
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 27 May 2022
Sec. Synthetic Biology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.919526
This article is part of the Research Topic Design and Construction of Microbial Cell Factories for the Production of Fuels and Chemicals View all 15 articles
Mogrol plays important roles in antihyperglycemic and antilipidemic through activating the AMP-activated protein kinase pathway. Although the synthesis pathway of mogrol in Siraitia grosvenorii has been clarified, few studies have focused on improving mogrol production. This study employed a modular engineerin g strategy to improve mogrol production in a yeast chassis cell. First, a de novo synthesis pathway of mogrol in Saccharomyces cerevisiae was constructed. Then, the metabolic flux of each synthetic module in mogrol metabolism was systematically optimized, including the enhancement of the precursor supply, inhibition of the sterol synthesis pathway using the Clustered Regularly Interspaced Short Palindromic Repeats Interference system (CRISPRi), and optimization of the expression and reduction system of P450 enzymes. Finally, the mogrol titer was increased to 9.1 μg/L, which was 455-fold higher than that of the original strain. The yeast strains engineered in this work can serve as the basis for creating an alternative way for mogrol production in place of extraction from S. grosvenorii.
Mogrol, a cucurbitane-type tetracyclic triterpenoid compound, is the aglycone involved in mogroside synthesis. Mogrosides are digested into mogrol in vivo (Chiu et al., 2013; Wang et al., 2019). Therefore, mogrol may be the main functional unit of mogrosides to exert pharmacological effects (Liu et al., 2018). Many studies have shown that mogrol can effectively reduce blood glucose to contribute to antihyperglycemia and antilipidemia by activating AMP-activated protein kinase (Harada et al., 2016; Wang H. et al., 2020; Liang et al., 2021). Further studies have identified that it could attenuate pulmonary fibrosis, memory impairment, neuroinflammation, and apoptosis (Chen et al., 2018; Wang J. et al., 2020; Liu et al., 2021). Additionally, mogrol can also suppress leukemic cell growth by inhibiting the extracellular signal-regulated kinase 1/2 (ERK1/2) and signal transducer and activator of transcription three pathways (STAT3) implicated in leukemia carcinogenesis (Liu et al., 2015). Therefore, mogrol has a good application prospect in medicine.
Mogrosides and mogrol are extracted from Siraitia grosvenorii fruit, but the yield is low (0.55%–0.65%) (EFSA Panel on Food Additives and Flavourings et al., 2019). Besides, the tedious extraction process, slow growth rate, and growth process of S. grosvenorii are vulnerable to environmental perturbations, further limiting the large-scale production efficiency of mogrol. Therefore, it is crucial to develop another method for efficiently synthesizing mogrol. Recently, the introduction of heterologous natural product biosynthesis pathways into microbial cell factories to achieve the high-efficiency synthesis of natural products is attracting the attention of researchers. The advantages of microbial synthesis of natural products include rapid growth rate, simple cell genetic manipulation, and product extraction process (Scalcinati et al., 2012; Kotopka et al., 2018; Li et al., 2018). Many natural products have been efficiently synthesized in microbial cell factories, such as protopanaxadiol (Zhao et al., 2016; Zhao et al., 2019), β-carotene (Araya et al., 2012; Bu et al., 2020), taxol (Engels et al., 2008; Ajikumar et al., 2010), and cannabinoids (Luo et al., 2019).
The biosynthesis pathway of mogrol has been extensively studied in recent years. Many studies have presumed that cucurbitadienol may be a precursor for mogrol synthesis. Therefore, a cucurbitadienol synthase gene (SgCS) from S. grosvenorii was heterologously expressed in Saccharomyces cerevisiae, and cucurbitadienol was detected in the fermentation broth, which realized the de novo biosynthesis of cucurbitadienol (Ta et al., 2012). However, the biosynthesis pathway from cucurbitadienol to mogrol has not been explored yet. Zhang et al. (2016) presumed that the enzyme CYP87D18 could catalyze the oxidation of cucurbitadienol at C-11 to generate 11-hydroxy cucurbitadienol, which could be a potential precursor to mogrol, but the following catalytic steps remained to be discovered. Therefore, researchers are committed to elucidating other pathways that could synthesize mogrol. Transcriptome and metabolomics are effective strategies to clarify the biosynthetic pathway of plant natural products (Tang et al., 2011; Dai et al., 2015). Itkin et al. (2016) screened a large number of candidate genes for mogrol biosynthesis according to the combination of genomic and transcriptomic profiling of S. grosvenorii and identified SQE (encoding squalene epoxidase), CDS (encoding cucurbitadienol synthase), EPH (encoding epoxide hydrolase), and CYP (encoding cytochrome P450-dependent monooxygenase) as the key genes for mogrol synthesis. Finally, Itkin et al. (2016) realized the de novo biosynthesis of mogrol in S. cerevisiae (Figure 1) and verified in vitro that UGT (glucosyltransferase) could catalyze mogrol to form mogrosides. However, the mogrol titer in their study was not quantified because it was still low.
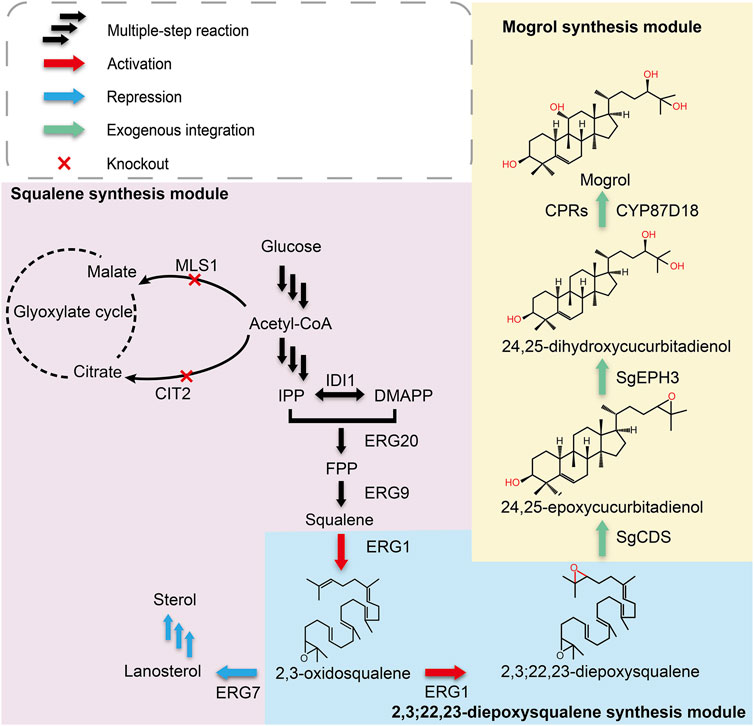
FIGURE 1. Biosynthetic pathway for mogrol production in S. cerevisiae. Single arrows represent one-step conversions, whereas triple arrows represent multiple steps. Overexpressed steps are shown as red arrows, suppressed steps are shown as blue arrows, and exogenous integrated steps are shown as green arrows. Abbreviations: IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; FPP, farnesyl; IDI1, isopentenyl diphosphate isomerase; ERG20, farnesyl pyrophosphate synthetase; ERG9, squalene synthase; ERG1, squalene epoxidase; ERG7, lanosterol synthase; MLS1, malate synthase; CIT2, citrate synthase; SgCDS, cucurbitadienol synthase; SgEPH3, epoxide hydrolase; CYP87D18, cytochrome P450 enzyme.
In this study, S. cerevisiae was engineered as a chassis cell for mogrol biosynthesis. A modular engineering strategy was proposed to improve the synthesis efficiency of mogrol in chassis cells. A de novo biosynthesis pathway of mogrol was constructed in yeast by heterologously expressing SgCDS, SgEPH3, CYP87D18, and AtCPR1 in strain Y4 (a high-yield squalene yeast constructed in our laboratory). MLS1 (malate synthase; GenBank ID: 855606) and CIT2 (citrate synthase; GenBank ID: 850361) were knocked out using the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRi) to enhance the precursor acetyl-CoA supply. The sterol synthesis pathway (essential for cell growth; Jordá and Puig, 2020) was repressed using CRISPRi to balance cell growth and production. The first step of the mogrol biosynthesis pathway was enhanced by overexpressing the ERG1 gene, an important rate-limiting enzyme in the ergosterol synthesis pathway. Subsequently, cytochrome P450 reductases (CPRs) from various sources were codon-optimized and coexpressed with CYP87D18 in the chassis cell to improve the catalysis efficiency of the last step of the mogrol biosynthesis pathway. Finally, by increasing the CYP87D18 expression levels and paring with different CPRs, mogrol production reached 9.1 μg/L, which was 455-fold higher than that of the original strain. Overall, this study constructed a yeast platform strain for bioproduction of mogrol by systems metabolic engineering strategies.
All chemicals were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China) unless otherwise specified. Mogrol standards were purchased from Sigma-Aldrich (St. Louis, MO, United States ). PrimeSTAR HS DNA polymerase used for target genes amplification was purchased from Takara (Japan). The GeneJET polymerase chain reaction (PCR) purification kit was purchased from Thermo Fisher Scientific (United States ), and SanPrep Spin Column & Collection Tube used for plasmid extraction was purchased from Sangon (Shanghai, China). The primers used in this study were synthesized by Genewiz (Suzhou, China). 5-Fluoroorotic acid (5-FOA), tyrosine, histidine, leucine, and uracil were purchased from Solarbio (Beijing, China).
