
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 08 June 2022
Sec. Industrial Biotechnology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.905110
Bacillus amyloliquefaciens is the dominant strain used to produce γ-polyglutamic acid from inulin, a non-grain raw material. B. amyloliquefaciens has a highly efficient tricarboxylic acid cycle metabolic flux and glutamate synthesis ability. These features confer great potential for the synthesis of glutamate derivatives. However, it is challenging to efficiently convert high levels of glutamate to a particular glutamate derivative. Here, we conducted a systematic study on the biosynthesis of L-ornithine by B. amyloliquefaciens using inulin. First, the polyglutamate synthase gene pgsBCA of B. amyloliquefaciens NB was knocked out to hinder polyglutamate synthesis, resulting in the accumulation of intracellular glutamate and ATP. Second, a modular engineering strategy was applied to coordinate the degradation pathway, precursor competition pathway, and L-ornithine synthesis pathway to prompt high levels of intracellular precursor glutamate for l-ornithine synthesis. In addition, the high-efficiency L-ornithine transporter was further screened and overexpressed to reduce the feedback inhibition of L-ornithine on the synthesis pathway. Combining these strategies with further fermentation optimizations, we achieved a final L-ornithine titer of 31.3 g/L from inulin. Overall, these strategies hold great potential for strengthening microbial synthesis of high value-added products derived from glutamate.
With the development of synthetic biology, chemical and material products are increasingly being obtained by microbial synthesis (McCarty and Ledesma-Amaro, 2019; Luo et al., 2021). These products are mainly obtained through microbial fermentation using food materials (glucose, sucrose, and starch) as substrates (Qiu et al., 2017; Jiang et al., 2019). The main reason is that food materials are easy to use, and their metabolic pathways have also been extensively studied. Nonetheless, the demand for various products continues to increase, and microbial fermentation using food materials has some shortcomings, i.e., competition with humans for grain resources (Chen et al., 2020). In recent years, research on the use of non-grain food materials for fermentation has been growing steadily. The use of non-grain food materials can make up for the shortage of raw food materials. Nevertheless, non-grain food materials are difficult to use, warranting the development of methods for efficient processing of these materials. To improve efficiency in the utilization of non-grain raw materials, we need to overcome limitations such as insufficient degrading enzymes and stress factor inhibition. The development of metabolic engineering and synthetic biology provides a means to solve these problems.
L-Ornithine is an essential non-protein amino acid widely used in the food, pharmaceutical, and chemical industries (Sheng et al., 2021). It is an essential part of the urea cycle and is widely used to treat liver diseases and burns. In recent years, the development of L-ornithine products (sweeteners, hepatoprotective drugs, and slimming products) has been gradually increasing, resulting in a rapidly growing demand for L-ornithine (Zajac et al., 2010). At present, L-ornithine obtained by chemical synthesis and enzymatic catalysis cannot meet the increasing demand. In addition, chemical synthesis has some drawbacks: chemically synthesized L-ornithine is a mixture of d-ornithine and l-ornithine (only L-ornithine has biological activity) and cannot be directly used in the pharmaceutical and food industries (Shu et al., 2018a). Fortunately, the rapid development of synthetic biology provides new ways for microorganisms to synthesize L-ornithine. Research on L-ornithine production by biological fermentation thus far has mainly focused on Corynebacterium glutamicum and Escherichia coli. Hwang et al. overexpressed the l-ornithine synthesis pathway genes (argCJBD) in an engineered strain of C. glutamicum (C.glu/ΔargF/ΔargR/Δprob), and the level of L-ornithine fermentation increased by 30% to 16.49 g/L (Hwang et al., 2008). Lee et al. overexpressed N-acetyl synthase (ArgA214) in E. coli and knocked out speF and proB, and the L-ornithine titer reached 13.2 g/L (Lee and Cho, 2006). Current research on the biosynthesis of L-ornithine mainly uses food materials for fermentation. With the increased demand for L-ornithine in the pharmaceutical and food industries, the production scale will inevitably need to expand. This will require more food materials for fermentation. Moreover, an insufficient supply of food materials for fermentation will undoubtedly become a significant problem.
Jerusalem artichoke, a non-grain food crop, is considered a potential energy source because of its high sugar content. Jerusalem artichoke contains the storage polysaccharide, inulin, which accounts for 85% of its dry weight (Li et al., 2014). This polysaccharide can be used with simple preprocessing. In recent years, Jerusalem artichoke has been used as substrate to produce products such as ethanol (Zhang et al., 2010; Lim et al., 2011), lactic acid (Ge et al., 2009), and 2,3-butanediol (Sun et al., 2009). In previous studies, we isolated and identified a bacterial strain from the root of Jerusalem artichoke, named B. amyloliquefaciens NB (Qiu et al., 2019b). This strain is deposited in the China Center for Type Culture Collection (China Center for Type Culture Collection), and the deposit number is CCTCC NO: M2016346. This strain can efficiently use the crude extract of Jerusalem artichoke (non-grain food material). Furthermore, B. amyloliquefaciens NB can efficiently synthesize poly-γ-glutamic acid (γ-PGA) without exogenous addition of glutamine (Sha et al., 2020a). Since γ-PGA is a glutamate polymer, B. amyloliquefaciens NB might have a highly efficient tricarboxylic acid cycle metabolic flux and glutamate synthesis ability. These characteristics highlight the potential for synthesizing glutamate derivatives from non-grain raw materials in B. amyloliquefaciens through the rational design of metabolic pathways.
In the present study, B. amyloliquefaciens NB was engineered for de novo biosynthesis of L-ornithine from inulin. Upon knocking out the γ-PGA synthase gene in B. amyloliquefaciens NB, intracellular glutamate and ATP levels were enhanced by 7and 4.5-fold, respectively. The glutamate degradation pathway, the precursor competition pathway, the L-ornithine synthesis pathway, and the L-ornithine transport process were further coordinated to prompt the high level of intracellular precursor glutamate into L-ornithine synthesis. Building upon these results, we successfully achieved a final titer of 31.3 g/L L-ornithine from non-food raw materials (inulin) in an optimized bioreactor system. Overall, this study aimed to provide a reference for improving L-ornithine synthesis and a strategy for the use of non-grain raw materials for microbial synthesis of value-added products derived from glutamate.
The bacterial strains and main plasmids used in this study are listed in Table 1. Escherichia coli DH5α was used as a recipient for plasmid construction. We used the dam− and dcm−-deficient host E. coli GM2163 to prepare the unmethylated plasmids (Sha et al., 2020a).
