- Department of Biomedical Engineering, School of Basic Medical Science, Central South University, Changsha, Hunan, China
Magnetic nanoparticles (MNPs) can be quantified based on their magnetic relaxation properties by volumetric magnetic biosensing strategies, for example, alternating current susceptometry. Volume-amplified magnetic nanoparticle detection assays (VAMNDAs) employ analyte-initiated nucleic acid amplification (NAA) reactions to increase the hydrodynamic size of MNP labels for magnetic sensing, achieving attomolar to picomolar detection limits. VAMNDAs offer rapid and user-friendly analysis of nucleic acid targets but present inherence defects determined by the chosen amplification reactions and sensing principles. In this mini-review, we summarize more than 30 VAMNDA publications and classify their detection models for NAA-induced MNP size increases, highlighting the performances of different linear, cascade, and exponential NAA strategies. For some NAA strategies that have not yet been reported in VAMNDA, we predicted their performances based on the reaction kinetics and feasible detection models. Finally, challenges and perspectives are given, which may hopefully inspire and guide future VAMNDA studies.
Introduction
Magnetic micron/nano-sized particles stand out in comparison to alternative nanomaterials due to their unique properties, for example, easy manipulation, biocompatibility, signal stability, and high contrast (against the inherently negligible magnetic susceptibilities of biological objects), allowing for extensive applications in biomedical engineering (Gloag et al., 2019; Moerland et al., 2019; Cheng et al., 2021). Magnetic nanoparticles (MNPs), including micron-sized particles consisting of nanocomposites, can be directly analyzed as labels and transducers by magnetic sensors, facilitating an ideal mix-and-read biosensing approach that is attractive for on-site analysis and point-of-care testing (Schrittwieser et al., 2016; Xianyu et al., 2018). Based on the sensing principle, MNP detection sensors can be classified as surface-based and volumetric sensors (Issadore et al., 2014). Surface-based MNP sensing (e.g., micro-Hall sensing and giant magnetoresistance sensing) detects the MNP’s magnetic stray field induced by an external magnetic field. Due to the fast decay of the magnetic stray field with distance, only MNPs located close to the surface of the sensing element can be detected with the requirement of separation steps to remove unbound MNPs. In contrast, volumetric magnetic sensing could measure the presence and/or the property (e.g., relaxivity) changes of MNPs dispersed in the entire suspension, which is simple and convenient (Lee et al., 2015), especially when combined with homogeneous reaction strategies. The hydrodynamic size changes of MNPs are related to several magnetic properties such as Brownian relaxation frequency and magnetic anisotropy, allowing the volume-amplified magnetic nanoparticle detection assay (VAMNDA) (Strömberg et al., 2008) based on different volumetric magnetic sensors.
Various nucleic acid amplification (NAA) strategies have been employed to increase the hydrodynamic volume of MNPs in VAMNDA. The hydrodynamic size increase of MNPs can be induced by analyte-initiated NAA in three detection models: (I) micrometer-sized single-stranded tandem amplicon coil-induced MNP aggregation, (II) amplicon monomer-mediated MNP linkage, and (III) double-stranded tandem amplicon chain-based MNP “hair growth,” as illustrated in Figure 1A i, ii, and iii, respectively. In VAMNDA, the NAA-induced hydrodynamic size changes of MNPs are usually analyzed by alternating current (AC) susceptometry (or equivalent methodologies), measuring the susceptibility of MNPs exposed to an AC magnetic field. For commonly used MNPs with diameters of 30–300 nm, the MNP’s relaxation process after switching off the external field is dominated by thermal rotational diffusion, that is, Brownian relaxation (Strömberg et al., 2007b). In AC susceptometry, for the simplest case, the Brownian relaxation frequency
