
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Bioeng. Biotechnol. , 08 February 2022
Sec. Synthetic Biology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.844405
This article is part of the Research Topic Fiat Lux! Light-Driven and Light-Controlled Synthetic Biological Parts, Devices, Systems and Processes View all 9 articles
Light switchable two-component protein dimerization systems offer versatile manipulation and dissection of cellular events in living systems. Over the past 20 years, the field has been driven by the discovery of photoreceptor-based interaction systems, the engineering of light-actuatable binder proteins, and the development of photoactivatable compounds as dimerization inducers. This perspective is to categorize mechanisms and design approaches of these dimerization systems, compare their advantages and limitations, and bridge them to emerging applications. Our goal is to identify new opportunities in combinatorial protein design that can address current engineering challenges and expand in vivo applications.
The control of molecular proximity by protein-protein interactions (PPIs) is a fundamental mechanism in biology. It is widely employed in signal transduction, regulation of gene expression, metabolism, and other processes. The ability to noninvasively control PPIs in vivo with sufficient sensitivity and specificity would have numerous biomedical applications, for example, the site- and time-specific activation of therapeutic immune cells in solid tumors. In chemogenetic and optogenetic approaches, PPIs are controlled by chemicals [e.g., chemically-induced dimerization (CID) (Spencer et al., 1993; Stanton et al., 2018)] and light [e.g., light-induced dimerization (Shimizu-Sato et al., 2002; Klewer and Wu, 2019)], respectively. Among those, photoswitchable PPI systems are superior in spatiotemporal resolution and are not limited by toxicity and delivery issues associated with chemical inducers. Compared with single-component actuators such as microbial opsins (Zhang et al., 2011), the PPI systems provide the flexibility to regulate diverse biological processes at various subcellular locations (Stanton et al., 2018; Klewer and Wu, 2019).
Photoswitchable PPI systems function via different mechanisms with specific applications. A typical example is photoswitchable heterodimerization of a photoreceptor and a dimerization protein, which are genetically fused to proteins of interest to control the proximity. Some commonly used systems include phytochrome (Shimizu-Sato et al., 2002; Chernov et al., 2017), cryptochrome (Kennedy et al., 2010; Losi et al., 2018), and light-oxygen-voltage (LOV)-sensing-based dimerization systems (Strickland et al., 2012; Guntas et al., 2015; Kawano et al., 2015; Pudasaini et al., 2015). For specific applications, critical parameters to consider are activation wavelength, light sensitivity, the affinity, specificity, kinetics, and reversibility of dimerization, and expression compatibility in host cells. Due to the limited understanding of phototransduction in these proteins, it has been challenging to rationally design them for particular needs. Another important example is photoswitchable protein binders, hereafter named opto-binders, which can target unmodified endogenous proteins. A first opto-binder was a split green fluorescent protein (GFP) antibody comprising two fragments genetically tagged with a photoswitchable heterodimerization system (Yu et al., 2019). Like custom antibodies, opto-binders with defined binding properties are needed for a growing list of endogenous targets and for different cell types. However, there lacks an efficient, generalizable approach for creating such binders against arbitrary targets. The last but not the least example is photocaged drug-controlled CID which is independent of a photoreceptor. This mechanism is exemplified by an emerging class of protein degradation drugs, known as photoswitchable proteolysis targeting chimeras (photoPROTACs) (Pfaff et al., 2019). Presumably, photo-uncaging of drugs can be used to simultaneously control their pharmacologic effects and the CID-regulated other outcomes. However, demonstrating the full potential of this actuation mechanism relies on the ability to create CID systems for a variety of drugs of interest.
The complex physicochemical properties and conformational dynamics of photoswitchable PPI systems impose challenges on the de novo design, but some successes were recently achieved by screening combinatorial protein libraries (Jang and Woolley, 2021). This perspective aims to build upon the initial success to identify new opportunities to break existing barriers in the protein design. It is worth noting that PPI systems have different degrees of complexity (e.g., homo- or hetero-assemblies with two or more protein components). Here, we focus on heterodimerization systems which in our opinion are the simplest but most convenient actuator device for engineering and applications. We will discuss the three major photoswitchable dimerization mechanisms, current challenges in their design, and then propose combinatorial approaches to address them.
A typical heterodimerization system comprises a photoreceptor protein or domain that undergoes a conformational switch between a light and a dark form and a dimerization binder recognizing the conformational change (Figure 1A). For actuation applications, photoreceptors and dimerization binders are fused to proteins of interest to exert the proximity control (Levskaya et al., 2009; Kennedy et al., 2010). Their applications have benefited from the availability of naturally occurring photoreceptors activated by a broad range of wavelengths from ultraviolet to near-infrared (NIR). These photoreceptors have been thoroughly discussed in the most recent reviews (Farahani et al., 2021; Manoilov et al., 2021; Seong and Lin, 2021) and are not the focus of this perspective. A broad selection of optical inputs enables advanced biological actuation such as controlling multiple cellular events with different wavelengths (Redchuk et al., 2017; Ochoa-Fernandez et al., 2020) and activating gene expression in deep tissues with NIR light (Kaberniuk et al., 2016). However, natural photoreceptors and dimerization binders often have extra domains irrelevant to photosensory and dimerization functions but with other important roles in their original hosts. In an actuator, they can cause unwanted interference to cell signaling and difficulties for mammalian expression or gene delivery due to the large gene sizes surpassing the packaging capacity of adeno-associated virus (Wu et al., 2010). Sometimes simply truncating them can disrupt the stability and function of the core photosensory and dimerization domains due to possible interdomain interactions (Bellini and Papiz, 2012). Additionally, the dimerization sometimes lacks adequate affinity or specificity, resulting in a low light activation sensitivity or a high dark activity. To overcome inherent limitations of the natural proteins, it is necessary to de novo engineer components of the systems such as dimerization binders.