For regular cloning and transformation experiments, E. coli and B. amyloliquefaciens strains were grown at 37°C in Luria-Bertani (LB) medium (10 g/L tryptone, 10 g/L yeast extract, and 5 g/L NaCl) containing the appropriate antibiotic. The composition of the initial fermentation medium was as follows: 80 g/L inulin, 40 g/L (NH4)2SO4, 20 g/L K2HPO4·3H2O, 2 g/L KH2PO4, 0.4 g/L MgSO4, 0.06 g/L MnSO4·H2O. The initial fermentation inoculum was 6%. For shake flask and batch fermentation, the cells were precultured in a 250 ml shake flask with 40 ml of seed culture and incubated at 37°C while shaking at 200 rpm for 12 h. For flask cultures, 2.5 ml of the seed culture was transferred into 500 ml flasks containing 50 ml of medium and cultured at 32°C and 200 rpm for 72 h. All fermentations in shake flasks were performed without the addition of exogenous sodium glutamate. During the fermentation process of L-ornithine, samples were collected periodically to evaluate the synthesis of L-ornithine and analyze bacterial growth by measuring optical density at 600 nm (OD600).
To obtain the optimal conditions for B. amyloliquefaciens to synthesize L-ornithine, we systematically optimized the fermentation conditions and medium composition in shake flasks. Some important factors (temperature, inoculum size, liquid volume, initial pH, carbon source, nitrogen source and metal ions) were selected to investigate their effects on L-ornithine synthesis. Firstly, 28°C, 30°C, 32°C, 34°C, 37°C and 40°C were selected to study the optimal temperature for L-ornithine fermentation. Secondly, we investigated the effect of the initial pH of the medium at 6.0, 7.0, 8.0, and 9.0 on L-ornithine synthesis, respectively. Then, the effects of inoculum size of 1%, 2%, 3%, 4%, 5%, and 6% on L-ornithine synthesis were investigated. Finally, the effects of adding 40 ml, 50 ml, 60 ml, 70 and 80 ml of fermentation broth to 500-ml shake flask on L-ornithine synthesis were investigated. In addition, the effects of the carbon source, nitrogen source and inorganic salt of the medium on l-ornithine synthesis were also investigated respectively. The Box–Behnken experimental design and response surface analysis were used to optimize the concentration of three key components in the L-ornithine fermentation medium: inulin, peptone, and MgSO4. The Statistica 7.0 software Experimental Design module was used to perform a quadratic polynomial regression fitting on the experimental data, and obtain the quadratic empirical equation model of L-ornithine on inulin, peptone, and MgSO4 (Y = 12.22 + 0.060A− 0.28B+ 0.17C + 0.087AB+ 0.027AC + 0.010BC − 1.64A2 − 1.29B2 − 1.35C2) (Y is the predicted value of L-ornithine production; A, B, and C are the coding values of inulin, peptone, and MgSO4, respectively) (Supplementary Tables S1–S3).
For batch fermentation in a 7.5 L fermenter, single colonies of engineered strains were picked and grown overnight at 32°C in LB medium, then inoculated at 1% (V/V) into 250 ml shake flasks containing 20 ml of seed medium for 12 h. Seed cultures were inoculated at 5% inoculum into a 7.5 L fermenter (BioFlo 115, New Brunswick Scientific, United States) with 4.5 L working volume. The stirring rate was set to 400 rpm, and the airflow was 1 vvm. To further improve the accumulation of L-ornithine, a fed-batch fermentation strategy was adopted. The feed solution with 20 g/L sodium glutamate were further fed into the fermenter at a constant flow rate during the fermentation period from 24 to 48 h.
The primers used in this study are listed in Supplementary Table S4. The genome of B. amyloliquefaciens (NC_017190.1) was extracted using a bacterial total DNA extraction kit (Code DC103-01, Vazyme, Nanjing, China). DNA fragments of argA (Gene ID: 947289), argB (Gene ID: 56457340), argC (Gene ID: 56457338), argD (ID: 12201583), and argE (Gene ID: 948456) were obtained from this genome using primer pairs ArgA-F/R, ArgB-F/R, ArgC-F/R, ArgD-F/R and ArgE-F/R, respectively. The plasmid pHY (containing the constitutive strong promoter PHpaII, p15A ori, CmR) was digested with restriction enzymes SalI/xhoI and purified by column (Code DC301-01, Vazyme, Nanjing, China). ArgA, ArgB, ArgC, ArgD, ArgE, ArgABCDE and ArgA-ArgE were ligated with the linearized pHY vector using ClonExpress II one-step cloning kit (Code C112-02-AB, Vazyme, Nanjing, China) to obtain recombinant plasmids pHY-ArgA, pHY-ArgB, pHY-ArgC, pHY-ArgD, pHY-ArgE, pHY-ArgABCDE and pHY-ArgA-ArgE, respectively. The genome of B. amyloliquefaciens LL3, C. glutamicum, E. coli MG1655 and B. subtilis 168 was extracted using a bacterial total DNA extraction kit (Code DC103-01, Vazyme, Nanjing, China). The genes encoding L-ornithine transporters BA. lysE, Cg. lysE, E. coli.lysE and BS. lysE were amplified from genomes of the different strains mentioned above with primer pairs BA. lysE-F/R, Cg. lysE-F/R, E. coli.lysE-F/R and BS. lysE-F/R, respectively. The recombinant plasmids pHY-BA. lysE, pHY-Cg. lysE, pHY-E. coli.lysE and pHY-BS. lysE were obtained by using the above-mentioned expression plasmid construction method. The constructed recombinant plasmids were verified by PCR and sanger sequencing.
The gene knockout method refers to previous research reports (Qiu et al., 2020b). To knock out argF, upstream and downstream fragments of argF (Gene ID: 56457344) were amplified from B. amyloliquefaciens genome (NC_017190.1) with primer pairs argFL-F/R and argFR-F/R, respectively. sgRNA of argF was designed using online software (http://cistrome.org/SSC/) and obtained by designing primers sgargF-F/R. The DNA fragments amplified above were ligated by overlapping PCR to obtain argFsgRNA-argFL-argFR. The fragment argFsgRNA-argFL-argFR was then ligated with the linearized pDR-uppsgRNA vector (linearized with restriction enzymes salI/xhoI) using ClonExpress II one-step cloning kit (Code C112-02-AB, Vazyme, Nanjing, China) to obtain recombinant plasmids pDR-argFsgRNA. The argI (Gene ID: 12203901), speF (Gene ID: 945297) and prob (Gene ID: 56457565) genes of B. amyloliquefaciens were knocked out using the above method to construct different engineered strains. The constructed recombinant plasmids were verified by colony PCR and sanger sequencing.
The transformation of wild-type B. amyloliquefaciens to B. amyloliquefaciens NB was performed using a modified high-osmolarity electroporation method (Sha et al., 2019). An overnight culture of B. amyloliquefaciens was diluted 100-fold in fresh medium (LB broth containing 0.5 M sorbitol) to prepare electrocompetent cells. When the OD600 of the culture reached 0.5, the NB cells were harvested by centrifugation at 4°C and 8,000 rpm for 10 min. After four washes in ice-cold electroporation medium (0.5 M sorbitol, 0.5 M mannitol, and 10% glycerol), the electrocompetent cells were suspended at a cell density of 1 × 1010 colony-forming units/mL.