The strains and plasmids used in the study are listed in Table 1. Escherichia coli JM109 was used for constructing recombinant plasmids. S. cerevisiae Y4 was employed as the original strain, a high-yield strain of squalene constructed previously. All strains constructed in this study are listed in Table 1. E. coli was cultured in Luria–Bertani medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) containing 100 mg/L ampicillin at 37°C and 220 rpm, and yeast strains were cultured in YPD medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L glucose) at 30°C and 220 rpm. Yeast transformants were selected on synthetic dextrose (SD-Ura) agar plates containing 6.9 g/L yeast nitrogen base without amino acids (YNB medium), 20 g/L glucose, 0.1 g/L amino acids (l-leucine, l-tryptophan, and l-histidine), and 20 g/L agar. Dimethyl sulfoxide with 1 g/ml 5-FOA was used for the counterselection of recombinants containing the URA3 marker. G418 sulfate (200 mg/L; Sigma) was supplemented into the medium as required.
SgCDS, SgEPH3, and CYP87D18 involved in mogrol synthesis were synthesized and codon-optimized by Genewiz. The CPRs coding AtCPR1 (GenBank ID: 828554) and AtCPR2 (GenBank ID: 829144) from Arabidopsis thaliana and SgCPR2 (GenBank ID: AYE89265.1) from S. grosvenorii (Zhao et al., 2018) were synthesized and codon-optimized by Genewiz. SrCPR1 from Stevia (Gold et al., 2018) was synthesized and codon-optimized by Genscript (Nanjing, China). CPRs coding CsCPR from Cucumber were provided by Prof. Sanwen Huang (Chinese Academy of Agricultural Sciences). All genetic modifications used the CRISPR-Cas9-mediated genome editing method and were performed as previously described (Gilbert et al., 2013). Genome integration loci (Reider Apel et al., 2017) and the corresponding single guide RNA (sgRNA) plasmids are listed in Supplementary Table S1. The target fragments were amplified using PCR, and the corresponding plasmid or S. cerevisiae genome was used as the template. The primers used in this experiment were synthesized by Genewiz. Overlaps (40–50 bp) of each adjacent fragment were used to implement homologous recombination in yeast. Yeast transformation was performed by the LiAc/SS carrier DNA/PEG method previously described (Gietz and Schiestl, 2007).
A green fluorescent protein (GFP) was fused with SgCDS, SgEPH3, CYP87D18, and AtCPR1, generating plasmids pY15-SgCDS-GFP, pY15-SgEPH3-GFP, pY15-CYP87D18-GFP, and pY15-AtCPR1-GFP, respectively, to determine the localization of heterologous enzymes involved in the biosynthesis pathway of mogrol. Most plant-derived P450 enzymes are located in the endoplasmic reticulum (ER) in S. cerevisiae (Arendt et al., 2017). An expression cassette PGAP-mCherry-Linker-SCE12-TADH1 was inserted into pY15-CYP87D18-GFP and pY15-AtCPR1-GFP, resulting in plasmids pY15-CYP87D18-GFP-mCherry-SEC12 and pY15-AtCPR1-GFP-mCherry-SEC12, respectively.
CYP and CPR genes, promoter PGAL1,10, and terminators TTDH3 and TCYC1 were amplified using PCR to construct pESC-G418 series vectors, and the corresponding plasmid or S. cerevisiae genome was used as the template. These fragments with a 40-bp homology arm were inserted into multiple cloning sites of pESC-G418 using the Gibson Assembly® Cloning Kit (NEB). Potential sgRNA targets were predicted at http://www.rgenome.net/cas-designer/ to design specific sgRNAs for corresponding targeted genes. All plasmids used in the study are listed in Table 2, and the corresponding primers are listed in Supplementary Table S2.
Gene localization of the mogrol biosynthesis pathway was observed in strain CEN-PK2-1C. Strain CEN-PK2-1C was transformed by plasmids pY15-SgCDS-GFP, pY15-SgEPH3-GFP, pY15-CYP87D18-GFP-mCherry-SEC12, and pY15-AtCPR1-GFP-mCherry-SEC12, thereby yielding strains LMOR001, LMOR002, LMOR003 and LMOR004. These strains were streaked onto an SD-Leu plate and cultured at 30°C for 2 days. Later, a single colony was picked into a 2 ml SD-Leu medium, cultured at 30°C and 220 rpm for 24 h, and washed twice with 1× phosphate-buffered saline (0.8% NaCl, 0.02% KCl, 0.144% Na2HPO4, and 0.024% KH2PO4, pH 7.4). Subsequently, 2-μL preparations were directly plated on slides. Strain LMOR001 culture was stained with Nile red solution in acetone (1:10, v/v; 1 mg/ml; Solarbio) and incubated for 60 min in the dark at 25°C to further confirm the specific distribution of SgCDS.
Fluorescence imaging was performed on a Nikon C-HGF microscope with a ×100 oil immersion objective. GFP fluorescence (excitation, 490 nm; emission, 530 nm), mCherry fluorescence (excitation, 580 nm; emission, 615 nm), and Nile red fluorescence (excitation, 561 nm; emission, 615 nm) were detected by microscopy. Image analysis was carried out on the Leica TCS SP8 software package and ImageJ (NIH).
The endogenous promoter of ERG7 and the gfp gene were inserted into plasmid pY13 to construct plasmid pY13-PERG7-GFP. Strain CEN-PK2-1C was transformed by plasmid pY13-PERG7-GFP to yield strain FMOR001. The mCherry protein shared the bidirectional promoter PGAL1,10 with the dcas9 protein on plasmid pML104-dCas9. The corresponding sgRNA was integrated into plasmid pML104-dCas9 to construct plasmid pML104-dCas9-mCherry-1/2/3. Strain CEN-PK2-1C was transformed by plasmids pY13-PERG7-GFP and pML104-dCas9, thereby yielding strains FMOR002, FMOR003, and FMOR004, respectively. These strains were streaked onto an SD-Ura-His (FMOR002, FMOR003 and FMOR004) or SD-His (FMOR001) plate and cultured at 30°C for 2 days. Later, a single colony was picked into a 2 ml SD-Ura-His or SD-His medium and cultured at 30°C and 220 rpm for 24 h. The 2% (v/v) seed cultures were inoculated into a 200 μL SD-Ura-His or SD-His medium in a 96-well fluorescent plate and cultured at 30°C for 72 h. Strains with plasmids pML104-dCas9 and pY13-PERG7-GFP were cultivated in the SD-Ura-His medium, and the strain with pY13-PERG7-GFP was cultivated in the SD-His medium. All strains containing plasmid pML104-dcas9 needed 2%, 4%, 6%, 8%, 10%, and 20% galactose at 0 h. GFP fluorescence (excitation, 490 nm; emission, 530 nm), mCherry fluorescence (excitation, 580 nm; emission, 615 nm), and OD600 of strains were detected using a Cytation microplate reader (BioTek). Eq. 1 was used to calculate the repression fold of GFP fluorescence (θ) by different sgRNAs.
where (GFP/OD)x is the relative fluorescence intensity of GFP in strain FMOR002, FMOR003 or FMOR004; (GFP/OD)0 is the relative fluorescence intensity of GFP in strain MOR001 (without GFP); and (GFP/OD)1 is the relative fluorescence intensity of GFP in strain FMOR001.
The strains were streaked onto a YPD solid plate and cultured at 30°C for 2 days. A single colony was picked into a 2 ml YPD medium at 30°C and 220 rpm for 24 h. The 1% (v/v) cultures were inoculated into a 250 ml flask containing 50 ml YPD medium. Strains containing plasmid pESC-G418 needed 200 mg/L G418 to add to the YPD medium. All flask fermentation results represented the mean ± standard deviation (SD) of three independent experiments.
A 1 ml fermentation culture was collected and broken by a high-pressure homogenizer (FastPrep-24 5G, United States ) and extracted twice with ethyl acetate to measure the mogrol and squalene concentrations. For squalene analysis (Paramasivan and Mutturi, 2017), the samples were filtered using a 0.45 μm organic phase filtration membrane and injected into a high-performance liquid chromatography instrument (Agilent 1260 series). The column was a C18 ODS column (5 μm, 250 × 4.6 mm; Thermo Fisher Scientific), and the column temperature was 40°C. The pump flow rate was 1.6 ml/min, and the mobile phase was 100% acetonitrile. The injection volume was fixed to 10 μL. For mogrol analysis, the samples extracted with ethyl acetate were resuspended in 100 μL methanol after evaporation. Mogrol was quantified using liquid chromatography mass spectrometry (LC–MS) according to Itkin et al. (2016).
