- 1School of Chemical and Biological Engineering, Seoul National University, Seoul, South Korea
- 2Institute of Molecular Biology and Genetics, Seoul National University, Seoul, South Korea
- 3Institute of Engineering Research, Seoul National University, Seoul, South Korea
- 4Bio-MAX/N-Bio Institute, Seoul National University, Seoul, South Korea
- 5Institute for Sustainable Development (ISD), Seoul National University, Seoul, South Korea
Several regiospecific enantiomers of hydroxy-(S)-equol (HE) were enzymatically synthesized from daidzein and genistein using consecutive reduction (four daidzein-to-equol–converting reductases) and oxidation (4-hydroxyphenylacetate 3-monooxygenase, HpaBC). Despite the natural occurrence of several HEs, most of them had not been studied owing to the lack of their preparation methods. Herein, the one-pot synthesis pathway of 6-hydroxyequol (6HE) was developed using HpaBC (EcHpaB) from Escherichia coli and (S)-equol-producing E. coli, previously developed by our group. Based on docking analysis of the substrate or products, a potential active site and several key residues for substrate binding were predicted to interpret the (S)-equol hydroxylation regioselectivity of EcHpaB. Through investigating mutations on the key residues, the T292A variant was verified to display specific mono-ortho-hydroxylation activity at C6 without further 3′-hydroxylation. In the consecutive oxidoreductive bioconversion using T292A, 0.95 mM 6HE could be synthesized from 1 mM daidzein, while 5HE and 3′HE were also prepared from genistein and 3′-hydroxydaidzein (3′HD or 3′-ODI), respectively. In the following efficacy tests, 3′HE and 6HE showed about 30∼200-fold higher EC50 than (S)-equol in both ERα and ERβ, and they did not have significant SERM efficacy except 6HE showing 10% lower β/α ratio response than that of 17β-estradiol. In DPPH radical scavenging assay, 3′HE showed the highest antioxidative activity among the examined isoflavone derivatives: more than 40% higher than the well-known 3′HD. In conclusion, we demonstrated that HEs could be produced efficiently and regioselectively through the one-pot bioconversion platform and evaluated estrogenic and antioxidative activities of each HE regio-isomer for the first time.
Introduction
Anaerobic equol-producing bacteria such as Slackia isoflavoniconvertens, Eggerthella sp. YY7918, and Lactococcus garvieae can convert daidzein (or genistein) into (S)-equol (or (-)-5-hydroxyequol) in the human intestine with a strict enantioselective manner (Uchiyama et al., 2007; Yokoyama and Suzuki, 2008; Matthies et al., 2012). When such gut bacterial metabolites reach their biological target organs or tissues, they function as a selective estrogen receptor modulator (SERM) based on their selective binding affinity for ERβ over ERα (Setchell et al., 2005). The functional phytoestrogen (S)-equol has been clinically verified to be effective against estrogen or testosterone-related health problems including women’s menopausal symptoms, ovarian/prostate cancers, osteoporosis, and even hair loss (Lund et al., 2004; Jackson et al., 2011). However, its metabolism in the human body has not been completely understood yet. R.J. Schwen et al. observed that (S)-equol in rats, monkeys, and humans is mainly metabolized through the 4′-glucuronide conjugated form or 7-sulfated form in minor (Schwen et al., 2012). Otherwise, most researchers believe that it is just diluted and eventually removed along with urine, but a study suggests that hydroxylation of equol in the liver might be another route to discard equol in the human body (Marrian and Haslewood, 1932; Rufer et al., 2006). According to the observation, equol is mainly hydroxylated at C-3′, six or 8 ortho-positions of its inherent hydroxyl groups by enzymes, possibly cytochrome P450s (CYPs) present in the liver microsome.
