
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 25 April 2022
Sec. Industrial Biotechnology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.900800
(R)-(+)-perillyl alcohol is a much valued supplemental compound with a wide range of agricultural and pharmacological characteristics. The aim of this study was to improve (R)-(+)-perillyl alcohol production using a whole-cell catalytic formula. In this study, we employed plasmids with varying copy numbers to identify an appropriate strain, strain 03. We demonstrated that low levels of alKL provided maximal biocatalyst stability. Upon determination of the optimal conditions, the (R)-(+)-perillyl alcohol yield reached 130 mg/L. For cofactor regeneration, we constructed strain 10, expressing FDH from Candida boidinii, and achieved (R)-(+)-perillyl alcohol production of 230 mg/L. As a result, 1.23 g/L (R)-(+)-perillyl alcohol was transformed in a 5 L fermenter. Our proposed method facilitates an alternative approach to the economical biosynthesis of (R)-(+)-perillyl alcohol.
(R)-(+)-limonene is a ubiquitous natural monocyclic monoterpene that accounts for over 90% of orange peel oil (Bicas et al., 2009). This makes it a readily available and affordable ingredient for fine chemical production (Berger et al., 1999). For the past few years, people considered using (R)-(+)-limonene to produce biological flavors. (R)-(+)-perillyl alcohol, one of its oxygenated derivatives, is a stable alcohol that is often used in anti-cancer drugs, such as colon, skin, head and neck (Gupta et al., 2005; Chen et al., 2006; Peffley et al., 2007; Yeruva et al., 2007; Das et al., 2010; Sobral et al., 2014).
In recent years, (R)-(+)-perillyl alcohol synthesis, particularly using microbial fermentation (MF) and enzymatic conversion (EC), has garnered much attention. The MF approach utilizes genetic engineering of the mevalonate pathway to simultaneously promote cell proliferation and (R)-(+)-perillyl alcohol synthesis. Until recently, it was reported that the content of glucose-derived (R)-(+)-perillyl alcohol, by artificially engineered Escherichia coli, was extremely low (87 mg/L) (Sun et al., 2021). This may be due to the complicated metabolic pathways, byproduct toxicity, and metabolic burden (Marmulla and Harder, 2014; Zhu et al., 2021). Thus, it is necessary to develop a new, simple, and efficient method for (R)-(+)-perillyl alcohol production.
EC methods include cell-free enzymes and whole cells (Li et al., 2020). Employing whole cells eliminate the need for cell-free enzyme purification and separation. Moreover, whole cells are more stable and resistant to environmental stimuli than free enzymes (Song et al., 2015; Yang et al., 2019). Despite limited investigations on the whole-cell bioconversion of (R)-(+)-limonene to (R)-(+)-perillyl alcohol, this approach offers much promise owing to the successful and efficient conversion of other limonene-derived monoterpenes using whole cell catalysts. Fortunately, the limonene-derived biotransformation of other monoterpenes have been reported in Penicillium sp. (Rottava et al., 2010; Badee et al., 2011; Molina et al., 2013), Aspergillus sp. (García-Carnelli et al., 2014), Fusarium oxysporum (Bicas et al., 2010a), Yarrowia lipolytica ATCC 18942 (Ferrara et al., 2013), Colletotrichum nymphaeae (Sales et al., 2018), Pseudomonas fluorescens (Chatterjee and Bhattacharyya, 2001; Bicas et al., 2008; Mirata et al., 2009; Soares-Castro et al., 2017) and Sphingobium sp. (Bicas et al., 2010b). (R)-(+)-α-terpineol synthesis from (R)-(+)-limonene using Sphingobium sp., with a Plackett-Burman design, followed by a Central Composite Design, gave a yield of 240 g/L after 96 h (Molina et al., 2019), which was the highest reported monoterpene yield. Mirata et al. (2009) also demonstrated that Pseudomonas putida DSM 12264 oxidizes the C7 site of limonene to produce (R)-(+)-perillic acid as the main product. Moreover, the perillic acid content was improved using in situ separation technology, thus, producing a yield of 31 g/L after 7 days. In addition, whole-cell reactions and subsequent purifications are relatively easy to perform, relative to the direct fermentation process (Oh et al., 2015).
In this study, we co-expressed p-cymene monooxygenase hydroxylase (cymAa) and p-cymene monoxygenase reductase (cymAb) in E. coli. Doing so, we converted (R)-(+)-limonene to (R)-(+)-perillyl alcohol (Figure 1A). Firstly, the catalytic activity was improved by expressing plasmids of varying copies. Moreover, alkL overexpression enhanced hydrophobic substrate uptake, whereas, low alkL level augmented whole cell stability. Next, we optimized biotransformation kinetics by altering parameters like initial pH, temperature, whole cell biocatalyst, and substrate concentration, to achieve the highest (R)-(+)-perillyl alcohol concentration. Employing the optimal conditions, along with formate dehydrogenase (FDH) for NADH regeneration, the strain 10 produced 1.23 g/L (R)-(+)-perillyl alcohol for 20 h in a 5 L fermenter. Our novel approach to (R)-(+)-perillyl alcohol synthesis was demonstrated to be highly efficient and green, with great potential for large-scale synthesis of monoterpenoids.
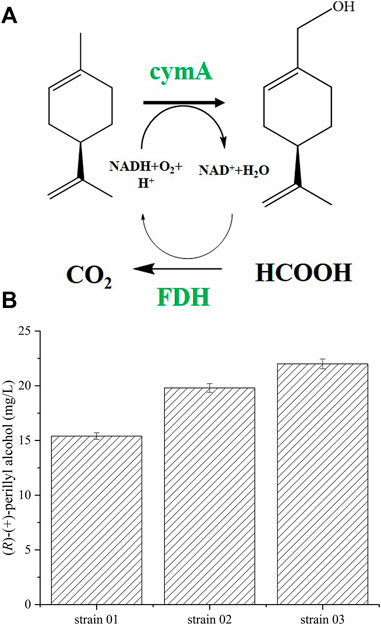
FIGURE 1. (R)-(+)-perillyl alcohol synthesis using whole cell biocatalysts. (A) (R)-(+)-perillyl alcohol biosynthesis from (R)-(+)-limonene, using a whole cell system. (B) (R)-(+)-perillyl alcohol production with different plasmids. Reaction conditions (20 ml): whole cell catalysts (OD600 = 30), 25 μl (R)-(+)- limonene, 8.0 ml dioctyl phthalate (DINP), and PBS buffer (50 mM, pH 7.4) at 30°C for 6 h. Data expressed as mean ± s.d. (n = 3). Abbreviations: CymAa, p-cymene monoxygenase hydroxylase, CymAb, p-cymene monoxygenase reductase, CymA, CymAa and CymAb, FDH, formate dehydrogenase.
