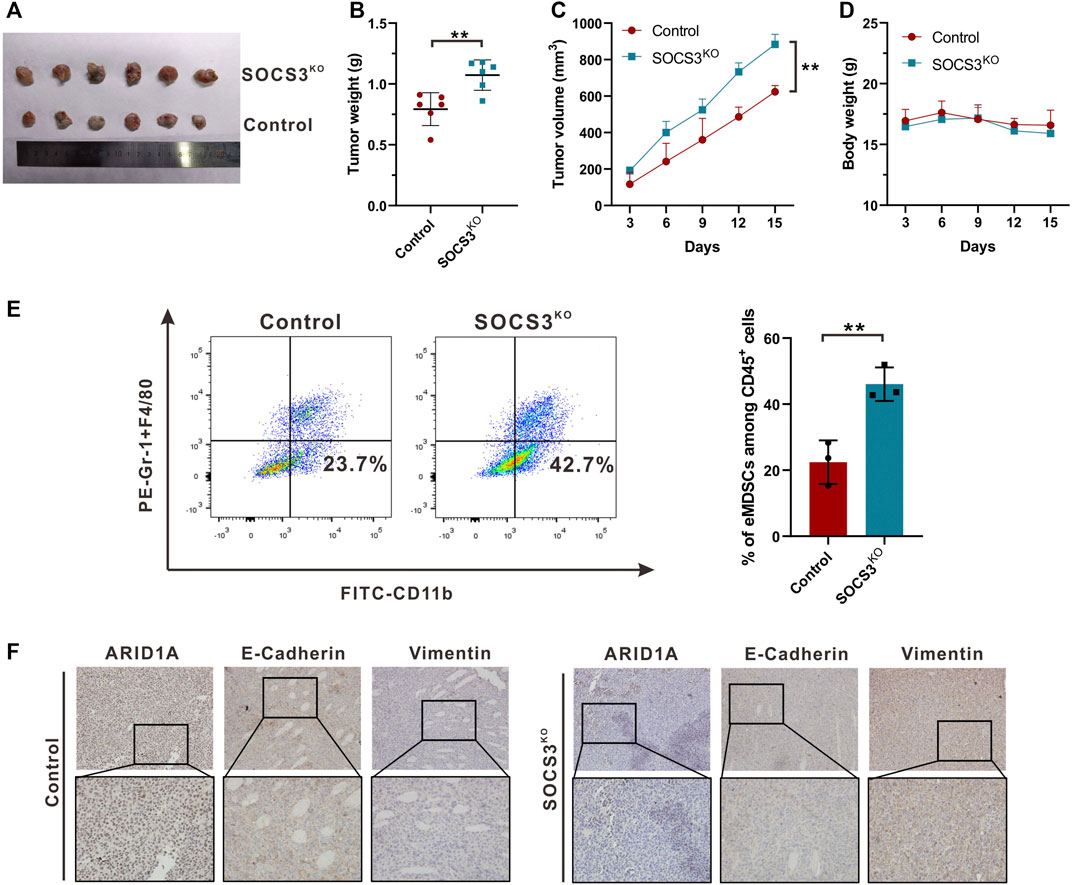
FIGURE 6. eMDSCs promote tumor growth via accelerating EMT process (A) Representative images for xenografts model in SOCS3fl/fl and SOCS3KO mice. (B,C) The weight and volume of tumors in the SOCS3KO group were significantly greater than those in the control group. (D) There was no discernible change in body weight between the SOCS3KO group and the negative control group. (E) The percentages of eMDSCs were detected by FCM in primary tumor tissues. (F) The images show immunohistochemistry staining for ARID1A, E-Cadherin, and Vimentin in xenografts. *p < 0.05, **p < 0.01.