The optical density of a sample was measured to determine cell growth. The OD600 of the diluted sample was measured using a spectrophotometer. The glutamate level was measured using Glutamate Content Assay Kit (Code MS 1906, Shanghai, China) from Shanghai Suoqiao Biological Co., Ltd. The detection principle of the kit is that the enzyme reagent can specifically recognize glutamate substrate from a mixture and catalyze it to produce a colored product, and the reaction product has a maximum absorption peak at a wavelength of 570 nm. The ATP content was determined using the ATP content kit (phosphomolybdic acid colorimetry) (Code G0815W, Suzhou, China) purchased from Suzhou Grace Biotechnology Co., Ltd. The creatine kinase in the kit can catalyze the ATP reaction with creatine to produce creatine phosphate, which is detected by the phosphomolybdic acid colorimetric method. Therefore, the ATP content can be calculated based on the maximum absorption peak of the reaction product at 700 nm. The L-ornithine content was detected through Chinard’s L-ornithine measurement method. Specifically, 6 mol/L H3PO4-glacial acetic acid (1/3, v/v) was used to prepare a 25 mg/ml ninhydrin solution as the coloring solution. After the coloring solution reacted with L-ornithine in a water bath at 100°C for 60 min, the absorption peak was measured at 510 nm (Shu et al., 2018a).
In an earlier study, we found that B. amyloliquefaciens NB can efficiently use inulin to synthesize PGA and has a highly-efficient tricarboxylic acid cycle metabolic flux and glutamate synthesis ability (Qiu et al., 2020a; Sha et al., 2020b). Therefore, we speculate that this strain may be optimal for synthesizing glutamate derivatives (L-ornithine). First, the polyglutamate synthase pgsBCA of B. amyloliquefaciens NB was knocked out to prevent the synthesis of γ-PGA from glutamate and release ATP for the synthesis of γ-PGA. After pgsBCA was knocked out, the colony morphology of B. amyloliquefaciens NB changed from wet to rough and almost no γ-PGA was detected in the fermentation broth of B. amyloliquefaciens NB (ΔpgsBCA), but 0.43 g/L of L-ornithine was obtained (Almost no L-ornithine was present in the fermentation broth of the original strain) (Supplementary Figures S1, S2). In addition, compared with B. amyloliquefaciens NB, the intracellular glutamate and ATP levels of B. amyloliquefaciens NB (ΔpgsBCA) increased by 7 times and 4.5 times, respectively (Figure 1). By blocking the synthesis of γ-PGA, the intracellular glutamate and ATP content of B. amyloliquefaciens NB (ΔpgsBCA) were increased to 531 μg/g and 3.2 μmol/g, respectively. In addition, the growth of B. amyloliquefaciens NB was also improved, and its OD600 increased from 3.2 to 4.3. These results demonstrated that B. amyloliquefaciens NB has certain advantages as an L-ornithine-producing strain. However, glutamate represents a critical node in many important metabolic pathways, and it is an essential intermediate of many products. For example, glutamate is a precursor of proline, L-ornithine, and arginine (Jiang et al., 2021; Tran et al., 2021). Therefore, achieving efficient glutamate conversion to L-ornithin in B. amyloliquefaciens is a fundamental challenge for obtaining a high-efficiency strain.
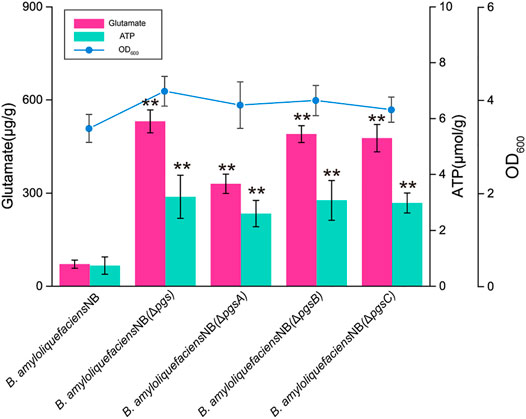
FIGURE 1. The lack of polyglutamate synthase (pgsBCA) improves glutamate and ATP concentrations and growth. Biomass and glutamate and ATP concentrations for B. amyloliquefaciens NB, B. amyloliquefaciens (ΔpgsBCA), B. amyloliquefaciens (ΔpgsA), B. amyloliquefaciens (ΔpgsB), and B. amyloliquefaciens (ΔpgsC) in shake flasks at 24 h. All data were the average of three independent studies with standard deviations. The ** and * indicate p < 0.01 and 0.05 relative to the control strain B. amyloliquefaciens NB, respectively.
Optimizing the L-ornithine synthesis pathway involves multiple metabolic pathways: the L-ornithine degradation pathway, the precursor competition pathway, and the L-ornithine synthesis pathway. These need to be coordinated to prompt the synthesis L-ornithine from glutamate. We divided these pathways into three modules, namely, module one (L-ornithine catabolism), module two (precursor competition), and module three (L-ornithine synthesis) (Figure 2A).
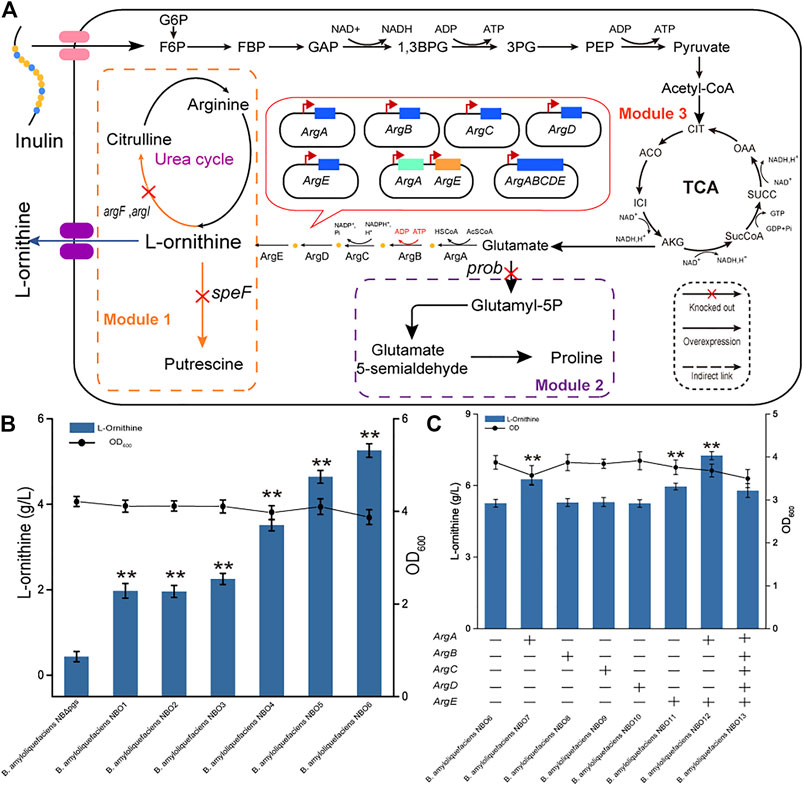
FIGURE 2. Effect of the L-ornithine synthesis module on putrescine production. (A) Schematic overview of L-ornithine catabolism pathway, precursor competition pathway, and synthesis pathways. The main pathway of l-ornithine catabolism is catalyzed by L-ornithine carbamoyltransferase and l-ornithine decarboxylase. Glutamate is a key starting material for L-ornithine synthesis. Blocking proline synthesis improves the supply of the precursor glutamate for L-ornithine synthesis. Some key genes in the L-ornithine synthesis pathway included amino-acid N-acetyltransferase (ArgA), acetylglutamate kinase (ArgB), N-acetyl-gamma-glutamyl-phosphate reductase (ArgC), acetylornithine aminotransferase (ArgD), and acetylornithine deacetylase (ArgE). G6P: Glucose-6-Phosphate; F6P: Frucose-6- Phosphate; GAP: Glyceraldedyde-3-phosphate; 1,3BPG: 1,3-Bisphospho-glyerate; 3 PG: 3-phosphoglycerate; PEP: Phosphoenolpyruvate (B) The L-ornithine titers of the recombinant strains lacking argF, argI, speF, and prob. All data were the average of three independent studies with standard deviations. The ** and * indicate p < 0.01 and 0.05 relative to the control strain B. amyloliquefaciens NB (ΔpgsBCA), respectively. (C) Overexpression of the L-ornithine synthase gene in B. amyloliquefaciens NBO6. “+” indicates that the relevant gene is overexpressed in the strain; “-” indicates that the relevant gene is not overexpressed in the strain. All data were the average of three independent studies with standard deviations. The ** and * indicate p < 0.01 and 0.05 relative to the control strain B. amyloliquefaciens NBO6, respectively. Co-expression of genes with noticeable effect to further improve the fermentation effect.