Phytochemicals comprising multiple phenol groups on their backbone are called ‘polyphenols’. Flavonoids or isoflavonoids are the representatives of the plant polyphenol, and their physiological activities against the human body vary along with the position or number of the hydroxyl groups on their aromatic backbone. For example, one of the major isoflavones in soybean, namely daidzein (7,4′-dihydroxyisoflavone), has moderate antioxidative activity; Also, ortho-dihydroxyisoflavone (ODI) derivatives such as 7,3′,4′-trihydroxyisoflavone (3′-hydroxydaidzein), 6,7,4′-trihydroxyisoflavone (6-hydroxydaidzein) and 7,8,4′-trihydroxyisoflavone (8-hydroxydaidzein) display superior antioxidative characters and unique biological functions caused by their actions on different signaling pathways such as anti-skin cancer (3′-hydroxydaidzein), anti-colon cancer, anti-adipogenesis (6-hydroxydaidzein), and anti-atopic dermatitis (8-hydroxydaidzein) pathways (Park et al., 2008; Park et al., 2010; Lee et al., 2011a; Lee et al., 2011b; Seo et al., 2013; Kim et al., 2014). In order to synthesize such hydroxylated polyphenols efficiently, scientists isolated several microbial enzymes and constructed the respective biocatalytic reaction systems. Those studies harnessed microbial cytochrome P450s, flavin-dependent monooxygenases (FMO), or tyrosinases in their natural or mutated forms for an efficient and regioselective production of plant polyphenols (Pandey et al., 2010; Lee et al., 2012; Lee et al., 2014; Lee S.-H. et al., 2016; Lee et al., 2019). In recent studies, a two-component FMO called HpaBC derived from E. coli or P. aeruginosa has been introduced as another potent candidate to hydroxylate aromatic chemicals. The two component FMO exhibited broad substrate specificities toward plant polyphenols so that naringenin and resveratrol were readily functionalized into eriodictyol (3′-hydroxynaringenin) and piceatannol (3-hydroxyresveratrol), respectively (Furuya and Kino, 2014; Lin and Yan, 2014).
Herein, we focused on the potential estrogenic and antioxidative activity of HEs which had not been studied due to the lack of proper synthetic methods. One study reported a single-step hydroxylation reaction of equol to 3′HE and 6HE, but the percent yields were quite low (<50%) probably due to the over-oxidation arising from adding too many HpaBC-expressing cells (Hashimoto et al., 2019). To overcome such limitations, herein, a set of polyphenol-hydroxylating enzymes from microbial monooxygenases including cytochrome P450, FMO and HpaBC was tested (Figure 1). Among the candidates, EcHpaBC (HpaBC from Escherichia coli) was selected for its relatively high activity toward (S)-equol. Then, site-directed mutagenesis was performed for the key residues identified by ligand-docking analysis to modulate its regioselectivity. Finally, regioselective synthesis of 6-hydroxy-(S)-equol from daidzein was achieved using recombinant equol-producing and equol-hydroxylating strains simultaneously as whole-cell catalysts.
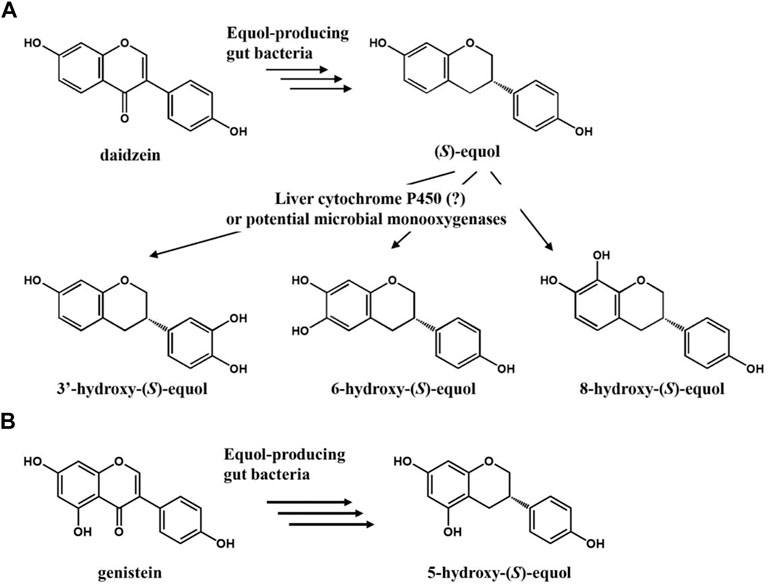
FIGURE 1. (A) Potential scheme of (S)-equol hydroxylation after metabolism of daidzein into (S)-equol by gut bacteria. (B) Metabolism of genistein into 5-hydroxy-(S)-equol by gut bacteria.
Other hydroxyequols including 3′-hydroxy-(S)-equol (3′HE) and 5-hydroxy-(S)-equol (5HE) were also prepared using the same recombinant equol-producing strain (Lee P.-G. et al., 2016; Lee S.-H. et al., 2016; Lee et al., 2017). The synthesized hydroxyequols as well as (S)-equol were subjected to the evaluation of their estrogenic activities using yeast two hybrid (Y2H) assay, which examined dose-responsive agonism of two ERs according to interactions with steroid receptor coactivator 1 (SRC1), and DPPH radical scavenging assay was used for the antioxidative efficacy test.