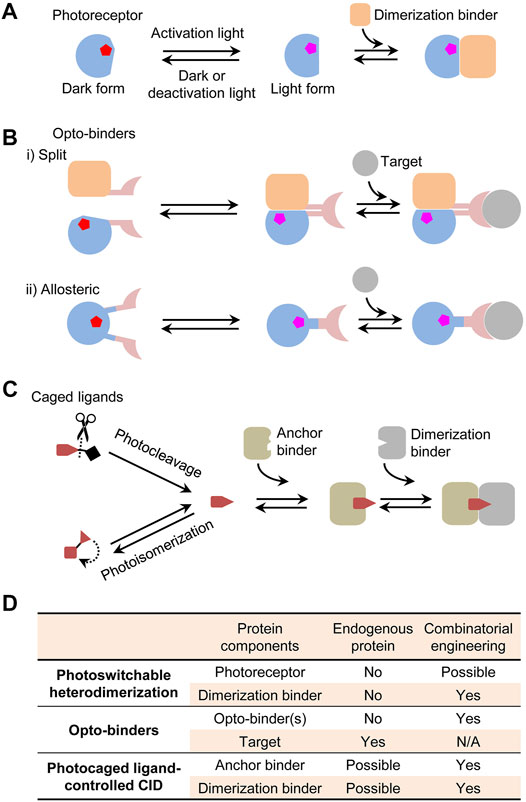
FIGURE 1. Photoswitchable protein dimerization mechanisms. (A) Photoswitchable heterodimerization. Pentagons indicate chromophores in different conformations (red or magenta). Although a dimerization binder might specifically recognize the light or dark form, only the binding to the light form is shown. (B) Opto-binders. Split and allosteric opto-binders function via the protein-fragment complementation and allosteric mechanisms, respectively. (C) Photocaged ligand-controlled CID. The uncaged ligand first binds to an anchor binder and then the anchor binder-ligand complex, not the free anchor binder, is recognized by a dimerization binder. (D) Comparison of protein components in the dimerization systems regarding whether they can be an endogenous protein and whether they can be engineered by a combinatorial approach. N/A, not applicable.
Some applications require controlling unmodified endogenous proteins rather than those genetically tagged with photoreceptors or dimerization binders. This became possible by using opto-binders derived from a binder of an endogenous target. Two types of opto-binders were demonstrated based on protein-fragment complementation and allosteric mechanisms (Figure 1B). The first type is similar to a protein-fragment complementation assay (Pelletier et al., 1998) in which, upon light activation, two fragments of a split binder tagged with a photoswitchable dimerization system are brought in close proximity to form a functional unit (Yu et al., 2019). An opto-binder can also be designed by inserting a photoreceptor domain into an allosteric site of a binder [e.g., nanobody (Gil et al., 2020; He et al., 2021), monobody (Carrasco-López et al., 2020; He et al., 2021), and affibody (Woloschuk et al., 2021)] to regulate the binding activity. By comparison, single-component allosteric opto-binders are easier to use than split opto-binders because it is more difficult to control expression levels and binding behaviors of two-component than single-component systems in cells. However, the design of the allostery in opto-binders had limited success and typically relied on a slow and costly trial-and-error process to select photoreceptors, binders, insertion sites, and linkers to create a chimeric fusion.
Some dimerization systems do not require a photoreceptor but instead use a photocaged ligand to control a CID system. Photo-uncaging of the ligand induces the dimerization of two proteins termed as anchor and dimerization binders (Figure 1C). This mechanism was first demonstrated with an engineered CID system induced by a photocleavage analogue of rapamycin (Karginov et al., 2011) and reviewed in the previous literature (Ankenbruck et al., 2018). For in vivo regulation, CID systems offer better flexibility than the photoreceptor-dependent dimerization because unmodified endogenous proteins can possibly serve as an anchor binder [e.g., ligand-induced G protein-coupled receptor–nanobody complex (Kruse et al., 2013)] or both an anchor and a dimerization binders [e.g., PROTAC-induced E3 ligase–target complexes (Sakamoto et al., 2001)] (Figure 1D). There could be a wide choice of photocaged CID inducers derived from natural or synthetic drugs, metabolites, or other molecules by introducing a photocleavable [e.g., coumarin and 2-nitrobenzyl derivatives (Klán et al., 2013)] or photoisomerizable moiety [e.g., azobenzenes (Beharry and Woolley, 2011)]. Compared with other dimerization mechanisms, a distinct advantage of these systems is the dual-control by chemical and light where a drug inducer can potentially exert both the pharmacological and CID-regulated effects. Although progress was made on designing ligands for some classes of proteins [e.g., photoPROTACs (Liu et al., 2020; Reynders et al., 2020)], a generalizable strategy is needed for creating CIDs for any arbitrary drug.