Table 1 lists the strains employed in our research. Bacterial cultures were grown in LB medium (per liter: 10 g tryptone, 5 g yeast extract and 10 g NaCl) with 10 g/L glucose during shake-flask fermentation. The bioreactor fermentation medium contained (per liter) 8.7 g glycerol, 10 g tryptone, 11 g yeast extract, 2.4 g (NH4)SO4, 2 g citric acid. H2O, 20.6 g Na2HPO4.12H2O, 5.22 g KH2PO4, and 0.12 g MgSO4. A feed solution (50% glycerol, 5 g/L yeast extract and 5 g/L tryptone) was utilized. We also introduced supplemental antibiotics (100 μg/ml ampicillin, or 50 μg/ml kanamycin) to maintain corresponding plasmids.
First, we codon optimized cymAa and cymAb from Pseudomonas putida (Sun et al., 2021). Next, they were cloned into pACYCDuet-2 using BamHI/SacI restriction sites to construct pACYCDuet-2-cymA. The pET28a(+)-cymA and pRSFDuet-1-cymA plasmids were constructed in a similar fashion. The alkL gene (GenBank: AJ245436.1) from Pseudomonas putida GPo1 and fdh gene (GenBank: AJ011046.2) were codon optimized by BGI and produced by GeneWiz (Suzhou, China), before cloning into PUC57. The alkL gene was assembled into the pACYCDuet-2 plasmid using C115 (Vazyme Biotech Co., Ltd., Nanjing, China) at the NcoI/SacI sites, thus, generating the pACYCDuet-2-alkL plasmid. The fdh gene from Candida boidinii was then ligated into pRSFDuet-1-cymA to create pRSFDuet-1-cymA-fdh. All primers are shown in Supplementary Table S1.
First, monoclonal strain was selected from the LB agar plate and cultured in a tube with 4 ml LB broth till the exponential phase. Next, seed cultures were inoculated into 500 ml baffled triangular flask carrying 100 ml LB and 10 g/L glucose at 37°C, 200 pm with an OD600 of 0.6–0.8, the temperature was set to 16°C, and induction was done with 0.2 mM IPTG for 20 h.
The recombinant E. coli were collected by centrifugation (8000 g for 10 min), PBS-rinsed two times (50 mM PBS buffer, pH 7.4), and resuspended to obtain a whole-cell catalyst, followed by storage at 4°C until further analysis.
All optimization experiments were performed in 250 ml flasks with 20 ml reaction broth, whole-cell catalysts (OD600 = 30), 25 μl (R)-(+)- limonene, 8.0 ml dioctyl phthalate (DINP), and 50 mM PBS buffer (pH 7.4). All reactions were carried out for 6 h at 30°C, with stirring at 200 rpm.
The variables examined included pH, reaction temperature, NADH, cell concentrations, and substrate concentrations. To optimize pH, experiments were conducted in several buffers (pH 5.8–8.0). Temperature optimization was done by monitoring whole-cell conversion activity between 16 and 37°C. To optimize NADH concentrations, 0, 1, and 2 mM NADH were examined. To establish optimal cell concentration, whole cell catalysts between OD600 10–70 were used. Moreover, following optimization of the above conditions, the effects of different substrate concentrations (1–40 g/L) were examined. Lastly, different ammonium formate concentrations (10–100 g/L) were assessed for optimal reaction results.
The engineered strain was cultured for 12 h at 37°C in 250 ml shake flask with 50 ml LB medium. Fed-batch fermentations were applicated in a 5 Lfermenter (ez-control, Applikon) containing 2 L fermentation broth at 0.5–2 vvm aeration and 400–800 rpm, respectively. Subsequently, the fermentation was inoculated with 5.0% (v/v) seed culture, and performed at 37°C and pH 7.0 with the addition of NH4OH (25%, v/v). When the OD600 reached 15 (initial glycerol was consumed), 0.2 mM IPTG was introduced and the temperature was set to 16°C for 20 h. The feeding speed was variable for the induction period with 50% glycerol supplement (1.5–2.0 g/L.h). Dissolved oxygen (DO) levels were not less than 20%. The recombinant E. coli were then collected via centrifugation (8000 g for 10 min), PBS-rinsed two times (50 mM, pH 7.4), and resuspended to obtain the whole-cell catalyst, followed by storage at 4°C.
Under optimized reaction conditions, we employed whole-cell biocatalyst to synthesize (R)-(+)-perillyl alcohol in a 5 L fermenter. The conversion mixture (1 L) included 20 g/L (R)-(+)-limonene, 40 g/L ammonium formate, recombinant cells (OD600 = 50), 40 ml dioctyl phthalate (DINP), and PBS buffer (50 mM, pH 7.4), with constant stirring at 20°C for 24 h. (R)-(+)-limonene and (R)-(+)-perillyl alcohol were detected every 4 h.
The whole-cell catalyst enzymatic activity was determined by adding 2 mM (R)-(+)-limonene and whole-cell catalyst (0.5 gCDW.L−1), followed by detection of (R)-(+)-perillyl alcohol concentration. The reaction (1 ml) was carried out in triplicate at 30°C for 10 min and 200 rpm (Cornelissen et al., 2011). Activities are presented in 1 UgCDW−1, with 1 UgCDW−1 = 1 μmol product min−1 gCDW−1.
The E. coli biomass was measured using OD600 on a spectrophotometer (Cary 50 UV-vis, Varian). Cell Dry weight (CDW) was calculated as follows: 1 OD600 = 0.323 gCDW.L−1. The Shimadzu GC-MS system (TQ8050) was employed to detect (R)-(+)-limonene, (R)-(+)-perillyl alcohol, and (R)-(+)-perillyl aldehyde, using the following conditions: 30 m DB-5MS column. (inside diameter 0.32 mm, film thickness 0.25 μm); temperature: held at 50°C, before ramp up 10°C/min to 250°C, followed by another hold at 250°C for 10 min. Highly pure helium at a linear velocity of 1 ml/min served as the carrier; the injector temperature was set to 250°C; a split ratio was adjusted to 1:10; ion source temperature was set to 230°C, and mass range was set to m/z 40–500. The fermentation broth was mixed with equal volume of ethyl acetate before spinning. The organic layer was further assessed using specific test methods and sample treatments, as reported earlier (Sun et al., 2021).
In a previous report, (R)-(+)-perillyl alcohol was obtained in an engineered E. coli by expressing cymA genes from glucose (Sun et al., 2021). Using this technique, strains 01, 02, and 03 were constructed (Table 1). In prior research, enzyme expression was balanced using plasmids with varying copy numbers (Hou et al., 2017). In this study, we employed three plasmids with distinct copy numbers: pRSFDuet-1 (100 copies per cell), pET28a (+) (40 copies per cell), and pACYCDuet-1 (10 copies per cell) (Rico et al., 2019). Next, we assessed their synthetic ability and enzymatic activity (Figure 1B and Supplementary Table S2). Relative to strain 03, the specific activities of cymA in strains 01 and 02 were 2.56 and 2.89 U gCDW−1, respectively, and they decreased by 32 and 24%. Based on our analysis, strain 03, constructed by pRSFDuet-1 (high copy number), provided the largest yield of (R)-(+)-perillyl alcohol at 22.0 mg/L (all units presented as the total fermentation volume, mg/Ltot).