Discussion
Growing evidence shows that crosstalk between cancer cells and the surrounding microenvironment appears to plays an important role in the recurrence and metastasis of various cancers (Wang et al., 2018; Hinshaw and Shevde, 2019; Lan et al., 2019; Zavros and Merchant, 2022). MDSCs are a prominent component of the tumor microenvironment and have extraordinary suppressive abilities, including the ability to limit T cell activation, induce NK cell anergy, and influence regulatory T cell accumulation (Ma et al., 2011; Lindau et al., 2013; Hegde et al., 2021). Aside from their impacts on immune responses, MDSCs also contribute to tumor development through a variety of non-immunological pathways, including angiogenesis support, tumor cell stemness promotion, EMT facilitation, and pre-metastatic niche creation (Safarzadeh et al., 2018; Wang et al., 2019; Tang et al., 2021). Since 2013, we have focused on CD33+ eMDSCs in human breast cancer tissues, and found they possess potent suppression on T cells proliferation and cytokine production (Yu et al., 2013). We demonstrated that cancer-derived interleukin-6 (IL-6) stimulates STAT3-dependent, nuclear factor-κB-mediated indoleamine 2,3-dioxygenase (IDO) upregulation in eMDSCs which triggers immunosuppressive effects of eMDSCs (Yu et al., 2014). Furthermore, we analyzed the immunosuppressive capacity of eMDSCs in 4T1 mammary tumor-bearing mice and found eMDSCs impaired T cell immunity significantly in SOCS3 deficiency-dependent manner by activating the JAK/STAT signaling pathway (Zhang et al., 2018). However, the mechanism that eMDSCs promote breast cancer recurrence and affect prognosis through EMT has not been fully understood.
In this study, using in-house tissue array and TCGA dataset, we demonstrated that the infiltration of eMDSCs within breast cancer tissues affected the prognosis of luminal A breast cancer patients which was negatively correlated with ARID1A expression in situ. Furthermore, we demonstrated that eMDSCs promoted the migration and invasion of luminal A breast cancer cells by accelerating EMT. Finally, we proposed a significant role of eMDSCs in promoting EMT of luminal A breast cancer cells by downregulating ARID1A which caused poor prognosis in luminal A breast cancer patients.
ARID1A is a highly conserved subunit of the BAF complex, which hydrolyzes ATP and uses the resulting energy to mobilize nucleosomes and alter accessibility of chromatin to transcriptional and coregulatory machineries (Clapier et al., 2017). Among the genetic abnormalities reported in ER+ breast cancer, mutations are commonly detected in genes encoding the subunits of the BAF chromatin remodeling complexes and ARID1A is the most frequently mutated one in BAF (Garraway and Lander, 2013). ARID1A depletion accelerates epithelial-mesenchymal transition, increases cancer stemness and promotes migration, invasion and angiogenesis in various cancers, including breast cancer, uterus cancer and pancreatic cancer by reducing the chromatin accessibility of target genes or influencing the post-transcriptional modification process (Wilson et al., 2019; Luo et al., 2020; Wang et al., 2020; Tomihara et al., 2021). We found that eMDSCs was negatively correlated with the expression of ARID1A protein and accelerated EMT progression in luminal A breast cancer patients, which suggest a possibility that eMDSCs promote breast cancer cell migration and invasion through down-regulation of ARID1A. The results of cellular experiments verified the authenticity of our idea.
The connection between immune cells and ARID1A had been reported (Shen et al., 2018; Li et al., 2019; Jung et al., 2021), but the regulatory mechanisms have not been fully understood. We found that eMDSCs significantly downregulated ARID1A expression in luminal A breast cancer but the mechanistic events involved in this process are worth more consideration and exploration. It is reported that MDSCs suppress CtBP2 in ovarian cancer cells by inducing miR-101 expression which enhanced cancer cell stemness and spreading potential (Cui et al., 2013). In our previous study, we found that some miRNAs were significantly upregulated in eMDSCs when co-cultured with breast cancer cell line 4T1 (Jiang et al., 2020). To find potential miRNA that may influence the ARID1A expression, we used the miRNA target prediction system “TargetScan” to find high-score potential miRNAs of ARID1A and mmu-miR-342-3p and mmu-miR-9-5p were selected (Supplementary Figure S5A). We validated the expression of the above miRNAs in EO771 cell and found that only mmu-miR-342-3p increased after co-cultured with eMDSCs (Supplementary Figure S5B). Moreover, ARID1A showed significantly downregulated in both mRNA and protein expression after transfected with mmu-miR-342-3p mimics (Supplementary Figure S5C,D). This discovery suggests that eMDSCs downregulated ARID1A expression via inducing mmu-miR-342-3p in EO771 cell and miRNA-related post-transcriptional regulation also participated in eMDSCs-induced ARID1A downregulation in luminal A breast cancer.
In summary, our study provided evidence for the tumor promoting function of eMDSCs in breast cancer. We constructed a 21-gene signature in the prediction of the infiltration of eMDSCs within breast cancer tissues. Meanwhile, eMDSCs significantly downregulate ARID1A expression and promote EMT in luminal A breast cancer. These findings shed light on the mechanisms by which eMDSCs could contribute to breast cancer metastasis in the primary tumor microenvironment. The eMDSCs-ARID1A axis was essential for luminal A type of breast cancer metastasis and could be a potential target for metastatic breast cancer therapies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Gene Expression Omnibus, GSE140092.