According to the mogrol synthetic pathway in plants, the initial step in mogrol biosynthesis is the epoxidation of 2, 3-oxidosqualene to form 2, 3; 22, 23-diepoxysqualene (the triterpenoid skeleton of mogrol). Then, 2, 3; 22, 23-diepoxysqualene is cycled into 24, 25-epoxycucurbiadienol by SgCDS. SgEPH3 catalyzes the unique hydroxylation of 24, 25-epoxycucurbiadienol to form trans-24, 25-dihydroxycucurbitadienol. Finally, CYP87D18 adds an oxygen atom to position C11 of 24, 25-dihydroxycucurbitadienol to form mogrol (Figure 1).
Therefore, to construct the de novo biosynthetic pathway of mogrol in S. cerevisiae, SgCDS, SgEPH3, and CYP87D18 from S. grosvenorii and the CPR gene AtCPR1 from A. thaliana were integrated into the chromosome of strain Y4 (a high-yield strain of squalene constructed previously), yielding strain MOR001. The expression of these genes was under the control of the galactose-inducible promoter PGAL1,10. Synthetic genes were first employed as templates and then the genes were amplified to investigate the expression and distribution of SgCDS, SgEPH3, CPY87D18, and AtCPR1. Genes were integrated into plasmids, and the gfp gene was fused downstream of them (Figure 2A). These plasmids were transformed into strain MOR001, thereby yielding strains LMOR001, LMOR002, LMOR003 and LMOR004. Comparing the fluorescent images of SgCDS to the bright-field images, the green fluorescent spots showed a point-like distribution. These apparent traits showed that these spots are like lipid drops (LDs), the primary storage sites for neutral lipids in cells (Hammer and Avalos, 2017). Then, cells were stained with Nile red, a dye that can directly stain LDs (Liu et al., 2020). The spots in Nile red images exactly overlapped with the green fluorescent spots. It was concluded that SgCDS was specifically localized in LDs (Figure 2B), which was similar to the cycloartenol synthase from Arabidopsis thaliana (Milla et al., 2002) and α-amyrin synthase from Malus × domestica (Yu et al., 2020). According to the merged results of the colocalization of AtCPR1 and ER, the localization of GFP fluorescence was consistent with sec12p, suggesting that AtCPR1 was an ER-localized protein in S. cerevisiae (Figure 2C). Besides, laser scanning confocal microscopy (LSCM) analysis revealed that the GFP fluorescence of SgEPH3 filled the entire cytoplasm of S. cerevisiae, showing that SgEPH3 was located in the cytoplasm (Figure 2D). Furthermore, the GFP fluorescence of CYP87D18 was weak, indicating that the enzyme activity of CYP87D18 was a bottleneck in the biosynthesis pathway of mogrol (Figure 2E). These results proved that heterologous SgCDS, SgEPH3, CPY87D18 and AtCPR1 genes were successfully expressed in strain MOR001.
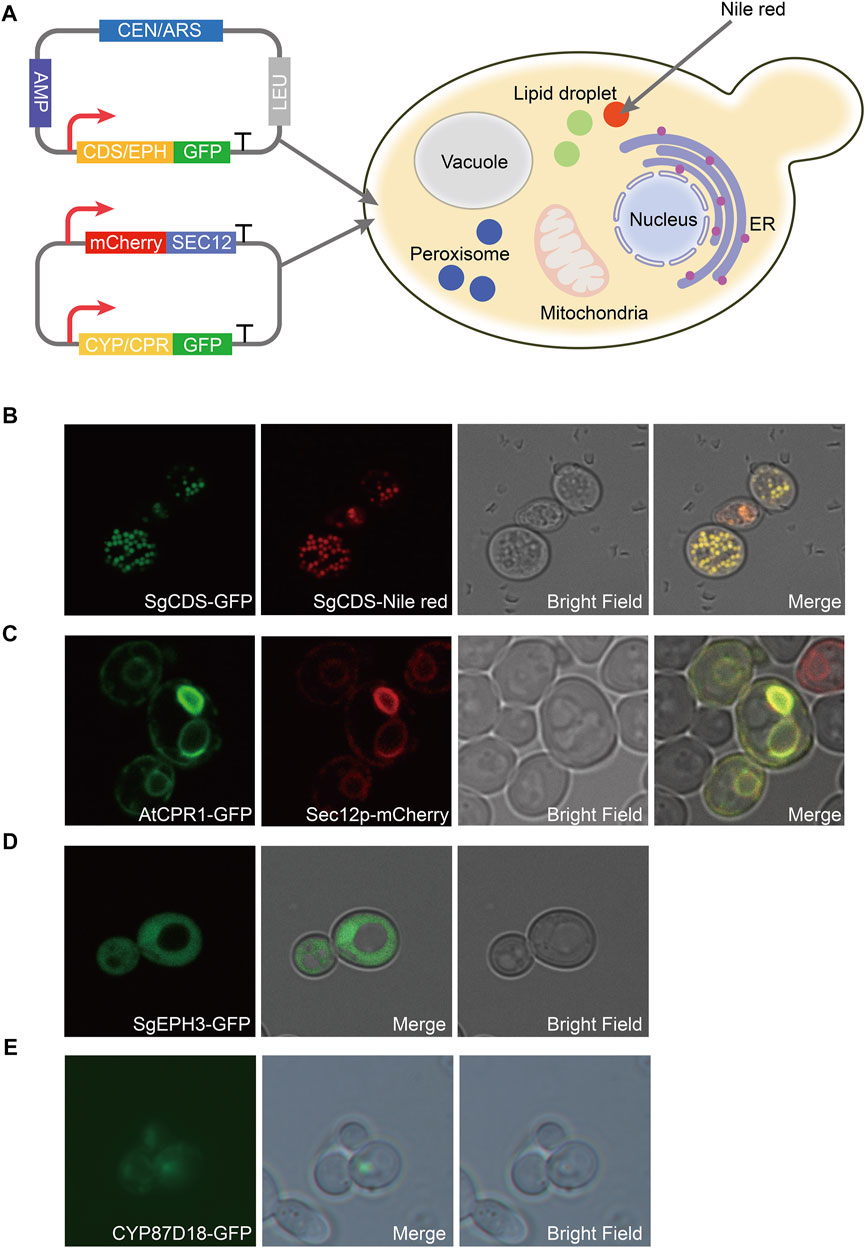
FIGURE 2. Distribution of enzymes involved in the synthesis pathway of mogrol. (A) Construction of plasmids pY15-GFP and pY15-GFP-mCherry. SEC12 was proven to be an ER-localized protein. GFP protein was used to visualize the localization of the target protein. mCherry protein was used to specifically mark the ER, and Nile red was used to show intracellular lipophilic regions (B–E) Distribution analysis of SgCDS, SgEPH, CYP87D18, and AtCPR1 by LSCM, respectively.
After strain MOR001 cultivation, a peak with a mass-to-charge ratio (m/z) value of 521.389 was detected using LC–MS in strain MOR001, in accordance with the m/z values of mogrol standard (Figures 3A,B). Mogrol could only be detected when the inducer galactose was added to the medium. These results demonstrated that the de novo biosynthetic pathway of mogrol was successfully constructed in strain MOR001. However, the mogrol titer in strain MOR001 was only 19.1 ng/L. The metabolic network was divided into three modules, including the squalene, 2, 3; 22, 23-diepoxysqualene, and mogrol synthesis modules, to synergistically regulate the metabolic network of mogrol (Figure 1). For the squalene synthesis module, the precursor acetyl-CoA supply was enhanced to facilitate squalene synthesis. In the cytoplasm, CIT2 and MLS1 involved in the glyoxylate pathway consumed intracellular acetyl-CoA. Therefore, CIT2 and MLS1 were knocked out to reduce the consumption of the precursor acetyl-CoA using the CRISPR–Cas9 system, generating strain MOR002. However, the mogrol titer in strain MOR002 only reached 25.9 ng/L, which was 1.3-fold higher than that of MOR001 (Figure 3D). These results indicated that the intracellular content of acetyl-CoA and squalene was sufficient to supply mogrol synthesis, which may be due to the high content of precursor squalene in the original strain Y4.
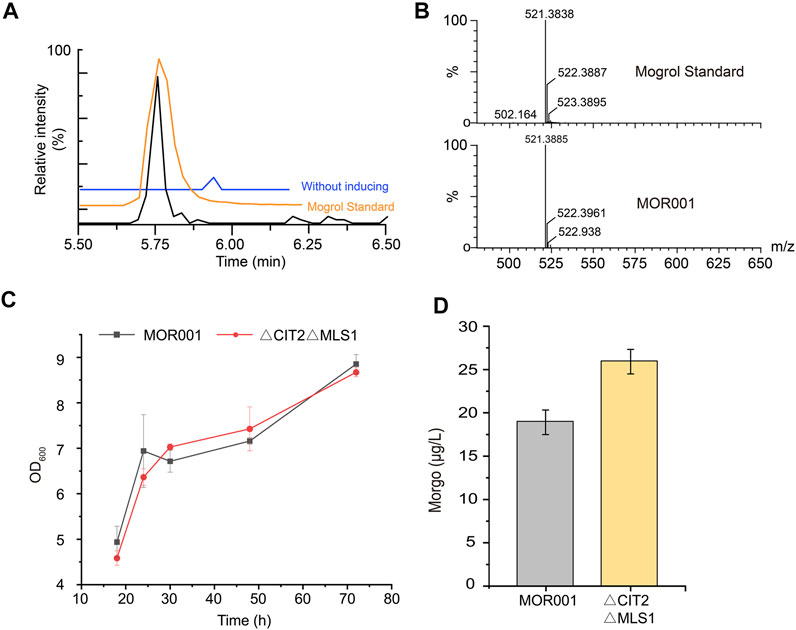
FIGURE 3. LC–MS analysis of mogrol produced by strain MOR001. (A,B) Chromatogram analysis (A) and mass spectrogram analysis (B) of MOR001 and mogrol standard. The blue line in (A) represents strain MOR001 in the YPD medium without induction with galactose (C,D) Cell growth curves (C) and mogrol titer (D) in engineered strain MOR002, in which MLS1 and CIT2 genes were knocked out. Error bars represent the SD of biological triplicates (n = 3).
