- 1Department of Electronics, Information and Bioengineering, Politecnico di Milano, Milano, Italy
- 2Department of Chemistry, Materials and Chemical Engineering “Giulio Natta”, Politecnico di Milano, Milano, Italy
- 3Department of Bioscience (DBS), University of Milan, Milano, Italy
- 4Institute of Photonics and Nanotechnology (IFN)-CNR and Department of Physics, Politecnico di Milano, Milano, Italy
Mesenchymal stem cells (MSCs) are known to be ideal candidates for clinical applications where not only regenerative potential but also immunomodulation ability is fundamental. Over the last years, increasing efforts have been put into the design and fabrication of 3D synthetic niches, conceived to emulate the native tissue microenvironment and aiming at efficiently controlling the MSC phenotype in vitro. In this panorama, our group patented an engineered microstructured scaffold, called Nichoid. It is fabricated through two-photon polymerization, a technique enabling the creation of 3D structures with control of scaffold geometry at the cell level and spatial resolution beyond the diffraction limit, down to 100 nm. The Nichoid’s capacity to maintain higher levels of stemness as compared to 2D substrates, with no need for adding exogenous soluble factors, has already been demonstrated in MSCs, neural precursors, and murine embryonic stem cells. In this work, we evaluated how three-dimensionality can influence the whole gene expression profile in rat MSCs. Our results show that at only 4 days from cell seeding, gene activation is affected in a significant way, since 654 genes appear to be differentially expressed (392 upregulated and 262 downregulated) between cells cultured in 3D Nichoids and in 2D controls. The functional enrichment analysis shows that differentially expressed genes are mainly enriched in pathways related to the actin cytoskeleton, extracellular matrix (ECM), and, in particular, cell adhesion molecules (CAMs), thus confirming the important role of cell morphology and adhesions in determining the MSC phenotype. In conclusion, our results suggest that the Nichoid, thanks to its exclusive architecture and 3D cell adhesion properties, is not only a useful tool for governing cell stemness but could also be a means for controlling immune-related MSC features specifically involved in cell migration.
1 Introduction
Traditionally, the most utilized substrates for cell culture have been treated, polystyrene or glass surfaces (Kapałczyńska et al., 2016), but since the environment provided by these systems is generally flat, cells grow forming monolayers, consequently impairing cell–cell and cell–ECM interactions and also modifying their morphology with respect to their native configuration. In this way, cells shape their cytoskeleton, sending different messages to the nucleus compared to the physiological state (Von der Mark et al., 1977; Petersen et al., 1992; Debnath and Brugge, 2005; Nelson and Bissell, 2006; Mahmud et al., 2009; Kilian et al., 2010), thus altering gene expression, protein synthesis, and other cell functions (Greiner et al., 2012; Santos et al., 2012). For these reasons, over the last few decades, three-dimensional (3D) systems for in vitro cell culture have gained increasing interest in several fields of biological research. It has been largely demonstrated that 3D culture conditions constitute a more realistic model as compared to bidimensional (2D) systems, since they create a more physiological microenvironment and, thus, promote cell responses that are more similar to the in vivo ones (Pampaloni et al., 2007; Edmondson et al., 2014; Shamir and Ewald, 2014).
One of the areas of greatest interest for the employment of 3D culture systems is stem cell research for tissue engineering and regenerative medicine, therapeutical approaches based on the use of stem cells to repair and regenerate damaged organs and tissues in place of resorting to allogenic transplantation (Heidary Rouchi and Mahdavi-Mazdeh, 2015). This is possible thanks to the self-renewal capacity of stem cells, the ability to maintain their stemness while dividing, and their ability to differentiate toward specific lineages under precise conditions (Kolios and Moodley, 2012; Dulak et al., 2015). The therapeutic potential of different stem cell types has been investigated, and adult stem cells and mesenchymal stem cells, in particular, have displayed the highest potential (Dulak et al., 2015; Liu, 2017). Mesenchymal stem cells (MSCs) are multipotent adult stem cells that can be easily isolated from the bone marrow (BM), adipose tissue, or umbilical cord and are able to differentiate into adipocytes, chondrocytes, osteoblasts, myocytes, smooth muscle cells, and neuron-like cells (Almalki and Agrawal, 2016; Liu, 2017; McKee and Chaudhry, 2017; Saidova and Vorobjev, 2020). Physiologically, MSCs and adult stem cells, in general, reside in specific “niches” which not only provide an anatomical location but also support self-renewal and stemness maintenance through biochemical and biophysical cues (Morrison and Spradling, 2008; Walker et al., 2008). In the last decade, increasing efforts have been made in designing and fabricating 3D synthetic niches, aiming to efficiently control MSCs’ fate in vitro and produce therapeutic cells on a large scale (Joddar and Ito, 2013). These scaffolds are conceived to emulate the native microenvironment by finely tuning the substrate physical properties, such as nanotopography (Dalby et al., 2007; Engel et al., 2009; McMurray et al., 2011), material stiffness (Engler et al., 2006; Khatiwala et al., 2009; Winer et al., 2009), and microgeometry (X. Li et al., 2012; Naito et al., 2011; Nerurkar et al., 2011). In the panorama of 3D cell culture systems for stem cell expansion, an innovative engineered substrate mimicking the native stem cell niche, called Nichoid, has recently been developed. It is based on an elementary and easily reproducible microarchitecture. Indeed, it is composed of a 3D interconnection of grids and columns able to create perfectly defined pores at the micrometric scale (Ricci et al., 2017). The main peculiarity of this scaffold is the use of two-photon polymerization (2PP) as a fabrication method, a technique enabling the creation of 3D structures based on a computer-generated model, with control of scaffold geometry at the cellular level (10 μm) and spatial resolution beyond the diffraction limit, up to 100 nm (Wu et al., 2006; Malinauskas et al., 2010; Nguyen and Narayan, 2017). It is known from the literature that isotropic cytoskeletal forces promote self-renewal and pluripotency, together with low extracellular loads and low oxygen concentration (McBeath et al., 2004; Guilak et al., 2009; Gilbert et al., 2010; Wan et al., 2010; Nava et al., 2012). The Nichoid substrate was conceived with a 3D microtopology capable of exerting an isotropic system of adhesion forces on cells while reducing cytoskeletal tension. The pore microgeometry is proven to promote stem cell homing inside the structure (Raimondi et al., 2013). Therefore, the fundamental property reproduced on cells by the Nichoid scaffold is the capability to interact mechanically with cells at the single-cell scale, thanks to the very high spatial resolution of the technique used for its microfabrication. The “niche effect” has been investigated by our group on murine embryonic stem cells (mESCs), human and rat bone marrow MSCs, and murine neural precursor cells (NPCs); all these studies have highlighted the Nichoid’s ability to maintain stemness and pluripotency genes switched on at higher levels compared to traditional flat substrates, with no need for adding exogenous soluble factors or feeder layers (Nava et al., 2016a; Nava et al., 2016b; Bonandrini et al., 2018; Carelli et al., 2020; Remuzzi et al., 2020; Rey et al., 2020). This phenomenon occurs thanks to the forces that the Nichoid provides to cells, thus inducing genetic reprogramming by controlling the cytoskeletal tension (Jacchetti et al., 2021). These results suggest that the Nichoid structure is able to emulate the native stem niche microenvironment in terms of self-renewal and stemness conservation, dictating stem cell fate.
In all the aforementioned studies, proliferation of stem cells inside the polymerized niches was also assessed and compared with that in bidimensional controls, demonstrating the ability of the 3D Nichoid to allow and promote cell expansion.
The aim of this work is to go beyond the examination of mesenchymal stemness, thus interrogating the whole transcriptome of these cells cultured inside the Nichoid, in order to deeper understand gene expression and, thus, better elucidate all MSCs’ physiological functions and therapeutic modes of action (Pittenger et al., 2019). We, thus, performed a complete transcriptome analysis, investigating gene modulation and all the biological functions that are modified by culturing MSCs on 3D Nichoids and comparing them with those of MSCs grown on the recently introduced 2D Nichoid, a substrate consisting of a single layer of polymerized grids that we chose as the bidimensional control since it not only allowed a spreading expansion of cells but also enabled us to evaluate how the pure 3D rearrangement of cells can influence their entire genomic profile.
2 Materials and methods
2.1 Nichoid fabrication
In this work, we compared Nichoids with two geometries: a single-level pattern (2D Nichoids) and a three-level pattern (3D Nichoids). 2D and 3D micro-patterned substrates were fabricated through 2PP (Raimondi et al., 2012) of a negative hybrid organic–inorganic photoresist named SZ2080 (Ovsianikov et al., 2008) and composed of a 2:8 molar ratio of zirconium propoxide (Sigma-Aldrich, United States) and methacryloxypropil trimethoxysilane (Sigma-Aldrich, United States) (Raimondi et al., 2012; Danilevičius et al., 2013; Ricci et al., 2017). Then, 1% concentration of Irg photoinitiator (Irgacure 369, 2-benzyl-2-dimethylamino-1-(4-morpholinophenyl)-butanone-1) was added to accelerate photopolymerization (Suzuki et al., 2012). A 20 µL drop of resist was deposited on a 12 mm-diameter round glass coverslip suitable for optical microscopy (Bio Optica, Italy) and allowed to harden to a semi-solid state; micropatterns were then fabricated directly on the coverslip. The pulsed laser source employed for fabrication is an ytterbium (Yb):KWY system based on a cavity-dumped oscillator in mode-locking with 1 MHz repetition rate, 1,030 nm wavelength (near-infrared, NIR), and 300 fs pulse duration. The laser beam was focused inside the sample through a 100× magnification oil-immersion microscope objective with 1.4 numerical aperture (NA) (plan apochromat, Carl Zeiss, Germany) and displacement along the three axes controlled by software (Automation 3200 CNC Operator Interface, Aerotech, United States). A spatial light modulator (SLM) was introduced in the setup to speed up the fabrication by splitting the beam into multiple parallel foci (Zandrini et al., 2019). The employed laser power was 100 mW, about 17 mW for each focus of the six-foci mask, and the writing speed was 3 mm/s. There was no slicing in the z-direction since it was a single line of 3 µm resolution, while the hatching parameter was 300 nm among two lines of 400 nm, leading to a final resolution of about 1 µm in the xy plane. At the end of the laser writing, the unpolymerized photoresist was removed by leaning the sample in a 50% (v/v) methyl isobutyl ketone and 50% (v/v) isopropyl alcohol solution (Sigma-Aldrich, United States) for 20 min (LaFratta et al., 2007; Malinauskas et al., 2013; Ricci et al., 2017).