As a precursor, L-ornithine could be converted into citrulline, putrescine, and other substances in the cell. For module 1, the argF and argI genes encoding L-ornithine carbamoyltransferase (Sander et al., 2019). were first knocked out to block the catabolism of L-ornithine to citrulline, thereby obtaining strains B. amyloliquefaciens NBO1 and NBO2. The fermentation results showed that blocking these two genes promoted the accumulation of L-ornithine; the L-ornithine titers of the NBO1 and NBO2 strains reached 1.97 g/L and 1.95 g/L, respectively. In addition, the speF gene, encoding L-ornithine decarboxylase (Bao et al., 2021), was knocked out to block the catabolism of L-ornithine to putrescine in B. amyloliquefaciens NBO3 [with argF and argI genes deleted in B. amyloliquefaciens NB (ΔpgsBCA)], thereby obtaining the B. amyloliquefaciens strain NBO4. The L-ornithine titer of NBO4 was further increased to 3.51 g/L (Figure 2B).
Glutamate is essential for L-ornithine synthesis and is a key starting material for other metabolic pathways (Xu et al., 2019). Therefore, the metabolic flow of glutamate directly affects the synthesis efficiency of L-ornithine. For module 2, the prob-encoded glutamate 5-kinase was knocked out to block proline synthesis, which competes with L-ornithine synthesis for the precursor glutamate. B. amyloliquefaciens NBO5 and NBO6 were obtained by knocking out the prob gene (ID: 56457565) in B. amyloliquefaciens NBO3 and NBO4, respectively. The results showed that the deletion of glutamate 5-kinase produced the most significant increase in the titer of L-ornithine. The titer of L-ornithine in B. amyloliquefaciens NBO5 and NBO6 reached 4.64 and 5.26 g/L, respectively (Figure 2B).
Previous studies have reported that the L-ornithine synthase cluster is an important factor limiting the efficient synthesis of L-ornithine. For example, the rate-limiting steps in the L-ornithine synthesis pathways of C. glutamicum and E. coli are N-acetylglutamate synthase encoded by argA and N-acetylglutamate kinase encoded by argB, respectively (Yoshida et al., 1979; Rajagopal et al., 1998). Higher yields can be obtained by overexpressing related genes using high-copy plasmid vectors, etc. Therefore, it is imperative to study the expression of L-ornithine synthase for efficient L-ornithine synthesis. For module 3, we studied the effects of the following enzymes on L-ornithine synthesis: amino-acid N-acetyltransferase (ArgA), acetylglutamate kinase (ArgB), N-acetyl-gamma-glutamyl-phosphate reductase (ArgC), acetylornithine aminotransferase (ArgD), and acetylornithine deacetylase (ArgE). These enzymes were expressed in strain B. amyloliquefaciens NBO6, respectively, resulting in engineering strains B. amyloliquefaciens NBO7, NBO8, NBO9, NBO10, and NBO11. The fermentation results of five recombinant strains and the control strain (B. amyloliquefaciens NBO6) showed that the overexpression of ArgA and ArgE contributed to L-ornithine synthesis. Subsequently, we co-overexpressed ArgA and ArgE in B. amyloliquefaciens NBO6 and found that the titer of L-ornithine of NBO12 was further improved, reaching 7.26 g/L. However, co-overexpression of ArgA, ArgB, ArgC, ArgD and ArgE in B. amyloliquefaciens NBO6 did not significantly improve L-ornithine production, and the production of B. amyloliquefaciens NBO13 was lower than that of NBO12. Overall, by optimizing the three modules, the production of L-ornithine synthesized by B. amyloliquefaciens using inulin as a substrate was increased almost 17-fold from 0.43 to 7.26 g/L (Figure 2C).
Products accumulated in the cell cause feedback inhibition on the activity of critical enzymes, which can have detrimental effects on cell growth (Luo et al., 2018). Therefore, accelerating the extracellular transport efficiency of l-ornithine is very important to enhance the extracellular accumulation of L-ornithine and alleviate its feedback inhibition on the synthesis pathway. First, the gene lysE (encoding the L-ornithine transporter) from different strains (E. coli, C. glutamicum, B. subtilis, and B. amyloliquefaciens) was overexpressed in NBO6 to screen high-efficiency transporters. The results indicated that L-ornithine production of the strains expressing the transporter was improved at different levels compared to the control strains (Figure 3). Among them, the overexpression of lysE from B. amyloliquefaciens (BA.lysE) resulted in a significant increase in L-ornithine titer and biomass. The titer of L-ornithine increased by 30% compared with NBO6, reaching 6.83 g/L. Subsequently, the lysE from B. amyloliquefaciens was further overexpressed in B. amyloliquefaciens NBO12 to enhance L-ornithine production, resulting in B. amyloliquefaciens NBO18. The L-ornithine titer of B. amyloliquefaciens NBO18 reached 8.6 g/L, representing a 20% increase compared with B. amyloliquefaciens NBO12 (Figure 3).
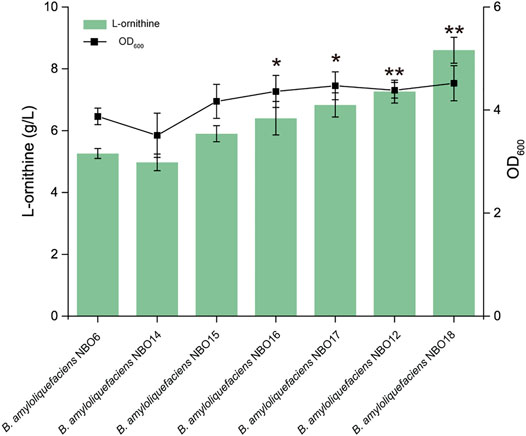
FIGURE 3. The expression of lysE increases L-ornithine synthesis and cell growth. The effect of the overexpression of four transporter genes on L-ornithine production and cell growth. Four L-ornithine transporters from different sources were screened, namely, E. coli, C. glutamicum, B. subtilis, and B. amyloliquefaciens. All data were the average of three independent studies with standard deviations. The ** and * indicate p < 0.01 and 0.05 relative to the control strain B. amyloliquefaciens NBO6, respectively.