In the 2, 3; 22, 23-diepoxysqualene synthesis module, Erg7p (lanosterol synthase) catalyzed the cyclization of (3S)-2, 3-oxidosqualene to lanosterol and further flowed into the synthesis pathway of ergosterol, one of the main components of the plasma membrane (Nes, 2011; Jordá and Puig, 2020). In general, 2,3-oxidosqualene prefers to be cyclized by ERG7 rather than oxidized into 2, 3; 22, 23-diepoxysqualene (Corey and Gross, 1967; Shan et al., 2005). Therefore, knocking out or inhibiting the activity of ERG7 is crucial for 2, 3; 22, 23-diepoxysqualene synthesis. However, directly blocking the metabolic flux to ergosterol would affect cell growth and require additional supplementation of ergosterol and Tween 80 in the medium to maintain cell growth, which was not cost-saving and conducive to industrial development (Itkin et al., 2016; Qiao et al., 2019).
The CRISPRi system is widely used to inhibit competing pathways in metabolic networks of various products (Gilbert et al., 2013; Jensen et al., 2017; Wu et al., 2018). Therefore, the CRISPRi system was used to inhibit ERG7 expression, thereby redirecting the metabolic flux toward mogrol biosynthesis, enabling the decoupling of cell growth and product synthesis. First, the inhibitory targets of the CRISPRi system on the promoter of ERG7 (PERG7) and their corresponding inhibitory effects were screened using the gfp reporter gene (Figure 4A). Control plasmid pY13-PERG7-GFP containing the reporter gene gfp was constructed to characterize the transcription strength of promoter PERG7. Three sgRNAs targeting different positions of the promoter PERG7 were inserted into plasmid pML104-dCas9-mCherry, which contained a tandem expression cassette dCas9-mCherry under the control of galactose-inducible promoter PGAL1,10, thereby obtaining plasmids pML104-dCas9-mCherry-1/2/3 (Figure 4B). The dCas9 expression level in plasmid pML104-dCas9-mCherry-1/2/3 was characterized according to the fluorescence intensity of mCherry. The control plasmid pY13-PERG7-GFP was transformed into the wild-type strain CEN-PK2-1C, yielding strain FMOR001. Plasmids pY13-PERG7-GFP and pML104-dCas9-mCherry-1/2/3 were cotransformed into the wild-type strain CEN-PK2-1C, thereby yielding strain FMOR002, FMOR003, and FMOR004 (Figure 4B). Finally, the fluorescence intensity of GFP and mCherry in strains FMOR001, FMOR002, FMOR003, and FMOR004 was measured. The results showed that when the inducer galactose concentration was 6 g/L, the repression fold of GFP fluorescence (θ) in strains FMOR002, FMOR003 and FMOR004 were 6.1-, 7.4-, and 3.5-fold, respectively (Figure 4C, Supplementary Figure S1). When the galactose concentration was >6 g/L, the repression fold of GFP fluorescence (θ) by different sgRNAs was gradually decreased (Figure 4C). In addition, the expression level of dCas9 in strains FMOR004 was higher than FMOR002 and FMOR003, but the repression fold of GFP fluorescence (θ) in strains FMOR004 were lowest (Figure 4D). This result indicating that sgRNA rather than the dCas9 protein was the most essential for the efficiency of the CRISPRi system.
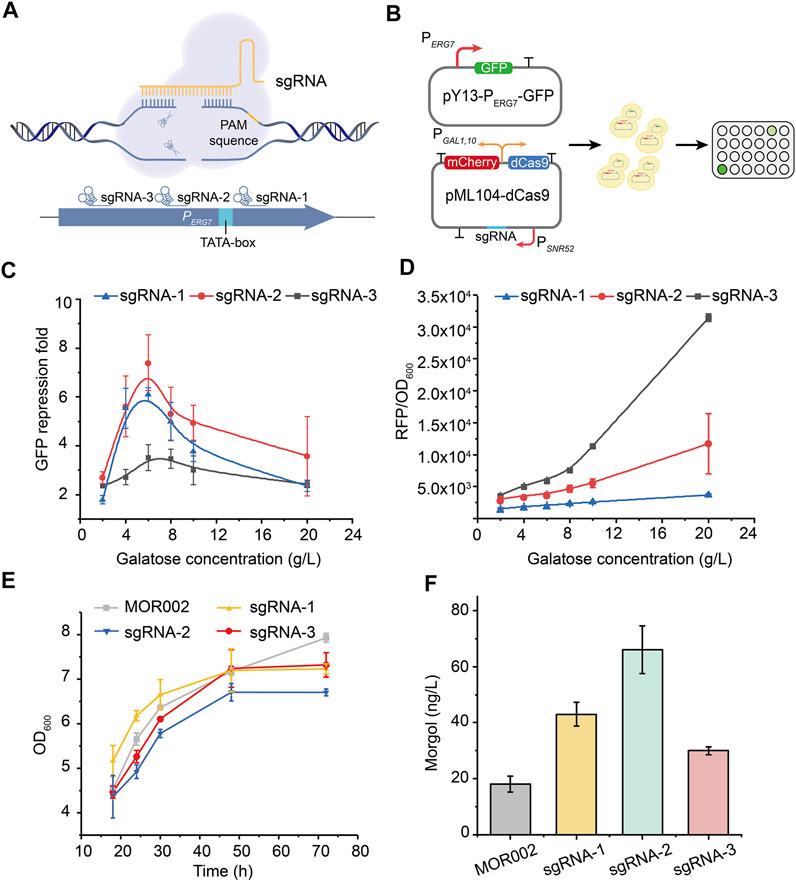
FIGURE 4. Repression of the lanosterol biosynthesis pathway by the CRISPRi system. (A) CRISPRi was employed to repress gene expression by an inactive Cas9 protein (dCas9), which can bind to a specific sequence by sgRNA and block the elongation of RNA polymerase. sgRNAs were chosen from the endogenous ERG7 promoter. (B) Construction of plasmids that characterize the feasibility of the CRISPRi system in yeast and the verification of fluorescence in a 96-well plate. (C) GFP fluorescence repression fold of different sgRNAs of promoter PERG7. (D) mCherry fluorescence intensity of dCas9 protein. (E) Cell growth curves of engineered strains in a 250-ml flask culture in a YPG medium (F) Mogrol production in engineered strains at 96 h. In (E) and (F), sgRNA-1 represented strain MOR003, sgRNA-2 represented strain MOR004, sgRNA-3 represented strain MOR005. Error bars represent the SD of biological triplicates (n = 3).
Subsequently, dcas9-sgRNA expression cassettes in plasmids pML104-dCas9-mCherry-1/2/3 were cloned and integrated into the genome of strain MOR002 to repress ERG7 expression, thereby yielding strains MOR003, MOR004, and MOR005, respectively. The growth of strains MOR003, MOR004, and MOR005 was similar to strain MOR002 (Figure 4E), demonstrating that the CRISPRi system could inhibit ERG7 expression without affecting cell growth. Meanwhile, the mogrol titer in strains MOR003, MOR004, and MOR005 reached 43, 66, and 30 ng/L, increased by 53%, 135%, and 7%, respectively, compared to that of MOR002 (Figure 4F), consistent with the GFP fluorescence repression fold of different sgRNAs.
