- 1School of Life Sciences and Medicine, Shandong University of Technology, Zibo, China
- 2State Key Lab of Microbial Technology, Shandong University, Qingdao, China
- 3CAS Key Laboratory of Biobased Materials, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao, China
- 4School of Chemical Engineering, University of Chinese Academy of Sciences, Beijing, China
Succinic acid, a dicarboxylic acid produced as an intermediate of the tricarboxylic acid (TCA) cycle, is one of the most important platform chemicals for the production of various high value-added derivatives. As traditional chemical synthesis processes suffer from nonrenewable resources and environment pollution, succinic acid biosynthesis has drawn increasing attention as a viable, more environmentally friendly alternative. To date, several metabolic engineering approaches have been utilized for constructing and optimizing succinic acid cell factories. In this review, different succinic acid biosynthesis pathways are summarized, with a focus on the key enzymes and metabolic engineering approaches, which mainly include redirecting carbon flux, balancing NADH/NAD+ ratios, and optimizing CO2 supplementation. Finally, future perspectives on the microbial production of succinic acid are discussed.
1 Introduction
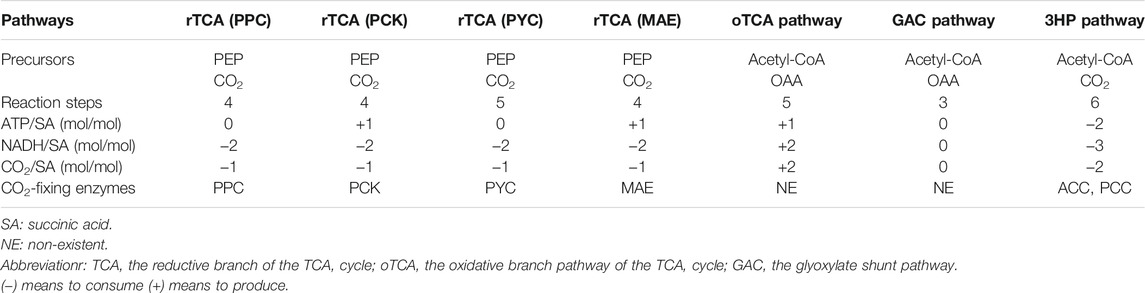
Succinic acid is a C4-dicarboxylic acid produced as a key intermediate of the tricarboxylic acid (TCA) cycle. As a widely investigated high-value chemical, it has numerous applications in the fields of agriculture, green solvents, pharmaceuticals, and biodegradable plastics. It can also be chemically converted into a variety of important industrial chemicals such as 1,4-butanediol, butadiene, and tetrahydrofuran. Succinic acid has been identified as one of the 12 value-added bio-based platform chemicals by the United States Department of Energy (DOE) (Werpy et al., 2004). The market potential of succinic acid and its direct derivatives is estimated to be as high as 245,000 tons per year, while the market size of succinic acid-based polymers is expected to be 25 million tons per year (Bozell and Petersen, 2010). In traditional chemical synthesis methods, maleic anhydride from petrochemical feedstocks serves as the key substrate for succinic acid, and Ni or Pd-based catalysts have been employed for the hydrogenation of maleic anhydride to succinic acid (Zhang et al., 2020). Although the conversion rate is high, many problems still exist, such as the complex operations required for synthesis, the high-energy consumption, and the harsh reaction conditions (Louasté and Eloutassi, 2020; Zhang et al., 2020). Therefore, much attention has focused on succinic acid biosynthesis, and to date, some efficient succinic acid bio-producers have been successfully applied for industrial purposes (Willke and Vorlop, 2004; Kequan et al., 2008). Compared with chemical processing, the raw materials of biosynthesis are more widely availableand lower in cost. Crude glycerol (Gao et al., 2016), food processing waste (Li et al., 2019), corn fiber (Chen et al., 2011), cassava root (Thuy et al., 2017), lignocellulose hydrolyzate (Zhang et al., 2020), maltose syrup, and other renewable biomass resources are potential sources of the raw materials for these reactions. Significantly, CO2 fixation and utilization processes are an integral part of succinic acid biosynthesis in almost all pathways (Table 1). Theoretically speaking, the biological production of 1 kg of succinic acid can fix at least 0.37 kg CO2, which presents an environmental benefit to the use of succinic acid biosynthesis upstream of the production of other chemicals (Wang et al., 2011).
Many anaerobic and facultative anaerobic microbes produce succinic acid as their fermentation end product. These succinic acid-producing strains can be divided into two categories, namely, the natural succinic acid producers (Table 2) and the metabolic engineering succinic acid producers (Table 3). The natural succinic acid producers mainly include Actinobacillus succinogenes (Thuy et al., 2017), Anaerobiospirillum succiniciproducens (Meynial-Salles et al., 2010), Mannheimia succiniciproducens (Lee et al., 2003), Basfia succiniciproducens (Choi et al., 2015) and Escherichia coli (Thakker et al., 2011). Among these strains, A. succinogenes and A. succiniciproducens were the first identified natural overproducers of succinic acid (Bretz and Kabasci, 2012). A. succinogenes, originally isolated from bovine rumen, is a Gram-negative, capnophilic, facultative anaerobic bacterium, and has been used as a succinic acid-producing chassis since 1981 by the Michigan Biotechnology Institute (MBI). A. succinogenes has a better tolerance of high concentrations of succinic acid than other strains (Wan et al., 2008) and can utilize extensive carbon sources, including glucose, xylose, glycerol, cellobiose, cheese whey (Wan et al., 2008), cane molasses (Liu et al., 2008), straw hydrolysate (Zheng et al., 2009), and crop stalk wastes. However, A. succinogenes also has some drawbacks as a succinic acid production platform, such as numerous auxotrophies and lack of useful genetic tools (Ahn et al., 2016). A. succiniciproducens is a Gram-negative and strictly anaerobic bacterium (Wang et al., 2011) that produces succinic acid and acetate as its major end products, while producing lactate and ethanol as minor products, depending on culture conditions (Lee et al., 1999). Compared with A. succinogenes, A. succiniciproducens shows only weak salt tolerance (Bretz and Kabasci, 2012), which partly hinders the wide application of this strain. M. succiniciproducens, also isolated from bovine rumen, is a facultatively anaerobic, mesophilic, non-motile, non-spore-forming, Gram-negative bacterium, and can utilize a wide variety of carbon sources, similar to A. succinogenes (Lee et al., 2002). This bacterium has an excellent CO2-fixing pathway and can produce succinic acid as a major end product through simple anaerobic fermentation. Nevertheless, this species also has some disadvantages, such as its pH sensitivity and auxotrophy for several amino acids and vitamins (Ahn et al., 2016). B. succiniciproducens, a new member of the family Pasteurellaceae, was isolated from the rumen of a German cow in 2008, and shows high similarity to M. succinciproducens (Ahn et al., 2016). In the 2,363 open reading frames (ORFs) of B. succiniciproducens DD1 and the 2,380 ORFs of M. succiniciproducens MBEL55 E, 2006 ORFs were found to be homologous (Kuhnert et al., 2010). This bacterium can export succinic acid as an end product and has been used as a succinic acid-producing chassis by the Succinity Company (a joint venture of BASF and Purac). Wild-type E. coli can also produce minor amounts of succinic acid under anaerobic conditions, whereas under aerobic conditions succinic acid is formed only as an intermediate of the TCA cycle unless the glyoxylate bypass is operating (Thakker et al., 2012). Table 2 shows a summary of bio-based succinic acid production by different wild-type strains with various fermentation substrates reported in selected papers. Among this list, A. succinogenes, B. succiniciproducens, and M. succiniciproducens are the most promising wild-type bacterial strains which have relatively high yield and productivity, but they still cannot satisfy industrial production requirements. Most wild-type strains show a tendency to degenerate (Lin et al., 2005b), pH sensitivity and auxotrophy, and they require a rich, complex medium for efficient growth. Thus, further optimization and improvement is needed before natural succinic acid producers can be used for industrial purposes.
Metabolic engineering strategies, however, is a promising way of generating succinic acid-producing strains, using not only natural succinic acid producers, but also unnatural producers, such as Corynebacterium glutamicum, Yarrowia lipolytica, Saccharomyces cerevisiae, and Pichia kudriavzevii (Table 3). Taking C. glutamicum as an example, the highest concentration of 152.2 g/L of succinic acid with a yield and productivity of 1.1 g/g glucose and 1.11 g/L/h, respectively, were achieved by the engineered strain S071 (Δldh, Δpta-ackA, ΔactA, ΔpoxB, pycP458, Δpck_Ptuf:Ms.pckG, Ptuf:ppc, ΔptsG)/pGEX4-NCgl0275 under anaerobic conditions. Compared with the initial strain, this engineered strain showed more than 30-fold increase in succinic acid production. (Chung et al., 2017). In the case of Y. lipolytica, the highest succinic acid concentration of 110.7 g/L was achieved, by the engineered strain Y. lipolytica PGC202 (ΔSdh5, ΔAch1, with pck from S. cerevisiae overexpression and SCS2 from Y. lipolytica overexpression) in fed-batch cultivation using glycerol without pH control. Compared with the initial strain, this engineered strain showed 4.3-fold increase and has the highest fermentative succinic acid titer achieved in yeast at low pH condition (Cui et al., 2017). For E. coli, metabolic engineering strategies could be used to improve its succinic acid biosynthesis ability. Wu et al. constructed an engineered strain E. coli K12 (Δpfl, ΔldhA, ΔptsG)/pMD19T-gadBC. This strain could produce 32.01 g/L succinic acid at pH 5.6, which was 2.6-fold higher than the initial strain (Wu et al., 2017). These metabolic engineering producers have shown high production efficiency, demonstrating the great potential of the production of industrially competitive bio-based succinic acid.
In this review, several different typical succinic acid biosynthetic pathways reported in recent years are summarized, and the main metabolic engineering strategies to improve the production efficiency are presented in detail. Additionally, the main challenges as well as the future perspectives for succinic acid biosynthesis are discussed.
2 Biosynthetic Pathway of Succinic Acid
To date, seven natural or artificial succinic acid biosynthesis pathways have been reported in the literature (Figure 1), including the reductive branch of the TCA cycle coupled to PEP carboxylase (PPC) pathway, the reductive branch of the TCA cycle coupled to PEP carboxykinase (PCK) pathway, the reductive branch of the TCA cycle coupled to pyruvate carboxylase (PYC) pathway, the reductive branch of the TCA cycle coupled to malic enzyme (MAE) pathway, the glyoxylate shunt pathway, the oxidative branch pathway of the TCA cycle, and the 3-hydroxypropionate cycle for the production of succinic acid. Each biosynthesis pathway has its own characteristics that determine the scope of their unique application (Table 1). The reductive branch pathways of the TCA cycle are the main succinic acid-producing approaches under anaerobic conditions (Zhu and Tang, 2017), while the other pathways can be effective under aerobic conditions. In the next subsections, these different pathways and their specific features are discussed.
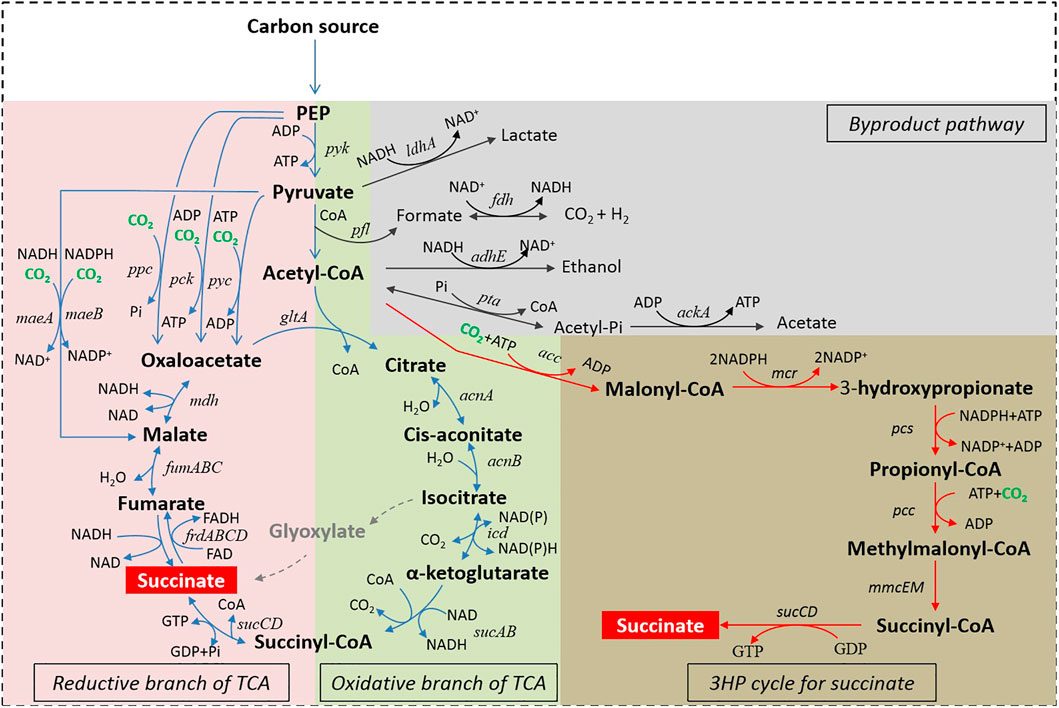
FIGURE 1. Succinic acid production biosynthetic pathways. Abbreviations: PEP, Phosphoenolpyruvic acid; ldhA, lactic dehydrogenase; pfl, pyruvate formate lyase; fdh, formate dehydrogenase; adhE, alcohol dehydrogenase; pta, phosphotransacetylase; ackA, acetate kinase; gltA, citrate synthetase; acnAB, aconitase; icd, isocitrate dehydrogenase; sucABCD, succinyl-CoA synthetase; frdABCD, succinate dehydrogenase; fumABC, fumarate hydratase; mdh, malate dehydrogenase; ppc, PEP carboxylase; pck, PEP carboxykinase; pyc, pyruvate carboxylase; pyk, pyruvate kinase; maeAB, malic enzyme; acc, acetyl-CoA carboxylase; mcr, malonyl-CoA reductase; pcc, propionyl-CoA carboxylase; pcs, propionyl-CoA synthase; mmcEM, methylmalonyl-CoA epimerase and mutase.