Earlier reports suggested that alkL enhances uptake and oxygenation of substrate, such as, alkanes, FAMEs, located in the inner membrane or in the cytosol (Julsing et al., 2012; Cornelissen et al., 2013; Grant et al., 2014). To elucidate the outcome of varying alkL levels on (R)-(+)-limonene conversion rates and product synthesis kinetics, recombinant E. coli BL21 (DE3), with different promoters, were used to construct three distinct strains (strain 04, strain 05, and strain 06) (Table 1 and Figure 2). Predictably, the alkL-negative strain 03 produced the least specific (R)-(+)-limonene hydroxylation activity (Figure 2B). In contrast, all alkL-positive strains exhibited markedly higher activities, and the level of activity corresponded to the level of alkL. AlkL-positive strain 06 (medium to high alkL expression) initially produced the maximum specific (R)-(+)-limonene hydroxylation activity at 12–14 U gDCW−1, followed by a rapid decrease. Conversely, strain 04, carrying a low alkL expression, produced relatively stable activity at 10 U gDCW−1 (Figure 2B), and maintained 45 mg/L (R)-(+)-perillyl alcohol, which reached 1.7 times higher than strain 06, with high alkL level (Figure 2C).
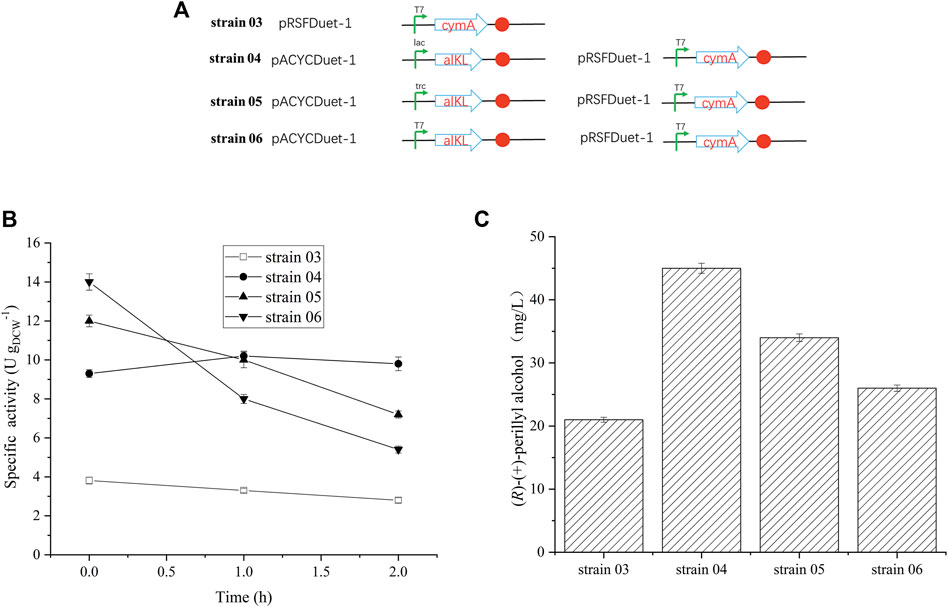
FIGURE 2. Effect of alkL protein expression on (R)-(+)-perillyl alcohol production. (A) Strains constructed by different promotors. (B) Whole-cell activity assays related to alkL expression. (C) Comparison of (R)-(+)-perillyl alcohol production in each strain. Reaction conditions (20 ml): whole cell catalysts (OD600 = 30), 25 μl (R)-(+)- limonene, 8.0 ml dioctyl phthalate (DINP), and PBS buffer (50 mM, pH 7.4) at 30°C for 6 h. Data expressed as mean ± s.d. (n = 3).
Recombinant E.coli. BL21 (DE3), E.coli. JM109 (DE3), E.coli Rosstta (DE3), and E.coli MG1655 (DE3), harboring both pRSFDuet-1-cymA and pACYCDet-2-lac-alkL, were examined, in terms of (R)-(+)-perillyl alcohol synthesis, in PBS buffer containing 1 g/L of (R)-(+)-limonene (Figure 3). The highest concentration was achieved by strain 09, yielding 54 mg/L (R)-(+)-perillyl alcohol, which was 1.2 times higher than strain 04. Thus, strain 09 was selected for subsequent investigations on (R)-(+)-perillyl alcohol synthesis.
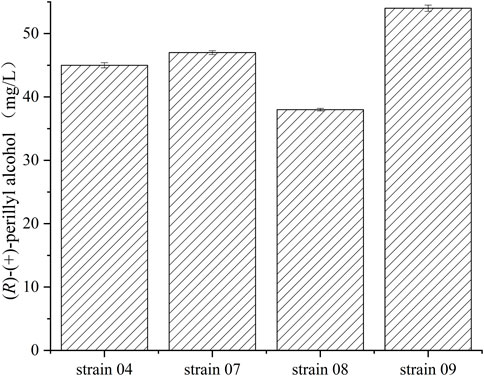
FIGURE 3. Various concentrations of (R)-(+)-perillyl alcohol production by different E. coli strains. Reaction conditions (20 ml): whole cell catalysts (OD600 = 30), 25 μl (R)-(+)- limonene, 8.0 ml dioctyl phthalate (DINP), and PBS buffer (50 mM, pH 7.4) at 30°C for 6 h. Data expressed as mean ± s.d. (n = 3).
We further optimized reaction conditions to produce the largest yield of (R)-(+)-perillyl alcohol from (R)-(+)-limonene. The assessed parameters included initial pH, temperature, NADH introduction, and whole-cell catalysts.
To augment (R)-(+)-perillyl alcohol synthesis, pH and temperature of whole cell catalysis were optimized in strain 09. We synthesized (R)-(+)-perillyl alcohol in different phosphate buffer (pH 5.8–8.0), and pH 7.4 produced the largest yield (Figure 4A). The examined temperatures were 16, 20, 25, 30, and 37°C, and the temperature the produced the most amount of (R)-(+)-perillyl alcohol was 20°C (Figure 4B). Taken together, the optimal conditions for (R)-(+)-perillyl alcohol synthesis in strain 09 were pH 7.4 and 20°C.