To further improve mogrol production, the metabolic flux of the mogrol synthesis module was engineered. First, the precursor 2, 3-oxidosqualene flow was “pulled” into the mogrol synthesis module rather than cell growth by overexpressing ERG1 (squalene epoxidase), which catalyzed squalene to 2, 3-oxidosqualene and 2, 3; 22, 23-diepoxysqualene. Another copy of ERG1 was integrated into the genome of strain MOR002, and GAL80 (a transcriptional factor) (Pilauri et al., 2005) was simultaneously knocked out, yielding strain MOR006. LC–MS results showed that the mogrol titer in strain MOR006 reached 48 ng/L, which showed a further 240% increase than that of MOR002 (Figure 5A). When another copy of ERG1 was integrated into the genome of strains MOR003, MOR004 and MOR005, the mogrol titer in the resulting strains MOR007, MOR008 and MOR009 were 13.9-, 33-, and 4.8-fold as high as that of strain MOR006, reached 670, 1600, and 230 ng/L, respectively (Figure 5A). These results indicated that only inhibiting the synthetic pathway of lanosterol and synergistically “pulling” the metabolic flux flow into the 2, 3; 22, 23-diepoxysqualene could maximize mogrol production. Subsequently, the copy number of ERG1 in strain MOR008 was further increased to improve mogrol production. When the copy number of ERG1 increased to three copies (MOR010), the mogrol titer was further increased to 5.67 μg/L. When the copy number of ERG1 reached four copies (MOR011), the mogrol titer decreased to 0.68 μg/L (Figure 5C). Finally, the titer of the intermediate metabolite squalene was also measured in strains MOR004, MOR008, MOR010, and MOR011. The results showed that the squalene titer in strains MOR008, MOR010 and MOR011 at 120 h reached 617, 1002.9, and 971.4 mg/L, which was 1.1-, 1.86- and 1.81-fold higher respectively, compared to that of the strain MOR004 (Figure 5D). The biomass of MOR008, MOR010 and MOR011 was significantly improved compared to MOR004, which may be due to squalene being an essential precursor for cells to synthesize the plasma membrane (Figure 5B). These results demonstrated that ERG1 increased mogrol production by “pulling” the metabolic flux toward the biosynthesis pathway of mogrol.
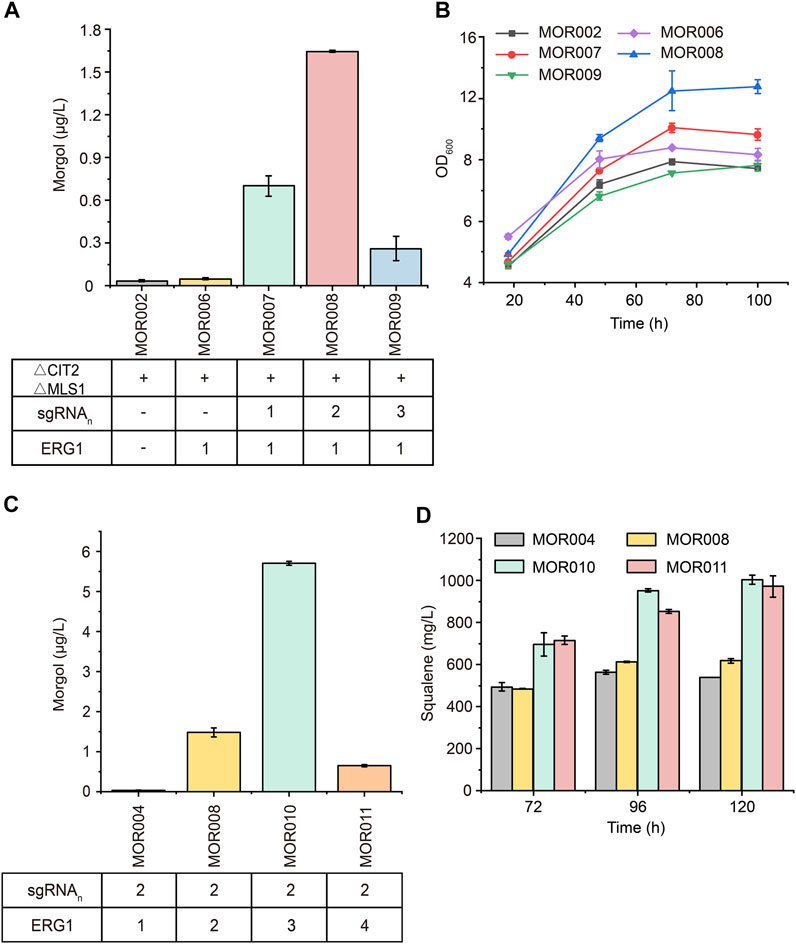
FIGURE 5. Effects of multicopy expression of ERG1 on mogrol. (A) Mogrol production in MOR007 to MOR009 compared to MOR002 and MOR006 (B) Cell growth curves of engineered strains that overexpressed ERG1 in YPD medium. (C,D) Mogrol (C) and squalene (D) production after multicopy genome integration of ERG1. Error bars represent the SD of biological triplicates (n = 3).
Most plant-derived P450 enzymes (CYP) are located in the ER and are often considered as the rate-limiting enzymes in the biosynthetic pathway (Hausjell et al., 2018). They rely on CPRs to obtain electrons from NAD(P)H for subsequent catalytic reactions (Sadeghi and Gilardi, 2013). Many studies showed that different sources of CPRs may affect CYP activity, and the compatibility of native CPRs with CYPs in heterologous hosts is often not optimal (Theron et al., 2019). Hence, screening CPRs from different sources is an effective strategy to increase the activity of P450 enzymes.
Therefore, CYP87D18 was first inserted into a high-copy plasmid pESC-G418 to test whether increasing its expression would improve mogrol production, yielding plasmid pESC-G418-CYP. Plasmid pESC-G418-CYP was transformed to MOR004 and MOR010 to construct strains MOR012 and MOR013, respectively. Compared to MOR004, CYP87D18 expression on a multicopy plasmid significantly increased mogrol production (1.05 μg/L; Figure 6A). This result revealed that increasing P450 enzyme expression was beneficial to mogrol synthesis, whereas it decreased in strain MOR013 (1.64 μg/L) compared to strain MOR010 (5.67 μg/L).
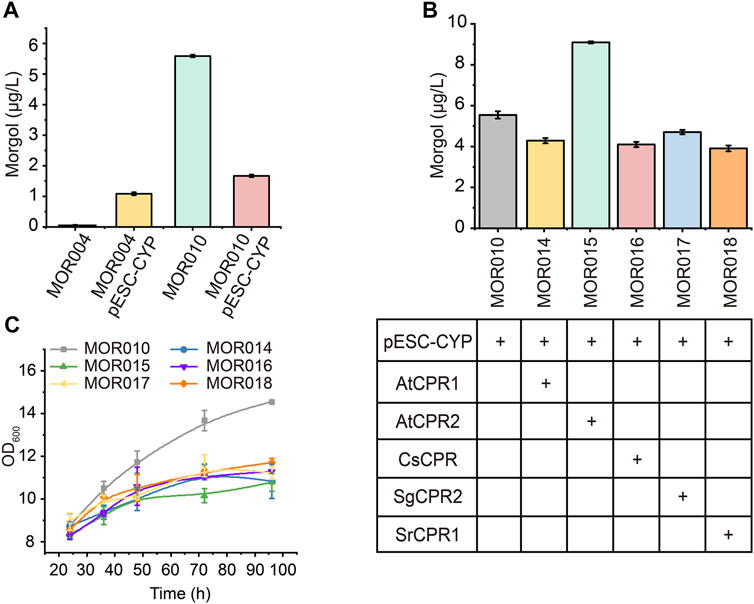
FIGURE 6. Combined optimization of CYPs-CPRs pairing to improve mogrol production. (A) Mogrol production by overexpressing CYP87D18 using a high-copy plasmid. (B,C) Mogrol production by optimizing CYPs-CPRs pairing (B) and cell growth curves of engineered strains (C) in a 250-ml flask. Error bars represent the SD of biological triplicates (n = 3).
Subsequently, CYP87D18 was coexpressed in combination with five different CPRs in plasmid pESC-G418, including AtCPR1 and AtCPR2 from A. thaliana, SgCPR2 from S. grosvenorii (Zhao et al., 2018), CsCPR from Cucumber, and SrCPR1 from Stevia (Gold et al., 2018). The resulting plasmids were transformed into strain MOR010, thereby obtaining strains MOR014, MOR015, MOR016, MOR017, and MOR018, respectively. Fermentation data showed that the mogrol titer in strain MOR015 reached 9.1 μg/L, which was 1.6-fold higher than that of the strain MOR010. In contrast, the mogrol titer in strains MOR014 MOR016, MOR017 and MOR018 was lower than that of MOR010 (Figure 6B). These results demonstrated that the selection of appropriate CPR is crucial for the catalytic efficiency of CYPs in heterologous hosts. Additionally, the biomass of these strains was only 70% of MOR010 (Figure 6C), which may be due to the additional burden on the organism caused by that the overexpression of the second recombinant protein (Hausjell et al., 2018).
As the aglycone of mogrosides, mogrol has a significant effect on reducing blood glucose and blood lipid. However, more studies focused on the biological properties of mogrol than on its biosynthesis. Additionally, the biosynthesis pathways for mogrol from other precursors have not been explored (Li et al., 2019; Qiao et al., 2019). S. cerevisiae is an ideal host for the heterologous synthesis of plant-derived natural products due to its ability to express P450 enzymes located in the ER. Therefore, this study aimed to construct a yeast cell factory for efficient mogrol production. First, a de novo synthesis pathway for mogrol was successfully constructed in S. cerevisiae. Subsequently, to improve the biosynthesis efficiency of the chassis cell, the metabolic network was divided into three modules. For the squalene synthesis module, the central metabolite acetyl-CoA flow was promoted into the squalene synthesis pathway by inhibiting its flow into the glyoxylate cycle. Surprisingly, in this study, directly knocking out CIT2 and MLS1 involved in the glyoxylate cycle would not affect cell growth (Figure 3C). However, Liu et al. (2019) knocked out CIT2 and MLS1 in a chassis cell of β-amyrin, resulting in cell growth inhibition. This may be caused by changes in the metabolic network of cells due to the introduction of different heterologous pathways. In addition, our results indicated that the content of the precursor squalene was sufficient for the synthesis of mogrol. Compared to the squalene synthesis module, the low metabolic fluxes of the 2, 3; 22, 23-diepoxysqualene synthesis and mogrol synthesis module may be the major rate-limiting steps for the synthesis of mogrol. Therefore, we subsequently enhanced the metabolic fluxes of 2, 3; 22, 23-diepoxysqualene synthesis module and mogrol synthesis module.