Results and Discussion
Screening of (S)-Equol-Hydroxylating Monooxygenases
Since no specific oxygenases responsible for hydroxylation of equol have been reported yet, we examined several microbial monooxygenase candidates including cytochrome P450, FMO, and HpaBC. Because of the unfavorable diphenolase activity, tyrosinase was excluded in this study (Lee et al., 2012). CYP102G4 is a self-sufficient microbial cytochrome P450 isolated from Streptomyces cattelya. Similar to other self-sufficient P450s, CYP102G4 directly consumes NAD(P)H as an electron source to activate heme, forming ‘Compound I’ Fe=O (IV) that finally breaks the substrate C-H bond for monooxygenation (Rittle and Green, 2010; Kim et al., 2018). In our previous study, the CYP102G4, was characterized to show a naturally high monooxygenation activity for various polyaromatic substrates including benzophenone and flavone owing to its wide cavity in the active site that enables bulky substrates to easily access the activated heme species (Kim et al., 2018). Whole-cells expressing CYP102G4, however, gave poor conversion of (S)-equol (ca. < 1%) with minor detection of 3′-hydroxy-(S)-equol (3′HE), 6-hydroxy-(S)-equol (6HE), and 8-hydroxy-(S)-equol (8HE) as reaction products (Figure 2 and Figure 3).
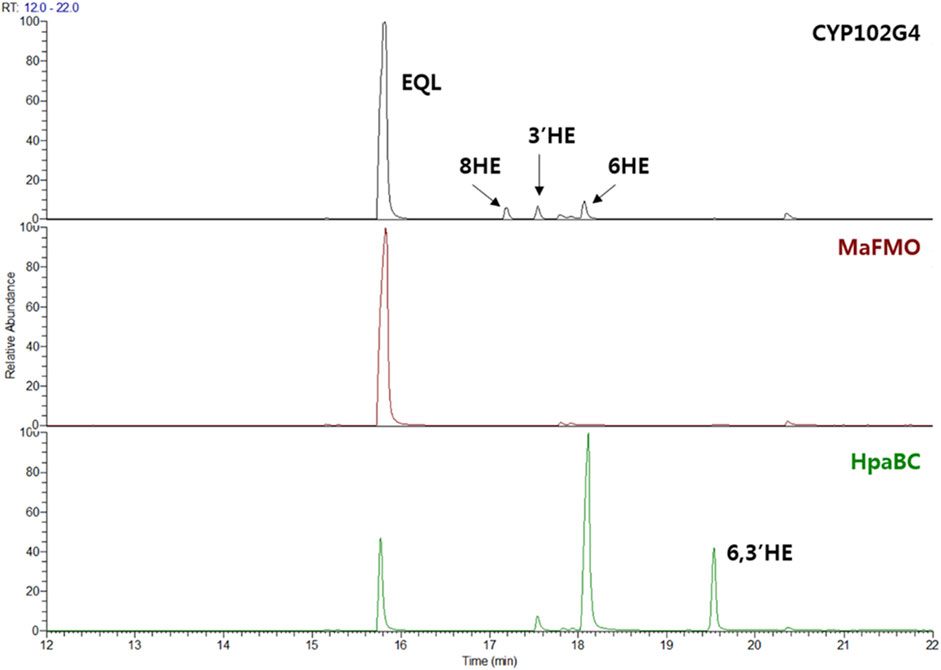
FIGURE 2. (S)-equol (EQL) monooxygenation assay using whole-cells expressing three microbial monooxygenases: CYP102G4, MaFMO, and EcHpaBC. 200 μM of (S)-equol was initially used for the whole-cell (OD600 = 10) biotransformation, and reaction extracts at 4 h were analyzed using GC-MS.