A strategy to design photoswitchable dimerization is to use protein binders to sense light-induced conformational change(s). However, photoreceptors exhibit function-associated structural dynamics (Genick et al., 1998; Takala et al., 2014), imposing a major challenge for the structure-based binder design. Some conformational states of these proteins are difficult to stabilize to produce immunogens for animal immunization. To date, the de novo engineering of binders to an existing photoreceptor mainly relies on in vitro screening of combinatorial protein libraries. In this approach, a critical step is to design and synthesize a high-quality combinatorial library (Figure 2A). Like natural antibody repertoires, a synthetic combinatorial library is designed from a shared protein scaffold, e.g., an immunoglobulin (Maynard and Georgiou, 2000; Muyldermans, 2013), non-immunoglobulin (Yu et al., 2017; Gebauer and Skerra, 2020), or de novo designed scaffold. The scaffold is introduced with rationally randomized binding site(s), for example, variable flexible loops like antibody complementarity-determining regions or residues on a relatively rigid binding surface. To obtain a high percentage of protein sequences in the library that might be correctly translated and stably folded, the randomization is usually guided by amino acid distributions found in experimentally validated natural [e.g., antibody repertoires (Moutel et al., 2016)] or engineered binder libraries. A trinucleotide phosphoramidite chemistry (Virnekäs et al., 1994) is ideally suited to control codon ratios at randomization sites. The combinatorial gene synthesis can generate a vastly diverse library (>109) with clones that can virtually bind to any given epitope. Such large libraries are suitable for protein display-based in vitro screening, e.g., phage (Smith, 1985), ribosome (Hanes and Plückthun, 1997), and mRNA display (Roberts and Szostak, 1997). An important advantage of the in vitro screening is that photosensitive reagents such as photoreceptors are easier to stabilize in light-conditioned assays. Based on our previous experience, an effective screening of a high-quality library can identify suitable conformation-selective binders even without further mutagenesis.
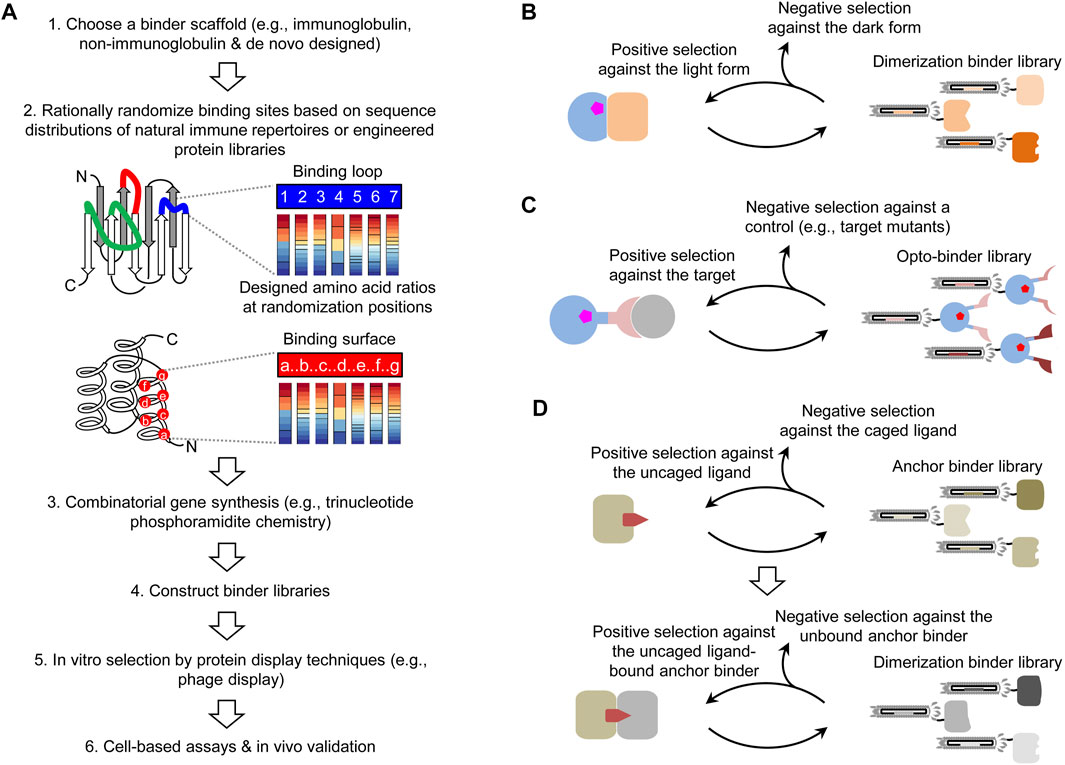
FIGURE 2. Combinatorial approaches for creating photoswitchable dimerization systems. (A) Pipeline of the design and screening of combinatorial binder libraries. (B) Combinatorial approach for obtaining dimerization binders in a photoswitchable heterodimerization system. (C) Selection of a combinatorial optobinder library constructed with a binder scaffold inserted with a photoreceptor for the allosteric regulation. The opto-binder library is applicable to arbitrary targets. (D) Generalizable, two-step approach for creating CID systems for an uncaged ligand.
The first application of combinatorial binder libraries is to select dimerization binders against the light or dark form of a photoreceptor to create heterodimerization systems. This approach can circumvent unavoidable constraints of natural dimerization binders and provide the flexibility to select the protein size and binding properties. Typically, dimerization binders are obtained by screening a combinatorial library against the light or dark form of a photoreceptor as the positive selection and the other form as the negative selection (Figure 2B). Candidates with various binding parameters are chosen or further engineered for specific applications. It might be possible to design combinatorial photoreceptor libraries (e.g., surface mutants), but it is necessary to assess how these mutations will impact the photoconversion, for example, by a deep mutational scanning analysis (Fowler and Fields, 2014).