2.1 Anaerobic Succinic Acid-Producing Pathways
The reductive branch of the TCA cycle coupled to PEP carboxylase (PPC) for succinic acid formation (rTCA (PPC) pathway), which exists in almost all microbes, is one of the most commonly used processes in bio-based succinic acid production. In this pathway, PEP carboxylase (PPC) converts PEP and CO2 into oxaloacetate, which is then converted into malate catalyzed by malate dehydrogenase (MDH). Then, malate is transformed into fumarate by fumarate hydratase (FumABC). Finally, fumarate and NADH are turned into succinic acid catalyzed by fumarate reductase (FrdABCD). Through this pathway, succinic acid is synthesized from PEP and CO2 at a stoichiometric ratio of 1:1. Among the components in this process, PPC (EC 4.1.1.31) plays a critical role in CO2 fixation. Taking E. coli as an example, PPC is produced during growth on glycolytic substrates (Keseler et al., 2011). PPC catalyzes the carboxylation of PEP with bicarbonate to form oxaloacetate and inorganic phosphate using Mg2+ as a cofactor (Millard et al., 1996; Kai et al., 1999). The main function of PPC is to replenish the oxaloacetate consumed by biosynthetic reactions, and under anaerobic fermentation conditions, it can direct a portion of PEP to succinic acid (Millard et al., 1996). Studies have shown that under a CO2 atmosphere, E. coli Δppc strains grow slower and more poorly than wild-type strains in glucose-based medium (40% reduction in biomass and more than 70% reduction in succinic acid production) (Millard et al., 1996; Ahn et al., 2020). PPC also exhibits high catalytic velocity (the enzymatic activity is 250 μmol/min/mg) and high substrate affinity (the Km value for HCO3- is 0.1 mM and the Km value for PEP is 0.19 mM), which is conducive to succinic acid biosynthesis (Millard et al., 1996; Kai et al., 1999; Tan et al., 2013). Millard et al. evaluated the effect of PPC on the amount of succinic acid produced in E. coli under control of the tac promoter. The engineered strain that overexpressed PPC showed a 3.5-fold increase in the concentration of succinic acid, from 3.0 g/L in a control culture to 10.7 g/L, and the yield showed a 3.75-fold increase, from 0.12 mol/mol glucose to 0.45 mol/mol glucose (Table 3) (Millard et al., 1996). There seemed to be a positive correlation between PPC activity and succinic acid production, but excessive PPC activity was detrimental to cell growth and succinic acid formation (Tan et al., 2013). Tan et al. found higher PPC activity along with higher succinic acid production, when the PPC activity was equal to or less than 0.47 U/mg protein (Tan et al., 2013). The reason may have been that the overexpression of PPC caused a shortage of PEP. PEP is also essential for the PEP-PTS (phosphoenolpyruvate-phosphotransferase) system, which has both a transport function and an extensive regulatory function. And the shortage of PEP will lead to a compromise in glucose uptake and cell growth (Gokarn et al., 2000). Therefore, the regulation of PPC expression was significant when using the PPC dependent biosynthesis pathway for succinic acid production.
The reductive branch of the TCA cycle coupled to PEP carboxykinase (PCK) for succinic acid formation (rTCA (PCK) pathway), is another representative succinic acid biosynthesis route that mainly exists in bacteria and fungi, especially in some natural succinic acid producers (Werf et al., 1997; Lee et al., 2006). This process is almost the same as the PPC-dependent pathway, except for the key CO2-fixation enzyme. In this pathway, the key CO2-fixation enzyme is PCK (EC 4.1.1.31) which can catalyze the carboxylation of PEP with bicarbonate and ADP to form oxaloacetate and ATP. Compared with PPC, the PCK catalytic process is accompanied by the production of energy, and as such it is more suitable for cell growth and succinic acid production (Zhu and Tang, 2017). However, the overexpression of PCK in E. coli at low CO2 concentrations in fact has no effect on succinic acid biosynthesis. PCK can be valuable for succinic acid production but only in the presence of high HCO3- concentrations (Millard et al., 1996; Kwon et al., 2006). There are two possible explanations for this result: 1) PCK has low HCO3- affinity (the Km value for HCO3- is 13 mM, 130 times larger than that of PPC, the Km value for PEP is 0.07 mM and the Km value for ADP is 0.05 mM); 2) PCK has low catalytic velocity (the enzymatic activity is 28 μmol/min/mg, which is just 11.2% of the PPC activity) (Krebs and Bridger, 1980; Zhang et al., 2009a; Tan et al., 2013). Previous studies have demonstrated that when 20 g/L NaHCO3 was added to a culture, the succinic acid titer in recombinant E. coli overexpressing PCK was 2.2-fold higher than a wild-type strain, from 6.4 g/L in a control culture to 14.2 g/L (Kwon et al., 2006). Kim et al. found that in wild-type E. coli K12 cells, PCK overexpression had no effect on succinic acid fermentation, but the overexpression of PCK in an engineered E. coli K12 Δppc strain was 6.5-fold higher than E. coli K12 Δppc, and rose from 3.1 g/L to 20.2 g/L (Table 3) (Millard et al., 1996). In addition, the PCK activity was closely related to succinic acid biosynthesis. Tan et al. reported that the succinic acid titer and yield were positively correlated with the PCK activity. A 16-fold increase in PCK activity resulted in a 57-fold increase in succinic acid titer (from 4 to 226 mM) in E. coli Suc-T108 (Tan et al., 2013). In some natural succinic acid producers, such as A. succiniciproducens (Millard et al., 1996), M. succiniciproducens (Hong et al., 2004; Lee et al., 2006), and A. succinogenes (Millard et al., 1996; Werf et al., 1997), PCK is mainly employed in CO2 fixation and cell growth instead of PPC. Taking M. succiniciproducens as an example, genes encoding PPC, PCK, or MAE were separately disrupted, and fermentation results showed that strains with PPC or MAE deletion did not show much effect on succinic acid productivity or cell growth, while the strain with PCK inactivation showed severe growth retardation and a remarkable decrease in succinic acid productivity (Lee et al., 2006). This result was significant because although PCK has a high Km value for HCO3-, it still plays a major role in succinic acid biosynthesis in M. succiniciproducens. Compared with M. succiniciproducens, the indispensable feature of PCK was not observed in wild-type E. coli. It might have been due to the differences in the internal concentration of HCO3- in these two organisms (Millard et al., 1996). These results also suggests that the optimization of a CO2 delivery system and the amelioration of the intracellular inorganic carbon concentration may be a good choice for improving the PCK depended succinic acid biosynthesis route (Xiao et al., 2017). Except for PCK, MDH was reported to be another rate-limiting enzyme for succinic acid production (Hong et al., 2004; Ahn et al., 2020). In M. succiniciproducens, MDH reduces oxaloacetate to malate using NADH as a cofactor, but it shows low specific activity and strong uncompetitive inhibition towards oxaloacetate (Ahn et al., 2020). In silico simulation suggested that MDH is instrumental in succinic acid production (Ahn et al., 2020). Jung et al. reported biochemical and structural analyses of various MDH variants and discovered a key amino acid residue (Gly-11) required for high kinetic efficiency in MDH. After screening, a valuable MDH mutant (MDH-G11Q) was obtained, which had high enzyme activity (2.9-fold increase in activity) and less substrate inhibition (6.4-fold increase in ki) compared with a wild type MDH. Furthermore, the application of this MDH-G11Q mutant in engineering bacteria led to a 1.4-fold increase in succinic acid production, indicating the significance of enzyme optimization in strain development (Choi et al., 2016; Ahn et al., 2020).
The reductive branch of the TCA cycle coupled to pyruvate carboxylase (PYC) for succinic acid formation (rTCA (PYC) pathway), is widespread in fungi, plants, as well as some bacteria (Modak and Kelly, 1995), but it is absent in Enterobacteriaceae (Gokarn et al., 2000). In this pathway, PEP is first converted into pyruvate by pyruvate kinase (PYK). Then PYC (EC 6.4.1.1) catalyzes the ATP-dependent carboxylation of pyruvate to produce oxaloacetate. Oxaloacetate is further converted into succinic acid through MDH, FumABC and FrdABCD. As the key CO2-fixation step, the reaction catalyzed by PYC also plays an anaplerotic role in the provision of oxaloacetate for maintaining the dynamic balance of the TCA cycle. In R. etli, the anaplerotic role of PYC has been shown to be essential for its normal growth (Jitrapakdee et al., 2008), and thus PYC can be considered to be one of the most significant regulatory enzymes in this pathway (Modak and Kelly, 1995). In most PYC catalytic reactions, acetyl-CoA acts as a positive allosteric activator (Jitrapakdee et al., 2008; Tong, 2013), except for PYC from L. lactis, which is regulated by the bacterial second messenger c-di-AMP (Choi et al., 2017). The PYC depended route is a promising tool for succinic acid biosynthesis. Yan et al. reconstructed this pathway in S. cerevisiae, and found that the engineered strain showed a 19.3-fold increase in succinic acid production in shaken flasks, rising from 0.32 g/L to 6.17 g/L (Yan et al., 2014). Chen et al. reported that the overexpression of PYC from R. oryzae in a S. cerevisiaeΔsdh2 strain resulted in a 1.2-fold increase in the concentration of succinic acid, from 0.709 g/L to 0.841 g/L (Table 3) (Chen et al., 2019). In addition, the functions of PYC, PPC and MAE on the carboxylation processes for high succinic acid productivity in E. coli were analyzed in silico. According to analysis results, carboxylation reactions catalyzed by PYC were the most suitable ones for obtain high productivity in E. coli. Furthermore, based on these analysis results, PYC from C. glutamicum ATCC13032 was overexpressed in E. coli ZJG13 and the final succinic acid production was 36.1 g/L, with a specific productivity and yield of 2.75 mmol g CDW−1 h−1 and 0.72 mol mol−1 glucose, respectively (Table 3) (Yang et al., 2014). Sánchez et al. overexpressed the L. lactis pyc gene in E. coli MG1655 (ΔadhE, ΔldhA) and found that a 25-fold increasein succinic acid production, from 0.6 g/L to 15.6 g/L, as well as a 6-fold increase in the yield, from 0.13 g/g glucose to 0.78 g/g glucose (Table 3) (Sánchez et al., 2005).
The reductive branch of the TCA cycle coupled to malic enzyme (MAE) for succinic acid formation (rTCA (MAE) pathway), is shorter than the PYC dependent pathway. In the MAE dependent pathway, pyruvate and CO2 are directly converted into malate by malic enzyme, and malate is further converted into succinic acid through FumABC and FrdABCD. There are two forms of malic enzymes: MaeA (EC 1.1.1.38) and MaeB (EC 1.1.1.40), which can use NADH and NADPH as electron donors, respectively. Stols et al. overexpressed MAE from A. suum in E. coli NZN111 to produce succinic acid, resulting in a 2.9-fold increase, from 2.45 g/L to 7.07 g/L (Table 3) (Stols et al., 1997). Kwon et al. cloned MaeA and MaeB, derived from E. coli W3110, and overexpressed them in E. coli W3110 under anaerobic conditions. They found that the enzyme activities of overexpressed MaeA and MaeB were 2.34 and 1.69 μmol min−1mgprotein−1, whereas their activities were only 0.82 and 0.01 μmol min−1mgprotein−1 in the wild-type W3110 strain, respectively (Kwon et al., 2007). Furthermore, the succinic acid yields were measured in different strains. MaeB overexpression resulted in a 2.4-fold increase in succinic acid production, to 15.43 mmol/L, compared with 6.41 mmol/L in wild-type E. coli W3110. However, MaeA overexpression resulted in just 6.69 mmol/L succinic acid, which was similar to a wild-type strain under the same anaerobic fermentation conditions (Kwon et al., 2007). In this study, there was a negative correlation between succinic acid production and the activities of MaeA and MaeB. The main reason for this result might have been related to the differences in the intracellular co-substrate availability of MaeA and MaeB. The intracellular concentration of NADPH was 146 μmol/L, which was 9.12-fold higher than the 16 μmol/L of NADH intracellular concentration (Bochner and Ames, 1982; Kwon et al., 2007). Correspondingly, NADPH-dependent MaeB showed an advantage over the NADH-dependent MaeA, which would encourage CO2 fixation by MaeB over by MaeA in wild-type E. coli (Kwon et al., 2007). In addition, the reaction catalyzed by MAE is reversible and this enzyme prefers to form pyruvate, which is unfavorable for succinic acid formation (Hong and Lee, 2002). The Km values of MAE for malate and pyruvate in E. coli are 0.4 and 16 mM, respectively. Similarly, the Km values of MAE in C. glutamicumare 3.8 mM for malate and 13.8 mM for pyruvate, respectively (Stols and Donnelly, 1997; Gourdon et al., 2000). This indicates that sufficiently high intracellular pyruvate concentrations may have a significant influence to overcome the unfavorable equilibrium of MAE catalytic processes. However, the wild-type strain is unable to accumulate a sufficiently high pyruvate concentration to allow MAE to be effective for producing excessive malate (Stols et al., 1997). Therefore, the enhancement of the PYK catalytic process, which can convert PEP and ADP into pyruvate and ATP, maybe a good choice for intracellular pyruvate enrichment. In some natural succinic acid producers, this reaction is also closely related to normal metabolic activity. Taking M. succiniciproducens as an example, Lee found that PYK operates by providing the pyruvate and ATP required for cell growth, as its pyruvate concentration increases with cell growth and remains constant after cell growth has stopped (Lee et al., 2006).