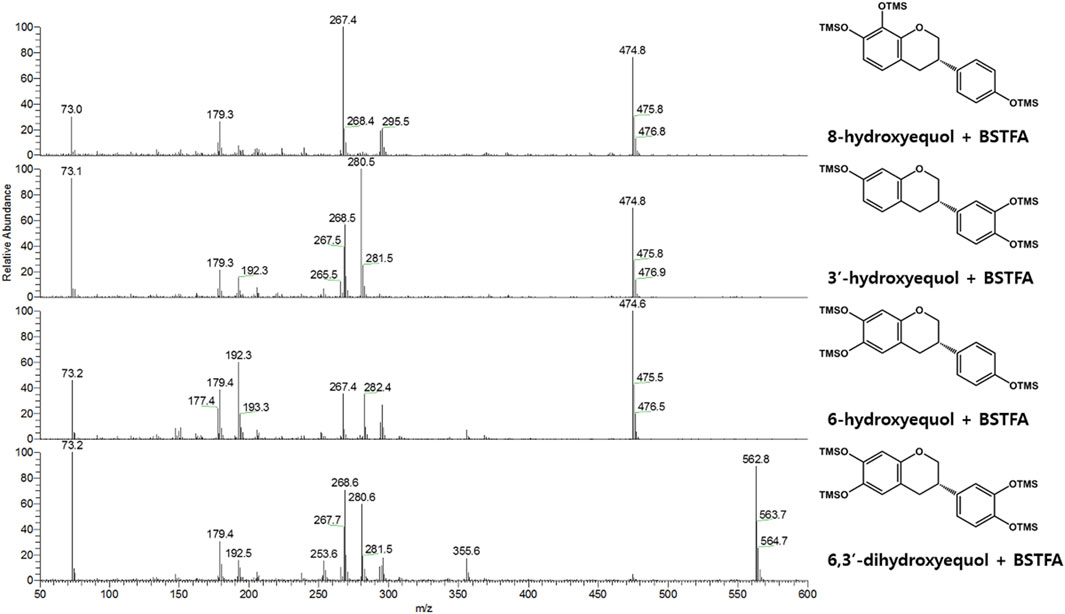
FIGURE 3. EI-mass spectra (BSTFA: N, O-bis(trimethylsilyl)trifluoroacetamide) of trimethylsilylated hydroxy-(S)-equols biosynthesized by CYP0102G4 and EcHpaBC. 3′-hydroxyequol and 6-hydroxyequol were detected in both enzyme reaction samples, while 8-hydroxyequol and 6,3′-dihydroxyequol were detected in the reaction extracts of CYP102G4 and EcHpaBC, respectively.
Next, flavin-containing monooxygenases (FMOs) are potent catalysts to prepare hydroxylated phytochemicals. In former studies, monooxygenation of daidzein, naringenin, and stilibene was achieved using FMOs from various microbial sources (Lee et al., 2014). We selected an FMO derived from Methylophaga aminisulfidivorans (MaFMO), which was mainly examined for hydroxylation of indole to form indigo, an aromatic natural dye (Han et al., 2008). Although MaFMO has been shown to exhibit superior activity for indole, no detectable monooxygenation activity for (S)-equol was found (Figure 2). Instead, we paid attention to a two-component FAD-dependent monooxygenase called EcHpaBC that was previously identified as a 4-hydroxyphenylacetate 3-monooxygenase in E. coli (Prieto and Garcia, 1994). The two-component FMO showed excellent hydroxylation activity for plant-derived polyphenols such as naringenin, resveratrol, and afzelechin with high efficiency (Lin and Yan, 2014; Jones et al., 2016). Interestingly, EcHpaBC-expressing whole cells resulted in remarkable consumption of (S)-equol (for OD600 = 10, ca. 79% conversion in 4 h) and generated three products with single or multiple hydroxyl group(s), which were finally identified as 6HE, 3′HE, and 6,3′-dihydroxy-(S)-equol (6,3′diHE) based on GC-MS analysis (Figure 2 and Figure 3). The EI-MS fragmentation patterns of the hydroxyequols (HEs) corresponded to those of previously identified products, and the fragment ions of m/z 280 and 295 generated from m/z 562 mother peak in the 6,3′diHE mass spectrum indicated the presence of hydroxyl groups at C3’ (B-ring) and C6 (A-ring), respectively (Rufer et al., 2006).
The unexpected high conversion rate of EcHpaBC from enterobacterium, E. coli, for (S)-equol led us to assume that the enzyme might be potentially involved in isoflavone metabolism in the human intestine as well as biodegradation of aromatic compounds by E. coli (Diaz et al., 2001). However, a BLAST search of EcHpaB for the human microbiome database (MetaQuery) revealed that the abundance and prevalence of HpaB in the human gut microbiome was only 0.089 and 33.19%, respectively, whereas those of daidzein reductase from Slackia isoflavoniconvertens were 0.26 and 100%, respectively. Even though several homologous HpaB proteins were observed in Proteobacteria including genus Escherichia, Klebsiella, Enterobacter, and Providencia with >50% sequence identity, the apparent low occurrence of HpaB might lead to infrequent detection of such HEs in the gut. Otherwise, generation of HEs might occur in the human liver via human cytochrome P450(CYP) enzymes, and the produced HEs would be estrogenic ligands such as (S)-equol or biologically active compounds with different roles from the parent compound (Rufer et al., 2006).