The 3D sample had the traditional Nichoid architecture (Ricci et al., 2017): 218 squared blocks made up of 5 × 5 (450 × 450 μm2) structures with a spacing of 15 μm (Supplementary Figures S1A, B). Each structure is composed of individual niches, the elementary unit of the structure, with size corresponding to 90 × 90 × 30 μm3. Inside, there were three “floors” of interconnected rods, with a graded spacing (10, 20, and 30 μm) in the transverse direction and a uniform spacing of 15 μm in the vertical direction. The 2D Nichoid had the same grid geometry, but since it was made up of only one “floor,” it had an overall thickness of 2 μm, resulting in a partial bi-dimensional scaffold (Supplementary Figures S1C, D) used as a control in all biological experiments.
2.2 Substrate preparation and cell seeding
To confine cells inside the surface covered by the polymerized structures and to avoid unwanted cell adhesion outside the Nichoid substrate, different strategies were implemented. First, Costar® 6-well plates with an ultra-low attachment surface (Corning Incorporated, United States) were used as supports for cell seeding both on 3D and 2D Nichoids in order to avoid cell adhesion and proliferation on the surface of the wells. Second, the diameter of the glass sample covered with the Nichoid is 8 mm. To avoid cell adhesion on the glass portion of the coverslip surrounding the patterned portion of the samples, the bottom of the culture wells was drilled to create a small hole, 7 mm in diameter, with respect to the diameter of the Nichoid-covered coverslips. Then, the glass samples covered with the Nichoid were glued to the bottom of the culture wells from below using the biocompatible LOCTITE AA 3321 glue (Henkel, Germany) (Supplementary Figure S2). The glass ring surrounding the Nichoid-covered area of the coverslip corresponded to the glued surface in this assembly. Finally, cells were suspended for seeding in 100 µL of culture medium only, that is, the exact volume needed to guarantee complete covering of the structures only and uniform distribution of cells. This procedure was mainly employed in the case of sample seeded for subsequent RNA extraction in order to limit the cell sample to cells expanded inside the Nichoid structures. For qualitative experiments, such as immunofluorescence image acquisitions, we only used the drop seeding technique, which nevertheless allowed us to ensure controlled disposition of cells on the polymerized structures.
Before seeding, all fabricated samples were sterilized by washing them twice with sterile deionized water, immersing in 70% ethanol for at least 30 min, washing again with sterile water, and letting them dry under a UV lamp (Raimondi et al., 2014).
Primary rat mesenchymal stem cells (rBMSCs), obtained from the bone marrow of Lewis rat and extracted at the Mario Negri Institute for Pharmacological Research (Milano, Italy) following the protocol presented by Zoja et al. (2012), were maintained in culture with an α-minimum essential medium (α-MEM, Gibco, Thermo Fisher Scientific, United States) supplemented by 20% fetal bovine serum (FBS) (EuroClone, Italy), 1% penicillin–streptomycin (EuroClone, Italy), 1% of L-glutamine 200 mM (EuroClone, Italy), and 100 mM of 1% sodium pyruvate (EuroClone, Italy). To avoid differentiation and stemness loss, cells were kept in culture for two passages only and then were detached by trypsin–EDTA solution 1X (EuroClone, Italy), resuspended in phosphate buffered saline (PBS, EuroClone, Italy), and counted using a cell counter (CytoSMART, Netherlands). A total of 3 × 104 cells were seeded on each substrate, and drop-seeded samples were incubated for 1 h to permit proper cell adhesion to the structures; the required volume of medium was then added inside each well.
2.3 Viability assay and morphology evaluation
To evaluate viability at the fourth day after seeding, live samples were incubated for 10 min with .5 µm Calcein-AM (Invitrogen, Thermo Fisher Scientific, United States) and .2 µM ethidium homodimer-1 (Invitrogen, Thermo Fisher Scientific, United States) diluted in the fresh culture medium. Calcein-AM distribution was also exploited for a morphological evaluation, and nuclei were stained with 1 µM DRAQ5™ (Thermo Fisher Scientific, United States).
Live fluorescence images were acquired by using the Nikon Ar1+ confocal microscope (Nikon, Japan), equipped with an incubator chamber and four wavelength diode lasers (λexcitation = 405/488/561/640 nm). Stained cells were imaged with a 60× immersion oil objective, with 1.4 NA and .13 working distance (WD). The pinhole was set to 1 Airy unit, and 1,024 × 1,024 pixel images were acquired as z-stack images. Cells grown on glass coverslips and 2D and 3D Nichoids were imaged with a 1 μm step, resulting in an acquisition depth of approximately 10 μm for the first two and a depth of approximately 40 μm for the latter.
After z-stack maximum projections, cellular and nuclear areas were measured through Fiji software v2.3.0/1.53f (Schindelin et al., 2012) by manually drawing cell and nucleus profiles.
2.4 RNA data collection
Total RNA was isolated at day 4 using the TRIzol reagent (Total RNA Isolation reagent, Invitrogen, Thermo Fisher Scientific, United States) following the standard protocol (Rio et al., 2010). The extracted RNA was quantified with the Qubit 2.0 Fluorometer (Invitrogen, Thermo Fisher Scientific, United States), and its quality and integrity were evaluated through the Agilent 2100 Bioanalyzer (Agilent Technology, United States). Complementary DNA (cDNA) libraries were prepared using the Universal Plus mRNA-Seq kit (Tecan Genomics, United States) and sequenced in a single-end 75 base pair mode on the NextSeq 500 platform (Illumina, IGA Technologies Services, Italy).
2.5 Raw data processing
The per-base quality of the sequenced reads contained in the fastq files was checked with FastQC v0.11.9 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). All samples were mapped to the mRatBN7.2 Rattus norvegicus reference genome (https://ftp.ensembl.org/pub/release-108/fasta/rattus_norvegicus/dna/), and the BAM files were sorted with STAR v2.7.0a (Dobin et al., 2013). Genes were annotated using standard Ensembl gene annotations, a high-quality annotation system used for vertebrate species that automatically indexes the genomic coordinates of each gene by plotting it onto genome assemblies (Aken et al., 2016). Gene expression levels were assessed by counting aligned reads with HTSeq v2.0.2 (Anders et al., 2015).
2.6 Differential expression and functional enrichment analyses
The DESeq2 package of R/Bioconductor (Love et al., 2014) was used to analyze the counts matrix produced by HTseq and to identify differentially expressed genes (DEGs) between rBMSCs grown inside the 3D and 2D Nichoids. As implemented in the DESeq2 package, size factors are estimated to normalize the read counts. Values of |log2(FoldChange)| > 1 and false discovery rate (FDR)-adjusted p value
A functional enrichment analysis to investigate the biological role of DEGs was conducted with the web-based tool g:Profiler (Raudvere et al., 2019) using the Gene Ontology (GO) database. The list of DEGs was ranked by decreasing log2(FC), and an ordered enrichment test was executed. g:Profiler performs Fisher’s exact test to compute the p value of the enrichment of a pathway, and multiple-test corrections are applied (Reimand et al., 2019). The cutoff value for significantly enriched pathways was fixed to Benjamini–Hochberg (BH) FDR-adjusted p value
Subsequently, the GSEA desktop application v4.1.0 (Mootha et al., 2003; Subramanian et al., 2005) was employed to perform a gene set permutation test with default parameters. It takes as input a pre-ranked list made up of all the available genes coming from the expression profile, without applying any cutoff, and uses a permutation-based test aiming to determine whether the genes included in a gene set fall at the top or at the bottom of the ranked list, rather than being randomly distributed within the list (Subramanian et al., 2005). The analysis was carried out with the H (hallmark) and the C2 (curated) gene sets of the Molecular Signatures Database (MSigDB) v7.2 (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp), retrieved and adapted to the Rattus norvegicus species through the msigdbr package of R. The number of permutations was settled at 1,000, and an FDR q-value < .25 was chosen as the cutoff for statistical significance.
For a graphical visualization of interactions, a protein–protein interaction network was built with Cytoscape software v3.9.1 (Shannon et al., 2003), and the STRING functional enrichment network plugin was used to generate clusters. As query terms for the network, genes belonging to the most enriched GSEA gene sets were selected.
2.7 Quantitative real-time PCR
For validation of selected genes through real-time PCR (RT-PCR), RNA was extracted from 3D and 2D Nichoids as previously described, treated with the DNA-free™ DNA removal kit (Invitrogen, Thermo Fisher Scientific, United States), and reverse transcribed into cDNA using the SuperScript™ IV VILO™ kit (Invitrogen, Thermo Fisher Scientific, United States). Real-time PCR amplification was performed with the StepOnePlus™ Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, United States) and the GoTaq® qPCR Master Mix (Promega, United States). The following primers (Thermo Fischer Scientific, United States) were designed with NCBI Primer-BLAST and Primer3Plus web tools and cross-checked with the Ensembl gene database: Vcam1 (forward: CTGTTTGCAGTCTCTCAAGC; reverse: AGTCTCCAATCTGAGCGAGC), Ncam1 (forward: GTATGATGCCAAAGAAGCCAACA; reverse: TGTCTTGAACTCAGTGGCTG), Selplg (forward: GGGGCTGGAACTTCTGAGAC; reverse: CCGTGGGTGCTAGCCG), and Efemp1 (forward: GCTCCCCGCAGGTATCTTTT; reverse: ATCGGTGCATTGCGTGTATG). Gapdh (forward: GGCAAGTTCAACGGCACAG; reverse: CGCCAGTAGACTCCACGAC) was chosen as the housekeeping gene to normalize samples and calculate the gene expression level following the
2.8 Immunofluorescence assay
For immunofluorescence (IF) staining, samples were washed in PBS (Sigma-Aldrich, United States) and fixed in 10% formalin (Bio Optica, Italy) for 15 min. After three washes in glycine (Sigma-Aldrich, United States) to reduce autofluorescence, cells were permeabilized with PBS–.25% Triton® X-100 (Sigma-Aldrich, United States) for 10 min and blocked in PBS–.1% TWEEN® 20 (Sigma-Aldrich, United States) + 2% FBS (EuroClone, Italy) for 4 h. Primary antibodies to stain YAP (rabbit monoclonal anti-YAP antibody, 1:100, #14074, Cell Signaling Technology, United States) and N-cadherin (mouse anti-cadherin N antibody, 1:200, #94622, Immunological Science, Italy) were introduced and incubated at 4°C overnight. Samples were then washed three times in PBS–.1% TWEEN® 20 and incubated with Alexa Fluor® 647 and Alexa Fluor® 488 (1:750, Abcam, UK) secondary antibodies for 45 min at room temperature. FITC-conjugated phalloidin (Sigma-Aldrich, United States) was added, where needed, to stain the actin cytoskeleton. After three additional washes in PBS–.1% TWEEN® 20, the nuclei were stained with Hoechst 33342 (1:500, Thermo Fisher Scientific, United States) and incubated for 10 min at room temperature. Finally, samples were mounted with Mowiol DABCO® (Sigma-Aldrich, United States) and inspected with a confocal microscope (FLUOVIEW FV10i, Olympus, Italy) equipped with four diode lasers (405, 473, 559, and 635 nm excitation wavelengths). A ×60 water immersion objective with 1.2 NA was used, and the pinhole was set to 1 Airy unit. Z-stack images of around 35 µm depth for 3D Nichoids and 10 µm depth for 2D Nichoids were acquired with a 1 µm step.