2.2 Aerobic Succinic Acid-Producing Pathways
Although high productivities and yields of succinic acid can be achieved through anaerobic biosynthesis pathways, some drawbacks still exist with using these pathways, such as their slow carbon throughput, limitation of NADH availability (2 mol of NADH are required for 1 mol succinic acid formation) and poor cell growth (Yang et al., 2014). One solution is to produce succinic acid under aerobic conditions, which can generate higher biomass and extensive energy with O2 as the electron acceptor (Lin et al., 2005a; 2005b; Yang et al., 2014). The oxidative branch pathway of the TCA cycle (oTCA pathway) is one of the candidate aerobic biosynthesis routes for succinic acid production. In this pathway, acetyl-CoA and oxaloacetate are converted into succinic acid through citrate synthetase (GltA), aconitase (AcnAB), isocitrate dehydrogenase (ICD), and Succinyl-CoA synthetase (SucABCD), accompanied by the synthesis of NADH at a stoichiometric ratio of 2:1. Although succinic acid can be generated by oTCA under aerobic conditions, most of it will be further converted in the TCA cycle or to biomass and cannot be excreted from the cells. Thus, the metabolic engineering studies are necessary to improve its yield. Taking E. coli as an example, succinic acid is a minor fermentation product under anaerobic conditions. Under aerobic conditions, however, succinic acid is only an intermediate of the TCA cycle and cannot be detected in the extracellular medium (Clark, 1989; Lin et al., 2005b). Correspondingly, acetate is the main byproduct under the same aerobic conditions (Lin et al., 2005b). In order to produce succinic acid as a major product aerobically, Lin et al. created a mutant E. coli HL27659k (pKK313) strain that overexpressed of S. vulgare pepc and had five enzymes inactivated: ΔsdhAB, Δ(ackA-pta), ΔpoxB, ΔiclR, andΔptsG. This mutant strain could use both the oxidative branch pathway of the TCA cycle and the glyoxylate shunt pathway to produce succinic acid under aerobic conditions, and its maximum theoretical succinic acid yield could reach 1.0 mol/mol glucose consumed (Lin et al., 2005c). Fed-batch fermentation results showed that this strain could produce 58.3 g/L of succinic acid under complete aerobic conditions with a yield of 0.94 mol/mol glucose consumed (Table 3) (Lin et al., 2005b; 2005d). Arikawa et al. constructed a Saccharomyces cerevisiae mutant with fumarate reductase (FRDS) and succinate dehydrogenase (SDH1) inactivated. However, under aerobic conditions, this mutant strain could synthesize succinic acid through the oxidative branch pathway of the TCA cycle and the succinic acid yield showed a 2.7-fold increase compared with the parental strain (Arikawa et al., 1998). S. cerevisiae exhibits a high tolerance to low pH values, which makes it superior for succinic acid production (Qiang et al., 2010). These results were significant and Lin et al. inferred the potential of the oTCA pathway for aerobic succinic acid biosynthesis (Lin et al., 2005b).
The glyoxylate shunt pathway (GAC pathway) is another aerobic succinic acid biosynthesis route. In this process, 2 mol of acetyl-CoA and 1 mol of oxalacetate are converted into 1 mol of malate and 1 mol of succinic acid by citrate synthetase (GltA), aconitase (AcnAB) and isocitrate lyase (AceAB). Compared with the oxidative branch pathway of the TCA cycle, this process bypasses the two oxidative steps where CO2 is released, and thus it is considered to be an atom-economic aerobic pathway (Lin et al., 2004). Additionally, malate is produced as the main byproduct in the glyoxylate shunt pathway. Malate can be catalyzed by malate dehydrogenase (MDH) to generate NADH and regeneration of oxaloacetate. Under anaerobic conditions, 1 mol of malate can also be further transformed into 1 mol of succinic acid by FumABC and FrdABCD by consuming 1 mol of NADH. Compared with the traditional rTCA pathway (formation of 1 mol of succinic acid requires 2 mol of NADH), the glyoxylate shunt pathway does not contain NADH consumption process. Therefore, its application will be beneficial for solving the limitation of NADH availability during succinic acid biosynthesis (Sánchez et al., 2005). Vemuri et al. found that in the absence of an additional electron acceptor under dual-phase conditions (an aerobic growth phase followed by an anaerobic production phase), the maximum theoretical succinic acid yield was up to 1.714 mol/mol glucose, when 28.6% of the carbon flows to the glyoxylate shunt pathway and 71.4% of the carbon flows to the reductive branch pathway of the TCA cycle. It is worth noting that the maximal yield of succinic acid cannot be achieved without an active glyoxylate shunt pathway (Vemuri et al., 2002). It should be noticed that C2 substrates like acetate or fatty acids are very prominent activators for the glyoxylate shunt pathway under aerobic conditions (Gui et al., 1996). The aceBAK operon, which encodes enzymes of the glyoxylate cycle, is controlled by IclR. Under aerobic conditions, acetate can induce aceBAK and reduce the repressor activity of IclR, resulting in activation of the glyoxylate cycle (Gui et al., 1996; Sánchez et al., 2005). Meanwhile, studies have also revealed that the disruption of iclR could dramatically induce the expression of the aceBAK operon even when growing on glucose (Gui et al., 1996; Lin et al., 2004; Hendrik et al., 2011). Thus, the inactivation of iclR is significant for activating the glyoxylate shunt pathway for succinic acid production (Sánchez et al., 2005). Lin et al. constructed an engineered E. coli HL27615k strain with mutations in the TCA cycle (ΔiclR, Δicd, and ΔsdhAB) and acetate pathways (ΔpoxB, and ΔackA-pta) for maximal aerobic succinic acid production through the glyoxylate shunt pathway. The results of aerobic batch fermentation showed that the succinic acid production reached 43 mM with a yield of 0.7, which was close to the maximum theoretical yield of 1 mol/mol glucose (Lin et al., 2004). Except for the absence of the iclR mutation, Zhu et al. demonstrated that the glyoxylate shunt pathway could also be activated by deleting other relevant genes in E. coli under anaerobic conditions. The key genes included ackA (a gene encoding an acetate kinase) and pta (a gene encoding a phosphotransacetylase). Metabolic flux analysis reflected that the carbon flow shunted to the glyoxylate pathway from oxalacetate in wild-type E. coli DY329 was 0 and 31% in the mutant YJ003 (ΔackA, Δpta, ΔldhA, and ΔpstG). In addition, the succinic acid production also showed a 6-fold increase, from 25.13 mM in DY329 to 150.78 mM in YJ003, implying the importance of the activation of the glyoxylate shunt pathway (Zhu et al., 2014).
The 3-hydroxypropionate cycle is one of the natural aerobic CO2-fixation pathways, and mainly exists in photosynthetic green nonsulfur bacteria (Herter et al., 2001). This cycle is complex, containing 16 enzymatic reaction steps that are catalyzed by 13 enzymes (Gong et al., 2016). Liu et al. introduced part of this cycle into recombinant E. coli for succinic acid production (3HP pathway) (Liu et al., 2020). In this process, acetyl-CoA is carboxylated by acetyl-CoA carboxylase (ACC) to generate malony-CoA. Malony-CoA is converted into propionyl-CoA by malonyl-CoA reductase (MCR) and propionyl-CoA synthase (PCS). Then propionyl-CoA is carboxylated by propionyl-CoA carboxylase (PCC) to generate (S)-methylmalonyl-CoA. Next, methylmalonyl-CoA epimerase (MmcE) and methylmalonyl-CoA mutase (MmcM) convert (S)-methylmalonyl-CoA into its isomer succinyl-CoA. Finally, succinyl-CoA is deesterificated by succinyl-CoA synthetase (SucCD) to generate succinic acid (Menendez et al., 1999; Liu et al., 2020). The conversion of 1 mol of acetyl-CoA into 1 mol of succinic acid via the 3-hydroxypropionate cycle can fix 2 mol of CO2, which was confirmed by isotope labeling experiments with NaH13CO3 (Liu et al., 2020). Compared with succinic acid production based on carboxylation of PEP or pyruvate, this route showed a higher CO2 fixation efficiency. Among these pathway, ACC (EC 6.4.1.2) and PCC (EC 6.4.1.3) are the key CO2-fixing and rate-limiting enzymes. Both ACC and PCC belong to a subgroup of biotin-dependent short-chain acyl-CoA carboxylases that contain biotin carboxyl carrier protein domain (BCCP), biotin carboxylase domain (BC), and carboxyltransferase domain (CT), and utilize a covalently bound biotin as a cofactor (Wongkittichote et al., 2017; Liu and Jiang, 2021). The CT domain determines the specificity of the substrate, and these proteins can even work with other BCCP and BC domains from different enzymes (Lombard and Moreira, 2012; Tong, 2013). To obtain PCC with high enzymatic activity, Liu et al. tested different CT subunits of the PCC homologs from diverse bacterial species and developed a direct evolution strategy to further optimize this protein. A highly active PCC mutant (PccBBS-N220I/I391T) was obtained, which showed a 94-fold increase in overall catalytic efficiency indicated by kcat/Km compared with a wild-type PCC. In addition, the application of this PCC mutant resulted in a 1.5-fold increase in succinic acid production compared to the engineered strain with wild-type PCC, which indicated the significance of this rate-limiting enzyme engineering strategy for microbial production (Liu et al., 2020). ACC acts as another CO2-fixing enzyme and ACC is essential for most living organism’s growth (Baba et al., 2006). The intracellular concentration of its carboxylation product (malonyl-CoA) is tightly regulated to be very low, leading to limited production of ACC-derived compounds (Zha et al., 2009; Liu et al., 2010). It has been reported that ACC is negatively regulated by AMP-activated serine/threonine protein kinase (Snf1) in S. cerevisiae when glucose is depleted. Moreover, the phosphorylation triggered by Snf1 at one or more serine residues results in the deactivation of ACC (Woods et al., 1994; Scott et al., 2002). Jin et al. identified a critical amino acid (Ser-1157) responsible for deactivation via phosphorylation and mutated it to an alanine. The in vitro activity results showed that this ACC mutant (ACC-S1157A) resulted in 9-fold higher specific activity relative to wild-type ACC (Choi and Silva, 2014). Shi et al. demonstrated that introduction of this S659A mutation in ACC-S1157A could also lead to an enhanced enzyme activity with 1.72-fold increase (Shi et al., 2014). Further utilization of these modified acetyl-CoA carboxylase will be beneficial for product biosynthesis built from malonyl-CoA, such as succinic acid (Choi and Silva, 2014). Except for ACC and PCC, malonyl-CoA reductase (MCR), a characteristic enzyme of the 3-hydroxypropionate cycle, is regarded as another rate-limiting enzyme (Fixation et al., 2002). MCR catalyzes a two-step NADPH-dependent reduction of malonyl-CoA to 3-hydroxypropionate, and malonate semialdehyde has been suggested to be the likely free intermediate (Fixation et al., 2002; Liu et al., 2013). Liu et al. demonstrated that MCR, a natural fusion protein of two short-chain dehydrogenase/reductases, has higher enzyme activity when dissected into two functionally individual fragments, including an MCR-C fragment (amino acids 550–1,219) and MCR-N fragment (amino acids 1–549) (Liu et al., 2013). Further studies indicated that the initial reduction of malonyl-CoA to malonate semialdehyde catalyzed by MCR-C was the rate-limiting step of MCR (Liu et al., 2013; Liu et al., 2016). Liu et al. developed a direct evolution strategy to further optimize MCR-C. A triple MCR-C mutant (N940V/K1106W/S1114R) was obtained, which showed a 5.54-fold increase in enzyme activity. Combined with fine tuning of the MCR-N expression levels and culture condition optimization, the 3-hydroxypropionate yield showed a 270-fold increase, which could relieve the pressure of being the rate-limiting step in succinic acid biosynthesis (Liu et al., 2016). Although some progress has been achieved in this succinic acid pathway, some problems are still worthy of attention. The main challenge is that significant amounts of ATP and reducing power (3ATP and 3NADPH) are consumed from one molecule acetyl-CoA to one molecule succinic acid. Using fatty acids as carbon sources or feeding electricity to bacteria to produce intracellular energy may be optional promising approaches (Chen et al., 2018; Liu et al., 2019).