Initial successes were demonstrated by screening libraries of small size proteins against a few photosensory domains. Combinatorial libraries of affibodies [∼6 kDa (kDa)] were screened by mRNA and ribosome display against a dark-form LOV2 domain of Avena Sativa phototropin 1 (AsLOV2) (Wang et al., 2016) and a light-form photosensory module of a Deinococcus radiodurans bacterial phytochrome (DrBphP) (Kuwasaki et al., 2021) to obtain the light-induced dissociation and dimerization, respectively. Other dimerization systems were obtained by phage display screening of a surface mutant library of an albumin-binding domain (∼5 kDa) against circularly permuted photoactive yellow protein (PYP) (Reis et al., 2018) and a cGMP phosphodiesterase-adenylate cyclase-FhlA (GAF) domain of Acryochloris marina cyanobacteriochrome (CBCR) (Jang et al., 2019). We recently demonstrated an efficient method, COMBINES-LID, by screening a nanobody library to create red light-switchable actuators (Huang et al., 2020). In this work, a widely used library of nanobodies (∼12–15 kDa, a diversity of 1.23–7.14 × 109) with a humanized scaffold was screened by phage display against the DrBphP photosensory module. Column-based biopanning was designed for the selection of both the conformational specificity and dimerization reversibility. The library screening was demonstrated to be highly efficient; in vivo actuators constructed with selected dimerization binder clones showed drastically improved light-induction specificity and a significantly lower dark activity than a similar natural Rhodopseudomonas palustris BphP1 (RpBphP1) system and its derivative. Although the protein display screening is efficient, it is necessary to validate the expression of in vitro selected binders by cell-based assays [e.g., yeast (Fields and Song, 1989) or mammalian-two hybrid (Lievens et al., 2009)]. Based on these results, we propose that this approach should be applicable to other photoreceptors and combinatorial libraries for designing actuators with different structural, optical, and biochemical properties.
Beyond standard binder libraries, it is possible to insert a photosensory domain into a binder scaffold to design a combinatorial opto-binder library. Photoreceptor-regulated allostery was engineered to control enzyme activities (Reynolds et al., 2011; Richter et al., 2016) and recently demonstrated in creating opto-binders. For opto-binders derived from nanobodies (Gil et al., 2020; He et al., 2021) and monobodies (Carrasco-López et al., 2020; He et al., 2021), a truncated AsLOV2 domain was inserted into specific loops away from their target binding sites, so the light-triggered unfolding of the Jɑ helix in AsLOV2 allosterically regulates the binding site conformation. In the affibody-based opto-binders (Woloschuk et al., 2021), the PYP was inserted into a loop connecting two helices important to the affibody stabilization and target binding, resulting in mutually exclusive folding of the affibody and PYP. Thus, the light-induced partial unfolding of PYP drives the affibody binding to its target. This approach of converting a known binder to opto-binders for the same target is error prone because the photoreceptor insertion tends to change or disrupt the binding to the target even without the allosteric regulation (Mathony and Niopek, 2021). Thus, the current opto-binder design from a known binder is relatively inefficient. Alternatively, we propose to directly screen a combinatorial opto-binder library against a target. Given that photoreceptor insertion and target binding sites are different in successful opto-binders, it is possible to use them as scaffolds and implement a similar binding site randomization protocol to generate combinatorial opto-binder libraries. If it is successful, this new approach will greatly facilitate the generation of opto-binders targeting arbitrary endogenous proteins, interactions, and modifications.
The screening of opto-binder libraries can be performed with established methods such as COMBINES-LID (Huang et al., 2020). For example, to select opto-binders targeting a specific epitope in a target, an activation light-illuminated phage library will pass through a negative and a positive selection columns preloaded with an epitope mutant and the wildtype target, respectively (Figure 2C). Binders captured in the positive selection column will be eluted by deactivation light illumination or dark relaxation to select reversible opto-binders. Because it is unknown whether opto-binders can be functionally displayed by protein display techniques, it is necessary to test the efficiency of protein folding and chromophore incorporation for different opto-binder scaffolds.
Besides the photoreceptor-dependent dimerization, the combinatorial screening is applicable to engineering CID. CID systems have different designs for specific applications, but considering wide interest in optopharmacology, we focus on those whose chemical inducers are photo-uncaged drugs targeting endogenous proteins. Here, one or two binder components in these systems are selected from combinatorial libraries. Compared with other CIDs, this type of system provides three switches: 1) the photo-uncaging of drugs, 2) the binding of drugs to endogenous targets, and 3) the drug-induced dimerization. Although such CID systems are yet to be reported, based on available methods for the CID engineering (Kang et al., 2019) and chemical synthesis (Klán et al., 2013), it is possible to design them for many intriguing applications. For example, to spatiotemporally regulate therapeutic T cells, a drug can be light activated at a tumor site to mediate multiple cell signaling events: e.g., targeting endogenous proteins such as a programmed death ligand-1 (PD-L1) to inhibit the checkpoint signaling of tumor-dwelling T cells (Zak et al., 2016) and dimerizing T-cell receptor signaling proteins to activate the T cells (Wu et al., 2015).