Fiji software v2.3.0/1.53f (Schindelin et al., 2012) was used for channel merge and fluorescence intensity quantification.
2.9 Statistical analysis
Data were reported as mean values ± standard deviations. The statistical analyses were performed with Student’s t-test, and a p value
3 Results
As a starting point, we carried out a comparison in terms of viability and morphology among cells cultured on traditional glass coverslips and both the 2D and 3D Nichoids.
rBMSC viability was assessed after 4 days of culture, the same time point of RNA extraction used for sequencing analysis. At least three areas per sample were acquired, and a strong predominance (close to 100%) of vital cells (in green) with respect to apoptotic cells (in red) was observed on all the used substrates (Figure 1A).
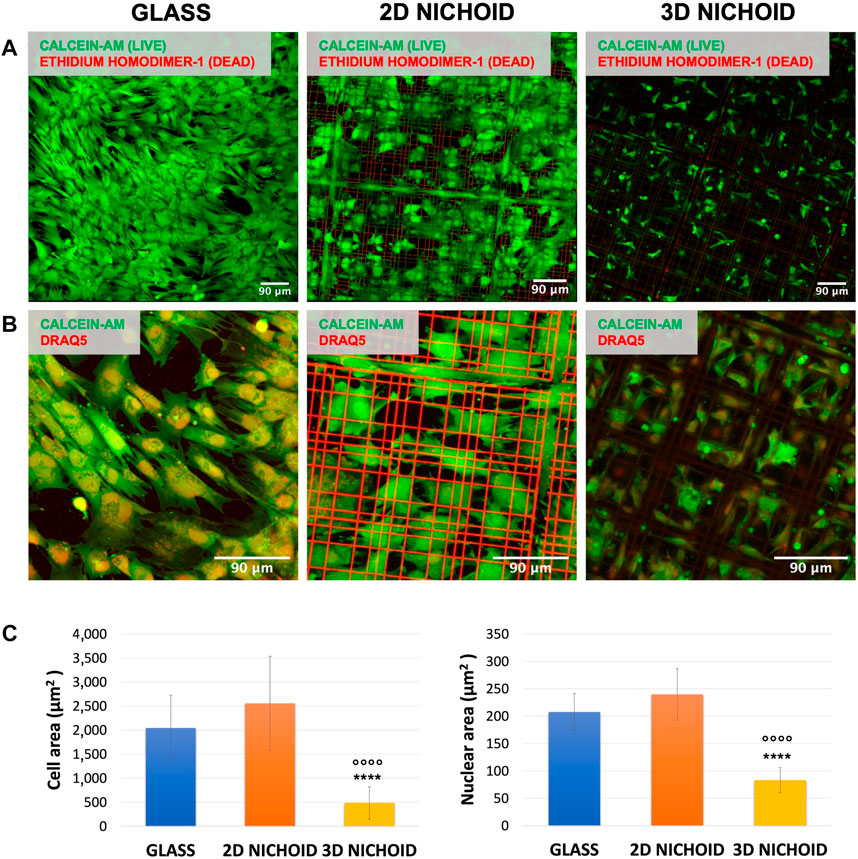
FIGURE 1. (A) Fluorescence images of live (green) and dead (red) rBMSCs cultured on glass coverslips and 2D and 3D Nichoids at day 4. Scale bar = 90 μm. (B) Fluorescence images of the cytoplasm (green) and nuclei (red) of rBMSCs cultured on glass coverslips and 2D and 3D Nichoids at day 4. Scale bar = 90 μm. (C) Quantification of the cellular and nuclear area in the three conditions. **** indicates p value
On the same samples, morphology analyses were performed, and the fluorescent dye DRAQ5 was added to also visualize the cell nucleus. Our observations revealed a variation between the three substrates in terms of cell shape. As visible in Figure 1B, cells cultured on 2D Nichoids were wide and spread, with morphology and dimension more comparable to those grown on glass coverslips, whereas in 3D Nichoids, cells tended to be more retained and confined. The quantification of cellular areas confirmed that cells inside the 3D environment are significantly smaller than those in both of the two flat substrates, whereas among the 2D Nichoid and glass coverslips, no discrepancy emerged. In addition, the quantification of nuclear areas highlighted a significant reduction in the dimensions of 3D-grown cells’ nuclei with respect to the other conditions (Figure 1C).
Based on these observations, we could state that rBMSCs cultured on 2D Nichoids had a bidimensional and spreading expansion compared to cells growing on glass coverslips. For this reason, we held it reasonable to employ a 2D Nichoid as a flat control in all subsequent experiments.
3.1 YAP nuclear/cytoplasmic localization
To corroborate the possibility of using a 2D Nichoid instead of a glass coverslip, we stained the mechano-transducer factor yes-associated protein (YAP) by immunofluorescence to compare its localization in bidimensional and tridimensional culture conditions. Immunofluorescence images (Figure 2A) clearly displayed that, just as it happened on glass coverslips (Remuzzi et al., 2020), in rBMSCs cultured on 2D Nichoids, YAP was almost exclusively localized in the nucleus, while it consistently translocated in the cytoplasm when cells were grown in a 3D environment. Indeed, on average, 93% of cells in 2D Nichoids retained YAP mostly inside the nucleus, whereas in 3D Nichoids, this percentage was around 12% (Figure 2B), and the YAP fluorescence level was significantly higher in the nuclei of cells cultured on bidimensional controls (Figure 2C).
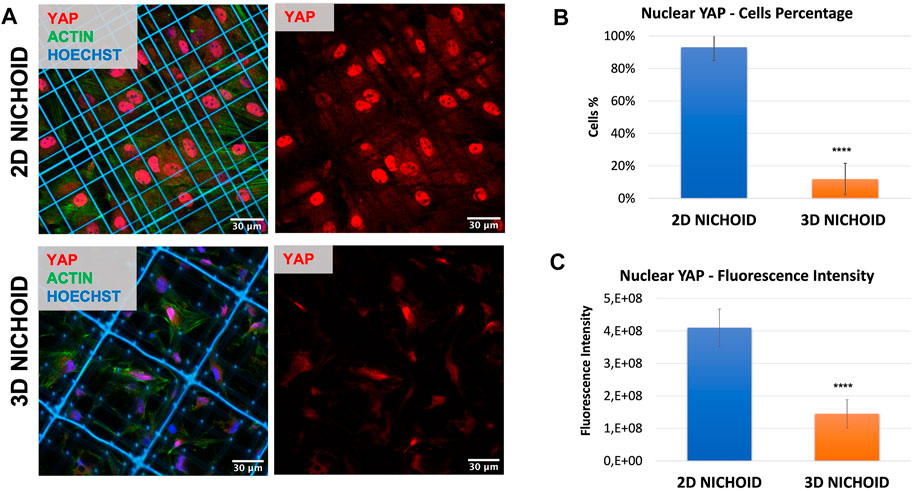
FIGURE 2. (A) Immunofluorescence images of rBMSCs with stained nuclei (blue), actin (green), and YAP (red) in 3D and 2D Nichoids. Scale bar = 30 µm. (B) Quantification of the rBMSC percentage of cells with YAP mainly localized in the nucleus; **** indicates p value
3.2 Differentially expressed genes identification and functional enrichment
To investigate how the previously observed morphological differences generated by Nichoid three-dimensionality translate into variations in terms of rBMSC gene expression, the profiling of the complete transcriptome of Nichoid cultures was performed. All genes resulting from the total RNA-seq are reported in Supplementary Table S1. Results from the bioinformatic pipeline showed that at 4 days from cell seeding, gene deregulation was affected in a significant way, since 654 genes appeared to be differentially expressed (392 upregulated and 262 downregulated) between 3D and 2D Nichoids, which are listed in Supplementary Table S2. All DEGs, defined as those genes that meet both |log2(FoldChange)| > 1 and false discovery rate (FDR)-adjusted p value
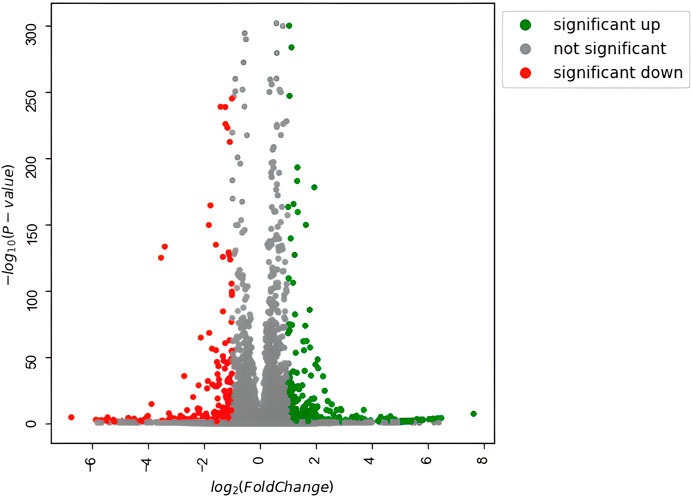
FIGURE 3. Volcano plot of differentially expressed genes. Significantly upregulated genes [adjusted p value
DEGs were successively used as input datasets for a biological enrichment analysis with the GO database to examine in which pathways they are mainly enriched. Interestingly, the highest dysregulated cellular components category has an extracellular matrix (GO:0031012), in particular, collagen-containing extracellular matrix (GO:0062023), apical plasma membrane (GO:0016324), apical part of the cell (GO:0045177), cell–cell junction (GO:0005911), actin cytoskeleton (GO:0015629), cluster of actin-based cell projections (GO:0098862), external side of the plasma membrane (GO:0009897), and cortical cytoskeleton (GO:0030863). The extracellular space appears to be dysregulated also in the biological functions category [extracellular matrix organization (GO:0030198) and extracellular structure organization (GO:0043062)], together with the regulation of cell adhesion (GO:0045785) and a series of pathways related to the muscle (GO:0033002; GO:0048659), bone (GO:0001503), cartilage (GO:0051216), and adipose tissue (GO:0045444) lineages. Moreover, in the molecular functions category, the top deregulated pathways are related to receptor activity (GO:0030546), cytokine activity (GO:0005125), cell adhesion molecule binding (GO:0050839), actin filament binding (GO:0051015), and integrin binding (GO:0005178) (Figure 4).