Ethics statement
The studies involving human participants were reviewed and approved by Medical Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Medical Ethics Committee of Tianjin Medical University Cancer Institute and Hospital.
Author contributions
JY, JL, and GC designed the research. GC, XL, CJ, and PL performed the research, data analyses, clinical information and samples for the study. LZ, DX, DW, and JL contributed to bioinformatic analysis. JY and GC wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (grant No. 81872143), Tianjin Research Innovation Project for Postgraduate Students (grant No.2021YJSB261), Tianjin Health Research Project (grant No. TJWJ2022QN013), National Science and Technology Support Program of China (grant No. 2018ZX09201015), and Project of Science and Technology of Tianjin (grant No. 18JCQNJC82700).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.973731/full#supplementary-material
References
Bild, A. H., Yao, G., Chang, J. T., Wang, Q., Potti, A., Chasse, D., et al. (2006). Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439, 353–357. doi:10.1038/nature04296
Bronte, V., Brandau, S., Chen, S. H., Colombo, M. P., Frey, A. B., Greten, T. F., et al. (2016). Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 7, 12150. doi:10.1038/ncomms12150
Carroll, J. S. (2016). EJE PRIZE 2016: Mechanisms of oestrogen receptor (ER) gene regulation in breast cancer. Eur. J. Endocrinol. 175, R41–R49. doi:10.1530/EJE-16-0124
Chanrion, M., Negre, V., Fontaine, H., Salvetat, N., Bibeau, F., Mac Grogan, G., et al. (2008). A gene expression signature that can predict the recurrence of tamoxifen-treated primary breast cancer. Clin. Cancer Res. 14, 1744–1752. doi:10.1158/1078-0432.Ccr-07-1833
Clapier, C. R., Iwasa, J., Cairns, B. R., and Peterson, C. L. (2017). Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422. doi:10.1038/nrm.2017.26
Clarke, R., Tyson, J. J., and Dixon, J. M. (2015). Endocrine resistance in breast cancer-An overview and update. Mol. Cell. Endocrinol. 418, 220–234. doi:10.1016/j.mce.2015.09.035
Cui, T. X., Kryczek, I., Zhao, L., Zhao, E., Kuick, R., Roh, M. H., et al. (2013). Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity 39, 611–621. doi:10.1016/j.immuni.2013.08.025
Dongre, A., and Weinberg, R. A. (2019). New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84. doi:10.1038/s41580-018-0080-4
Erin, N., Grahovac, J., Brozovic, A., and Efferth, T. (2020). Tumor microenvironment and epithelial mesenchymal transition as targets to overcome tumor multidrug resistance. Drug resist. Updat. 53, 100715. doi:10.1016/j.drup.2020.100715
Garraway, L. A., and Lander, E. S. (2013). Lessons from the cancer genome. Cell 153, 17–37. doi:10.1016/j.cell.2013.03.002
Gyorffy, B., Lanczky, A., Eklund, A. C., Denkert, C., Budczies, J., Li, Q., et al. (2010). An online survival analysis tool to rapidly assess the effect of 22, 277 genes on breast cancer prognosis using microarray data of 1, 809 patients. Breast Cancer Res. Treat. 123, 725–731. doi:10.1007/s10549-009-0674-9
Hegde, S., Leader, A. M., and Merad, M. (2021). MDSC: Markers, development, states, and unaddressed complexity. Immunity 54, 875–884. doi:10.1016/j.immuni.2021.04.004
Hinshaw, D. C., and Shevde, L. A. (2019). The tumor microenvironment innately modulates cancer progression. Cancer Res. 79, 4557–4566. doi:10.1158/0008-5472.Can-18-3962
Huang, R. Y., Guilford, P., and Thiery, J. P. (2012). Early events in cell adhesion and polarity during epithelial-mesenchymal transition. J. Cell Sci. 125, 4417–4422. doi:10.1242/jcs.099697
Jiang, M., Chen, J., Zhang, W., Zhang, R., Ye, Y., Liu, P., et al. (2017). Interleukin-6 trans-signaling pathway promotes immunosuppressive myeloid-derived suppressor cells via suppression of suppressor of cytokine signaling 3 in breast cancer. Front. Immunol. 8, 1840. doi:10.3389/fimmu.2017.01840