3 Metabolic Engineering for Enhancing Succinic Acid Biosynthesis
Up to now, the bio-based succinic acid synthetic process has become increasingly mature. There are several strategies to enhance succinic acid biosynthesis, which mainly include redirecting carbon flux, balancing the redox ratio (NADH/NAD+) and optimizing CO2 supplementation. And the different strategies will be discussed in more details in the later chapters.
3.1 Redirecting Carbon Flux
Whether under anaerobic or aerobic conditions, succinic acid is not the main product of many microbes. In general, microbes preferentially accumulate substances such as acetate, lactate, and ethanol etc. These by-product pathways compete with the succinic acid pathway for ATP or reducing power. Therefore, it is necessary to redirect the carbon flux from these competitive pathways into succinic acid production (Li et al., 2020). The redirection methods often used mainly include blocking these competitive pathways and regulating succinic acid biosynthesis pathways.
3.1.1 Blocking Competitive Pathways
As the main competing by-products, acetate, lactate, and ethanol are the targets of metabolic regulation, and deletion of the genes related to their production is usually performed. Specifically, single deletion of ldhA (encoding lactate dehydrogenase), which can convert pyruvate and NADH into lactate and NAD+ in E. coli, led to a 1.6-fold increase in succinic acid yield under anaerobic conditions, from 0.13 to 0.21 mol/mol glucose. Meanwhile, the lactate concentration of this ΔldhA mutant was 92% lower than a wild-type strain, dropping from 214 to 17 mM (Zhang et al., 2009b). Single deletion of adhE (encoding alcohol dehydrogenase), which converts acetyl-CoA and NADH into ethanol and NAD+, resulted in a 1.125-fold increase in succinic acid yield. Correspondingly, the ethanol concentration of this ΔadhE mutant was 0, compared with 133 mM in a wild-type strain (Zhang et al., 2009b). Zhang et al. constructed an E. coli ΔldhAΔadhE double mutant strain, which showed a 1.77-fold increase in succinic acid yield, and lactate and ethanol were not detected in its fermentation broth (Zhang et al., 2009b). There are two major acetate-producing pathways in E. coli, respectively through the enzyme pyruvate oxidase (encoded by poxB) and acetate kinase/phosphotransacetylase (encoded by ackA-pta). Pyruvate oxidase catalyzes the decarboxylation of pyruvate to acetate and CO2, and is more active in the stationary stage of cell growth (Mey et al., 2007; Dittrich, 2010). Ahmed et al. demonstrated that PoxB was useful for overall metabolism functioning and single deletion of poxB led to a decrease of 24% in biomass. Thus, they suggested that employing poxB served as a method to decrease acetate production (Abdel-Hamid et al., 2001). The ackA-pta pathway, which is more active in the exponential phases of cell growth, involves two consecutive reactions. PTA converts acetyl-CoA and Pi into acetyl phosphate and CoA. AckA then converts acetyl phosphate and ADP into acetate and ATP (Zhu et al., 2014). It was demonstrated that pta mutants grow more slowly than wild-type strains. The specific rate of growth of E. coli MG1655 Δpta was 9.3% lower than the parent strain under aerobic conditions and 37.2% lower under anaerobic conditions (Schütze et al., 2020). This may have been due to pyruvate accumulation, acetyl-P accumulation or a redox imbalance in pta mutants (Wolfe, 2005). Wolfe reported that pta mutants or ackA-pta mutants did not accumulate extracellular acetate, and ackA mutants could accumulate small amounts of acetate (Wolfe, 2005). Compared to a particular mutation, the combination of mutations was proved more useful for enhancing succinic acid production in many cases. Zhang et al. constructed a ΔldhAΔadhEΔackA triple mutant, which showed a 3.2-fold increase in succinic acid concentration and a 5.4-fold increase in yield (Zhang et al., 2009b). Lu et al. constructed a E. coli JH208 (ΔldhA, ΔpflB, ΔadhE, ΔpoxB, ΔackA, ΔcscR) mutant strain, and found that this mutant could produce 48.46 g/L succinic acid in 46 h with the productivity of 1.01 g/L/h and the yield of 0.83 g/g sucrose. Compared with the initial strain, the production efficiency of succinic acid has been improved 72% (Lu et al., 2015). Olajuyin et al. constructed an E. coli K-12 (ΔldhA, ΔpflB, ΔpoxB, Δpta-ackA) mutant for succinic acid production, the concentration and molar yield of succinic acid were respectively 22.40 ± 0.12 g/L and 1.13 ± 0.02 mol/mol total sugar after 72 h dual phase fermentation, and the final concentration of succinic acid was 6.2-fold higher than the wild-type strain (Olajuyin et al., 2016). These results indicated the necessity of blocking competitive pathways for improving succinic acid yield.
3.1.2 Regulating Succinic Acid Pathways
Direct overexpression of important enzymes (mentioned in Section 2) is the most frequently used method for enhancing the production of the succinic acid pathways. However, biosynthetic processes are usually regulated by multiple genes, and positive expected results are difficult to obtain by simple genetic engineering. In recent years, investigation of the global regulation of gene expression has been shown to be effective for solving the limitations of classical metabolic engineering approaches (Chong et al., 2014; Zhu and Tang, 2017). Some global regulators are good targets for improving the succinic acid biosynthesis process, such as ArcA, Crp, Cya and Cra. Among them, the Arc (anoxic respiration control) system composed of ArcA and ArcB regulates gene expression in response to redox conditions (Liu and Wulf, 2004; Sun et al., 2020). As reported, ArcA, when phosphorylated, represses the expressions of the genes involved in the TCA cycle and the glyoxylate shunt genes (Shimizu, 2013). Knocking out of ArcA did not affect the growth rate and the glucose uptake rate, but increased the TCA cycle activity by over 60% under aerobic conditions. It is significant for the oTCA pathway and the glyoxylate shunt pathway (Perrenoud and Sauer, 2005). Crp (cyclic AMP receptor protein) is the catabolite repressor that is activated by adenylate cyclase (Cya)-synthesized cAMP (Zhang et al., 2014). Crp/cAMP is a positive transcriptional regulator for mdh, ptsG, pckA, sdhABCD, acnAB, gltA (Perrenoud and Sauer, 2005; Wilfredo et al., 2021). Research showed that knocking out of Crp or Cya would decrease the growth rate, the glucose consumption rate and reduce the metabolic flux from PEP to oxaloacetate (Perrenoud and Sauer, 2005). Catabolite repressor/activator (Cra), also known as FruR, is an important global transcription factor, and it plays a key role in balancing the levels of the genes involved in carbon metabolism in E. coli (Akira, 2000; Ishihama, 2010; Shimada et al., 2011). Cra can activate genes related to the tricarboxylic acid cycle, the glyoxylate shunt pathway (Cozzone and El-Mansi, 2005; Dayanidhi et al., 2008; Shimada et al., 2011), and negatively affects genes related with the ED and glycolytic pathways (Ramseier, 1996; Sarkar et al., 2008; Ramseier et al., 2010). In a study from Zhu et al., Cra was first engineered for achieving high product concentration through error-prone PCR. After screening and mutation site integration, a high succinic acid producing mutant strain (Cra-R57K/A58G/G59Q/R60Q/S75H/T76Y/D148I/R149I), was obtained. This strain (E. coli W1485 (ΔpflAB, ΔldhA, ΔptsG)/pTrc-mutant cra) produced succinic acid to a concentration of 79.8 g/L, which was 22.8% higher than a control strain (E. coli W1485 (ΔpflAB, ΔldhA, ΔptsG)/pTrc) (Zhu et al., 2016). Further studies demonstrated that the significant increase of succinic acid in a Cra mutant may be caused by the activation of PEP carboxylation, reductive branches of the TCA cycle and the glyoxylate pathway (Zhu et al., 2016). Meanwhile, Li et al. first reported a positive correlation between succinic acid production and the affinity of Cra for its effector fructose-1,6-bisphosphate (FBP) in E. coli (Wei et al., 2016). To heighten this affinity, a semi-rational strategy based on computer-assisted virtual saturation mutagenesis was designed, and a Cra mutant (D101R/D148R/G274R) was obtained. Compared with wild-type Cra (the Kd of Cra-FBP was 1,360 ± 6.2 nM), the triple mutant Cra showed a high affinity with its effector fructose-1,6-bisphosphate (FBP) (the Kd of Cra-FBP was 154.2 ± 4.3 nM). Further experimental results indicated that this enhanced affinity increased the activation of succinic acid biosynthetic pathways, especially pck and aceB, and led to a succinic acid concentration of 92.7 g/L in engineered strain (E. coli W1485 (ΔpflAB, ΔldhA, ΔptsG)/pTrc99A-cra D101R/D148R/G274R), which was 34% higher than a control strain (E. coli W1485 (ΔpflAB, ΔldhA, ΔptsG)/pTrc99A) (Wei et al., 2016). The advantage of using global transcription factors is that they can regulate the expression of multiple pathway-related genes (Wu et al., 2007; Wei et al., 2016). In conclusion, these results demonstrate the great potentials of global transcription factors combined with the traditional metabolic engineering strategies in succinic acid production process, and this strategy can also be used as a reference for other valuable chemical biosynthetic pathways.
3.2 Balancing the Redox Ratio (NADH/NAD+)
The intracellular redox ratio is closely related to metabolite profiles, membrane transport, microbial fitness and cellular functions (Berríos-Rivera et al., 2004; Singh et al., 2009). The redox ratio is mainly reflected by the intracellular NADH/NAD+ ratio, which is influenced by several factors, such as the physiological state of a given strain, the oxidation state of carbon sources, and the expression levels of NADH/NAD+-related enzymes (such as formate dehydrogenase, membrane-bound transhydrogenase, NAD+ dehydrogenase, terminal oxidases and lactic dehydrogenase) (Lin et al., 2005a; Singh et al., 2009). In microbes, both NADH and NAD+ are kept in balance. NADH is oxidized to NAD+ through the oxidative phosphorylation process or fermentation reactions. NADH provides the reducing power for reductive product formation and NAD+ serves as an electron acceptor during substrate degradation (Li et al., 2017). In addition, the appropriate NADH/NAD+ ratio is a major prerequisite of many biosynthetic processes, and it is also crucial for succinic acid production (Zhang Y. et al., 2009; Li et al., 2017).
Taking E. coli as an example, lactate dehydrogenase (encoded by ldhA), which drives the formation of lactate, and pyruvate formate lyase (encoded by pflB), which drives the formation of formate and acetyl-CoA derivants (acetate, ethanol), were deleted to redirect the carbon flux towards succinic acid under anaerobic conditions (Singh et al., 2010). This mutant was termed E. coli NZN111, and it has been considered an excellent candidate as a succinic acid producer (Hui et al., 2009). However, E. coli NZN111 lost its ability to ferment glucose anaerobically and accumulates high levels of pyruvate and NADH intracellularly (Wu et al., 2007; Ma et al., 2013). It has been reported that E. coli NZN111 growth was impaired mainly due to the anomalously high NADH/NAD+ ratio in vivo, which was shown to be three times higher than a wild-type E. coli strain (Singh et al., 2009; Singh et al., 2010). To reduce this NADH/NAD+ ratio, an effective genomic library-based screening approach was employed, and genes including pfkB (encoding phosphofructokinase II), marA (encoding DNA-binding transcriptional repressor), and moaE (encoding the subunit of molybdopterin synthase), were shown to be significant for balancing the redox ratio. Overexpression of these genes was beneficial for succinic acid production, especially pfkB, and led to a 7.8-fold increase in M9+10 g/L glucose medium under microaerobic conditions (that means controlled dosing of small amount of air or oxygen into reactor).