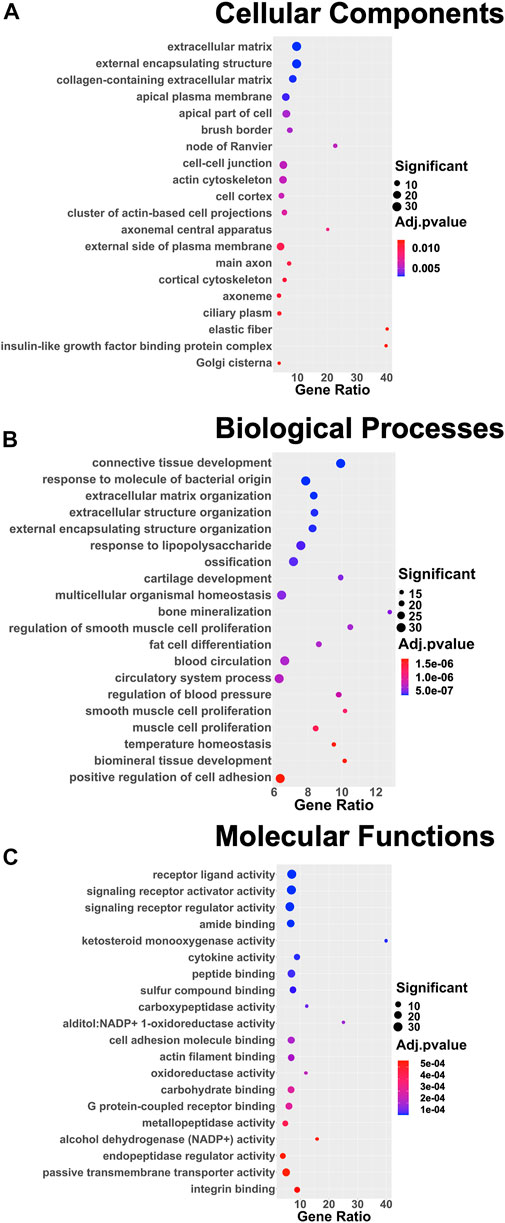
FIGURE 4. Dot plots of the 20 most enriched terms of the three different GO categories: (A) cellular components, (B) biological processes, (C) and molecular functions. The color of the dot represents the adjusted p value according to the color scale on the right side of the plots; the dimension of the dot represents the number of significant DEGs involved in the pathway; and the position of the dot on the x-axis represents the fraction of DEGs on the total number of genes belonging to the pathway.
For a deeper enrichment investigation, we exploited the GSEA approach. This analysis also showed, among all the aforementioned pathways, significant and marked modifications in the expression of genes related to cell adhesion molecules and the extracellular matrix. Indeed, the KEGG_CELL_ADHESION_MOLECULES_CAMS and BOWIE_RESPONSE_TO_EXTRACELLULAR_MATRIX gene sets had positive enrichment scores, ES = .51 and ES = .77, respectively, meaning that they were significantly (FDR q-value = .24 and FDR q-value = .17) overrepresented at the top of the ranked list of genes in the expression dataset. This is also graphically visible by the majority of black lines belonging to the gene set moved to the left. Interestingly, other positively and significantly enriched gene sets were REACTOME_INTERFERON_GAMMA_SIGNALING (ES = .56; FDR q-value = .14) and HALLMARK_INTERFERON_GAMMA_RESPONSE (ES = .44; FDR q-value = .002) (Figure 5A).
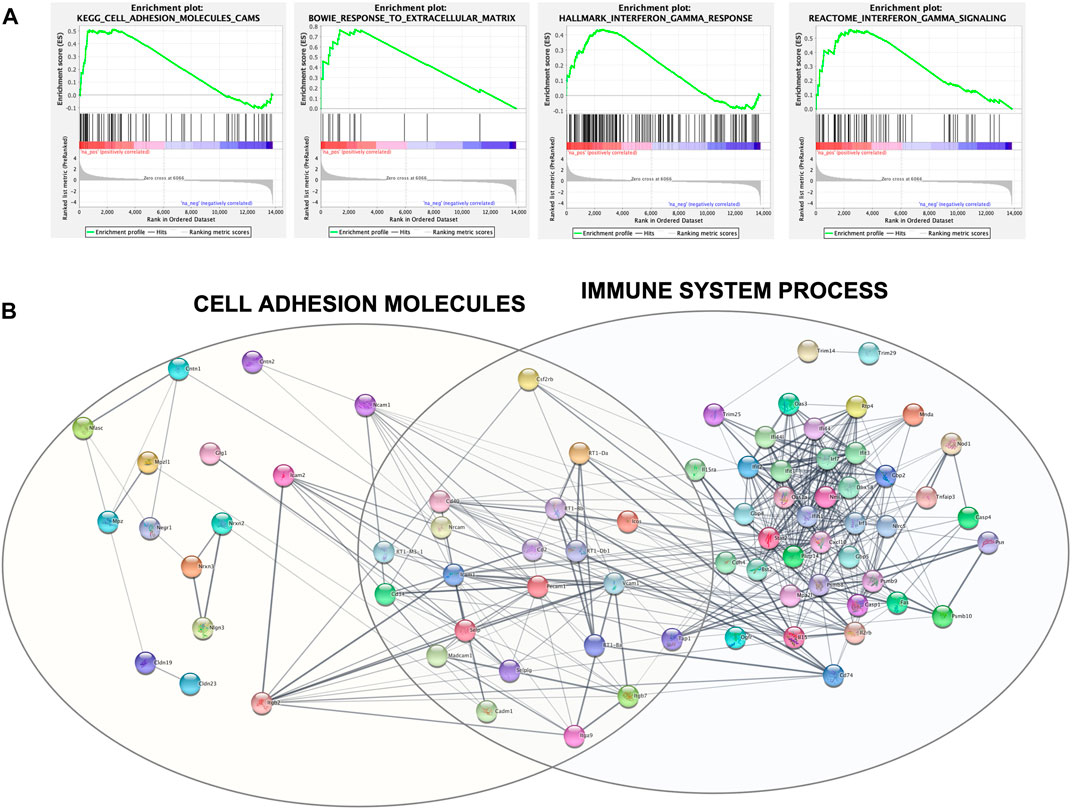
FIGURE 5. (A) GSEA output of the four mainly enriched gene sets. Each vertical black line corresponds to a gene, and its shift to the left indicates its positive enrichment. (B) Protein–protein interaction network built with Cytoscape and clustered with the STRING functional enrichment network plugin.
The set of genes belonging to these four significantly enriched gene sets was used as query terms for network construction to investigate potential connections. The network was divided into several clusters; the main two are shown in Figure 5B and include core genes with the highest rank metric scores of CAMs and IFN-
3.3 Cell adhesion molecules
We then focused our attention on adhesion molecules. Therefore, the expression of four upregulated genes belonging to enriched pathways was analyzed through RT-PCR, which confirmed the overexpression of Selplg, Vcam1, Ncam1, and Efemp1 detected from the sequencing in the 3D Nichoid with respect to the 2D Nichoid (Figure 6A).
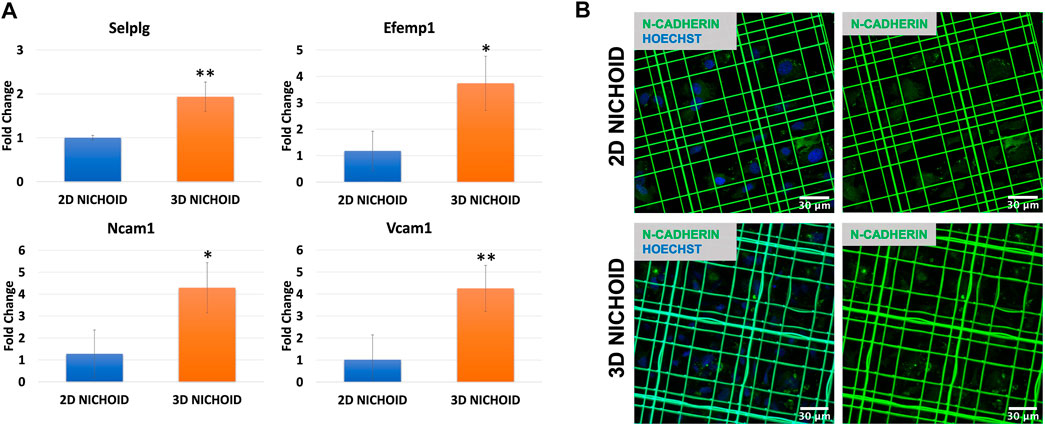
FIGURE 6. (A) Gene expression quantification through real-time PCR of Selplg (n = 3), Efemp1 (n = 4), Ncam1 (n = 3), and Vcam1 (n = 4) in rBMSCs grown in 3D and 2D Nichoids for 4 days. Gapdh was used as the housekeeping gene. Data are represented as mean ± SD; * indicates p value
To also evaluate homotypic cell–cell adhesions, we stained N-cadherin, a calcium-dependent cell adhesion protein that is predominantly expressed by MSCs (Hatta and Takeichi, 1986; Wuchter et al., 2007). Immunofluorescence images display a slight increase, although not significant, in the synthesis of this protein in rBMSCs grown in 3D Nichoids compared to bidimensional expansion. More interestingly, images showed a different localization of the proteins inside cells: in rBMSCs grown in 2D Nichoids, N-cadherin appears to be homogeneously distributed in the cytoplasm, whereas in 3D Nichoids, it is more localized in spots and distant from the nucleus in areas of cell–cell contacts (Figure 6B).