The fermentation process and medium components are critical for strain growth and for the synthesis of the desired products (Luo et al., 2020). Therefore, it is essential to optimize the fermentation process and medium composition to further enhance the accumulation of L-ornithine. First, the inoculum volume, liquid volume, temperature, and pH of the B. amyloliquefaciens NBO18 strain were optimized. The fermentation results showed that the inoculum volume, liquid volume, temperature, and pH were 5%, 10%, 32°C, and 7.0, respectively (Supplementary Figure S3). In addition, the composition of the fermentation medium was further optimized by combining single factor and response surface experiments (Supplementary Figure S3), and the best carbon source, nitrogen source, and metal ions were determined, which were 120 g/L of inulin, 60 g/L of peptone, and 0.4 g/L of MgSO4, respectively. The titer of L-ornithine of B. amyloliquefaciens NBO18 under optimal fermentation conditions reached 12.6 g/L, representing a 31.7% increase (Figure 4).
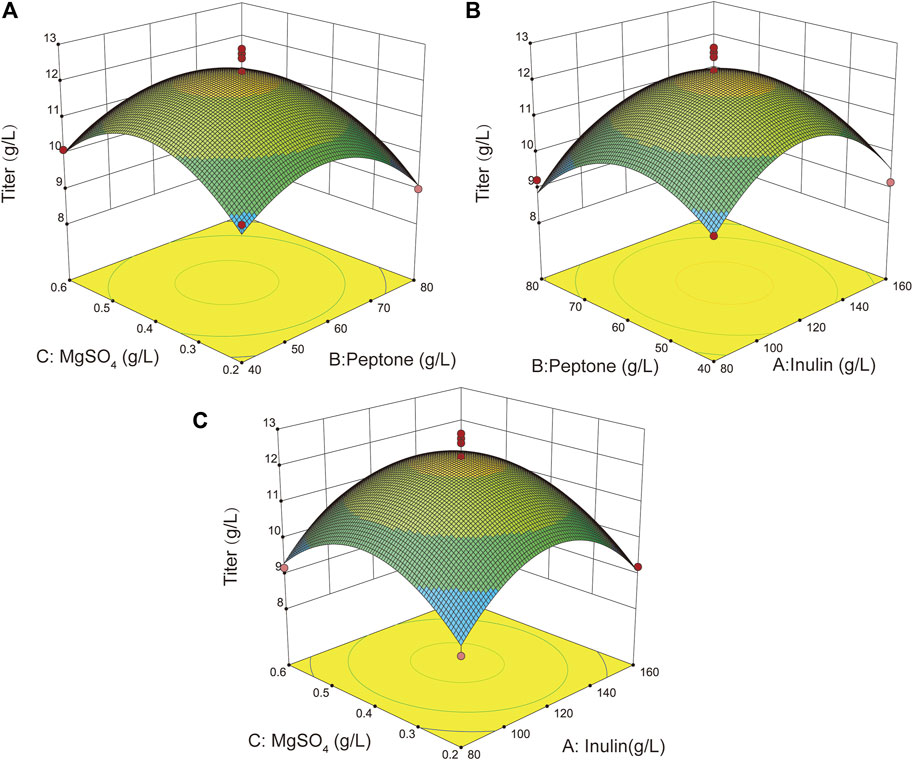
FIGURE 4. The effect of different fermentation conditions on L-ornithine production and response surface optimization results. Three-factor and three-level response surface optimization for three major components (inulin, peptone, and MgSO4) was done. (A) The response surface plot showed the effects of MgSO4 and peptone on l-ornithine production. (B) The response surface plot showed the effects of inulin and peptone on L-ornithine production. (C) The response surface plot showed the effects of inulin and MgSO4 on L-ornithine production.
To maximize L-ornithine production by B. amyloliquefaciens NBO18, we cultured NBO18 in 7.5 L batch bioreactors (BioFlo 115, New Brunswick Scientific, United States) based on the optimized fermentation conditions above, resulting in titers of 14.5 g/L L-ornithine (Figure 5A). To further improve L-ornithine production, 20 g/L of the precursor sodium glutamate was added to L-ornithine batch fermentation, and 19.3 g/L of L-ornithine was obtained (Figure 5B). However, the efficiency of sodium glutamate conversion into L-ornithine was very low, which may explain why the high concentration of sodium glutamate had a negative effect on strain metabolism. Thus, the sodium glutamate feeding strategy was adopted, and sodium glutamate was added to the fermenter at a constant flow rate of 0.5 ml/min during a fermentation period of 24–48 h (the final supplement amount of sodium glutamate was about 20 g/L). Finally, the titer of L-ornithine produced by B. amyloliquefaciens NBO18 reached 31.3 g/L, and the yield of L-ornithine was 0.22 g/g (L-ornithine/(inulin + glutamate)) (Figure 5C).
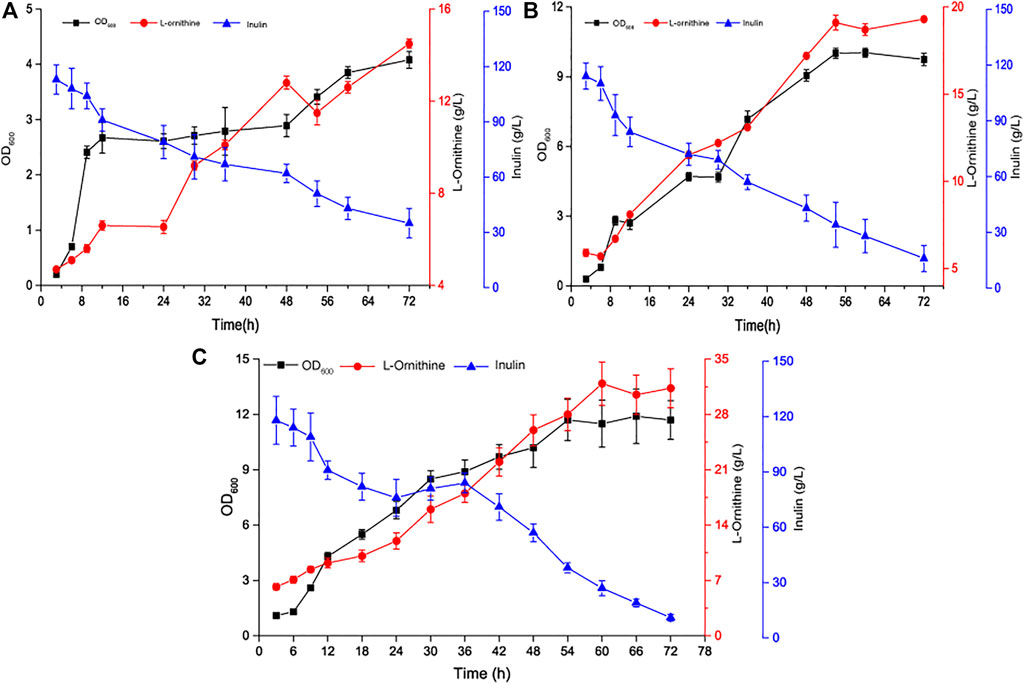
FIGURE 5. The time curve of L-ornithine fermentation in a 7.5 L fermenter. (A) Time profile of single-batch L-ornithine fermentation using inulin as the carbon source in a 7.5 L fermenter. (B) Time profile of single-batch L-ornithine fermentation using inulin and sodium glutamate as the carbon source in a 7.5 L fermenter. (C) Time profile of fed-batch L-ornithine fermentation in a 7.5 L fermenter. Fermentation conditions are as follows: the working volume is 3.5-L, the stirring rate is 400 rpm, the volume air per volume is 1 vvm, and the inoculation size is 6%.