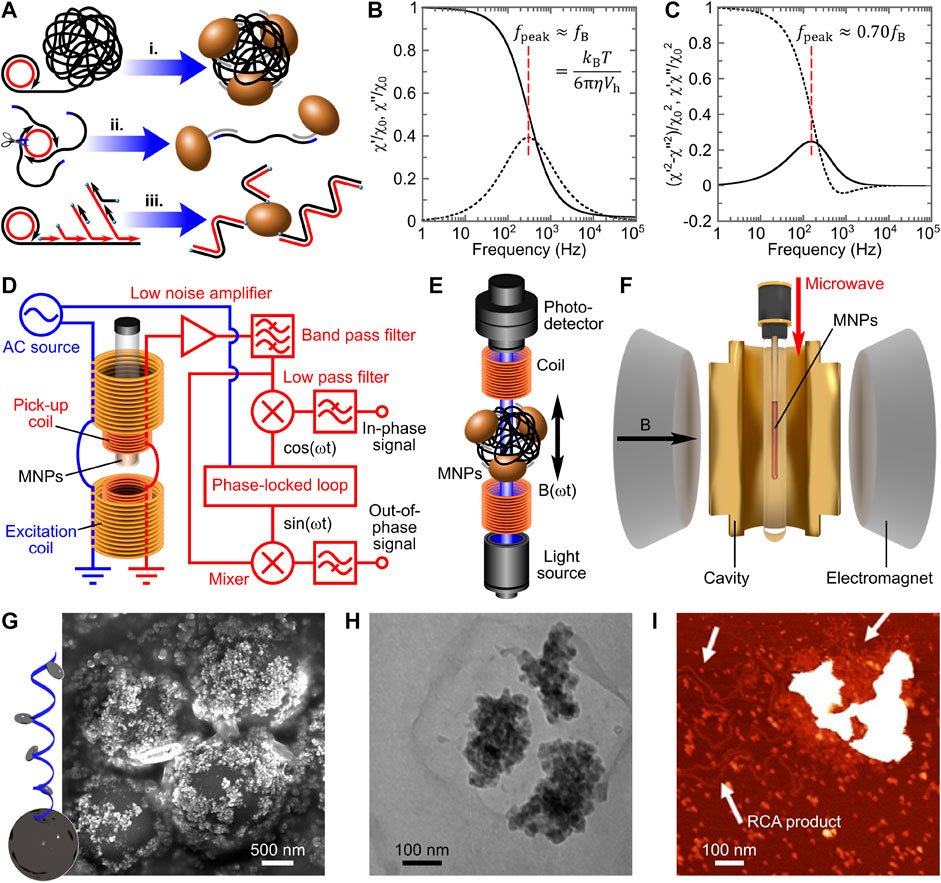
FIGURE 1. The basic principle of volume-amplified magnetic nanoparticle detection assays. (A) Three typical detection models for the nucleic acid amplification-based hydrodynamic size increase of MNPs, instanced by conventional, nicking-enhanced, and hyperbranched rolling circle amplification strategies, respectively. (B) Spectra of normalized in-phase and out-of-phase magnetic susceptibility (
Rolling circle amplification
Owing to its simplicity, robustness, and high efficiency, rolling circle amplification (RCA) is the most widely adopted isothermal NAA in biomedical engineering (Ali et al., 2014; Soares et al., 2021; Yao et al., 2021). RCA is the most effective linear amplification due to (I) the conformational stresses that facilitate the strand displacement (Joffroy et al., 2018) and (II) the high polymerizing processivity of the polymerase (whereas in linear template-based amplification reactions, polymerases undergo cycles of diffusion-polymerization-dissociation). In phi29 polymerase-based RCA, ca. 105 nt-long single-stranded tandem amplicons (about 103 times amplification) can be synthesized in 1 h (Banér et al., 1998). Moreover, a single-nucleotide specific process, that is, padlock probe ligation, is usually employed to prepare circular templates for the following RCA, offering the whole reaction system an ability of single-nucleotide discrimination (Nilsson et al., 2000). RCA amplicons form micrometer-sized DNA coils in the aqueous solution, which can be quantified by VAMNDA model I (Figure 1A, i). Figure 1G shows a representative scanning electron microscopy micrograph of RCA coil-aggregated MNPs on the surface of microbeads, demonstrating the formation of micrometer-sized MNP aggregates (Tian et al., 2018b). RCA coil-aggregated MNPs have dramatically increased hydrodynamic volumes characterized by a
For RCA-based end-point VAMNDA, where an additional hybridization step is required to anneal probe-modified MNPs onto the coils, 1–10 pM, LODs were reported with total assay times around 2 h (including 1 h of RCA) and dynamic detection ranges of approximately two orders of magnitude (Donolato et al., 2015b; Bejhed et al., 2015; Blomgren et al., 2018; Sepehri et al., 2018; Tian et al., 2019b). By using a microfluidic sample handling system with a simultaneous differential sensor, Sepehri et al. improved the picomolar LOD to 45 fM, being the most sensitive conventional RCA-based VAMNDA (Sepehri et al., 2019). However, in the end-point detection format, MNPs can probably be stopped at the surface of RCA coils due to the multivalent binding reaction, resulting in a limited aggregation effect (Zardán Gómez de la Torre et al., 2010). Transmission electron microscopy and atomic force microscopy studies showed that only a few particles were bound to each RCA coil (Figures 1H, I), suggesting potentially higher sensitivity of the real-time MNP aggregation-based assay (Akhtar et al., 2010; Oropesa-Nuñez et al., 2020). In order to increase the MNP-to-coil ratio and shorten the total assay time, real-time VAMNDAs were presented by performing RCA and MNP hybridization simultaneously with magnetic incubation (i.e., applying magnetic actuation to enhance the binding kinetics) and optomagnetic phase lag sensing, which could achieve an LOD of 0.3 pM. within 90 min (Tian et al., 2019a; Minero et al., 2020).