To create such systems, a critical need is to design CID systems for any given drug. Recent advances were made by selecting dimerization binders from phage-displayed libraries of antigen-binding fragments (Fabs) (Hill et al., 2018) and nano-CLostridial antibody mimetic proteins (nanoCLAMPs) (Guo et al., 2021) against an existing drug–target complex. However, this method is limited to structure-defined drug–target complexes and cannot be applied to other drugs with unknown or unstable drug–target complexes. To address this limitation, we recently developed a two-step screening method (COMBINES-CID) to sequentially select anchor and dimerization binders from a combinatorial nanobody library (Figure 2D) (Kang et al., 2019). As a proof-of-concept, we chose a challenging drug inducer, cannabidiol (CBD) with low molecular weight and high hydrophobicity, and obtained CIDs with high ligand specificity. We have successfully applied this method to other combinatorial libraries and drug inducers (unpublished). To obtain CIDs only induced by photo-uncaged not caged drugs and without drug-independent auto-dimerization, the dimerization binder screening requires the negative selection using unbound anchor binders and caged drugs as controls. Presumably, the proposed approach can be integrated with a variety of photo-caged drugs to achieve the flexible, multimodal actuation.
The proposed combinatorial approaches will leverage ongoing efforts on designing photoswitchable dimerization systems for in vivo actuation needs. They can potentially generate functional data for large-scale protein sequences which are useful for developing deep learning methods (Cong et al., 2019; Humphreys et al., 2021) to improve computational PPI design. Beyond traditional screening methods, other technologies enabling the “library-by-library” screening [e.g., SMI-seq (Gu et al., 2014)] can facilitate the engineering of complex dimerization surfaces. Besides using photoreceptors to create actuators, other sensory proteins that convert chemical or physical signals to conformational changes [e.g., covalent modifications (Hoppmann et al., 2014), electric field (Sasaki et al., 2006), heat (Caterina et al., 1997), and mechanical force (Liedtke et al., 2003)] can also be used as targets for dimerization binder selection to create switchable dimerization systems. Overall, the methodology innovation and the sensory protein discovery will significantly expand the biosensor toolkit and revolutionize in vivo applications.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
LG, XZ, SK, and YP conceptualized the manuscript. LG and XZ wrote the manuscript with input from other authors.
This work was supported by the grants from the National Institutes of Health (R35GM128918, R21DA051194, and R21DA051555 to LG).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ankenbruck, N., Courtney, T., Naro, Y., and Deiters, A. (2018). Optochemical Control of Biological Processes in Cells and Animals. Angew. Chem. Int. Ed. 57, 2768–2798. doi:10.1002/anie.201700171
Beharry, A. A., and Woolley, G. A. (2011). Azobenzene Photoswitches for Biomolecules. Chem. Soc. Rev. 40, 4422–4437. doi:10.1039/c1cs15023e
Bellini, D., and Papiz, M. Z. (2012). Structure of a Bacteriophytochrome and Light-Stimulated Protomer Swapping with a Gene Repressor. Structure 20, 1436–1446. doi:10.1016/j.str.2012.06.002
Carrasco-López, C., Zhao, E. M., Gil, A. A., Alam, N., Toettcher, J. E., and Avalos, J. L. (2020). Development of Light-Responsive Protein Binding in the Monobody Non-immunoglobulin Scaffold. Nat. Commun. 11, 4045. doi:10.1038/s41467-020-17837-7
Caterina, M. J., Schumacher, M. A., Tominaga, M., Rosen, T. A., Levine, J. D., and Julius, D. (1997). The Capsaicin Receptor: a Heat-Activated Ion Channel in the Pain Pathway. Nature 389, 816–824. doi:10.1038/39807
Chernov, K. G., Redchuk, T. A., Omelina, E. S., and Verkhusha, V. V. (2017). Near-Infrared Fluorescent Proteins, Biosensors, and Optogenetic Tools Engineered from Phytochromes. Chem. Rev. 117, 6423–6446. doi:10.1021/acs.chemrev.6b00700
Cong, Q., Anishchenko, I., Ovchinnikov, S., and Baker, D. (2019). Protein Interaction Networks Revealed by Proteome Coevolution. Science 365, 185–189. doi:10.1126/science.aaw6718
Farahani, P. E., Reed, E. H., Underhill, E. J., Aoki, K., and Toettcher, J. E. (2021). Signaling, Deconstructed: Using Optogenetics to Dissect and Direct Information Flow in Biological Systems. Annu. Rev. Biomed. Eng. 23, 61–87. doi:10.1146/annurev-bioeng-083120-111648