Conversely, deficiency in NADH has a negative impact on succinic acid biosynthesis as well. Except for the glyoxylate shunt pathway, conversion of phosphoenolpyruvate, pyruvate or acetyl-CoA to one molecule of succinic acid requires a minimum of two molecules of reducing equivalents (Lin et al., 2005c). However, only two molecules of NADH can be obtained from one molecule of glucose through the glycolytic pathway. Thus, the shortage of NADH limits the succinic acid production to a theoretical yield of 1 mol/mol glucose (Zhu et al., 2014). One solution is to use the glyoxylate shunt pathway (mentioned in section 2), which has a theoretical yield of 1.7 mol/mol glucose (Vemuri et al., 2002). Another solution is to provide more NADH. NAD+-dependent formate dehydrogenase can convert one molecule of formate into one molecule of CO2 and NADH. In this reaction, formate is one of the main by-products produced by succinic acid producers, while CO2 and NADH are necessary for succinic acid synthesis. When using glucose as a carbon source, the expression of formate dehydrogenase can double the maximum yield of NADH, from 2 to 4 mol NADH/mol glucose consumed (Berríos-Rivera et al., 2004). In Grant et al., fdh (encoding NAD+-dependent formate dehydrogenase) from C. boidiniiwas and pycA (encoding pyruvate carboxylase) from L. lactis were co-expressed in E. coli MG1655 (ΔadhE, ΔldhA, ΔiclR, Δack-pta) under the control of the same promoters (Thakker et al., 2011). Compared with the overexpression of pyc alone, this co-expression strain had better succinic acid production capacity. The succinic acid yield at 24 h showed a 1.9-fold increase, from 176 to 334 mM, and the succinic acid productivity showed a 2-fold increase, from 1 to 2 g/L/h. Meanwhile the byproduct formate concentration showed a significant decrease, from 17 mM to 0–3 mM. This indicated that higher NADH availability conditions significantly changed the final metabolite concentration pattern and promoted an increase in the contents of succinic acid. Additionally, providing carbon sources with different oxidation states is also an optional method. Sorbitol, for instance, has a higher NADH maximum theoretical yield (3 mol NADH/mol sorbitol) than glucose (Berríos-Rivera et al., 2004). It has been reported that using sorbitol can also generate higher yield and productivity of succinic acid (Chatterjee et al., 2001).
3.3 Optimizing CO2 Supplementation
In succinic acid biosynthesis, HCO3- is the major carboxylation substrate. During fermentation processes, CO2 that is dissolved in the liquid medium, first crosses the cell membrane through passive diffusion, and is then converted into HCO3- in the cytoplasm to participate in carboxylation (Lu et al., 2009). However, the low concentration of CO2 in the growth medium is the critical limiting step in succinic acid production (Zhu et al., 2015). To date, several strategies have been attempted to improve succinic acid production by optimizing CO2 supplementation and several satisfying results have been achieved.
3.3.1 Promoting the Intracellular Conversion of HCO3- and CO2
Under anaerobic fermentation conditions, both CO2 or HCO3- can serve as a carboxylation substrate for PEP carboxykinase (PCK), but PCK prefers to use CO2, and its catalytic velocity with CO2 was shown to be 7.6-fold higher than that with HCO3- (Cotelesage et al., 2007). Therefore, promoting the intracellular interconversion of HCO3- and CO2 is a way to improve its catalytic efficiency. Carbonic anhydrase (CA), which is an important component of the carboxysome, efficiently converts HCO3- to CO2 (Cotelesage et al., 2007). For improving the carboxylation velocity of PCK, Xiao et al. introduced the CA gene from Synechococcus sp. PCC7002 into E. coli Suc-T110 Δppc. Due to the increase of the local CO2 concentration, the carboxylation rate of PCK showed a 1.6-fold increase from 2.46 to 3.92 μmol/min/mg protein. As a result of CA gene overexpression, the succinic acid titer at 36 h showed a 35% increase as well (Xiao et al., 2017). These results demonstrated that carbonic anhydrase was useful for improving succinic acid productivity.
3.3.2 Enhancing HCO3- Transmembrane Transport
As the major carboxylation substrate, HCO3- can also cross the cell membrane through passive diffusion, but the rate is very slow, limiting its direct utilization (Badger and Price, 2003). One feasible strategy to increase intracellular HCO3- concentration is to use bicarbonate transporters. Among these, SbtA and BicA have been shown to be more efficient (Price et al., 2004). BicA, which was identified from Synechococcus sp. PCC7002, is a Na+-dependent transporter, and it has low transport affinity but a high flux rate (Price et al., 2004). SbtA-mediated transport, which was identified from Synechocystis PCC6803, is also Na+-dependent. Meanwhile, it can be induced by low CO2 levels and shows a relatively high affinity for HCO3- (Shibata et al., 2002; Zhu et al., 2015). To optimize succinic acid production, Xiao et al. overexpressed the bicA and pck genes in E. coli, and reported that the final cell density and succinic acid titers at 36 h were 29 and 22% higher than those of control strains, respectively (Xiao et al., 2017). Zhu et al. contrasted SbtA and BicA in E. coli AFP111, and demonstrated that overexpression of sbtA or/and bicA genes showed a positive effect on CO2 absorption but a negative effect on succinic acid biosynthesis. Meanwhile, they found that the succinic acid concentration was improved only when bicarbonate transporters and carboxylase (PCK or PPC) were co-expressed. Among these, co-overexpression of the sbtA and pck genes was superior, and led to a 15% increase in succinic production (Zhu et al., 2015). For further optimization, Yu et al. balanced HCO3- transport and fixation to maximize succinic acid titer. They demonstrated that the highest succinic acid production was obtained when the sbtA, bicA, and ppc genes were co-expressed under the control of the weak P4 promoter and the pck gene under the control of the strong P19 promoter. This led to a 1.4-fold increase in succinic acid production from 65 to 89.4 g/L, and a 1.3-fold increase in succinic acid yield from 0.98 to 1.27 mol/mol glucose (Yu et al., 2016). All of these results indicated the necessity of CO2 supplementation in succinic acid biosynthesis.
4 Conclusion and Perspectives
Succinic acid is an important four-carbon building-block chemical that can be used as the precursor of numerous products, including biodegradable plastics, feed additives, green solvents, and pharmaceutical products. As an alternative to environmentally unfriendly traditional method, biosynthesis is a promising means for succinic acid production and has become increasingly mature as well. This review summarized different succinic acid biosynthesis pathways, key enzymes, and the metabolic engineering approaches, particularly those developed for adjusting the carbon flux, balancing the redox ratio, and CO2 supplementation for succinic acid production. It needs to be clear that one or more strategies have been applied jointly to obtain high performance strains in practical application. To date, some efficient succinic acid producers have been obtained and applied in industry, but several problems still exist, such as the low robustness of engineered bacteria or the high cost of downstream product recovery. Thus, increasing attention should be paid to the following prospects in future research.
Firstly, an economical fermentation process should be further explored. Most succinic acid producing strains prefer neutral pH conditions, but succinic acid formation acidifies the medium. To keep a neutral pH, a common method is the conversion of succinic acid into succinate by titration with bases during fermentation. However, generation of salts increases the difficulty and cost of post-processing for succinic acid production. Compared with feedstock costs and some other upstream costs, the downstream costs are also considerable, and represent about 30–40% of the total production costs (López-Garzón and Straathof, 2014). One solution is to develop low pH (below or close to the pKa) fermentation strategies, which can minimize the consumption of bases, reduce the generation of salts and avoid re-conversion of succinate into succinic acid. However, the highest succinic acid concentration has so far been achieved at neutral pH, not at low pH. The main reason is the low tolerance to low pH of most succinic acid producers, such as C. glutamicum, E. coli, M. succiniciproducens, B. succiniciproducens and A. succinogenes (Ahn et al., 2016). Therefore, to reduce the overall production costs, development of a competitive acid-tolerant strain is critical for enabling bio-based production of succinic acid. Meanwhile, more attention should be paid to revealing acid-tolerant mechanisms, which are meaningful for the production of any carboxylic acid.
Furthermore, efficient biosynthesis process should be further explored. Making a detailed analyze and comparison of the seven succinic acid biosynthetic pathways is very important for understanding and designing greater efficiency biosynthesis pathway. And no matter which pathway is selected, the metabolic process for its production is a multigene pathway. Key enzyme overexpression is the traditional genetic engineering strategy used to overcome this, but the enzyme expression level in a multistep pathway is not simply “the more the better”. Liu et al. demonstrated the importance of metabolic balancing in a multistep biosynthetic pathway (Liu et al., 2016). Meanwhile, plasmid-based multigene over-expression systems can also lead to genetic instability and high cellular burdens. Thus, a functional balance between enzymes and the improvement of host stability in succinic acid biosynthetic processes should be considered. Some strategies are valuable, such as chromosomal integration, promoters or RBS engineering, microbial host engineering, and dynamic control of the pathway of interest.
Author Contributions
XF and XL developed the idea for the review. XL and XF wrote the manuscript. XF, GZ, SS, CF, and PX revised this manuscript. All authors read and approved the final manuscript.
Funding
This study was financially supported by the Natural Science Foundation of Shandong Province (ZR2021QB176 and ZR2019QB015), National Natural Science Foundation of China (31800081), State Key Laboratory of Microbial Technology Open Projects Fund (Project No. M2021-15), and start-up fund from Shandong University of Technology (to XL).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdel-Hamid, A. M., Attwood, M. M., and Guest, J. R. (2001). Pyruvate Oxidase Contributes to the Aerobic Growth Efficiency of Escherichia coli. Microbiology 147, 1483–1498. doi:10.1099/00221287-147-6-1483
Ahn, J. H., Bang, J., Kim, W. J., and Lee, S. Y. (2017). Formic Acid as a Secondary Substrate for Succinic Acid Production by Metabolically engineeredMannheimia Succiniciproducens. Biotechnol. Bioeng. 114, 2837–2847. doi:10.1002/bit.26435
Ahn, J. H., Jang, Y.-S., and Lee, S. Y. (2016). Production of Succinic Acid by Metabolically Engineered Microorganisms. Curr. Opin. Biotechnol. 42, 54–66. doi:10.1016/j.copbio.2016.02.034