Due to the steric hindrance and electrostatic repulsion, real-time RCA can hardly achieve the theoretical highest sensitivity (Zardán Gómez de la Torre et al., 2010). Nicking-enhanced RCA (NickRCA) performs nicking reactions during RCA, generating single-stranded amplicon monomers instead of amplicon coils (Li et al., 2010). Moreover, NickRCA allows several polymerases to act simultaneously on one single circular template, which further improves the amplification efficiency (Tian et al., 2019a). Amplicon monomers can bridge MNPs for turn-on measurement (detection model II, Figure 1A, ii). By real-time optomagnetic sensing of MNP phase lag in response to an AC field, NickRCA achieved an LOD of 15 fM target DNA with a total assay time of ca. 100 min (Tian et al., 2019a), which is the lowest LOD obtained by linear amplification-based VAMNDAs. As a comparison, optomagnetic analysis of synthetic DNA monomers (without NAA) presented an LOD of 50 pM. (Mezger et al., 2015), suggesting amplification of NickRCA of ca. 3 × 103 times.
Linear RCA-based VAMNDAs have been demonstrated for the quantification of bacterial/viral sequences (representing, e.g., influenza virus), discrimination of drug-resistance single-nucleotide mutations of Mycobacterium tuberculosis, and biplex detection of Vibrio cholerae and Escherichia coli. However, LODs at a 10−14 M range for nucleic acid targets are still inferior for many clinical applications (e.g., virus detection), suggesting the utilization of more efficient NAA strategies.
RCA-based cascade amplification
A cascade amplification strategy consists of tandemly performed amplification reactions, where the product of one amplification reaction is the trigger, primer, or template of the subsequent amplification reaction. Cascade amplification strategies are versatile to involve various tool enzymes at the expense of complicated strategy design (e.g., polymerases with exonucleolytic activity can digest the intermediate amplicons), especially for the one-pot homogeneous cascade amplification strategies that are preferred for VAMNDA. By stepwise performing
Circle-to-circle amplification (C2CA, illustrated in Figure 2A) is a representative cascade amplification that converts RCA amplicon coils into multiple circular monomers (through endonuclease monomerization followed by ligation) as templates for subsequent RCA (Dahl et al., 2004). The ideal gain of C2CA (i.e., the quantity of generated amplicon) is
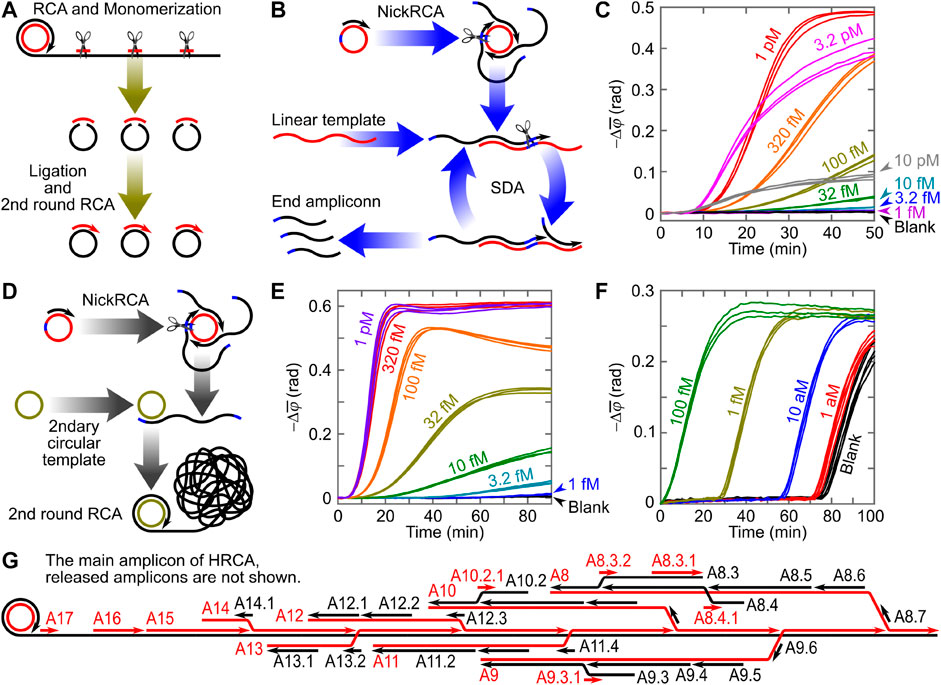
FIGURE 2. (A) Schematic illustration of conventional C2CA. (B,C) Schematic illustration and time-resolved optomagnetic signals (phase lag increases) of NECA-based VAMNDA. (D,E) Schematic illustration and time-resolved optomagnetic signals of HC2CA-based VAMNDA. (F) Time-resolved optomagnetic signals of PG-RCA-based VAMNDA. Panels (C,E,F) are adapted with permission from Tian et al. (2020a, 2020b, 2020c), Copyright 2020, Elsevier and Oxford University Press. (G) Illustration of HRCA processes on the RCA coil (amplicons that have already been released from the RCA coil are not shown). A8 means the eighth amplicon produced on the RCA coil (A1-A7 have been released), and A8.3 means the third amplicon produced on A8 (A8.1 and A8.2 have been released).