Fields, S., and Song, O.-K. (1989). A Novel Genetic System to Detect Protein-Protein Interactions. Nature 340, 245–246. doi:10.1038/340245a0
Fowler, D. M., and Fields, S. (2014). Deep Mutational Scanning: a New Style of Protein Science. Nat. Methods 11, 801–807. doi:10.1038/nmeth.3027
Gebauer, M., and Skerra, A. (2020). Engineered Protein Scaffolds as Next-Generation Therapeutics. Annu. Rev. Pharmacol. Toxicol. 60, 391–415. doi:10.1146/annurev-pharmtox-010818-021118
Genick, U. K., Soltis, S. M., Kuhn, P., Canestrelli, I. L., and Getzoff, E. D. (1998). Structure at 0.85 Å Resolution of an Early Protein Photocycle Intermediate. Nature 392, 206–209. doi:10.1038/32462
Gil, A. A., Carrasco-López, C., Zhu, L., Zhao, E. M., Ravindran, P. T., Wilson, M. Z., et al. (2020). Optogenetic Control of Protein Binding Using Light-Switchable Nanobodies. Nat. Commun. 11, 4044. doi:10.1038/s41467-020-17836-8
Gu, L., Li, C., Aach, J., Hill, D. E., Vidal, M., and Church, G. M. (2014). Multiplex Single-Molecule Interaction Profiling of DNA-Barcoded Proteins. Nature 515, 554–557. doi:10.1038/nature13761
Guntas, G., Hallett, R. A., Zimmerman, S. P., Williams, T., Yumerefendi, H., Bear, J. E., et al. (2015). Engineering an Improved Light-Induced Dimer (iLID) for Controlling the Localization and Activity of Signaling Proteins. Proc. Natl. Acad. Sci. USA 112, 112–117. doi:10.1073/pnas.1417910112
Guo, Z., Smutok, O., Johnston, W. A., Walden, P., Ungerer, J. P. J., Peat, T. S., et al. (2021). Design of a Methotrexate-Controlled Chemical Dimerization System and its Use in Bio-Electronic Devices. Nat. Commun. 12, 7137. doi:10.1038/s41467-021-27184-w
Hanes, J., and Pluckthun, A. (1997). In Vitro selection and Evolution of Functional Proteins by Using Ribosome Display. Proc. Natl. Acad. Sci. 94, 4937–4942. doi:10.1073/pnas.94.10.4937
He, L., Tan, P., Huang, Y., and Zhou, Y. (2021). Design of Smart Antibody Mimetics with Photosensitive Switches. Adv. Biol. (Weinh) 5, e2000541. doi:10.1002/adbi.202000541
Hill, Z. B., Martinko, A. J., Nguyen, D. P., and Wells, J. A. (2018). Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications. Nat. Chem. Biol. 14, 112–117. doi:10.1038/nchembio.2529
Hoppmann, C., Lacey, V. K., Louie, G. V., Wei, J., Noel, J. P., and Wang, L. (2014). Genetically Encoding Photoswitchable Click Amino Acids inEscherichia Coliand Mammalian Cells. Angew. Chem. 126, 4013–4017. doi:10.1002/ange.201400001
Huang, Z., Li, Z., Zhang, X., Kang, S., Dong, R., Sun, L., et al. (2020). Creating Red Light-Switchable Protein Dimerization Systems as Genetically Encoded Actuators with High Specificity. ACS Synth. Biol. 9, 3322–3333. doi:10.1021/acssynbio.0c00397
Humphreys, I. R., Pei, J., Baek, M., Krishnakumar, A., Anishchenko, I., Ovchinnikov, S., et al. (2021). Computed Structures of Core Eukaryotic Protein Complexes. Science 374, eabm4805. doi:10.1126/science.abm4805
Jang, J., McDonald, S., Uppalapati, M., and Woolley, G. A. (2019). Green, orange, Red, and Far-Red Optogenetic Tools Derived from Cyanobacteriochromes. bioRxiv, 769422. doi:10.1101/769422
Jang, J., and Woolley, G. A. (2021). Directed Evolution Approaches for Optogenetic Tool Development. Biochem. Soc. Trans. 49, 2737–2748. doi:10.1042/BST20210700
Kaberniuk, A. A., Shemetov, A. A., and Verkhusha, V. V. (2016). A Bacterial Phytochrome-Based Optogenetic System Controllable with Near-Infrared Light. Nat. Methods 13, 591–597. doi:10.1038/nmeth.3864
Kang, S., Davidsen, K., Gomez-Castillo, L., Jiang, H., Fu, X., Li, Z., et al. (2019). COMBINES-CID: An Efficient Method for De Novo Engineering of Highly Specific Chemically Induced Protein Dimerization Systems. J. Am. Chem. Soc. 141, 10948–10952. doi:10.1021/jacs.9b03522
Karginov, A. V., Zou, Y., Shirvanyants, D., Kota, P., Dokholyan, N. V., Young, D. D., et al. (2011). Light Regulation of Protein Dimerization and Kinase Activity in Living Cells Using Photocaged Rapamycin and Engineered FKBP. J. Am. Chem. Soc. 133, 420–423. doi:10.1021/ja109630v
Kawano, F., Suzuki, H., Furuya, A., and Sato, M. (2015). Engineered Pairs of Distinct Photoswitches for Optogenetic Control of Cellular Proteins. Nat. Commun. 6, 6256. doi:10.1038/ncomms7256
Kennedy, M. J., Hughes, R. M., Peteya, L. A., Schwartz, J. W., Ehlers, M. D., and Tucker, C. L. (2010). Rapid Blue-Light-Mediated Induction of Protein Interactions in Living Cells. Nat. Methods 7, 973–975. doi:10.1038/nmeth.1524
Klán, P., Šolomek, T., Bochet, C. G., Blanc, A., Givens, R., Rubina, M., et al. (2013). Photoremovable Protecting Groups in Chemistry and Biology: Reaction Mechanisms and Efficacy. Chem. Rev. 113, 119–191. doi:10.1021/cr300177k