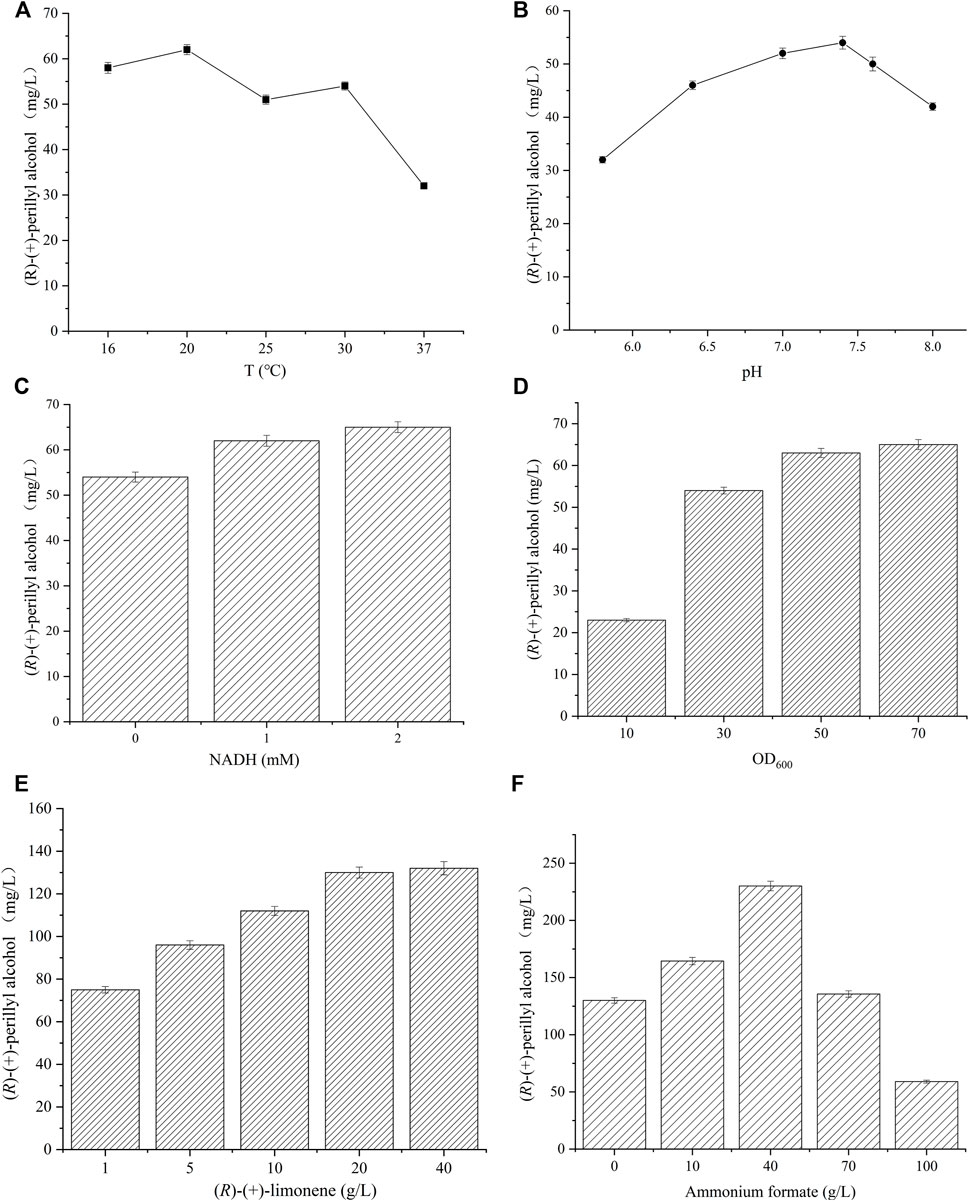
FIGURE 4. Reaction condition optimization for (R)-(+)-perillyl alcohol synthesis. The effect of (A) varying pHs and (B) temperatures on (R)-(+)-perillyl alcohol synthesis. Reaction conditions (20 ml) for different pHs: whole cell catalysts (OD600 = 30), 25 μl (R)-(+)- limonene, 8.0 ml dioctyl phthalate (DINP), and PBS buffer (50 mM, pH 5.8–8.0) at 30°C for 6 h. Reaction conditions (20 ml) for different temperatures: whole cell catalysts (OD600 = 30), 25 μl (R)-(+)-limonene, 8.0 ml dioctyl phthalate (DINP), and PBS buffer (50 mM, pH 7.4) at 16–37°C for 6 h. (C) Effects of NADH addition on (R)-(+)-perillyl alcohol production. Reaction conditions (20 ml): whole cell catalysts (OD600 = 30), 25 μl (R)-(+)-limonene, 8.0 ml dioctyl phthalate (DINP), NADH (0–2 mM), and PBS buffer (50 mM, pH 7.4) at 30°C for 6 h. (D) Effect of biocatalysts concentrations on (R)-(+)-perillyl alcohol production. Reaction conditions (20 ml): whole cell catalysts (OD600 = 10, 30, 50, 70), 25 μl (R)-(+)-limonene, 8.0 ml dioctyl phthalate (DINP), and PBS buffer (50 mM, pH 7.4) at 30°C for 6 h. (E) Whole cell biotransformation of (R)-(+)-perillyl alcohol at high (R)-(+)-limonene concentration. Reaction conditions (20 ml): whole cell catalysts (OD600 = 50), (R)-(+)-limonene (1, 5, 10, 20, 40 g/L), 8.0 ml dioctyl phthalate (DINP), 2 mM NADH, and PBS buffer (50 mM, pH 7.4) at 30°C for 6 h. (F) (R)-(+)-perillyl alcohol production with different ammonium formate concentrations. Reaction conditions (20 ml): whole-cell catalysts (OD600 = 50), 500 μl (R)-(+)-limonene, 8.0 ml dioctyl phthalate (DINP), ammonium formate (0, 10, 40, 70, 100 g/L), and PBS buffer (50 mM, pH 7.4) at 30°C for 6 h. Data expressed as mean ± s.d. (n = 3).
It is possible that the reductase portion of cymA transfers electrons from NADH to the hydroxylase subunit (Figure 1A), using flavin and ferredoxin moieties (Eaton, 1997; Dutta et al., 2010; Dutta et al., 2012). Thus, NADH is a necessary element for the production of (R)-(+)-perillyl alcohol from (R)-(+)-limonene (Figure 1A). The optimal NADH, according to our results, was 2 mM, which produced 65 mg/L of (R)-(+)-perillyl alcohol (Figure 4C).
An increasing amount of whole cell (OD600 of 10, 30, 50, 70) was introduced to identify the optimal whole cell concentration for efficient (R)-(+)-perillyl alcohol production. Based on our analysis, cell concentration with an OD600 value of 50 markedly enhanced (R)-(+)-perillyl alcohol synthesis, whereas, cell concentration of 70 did not alter the titer significantly (Figure 4D). Moreover, the influence of cell densities exceeding OD600 value of 50 was unremarkable, likely due to the lack of dissolved oxygen, which is a critical step in (R)-(+)-perillyl alcohol synthesis.
Based on the above findings, we synthesized 75 mg/L (R)-(+)-perillyl alcohol in strain 09, under the proper conditions (Figure 4E). Given that excess substrate can vastly improve volumetric productivity, we investigated different substrate concentrations varying from 1 to 40 g/L (Figure 4E). With 20 g/L (R)-(+)-limonene, the (R)-(+)-perillyl alcohol production was 130 mg/L, which was 1.7 times higher than the production from 1 g/L (R)-(+)-limonene.In addition, (R)-(+)-perillyl alcohol accumulation was not obvious, when the substrate exceeded 20 g/L.