Jiang, M., Zhang, W., Zhang, R., Liu, P., Ye, Y., Yu, W., et al. (2020). Cancer exosome-derived miR-9 and miR-181a promote the development of early-stage MDSCs via interfering with SOCS3 and PIAS3 respectively in breast cancer. Oncogene 39, 4681–4694. doi:10.1038/s41388-020-1322-4
Jung, U. S., Min, K. W., Kim, D. H., Kwon, M. J., Park, H., and Jang, H. S. (2021). Suppression of ARID1A associated with decreased CD8 T cells improves cell survival of ovarian clear cell carcinoma. J. Gynecol. Oncol. 32, e3. doi:10.3802/jgo.2021.32.e3
Lamouille, S., Xu, J., and Derynck, R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196. doi:10.1038/nrm3758
Lan, J., Sun, L., Xu, F., Liu, L., Hu, F., Song, D., et al. (2019). M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 79, 146–158. doi:10.1158/0008-5472.Can-18-0014
Li, J., Lenferink, A. E., Deng, Y., Collins, C., Cui, Q., Purisima, E. O., et al. (2010). Identification of high-quality cancer prognostic markers and metastasis network modules. Nat. Commun. 1, 34. doi:10.1038/ncomms1033
Li, L., Li, M., Jiang, Z., and Wang, X. (2019). ARID1A mutations are associated with increased immune activity in gastrointestinal cancer. Cells 8, 678. doi:10.3390/cells8070678
Lindau, D., Gielen, P., Kroesen, M., Wesseling, P., and Adema, G. J. (2013). The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology 138, 105–115. doi:10.1111/imm.12036
Luo, Q., Wu, X., Chang, W., Zhao, P., Nan, Y., Zhu, X., et al. (2020). ARID1A prevents squamous cell carcinoma initiation and chemoresistance by antagonizing pRb/E2F1/c-Myc-mediated cancer stemness. Cell Death Differ. 27, 1981–1997. doi:10.1038/s41418-019-0475-6
Ma, G., Pan, P. Y., Eisenstein, S., Divino, C. M., Lowell, C. A., Takai, T., et al. (2011). Paired immunoglobin-like receptor-B regulates the suppressive function and fate of myeloid-derived suppressor cells. Immunity 34, 385–395. doi:10.1016/j.immuni.2011.02.004
Mittal, V. (2018). Epithelial mesenchymal transition in tumor metastasis. Annu. Rev. Pathol. Mech. Dis. 13, 395–412. doi:10.1146/annurev-pathol-020117-043854
Ning, J., Ye, Y., Bu, D., Zhao, G., Song, T., Liu, P., et al. (2021). Imbalance of TGF-β1/BMP-7 pathways induced by M2-polarized macrophages promotes hepatocellular carcinoma aggressiveness. Mol. Ther. 29, 2067–2087. doi:10.1016/j.ymthe.2021.02.016
Onitilo, A. A., Engel, J. M., Greenlee, R. T., and Mukesh, B. N. (2009). Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin. Med. Res. 7, 4–13. doi:10.3121/cmr.2008.825
Pastushenko, I., and Blanpain, C. (2019). EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29, 212–226. doi:10.1016/j.tcb.2018.12.001
Razavi, P., Chang, M. T., Xu, G., Bandlamudi, C., Ross, D. S., Vasan, N., et al. (2018). The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell 34, 427–438. doi:10.1016/j.ccell.2018.08.008
Safarzadeh, E., Orangi, M., Mohammadi, H., Babaie, F., and Baradaran, B. (2018). Myeloid-derived suppressor cells: Important contributors to tumor progression and metastasis. J. Cell. Physiol. 233, 3024–3036. doi:10.1002/jcp.26075
Shen, J., Ju, Z., Zhao, W., Wang, L., Peng, Y., Ge, Z., et al. (2018). ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 24, 556–562. doi:10.1038/s41591-018-0012-z
Shibue, T., and Weinberg, R. A. (2017). EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 14, 611–629. doi:10.1038/nrclinonc.2017.44
Sørensen, K. P., Thomassen, M., Tan, Q., Bak, M., Cold, S., Burton, M., et al. (2013). Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res. Treat. 142, 529–536. doi:10.1007/s10549-013-2776-7
Tang, H., Li, H., and Sun, Z. (2021). Targeting myeloid-derived suppressor cells for cancer therapy. Cancer Biol. Med. 18, 992–1009. doi:10.20892/j.issn.2095-3941.2020.0806