For homogeneous cascade amplification, nicking-assisted on- and off-loop enzymatic cascade amplification (NECA, illustrated in Figure 2B) comprises a NickRCA to generate primers of a subsequent strand-displacement amplification (SDA) for VAMNDA model II (Tian et al., 2020a). Both NickRCA and SDA were designed with the same template sequence: a padlock probe ligation process was employed to produce the circular template of NickRCA, leaving unligated padlock probe molecules in the suspension served as the template of SDA. Before all the templates are occupied, the ideal gain of NECA follows a quadratic function,
Homogeneous C2CA (HC2CA, illustrated in Figure 2D) employs NickRCA to produce primers for the subsequent RCA, combining two amplification reactions in a one-pot homogeneous reaction to eliminate additional monomerization and ligation steps (Tian et al., 2020b). However, the secondary circular template competes with the detection probe. Thus, the RCA coils can hardly be monitored before all circular templates are occupied (either by a primer or by an amplicon coil), indicated by a dead time during which no signal can be detected:
Exponential amplification
An exponential amplification reaction, for example, the polymerase chain reaction (PCR), generates amplicons serving as the trigger, primer, or template of the reaction itself. NickRCA can react exponentially if the circular templates are added as reagents (instead of being prepared by padlock probe ligation), which is known as primer-generation RCA (PG-RCA) (Murakami et al., 2009). PG-RCA amplicons can trigger the formation of MNP dimers for detection model II, obtaining an attomolar LOD for a synthetic Dengue virus sequence (Figure 2F) (Tian et al., 2020c). However, the rapid generation of amplicon monomers requires real-time MNP binding. Otherwise, MNPs could be saturated by an excess amount of amplicons without a significant hydrodynamic size increase. Considering that some exponential NAAs include either high-temperature processes or molecular crowding agents, real-time VAMNDAs based on these NAAs are challenging. This may explain why PCR, EXPAR (exponential amplification reaction), and RPA (recombinase polymerase amplification) have not been reported in VAMNDA so far.
Loop-mediated isothermal amplification (LAMP) produces double-stranded amplicons of different lengths. By using biotinylated primers, LAMP amplicons can attach to the streptavidin-modified MNPs for VAMNDA model III, which can be achieved by either an end-point MNP binding or a real-time on-particle amplification, resulting in attomolar LODs for different synthetic (e.g., a Zika virus sequence) and real (e.g., Newcastle disease virus) targets within 30 min (Tian et al., 2016b, 2016c; Minero et al., 2017). Except for the amplicon-based detection, precipitation of Mg2P2O7 (a by-product of LAMP) onto MNPs can also be utilized for VAMNDA, providing a sub-femtomolar LOD by using a ferromagnetic resonance spectrometer (Tian et al., 2018a). Similar to LAMP, hyperbranched RCA (HRCA) is an exponential amplification strategy producing double-stranded amplicons of different lengths. HRCA employs a pair of primers to trigger SDA on the RCA coil (Figure 2G) with a gain of
Challenges and perspectives
Existing VAMNDA models present inherence defects: model I requires long single-stranded amplicons that can hardly be produced with high efficiency; model II has to discriminate MNP dimers from a background of individual MNPs; model III is limited by the length distribution of amplicons and cannot distinguish false-positive amplicons. Moreover, the sensitivity of VAMNDA is determined mainly by the amplification efficiency of the NAA strategy, implying that exponential NAAs are preferred for lower LODs. However, due to the mispriming, ab initio DNA synthesis, and polymerase side-reactions, nonspecific synthesis of false-positive products is inevitable in highly effective NAA reactions, especially the exponential ones (Zyrina and Antipova, 2021). A CRISPR/Cas12a-based internal negative control system was reported in combination with VAMNDA, which could indicate the onset of nonspecific amplification (Tian et al., 2020c). Nevertheless, such a warning system cannot suppress nonspecific amplification. We expect that the problem of nonspecific synthesis can hopefully be solved in the future by applying highly accurate and programmable techniques such as DNA logic gates.
For the purpose of system automation and miniaturization, one-pot homogeneous reactions are preferred for VAMNDA. However, clinical applications of NAA strategies usually consist of a series of processes such as analyte extraction and purification. The lab-on-a-disc technique can integrate multi-step assays into a chip on which density gradient centrifugation and centrifugo-pneumatic valving can be processed, facilitating a compact and fully automated sample-to-answer biosensor (Uddin et al., 2018). In another RCA-based VAMNDA study (Garbarino et al., 2019), all the detection processes, including target capture, padlock probe ligation, molecular amplification, and optomagnetic detection, were integrated into a microfluidic chip containing three connected reaction chambers, below which a motorized permanent magnet was positioned to move analytes (captured by magnetic microparticles) along the fluid channel.