4 Discussion
Mesenchymal stem cells appear to be among the most promising stem cell sources for cell-based regenerative therapies, thanks mainly to their ease of extraction from several tissues and their differentiation potential toward a wide cohort of end-stage lineages (Han et al., 2019). Nevertheless, beyond their potential for differentiation and tissue regeneration, it has been demonstrated that MSCs have immunomodulatory and anti-inflammatory properties exercised through cytokine secretion and trophic activity (Zhao et al., 2016; Weiss and Dahlke, 2019; Song et al., 2020); they express a large number of genes encoding for a variety of regulatory proteins involved not only in mesoderm specification but also in other biological processes including inflammation and immune activation, cell motility, and communication (Phinney et al., 2006; Phinney, 2007; Phinney, 2009; Cruz-Barrera et al., 2020). Furthermore, it has been demonstrated that among the mostly expressed transcripts, there are also those encoding for structural and functional extracellular matrix (ECM) proteins, which may contribute to MSCs’ clinical regenerative potential, immune modulatory effects, and anti-inflammatory effects (J. Ren et al., 2011). These features make MSCs an ideal source for autologous cell-based treatments since their trophism could be even more significant for their therapeutic potential than their pluripotency (Phinney, 2007; Lukomska et al., 2019).
However, to enable these cells to maintain all their characteristics and properties, it is of fundamental relevance to expand them in an appropriate environment where they can replicate in vitro the correct disposition and conformation they possess in vivo through the restoration of their physiological shape and the proper interactions with both the ECM and surrounding cells. In order to model a more physiological environment than the one that can be obtained with a simple slide or plate, we employed the Nichoid, a 3D scaffold designed to replicate the architectural microenvironment of the native stem niche. It is fabricated through a two-photon laser polymerization technique, the only one capable of achieving a defined structure at the sub-micrometric level, which is essential to ensure control over MSC responses at the individual cell scale.
Cells cultured on Nichoid samples were first examined to assess morphological and viability differences between three culture conditions. In 2D Nichoids, cells adhered to the underlying glass, and they showed a flattened and spread shape, correlated to an anisotropic tensional state, with an area comparable to that of cells cultured on traditional glass coverslips. On the contrary, in 3D Nichoids, the cytoplasm of cells was distributed in a more confined space, and evidence from several sources in the literature shows that this morphology is linked to reduced cytoskeletal tensions on the nucleus (Jacchetti et al., 2021) and to a greater cell multipotency (Nava et al., 2012; Saei Arezoumand et al., 2017). Despite these morphological differences, the viability assay did not show a significant difference, confirming the biocompatibility of the employed resin and demonstrating that the scaffold material in both 3D and 2D substrates does not impact rBMSC viability. These inspections allowed substituting the standard glass coverslip as the bidimensional control with the 2D Nichoid in all experiments, allowing us to determine the mere architectural effect of the scaffold tridimensionality on rBMSC gene expression and protein localization, separating it from the influence of the material properties relevant to the scaffold. In support of this hypothesis, our immunofluorescence results confirmed the same behavior observed by Remuzzi et al. (2020), where an increased percentage of the cytoplasmic fraction of YAP in rBMSCs cultured in 3D Nichoids was appreciable with respect to rBMSCs grown in monolayers on flat controls. In particular, this occurred despite differences in the number of cells seeded, time points, and, above all, substrate used as the flat control, which, in our case, was the 2D Nichoid in place of glass coverslips.
To investigate the consequent variation in gene expression inside the two different culture substrates, the transcriptome of rBMSCs cultured on Nichoids was sequenced and analyzed, aiming at determining the “niche effect” provided by the engineered cues of the 3D Nichoid architecture on the whole stem cell genetic response. For this purpose, the total RNA extracted from rBMSCs cultured on 2D and 3D Nichoids was profiled for the first time using next-generation sequencing technology, which allows for a more straightforward and affordable analysis of gene regulation compared to microarrays and other sequencing techniques with a higher sensitivity in detecting differentially expressed genes (J. Li et al., 2016; Marguerat and Bähler, 2010).
The measured differential expression confirmed that the morphological differences induced by the two substrates are enough to lead to a significant genetic reprogramming of cellular processes, and the experimental design in use leads to the conclusion that these results are attributable exclusively to the three-dimensionality of the culture substrate.
To understand in which pathways they were mainly involved, the identified DEGs were subsequently subjected to functional enrichment for GO annotation with g:Profiler. The list of the top 20 affected pathways in the three categories seems to suggest that there was a deregulation in the entire mechano-transduction chain, starting from the extracellular matrix, moving through cell adhesions, and, by means of a reorganization of the actin cytoskeleton, reaching the nucleus. The modifications in this chain influenced biological processes and molecular functions of rBMSCs that were different between the 3D and 2D Nichoids, which mainly involved differentiation toward different lineages, receptors, and cytokine activity. The GSEA further underlined the discrepancy between the two culture systems in the expression of genes encoding for cell adhesion molecules and in the response to the ECM. This analysis revealed that two gene sets related to IFN-
Following the results obtained from the enrichment analyses, we focused our attention on this gene set, confirming the upregulation of Selplg, Vcam1, Ncam1, and Efemp1 in the 3D Nichoid with respect to the 2D Nichoid via RT-PCR experiments. Selplg encodes for the P-selectin glycoprotein ligand-1, which is part of the adhesion molecules involved in the regulation of leukocyte migration in response to inflammatory stimuli (Hirata et al., 2000; Hidalgo et al., 2007); interestingly, it has been shown how MSCs expressing this gene can rapidly move to the site of inflammation, exerting a superior anti-inflammatory effect (Levy et al., 2013; Liao et al., 2016). In addition to being an MSC marker, Vcam1 (vascular cell adhesion molecule 1) is recognized to play a critical role in MSC-related immunosuppression, and its expression is induced by the simultaneous presence of IFN-
The upregulation of Vcam1 in 3D Nichoids could also be related to a higher number of cell–cell contacts mediated by N-cadherin that we investigated through immunofluorescence staining. It has been demonstrated that cell–cell adhesion mediated by N-cadherin promotes the expression of Vcam1 by activating the nuclear factor κB (NF-κB) pathway via the platelet-derived growth factor receptor beta (PDGFRβ) (Aomatsu et al., 2014; Chosa and Ishisaki, 2018). Our results show that this contact protein was slightly more expressed in 3D Nichoids, but above all, it was more aggregated; this suggests that the 3D structure of the scaffold promoted MSC-homotypic cell–cell adhesion among neighboring cells.
Cell–cell adhesions through N-cadherin also underlay the translocation of the mechano-transduction factor YAP from the nucleus to the cytoplasm. YAP, together with the transcriptional co-activator with a PDZ-binding motif (TAZ), is a key driver of stem cell behavior since it responds to physical stimuli, such as ECM stiffness, cell geometry, or mechanical forces of the cytoskeleton, with specific transcriptional programs (Donnaloja et al., 2020; Y. Li et al., 2021; Piccolo et al., 2014). As a matter of fact, the spatial organization of focal adhesions in cells inside the 3D Nichoid modifies the transmission of forces to the nucleus, determining weaker forces than in spreading cells and, thus, affecting the nuclear import of signaling molecules (Jacchetti et al., 2021). In addition to the effect of focal adhesions, the presence of N-cadherin, which is involved in mechano-transduction, affects the perception of the microenvironment stiffness, reducing the contractile state of the cell and subsequently YAP translocation (Cosgrove et al., 2016; Qin et al., 2020; Zhang et al., 2021).
YAP is a fundamental downstream effector of the Hippo pathway, which plays a role in several mechanisms such as development, stem cell self-renewal and differentiation, regeneration, immune modulation, and cancer (Moya and Halder, 2019; Dey et al., 2020; LeBlanc et al., 2021). Among target genes of activated YAP/TAZ, in our transcriptome analysis, we found both Snai2 (zinc finger protein SNAI2) and Jag1 (protein jagged-1) to be overexpressed in 3D Nichoids. It has been proven that Snai2 is a transcription factor included among the major upregulators for BM–MSC maintenance, fate decision, cell adhesion, and cell structure regulation (Sánchez-Luis et al., 2020; Wei et al., 2020). In turn, Snai2 can bind to Cxcl12 (C-X-C motif chemokine 12) promoters; Cxcl12, which we also found slightly over-expressed, is critical to retain both MSCs and hematopoietic stem cells in the bone marrow niche and also for hematopoietic stem and progenitor cells’ quiescent state maintenance (Roson-Burgo et al., 2016). Interestingly, Giri et al. (2020) also demonstrated that BM–MSC-secreted Cxcl12, cooperating with CCL2 (C-C motif chemokine 2), exercised an anti-inflammatory capacity in toxic colitis by upregulating IL10 expression in CCR2+ macrophages. Furthermore, Jag1, which is part of the Notch pathway, has been shown to be a potent pro-angiogenic regulator (Benedito et al., 2009).
To conclude, our results demonstrated how restoring both the correct morphology of the stem cells and the correct adhesions in an in vitro culture system is important to preserve the native cell functionality. It is known that adhesion molecules are the essential initial point of a mechano-transduction chain that starts from the membrane and, through the cytoskeletal organization and tension, reaches the nucleus, conditioning several biological aspects of the cell, such as transcription. In this context, the Nichoid proved to be able to perform these tasks and, thus, demonstrates, thanks to its peculiar capability to restore 3D cell adhesions, especially those among neighboring cells, to be not only an ideal substrate for MSC expansion and stemness maintenance but also a means for controlling other specific MSC features such as immunomodulation, thus supporting its potential for clinical application in immune-based diseases.
Data availability statement
The data presented in the study are deposited in the GEO repository, accession number GSE199846: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE199846.
Author contributions
CT, SO, and EJ designed the experiments and analyzed and interpreted the data. CT fabricated both the 3D and 2D microstructured substrates. CT and SO performed cell cultures, viability and morphological assays, RNA extraction, and RT-PCR experiments. CT, EJ, CM, and FD performed immunofluorescence experiments and confocal image acquisition. CT and PP performed RNA-seq raw data processing, differential expression, and functional enrichment analyses. RO and GC provided scientific supervision on the 2PP fabrication process. MR, SB, and SC supervised the work, revised the manuscript, and provided financial support. All authors read and approved the final version of the article.