NADH addition can obviously increase (R)-(+)-perillyl alcohol yield (Figure 4C). Since NADH is costly, we integrated a NADH generating pathway, such as, formate dehydrogenase (FDH) for the regeneration of NADH (Liu et al., 2019; Chen et al., 2021). FDH from Candida boidinii was expressed and strain 10 was constructed. NADH regeneration was achieved by adding different concentrations of ammonium formate. When 40 g/L ammonium formate was added, the (R)-(+)-perillyl alcohol production was 230 mg/L (Figure 4F). Conversely, when ammonium formate exceeded 40 g/L, the product yield decreased rapidly, likely due to enzyme inactivation in presence of excess ammonium formate.
Using the optimal parameters from aforementioned experiments (i.e., pH, cell density, and fdh gene expression), recombinant E. coli strain 10 was cultivated in a 5 L bioreactor. The biomass was obtained via centrifugation and employed for the biotransformation of (R)-(+)-limonene to (R)-(+)-perillyl alcohol. The reaction was conducted as follows: 20°C with 300–800 rpm and 0.5 vvm; pH of 7.4; and recombinant cell concentration OD600 of 50. The (R)-(+)-perillyl alcohol synthesis yield is shown in Figure 5. In brief, (R)-(+)-perillyl alcohol yield rose drastically between 0 and 12 h, then started to decreased between 12 and 20 h, and the titer reduced even more after 20 h. Our largest (R)-(+)-perillyl alcohol yield was 1.23 g/L by using strain 10. The loss of substrate limonene is partly oxidized to (R)-(+)-perillyl alcohol, partly oxidized to other by-products such as perillyl aldehyde, and partly evaporated throughout the reaction process.
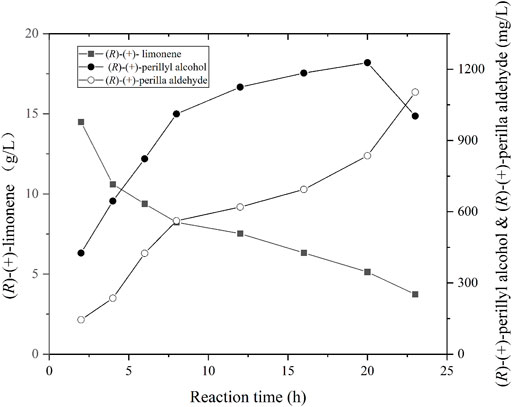
FIGURE 5. Time course of (R)-(+)-limonene hydroxylation using whole cell of strain 10. Reaction conditions (1 L): whole cell catalysts (OD600 = 50), 25 ml (R)-(+)-limonene, 40 g ammonium formate, 400 ml dioctyl phthalate (DINP), and PBS buffer (50 mM, pH 7.4) at 20°C for 24 h.
Whole-cell biocatalyst is commonly used to produce multiple compounds (Goldberg et al., 2007; Fukuda et al., 2008; Hou et al., 2015). The reasons for this are multifold. Whole-cell catalyst offers enhanced operability and economy, relative to extracted enzyme. Moreover, it provides better stability and can be repeatedly used in bioconversions, without loss of activity (Richter et al., 2010). Furthermore, whole-cell catalyst is more tolerant to substrate and provides higher product yield than extracted enzyme (Li et al., 2018).
During whole-cell biotransformation, low activity enzyme negatively affects (R)-(+)-perillyl alcohol production. Hence, it is crucial to balance enzyme levels during biosynthetic reactions. In this study, we employed varying plasmid copy numbers to regulate enzyme levels, as was previously reported (Wu et al., 2013; Hou et al., 2017). CymA overexpression in pRSFDuet-1 (high copy number) yielded high enzymatic activity and enhanced (R)-(+)-perillyl alcohol synthesis in strain 03 (Rico et al., 2019). (R)-(+)-perillyl alcohol synthesis by strain 01 (pACYCDet-2-cymA) and strain 02 (pET28a (+)-cymA) was slightly lower than strain 3 (Figure 2B and Supplementary Table S2). CymA carries the genes cymAa and cymAb, which encode the two subunits (hydroxylase subunit and reductase subunit) of p-cymene monooxygenase (Eaton, 1997). It is possible that the hydroxylase subunit cymAa was strongly expressed in strain 03 with the high copy plasmid.
We also demonstrated that different alkL expressions were constructed in strains 04, 05, and 06. Strain 04, producing the lowest alkL levels, maintained the stability of enzymatic activity for 2 h (Figure 2B). Strain 06, on the other hand, produced the highest alkL levels, which lead to whole-cell biocatalyst toxicity and reduced production (Figure 2B). In contrast, two-liquid phase biotransformation with whole cell catalyst (strain 04), featuring low alkL levels, enhanced biocatalyst viability and stability, thereby enhancing product titers. Julsing et al. (2012) similarly reported that well-regulated alkL production and low alkBGT expression facilitates a 4.3 times increase in product titers in a two-liquid phase bioprocess. These evidences suggest the critical role of a balance between biocatalyst stability and maximal titiers in determining hydroxylation efficiency of whole-cell-based reactions. Here, we achieved remarkable enhancement of biocatalyst stability via fine-tuning alkL expression, and thus, maximizing (R)-(+)-perillyl alcohol production.
One of the important factors of (R)-(+)-perillyl alcohol production via biotransformation is reaction condition. We established the optimal pH and temperature of whole-cell biocatalyst to be pH 7.4 and 20°C (Figures 4A,B). In addition, our (R)-(+)-perillyl alcohol production was 130 mg/L, when 50 OD600 whole-cell catalyst and 20 g/L substrate concentration was employed (Figures 4D,E). In prior studies, (R)-(+)-perillyl alcohol was shown to be crucial for whole-cell biotransformation at elevated substrate concentration. Limonene causes E. coli growth reduction during fermentation at concentrations between 1.05 and 3.36 g/L (Dunlop et al., 2011; Chubukov et al., 2015; Edgar et al., 2017). It is possible that the inhibitory concentration of limonene on cells was increased by using high concentration whole cell catalysis and, thus, the yield was effectively augmented. Owing to its remarkable tolerance to elevated substrate concentration, our developed biocatalyst became efficient in whole cell bioconversion.
The representative cofactor NADH plays a central role in the conversion of (R)-(+)-limonene to (R)-(+)-perillyl alcohol (Agullo et al., 2017). (R)-(+)-perillyl alcohol synthesis was augmented by 1.2 times with 2 mM NADH, as opposed to no NADH addition (Figure 4C). NADH regeneration, based on formate dehydrogenase, is highly developed (Alissandratos et al., 2014). Given that NADH is expensive ($260 per gram) (Britton et al., 2018), we expressed fdh from Candida boidinii to construct strain 10. Recently, 230 mg/L (R)-(+)-perillyl alcohol was produced using whole cells (OD600 = 50) of strain 10 during batch biotransformation. FDH is also used in the synthesis of multiple other compounds (L-gulose, aminobutyric acid) to achieve NADH regeneration (Chen et al., 2021; Zhang et al., 2021). At present, the formate dehydrogenase activity is not very strong, therefore, it is necessary to screen other dehydrogenases (GDH) or modify enzymes (Zhu et al., 2017).