For homogeneous VAMNDAs without any washing/separation steps, the multiplex sensing can be realized by utilizing MNPs showing distinct relaxivities. As reviewed in a previous section, MNPs of different hydrodynamic volumes can be distinguished based on their characteristic
Portable and ease-of-use magnetic biosensing systems based on the VAMNDA concept have been commercialized for clinical applications, for example, one-drop-of-blood quantitative virus tests (blusense-diagnostics.com). However, these commercialized biosensors were realized based on the (target antibody-induced) immuno-agglutination but not NAA, which could probably be explained by the lack of reliable and sensitive homogeneous isothermal amplification strategies with proper controls of background amplification. Nevertheless, some NAA-based VAMNDAs were verified by testing clinical samples such as virus specimens (vaccine and tissue) (Tian et al., 2016b) and bacterium specimens (urine) (Mezger et al., 2015), implying potential applications in diagnosis.
Conclusion
Following the trends of nanoscience, magnetics, and sensing techniques over the last decade, VAMNDAs have undergone a pronounced evolution toward point-of-care testing. However, despite the rapidly improved biosensing performances, technical challenges remain. We herein provide an overview of VAMNDA and introduce the concepts and performances of different NAA strategies employed in this technique. Thereafter, limitations and prospects toward point-of-care applications are discussed. We hope that this mini-review will motivate studies to help solve the current limitations that are preventing VAMNDA from clinical applications.
Author contributions
ZH: conceptualization, supervision, manuscript review and editing. JL and HZ: data analysis, manuscript review and editing. BT: project administration, funding acquisition, and original draft.
Funding
This work was financially supported by the National Natural Science Foundation of China (no. 32101214), the Hunan Provincial Natural Science Foundation of China (no. 2022JJ40582), and the Fundamental Research Funds for the Central Universities of Central South University (no. 202045006).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akhtar, S., Strömberg, M., Zardán Gómez de la Torre, T., Russell, C., Gunnarsson, K., Nilsson, M., et al. (2010). Real-space transmission electron microscopy investigations of attachment of functionalized magnetic nanoparticles to DNA-coils acting as a biosensor. J. Phys. Chem. B 114, 13255–13262. doi:10.1021/jp105756b
Ali, M. M., Li, F., Zhang, Z., Zhang, K., Kang, D. K., Ankrum, J. A., et al. (2014). Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 43, 3324–3341. doi:10.1039/c3cs60439j
Banér, J., Nilsson, M., Mendel-Hartvig, M., and Landegren, U. (1998). Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 26, 5073–5078. doi:10.1093/nar/26.22.5073
Bejhed, R. S., Zardán Gómez de la Torre, T., Donolato, M., Hansen, M. F., Svedlindh, P., and Strömberg, M. (2015). Turn-on optomagnetic bacterial DNA sequence detection using volume-amplified magnetic nanobeads. Biosens. Bioelectron. 66, 405–411. doi:10.1016/j.bios.2014.11.048
Blomgren, J., Ahrentorp, F., Ilver, D., Jonasson, C., Sepehri, S., Kalaboukhov, A., et al. (2018). Development of a sensitive induction-based magnetic nanoparticle biodetection method. Nanomaterials 8, 887. doi:10.3390/nano8110887
Cheng, H. W., Tsao, H. Y., Chiang, C. S., and Chen, S. Y. (2021). Advances in magnetic nanoparticle-mediated cancer immune-theranostics. Adv. Healthc. Mat. 10, 2001451. doi:10.1002/adhm.202001451
Cole, K. S., and Cole, R. H. (1941). Dispersion and absorption in dielectrics I. Alternating current characteristics. J. Chem. Phys. 9, 341–351. doi:10.1063/1.1750906
Dahl, F., Banér, J., Gullberg, M., Mendel-Hartvig, M., Landegren, U., and Nilsson, M. (2004). Circle-to-circle amplification for precise and sensitive DNA analysis. Proc. Natl. Acad. Sci. U. S. A. 101, 4548–4553. doi:10.1073/pnas.0400834101