Ahn, J. H., Seo, H., Park, W., Seok, J., Lee, J. A., Kim, W. J., et al. (2020). Enhanced Succinic Acid Production by Mannheimia Employing Optimal Malate Dehydrogenase. Nat. Commun. 11, 1970–1982. doi:10.1038/s41467-020-15839-z
Akira, I. (2000). Functional Modulation of Escherichia coli RNA Polymerase. Annu. Rev. Microbiol. 54, 499–518. doi:10.1146/annurev.micro.54.1.499
Arikawa, Y., Kuroyanagi, T., Shimosaka, M., Muratsubaki, H., Enomoto, K., Kodaira, R., et al. (1998). Effect of Gene Disruptions of the TCA Cycle on Production of Succinic Acid in Saccharomyces cerevisiae. J. Biosci. Bioeng. 87, 28–36. doi:10.1016/S1389-1723(99)80004-8
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 In-Frame, Single-Gene Knockout Mutants: the Keio Collection. Mol. Syst. Biol. 2, 2006–0008. doi:10.1038/msb4100050
Badger, M. R., and Price, G. D. (2003). CO2 Concentrating Mechanisms in Cyanobacteria: Molecular Components, Their Diversity and Evolution. J. Exp. Bot. 54, 609–622. doi:10.1093/jxb/erg076
Becker, J., Reinefeld, J., Stellmacher, R., Schäfer, R., Lange, A., Meyer, H., et al. (2013). Systems-wide Analysis and Engineering of Metabolic Pathway Fluxes in Bio-Succinate Producing Basfia Succiniciproducens. Biotechnol. Bioeng. 110, 3013–3023. doi:10.1002/bit.24963
Berríos-Rivera, S. J., Sánchez, A. M., Bennett, G. N., and San, K. Y. (2004). Effect of Different Levels of NADH Availability on Metabolite Distribution in Escherichia coli Fermentation in Minimal and Complex media. Appl. Microbiol. Biotechnol. 65, 426–432. doi:10.1007/s00253-004-1609-3
Bochner, B. R., and Ames, B. N. (1982). Complete Analysis of Cellular Nucleotides by Two-Dimensional Thin Layer Chromatography. J. Biol. Chem. 257, 9759–9769. doi:10.1016/s0021-9258(18)34138-3
Bozell, J. J., and Petersen, G. R. (2010). Technology Development for the Production of Biobased Products from Biorefinery Carbohydrates-The US Department of Energy's "Top 10" Revisited. Green. Chem. 12, 539–554. doi:10.1039/B922014C
Bretz, K., and Kabasci, S. (2012). Feed-control Development for Succinic Acid Production with Anaerobiospirillum Succiniciproducens. Biotechnol. Bioeng. 109, 1187–1192. doi:10.1002/bit.24387
Chatterjee, R., Millard, C. S., Champion, K., Clark, D. P., and Donnelly, M. I. (2001). Mutation of the ptsG Gene Results in Increased Production of Succinate in Fermentation of Glucose by Escherichia coli. Appl. Environ. Microbiol. 67, 148–154. doi:10.1128/AEM.67.1.148-154.2001
Chen, K.-Q., Li, J., Ma, J.-F., Jiang, M., Wei, P., Liu, Z.-M., et al. (2011). Succinic Acid Production by Actinobacillus Succinogenes Using Hydrolysates of Spent Yeast Cells and Corn Fiber. Bioresour. Technol. 102, 1704–1708. doi:10.1016/j.biortech.2010.08.011
Chen, X., Cao, Y., Li, F., Tian, Y., and Song, H. (2018). Enzyme-Assisted Microbial Electrosynthesis of Poly(3-Hydroxybutyrate) via CO2 Bioreduction by Engineered Ralstonia Eutropha. ACS Catal. 8, 4429–4437. doi:10.1021/acscatal.8b00226
Chen, Y., Cheng, H., Ningna, L. I., Ke, S. U., Jiayu, L. I., Xia, H., et al. (2019). Effects of Pyruvate Carboxylase on Succinic Acid Accumulation in Saccharomyces cerevisiae. Food Ferment. Industries 45, 38–44. doi:10.13995/j.cnki.11-1802/ts.018736
Choi, J. W., and Da Silva, N. A. (2014). Improving Polyketide and Fatty Acid Synthesis by Engineering of the Yeast Acetyl-CoA Carboxylase. J. Biotechnol. 187, 56–59. doi:10.1016/j.jbiotec.2014.07.430
Choi, P. H., Vu, T. M. N., Pham, H. T., Woodward, J. J., Turner, M. S., and Tong, L. (2017). Structural and Functional Studies of Pyruvate Carboxylase Regulation by Cyclic Di-AMP in Lactic Acid Bacteria. Proc. Natl. Acad. Sci. U S A. 114, E7226–E7235. doi:10.1073/pnas.1704756114
Choi, S., Song, C. W., Shin, J. H., and Lee, S. Y. (2015). Biorefineries for the Production of Top Building Block Chemicals and Their Derivatives. Metab. Eng. 28, 223–239. doi:10.1016/j.ymben.2014.12.007
Choi, S., Song, H., Lim, S. W., Kim, T. Y., Ahn, J. H., Lee, J. W., et al. (2016). Highly Selective Production of Succinic Acid by Metabolically engineeredMannheimia Succiniciproducensand its Efficient Purification. Biotechnol. Bioeng. 113, 2168–2177. doi:10.1002/bit.25988
Chong, H., Geng, H., Zhang, H., Song, H., Huang, L., and Jiang, R. (2014). EnhancingE. Coliisobutanol Tolerance through Engineering its Global Transcription Factor cAMP Receptor Protein (CRP). Biotechnol. Bioeng. 111, 700–708. doi:10.1002/bit.25134
Chung, S.-C., Park, J.-S., Yun, J., and Park, J. H. (2017). Improvement of Succinate Production by Release of End-Product Inhibition in Corynebacterium Glutamicum. Metab. Eng. 40, 157–164. doi:10.1016/j.ymben.2017.02.004
Clark, D. P. (1989). The Fermentation Pathways ofEscherichia Coli. FEMS Microbiol. Lett. 63, 223–234. doi:10.1111/j.1574-6968.1989.tb03398.x
Cotelesage, J. J. H., Puttick, J., Goldie, H., Rajabi, B., Novakovski, B., and Delbaere, L. T. J. (2007). How Does an Enzyme Recognize CO2? Int. J. Biochem. Cel Biol. 39, 1204–1210. doi:10.1016/j.biocel.2007.03.015
Cozzone, A. J., and El-Mansi, M. (2005). Control of Isocitrate Dehydrogenase Catalytic Activity by Protein Phosphorylation in Escherichia coli. J. Mol. Microbiol. Biotechnol. 9, 132–146. doi:10.1159/000089642
Cui, Z., Gao, C., Li, J., Hou, J., Lin, C. S. K., and Qi, Q. (2017). Engineering of Unconventional Yeast Yarrowia Lipolytica for Efficient Succinic Acid Production from Glycerol at Low pH. Metab. Eng. 42, 126–133. doi:10.1016/j.ymben.2017.06.007
Dayanidhi, S., Siddiquee, K. A., Araúzo-Bravo, M. J., Oba, T., and Shimizu, K. (2008). Effect of Cra Gene Knockout Together with Edd and iclR Genes Knockout on the Metabolism in Escherichia coli. Arch. Microbiol. 190, 559–571. doi:10.1007/s00203-008-0406-2
De Mey, M., De Maeseneire, S., Soetaert, W., and Vandamme, E. (2007). Minimizing Acetate Formation in E. coli Fermentations. J. Ind. Microbiol. Biotechnol. 34, 689–700. doi:10.1007/s10295-007-0244-2
Dittrich, C. R., Vadali, R. V., Bennett, G. N., and San, K. Y. (2010). Redistribution of Metabolic Fluxes in the Central Aerobic Metabolic Pathway of E. coli Mutant Strains with Deletion of the ackA-Pta and poxB Pathways for the Synthesis of Isoamyl Acetate. Biotechnol. Prog. 21, 627–631. doi:10.1021/bp049730r
Fixation, A. C., Hügler, M., Menendez, C., Schägger, H., and Fuchs, G. (2002). Malonyl-coenzyme A Reductase from Chloroflexus Aurantiacus, a Key Enzyme of the 3-hydroxypropionate Cycle for Autotrophic CO(2) Fixation. J. Bacteriol. 184, 2404–2410. doi:10.1128/JB.184.9.2404–2410.2002
Gao, C., Yang, X., Wang, H., Rivero, C. P., Li, C., Cui, Z., et al. (2016). Robust Succinic Acid Production from Crude Glycerol Using Engineered Yarrowia Lipolytica. Biotechnol. Biofuels 9, 179–190. doi:10.1186/s13068-016-0597-8
Gokarn, R. R., Eiteman, M. A., and Altman, E. (1998). Expression of Pyruvate Carboxylase Enhances Succinate Production in Escherichia coli without Affecting Glucose Uptake. Biotechnol. Lett. 20, 795–798. doi:10.1023/B:BILE.0000015925.52287.1f
Gokarn, R. R., Eiteman, M. A., and Altman, E. (2000). Metabolic Analysis of Escherichia coli in the Presence and Absence of the Carboxylating Enzymes Phosphoenolpyruvate Carboxylase and Pyruvate Carboxylase. Appl. Environ. Microbiol. 66, 1844–1850. doi:10.1128/AEM.66.5.1844-1850.2000
Gong, F., Cai, Z., and Li, Y. (2016). Synthetic Biology for CO2 Fixation. Sci. China Life Sci. 59, 1106–1114. doi:10.1007/s11427-016-0304-2
Gourdon, P., Baucher, M.-F., Lindley, N. D., and Guyonvarch, A. (2000). Cloning of the Malic Enzyme Gene from Corynebacterium Glutamicum and Role of the Enzyme in Lactate Metabolism. Appl. Environ. Microbiol. 66, 2981–2987. doi:10.1128/aem.66.7.2981-2987.2000
Guettler, M. V., Jain, M. K., and Rumler, D. (1996). Method for Making Succinic Acid, Bacterial Variants for Use in the Process, and Methods for Obtaining Variants. United States Patent. Available at http://www.google.com/patents/US5723322.
Gui, L., Sunnarborg, A., Pan, B., and Laporte, D. C. (1996). Autoregulation of iclR, the Gene Encoding the Repressor of the Glyoxylate Bypass Operon. J. Bacteriol. 178, 321–324. doi:10.1128/jb.178.1.321-324.1996
Hendrik, W., Joeri, B., Helena, M., Jo, M., Marjan, D. M., Maria, R., et al. (2011). Effect of iclR and arcA Knockouts on Biomass Formation and Metabolic Fluxes in Escherichia coli K12 and its Implications on Understanding the Metabolism of Escherichia coli BL21 (DE3). Bmc Microbiol. 11, 1–17. doi:10.1186/1471-2180-11-70
Herter, S., Farfsing, J., Gad'On, N., Rieder, C., Eisenreich, W., Bacher, A., et al. (2001). Autotrophic CO 2 Fixation by Chloroflexus Aurantiacus : Study of Glyoxylate Formation and Assimilation via the 3-Hydroxypropionate Cycle. J. Bacteriol. 183, 4305–4316. doi:10.1128/jb.183.14.4305-4316.2001
Hong, S. H., and Lee, S. Y. (2002). Importance of Redox Balance on the Production of Succinic Acid by Metabolically Engineered Escherichia coli. Appl. Microbiol. Biotechnol. 58, 286–290. doi:10.1007/s00253-001-0899-y
Hong, S. H., Kim, J. S., Lee, S. Y., In, Y. H., Choi, S. S., Rih, J.-K., et al. (2004). The Genome Sequence of the Capnophilic Rumen Bacterium Mannheimia Succiniciproducens. Nat. Biotechnol. 22, 1275–1281. doi:10.1038/nbt1010
Hui, W., Zhimin, L., Li, Z., and Qin, Y. (2009). Enhanced Anaerobic Succinic Acid Production by Escherichia coli NZN111 Aerobically Grown on Gluconeogenic Carbon Sources. Enzyme Microb. Technol. 44, 165–169. doi:10.1016/j.jbiotec.2008.07.964
Ishihama, A. (2010). Prokaryotic Genome Regulation: Multifactor Promoters, Multitarget Regulators and Hierarchic Networks. FEMS Microbiol. Rev. 34, 628–645. doi:10.1111/j.1574-6976.2010.00227.x
Jiang, M., Liu, S.-w., Ma, J.-f., Chen, K.-q., Yu, L., Yue, F.-f., et al. (2010). Effect of Growth Phase Feeding Strategies on Succinate Production by Metabolically Engineered Escherichia coli. Appl. Environ. Microbiol. 76, 1298–1300. doi:10.1128/AEM.02190-09
Jitrapakdee, S., St Maurice, M., Rayment, I., Cleland, W. W., Wallace, J. C., and Attwood, P. V. (2008). Structure, Mechanism and Regulation of Pyruvate Carboxylase. Biochem. J. 413, 369–387. doi:10.1042/bj20080709
Kai, Y., Matsumura, H., Inoue, T., Terada, K., Nagara, Y., Yoshinaga, T., et al. (1999). Three-dimensional Structure of Phosphoenolpyruvate Carboxylase: a Proposed Mechanism for Allosteric Inhibition. Proc. Natl. Acad. Sci. 96, 823–828. doi:10.1073/pnas.96.3.823
Kequan, C., Min, J., Ping, W., Jiaming, Y., and Hao, W. (2008). Succinic Acid Production from Acid Hydrolysate of Corn Fiber by Actinobacillus Succinogenes. Appl. Biochem. Biotechnol. 160, 477–485. doi:10.1016/j.biortech.2007.03.044
Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muñiz-Rascado, L., et al. (2011). EcoCyc: a Comprehensive Database of Escherichia coli Biology. Nucleic Acids Res. 39, D583–D590. doi:10.1093/nar/gkq1143
Kim, D. Y., Yim, S. C., Lee, P. C., Lee, W. G., Lee, S. Y., and Chang, H. N. (2004). Batch and Continuous Fermentation of Succinic Acid from wood Hydrolysate by Mannheimia Succiniciproducens MBEL55E. Enzyme Microb. Technol. 35, 648–653. doi:10.1016/j.enzmictec.2004.08.018
Krebs, A., and Bridger, W. A. (1980). The Kinetic Properties of Phosphoenolpyruvate Carboxykinase of Escherichia coli. Can. J. Biochem. 58, 309–318. doi:10.1139/o80-041