Klewer, L., and Wu, Y. W. (2019). Light‐Induced Dimerization Approaches to Control Cellular Processes. Chem. Eur. J. 25, 12452–12463. doi:10.1002/chem.201900562
Kruse, A. C., Ring, A. M., Manglik, A., Hu, J., Hu, K., Eitel, K., et al. (2013). Activation and Allosteric Modulation of a Muscarinic Acetylcholine Receptor. Nature 504, 101–106. doi:10.1038/nature12735
Kuwasaki, Y., Suzuki, K., Yu, G., Kakihara, Y., Nishiwaki, M., Fushimi, K., et al. (2021). A Semi-synthetic Red Light Photoswitch for Optogenetic Control of Protein Activity. Research Square Platform LLC. doi:10.21203/rs.3.rs-450425/v1
Levskaya, A., Weiner, O. D., Lim, W. A., and Voigt, C. A. (2009). Spatiotemporal Control of Cell Signalling Using a Light-Switchable Protein Interaction. Nature 461, 997–1001. doi:10.1038/nature08446
Liedtke, W., Tobin, D. M., Bargmann, C. I., and Friedman, J. M. (2003). Mammalian TRPV4 (VR-OAC) Directs Behavioral Responses to Osmotic and Mechanical Stimuli in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 100 (Suppl. 2), 14531–14536. doi:10.1073/pnas.2235619100
Lievens, S., Lemmens, I., and Tavernier, J. (2009). Mammalian Two-Hybrids Come of Age. Trends Biochem. Sci. 34, 579–588. doi:10.1016/j.tibs.2009.06.009
Liu, J., Chen, H., Ma, L., He, Z., Wang, D., Liu, Y., et al. (2020). Light-induced Control of Protein Destruction by Opto-PROTAC. Sci. Adv. 6, eaay5154. doi:10.1126/sciadv.aay5154
Losi, A., Gardner, K. H., and Möglich, A. (2018). Blue-Light Receptors for Optogenetics. Chem. Rev. 118, 10659–10709. doi:10.1021/acs.chemrev.8b00163
Manoilov, K. Y., Verkhusha, V. V., and Shcherbakova, D. M. (2021). A Guide to the Optogenetic Regulation of Endogenous Molecules. Nat. Methods 18, 1027–1037. doi:10.1038/s41592-021-01240-1
Mathony, J., and Niopek, D. (2021). Enlightening Allostery: Designing Switchable Proteins by Photoreceptor Fusion. Adv. Biol. (Weinh) 5, e2000181. doi:10.1002/adbi.202000181
Maynard, J., and Georgiou, G. (2000). Antibody Engineering. Annu. Rev. Biomed. Eng. 2, 339–376. doi:10.1146/annurev.bioeng.2.1.339
Moutel, S., Bery, N., Bernard, V., Keller, L., Lemesre, E., de Marco, A., et al. (2016). NaLi-H1: A Universal Synthetic Library of Humanized Nanobodies Providing Highly Functional Antibodies and Intrabodies. Elife 5, e16228. doi:10.7554/eLife.16228
Muyldermans, S. (2013). Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 82, 775–797. doi:10.1146/annurev-biochem-063011-092449
Ochoa-Fernandez, R., Abel, N. B., Wieland, F.-G., Schlegel, J., Koch, L.-A., Miller, J. B., et al. (2020). Optogenetic Control of Gene Expression in Plants in the Presence of Ambient white Light. Nat. Methods 17, 717–725. doi:10.1038/s41592-020-0868-y
Pelletier, J. N., Campbell-Valois, F. X., and Michnick, S. W. (1998). Oligomerization Domain-Directed Reassembly of Active Dihydrofolate Reductase from Rationally Designed Fragments. Proc. Natl. Acad. Sci. 95, 12141–12146. doi:10.1073/pnas.95.21.12141
Pfaff, P., Samarasinghe, K. T. G., Crews, C. M., and Carreira, E. M. (2019). Reversible Spatiotemporal Control of Induced Protein Degradation by Bistable PhotoPROTACs. ACS Cent. Sci. 5, 1682–1690. doi:10.1021/acscentsci.9b00713
Pudasaini, A., El-Arab, K. K., and Zoltowski, B. D. (2015). LOV-based Optogenetic Devices: Light-Driven Modules to Impart Photoregulated Control of Cellular Signaling. Front. Mol. Biosci. 2, 18. doi:10.3389/fmolb.2015.00018
Redchuk, T. A., Omelina, E. S., Chernov, K. G., and Verkhusha, V. V. (2017). Near-infrared Optogenetic Pair for Protein Regulation and Spectral Multiplexing. Nat. Chem. Biol. 13, 633–639. doi:10.1038/nchembio.2343
Reis, J. M., Xu, X., McDonald, S., Woloschuk, R. M., Jaikaran, A. S. I., Vizeacoumar, F. S., et al. (2018). Discovering Selective Binders for Photoswitchable Proteins Using Phage Display. ACS Synth. Biol. 7, 2355–2364. doi:10.1021/acssynbio.8b00123
Reynders, M., Matsuura, B. S., Bérouti, M., Simoneschi, D., Marzio, A., Pagano, M., et al. (2020). PHOTACs Enable Optical Control of Protein Degradation. Sci. Adv. 6 (8), eaay5064. doi:10.1126/sciadv.aay5064
Reynolds, K. A., McLaughlin, R. N., and Ranganathan, R. (2011). Hot Spots for Allosteric Regulation on Protein Surfaces. Cell 147, 1564–1575. doi:10.1016/j.cell.2011.10.049
Richter, F., Fonfara, I., Bouazza, B., Schumacher, C. H., Bratovič, M., Charpentier, E., et al. (2016). Engineering of Temperature- and Light-Switchable Cas9 Variants. Nucleic Acids Res. 44, 10003–10014. doi:10.1093/nar/gkw930