Donolato, M., Antunes, P., Bejhed, R. S., Zardán Gómez de la Torre, T., Østerberg, F. W., Strömberg, M., et al. (2015a). Novel readout method for molecular diagnostic assays based on optical measurements of magnetic nanobead dynamics. Anal. Chem. 87, 1622–1629. doi:10.1021/ac503191v
Donolato, M., Antunes, P., Zardán Gómez de la Torre, T., Hwu, E. T., Chen, C. H., Burger, R., et al. (2015b). Quantification of rolling circle amplified DNA using magnetic nanobeads and a Blu-ray optical pick-up unit. Biosens. Bioelectron. 67, 649–655. doi:10.1016/j.bios.2014.09.097
Fock, J., Jonasson, C., Johansson, C., and Hansen, M. F. (2017a). Characterization of fine particles using optomagnetic measurements. Phys. Chem. Chem. Phys. 19, 8802–8814. doi:10.1039/c6cp08749c
Fock, J., Parmvi, M., Strömberg, M., Svedlindh, P., Donolato, M., and Hansen, M. F. (2017b). Comparison of optomagnetic and AC susceptibility readouts in a magnetic nanoparticle agglutination assay for detection of C-reactive protein. Biosens. Bioelectron. 88, 94–100. doi:10.1016/j.bios.2016.07.088
Garbarino, F., Minero, G. A. S., Rizzi, G., Fock, J., and Hansen, M. F. (2019). Integration of rolling circle amplification and optomagnetic detection on a polymer chip. Biosens. Bioelectron. 142, 111485. doi:10.1016/j.bios.2019.111485
Gloag, L., Mehdipour, M., Chen, D., Tilley, R. D., and Gooding, J. J. (2019). Advances in the application of magnetic nanoparticles for sensing. Adv. Mat. 31, 1904385. doi:10.1002/adma.201904385
Göransson, J., Zardán Gómez de la Torre, T., Strömberg, M., Russell, C., Svedlindh, P., Strømme, M., et al. (2010). Sensitive detection of bacterial DNA by magnetic nanoparticles. Anal. Chem. 82, 9138–9140. doi:10.1021/ac102133e
Issadore, D., Park, Y. I., Shao, H., Min, C., Lee, K., Liong, M., et al. (2014). Magnetic sensing technology for molecular analyses. Lab. Chip 14, 2385. doi:10.1039/c4lc00314d
Joffroy, B., Uca, Y. O., Prešern, D., Doye, J. P. K., and Schmidt, T. L. (2018). Rolling circle amplification shows a sinusoidal template length-dependent amplification bias. Nucleic Acids Res. 46, 538–545. doi:10.1093/nar/gkx1238
Lee, H., Shin, T. H., Cheon, J., and Weissleder, R. (2015). Recent developments in magnetic diagnostic systems. Chem. Rev. 115, 10690–10724. doi:10.1021/cr500698d
Li, J., Deng, T., Chu, X., Yang, R., Jiang, J., Shen, G., et al. (2010). Rolling circle amplification combined with gold nanoparticle aggregates for highly sensitive identification of single-nucleotide polymorphisms. Anal. Chem. 82, 2811–2816. doi:10.1021/ac100336n
Mezger, A., Fock, J., Antunes, P., Østerberg, F. W., Boisen, A., Nilsson, M., et al. (2015). Scalable DNA-based magnetic nanoparticle agglutination assay for bacterial detection in patient samples. ACS Nano 9, 7374–7382. doi:10.1021/acsnano.5b02379
Minero, G. A. S., Nogueira, C., Rizzi, G., Tian, B., Fock, J., Donolato, M., et al. (2017). Sequence-specific validation of LAMP amplicons in real-time optomagnetic detection of Dengue serotype 2 synthetic DNA. Analyst 142, 3441–3450. doi:10.1039/c7an01023k
Minero, G. A. S., Bagnasco, M., Fock, J., Tian, B., Garbarino, F., and Hansen, M. F. (2020). Automated on-chip analysis of tuberculosis drug-resistance mutation with integrated DNA ligation and amplification. Anal. Bioanal. Chem. 412, 2705–2710. doi:10.1007/s00216-020-02568-x
Moerland, C. P., Van IJzendoorn, L. J., and Prins, M. W. J. (2019). Rotating magnetic particles for lab-on-chip applications-a comprehensive review. Lab. Chip 19, 919–933. doi:10.1039/C8LC01323C
Murakami, T., Sumaoka, J., and Komiyama, M. (2009). Sensitive isothermal detection of nucleic-acid sequence by primer generation-rolling circle amplification. Nucleic Acids Res. 37, e19. doi:10.1093/nar/gkn1014
Nilsson, M., Barbany, G., Antson, D. O., Gertow, K., and Landegren, U. (2000). Enhanced detection and distinction of RNA by enzymatic probe ligation. Nat. Biotechnol. 18, 791–793. doi:10.1038/77367