Funding
Funded by the European Union grant ERC-AdG-2021 project BEACONSANDEGG; G.A. nr 101053122. Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the ERC. Neither the European Union nor the granting authority can be held responsible for them. Funded also by an unrestricted grant from “Fondazione Romeo ed Enrica Invernizzi ” to Stefano Biffo.
Acknowledgments
The authors wish to thank Joy Atoe for generating part of the data in the context of her master’s thesis.
Conflict of interest
MTR, RO, and GC are co-founders of a university spin-off company, MOAB srl, and hold shares.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.945474/full#supplementary-material
References
Aken, B. L., Ayling, S., Barrell, D., Clarke, L., Curwen, V., Fairley, S., et al. (2016). The Ensembl gene annotation system. Database J. Biol. Databases Curation 2016, baw093. doi:10.1093/database/baw093
Almalki, S. G., and Agrawal, D. K. (2016). Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation 92 (1–2), 41–51. doi:10.1016/j.diff.2016.02.005
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 31 (2), 166–169. doi:10.1093/bioinformatics/btu638
Aomatsu, E., Chosa, N., Nishihira, S., Sugiyama, Y., Miura, H., and Ishisaki, A. (2014). Cell-cell adhesion through N-cadherin enhances VCAM-1 expression via PDGFRβ in a ligand-independent manner in mesenchymal stem cells. Int. J. Mol. Med. 33 (3), 565–572. doi:10.3892/ijmm.2013.1607
Benedito, R., Roca, C., Sörensen, I., Adams, S., Gossler, A., Fruttiger, M., et al. (2009). The Notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell 137 (6), 1124–1135. doi:10.1016/j.cell.2009.03.025
Bonandrini, B., Figliuzzi, M., Conti, S., Zandrini, T., Osellame, R., Cerullo, G., et al. (2018). Effect of the 3D artificial nichoid on the morphology and mechanobiological response of mesenchymal stem cells cultured in vitro. Cells 9, 1873–1928. doi:10.3390/cells9081873
Carelli, S., Giallongo, T., Rey, F., Barzaghini, B., Zandrini, T., Pulcinelli, A., et al. (2020). Neural precursors cells expanded in a 3D micro-engineered niche present enhanced therapeutic efficacy in vivo. Nanotheranostics 5 (1), 8–26. doi:10.7150/ntno.50633
Chosa, N., and Ishisaki, A. (2018). Two novel mechanisms for maintenance of stemness in mesenchymal stem cells: SCRG1/BST1 axis and cell–cell adhesion through N-cadherin. Jpn. Dent. Sci. Rev. 54 (1), 37–44. doi:10.1016/j.jdsr.2017.10.001
Cosgrove, B. D., Mui, K. L., Driscoll, T. P., Caliari, S. R., Mehta, K. D., Assoian, R. K., et al. (2016). N-Cadherin adhesive interactions modulate matrix mechanosensing and fate commitment of mesenchymal stem cells. Nat. Mater 15, 1297–1306. doi:10.1038/nmat4725
Cruz-Barrera, M., Flórez-Zapata, N., Lemus-Diaz, N., Medina, C., Galindo, C. C., González-Acero, L. X., et al. (2020). Integrated analysis of transcriptome and secretome from umbilical cord mesenchymal stromal cells reveal new mechanisms for the modulation of inflammation and immune activation. Front. Immunol. 11, 575488–575519. doi:10.3389/fimmu.2020.575488
Dalby, M. J., Gadegaard, N., Tare, R., Andar, A., Riehle, M. O., Herzyk, P., et al. (2007). The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 6 (12), 997–1003. doi:10.1038/nmat2013
Danilevičius, P., Rekštyte, S., Balčiunas, E., Kraniauskas, A., Širmenis, R., Baltriukiene, D., et al. (2013). Laser 3D micro/nanofabrication of polymers for tissue engineering applications. Opt. Laser Technol. 45 (1), 518–524. doi:10.1016/j.optlastec.2012.05.038
Debnath, J., and Brugge, J. S. (2005). Modelling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer 5 (9), 675–688. doi:10.1038/nrc1695
Dey, A., Varelas, X., and Guan, K.-L. (2020). Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 19 (7), 480–494. doi:10.1038/s41573-020-0070-z
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29 (1), 15–21. doi:10.1093/bioinformatics/bts635
Donnaloja, F., Carnevali, F., Jacchetti, E., and Raimondi, M. T. (2020). Lamin A/C mechanotransduction in laminopathies. Cells 9 (5), 1306. doi:10.3390/cells9051306
Dulak, J., Szade, K., Szade, A., Nowak, W., and Józkowicz, A. (2015). Adult stem cells: Hopes and hypes of regenerative medicine. Acta Biochim. Pol. 62 (3), 329–337. doi:10.18388/abp.2015_1023
Edmondson, R., Broglie, J. J., Adcock, A. F., and Yang, L. (2014). Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 12 (4), 207–218. doi:10.1089/adt.2014.573
Engel, E., Martínez, E., Mills, C. A., Funes, M., Planell, J. A., and Samitier, J. (2009). Mesenchymal stem cell differentiation on microstructured poly (methyl methacrylate) substrates. Ann. Anat. 191 (1), 136–144. doi:10.1016/j.aanat.2008.07.013
Engler, A. J., Sen, S., Sweeney, H. L., and Discher, D. E. (2006). Matrix elasticity directs stem cell lineage specification. Cell 126 (4), 677–689. doi:10.1016/j.cell.2006.06.044
Gilbert, P. M., Havenstrite, K. L., Magnusson, K. E. G., Sacco, A., Leonardi, N. A., Kraft, P., et al. (2010). Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 329 (5995), 1078–1081. doi:10.1126/science.1191035
Giri, J., Das, R., Nylen, E., Chinnadurai, R., and Galipeau, J. (2020). CCL2 and CXCL12 derived from mesenchymal stromal cells cooperatively polarize IL-10+ tissue macrophages to mitigate gut injury. Cell Rep. 30 (6), 1923–1934. doi:10.1016/j.celrep.2020.01.047
Greiner, A. M., Richter, B., and Bastmeyer, M. (2012). Micro-engineered 3D scaffolds for cell culture studies. Macromol. Biosci. 12 (10), 1301–1314. doi:10.1002/mabi.201200132
Guilak, F., Cohen, D. M., Estes, B. T., Gimble, J. M., Liedtke, W., and Chen, C. S. (2009). Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell 5 (1), 17–26. doi:10.1016/j.stem.2009.06.016
Han, Y., Li, X., Zhang, Y., Han, Y., Chang, F., and Ding, J. (2019). Mesenchymal stem cells for regenerative medicine. Cells 8 (8), 886. doi:10.3390/cells8080886
Hatta, K., and Takeichi, M. (1986). Expression of N-cadherin adhesion molecules associated with early morphogenetic events in chick development. Nature 324, 447–449. doi:10.1038/320447a0
Heidary Rouchi, A., and Mahdavi-Mazdeh, M. (2015). Regenerative medicine in organ and tissue transplantation: Shortly and practically achievable? Int. J. Organ Transplant. Med. 6 (3), 93–98.
Hidalgo, A., Peired, A. J., Wild, M., Vestweber, D., and Frenette, P. S. (2007). Complete identification of E-selectin ligands on neutrophils reveals distinct functions of PSGL-1, ESL-1, and CD44. Immunity 26 (4), 477–489. doi:10.1016/j.immuni.2007.03.011
Hirata, T., Merrill-Skoloff, G., Aab, M., Yang, J., Furie, B. C., and Furie, B. (2000). P-selectin glycoprotein ligand 1 (Psgl-1) is a physiological ligand for E-selectin in mediating T helper 1 lymphocyte migration. J. Exp. Med. 192 (11), 1669–1676. doi:10.1084/jem.192.11.1669
Jacchetti, E., Nasehi, R., Boeri, L., Parodi, V., Negro, A., Albani, D., et al. (2021). The nuclear import of the transcription factor MyoD is reduced in mesenchymal stem cells grown in a 3D micro-engineered niche. Sci. Rep. 11 (1), 3021–3119. doi:10.1038/s41598-021-81920-2
Joddar, B., and Ito, Y. (2013). Artificial niche substrates for embryonic and induced pluripotent stem cell cultures. J. Biotechnol. 168 (2), 218–228. doi:10.1016/j.jbiotec.2013.04.021
Kapałczyńska, M., Kolenda, T., Przybyła, W., Zajączkowska, M., Teresiak, A., Filas, V., et al. (2016). 2D and 3D cell cultures – a comparison of different types of cancer cell cultures. Archives Med. Sci. 14 (4), 910–919. doi:10.5114/aoms.2016.63743
Khatiwala, C. B., Kim, P. D., Peyton, S. R., and Putnam, A. J. (2009). ECM compliance regulates osteogenesis by influencing MAPK signaling downstream of RhoA and ROCK. J. Bone Mineral Res. 24 (5), 886–898. doi:10.1359/jbmr.081240
Kilian, K. A., Bugarija, B., Lahn, B. T., and Mrksich, M. (2010). Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. U. S. A. 107 (11), 4872–4877. doi:10.1073/pnas.0903269107
Kim, D. S., Jang, I. K., Lee, M. W., Ko, Y. J., Lee, D. H., Lee, J. W., et al. (2018). Enhanced immunosuppressive properties of human mesenchymal stem cells primed by interferon-γ. EBioMedicine 28, 261–273. doi:10.1016/j.ebiom.2018.01.002
Klinker, M. W., Marklein, R. A., Lo Surdo, J. L., Wei, C. H., and Bauer, S. R. (2017). Morphological features of IFN-γ-stimulated mesenchymal stromal cells predict overall immunosuppressive capacity. Proc. Natl. Acad. Sci. U. S. A. 114 (13), E2598–E2607. doi:10.1073/pnas.1617933114
Kolios, G., and Moodley, Y. (2012). Introduction to stem cells and regenerative medicine. Respiration 85 (1), 3–10. doi:10.1159/000345615
LaFratta, C. N., Fourkas, J. T., Baldacchini, T., and Farrer, R. A. (2007). Multiphoton fabrication. Angew. Chem. - Int. Ed. 46, 6238–6258. doi:10.1002/anie.200603995
LeBlanc, L., Ramirez, N., and Kim, J. (2021). Context-dependent roles of YAP/TAZ in stem cell fates and cancer. Cell. Mol. Life Sci. 78 (9), 4201–4219. doi:10.1007/s00018-021-03781-2