In 2, 3; 22, 23-diepoxysqualene synthesis module, ERG1 was the only enzyme involved. ERG1 is a key rate-limiting enzyme in the sterol biosynthetic pathway. When the intracellular concentration of lanosterol is high, its activity will be feedback-inhibited, thereby maintaining the homeostasis of sterols. Therefore, reducing the intracellular sterol content is crucial for maintaining the catalytic activity of ERG1. Inhibiting ERG7 (related to sterol synthesis pathway) expression will promote ERG1 to catalyze the synthesis of 2, 3; 22, 23-diepoxysqualene from squalene (Xu et al., 2004; Zhao et al., 2019). The traditional methods of inhibiting ERG7 expression were mainly by replacing the native PERG7 promoter with an inducible promoter, such as methionine-inhibited promoter MET3 (Baadhe et al., 2013) and a copper-inhibited promoter CTR3 (Paddon et al., 2013; Rodriguez et al., 2016). However, adding methionine and copper ions will increase production costs, and copper ions are toxic to yeast. Therefore, the CRISPR-dCas9 system was employed to inhibit ERG7 expression. Three sgRNAs that inhibited ERG7 expression in different strengths were screened. sgRNA-2 had the best inhibitory effect on ERG7 expression, probably because its target was closest to the TATA-box of PERG7 (Figure 3). Moreover, the mogrol titer gradually increased with the inhibited effect of sgRNA. These results confirmed that reducing the metabolic flux of the sterol pathway is an effective way to increase mogrol production. More importantly, compared with other modules, the yield of mogrol was improved most obviously by adjusting this module. Furthermore, although ERG1 could catalyze the synthesis of 2, 3; 22, 23-diepoxysqualene from squalene, its activity may be low due to 2, 3; 22, 23-diepoxysqualene is not a known precursor for a metabolite in S. cerevisiae. Therefore, the activity of ERG1 is likely a major bottleneck for mogrol biosynthesis in S. cerevisiae. In future work, screening for a more efficient squalene epoxidase, or using enzyme engineering techniques to enhance the catalytic activity of ERG1, is necessary to increase the production of mogrol.
In mogrol synthesis module, the critical engineering step for mogrol overproduction was the low catalytic efficiency of P450 catalytic system. P450 enzymes are generally considered to be the rate-limiting step for triterpene synthesis (Li et al., 2021). In this study, CYP87D18 expression in MOR001 was weak. Therefore, for the mogrol synthesis module, CYP87D18 was expressed on the high-copy plasmid in strain MOR004, resulting in a 12-fold mogrol production. This result demonstrated that CYP87D18 might be a rate-limiting factor for improving the mogrol titer. While overexpressing CYP87D18 in strain MOR010, mogrol production decreased. This may be due to an imbalance of the cofactor NADPH in the cytoplasm, as Erg9p (Paramasivan and Mutturi, 2017), Erg1p (Ruckenstuhl et al., 2007) and P450 enzymes will consume NADPH for catalytic reactions. If the expression of the P450 enzyme and Erg1p increases, it may lead to an insufficient supply of intracellular NADPH. Most eukaryotic P450s are complex membrane proteins and require their redox partner CPRs for electron transfer (Jiang et al., 2021). Besides, a suitable CYP-CPR pairing is usually beneficial for the catalytic efficiency of P450 enzymes (Chen et al., 2021). The electron transfer efficiency between them will further affect the subsequent catalytic ability of P450 enzymes. Theron et al. (2019) found that a coexpressed CPR from Ustilago maydis (UmCPR) with CYP53B1 resulted in a six-to seven-fold p-hydroxybenzoic acid volumetric yield than that of the endogenous CPR. Additionally, Zhu et al. (2018) introduced a novel CPR reduction system to the metabolic network of 11-oxo-β-amyrin and glycyrrhetinic acid (GA), and the 11-oxo-β-amyrin and GA titer increased by 1422- and 946.5-fold, which reached 108.1 and 18.9 mg/L, respectively. CPRs from different sources have different catalytic efficiencies for P450 enzymes, which will further affect the production synthesis. In this study, by selecting CPR from different sources, AtCPR2 from A. thaliana had the highest electron transfer efficiency among the candidate CPRs, and the mogrol titer was 1.6-fold higher compared to that of MOR010. Meanwhile, we found that expressing CYP and CPRs through a high-copy plasmid resulted in a reduction of biomass. When the reaction ratio of CYP and CPR was not optimal, the excessive electron transport might lead to the release of reactive oxygen species (ROS) and cause cell damage (Zhao et al., 2017; Lin et al., 2022).
Though we significantly improved the mogrol titer by optimizing the expression of key genes of the mogrol metabolic network and improving the catalytic activity of P450 enzyme, the mogrol titer was still very low. We speculated that the activity of squalene epoxidase ERG1 and P450 enzyme might be the main rate-limiting steps that limited the synthesis of mogrol. Generally, squalene epoxidase only has the function of cyclizing squalene to form 2, 3-oxidosqualene. However, Itkin et al. (2016) found that squalene epoxidase derived from S. grosvenorii could further catalyze the synthesis of 2, 3; 22, 23-diepoxysqualene from 2, 3-oxidosqualene. ERG1 could perform the same function in yeast, but this catalytic step may be a side reaction because 2, 3; 22, 23-diepoxysqualene is not a known precursor for a metabolite in S. cerevisiae. Moreover, the enzymatic activity of P450 enzyme is the rate-limiting step that restricts the efficient synthesis of most terpenoids in S. cerevisiae (Hausjell et al., 2018; Jiang et al., 2021). Herein we improved the catalytic activity of P450 enzymes by paring it with a most suitable CPR, and we will carry out directed evolution of P450 enzyme to further improve the catalytic performance in the future. The titers of various hydrophobic terpenoids were improved through compartmentalizing their metabolite pathways into LDs, such as lycopene (Ma et al., 2019), α-amyrin (Yu et al., 2020), and ginsenosides (Shi et al., 2021). Therefore, compartmentalizing ERG1 and the enzymes involved in mogrol synthesis module into LDs may be an effective approach to increase the mogrol production. In summary, we achieved the bioproduction of mogrol by the engineered yeast cell factories, and the used metabolic engineering strategies may be useful for the bioproduction of the other natural products in yeast.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
SW and LL conceived this project and designed the experiments. SW performed experiments. SW wrote the manuscript. XX, XL, YL, JL, GD, and LL edited the manuscript.