Sufficient oxygen is required for the hydroxylation of monoxygenase, but not NADH (Li et al., 2002). This was supplied with aeration in the bioreactor. Finally, 20 g/L(R)-(+)-limonene was catalyzed to produce 1.23 g/L (R)-(+)-perillyl alcohol, with a substrate conversion rate 6% (Figure 5). In two-liquid phase bioconversion, 20–30% (R)-(+)-limonene was lost via evaporation, and the remaining 70–80% was oxidized (Figure 5). The loss of (R)-(+)-limonene was increased with the prolongation of conversion time. Hence, excess substrate was sacrificed. The production was not very obvious with the addition of 5 g/L (R)-(+)-limonene at 12 h (date not known). It is possible that (R)-(+)-perillyl alcohol was converted to (R)-(+)-perilla aldehyde (Figure 5). CymA converts p-cymene to a combination of p-cumic alcohol and p-cumic aldehyde (Eaton, 1997). So, it is likely that there were (R)-(+)-perillyl alcohol and (R)-(+)-perilla aldehyde mixtures in the fermentation products. In order to further enhance production, directed evolution of cymA or deletion of alcohol dehydrogenase [conversion of (R)-(+)-perillyl alcohol into (R)-(+)-perilla aldehyde] was carried out.
Here, we demonstrated that recombinant E. coli expressing cymA can synthesize (R)-(+)-perillyl alcohol, with whole cell catalysis. High plasmid copy number expressing cymA is highly beneficial to (R)-(+)-perillyl alcohol production, and low alkL levels promote whole cell stability. We optimized catalytic conditions, as well as cell and substrate concentration, to further enhance (R)-(+)-perillyl alcohol production. Simultaneously, we also achieved NADH cofactor regeneration. Finally, (R)-(+)-perillyl alcohol production in strain 10 was 1.23 g/L in a 5 L bioreactor, and improved by 82 times, compared to strain 01 (15 mg/L). The whole cell catalysis approach could be useful for various valuable chemicals and pharmaceuticals.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
CS: Data analysis, manuscript writing. RZ: Methodology. CX: Investigation, review and editing. All authors read and approved the manuscript.
This work was supported by the National Natural Science of China (grant number 21878166, 22007091), and the Taishan Scholar Project of Shandong Province, China (grant number ts201511033).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors appreciate the contribution of the Yujin Cao for his advice on strain design.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.900800/full#supplementary-material
CymAa, p-cymene monoxygenase hydroxylase; CymAb, p-cymene monoxygenase reductase; CymA, p-cymene monoxygenase hydroxylase and p-cymene monoxygenase reductase; FDH, formate dehydrogenase; alkL, outer membrane protein; GDH, glucose 1-dehydrogenase.
Agulló, L., Romero-Silva, M. J., Domenech, M., and Seeger, M. (2017). p-Cymene Promotes its Catabolism through the P-Cymene and the P-Cumate Pathways, Activates a Stress Response and Reduces the Biofilm Formation in Burkholderia Xenovorans LB400. PLoS One 12 (1), e0169544. doi:10.1371/journal.pone.0169544
Alissandratos, A., Kim, H.-K., and Easton, C. J. (2014). Formate Production through Carbon Dioxide Hydrogenation with Recombinant Whole Cell Biocatalysts. Bioresour. Tech. 164, 7–11. doi:10.1016/j.biortech.2014.04.064
Badee, A. Z. M., Helmy, S. A., and Morsy, N. F. S. (2011). Utilisation of orange Peel in the Production of α-terpineol by Penicillium digitatum (NRRL 1202). Food Chem. 126 (3), 849–854. doi:10.1016/j.foodchem.2010.11.046
Berger, R. G., De Bont, J. A. M., Eggink, G., Da Fonseca, M. M., Gehrke, M., Gros, J.-B., et al. (1999). “Biotransformations in the Flavour Industry,” in Current Topics in Flavours and Fragrances: Towards a New Millennium of Discovery. Editor KAD Swift (Dordrecht: Springer Netherlands), 139–170. doi:10.1007/978-94-011-4022-5_8
Bicas, J. L., de Quadros, C. P., Néri-Numa, I. A., and Pastore, G. M. (2010a). Integrated Process for Co-production of Alkaline Lipase and (R)-(+)-α-terpineol by Fusarium Oxysporum. Food Chem. 120 (2), 452–456. doi:10.1016/j.foodchem.2009.10.037
Bicas, J. L., Dionísio, A. P., and Pastore, G. M. (2009). Bio-oxidation of Terpenes: An Approach for the Flavor Industry. Chem. Rev. 109 (9), 4518–4531. doi:10.1021/cr800190y
Bicas, J. L., Fontanille, P., Pastore, G. M., and Larroche, C. (2010b). A Bioprocess for the Production of High Concentrations of (R)-(+)-α-terpineol from (R)-(+)-limonene. Process Biochem. 45 (4), 481–486. doi:10.1016/j.procbio.2009.11.007
Bicas, J. L., Fontanille, P., Pastore, G. M., and Larroche, C. (2008). Characterization of Monoterpene Biotransformation in Two Pseudomonads. J. Appl. Microbiol. 105 (6), 1991–2001. doi:10.1111/j.1365-2672.2008.03923.x
Britton, J., Majumdar, S., and Weiss, G. A. (2018). Continuous Flow Biocatalysis. Chem. Soc. Rev. 47 (15), 5891–5918. doi:10.1039/c7cs00906b
Chatterjee, T., and Bhattacharyya, D. K. (2001). Biotransformation of Limonene by Pseudomonas Putida. Appl. Microbiol. Biotechnol. 55 (5), 541–546. doi:10.1007/s002530000538
Chen, J., Lu, M., Jing, Y., and Dong, J. (2006). The Synthesis of L-Carvone and Limonene Derivatives with Increased Antiproliferative Effect and Activation of ERK Pathway in Prostate Cancer Cells. Bioorg. Med. Chem. 14 (19), 6539–6547. doi:10.1016/j.bmc.2006.06.013
Chen, J., Zhu, R., Zhou, J., Yang, T., Zhang, X., Xu, M., et al. (2021). Efficient Single Whole-Cell Biotransformation for L-2-Aminobutyric Acid Production through Engineering of Leucine Dehydrogenase Combined with Expression Regulation. Bioresour. Tech. 326, 124665. doi:10.1016/j.biortech.2021.124665