Thomas, C., and Gustafsson, J. A. (2015). Estrogen receptor mutations and functional consequences for breast cancer. Trends Endocrinol. Metab. 26, 467–476. doi:10.1016/j.tem.2015.06.007
Thomas, C., and Gustafsson, J. A. (2011). The different roles of ER subtypes in cancer biology and therapy. Nat. Rev. Cancer 11, 597–608. doi:10.1038/nrc3093
Tomihara, H., Carbone, F., Perelli, L., Huang, J. K., Soeung, M., Rose, J. L., et al. (2021). Loss of ARID1A promotes epithelial-mesenchymal transition and sensitizes pancreatic tumors to proteotoxic stress. Cancer Res. 81, 332–343. doi:10.1158/0008-5472.CAN-19-3922
Wang, N., Liu, W., Zheng, Y., Wang, S., Yang, B., Li, M., et al. (2018). CXCL1 derived from tumor-associated macrophages promotes breast cancer metastasis via activating NF-κB/SOX4 signaling. Cell Death Dis. 9, 880. doi:10.1038/s41419-018-0876-3
Wang, T., Gao, X., Zhou, K., Jiang, T., Gao, S., Liu, P., et al. (2020). Role of ARID1A in epithelialmesenchymal transition in breast cancer and its effect on cell sensitivity to 5FU. Int. J. Mol. Med. 46, 1683–1694. doi:10.3892/ijmm.2020.4727
Wang, Y., Ding, Y., Guo, N., and Wang, S. (2019). MDSCs: key criminals of tumor pre-metastatic niche formation. Front. Immunol. 10, 172. doi:10.3389/fimmu.2019.00172
Wilson, M. R., Reske, J. J., Holladay, J., Wilber, G. E., Rhodes, M., Koeman, J., et al. (2019). ARID1A and PI3-kinase pathway mutations in the endometrium drive epithelial transdifferentiation and collective invasion. Nat. Commun. 10, 3554. doi:10.1038/s41467-019-11403-6
Yasukawa, H., Ohishi, M., Mori, H., Murakami, M., Chinen, T., Aki, D., et al. (2003). IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat. Immunol. 4, 551–556. doi:10.1038/ni938
Yu, J., Du, W., Yan, F., Wang, Y., Li, H., Cao, S., et al. (2013). Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J. I. 190, 3783–3797. doi:10.4049/jimmunol.1201449
Yu, J., Wang, Y., Yan, F., Zhang, P., Li, H., Zhao, H., et al. (2014). Noncanonical NF-κB activation mediates STAT3-stimulated IDO upregulation in myeloid-derived suppressor cells in breast cancer. J. I. 193, 2574–2586. doi:10.4049/jimmunol.1400833
Zavros, Y., and Merchant, J. L. (2022). The immune microenvironment in gastric adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 19, 451–467. doi:10.1038/s41575-022-00591-0
Zhang, W., Jiang, M., Chen, J., Zhang, R., Ye, Y., Liu, P., et al. (2018). SOCS3 suppression promoted the recruitment of CD11b+Gr-1-F4/80-MHCII- early-stage myeloid-derived suppressor cells and accelerated interleukin-6-related tumor invasion via affecting myeloid differentiation in breast cancer. Front. Immunol. 9, 1699. doi:10.3389/fimmu.2018.01699
Keywords: breast cancer, early-stage myeloid-derived suppressor cells, ARID1A, epithelial-mesenchymal transition, prognostic factor
Citation: Chen G, Li X, Ji C, Liu P, Zhou L, Xu D, Wang D, Li J and Yu J (2022) Early myeloid-derived suppressor cells accelerate epithelial-mesenchymal transition by downregulating ARID1A in luminal A breast cancer. Front. Bioeng. Biotechnol. 10:973731. doi: 10.3389/fbioe.2022.973731
Received: 20 June 2022; Accepted: 03 October 2022;
Published: 18 October 2022.
Edited by:
Shengjun Wang, Jiangsu University Affiliated People’s Hospital, ChinaReviewed by:
Adeleye Oluwatosin Adeshakin, St. Jude Children’s Research Hospital, United StatesCheng Yang, Fudan University, China
Copyright © 2022 Chen, Li, Ji, Liu, Zhou, Xu, Wang, Li and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinpu Yu, anl1QHRtdS5lZHUuY24=; Jie Li, amllbGlAaGl0LmVkdS5jbg==
†These authors have contributed equally to this work
 Guidong Chen
Guidong Chen Xingchen Li
Xingchen Li Chenyan Ji1†
Chenyan Ji1† Dong Wang
Dong Wang Jie Li
Jie Li Jinpu Yu
Jinpu Yu