This work was financially supported by the National Natural Science Foundation of China (32021005, 32070085, 31871784), National Key Research and Development Program of China (2020YFA0908300), and the Fundamental Research Funds for the Central Universities (JUSRP52019A, JUSRP121010, JUSRP221013).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Guanjun Tao and Tian Yang for helping us to detect mogrol by LC-MS. We would like to express our gratitude to Prof. Sanyang Li for providing the gene CsCPR.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.919526/full#supplementary-material
EFSA Panel on Food Additives and Flavourings Younes, M., Aquilina, G., Engel, K. H., Fowler, P., Frutos Fernandez, M. J., et al. (2019). Safety of Use of Monk Fruit Extract as a Food Additive in Different Food Categories. EFSA. J. 17, e05921. doi:10.2903/j.efsa.2019.5921
Ajikumar, P. K., Xiao, W.-H., Tyo, K. E. J., Wang, Y., Simeon, F., Leonard, E., et al. (2010). Isoprenoid Pathway Optimization for Taxol Precursor Overproduction in Escherichia coli. Science 330, 70–74. doi:10.1126/science.1191652
Araya-Garay, J. M., Feijoo-Siota, L., Rosa-dos-Santos, F., Veiga-Crespo, P., and Villa, T. G. (2012). Construction of New Pichia pastoris X-33 Strains for Production of Lycopene and β-carotene. Appl. Microbiol. Biotechnol. 93, 2483–2492. doi:10.1007/s00253-011-3764-7
Arendt, P., Miettinen, K., Pollier, J., De Rycke, R., Callewaert, N., and Goossens, A. (2017). An Endoplasmic Reticulum-Engineered Yeast Platform for Overproduction of Triterpenoids. Metab. Eng. 40, 165–175. doi:10.1016/j.ymben.2017.02.007
Baadhe, R. R., Mekala, N. K., Parcha, S. R., and Prameela Devi, Y. (2013). Combination ofERG9Repression and Enzyme Fusion Technology for Improved Production of Amorphadiene inSaccharomyces Cerevisiae. J. Anal. Methods Chem. 2013, 1–8. doi:10.1155/2013/140469
Bu, X., Lin, J.-Y., Cheng, J., Yang, D., Duan, C.-Q., Koffas, M., et al. (2020). Engineering Endogenous ABC Transporter with Improving ATP Supply and Membrane Flexibility Enhances the Secretion of β-carotene in Saccharomyces cerevisiae. Biotechnol. Biofuels. 13, 168. doi:10.1186/s13068-020-01809-6
Chen, G., Liu, C., Meng, G., Zhang, C., Chen, F., Tang, S., et al. (2018). Neuroprotective Effect of Mogrol against Aβ1-42-Induced Memory Impairment Neuroinflammation and Apoptosis in Mice. J. Pharm. Pharmacol. 71, 869–877. doi:10.1111/jphp.13056
Chen, K., Liu, C., Shao, M., Xu, Z., Yang, T., and Rao, Z. (2021). Enhancing the Biotransformation Efficiency of Human CYP17A1 in Pichia pastoris by Co-expressing CPR and Glucose-6-Phosphate Dehydrogenase Simultaneously. Syst Microbiol Biomanuf. doi:10.1007/s43393-021-00063-7
Chiu, C.-H., Wang, R., Lee, C.-C., Lo, Y.-C., and Lu, T.-J. (2013). Biotransformation of Mogrosides from Siraitia Grosvenorii Swingle by Saccharomyces cerevisiae. J. Agric. Food Chem. 61, 7127–7134. doi:10.1021/jf402058p
Corey, E. J., and Gross, S. K. (1967). Formation of Sterols by the Action of 2,3-Oxidosqualene-Sterol Cyclase on the Factitious Substrates, 2,3,: 22,23-dioxidosqualene and 2,3-Oxido-22,23-Dihydrosqualene. J. Am. Chem. Soc. 89, 4561–4562. doi:10.1021/ja00993a079
Dai, L., Liu, C., Zhu, Y., Zhang, J., Men, Y., Zeng, Y., et al. (2015). Functional Characterization of Cucurbitadienol Synthase and Triterpene Glycosyltransferase Involved in Biosynthesis of Mogrosides from Siraitia Grosvenorii. Plant Cell Physiol. 56, 1172–1182. doi:10.1093/pcp/pcv043
Engels, B., Dahm, P., and Jennewein, S. (2008). Metabolic Engineering of Taxadiene Biosynthesis in Yeast as a First Step towards Taxol (Paclitaxel) Production. Metab. Eng. 10, 201–206. doi:10.1016/j.ymben.2008.03.001
Gietz, R. D., and Schiestl, R. H. (2007). High-efficiency Yeast Transformation Using the LiAc/SS Carrier DNA/PEG Method. Nat. Protoc. 2, 31–34. doi:10.1038/nprot.2007.13
Gilbert, L. A., Larson, M. H., Morsut, L., Liu, Z., Brar, G. A., Torres, S. E., et al. (2013). CRISPR-mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 154, 442–451. doi:10.1016/j.cell.2013.06.044
Gold, N. D., Fossati, E., Hansen, C. C., Difalco, M., Douchin, V., and Martin, V. J. J. (2018). A Combinatorial Approach to Study Cytochrome P450 Enzymes for De Novo Production of Steviol Glucosides in Baker's Yeast. ACS Synth. Biol. 7, 2918–2929. doi:10.1021/acssynbio.8b00470
Hammer, S. K., and Avalos, J. L. (2017). Harnessing Yeast Organelles for Metabolic Engineering. Nat. Chem. Biol. 13, 823–832. doi:10.1038/nchembio.2429
Harada, N., Ishihara, M., Horiuchi, H., Ito, Y., Tabata, H., Suzuki, Y. A., et al. (2016). Mogrol Derived from Siraitia Grosvenorii Mogrosides Suppresses 3T3-L1 Adipocyte Differentiation by Reducing cAMP-Response Element-Binding Protein Phosphorylation and Increasing AMP-Activated Protein Kinase Phosphorylation. PLoS One 11, e0162252. doi:10.1371/journal.pone.0162252
Hausjell, J., Halbwirth, H., and Spadiut, O. (2018). Recombinant Production of Eukaryotic Cytochrome P450s in Microbial Cell Factories. Biosci. Rep. 38, BSR20171290. doi:10.1042/BSR20171290
Itkin, M., Davidovich-Rikanati, R., Cohen, S., Portnoy, V., Doron-Faigenboim, A., Oren, E., et al. (2016). The Biosynthetic Pathway of the Nonsugar, High-Intensity Sweetener Mogroside V from Siraitia Grosvenorii. Proc. Natl. Acad. Sci. U.S.A. 113, e7619–e7628. doi:10.1073/pnas.1604828113
Jensen, E. D., Ferreira, R., Jakočiūnas, T., Arsovska, D., Zhang, J., Ding, L., et al. (2017). Transcriptional Reprogramming in Yeast Using dCas9 and Combinatorial gRNA Strategies. Microb. Cell Fact. 16, 46. doi:10.1186/s12934-017-0664-2
Jiang, L., Huang, L., Cai, J., Xu, Z., and Lian, J. (2021). Functional Expression of Eukaryotic Cytochrome P450s in Yeast. Biotechnol. Bioeng. 118, 1050–1065. doi:10.1002/bit.27630
Jordá, T., and Puig, S. (2020). Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae. Genes 11, 795. doi:10.3390/genes11070795
Kotopka, B. J., Li, Y., and Smolke, C. D. (2018). Synthetic Biology Strategies toward Heterologous Phytochemical Production. Nat. Prod. Rep. 35, 902–920. doi:10.1039/c8np00028j
Li, S., Li, Y., and Smolke, C. D. (2018). Strategies for Microbial Synthesis of High-Value Phytochemicals. Nat. Chem. 10, 395–404. doi:10.1038/s41557-018-0013-z
Li, D., Ma, Y., Zhou, Y., Gou, J., Zhong, Y., Zhao, L., et al. (2019). A Structural and Data-Driven Approach to Engineering a Plant Cytochrome P450 Enzyme. Sci. China Life Sci. 62, 873–882. doi:10.1007/s11427-019-9538-3
Li, X., Wang, Y., Fan, Z., Wang, Y., Wang, P., Yan, X., et al. (2021). High-level Sustainable Production of the Characteristic Protopanaxatriol-type Saponins from Panax Species in Engineered Saccharomyces cerevisiae. Metab. Eng. 66, 87–97. doi:10.1016/j.ymben.2021.04.006
Liang, H., Cheng, R., Wang, J., Xie, H., Li, R., Shimizu, K., et al. (2021). Mogrol, an Aglycone of Mogrosides, Attenuates Ulcerative Colitis by Promoting AMPK Activation. Phytomedicine 81, 153427. doi:10.1016/j.phymed.2020.153427
Lin, N.-X., Xu, Y., and Yu, X.-W. (2022). Overview of Yeast Environmental Stress Response Pathways and the Development of Tolerant Yeasts. Syst. Microbiol. Biomanuf. 2, 232–245. doi:10.1007/s43393-021-00058-4
Liu, C., Dai, L., Liu, Y., Dou, D., Sun, Y., and Ma, L. (2018). Pharmacological Activities of Mogrosides. Future Med. Chem. 10, 845–850. doi:10.4155/fmc-2017-0255
Liu, C., Zeng, Y., Dai, L. H., Cai, T. Y., Zhu, Y. M., Dou, D. Q., et al. (2015). Mogrol Represents a Novel Leukemia Therapeutic, via ERK and STAT3 Inhibition. Am. J. Cancer Res. 5, 1308–1318.