Chubukov, V., Mingardon, F., Schackwitz, W., Baidoo, E. E. K., Alonso-Gutierrez, J., Hu, Q., et al. (2015). Acute Limonene Toxicity in Escherichia coli Is Caused by Limonene Hydroperoxide and Alleviated by a Point Mutation in Alkyl Hydroperoxidase AhpC. Appl. Environ. Microbiol. 81 (14), 4690–4696. doi:10.1128/aem.01102-15
Cornelissen, S., Julsing, M. K., Volmer, J., Riechert, O., Schmid, A., and Bühler, B. (2013). Whole-cell-based CYP153A6-Catalyzed (S)-limonene Hydroxylation Efficiency Depends on Host Background and Profits from Monoterpene Uptake via AlkL. Biotechnol. Bioeng. 110 (5), 1282–1292. doi:10.1002/bit.24801
Cornelissen, S., Liu, S., Deshmukh, A. T., Schmid, A., and Bühler, B. (2011). Cell Physiology rather Than Enzyme Kinetics Can Determine the Efficiency of Cytochrome P450-Catalyzed C-H-Oxyfunctionalization. J. Ind. Microbiol. Biotechnol. 38 (9), 1359–1370. doi:10.1007/s10295-010-0919-y
Das, B. C., Mahalingam, S. M., Panda, L., Wang, B., Campbell, P. D., and Evans, T. (2010). Design and Synthesis of Potential New Apoptosis Agents: Hybrid Compounds Containing Perillyl Alcohol and New Constrained Retinoids. Tetrahedron Lett. 51 (11), 1462–1466. doi:10.1016/j.tetlet.2010.01.003
Dunlop, M. J., Dossani, Z. Y., Szmidt, H. L., Chu, H. C., Lee, T. S., Keasling, J. D., et al. (2011). Engineering Microbial Biofuel Tolerance and export Using Efflux Pumps. Mol. Syst. Biol. 7, 487. doi:10.1038/msb.2011.21
Dutta, T. K., Chakraborty, J., Roy, M., Ghosal, D., Khara, P., and Gunsalus, I. C. (2010). Cloning and Characterization of a P-Cymene Monooxygenase from Pseudomonas Chlororaphis Subsp. Aureofaciens. Res. Microbiol. 161 (10), 876–882. doi:10.1016/j.resmic.2010.10.008
Dutta, T. K., Dutta, A., Chakraborty, J., Sarkar, J., Pal Chowdhury, P., and Gunsalus, I. C. (2012). Purification and Properties of Reductase of the Three-Component P-Cymene Methyl Hydroxylase from Pseudomonas Chlororaphis Subsp. Aureofaciens. Process Biochem. 47 (8), 1263–1267. doi:10.1016/j.procbio.2012.04.021
Eaton, R. W. (1997). p-Cymene Catabolic Pathway in Pseudomonas Putida F1: Cloning and Characterization of DNA Encoding Conversion of P-Cymene to P-Cumate. J. Bacteriol. 179 (10), 3171–3180. doi:10.1128/jb.179.10.3171-3180.1997
Edgar, S., Li, F.-S., Qiao, K., Weng, J.-K., and Stephanopoulos, G. (2017). Engineering of Taxadiene Synthase for Improved Selectivity and Yield of a Key Taxol Biosynthetic Intermediate. ACS Synth. Biol. 6 (2), 201–205. doi:10.1021/acssynbio.6b00206
Ferrara, M. A., Almeida, D. S., Siani, A. C., Lucchetti, L., Lacerda, P. S. B., Freitas, A., et al. (2013). Bioconversion of (R)-(+)-limonene to Perillic Acid by the Yeast Yarrowia Lipolytica. Braz. J. Microbiol. 44 (4), 1075–1080. doi:10.1590/s1517-83822014005000008
Fukuda, H., Hama, S., Tamalampudi, S., and Noda, H. (2008). Whole-cell Biocatalysts for Biodiesel Fuel Production. Trends Biotechnol. 26 (12), 668–673. doi:10.1016/j.tibtech.2008.08.001
García-Carnelli, C., Rodríguez, P., Heinzen, H., and Pilar Menéndez, P. (2014). Influence of Culture Conditions on the Biotransformation of (+)-Limonene by Aspergillus niger. Z. für Naturforschung C 69 (1-2), 61–67. doi:10.5560/znc.2013-0048
Goldberg, K., Schroer, K., Lütz, S., and Liese, A. (2007). Biocatalytic Ketone Reduction-A Powerful Tool for the Production of Chiral Alcohols-Part I: Processes with Isolated Enzymes. Appl. Microbiol. Biotechnol. 76 (2), 237–248. doi:10.1007/s00253-007-1002-0
Grant, C., Deszcz, D., Wei, Y. C., Martínez-Torres, R. J., Morris, P., Folliard, T., et al. (2014). Identification and Use of an Alkane Transporter Plug-In for Applications in Biocatalysis and Whole-Cell Biosensing of Alkanes. Sci. Rep. 4 (1), 5844–5849. doi:10.1038/srep05844
Gupta, A., Stratton, S. P., and Myrdal, P. B. (2005). An HPLC Method for Quantitation of Perillyl Alcohol in a Topical Pharmaceutical Cream Formulation. J. Pharm. Biomed. Anal. 37 (3), 447–452. doi:10.1016/j.jpba.2004.02.039
Hou, Y., Hossain, G. S., Li, J., Shin, H.-D., Du, G., Chen, J., et al. (2017). Metabolic Engineering of Cofactor Flavin Adenine Dinucleotide (FAD) Synthesis and Regeneration inEscherichia Colifor Production of α-keto Acids. Biotechnol. Bioeng. 114 (9), 1928–1936. doi:10.1002/bit.26336
Hou, Y., Hossain, G. S., Li, J., Shin, H.-d., Liu, L., and Du, G. (2015). Production of Phenylpyruvic Acid from L-Phenylalanine Using an L-Amino Acid Deaminase from Proteus mirabilis: Comparison of Enzymatic and Whole-Cell Biotransformation Approaches. Appl. Microbiol. Biotechnol. 99 (20), 8391–8402. doi:10.1007/s00253-015-6757-0
Julsing, M. K., Schrewe, M., Cornelissen, S., Hermann, I., Schmid, A., and Bühler, B. (2012). Outer Membrane Protein AlkL Boosts Biocatalytic Oxyfunctionalization of Hydrophobic Substrates in Escherichia coli. Escherichia Coliappl Environ. Microbiol. 78 (16), 5724–5733. doi:10.1128/aem.00949-12
Li, H., Lu, X., Chen, K., Yang, J., Zhang, A., Wang, X., et al. (2018). β-Alanine Production Using Whole-Cell Biocatalysts in Recombinant Escherichia coli. Escherichia coliMolecular Catal. 449, 93–98. doi:10.1016/j.mcat.2018.02.008
Li, Y., Liu, S., and You, C. (2020). Permeabilized Escherichia coli Whole Cells Containing Co‐Expressed Two Thermophilic Enzymes Facilitate the Synthesis of Scyllo ‐Inositol from Myo ‐Inositol. Biotechnol. J. 15 (2), 1900191. doi:10.1002/biot.201900191
Li, Z., van Beilen, J. B., Duetz, W. A., Schmid, A., de Raadt, A., Griengl, H., et al. (2002). Oxidative Biotransformations Using Oxygenases. Curr. Opin. Chem. Biol. 6 (2), 136–144. doi:10.1016/s1367-5931(02)00296-x