Roberts, R. W., and Szostak, J. W. (1997). RNA-peptide Fusions for the In Vitro Selection of Peptides and Proteins. Proc. Natl. Acad. Sci. 94, 12297–12302. doi:10.1073/pnas.94.23.12297
Sakamoto, K. M., Kim, K. B., and Kumagai, A. (2001). Protacs: Chimeric Molecules that Target Proteins to the Skp1–Cullin–F Box Complex for Ubiquitination and Degradation. Proc. Natl. Acad. Sci. U S A. 98 (15), 8554–8559. Available at: https://www.pnas.org/content/98/15/8554.short. doi:10.1073/pnas.141230798
Sasaki, M., Takagi, M., and Okamura, Y. (2006). A Voltage Sensor-Domain Protein Is a Voltage-Gated Proton Channel. Science 312, 589–592. doi:10.1126/science.1122352
Seong, J., and Lin, M. Z. (2021). Optobiochemistry: Genetically Encoded Control of Protein Activity by Light. Annu. Rev. Biochem. 90, 475–501. doi:10.1146/annurev-biochem-072420-112431
Shimizu-Sato, S., Huq, E., Tepperman, J. M., and Quail, P. H. (2002). A Light-Switchable Gene Promoter System. Nat. Biotechnol. 20, 1041–1044. doi:10.1038/nbt734
Smith, G. P. (1985). Filamentous Fusion Phage: Novel Expression Vectors that Display Cloned Antigens on the Virion Surface. Science 228, 1315–1317. doi:10.1126/science.4001944
Spencer, D. M., Wandless, T. J., Schreiber, S. L., and Crabtree, G. R. (1993). Controlling Signal Transduction with Synthetic Ligands. Science 262, 1019–1024. doi:10.1126/science.7694365
Stanton, B. Z., Chory, E. J., and Crabtree, G. R. (2018). Chemically Induced Proximity in Biology and Medicine. Science 359, eaao5902. doi:10.1126/science.aao5902
Strickland, D., Lin, Y., Wagner, E., Hope, C. M., Zayner, J., Antoniou, C., et al. (2012). TULIPs: Tunable, Light-Controlled Interacting Protein Tags for Cell Biology. Nat. Methods 9, 379–384. doi:10.1038/nmeth.1904
Takala, H., Björling, A., Berntsson, O., Lehtivuori, H., Niebling, S., Hoernke, M., et al. (2014). Signal Amplification and Transduction in Phytochrome Photosensors. Nature 509, 245–248. doi:10.1038/nature13310
Virnekäs, B., Ge, L., Plückthun, A., Schneider, K. C., Wellnhofer, G., and Moroney, S. E. (1994). Trinucleotide Phosphoramidites: Ideal Reagents for the Synthesis of Mixed Oligonucleotides for Random Mutagenesis. Nucleic Acids Res. 22, 5600–5607. doi:10.1093/nar/22.25.5600
Wang, H., Vilela, M., Winkler, A., Tarnawski, M., Schlichting, I., Yumerefendi, H., et al. (2016). LOVTRAP: an Optogenetic System for Photoinduced Protein Dissociation. Nat. Methods 13, 755–758. doi:10.1038/nmeth.3926
Woloschuk, R. M., Reed, P. M. M., Jaikaran, A. S. I., Demmans, K. Z., Youn, J., Kanelis, V., et al. (2021). Structure‐based Design of a Photoswitchable Affibody Scaffold. Protein Sci. 30, 2359–2372. doi:10.1002/pro.4196
Wu, C.-Y., Roybal, K. T., Puchner, E. M., Onuffer, J., and Lim, W. A. (2015). Remote Control of Therapeutic T Cells through a Small Molecule-Gated Chimeric Receptor. Science 350, aab4077. doi:10.1126/science.aab4077
Wu, Z., Yang, H., and Colosi, P. (2010). Effect of Genome Size on AAV Vector Packaging. Mol. Ther. 18, 80–86. doi:10.1038/mt.2009.255
Yu, D., Lee, H., Hong, J., Jung, H., Jo, Y., Oh, B.-H., et al. (2019). Optogenetic Activation of Intracellular Antibodies for Direct Modulation of Endogenous Proteins. Nat. Methods 16, 1095–1100. doi:10.1038/s41592-019-0592-7
Yu, X., Yang, Y.-P., Dikici, E., Deo, S. K., and Daunert, S. (2017). Beyond Antibodies as Binding Partners: The Role of Antibody Mimetics in Bioanalysis. Annu. Rev. Anal. Chem. 10, 293–320. doi:10.1146/annurev-anchem-061516-045205
Zak, K. M., Grudnik, P., Guzik, K., Zieba, B. J., Musielak, B., Dömling, A., et al. (2016). Structural Basis for Small Molecule Targeting of the Programmed Death Ligand 1 (PD-L1). Oncotarget 7, 30323–30335. doi:10.18632/oncotarget.8730
Keywords: combinatorial protein library, light induced dimerization, chemically induced dimerization, photoreceptor, optogenetics, actuator, opto-binder, nanobody
Citation: Zhang X, Pan Y, Kang S and Gu L (2022) Combinatorial Approaches for Efficient Design of Photoswitchable Protein-Protein Interactions as In Vivo Actuators. Front. Bioeng. Biotechnol. 10:844405. doi: 10.3389/fbioe.2022.844405
Received: 28 December 2021; Accepted: 20 January 2022;
Published: 08 February 2022.
Edited by:
Pasquale Stano, University of Salento, ItalyReviewed by:
Brian D. Zoltowski, Southern Methodist University, United StatesCopyright © 2022 Zhang, Pan, Kang and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangcai Gu, Z3VsY0B1dy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.