Kuhnert, P., Scholten, E., Haefner, S., Mayor, D., and Frey, J. (2010). Basfia Succiniciproducens Gen. nov., Sp. nov., a New Member of the Family Pasteurellaceae Isolated from Bovine Rumen. Int. J. Syst. Evol. Microbiol. 60, 44–50. doi:10.1099/ijs.0.011809-0
Kwon, Y.-D., Kwon, O.-H., Lee, H.-S., and Kim, P. (2007). The Effect of NADP-dependent Malic Enzyme Expression and Anaerobic C4 Metabolism in Escherichia coli Compared with Other Anaplerotic Enzymes. J. Appl. Microbiol. 103, 2340–2345. doi:10.1111/j.1365-2672.2007.03485.x
Kwon, Y. D., Lee, S. Y., and Kim, P. (2006). Influence of Gluconeogenic Phosphoenolpyruvate Carboxykinase (PCK) Expression on Succinic Acid Fermentation in Escherichia coli under High Bicarbonate Condition. J. Microbiol. Biotechnol. 16, 1448–1452. doi:10.1007/s10295-006-0128-x
Lee, J. W., Yi, J., Kim, T. Y., Choi, S., Ahn, J. H., Song, H., et al. (2016). Homo-succinic Acid Production by Metabolically Engineered Mannheimia Succiniciproducens. Metab. Eng. 38, 409–417. doi:10.1016/j.ymben.2016.10.004
Lee, P. C., Lee, S. Y., Hong, S. H., and Chang, H. N. (2003). Batch and Continuous Cultures of Mannheimia Succiniciproducens MBEL55E for the Production of Succinic Acid from Whey and Corn Steep Liquor. Bioproc. Biosyst. Eng. 26, 63–67. doi:10.1007/s00449-003-0341-1
Lee, P. C., Lee, W. G., Kwon, S., Lee, S. Y., and Chang, H. N. (1999). Succinic Acid Production by Anaerobiospirillum Succiniciproducens: Effects of the H2/CO2 Supply and Glucose Concentration. Enzyme Microb. Technol. 24, 549–554. doi:10.1016/S0141-0229(98)00156-2
Lee, S., Hong, H., Chang, P., Lee, S. Y., Hong, S. H., and Chang, H. N. (2002). Isolation and Characterization of a New Succinic Acid-Producing Bacterium, Mannheimia Succiniciproducens MBEL55E, from Bovine Rumen. Appl. Microbiol. Biotechnol. 58, 663–668. doi:10.1007/s00253-002-0935-6
Lee, S. J., Song, H., and Lee, S. Y. (2006). Genome-based Metabolic Engineering of Mannheimia Succiniciproducens for Succinic Acid Production. Appl. Environ. Microbiol. 72, 1939–1948. doi:10.1128/aem.72.3.1939-1948.2006
Li, C., Ong, K. L., Yang, X., and Lin, C. S. K. (2019). Bio-refinery of Waste Streams for green and Efficient Succinic Acid Production by Engineered Yarrowia Lipolytica without pH Control. Chem. Eng. J. 371, 804–812. doi:10.1016/j.cej.2019.04.092
Li, J., Li, Y., Cui, Z., Liang, Q., and Qi, Q. (2017). Enhancement of Succinate Yield by Manipulating NADH/NAD+ Ratio and ATP Generation. Appl. Microbiol. Biotechnol. 101, 3153–3161. doi:10.1007/s00253-017-8127-6
Li, M., Hou, F., Wu, T., Jiang, X., Li, F., Liu, H., et al. (2020). Recent Advances of Metabolic Engineering Strategies in Natural Isoprenoid Production Using Cell Factories. Nat. Prod. Rep. 37, 80–99. doi:10.1039/C9NP00016J
Li, Q., Yang, M., Wang, D., Li, W., Wu, Y., Zhang, Y., et al. (2010). Efficient Conversion of Crop Stalk Wastes into Succinic Acid Production by Actinobacillus Succinogenes. Bioresour. Technol. 101, 3292–3294. doi:10.1016/j.biortech.2009.12.064
Lin, H., Bennett, G. N., and San, K.-Y. (2005c). Effect of Carbon Sources Differing in Oxidation State and Transport Route on Succinate Production in Metabolically Engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 32, 87–93. doi:10.1007/s10295-005-0206-5
Lin, H., Bennett, G. N., and San, K.-Y. (2005d). Fed-batch Culture of a Metabolically engineeredEscherichia Coli Strain Designed for High-Level Succinate Production and Yield under Aerobic Conditions. Biotechnol. Bioeng. 90, 775–779. doi:10.1002/bit.20458
Lin, H., Bennett, G. N., and San, K.-Y. (2004). Genetic Reconstruction of the Aerobic central Metabolism inEscherichia Coli for the Absolute Aerobic Production of Succinate. Biotechnol. Bioeng. 89, 148–156. doi:10.1002/bit.20298
Lin, H., Bennett, G. N., and San, K.-Y. (2005a). Metabolic Engineering of Aerobic Succinate Production Systems in Escherichia coli to Improve Process Productivity and Achieve the Maximum Theoretical Succinate Yield. Metab. Eng. 7, 116–127. doi:10.1016/j.ymben.2004.10.003
Lin, H., San, K.-Y., and Bennett, G. N. (2005b). Effect of Sorghum Vulgare Phosphoenolpyruvate Carboxylase and Lactococcus Lactis Pyruvate Carboxylase Coexpression on Succinate Production in Mutant Strains of Escherichia coli. Appl. Microbiol. Biotechnol. 67, 515–523. doi:10.1007/s00253-004-1789-x
Litsanov, B., Brocker, M., and Bott, M. (2012). Toward Homosuccinate Fermentation: Metabolic Engineering of Corynebacterium Glutamicum for Anaerobic Production of Succinate from Glucose and Formate. Appl. Environ. Microbiol. 78, 3325–3337. doi:10.1128/AEM.07790-11
Liu, Y., and Jiang, H. (2011). Directed Evolution of Propionyl-CoA Carboxylase for Succinate Biosynthesis. Trends Biotechnol. 39, 330–331. doi:10.1016/j.tibtech.2021.02.006
Liu, B., Xiang, S., Zhao, G., Wang, B., Ma, Y., Liu, W., et al. (2019). Efficient Production of 3-hydroxypropionate from Fatty Acids Feedstock in Escherichia coli. Metab. Eng. 51, 121–130. doi:10.1016/j.ymben.2018.10.003
Liu, C., Wang, Q., Xian, M., Ding, Y., and Zhao, G. (2013). Dissection of Malonyl-Coenzyme A Reductase of Chloroflexus Aurantiacus Results in Enzyme Activity Improvement. Plos One 8, e75554–8. doi:10.1371/journal.pone.0075554
Liu, C., Ding, Y., Zhang, R., Liu, H., Xian, M., and Zhao, G. (2016). Functional Balance between Enzymes in Malonyl-CoA Pathway for 3-hydroxypropionate Biosynthesis. Metab. Eng. 34, 104–111. doi:10.1016/j.ymben.2016.01.001i
Liu, T., Vora, H., and Khosla, C. (2010). Quantitative Analysis and Engineering of Fatty Acid Biosynthesis in E. coli. Metab. Eng. 12, 378–386. doi:10.1016/j.ymben.2010.02.003
Liu, X., and De Wulf, P. (2004). Probing the ArcA-P Modulon of Escherichia coli by Whole Genome Transcriptional Analysis and Sequence Recognition Profiling. J. Biol. Chem. 279, 12588–12597. doi:10.1074/jbc.M313454200
Liu, X., Feng, X., Ding, Y., Gao, W., Xian, M., Wang, J., et al. (2020). Characterization and Directed Evolution of Propionyl-CoA Carboxylase and its Application in Succinate Biosynthetic Pathway with Two CO2 Fixation Reactions. Metab. Eng. 62, 42–50. doi:10.1016/j.ymben.2020.08.012
Liu, Y.-P., Zheng, P., Sun, Z.-H., Ni, Y., Dong, J.-J., and Zhu, L.-L. (2008). Economical Succinic Acid Production from Cane Molasses by Actinobacillus Succinogenes. Bioresour. Technol. 99, 1736–1742. doi:10.1016/j.biortech.2007.03.044
Lombard, J., and Moreira, D. (2012). Early Evolution of the Biotin-dependent Carboxylase Family. BMC Evol. Biol. 11, 232. doi:10.1186/1471-2148-11-232
López-Garzón, C., and Straathof, A. (2014). Recovery of Carboxylic Acids Produced by Fermentation. Biotechnol. Adv. 32, 873–904. doi:10.1016/j.biotechadv.2014.04.002
Louasté, B., and Eloutassi, N. (2020). Succinic Acid Production from Whey and Lactose by Actinobacillus Succinogenes 130Z in Batch Fermentation. Biotechnol. Rep. (Amst) 27, e00481–486. doi:10.1016/j.btre.2020.e00481
Lu, C. D., Xiong, Y. Y., Zhao, J. F., and Wang, J. H. (2015). Fermentation Production of Succinic Acid from Sucrose by Engineered Escherichia Coli JH208. Amr 1088, 503–506. doi:10.4028/www.scientific.net/AMR.1088.503
Lu, S., Eiteman, M. A., and Altman, E. (2009). Effect of CO2 on Succinate Production in Dual-phase Escherichia coli Fermentations. J. Biotechnol. 143, 213–223. doi:10.1016/j.jbiotec.2009.07.012
Ma, J., Gou, D., Liang, L., Liu, R., Chen, X., Zhang, C., et al. (2013). Enhancement of Succinate Production by Metabolically Engineered Escherichia coli with Co-expression of Nicotinic Acid Phosphoribosyltransferase and Pyruvate Carboxylase. Appl. Microbiol. Biotechnol. 97, 6739–6747. doi:10.1007/s00253-013-4910-1
Menendez, C., Bauer, Z., Huber, H., Nasser, G. o., Stetter, K.-O., Fuchs, G., et al. (1999). Presence of Acetyl Coenzyme A (Coa) Carboxylase and Propionyl-Coa Carboxylase in Autotrophic Crenarchaeota and Indication for Operation of a 3-Hydroxypropionate Cycle in Autotrophic Carbon Fixation. J. Bacteriol. 181, 1088–1098. doi:10.1128/JB.181.4.1088-1098.1999
Meynial-Salles, I., Dorotyn, S., and Soucaille, P. (2010). A New Process for the Continuous Production of Succinic Acid from Glucose at High Yield, Titer, and Productivity. Biotechnol. Bioeng. 99, 129–135. doi:10.1002/bit.21521
Michael, F. A. B., Ali, M., Davinia, S., Holly, S., Brenna, A. B., Nancy, D., et al. (2015). Continuous Succinic Acid Production by Actinobacillus succinogenes on Xylose-Enriched Hydrolysate. Bioresour. Technol. 8, 181–198. doi:10.1186/s13068-015-0363-3
Millard, C. S., Chao, Y. P., Liao, J. C., and Donnelly, M. I. (1996). Enhanced Production of Succinic Acid by Overexpression of Phosphoenolpyruvate Carboxylase in Escherichia coli. Appl. Environ. Microbiol. 62, 1808–1810. doi:10.1128/aem.62.5.1808-1810.1996
Modak, H. V., and Kelly, D. J. (1995). Acetyl-CoA-dependent Pyruvate Carboxylase from the Photosynthetic Bacterium Rhodobacter Capsulatus: Rapid and Efficient Purification Using Dye-Ligand Affinity Chromatography. Microbiology 141, 2619–2628. doi:10.1099/13500872-141-10-2619
Okino, S., Inui, M., and Yukawa, H. (2005). Production of Organic Acids by Corynebacterium Glutamicum under Oxygen Deprivation. Appl. Microbiol. Biotechnol. 68, 475–480. doi:10.1007/s00253-005-1900-y
Okino, S., Noburyu, R., Suda, M., Jojima, T., Inui, M., and Yukawa, H. (2008). An Efficient Succinic Acid Production Process in a Metabolically Engineered Corynebacterium Glutamicum Strain. Appl. Microbiol. Biotechnol. 81, 459–464. doi:10.1007/s00253-008-1668-y
Olajuyin, A. M., Yang, M., Liu, Y., Mu, T., Tian, J., Adaramoye, O. A., et al. (2016). Efficient Production of Succinic Acid from Palmaria Palmata Hydrolysate by Metabolically Engineered Escherichia coli. Bioresour. Technol. 214, 653–659. doi:10.1016/j.biortech.2016.04.117
Pateraki, C., Patsalou, M., Vlysidis, A., Kopsahelis, N., Webb, C., Koutinas, A. A., et al. (2016). Actinobacillus Succinogenes : Advances on Succinic Acid Production and Prospects for Development of Integrated Biorefineries. Biochem. Eng. J. 112, 285–303. doi:10.1016/j.bej.2016.04.005
Perrenoud, A., and Sauer, U. (2005). Impact of Global Transcriptional Regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on Glucose Catabolism in Escherichia coli. J. Bacteriol. 187, 3171–3179. doi:10.1128/JB.187.9.3171-3179.2005
Price, G. D., Woodger, F. J., Badger, M. R., Howitt, S. M., and Tucker, L. (2004). Identification of a SulP-type Bicarbonate Transporter in marine Cyanobacteria. Proc. Natl. Acad. Sci. 101, 18228–18233. doi:10.1073/pnas.0405211101
Qiang, l., Dan, W., Yong, W., and Wangliang, L. (2010). One Step Recovery of Succinic Acid from Fermentation Broths by Crystallization. Separat. Purif. Technol. 72, 294–300. doi:10.1016/j.seppur.2010.02.021
Ramseier, T. M., Bledig, S., Michotey, V., Feghali, R., and Saier, M. H. (2010). The Global Regulatory Protein FruR Modulates the Direction of Carbon Flow in Escherichia coli. Mol. Microbiol. 16, 1157–1169. doi:10.1111/j.1365-2958.1995.tb02339.x
Ramseier, T. M. (1996). Cra and the Control of Carbon Flux via Metabolic Pathways. Res. Microbiol. 147, 489–493. doi:10.1016/0923-2508(96)84003-4
Sánchez, A. M., Bennett, G. N., and San, K. Y. (2005). Efficient Succinic Acid Production from Glucose through Overexpression of Pyruvate Carboxylase in an Escherichia coli Alcohol Dehydrogenase and Lactate Dehydrogenase Mutant. Biotechnol. Prog. 21, 358–365. doi:10.1021/bp049676e