Liu, S., Zhang, X., Liu, F., Xu, M., Yang, T., Long, M., et al. (2019). Designing of a Cofactor Self-Sufficient Whole-Cell Biocatalyst System for Production of 1,2-amino Alcohols from Epoxides. ACS Synth. Biol. 8 (4), 734–743. doi:10.1021/acssynbio.8b00364
Marmulla, R., and Harder, J. (2014). Microbial Monoterpene Transformationsâ€"a Review. Front. Microbiol. 5, 346. doi:10.3389/fmicb.2014.00346
Mirata, M. A., Heerd, D., and Schrader, J. (2009). Integrated Bioprocess for the Oxidation of Limonene to Perillic Acid with Pseudomonas Putida DSM 12264. Process Biochem. 44 (7), 764–771. doi:10.1016/j.procbio.2009.03.013
Molina, G., Pessôa, M. G., Bicas, J. L., Fontanille, P., Larroche, C., and Pastore, G. M. (2019). Optimization of Limonene Biotransformation for the Production of Bulk Amounts of α-terpineol. Bioresour. Tech. 294, 122180. doi:10.1016/j.biortech.2019.122180
Molina, G., Pinheiro, D. M., Pimentel, M. R., dos Ssanros, R., and Pastore, G. M. (2013). Monoterpene Bioconversion for the Production of Aroma Compounds by Fungi Isolated from Brazilian Fruits. Food Sci. Biotechnol. 22 (4), 999–1006. doi:10.1007/s10068-013-0176-8
Oh, Y. H., Kang, K.-H., Kwon, M. J., Choi, J. W., Joo, J. C., Lee, S. H., et al. (2015). Development of Engineered Escherichia coli Whole-Cell Biocatalysts for High-Level Conversion of L-Lysine into Cadaverine. J. Ind. Microbiol. Biotechnol. 42 (11), 1481–1491. doi:10.1007/s10295-015-1678-6
Peffley, D. M., Sharma, C., Hentosh, P., and Buechler, R. D. (2007). Perillyl Alcohol and Genistein Differentially Regulate PKB/Akt and 4E-BP1 Phosphorylation as Well as eIF4E/eIF4G Interactions in Human Tumor Cells. Arch. Biochem. Biophys. 465 (1), 266–273. doi:10.1016/j.abb.2007.05.022
Richter, N., Neumann, M., Liese, A., Wohlgemuth, R., Weckbecker, A., Eggert, T., et al. (2010). Characterization of a Whole-Cell Catalyst Co-expressing Glycerol Dehydrogenase and Glucose Dehydrogenase and its Application in the Synthesis of L-Glyceraldehyde. Biotechnol. Bioeng. 106 (4), 541–552. doi:10.1002/bit.22714
Rico, J., Duquesne, K., Petit, J. L., Mariage, A., Darii, E., Peruch, F., et al. (2019). Exploring Natural Biodiversity to Expand Access to Microbial Terpene Synthesis. Microb. Cel Fact 18 (1), 23–10. doi:10.1186/s12934-019-1074-4
Rottava, I., Toniazzo, G., Cortina, P. F., Martello, E., Grando, C. E., Lerin, L. A., et al. (2010). Screening of Microorganisms for Bioconversion of (−)-β-Pinene and (R)-(+)-limonene to α-terpineol. LWT - Food Sci. Tech. 43 (7), 1128–1131. doi:10.1016/j.lwt.2010.03.001
Sales, A., Afonso, L. F., Americo, J. A., de Freitas Rebelo, M., Pastore, G. M., and Bicas, J. L. (2018). Monoterpene Biotransformation by Colletotrichum Species. Biotechnol. Lett. 40 (3), 561–567. doi:10.1007/s10529-017-2503-2
Soares-Castro, P., Montenegro-Silva, P., Heipieper, H. J., and Santos, P. M. (2017). Functional Characterization of a 28-Kilobase Catabolic Island from Pseudomonas Sp. Strain M1 Involved in Biotransformation Of-Myrcene and Related Plant-Derived Volatiles. Appl. Environ. Microbiol. 83 (9), 4. doi:10.1128/AEM.03112-16
Sobral, M. V., Xavier, A. L., Lima, T. C., and de Sousa, D. P. (2014). Antitumor Activity of Monoterpenes Found in Essential Oils. Scientific World J. 2014, 1–35. doi:10.1155/2014/953451
Song, Y., Li, J., Shin, H.-d., Du, G., Liu, L., and Chen, J. (2015). One-step Biosynthesis of α-ketoisocaproate from L-Leucine by an Escherichia coli Whole-Cell Biocatalyst Expressing an L-Amino Acid Deaminase from Proteus Vulgaris. Sci. Rep. 5 (1), 12614. doi:10.1038/srep12614
Sun, C., Dong, X., Zhang, R., and Xie, C. (2021). Effectiveness of Recombinant Escherichia coli on the Production of (R)-(+)-perillyl Alcohol. BMC Biotechnol. 21 (1), 3. doi:10.1186/s12896-020-00662-7
Wu, J., Du, G., Zhou, J., and Chen, J. (2013). Metabolic Engineering of Escherichia coli for (2S)-Pinocembrin Production from Glucose by a Modular Metabolic Strategy. Metab. Eng. 16, 48–55. doi:10.1016/j.ymben.2012.11.009
Yang, J., Zhang, T., Tian, C., Zhu, Y., Zeng, Y., Men, Y., et al. (2019). Multi-enzyme Systems and Recombinant Cells for Synthesis of Valuable Saccharides: Advances and Perspectives. Biotechnol. Adv. 37 (7), 107406. doi:10.1016/j.biotechadv.2019.06.005
Yeruva, L., Pierre, K. J., Elegbede, A., Wang, R. C., and Carper, S. W. (2007). Perillyl Alcohol and Perillic Acid Induced Cell Cycle Arrest and Apoptosis in Non Small Cell Lung Cancer Cells. Cancer Lett. 257 (2), 216–226. doi:10.1016/j.canlet.2007.07.020
Zhang, B., Bian, L., Huang, P., Zhao, L., Chen, Y., and Wu, X. (2021). Efficient Whole-Cell Biosynthesis of L-Gulose by Coupling Mannitol-1-Dehydrogenase with NADH Oxidase. Enzyme Microb. Tech. 148, 109815. doi:10.1016/j.enzmictec.2021.109815
Zhu, K., Kong, J., Zhao, B., Rong, L., Liu, S., Lu, Z., et al. (2021). Metabolic Engineering of Microbes for Monoterpenoid Production. Biotechnol. Adv. 53, 107837. doi:10.1016/j.biotechadv.2021.107837
Keywords: whole cell catalysis, (R)-(+)-perillyl alcohol, NADH regeneration, alkL, escherichia coli
Citation: Sun C, Zhang R and Xie C (2022) Efficient Synthesis of (R)-(+)-Perillyl Alcohol From (R)-(+)-Limonene Using Engineered Escherichia coli Whole Cell Biocatalyst. Front. Bioeng. Biotechnol. 10:900800. doi: 10.3389/fbioe.2022.900800
Received: 21 March 2022; Accepted: 04 April 2022;
Published: 25 April 2022.
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaCopyright © 2022 Sun, Zhang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rubing Zhang, emhhbmdyYkBxaWJlYnQuYWMuY24=; Congxia Xie, eGllY29uZ3hpYUAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.