Using non-grain raw materials to synthesize biochemical products is very difficult due to the inefficiency of their bio-utilization. Although much work has been done to solve this issue, the current efficiency of synthesizing biochemical products from non-grain raw materials is lower than that of synthesizing biochemical products from food raw material substrates such as glucose (Qiu et al., 2019a; Becker and Wittmann, 2019). Here, we systematically studied the biosynthesis of L-ornithine by B. amyloliquefaciens using non-grain food materials. First, we demonstrated the feasibility of fermenting L-ornithine from Jerusalem artichoke by analyzing intracellular glutamate and ATP levels. Then, modular engineering and carrier engineering were applied to prompt high levels of intracellular precursor glutamate conversion into L-ornithine. This enabled B. amyloliquefaciens to efficiently produce L-ornithine from Jerusalem artichoke without glutamate supplementation. Combining these strategies with an optimized fermentation process, we successfully achieved a final titer of 31.3 g/L L-ornithine. We anticipate that these strategies should be widely applicable in the microbial synthesis of value-added glutamate derivatives by B. amyloliquefaciens using non-grain food materials.
To our knowledge, this is the first report of the use of non-grain food materials to produce L-ornithine using B. amyloliquefaciens. In previous studies, a small amount of L-ornithine was synthesized by model strains (E. coli, C. glutamicum) via fermentation of food raw materials (glucose and starch) (Lee and Cho, 2006; Wu et al., 2020). Compared with these model strains, B. amyloliquefaciens NB is advantageous since it serves as a cell factory for L-ornithine synthesis. We found that the intracellular glutamate content of B. amyloliquefaciens was significantly increased several times by blocking the PGA synthesis pathway. This demonstrates that the strain has an efficient glutamate synthesis flux and provides a sufficient precursor supply for L-ornithine synthesis. In addition, we compared the effects of different carbon sources on L-ornithine synthesis and found that inulin as a non-grain raw material was the dominant carbon source for L-ornithine synthesis compared with glucose, fructose, and other carbon sources. Overall, these results demonstrated that B. amyloliquefaciens could be optimal for the synthesis of glutamate derivatives from non-grain inulin. However, the synthesis of L-ornithine from inulin in this study requires a large amount of peptone and glutamate supplementation, which leads to an increase in the cost of producing L-ornithine. Therefore, it is necessary to rationally regulate the nitrogen metabolism pathway of B. amyloliquefaciens to improve its utilization efficiency of cheap nitrogen sources in the future.
Coordinated optimization of multiple pathways is essential for constructing efficient cell factories (Gong et al., 2020; Zhou et al., 2021). Most studies on the construction of L-ornithine cell factories thus far have focused on enhancing L-ornithine synthesis pathways but rarely on regulating the overall L-ornithine synthesis pathway (Shu et al., 2018b; Zhang et al., 2018). Although implementing these strategies improved L-ornithine production, local regulation of the metabolic pathway will cause an imbalance of the metabolic network limiting target product production (Wu et al., 2020). In this study, we systematically investigated and coordinated the optimization of the L-ornithine degradation pathway, precursor competing pathway, L-ornithine synthesis pathway and L-ornithine transport pathway. This enabled the production of a strain that efficiently utilized inulin to synthesize L-ornithine. In addition, the L-ornithine fermentation process was systematically optimized to further improve L-ornithine synthesis efficiency. Finally, the titer of L-ornithine increased from 0.43 to 31.3 g/L. These results demonstrate that systematically optimizing the metabolic network of strains is invaluable for efficient synthesis of target products. In addition, the strategies employed in this study could prove useful for constructing high-efficiency cell factories of glutamate and its related products.
Efficient utilization of non-grain raw materials is a crucial challenge, hampering efficient synthesis of target products by microbial strains (Kamimura et al., 2019). Surprisingly, we found that the utilization efficiency of inulin by microorganisms was significantly higher than that of other non-grain materials such as cellulose, hemicellulose, and lignin (Shu et al., 2018b; Cai et al., 2021). This may be because B. amyloliquefaciens NB has a highly active inulin degrading enzyme, enabling efficient inulin degradation into fermentable monosaccharides (glucose and fructose) (Qiu et al., 2019a). Therefore, the inulin utilization module pathway from B. amyloliquefaciens NB could be designed in model microbial cells to achieve efficient synthesis of target products using inulin non-food raw materials. Nonetheless, the conversion rate of L-ornithine synthesized by B. amyloliquefaciens from non-grain raw materials was lower than that of a model strain using food grain as the raw material and a theoretical conversion rate (Sheng et al., 2021). The main reason may be that the mechanism for efficient inulin utilization remains unclear. For example, key factors affecting strain metabolism and growth remain unclear. Furthermore, the mechanism for coordinated utilization of fructose and glucose from inulin has not been elucidated. Therefore, the effects of these factors on strain metabolism should be further analyzed. Moreover, the conversion rate of inulin into target products requires improvement in future studies, since this will play a vital role in the utilization efficiency of other complex carbon sources.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
YZ and ZL conceived the study; YZ, YH, YY, SD and FP preformed the study and analyzed the experiments; ZL and YZ wrote the manuscript with editing from SL and HX, ZL, and YZ revised the manuscript according to reviewer suggestions; all authors read and approved the manuscript.