Levy, O., Zhao, W., Mortensen, L. J., LeBlanc, S., Tsang, K., Fu, M., et al. (2013). MRNA-engineered mesenchymal stem cells for targeted delivery of interleukin-10 to sites of inflammation. Blood 122 (14), e23–e32. doi:10.1182/blood-2013-04-495119
Li, J., Hou, R., Niu, X., Liu, R., Wang, Q., Wang, C., et al. (2016). Comparison of microarray and RNA-Seq analysis of mRNA expression in dermal mesenchymal stem cells. Biotechnol. Lett. 38 (1), 33–41. doi:10.1007/s10529-015-1963-5
Li, X., Liu, H., Niu, X., Yu, B., Fan, Y., Feng, Q., et al. (2012). The use of carbon nanotubes to induce osteogenic differentiation of human adipose-derived MSCs in vitro and ectopic bone formation in vivo. Biomaterials 33 (19), 4818–4827. doi:10.1016/j.biomaterials.2012.03.045
Li, Y., Wang, J., and Zhong, W. (2021). Regulation and mechanism of YAP/TAZ in the mechanical microenvironment of stem cells (Review). Mol. Med. Rep. 24 (1), 506–511. doi:10.3892/mmr.2021.12145
Liao, W., Pham, V., Liu, L., Riazifar, M., Pone, E. J., Zhang, S. X., et al. (2016). Mesenchymal stem cells engineered to express selectin ligands and IL-10 exert enhanced therapeutic efficacy in murine experimental autoimmune encephalomyelitis. Biomaterials 77, 87–97. doi:10.1016/j.biomaterials.2015.11.005
Liu, T. M. (2017). Stemness of mesenchymal stem cells. J. Stem Cell Ther. Transplant. 1 (1), 071–073. doi:10.29328/journal.jsctt.1001008
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550–621. doi:10.1186/s13059-014-0550-8
Lukomska, B., Stanaszek, L., Zuba-Surma, E., Legosz, P., Sarzynska, S., and Drela, K. (2019). Challenges and controversies in human mesenchymal stem cell therapy. Stem Cells Int. 2019, 1–10. doi:10.1155/2019/9628536
Mahmud, G., Campbell, C. J., Bishop, K. J. M., Komarova, Y. A., Chaga, O., Soh, S., et al. (2009). Directing cell motions on micropatterned ratchets. Nat. Phys. 5 (8), 606–612. doi:10.1038/nphys1306
Malinauskas, M., Danilevicius, P., Baltriukien, D., Purlys, V., Paipulas, D., Bukelskien, V., et al. (2010). 3D aritificial polymeric scaffolds for stem cell growth fabricated by femtosecond laser. Lithuanian J. Phys. 50(1), 75–82. doi:10.3952/lithjphys.50121
Malinauskas, M., Farsari, M., Piskarskas, A., and Juodkazis, S. (2013). Ultrafast laser nanostructuring of photopolymers: A decade of advances. Phys. Rep. 533 (1), 1–31. doi:10.1016/j.physrep.2013.07.005
Marguerat, S., and Bähler, J. (2010). RNA-seq: From technology to biology. Cell. Mol. Life Sci. 67 (4), 569–579. doi:10.1007/s00018-009-0180-6
McBeath, R., Pirone, D. M., Nelson, C. M., Bhadriraju, K., and Chen, C. S. (2004). Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 6 (4), 483–495. doi:10.1016/S1534-5807(04)00075-9
McKee, C., and Chaudhry, G. R. (2017). Advances and challenges in stem cell culture. Colloids Surfaces B Biointerfaces 159, 62–77. doi:10.1016/j.colsurfb.2017.07.051
McMurray, R. J., Gadegaard, N., Tsimbouri, P. M., Burgess, K. V., McNamara, L. E., Tare, R., et al. (2011). Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat. Mater. 10 (8), 637–644. doi:10.1038/nmat3058
Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., et al. (2003). PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34 (3), 267–273. doi:10.1038/ng1180
Morrison, S. J., and Spradling, A. C. (2008). Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell 132 (4), 598–611. doi:10.1016/j.cell.2008.01.038
Moya, I. M., and Halder, G. (2019). Hippo–YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 20 (4), 211–226. doi:10.1038/s41580-018-0086-y
Murphy, K. C., Whitehead, J., Falahee, P. C., Zhou, D., Simon, S. I., and Leach, J. K. (2017). Multifactorial experimental design to optimize the anti- inflammatory and proangiogenic potential of mesenchymal stem cell spheroids. Stem Cells 35 (6), 1493–1504. doi:10.1002/stem.2606
Naito, H., Dohi, Y., Zimmermann, W. H., Tojo, T., Takasawa, S., Eschenhagen, T., et al. (2011). The effect of mesenchymal stem cell osteoblastic differentiation on the mechanical properties of engineered bone-like tissue. Tissue Eng. - Part A 17 (17–18), 2321–2329. doi:10.1089/ten.tea.2011.0099
Nava, M. M., Maggio, N., Di Zandrini, T., Cerullo, G., Osellame, R., Martin, I., et al. (2016a). Synthetic niche substrates engineered via two-photon laser polymerization for the expansion of human mesenchymal stromal cells. J. Tissue Eng. Regen. Med. 11, 2836. doi:10.1002/term
Nava, M. M., Piuma, A., Figliuzzi, M., Cattaneo, I., Bonandrini, B., Zandrini, T., et al. (2016b). Two-photon polymerized “nichoid” substrates maintain function of pluripotent stem cells when expanded under feeder-free conditions. Stem Cell Res. Ther. 7, 132. doi:10.1186/s13287-016-0387-z
Nava, M. M., Raimondi, M. T., and Pietrabissa, R. (2012). Controlling self-renewal and differentiation of stem cells via mechanical cues. J. Biomed. Biotechnol. 2012, 1–12. doi:10.1155/2012/797410
Nelson, C. M., and Bissell, M. J. (2006). Of extracellular matrix, scaffolds, and signaling: Tissue architecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 22 (1), 287–309. doi:10.1146/annurev.cellbio.22.010305.104315
Nerurkar, N. L., Han, W., Mauck, R. L., and Elliott, D. M. (2011). Homologous structure-function relationships between native fibrocartilage and tissue engineered from MSC-seeded nanofibrous scaffolds. Biomaterials 32 (2), 461–468. doi:10.1016/j.biomaterials.2010.09.015
Nguyen, A. K., and Narayan, R. J. (2017). Two-photon polymerization for biological applications. Biochem. Pharmacol. 20 (6), 314–322. doi:10.1016/j.mattod.2017.06.004
Ovsianikov, A., Viertl, J., Chichkov, B., Oubaha, M., Maccraith, B., Giakoumaki, A., et al. (2008). Ultra-low shrinkage hybrid photosensitive material for two-photon polymerization microfabrication. ACS Nano 2, 2257. doi:10.1021/nn800451w
Pampaloni, F., Reynaud, E. G., and Stelzer, E. H. K. (2007). The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 8 (10), 839–845. doi:10.1038/nrm2236
Petersen, O. W., Ronnov-Jessen, L., Howlett, A. R., and Bissell, M. J. (1992). Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 89 (19), 9064–9068. doi:10.1073/pnas.89.19.9064
Phinney, D. G. (2009). A Sage view of mesenchymal stem cells. Int. J. Stem Cells 2 (1), 1–10. doi:10.15283/ijsc.2009.2.1.1
Phinney, D. G. (2007). Biochemical heterogeneity of mesenchymal stem cell populations: Clues to their therapeutic efficacy. Cell Cycle 6 (23), 2884–2889. doi:10.4161/cc.6.23.5095
Phinney, D. G., Hill, K., Michelson, C., DuTreil, M., Hughes, C., Humphries, S., et al. (2006). Biological activities encoded by the murine mesenchymal stem cell transcriptome provide a basis for their developmental potential and broad therapeutic efficacy. Stem Cells 24 (1), 186–198. doi:10.1634/stemcells.2004-0236
Piccolo, S., Dupont, S., and Cordenonsi, M. (2014). The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 94 (4), 1287–1312. doi:10.1152/physrev.00005.2014
Pittenger, M. F., Discher, D. E., Péault, B. M., Phinney, D. G., Hare, J. M., and Caplan, A. I. (2019). Mesenchymal stem cell perspective: cell biology to clinical progress. Npj Regen. Med. 4 (1), 22. doi:10.1038/s41536-019-0083-6
Qin, E. C., Ahmed, S. T., Sehgal, P., Vu, V. H., Kong, H., and Leckband, D. E. (2020). Comparative effects of N-cadherin protein and peptide fragments on mesenchymal stem cell mechanotransduction and paracrine function. Biomaterials 239, 119846. doi:10.1016/j.biomaterials.2020.119846
Raimondi, M. T., Eaton, S. M., Laganà, M., Aprile, V., Nava, M. M., Cerullo, G., et al. (2013). Three-dimensional structural niches engineered via two-photon laser polymerization promote stem cell homing. Acta Biomater. 9 (1), 4579–4584. doi:10.1016/j.actbio.2012.08.022
Raimondi, M. T., Eaton, S. M., Nava, M. M., Laganà, M., Cerullo, G., and Osellame, R. (2012). Two-photon laser polymerization: From fundamentals to biomedical application in tissue engineering and regenerative medicine. J. Appl. Biomaterials Funct. Mater. 10 (1), 56–66. doi:10.5301/jabfm.2012.9278
Raimondi, M. T., Nava, M. M., Eaton, S. M., Bernasconi, A., Vishnubhatla, K. C., Cerullo, G., et al. (2014). Optimization of femtosecond laser polymerized structural niches to control mesenchymal stromal cell fate in culture. Micromachines 2, 341–358. doi:10.3390/mi5020341
Raudvere, U., Kolberg, L., Kuzmin, I., Arak, T., Adler, P., Peterson, H., et al. (2019). G:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47 (W1), W191–W198. doi:10.1093/nar/gkz369
Reimand, J., Isserlin, R., Voisin, V., Kucera, M., Tannus-Lopes, C., Rostamianfar, A., et al. (2019). Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 14 (2), 482–517. doi:10.1038/s41596-018-0103-9
Remuzzi, A., Bonandrini, B., Tironi, M., Longaretti, L., Figliuzzi, M., Conti, S., et al. (2020). Effect of the 3D artificial nichoid on the morphology and mechanobiological response of mesenchymal stem cells cultured in vitro. Cells 9 (8), 1873. doi:10.3390/cells9081873