Oropesa-Nuñez, R., Zardán Gómez de la Torre, T., Stopfel, H., Svedlindh, P., Strömberg, M., and Gunnarsson, K. (2020). Insights into the formation of DNA-magnetic nanoparticle hybrid structures: Correlations between morphological characterization and output from magnetic biosensor measurements. ACS Sens. 5, 3510–3519. doi:10.1021/acssensors.0c01623
Østerberg, F. W., Rizzi, G., and Hansen, M. F. (2013a). On-chip measurements of Brownian relaxation of magnetic beads with diameters from 10 nm to 250 nm. J. Appl. Phys. 113, 154507. doi:10.1063/1.4802657
Østerberg, F. W., Rizzi, G., Zardán Gómez de la Torre, T., Strömberg, M., Strømme, M., Svedlindh, P., et al. (2013b). Measurements of Brownian relaxation of magnetic nanobeads using planar Hall effect bridge sensors. Biosens. Bioelectron. 40, 147–152. doi:10.1016/j.bios.2012.07.002
Østerberg, F. W., Rizzi, G., Donolato, M., Bejhed, R. S., Mezger, A., Strömberg, M., et al. (2014). On-chip detection of rolling circle amplified DNA molecules from Bacillus globigii spores and Vibrio cholerae. Small 10, 2877–2882. doi:10.1002/smll.201303325
Schrittwieser, S., Pelaz, B., Parak, W. J., Lentijo-Mozo, S., Soulantica, K., Dieckhoff, J., et al. (2016). Homogeneous biosensing based on magnetic particle labels. Sensors Switz. 16, 828. doi:10.3390/s16060828
Sepehri, S., Eriksson, E., Kalaboukhov, A., Zardán Gómez de la Torre, T., Kustanovich, K., Jesorka, A., et al. (2018). Volume-amplified magnetic bioassay integrated with microfluidic sample handling and high-Tc SQUID magnetic readout. Apl. Bioeng. 2, 016102. doi:10.1063/1.4999713
Sepehri, S., Zardán Gómez de la Torre, T., Schneiderman, J. F., Blomgren, J., Jesorka, A., Johansson, C., et al. (2019). Homogeneous differential magnetic assay. ACS Sens. 4, 2381–2388. doi:10.1021/acssensors.9b00969
Soares, R. R. G., Madaboosi, N., and Nilsson, M. (2021). Rolling circle amplification in integrated microsystems: An uncut gem toward massively multiplexed pathogen diagnostics and genotyping. Acc. Chem. Res. 54, 3979–3990. doi:10.1021/acs.accounts.1c00438
Strömberg, M., Gunnarsson, K., Johansson, H., Nilsson, M., Svedlindh, P., and Strømme, M. (2007a). Interbead interactions within oligonucleotide functionalized ferrofluids suitable for magnetic biosensor applications. J. Phys. D. Appl. Phys. 40, 1320–1330. doi:10.1088/0022-3727/40/5/002
Strömberg, M., Gunnarsson, K., Valizadeh, S., Svedlindh, P., and Strømme, M. (2007b). Aging phenomena in ferrofluids suitable for magnetic biosensor applications. J. Appl. Phys. 101, 023911. doi:10.1063/1.2424522
Strömberg, M., Göransson, J., Gunnarsson, K., Nilsson, M., Svedlindh, P., and Strømme, M. (2008). Sensitive molecular diagnostics using volume-amplified magnetic nanobeads. Nano Lett. 8, 816–821. doi:10.1021/nl072760e
Strömberg, M., Zardán Gómez de la Torre, T., Göransson, J., Gunnarsson, K., Nilsson, M., Svedlindh, P., et al. (2009). Multiplex detection of DNA sequences using the volume-amplified magnetic nanobead detection assay. Anal. Chem. 81, 3398–3406. doi:10.1021/ac900561r
Strömberg, M., Zardán Gómez de la Torre, T., Nilsson, M., Svedlindh, P., and Strømme, M. (2014). A magnetic nanobead-based bioassay provides sensitive detection of single- and biplex bacterial DNA using a portable AC susceptometer. Biotechnol. J. 9, 137–145. doi:10.1002/biot.201300348
Tian, B., Bejhed, R. S., Svedlindh, P., and Strömberg, M. (2016a). Blu-ray optomagnetic measurement based competitive immunoassay for Salmonella detection. Biosens. Bioelectron. 77, 32–39. doi:10.1016/j.bios.2015.08.070
Tian, B., Ma, J., Zardán Gómez de la Torre, T., Bálint, Á., Donolato, M., Hansen, M. F., et al. (2016b). Rapid Newcastle disease virus detection based on loop-mediated isothermal amplification and optomagnetic readout. ACS Sens. 1, 1228–1234. doi:10.1021/acssensors.6b00379
Tian, B., Qiu, Z., Ma, J., Zardán Gómez de la Torre, T., Johansson, C., Svedlindh, P., et al. (2016c). Attomolar Zika virus oligonucleotide detection based on loop-mediated isothermal amplification and AC susceptometry. Biosens. Bioelectron. 86, 420–425. doi:10.1016/j.bios.2016.06.085