This work was financially supported by the National Key Research and Development Program of China (2021YFC2101700), National Natural Science Foundation of China (22108122), Natural Science Foundation of Jiangsu Province (BK20200692), Jiangsu Agriculture Science and Technology Innovation Fund (JASTIF) (CX (20)3049), China Postdoctoral Science Foundation (2020M671466), Natural Science Foundation of the Jiangsu Higher Education Institutions of China (20KJB530016), and Jiangsu Postdoctoral Research Foundation (2020Z115).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.905110/full#supplementary-material
Bao, X., Wang, F., Yang, R., Zhang, Y., Fu, L., and Wang, Y. (2021). Ornithine Decarboxylation System of Shewanella Baltica Regulates Putrescine Production and Acid Resistance. J. Food Prot. 84 (2), 303–309. doi:10.4315/jfp-20-227
Becker, J., and Wittmann, C. (2019). A Field of Dreams: Lignin Valorization into Chemicals, Materials, Fuels, and Health-Care Products. Biotechnol. Adv. 37 (6), 107360. doi:10.1016/j.biotechadv.2019.02.016
Cai, C., Xu, Z., Zhou, H., Chen, S., and Jin, M. (2021). Valorization of Lignin Components into Gallate by Integrated Biological Hydroxylation, O-Demethylation, and Aryl Side-Chain Oxidation. Sci. Adv. 7 (36), eabg4585. doi:10.1126/sciadv.abg4585
Chen, F. Y.-H., Jung, H.-W., Tsuei, C.-Y., and Liao, J. C. (2020). Converting Escherichia coli to a Synthetic Methylotroph Growing Solely on Methanol. Cell 182 (4), 933–946. doi:10.1016/j.cell.2020.07.010
Ge, X.-Y., Qian, H., and Zhang, W.-G. (2009). Improvement of L-Lactic Acid Production from Jerusalem Artichoke Tubers by Mixed Culture of Aspergillus niger and Lactobacillus Sp. Bioresour. Technol. 100 (5), 1872–1874. doi:10.1016/j.biortech.2008.09.049
Gong, Z., Wang, H., Tang, J., Bi, C., Li, Q., and Zhang, X. (2020). Coordinated Expression of Astaxanthin Biosynthesis Genes for Improved Astaxanthin Production in Escherichia coli. J. Agric. Food Chem. 68 (50), 14917–14927. doi:10.1021/acs.jafc.0c05379
Hwang, J. H., Hwang, G. H., and Cho, J. Y. (2008). Effect of Increased Glutamate Availability on L-Ornithine Production in Corynebacterium Glutamicum. J. Microbiol. Biotechnol. 18 (4), 704–710. doi:10.1007/s10295-007-0288-3
Jiang, Y., Wu, R., Zhou, J., He, A., Xu, J., Xin, F., et al. (2019). Recent Advances of Biofuels and Biochemicals Production from Sustainable Resources Using Co-cultivation Systems. Biotechnol. Biofuels 12, 155. doi:10.1186/s13068-019-1495-7
Jiang, Y., Sheng, Q., Wu, X.-Y., Ye, B.-C., and Zhang, B. (2021). l-Arginine Production in Corynebacterium Glutamicum: Manipulation and Optimization of the Metabolic Process. Crit. Rev. Biotechnol. 41 (2), 172–185. doi:10.1080/07388551.2020.1844625
Kamimura, N., Sakamoto, S., Mitsuda, N., Masai, E., and Kajita, S. (2019). Advances in Microbial Lignin Degradation and its Applications. Curr. Opin. Biotechnol. 56, 179–186. doi:10.1016/j.copbio.2018.11.011
Lee, Y.-J., and Cho, J.-Y. (2006). Genetic Manipulation of a Primary Metabolic Pathway for L-Ornithine Production in Escherichia coli. Biotechnol. Lett. 28 (22), 1849–1856. doi:10.1007/s10529-006-9163-y
Li, L., Chen, C., Li, K., Wang, Y., Gao, C., Ma, C., et al. (2014). Efficient Simultaneous Saccharification and Fermentation of Inulin to 2, 3-butanediol by Thermophilic Bacillus Licheniformis ATCC 14580. Appl. Environ. Microbiol. 80 (20), 6458–6464. doi:10.1128/aem.01802-14
Lim, S.-H., Ryu, J.-M., Lee, H., Jeon, J. H., Sok, D.-E., and Choi, E.-S. (2011). Ethanol Fermentation from Jerusalem Artichoke Powder Using Saccharomyces cerevisiae KCCM50549 without Pretreatment for Inulin Hydrolysis. Bioresour. Technol. 102 (2), 2109–2111. doi:10.1016/j.biortech.2010.08.044
Luo, Z., Liu, S., Du, G., Xu, S., Zhou, J., and Chen, J. (2018). Enhanced Pyruvate Production in Candida Glabrata by Carrier Engineering. Biotechnol. Bioeng. 115 (2), 473–482. doi:10.1002/bit.26477
Luo, Z., Liu, N., Lazar, Z., Chatzivasileiou, A., Ward, V., Chen, J., et al. (2020). Enhancing Isoprenoid Synthesis in Yarrowia Lipolytica by Expressing the Isopentenol Utilization Pathway and Modulating Intracellular Hydrophobicity. Metab. Eng. 61, 344–351. doi:10.1016/j.ymben.2020.07.010
Luo, Z., Yu, S., Zeng, W., and Zhou, J. (2021). Comparative Analysis of the Chemical and Biochemical Synthesis of Keto Acids. Biotechnol. Adv. 47, 107706. doi:10.1016/j.biotechadv.2021.107706
McCarty, N. S., and Ledesma-Amaro, R. (2019). Synthetic Biology Tools to Engineer Microbial Communities for Biotechnology. Trends Biotechnol. 37 (2), 181–197. doi:10.1016/j.tibtech.2018.11.002
Qiu, Y., Sha, Y., Zhang, Y., Xu, Z., Li, S., Lei, P., et al. (2017). Development of Jerusalem Artichoke Resource for Efficient One-step Fermentation of Poly-(γ-Glutamic Acid) Using a Novel Strain Bacillus Amyloliquefaciens NX-2S. Bioresour. Technol. 239, 197–203. doi:10.1016/j.biortech.2017.05.005
Qiu, Y., Zhu, Y., Zhan, Y., Zhang, Y., Sha, Y., Zhan, Y., et al. (2019a). Systematic Unravelling of the Inulin Hydrolase from Bacillus Amyloliquefaciens for Efficient Conversion of Inulin to Poly-(γ-Glutamic Acid). Biotechnol. Biofuels 12, 145. doi:10.1186/s13068-019-1485-9
Qiu, Y., Zhu, Y., Zhang, Y., Sha, Y., Xu, Z., Li, S., et al. (2019b). Characterization of a Regulator pgsR on Endogenous Plasmid p2Sip and its Complementation for Poly(γ-Glutamic Acid) Accumulation in Bacillus Amyloliquefaciens. J. Agric. Food Chem. 67 (13), 3711–3722. doi:10.1021/acs.jafc.9b00332
Qiu, Y., Zhu, Y., Sha, Y., Lei, P., Luo, Z., Feng, X., et al. (2020a). Development of a Robust Bacillus Amyloliquefaciens Cell Factory for Efficient Poly(γ-Glutamic Acid) Production from Jerusalem Artichoke. ACS Sustain. Chem. Eng. 8 (26), 9763–9774. doi:10.1021/acssuschemeng.0c02107