Sánchez, A. M., Bennett, G. N., and San, K.-Y. (2005). Novel Pathway Engineering Design of the Anaerobic central Metabolic Pathway in Escherichia coli to Increase Succinate Yield and Productivity. Metab. Eng. 7, 229–239. doi:10.1016/j.ymben.2005.03.001
Sarkar, D., Siddiquee, K. A. Z., Araúzo-Bravo, M. J., Oba, T., and Shimizu, K. (2008). Effect of Cra Gene Knockout Together with Edd and iclR Genes Knockout on the Metabolism in Escherichia coli. Arch. Microbiol. 190, 559–571. doi:10.1007/s00203-008-0406-2
Schütze, A., Benndorf, D., Püttker, S., Kohrs, F., and Bettenbrock, K. (2020). The Impact of ackA, Pta, and ackA-Pta Mutations on Growth, Gene Expression and Protein Acetylation in Escherichia coli K-12. Front. Microbiol. 11, 233. doi:10.3389/fmicb.2020.00233
Scott, J. W., Norman, D. G., Hawley, S. A., Kontogiannis, L., and Hardie, D. G. (2002). Protein Kinase Substrate Recognition Studied Using the Recombinant Catalytic Domain of AMP-Activated Protein Kinase and a Model Substrate. J. Mol. Biol. 317, 309–323. doi:10.1006/jmbi.2001.5316
Shi, S., Chen, Y., Siewers, V., and Nielsen, J. (2014). Improving Production of Malonyl Coenzyme A-Derived Metabolites by Abolishing Snf1-dependent Regulation of Acc1. mBio 5, e01130–14. doi:10.1128/mBio.01130-14
Shibata, M., Katoh, H., Sonoda, M., Ohkawa, H., Shimoyama, M., Fukuzawa, H., et al. (2002). Genes Essential to Sodium-dependent Bicarbonate Transport in Cyanobacteria: Function and Phylogenetic Analysis. J. Biol. Chem. 277, 18658–18664. doi:10.1074/jbc.M112468200
Shimada, T., Yamamoto, K., and Ishihama, A. (2011). Novel Members of the Cra Regulon Involved in Carbon Metabolism in Escherichia coli. J. Bacteriol. 193, 649–659. doi:10.1128/JB.01214-10
Shimizu, K. (2013). Metabolic Regulation of a Bacterial Cell System with Emphasis on Escherichia coli Metabolism. ISRN Biochem. 2013, 645983. doi:10.1155/2013/645983
Singh, A., Cher Soh, K., Hatzimanikatis, V., and Gill, R. T. (2010). Manipulating Redox and ATP Balancing for Improved Production of Succinate in E. coli. Metab. Eng. 13, 76–81. doi:10.1016/j.ymben.2010.10.006
Singh, A., Lynch, M. D., and Gill, R. T. (2009). Genes Restoring Redox Balance in Fermentation-Deficient E. coli NZN111. Metab. Eng. 11, 347–354. doi:10.1016/j.ymben.2009.07.002
Stols, L., and Donnelly, M. I. (1997). Production of Succinic Acid through Overexpression of NAD(+)-dependent Malic Enzyme in an Escherichia coli Mutant. Appl. Environ. Microbiol. 63, 2695–2701. doi:10.1128/aem.63.7.2695-2701.1997
Stols, L., Kulkarni, G., Harris, B. G., and Donnelly, M. I. (1997). Expression of Ascaris suum Malic Enzyme in a Mutant Escherichia coli Allows Production of Succinic Acid from Glucose. Appl. Biochem. Biotechnol. 63, 153–158. doi:10.1007/978-1-4612-2312-2_15
Sun, H., Song, Y., Chen, F., Zhou, C., and Feng, L. (2020). An ArcA-Modulated Small RNA in Pathogenic Escherichia coli K1. Front. Microbiol. 11, 1–13. doi:10.3389/fmicb.2020.574833
Tan, Z., Zhu, X., Chen, J., Li, Q., and Zhang, X. (2013). Activating Phosphoenolpyruvate Carboxylase and Phosphoenolpyruvate Carboxykinase in Combination for Improvement of Succinate Production. Appl. Environ. Microbiol. 79, 4838–4844. doi:10.1128/AEM.00826-13
Thakker, C., Martínez, I., San, K.-Y., and Bennett, G. N. (2012). Succinate Production in Escherichia coli. Biotechnol. J. 7 (2), 213–224. doi:10.1002/biot.201100061
Thakker, C., Zhu, J., San, K.-Y., and Bennett, G. (2011). Heterologous Pyc Gene Expression under Various Natural and Engineered Promoters in Escherichia coli for Improved Succinate Production. J. Biotechnol. 155, 236–243. doi:10.1016/j.jbiotec.2011.05.001
Thuy, N. T. H., Kongkaew, A., Flood, A., and Boontawan, A. (2017). Fermentation and Crystallization of Succinic Acid from Actinobacillus Succinogenes ATCC55618 Using Fresh Cassava Root as the Main Substrate. Bioresour. Technol. 233, 342–352. doi:10.1016/j.biortech.2017.02.114
Tong, L. (2013). Structure and Function of Biotin-dependent Carboxylases. Cell. Mol. Life Sci. 70, 863–891. doi:10.1007/s00018-012-1096-0
Vemuri, G. N., Eiteman, M. A., and Altman, E. (2002). Effects of Growth Mode and Pyruvate Carboxylase on Succinic Acid Production by Metabolically Engineered Strains of Escherichia coli. Appl. Environ. Microbiol. 68, 1715–1727. doi:10.1128/AEM.68.4.1715-1727.2002
Wan, C., Li, Y., Shahbazi, A., and Xiu, S. (2008). Succinic Acid Production from Cheese Whey Using Actinobacillus Succinogenes 130 Z. Appl. Biochem. Biotechnol. 145, 111–119. doi:10.1007/s12010-007-8031-0
Wang, D., Li, Q., Song, Z., Zhou, W., Su, Z., and Xing, J. (2011). High Cell Density Fermentation via a Metabolically Engineered Escherichia coli for the Enhanced Production of Succinic Acid. J. Chem. Technol. Biotechnol. 86, 512–518. doi:10.1002/jctb.2543
Wei, L.-N., Zhu, L.-W., and Tang, Y.-J. (2016). Succinate Production Positively Correlates with the Affinity of the Global Transcription Factor Cra for its Effector FBP in Escherichia coli. Biotechnol. Biofuels 9, 264–279. doi:10.1186/s13068-016-0679-7
Werf, M. J. V. D., Guettler, M. V., Jain, M. K., and Zeikus, J. G. (1997). Environmental and Physiological Factors Affecting the Succinate Product Ratio during Carbohydrate Fermentation by Actinobacillus Sp. 130Z. Arch. Microbiol. 167, 332–342. doi:10.1007/s002030050452
Werpy, T., Petersen, G., Aden, A., Bozell, J. J., and Jones, S. (2004). Top Value Added Chemicals from Biomass. Nato Adv. Sci. Institutes. doi:10.1126/science.1146356
Wilfredo, E., James, K., Levani, Z., Alexandre, E., White, M. A., Gribenko, A. V., et al. (2021). Signal Transmission in Escherichia coli Cyclic AMP Receptor Protein for Survival in Extreme Acidic Conditions. Biochemistry 60, 2987–3006. doi:10.1021/acs.biochem.1c00388
Wolfe, A. J. (2005). The Acetate Switch. Microbiol. Mol. Biol. Rev. 69, 12–50. doi:10.1128/mmbr.69.1.12-50.2005
Wongkittichote, P., Ah Mew, N., and Chapman, K. A. (2017). Propionyl-CoA Carboxylase - A Review. Mol. Genet. Metab. 122, 145–152. doi:10.1016/j.ymgme.2017.10.002
Woods, A., Munday, M. R., Scott, J., Yang, X., Carlson, M., and Carling, D. (1994). Yeast SNF1 Is Functionally Related to Mammalian AMP-Activated Protein Kinase and Regulates Acetyl-CoA Carboxylase In Vivo. J. Biol. Chem. 269, 19509–19515. doi:10.1016/S0021-9258(17)32198-1
Wu, H., Li, Z.-m., Zhou, L., and Ye, Q. (2007). Improved Succinic Acid Production in the Anaerobic Culture of an Escherichia coli pflB ldhA Double Mutant as a Result of Enhanced Anaplerotic Activities in the Preceding Aerobic Culture. Appl. Environ. Microbiol. 73, 7837–7843. doi:10.1128/AEM.01546-07
Wu, M., Li, X., Guo, S., Lemma, W. D., Zhang, W., Ma, J., et al. (2017). Enhanced Succinic Acid Production under Acidic Conditions by Introduction of Glutamate Decarboxylase System in E. coli AFP111. Bioproc. Biosyst Eng 40, 549–557. doi:10.1007/s00449-016-1720-8
Xiao, M., Zhu, X., Bi, C., Ma, Y., and Zhang, X. (2017). Improving Succinate Productivity by Engineering a Cyanobacterial CO2 Concentrating System (CCM) in Escherichia coli. Biotechnol. J. 12, 1–7. doi:10.1002/biot.201700199
Yan, D., Wang, C., Zhou, J., Liu, Y., Yang, M., and Xing, J. (2014). Construction of Reductive Pathway in Saccharomyces cerevisiae for Effective Succinic Acid Fermentation at Low pH Value. Bioresour. Technol. 156, 232–239. doi:10.1016/j.biortech.2014.01.053
Yang, J., Wang, Z., Zhu, N., Wang, B., Chen, T., and Zhao, X. (2014). Metabolic Engineering of Escherichia coli and In Silico Comparing of Carboxylation Pathways for High Succinate Productivity under Aerobic Conditions. Microbiol. Res. 169, 432–440. doi:10.1016/j.micres.2013.09.002
Yu, J.-H., Zhu, L.-W., Xia, S.-T., Li, H.-M., Tang, Y.-L., Liang, X.-H., et al. (2016). Combinatorial Optimization of CO2transport and Fixation to Improve Succinate Production by Promoter Engineering. Biotechnol. Bioeng. 113, 1531–1541. doi:10.1002/bit.25927
Yuzbashev, T. V., Yuzbasheva, E. Y., Sobolevskaya, T. I., Laptev, I. A., Vybornaya, T. V., Larina, A. S., et al. (2010). Production of Succinic Acid at Low pH by a Recombinant Strain of the Aerobic Yeast Yarrowia Lipolytica. Biotechnol. Bioeng. 107, 673–682. doi:10.1002/bit.22859
Zha, W., Rubin-Pitel, S. B., Shao, Z., and Zhao, H. (2009). Improving Cellular Malonyl-CoA Level in Escherichia coli via Metabolic Engineering. Metab. Eng. 11, 192–198. doi:10.1016/j.ymben.2009.01.005
Zhang, X., Jantama, K., Moore, J. C., Jarboe, L. R., Shanmugam, K. T., and Ingram, L. O. (2009a). Metabolic Evolution of Energy-Conserving Pathways for Succinate Production in Escherichia coli. Proc. Natl. Acad. Sci. 106, 20180–20185. doi:10.1073/pnas.0905396106
Zhang, X., Jantama, K., Shanmugam, K. T., and Ingram, L. O. (2009b). Reengineering Escherichia coli for Succinate Production in Mineral Salts Medium. Appl. Environ. Microbiol. 75, 7807–7813. doi:10.1128/AEM.01758-09
Zhang, Y., Huang, Z., Du, C., Li, Y., and Cao, Z. a. (2009c). Introduction of an NADH Regeneration System into Klebsiella Oxytoca Leads to an Enhanced Oxidative and Reductive Metabolism of Glycerol. Metab. Eng. 11, 101–106. doi:10.1016/j.ymben.2008.11.001
Zhang, Y., Qiu, X., Chen, C., Yu, Z., and Hong, H. (2020). Recent Progress in Microbial Production of Succinic Acid. CIESC J. 71, 1964–1975. doi:10.11949/0438-1157.20191430
Zhang, Z., Aboulwafa, M., and Saier, M. H. (2014). Regulation of Crp Gene Expression by the Catabolite Repressor/Activator, Cra, in Escherichia coli. J. Mol. Microbiol. Biotechnol. 24, 135–141. doi:10.1159/000362722
Zheng, P., Dong, J.-J., Sun, Z.-H., Ni, Y., and Fang, L. (2009). Fermentative Production of Succinic Acid from Straw Hydrolysate by Actinobacillus Succinogenes. Bioresour. Technol. 100, 2425–2429. doi:10.1016/j.biortech.2008.11.043
Zhu, L.-W., and Tang, Y.-J. (2017). Current Advances of Succinate Biosynthesis in Metabolically Engineered Escherichia coli. Biotechnol. Adv. 35, 1040–1048. doi:10.1016/j.biotechadv.2017.09.007
Zhu, L.-W., Xia, S.-T., Wei, L.-N., Li, H.-M., Yuan, Z.-P., and Tang, Y.-J. (2016). Enhancing Succinic Acid Biosynthesis in Escherichia coli by Engineering its Global Transcription Factor, Catabolite Repressor/activator (Cra). Sci. Rep. 6, 36526–36537. doi:10.1038/srep36526
Zhu, L.-W., Zhang, L., Wei, L.-N., Li, H.-M., Yuan, Z.-P., Chen, T., et al. (2015). Collaborative Regulation of CO2 Transport and Fixation during Succinate Production in Escherichia coli. Sci. Rep. 5, 17321–17333. doi:10.1038/srep17321
Keywords: succinic acid, biosynthesis pathways, metabolic engineering, CO2 fixation, NADH/NAD + ratio
Citation: Liu X, Zhao G, Sun S, Fan C, Feng X and Xiong P (2022) Biosynthetic Pathway and Metabolic Engineering of Succinic Acid. Front. Bioeng. Biotechnol. 10:843887. doi: 10.3389/fbioe.2022.843887
Received: 27 December 2021; Accepted: 16 February 2022;
Published: 08 March 2022.
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaReviewed by:
Jiangfeng Ma, Nanjing Tech University, ChinaKatja Bettenbrock, Max Planck Society, Germany
Copyright © 2022 Liu, Zhao, Sun, Fan, Feng and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinjun Feng, ZmVuZ3hqQHFpYmVidC5hYy5jbg==; Peng Xiong, eGlvbmdwQHNkdXQuZWR1LmNu
 Xiutao Liu
Xiutao Liu Guang Zhao
Guang Zhao Shengjie Sun1
Shengjie Sun1 Xinjun Feng
Xinjun Feng