Tian, B., Ma, J., Qiu, Z., Zardán Gómez de la Torre, T., Donolato, M., Hansen, M. F., et al. (2017). Optomagnetic detection of MicroRNA based on duplex-specific nuclease-assisted target recycling and multilayer core-satellite magnetic superstructures. ACS Nano 11, 1798–1806. doi:10.1021/acsnano.6b07763
Tian, B., Liao, X., Svedlindh, P., Strömberg, M., and Wetterskog, E. (2018a). Ferromagnetic resonance biosensor for homogeneous and volumetric detection of DNA. ACS Sens. 3, 1093–1101. doi:10.1021/acssensors.8b00048
Tian, B., Qiu, Z., Ma, J., Donolato, M., Hansen, M. F., Svedlindh, P., et al. (2018b). On-particle rolling circle amplification-based core-satellite magnetic superstructures for MicroRNA detection. ACS Appl. Mat. Interfaces 10, 2957–2964. doi:10.1021/acsami.7b16293
Tian, B., Fock, J., Minero, G. A. S., Garbarino, F., and Hansen, M. F. (2019a). Ultrasensitive real-time rolling circle amplification detection enhanced by nicking-induced tandem-acting polymerases. Anal. Chem. 91, 10102–10109. doi:10.1021/acs.analchem.9b02073
Tian, B., Han, Y., Fock, J., Strömberg, M., Leifer, K., and Hansen, M. F. (2019b). Self-assembled magnetic nanoparticle–graphene oxide nanotag for optomagnetic detection of DNA. ACS Appl. Nano Mat. 2, 1683–1690. doi:10.1021/acsanm.9b00127
Tian, B., Fock, J., Minero, G. A. S., and Hansen, M. F. (2020a). Nicking-assisted on-loop and off-loop enzymatic cascade amplification for optomagnetic detection of a highly conserved dengue virus sequence. Biosens. Bioelectron. 160, 112219. doi:10.1016/j.bios.2020.112219
Tian, B., Gao, F., Fock, J., Dufva, M., and Hansen, M. F. (2020b). Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 165, 112356. doi:10.1016/j.bios.2020.112356
Tian, B., Minero, G. A. S., Fock, J., Dufva, M., and Hansen, M. F. (2020c). CRISPR-Cas12a based internal negative control for nonspecific products of exponential rolling circle amplification. Nucleic Acids Res. 48, e30. doi:10.1093/nar/gkaa017
Uddin, R., Donolato, M., Hwu, E. T., Hansen, M. F., and Boisen, A. (2018). Combined detection of C-reactive protein and PBMC quantification from whole blood in an integrated lab-on-a-disc microfluidic platform. Sensors Actuators B Chem. 272, 634–642. doi:10.1016/j.snb.2018.07.015
Xianyu, Y., Wang, Q., and Chen, Y. (2018). Magnetic particles-enabled biosensors for point-of-care testing. TrAC Trends Anal. Chem. 106, 213–224. doi:10.1016/j.trac.2018.07.010
Xiao, X., Yuan, C., Li, T., Fock, J., Svedlindh, P., and Tian, B. (2022). Optomagnetic biosensors: Volumetric sensing based on magnetic actuation-induced optical modulations. Biosens. Bioelectron. 215, 114560. doi:10.1016/j.bios.2022.114560
Yao, C., Zhang, R., Tang, J., and Yang, D. (2021). Rolling circle amplification (RCA)-based DNA hydrogel. Nat. Protoc. 16, 5460–5483. doi:10.1038/s41596-021-00621-2
Zardán Gómez de la Torre, T., Strömberg, M., Russell, C., Göransson, J., Nilsson, M., Svedlindh, P., et al. (2010). Investigation of immobilization of functionalized magnetic nanobeads in rolling circle amplified DNA coils. J. Phys. Chem. B 114, 3707–3713. doi:10.1021/jp911251k
Zardán Gómez de la Torre, T., Ke, R., Mezger, A., Svedlindh, P., Strømme, M., and Nilsson, M. (2012). Sensitive detection of spores using volume-amplified magnetic nanobeads. Small 8, 2174–2177. doi:10.1002/smll.201102632
Keywords: magnetic biosensing, molecular amplification, magnetic nanoparticles, Brownian relaxation, homogeneous assays, volumetric sensing
Citation: Huang Z, Li J, Zhong H and Tian B (2022) Nucleic acid amplification strategies for volume-amplified magnetic nanoparticle detection assay. Front. Bioeng. Biotechnol. 10:939807. doi: 10.3389/fbioe.2022.939807
Received: 09 May 2022; Accepted: 11 July 2022;
Published: 10 August 2022.
Edited by:
Zhiyang Li, Nanjing Drum Tower Hospital, ChinaReviewed by:
Alfredo De La Escosura-Muñiz, University of Oviedo, SpainBingjie Zou, China Pharmaceutical University, China
Copyright © 2022 Huang, Li, Zhong and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Tian, tianbo@csu.edu.cn
 Zhongchao Huang
Zhongchao Huang