Qiu, Y., Zhu, Y., Sha, Y., Lei, P., and Xu, H. (2020b). Development of a Robust Bacillus Amyloliquefaciens Cell Factory for Efficient Poly(γ-Glutamic Acid) Production from Jerusalem Artichoke. ACS Sustain. Chem. Eng. 8, 9763–9774. doi:10.1021/acssuschemeng.0c02107
Rajagopal, B. S., DePonte, J., Tuchman, M., and Malamy, M. H. (1998). Use of Inducible Feedback-Resistant N -Acetylglutamate Synthetase ( argA ) Genes for Enhanced Arginine Biosynthesis by Genetically Engineered Escherichia coli K-12 Strains. Appl. Environ. Microbiol. 64 (5), 1805–1811. doi:10.1128/aem.64.5.1805-1811.1998
Sander, T., Wang, C. Y., Glatter, T., and Link, H. (2019). CRISPRi-based Downregulation of Transcriptional Feedback Improves Growth and Metabolism of Arginine Overproducing E. coli. ACS Synth. Biol. 8 (9), 1983–1990. doi:10.1021/acssynbio.9b00183
Sha, Y., Zhang, Y., Qiu, Y., Xu, Z., Li, S., Feng, X., et al. (2019). Efficient Biosynthesis of Low-Molecular-Weight Poly-γ-Glutamic Acid by Stable Overexpression of PgdS Hydrolase in Bacillus Amyloliquefaciens NB. J. Agric. Food Chem. 67 (1), 282–290. doi:10.1021/acs.jafc.8b05485
Sha, Y., Qiu, Y., Zhu, Y., Sun, T., Luo, Z., Gao, J., et al. (2020a). CRISPRi-Based Dynamic Regulation of Hydrolase for the Synthesis of Poly-γ-Glutamic Acid with Variable Molecular Weights. ACS Synth. Biol. 9 (9), 2450–2459. doi:10.1021/acssynbio.0c00207
Sha, Y., Huang, Y., Zhu, Y., Sun, T., Luo, Z., Qiu, Y., et al. (2020b). Efficient Biosynthesis of Low-Molecular-Weight Poly-γ-Glutamic Acid Based on Stereochemistry Regulation in Bacillus Amyloliquefaciens. ACS Synth. Biol. 9 (6), 1395–1405. doi:10.1021/acssynbio.0c00080
Sheng, Q., Wu, X., Jiang, Y., Li, Z., Wang, F., and Zhang, B. (2021). Highly Efficient Biosynthesis of L-Ornithine from Mannitol by Using Recombinant Corynebacterium Glutamicum. Bioresour. Technol. 327, 124799. doi:10.1016/j.biortech.2021.124799
Shu, Q., Xu, M., Li, J., Yang, T., Zhang, X., Xu, Z., et al. (2018a). Improved L-Ornithine Production in Corynebacterium Crenatum by Introducing an Artificial Linear Transacetylation Pathway. J. Ind. Microbiol. Biotechnol. 45 (6), 393–404. doi:10.1007/s10295-018-2037-1
Shu, Q., Xu, M., Li, J., Zhang, X., Yang, T., Xu, Z., et al. (2018b). Producing L-Ornithine by Heterologous Expression of N-Acetyl-L-Ornithine Deacetylase in Corynebacterium Crenatum. Chin. Biotechnol. 38 (7), 29–39. doi:10.13523/j.cb.20180705
Sun, L.-H., Wang, X.-D., Dai, J.-Y., and Xiu, Z.-L. (2009). Microbial Production of 2, 3-butanediol from Jerusalem Artichoke Tubers by Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 82 (5), 847–852. doi:10.1007/s00253-008-1823-5
Tran, D. H., Kesavan, R., Rion, H., Soflaee, M. H., Solmonson, A., Bezwada, D., et al. (2021). Mitochondrial NADP+ is Essential for Proline Biosynthesis during Cell Growth. Nat. Metab. 3 (4), 571–585. doi:10.1038/s42255-021-00374-y
Wu, X.-Y., Guo, X.-Y., Zhang, B., Jiang, Y., and Ye, B.-C. (2020). Recent Advances of L-Ornithine Biosynthesis in Metabolically Engineered Corynebacterium Glutamicum. Front. Bioeng. Biotechnol. 7, 12. doi:10.3389/fbioe.2019.00440
Xu, G., Zha, J., Cheng, H., Ibrahim, M. H. A., Yang, F., Dalton, H., et al. (2019). Engineering Corynebacterium Glutamicum for the De Novo Biosynthesis of Tailored Poly-γ-Glutamic Acid. Metab. Eng. 56, 39–49. doi:10.1016/j.ymben.2019.08.011
Yoshida, H., Araki, K., and Nakayama, K. (1979). Fermentative Production of L-Arginine. II. Mechanism of L-Arginine Production by L-Arginine-Producing Mutants of Corynebacterium Glutamicum. Agric. Biol. Chem. 43 (1), 105–111. doi:10.1271/bbb1961.43.105
Zajac, A., Poprzęcki, S., Żebrowska, A., Chalimoniuk, M., and Langfort, J. (2010). Arginine and Ornithine Supplementation Increases Growth Hormone and Insulin-like Growth Factor-1 Serum Levels after Heavy-Resistance Exercise in Strength-Trained Athletes. J. Strength Cond. Res. 24 (4), 1082–1090. doi:10.1519/JSC.0b013e3181d321ff
Zhang, T., Chi, Z., Zhao, C. H., Chi, Z. M., and Gong, F. (2010). Bioethanol Production from Hydrolysates of Inulin and the Tuber Meal of Jerusalem Artichoke by Saccharomyces Sp. W0. Bioresour. Technol. 101 (21), 8166–8170. doi:10.1016/j.biortech.2010.06.013
Zhang, B., Ren, L.-Q., Yu, M., Zhou, Y., and Ye, B.-C. (2018). Enhanced L-Ornithine Production by Systematic Manipulation of L-Ornithine Metabolism in Engineered Corynebacterium Glutamicum S9114. Bioresour. Technol. 250, 60–68. doi:10.1016/j.biortech.2017.11.017
Zhou, S., Yuan, S.-F., Nair, P. H., Alper, H. S., Deng, Y., and Zhou, J. (2021). Development of a Growth Coupled and Multi-Layered Dynamic Regulation Network Balancing Malonyl-CoA Node to Enhance (2S)-Naringenin Biosynthesis in Escherichia coli. Metab. Eng. 67, 41–52. doi:10.1016/j.ymben.2021.05.007
Keywords: Bacillus amyloliquefaciens, L-Ornithine, L-ornithine transporter, non-grain raw materials, modular metabolic engineering
Citation: Zhu Y, Hu Y, Yan Y, Du S, Pan F, Li S, Xu H and Luo Z (2022) Metabolic Engineering of Bacillus amyloliquefaciens to Efficiently Synthesize L-Ornithine From Inulin. Front. Bioeng. Biotechnol. 10:905110. doi: 10.3389/fbioe.2022.905110
Received: 26 March 2022; Accepted: 10 May 2022;
Published: 08 June 2022.
Edited by:
Zhi-Qiang Liu, Zhejiang University of Technology, ChinaReviewed by:
Mingfeng Cao, Xiamen University, ChinaCopyright © 2022 Zhu, Hu, Yan, Du, Pan, Li, Xu and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhengshan Luo, bHVvenNAbmp0ZWNoLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.