Liu, H., Fan, J., Wang, C., Li, C., and Zhou, X. (2019). Enhanced β-Amyrin Synthesis in Saccharomyces cerevisiae by Coupling an Optimal Acetyl-CoA Supply Pathway. J. Agric. Food Chem. 67, 3723–3732. doi:10.1021/acs.jafc.9b00653
Liu, G.-S., Li, T., Zhou, W., Jiang, M., Tao, X.-Y., Liu, M., et al. (2020). The Yeast Peroxisome: A Dynamic Storage Depot and Subcellular Factory for Squalene Overproduction. Metab. Eng. 57, 151–161. doi:10.1016/j.ymben.2019.11.001
Liu, B., Yang, J., Hao, J., Xie, H., Shimizu, K., Li, R., et al. (2021). Natural Product Mogrol Attenuates Bleomycin-Induced Pulmonary Fibrosis Development through Promoting AMPK Activation. J. Funct. Foods 77, 104280. doi:10.1016/j.jff.2020.104280
Luo, X., Reiter, M. A., d’Espaux, L., Wong, J., Denby, C. M., Lechner, A., et al. (2019). Complete Biosynthesis of Cannabinoids and Their Unnatural Analogues in Yeast. Nature 567, 123–126. doi:10.1038/s41586-019-0978-9
Ma, T., Shi, B., Ye, Z., Li, X., Liu, M., Chen, Y., et al. (2019). Lipid Engineering Combined with Systematic Metabolic Engineering of Saccharomyces cerevisiae for High-Yield Production of Lycopene. Metab. Eng. 52, 134–142. doi:10.1016/j.ymben.2018.11.009
Milla, P., Viola, F., Bosso, S. O., Rocco, F., Cattel, L., Joubert, B. M., et al. (2002). Subcellular Localization of Oxidosqualene Cyclases from Arabidopsis thaliana, Trypanosoma Cruzi, and Pneumocystis Carinii Expressed in Yeast. Lipids 37, 1171–1176. doi:10.1007/s11745-002-1017-9
Nes, W. D. (2011). Biosynthesis of Cholesterol and Other Sterols. Chem. Rev. 111, 6423–6451. doi:10.1021/cr200021m
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., Mcphee, D., et al. (2013). High-level Semi-synthetic Production of the Potent Antimalarial Artemisinin. Nature 496, 528–532. doi:10.1038/nature12051
Paramasivan, K., and Mutturi, S. (2017). Regeneration of NADPH Coupled with HMG-CoA Reductase Activity Increases Squalene Synthesis in Saccharomyces cerevisiae. J. Agric. Food Chem. 65, 8162–8170. doi:10.1021/acs.jafc.7b02945
Pilauri, V., Bewley, M., Diep, C., and Hopper, J. (2005). Gal80 Dimerization and the Yeast GAL Gene Switch. Genetics 169, 1903–1914. doi:10.1534/genetics.104.036723
Qiao, J., Luo, Z., Cui, S., Zhao, H., Tang, Q., Mo, C., et al. (2019). Modification of Isoprene Synthesis to Enable Production of Curcurbitadienol Synthesis in Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 46, 147–157. doi:10.1007/s10295-018-2116-3
Reider Apel, A., D'espaux, L., Wehrs, M., Sachs, D., Li, R. A., Tong, G. J., et al. (2017). A Cas9-Based Toolkit to Program Gene Expression in Saccharomyces Cerevisiae. Nucleic. Acids. Res. 45, 496–508. doi:10.1093/nar/gkw1023
Rodriguez, S., Denby, C. M., Van Vu, T., Baidoo, E. E. K., Wang, G., and Keasling, J. D. (2016). ATP Citrate Lyase Mediated Cytosolic Acetyl-CoA Biosynthesis Increases Mevalonate Production in Saccharomyces cerevisiae. Microb. Cell Fact. 15, 48. doi:10.1186/s12934-016-0447-1
Ruckenstuhl, C., Lang, S., Poschenel, A., Eidenberger, A., Baral, P. K., Kohút, P., et al. (2007). Characterization of Squalene Epoxidase of Saccharomyces cerevisiae by Applying Terbinafine-Sensitive Variants. Antimicrob. Agents Chemother. 51, 275–284. doi:10.1128/AAC.00988-06
Sadeghi, S. J., and Gilardi, G. (2013). Chimeric P450 Enzymes: Activity of Artificial Redox Fusions Driven by Different Reductases for Biotechnological Applications. Biotechnol. Appl. Biochem. 60, 102–110. doi:10.1002/bab.1086
Scalcinati, G., Knuf, C., Partow, S., Chen, Y., Maury, J., Schalk, M., et al. (2012). Dynamic Control of Gene Expression in Saccharomyces cerevisiae Engineered for the Production of Plant Sesquitepene α-santalene in a Fed-Batch Mode. Metab. Eng. 14, 91–103. doi:10.1016/j.ymben.2012.01.007
Shan, H., Segura, M. J. R., Wilson, W. K., Lodeiro, S., and Matsuda, S. P. T. (2005). Enzymatic Cyclization of Dioxidosqualene to Heterocyclic Triterpenes. J. Am. Chem. Soc. 127, 18008–18009. doi:10.1021/ja055822g
Shi, Y., Wang, D., Li, R., Huang, L., Dai, Z., and Zhang, X. (2021). Engineering Yeast Subcellular Compartments for Increased Production of the Lipophilic Natural Products Ginsenosides. Metab. Eng. 67, 104–111. doi:10.1016/j.ymben.2021.06.002
Ta, M. T., Kapterian, T. S., Fei, W., Du, X., Brown, A. J., Dawes, I. W., et al. (2012). Accumulation of Squalene is Associated with the Clustering of Lipid Droplets. FEBS J. 279, 4231–4244. doi:10.1111/febs.12015
Tang, Q., Ma, X., Mo, C., Wilson, I. W., Song, C., Zhao, H., et al. (2011). An Efficient Approach to Finding Siraitia Grosvenorii Triterpene Biosynthetic Genes by RNA-Seq and Digital Gene Expression Analysis. BMC Genomics 12, 343. doi:10.1186/1471-2164-12-343
Theron, C. W., Labuschagné, M., Albertyn, J., and Smit, M. S. (2019). Heterologous Coexpression of the Benzoate‐para‐hydroxylase CYP53B1 with Different Cytochrome P450 Reductases in Various Yeasts. Microb. Biotechnol. 12, 1126–1138. doi:10.1111/1751-7915.13321
Wang, H. T., Yang, J. T., Chen, K. I., Wang, T. Y., Lu, T. J., and Cheng, K. C. (2019). Hydrolyzation of Mogrosides: Immobilized β‐glucosidase for Mogrosides Deglycosylation from Lo Han Kuo. Food Sci. Nutr. 7, 834–843. doi:10.1002/fsn3.932
Wang, H., Meng, G.-L., Zhang, C.-T., Wang, H., Hu, M., Long, Y., et al. (2020a). Mogrol Attenuates Lipopolysaccharide (LPS)-induced Memory Impairment and Neuroinflammatory Responses in Mice. J. Asian Nat. Prod. Res. 22, 864–878. doi:10.1080/10286020.2019.1642878
Wang, J., Liu, J., Xie, Z., Li, J., Li, J., and Hu, L. (2020b). Design, Synthesis and Biological Evaluation of Mogrol Derivatives as a Novel Class of AMPKα2β1γ1 Activators. Bioorg. Med. Chem. Lett. 30, 126790. doi:10.1016/j.bmcl.2019.126790
Wu, Y., Chen, T., Liu, Y., Lv, X., Li, J., Du, G., et al. (2018). CRISPRi Allows Optimal Temporal Control of N-Acetylglucosamine Bioproduction by a Dynamic Coordination of Glucose and Xylose Metabolism in Bacillus Subtilis. Metab. Eng. 49, 232–241. doi:10.1016/j.ymben.2018.08.012
Xu, R., Fazio, G. C., and Matsuda, S. P. T. (2004). On the Origins of Triterpenoid Skeletal Diversity. Phytochemistry 65, 261–291. doi:10.1016/j.phytochem.2003.11.014
Yu, Y., Rasool, A., Liu, H., Lv, B., Chang, P., Song, H., et al. (2020). Engineering Saccharomyces cerevisiae for High Yield Production of α-amyrin via Synergistic Remodeling of α-amyrin Synthase and Expanding the Storage Pool. Metab. Eng. 62, 72–83. doi:10.1016/j.ymben.2020.08.010
Zhang, J., Dai, L., Yang, J., Liu, C., Men, Y., Zeng, Y., et al. (2016). Oxidation of Cucurbitadienol Catalyzed by CYP87D18 in the Biosynthesis of Mogrosides fromSiraitia Grosvenorii. Plant Cell Physiol. 57, 1000–1007. doi:10.1093/pcp/pcw038
Zhao, F., Bai, P., Liu, T., Li, D., Zhang, X., Lu, W., et al. (2016). Optimization of a Cytochrome P450 Oxidation System for Enhancing Protopanaxadiol Production inSaccharomyces Cerevisiae. Biotechnol. Bioeng. 113, 1787–1795. doi:10.1002/bit.25934
Zhao, F., Du, Y., Bai, P., Liu, J., Lu, W., and Yuan, Y. (2017). Enhancing Saccharomyces cerevisiae Reactive Oxygen Species and Ethanol Stress Tolerance for High-Level Production of Protopanoxadiol. Bioresour. Technol. 227, 308–316. doi:10.1016/j.biortech.2016.12.061
Zhao, H., Wang, J., Tang, Q., Mo, C., Guo, J., Chen, T., et al. (2018). Functional Expression of Two NADPH-Cytochrome P450 Reductases from Siraitia Grosvenorii. Int. J. Biol. Macromol. 120, 1515–1524. doi:10.1016/j.ijbiomac.2018.09.128
Zhao, F., Bai, P., Nan, W., Li, D., Zhang, C., Lu, C., et al. (2019). A Modular Engineering Strategy for High‐level Production of Protopanaxadiol from Ethanol by Saccharomyces cerevisiae. AICHE J. 65, 866–874. doi:10.1002/aic.16502
Keywords: Siraitia grosvenorii, Mogrol, S. cerevisiae, CRISPRi, Metabolic Engineering
Citation: Wang S, Xu X, Lv X, Liu Y, Li J, Du G and Liu L (2022) Construction and Optimization of the de novo Biosynthesis Pathway of Mogrol in Saccharomyces Cerevisiae. Front. Bioeng. Biotechnol. 10:919526. doi: 10.3389/fbioe.2022.919526
Received: 13 April 2022; Accepted: 26 April 2022;
Published: 27 May 2022.
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaReviewed by:
Jian Zha, Shaanxi University of Science and Technology, ChinaCopyright © 2022 Wang, Xu, Lv, Liu, Li, Du and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Long Liu, bG9uZ2xpdUBqaWFuZ25hbi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.