Ren, G., Zhao, X., Zhang, L., Zhang, J., L’Huillier, A., Ling, W., et al. (2010). Inflammatory cytokine-induced intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 in mesenchymal stem cells are critical for immunosuppression. J. Immunol. 184 (5), 2321–2328. doi:10.4049/jimmunol.0902023
Ren, J., Jin, P., Sabatino, M., Balakumaran, A., Feng, J., Kuznetsov, S. A., et al. (2011). Global transcriptome analysis of human bone marrow stromal cells (BMSC) reveals proliferative, mobile and interactive cells that produce abundant extracellular matrix proteins, some of which may affect BMSC potency. Cytotherapy 13 (6), 661–674. doi:10.3109/14653249.2010.548379
Rey, F., Pandini, C., Barzaghini, B., Messa, L., Giallongo, T., Pansarasa, O., et al. (2020). Dissecting the effect of a 3D microscaffold on the transcriptome of neural stem cells with computational approaches: A focus on mechanotransduction. Int. J. Mol. Sci. 21 (18), 6775–6837. doi:10.3390/ijms21186775
Ricci, D., Nava, M. M., Zandrini, T., Cerullo, G., Raimondi, M. T., and Osellame, R. (2017). Scaling-up techniques for the nanofabrication of cell culture substrates via two-photon polymerization for industrial-scale expansion of stem cells. Materials 10, 66. doi:10.3390/ma10010066
Rio, D. C., Ares, M., Hannon, G. J., and Nilsen, T. W. (2010). Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 5 (6), pdb.prot5439–4. doi:10.1101/pdb.prot5439
Roson-Burgo, B., Sanchez-Guijo, F., Del Cañizo, C., and De Las Rivas, J. (2016). Insights into the human mesenchymal stromal/stem cell identity through integrative transcriptomic profiling. BMC Genomics 17 (1), 944–1027. doi:10.1186/s12864-016-3230-0
Saei Arezoumand, K., Alizadeh, E., Pilehvar-Soltanahmadi, Y., Esmaeillou, M., and Zarghami, N. (2017). An overview on different strategies for the stemness maintenance of MSCs. Artif. Cells, Nanomedicine Biotechnol. 45 (7), 1255–1271. doi:10.1080/21691401.2016.1246452
Saidova, A. A., and Vorobjev, I. A. (2020). Lineage commitment, signaling pathways, and the cytoskeleton systems in mesenchymal stem cells. Tissue Eng. - Part B Rev. 26 (1), 13–25. doi:10.1089/ten.teb.2019.0250
Sánchez-Luis, E., Joaquín-García, A., Campos-Laborie, F. J., Sánchez-Guijo, F., and De las Rivas, J. (2020). Deciphering master gene regulators and associated networks of human mesenchymal stromal cells. Biomolecules 10 (4), 557–617. doi:10.3390/biom10040557
Santos, E., Hernández, R. M., Pedraz, J. L., and Orive, G. (2012). Novel advances in the design of three-dimensional bio-scaffolds to control cell fate: Translation from 2D to 3D. Trends Biotechnol. 30 (6), 331–341. doi:10.1016/j.tibtech.2012.03.005
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9 (7), 676–682. doi:10.1038/nmeth.2019
Shamir, E. R., and Ewald, A. J. (2014). Three-dimensional organotypic culture: Experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol. 15 (10), 647–664. doi:10.1038/nrm3873
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (2498), 2498–2504. doi:10.1101/gr.1239303
Shi, Y., Xia, Y. Y., Wang, L., Liu, R., Khoo, K. S., and Feng, Z. W. (2012). Neural cell adhesion molecule modulates mesenchymal stromal cell migration via activation of MAPK/ERK signaling. Exp. Cell Res. 318 (17), 2257–2267. doi:10.1016/j.yexcr.2012.05.029
Song, N., Scholtemeijer, M., and Shah, K. (2020). Mesenchymal stem cell immunomodulation: Mechanisms and therapeutic potential. Trends Pharmacol. Sci. 41 (9), 653–664. doi:10.1016/j.tips.2020.06.009
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102 (43), 15545–15550. doi:10.1073/pnas.0506580102
Suzuki, T., Morikawa, J., Hashimoto, T., Buividas, R., Gervinskas, G., Paipulas, D., et al. (2012). Thermal and optical properties of sol-gel and SU-8 resists. Proc. SPIE - Int. Soc. Opt. Eng. 8249, 1–9. doi:10.1117/12.907028
von der Mark, K., Gauss, V., von der Mark, H., and Müller, P. (1977). Relationship between cell shape and type of collagen synthesised as chondrocytes lose their cartilage phenotype in culture. Nature 267, 531. doi:10.1038/267531a0
Vukovic, J., Ruitenberg, M. J., Roet, K., Franssen, E., Arulpragasam, A., Sasaki, T., et al. (2009). The glycoprotein fibulin-3 regulates morphology and motility of olfactory ensheathing cells in vitro. Glia 57 (4), 424–443. doi:10.1002/glia.20771
Walker, M., Patel, K., and Stappenbeck, T. (2008). The stem cell niche. J. Pathology, 231–241. doi:10.1002/path
Wan, L. Q., Kang, S. M., Eng, G., Grayson, W. L., Lu, X. L., Huo, B., et al. (2010). Geometric control of human stem cell morphology and differentiation. Integr. Biol. 2 (7–8), 346–353. doi:10.1039/c0ib00016g
Wei, Q., Nakahara, F., Asada, N., Zhang, D., Gao, X., Xu, C., et al. (2020). Snai2 maintains bone marrow niche cells by repressing osteopontin expression. Dev. Cell 53 (5), 503–513. doi:10.1016/j.devcel.2020.04.012
Weiss, A. R. R., and Dahlke, M. H. (2019). Immunomodulation by mesenchymal stem cells (MSCs): Mechanisms of action of living, apoptotic, and dead MSCs. Front. Immunol. 10, 1191–1210. doi:10.3389/fimmu.2019.01191
Winer, J. P., Janmey, P. A., McCormick, M. E., and Funaki, M. (2009). Bone marrow-derived human mesenchymal stem cells become quiescent on soft substrates but remain responsive to chemical or mechanical stimuli. Tissue Eng. - Part A 15 (1), 147–154. doi:10.1089/ten.tea.2007.0388
Wu, S., Serbin, J., and Gu, M. (2006). Two-photon polymerisation for three-dimensional micro-fabrication. J. Photochem. Photobiol. A Chem. 181 (1), 1–11. doi:10.1016/j.jphotochem.2006.03.004
Wuchter, P., Boda-Heggemann, J., Straub, B. K., Grund, C., Kuhn, C., Krause, U., et al. (2007). Processus and recessus adhaerentes: Giant adherens cell junction systems connect and attract human mesenchymal stem cells. Cell Tissue Res. 328 (3), 499–514. doi:10.1007/s00441-007-0379-5
Xie, A. W., Zacharias, N. A., Binder, B. Y. K., and Murphy, W. L. (2021). Controlled aggregation enhances immunomodulatory potential of mesenchymal stromal cell aggregates. Stem Cells Transl. Med. 10 (8), 1184–1201. doi:10.1002/sctm.19-0414
Ylostalo, J. ., Bartsoh, T. ., Toblow, A., and Prockop, D. (2014). Unique characteristics of human mesenchymal stromal/progenitor cells pre-activated in 3-dimensional cultures under different conditions. Cytotherapy 16 (11), 1486–1500. doi:10.1016/j.jcyt.2014.07.010
Zandrini, T., Shan, O., Parodi, V., Cerullo, G., Raimondi, M. T., and Osellame, R. (2019). Multi-foci laser microfabrication of 3D polymeric scaffolds for stem cell expansion in regenerative medicine. Sci. Rep. 9, 11761–11769. doi:10.1038/s41598-019-48080-w
Zhang, C., Zhu, H., Ren, X., Gao, B., Cheng, B., Liu, S., et al. (2021). Mechanics-driven nuclear localization of YAP can be reversed by N-cadherin ligation in mesenchymal stem cells. Nat. Commun. 12 (1), 6229. doi:10.1038/s41467-021-26454-x
Zhao, Q., Ren, H., and Han, Z. (2016). Mesenchymal stem cells: Immunomodulatory capability and clinical potential in immune diseases. J. Cell. Immunother. 2 (1), 3–20. doi:10.1016/j.jocit.2014.12.001
Zoja, C., Garcia, P. B., Rota, C., Conti, S., Gagliardini, E., Corna, D., et al. (2012). Mesenchymal stem cell therapy promotes renal repair by limiting glomerular podocyte and progenitor cell dysfunction in adriamycin-induced nephropathy. Am. J. Physiology - Ren. Physiology 303 (9), 1370–1381. doi:10.1152/ajprenal.00057.2012
Keywords: mechanobiology, mesenchymal stem cells, synthetic, niche, 3D cell culture, transcriptomic analysis, cell adhesion molecules, bioengineering, immunemodulation
Citation: Testa C, Oliveto S, Jacchetti E, Donnaloja F, Martinelli C, Pinoli P, Osellame R, Cerullo G, Ceri S, Biffo S and Raimondi MT (2023) Whole transcriptomic analysis of mesenchymal stem cells cultured in Nichoid micro-scaffolds. Front. Bioeng. Biotechnol. 10:945474. doi: 10.3389/fbioe.2022.945474
Received: 16 May 2022; Accepted: 15 December 2022;
Published: 06 January 2023.
Edited by:
Sandra Camarero-Espinosa, Polymat, SpainReviewed by:
Gonzalo Riadi, University of Talca, ChileAngelo Accardo, Delft University of Technology, Netherlands
Copyright © 2023 Testa, Oliveto, Jacchetti, Donnaloja, Martinelli, Pinoli, Osellame, Cerullo, Ceri, Biffo and Raimondi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carolina Testa, Y2Fyb2xpbmEudGVzdGFAcG9saW1pLml0; Manuela T. Raimondi, bWFudWVsYS5yYWltb25kaUBwb2xpbWkuaXQ=
†These authors share senior authorship
 Carolina Testa
Carolina Testa Stefania Oliveto
Stefania Oliveto Emanuela Jacchetti
Emanuela Jacchetti Francesca Donnaloja
Francesca Donnaloja Chiara Martinelli
Chiara Martinelli Pietro Pinoli
Pietro Pinoli Roberto Osellame4
Roberto Osellame4 Giulio Cerullo
Giulio Cerullo Stefano Ceri
Stefano Ceri Stefano Biffo
Stefano Biffo Manuela T. Raimondi
Manuela T. Raimondi