Modulation of Regioselectivity of HpaB
According to the EcHpaBC-WT whole-cell reaction profile, EcHpaBC catalyzed (S)-equol into 6HE and 3′HE as major and minor products, respectively, and then, the mono-hydroxylated HEs were subjected to consecutive second hydroxylation, being converted into 6,3′diHE. To interpret the regioselectivity of EcHpaB, biochemical regiospecific analysis of the reaction products was carried out using protein–ligand docking analysis. The two-component HpaBC consists of HpaB, a catalytic component containing flavin adenine dinucleotide (FAD) as a prosthetic group, and HpaC, a NAD(P)H:flavin oxidoreductase component that regenerates FADH2 for complete oxidation of the substrate and reduction of molecular oxygen by HpaB (Kim et al., 2007). In a recent report, the well-known broad substrate specificity of EcHpaB toward various aromatic phytochemicals was explained by identifying the crystal structure (PDB code: 6QYI) (Deng et al., 2020). As proposed earlier, FAD(H2) freely associates and dissociates in between HpaB and HpaC to complete a cycle of the catalysis, explaining a loosely bound FAD in the active site of EcHpaB (Kim et al., 2007; Deng et al., 2020). Arg164 and Arg333 stabilize diphosphate and adenine moieties, respectively, by constructing a substrate binding site in a groove of EcHpaB. The possible substrate binding site on the re face (over flavin) of FAD, partially exposed to the protein surface, was predicted from the binding mode of 4-hydrophenylacetate in Thermus thermophilus HB8 HpaB (PDB code: 2YYI) (Kim et al., 2007). Based on the hypothesis that (S)-equol would bind to the same re face of FAD, energetically minimized (S)-equol was docked into the substrate binding region, which resulted in highly probable two binding modes of (S)-equol for 3′ and 6-hydroxylation with binding energy -7.6 and -7.4 kcal/mol, respectively (Figure 4). This computational prediction intuitively explains the production of both 3′HE and 6HE by EcHpaBC. Moreover, the generation of another major product of EcHpaBC, 6,3′diHE, could be rationalized by assuming a potential binding of 6HE in the binding pocket for the second hydroxylation at C-3′ and vice versa with low probability (Supplementary Figure S1). In the predicted bindings, virtual interactions between the bound substrate and enzyme residues were observed at R113, Y117, H155, I157, T292, and Y301. Among them, R113 and Y117 are the key residues to possibly stabilize 4′- or 7-OH of (S)-equol by hydrogen bondings in the respective two binding modes. The other residues might be involved in hydrophobic interactions with the aromatic substrate backbone.
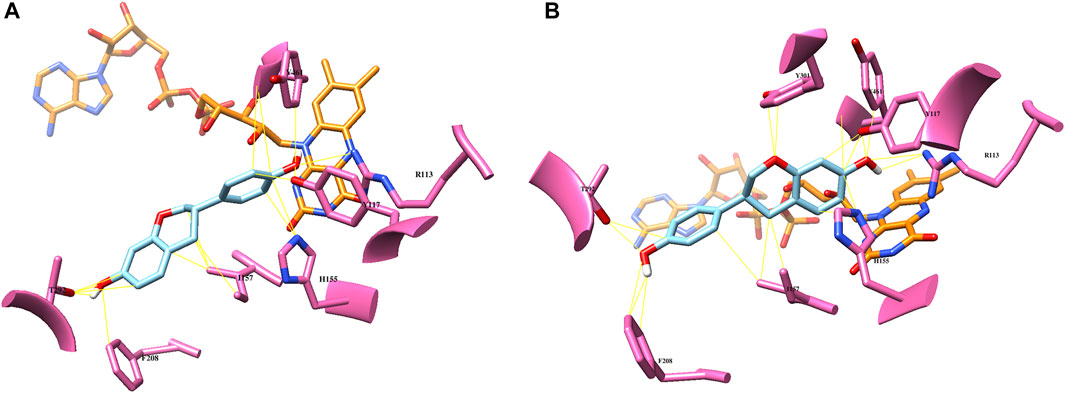
FIGURE 4. Computational docking analysis of (S)-equol (cyan) of the crystal structure of EcHpaB (PDB code: 6QYI) (A): binding mode of 3′-hydroxylation (B): binding mode of 6-hydroxylation. Potential substrate binding residues are highlighted with pink, and the FAD cofactor is shown as orange.
Site-Directed Mutagenesis of HpaB to Enhance Its Regioselectivity
Given the possible binding mode of (S)-equol to EcHpaB, we investigated the role of several active site residues that might determine the regioselectivity of EcHpaB toward (S)-equol. Because the native regioselectivity of EcHpaB favored 6-hydroxylation of (S)-equol, to generate 6HE as a sole product, we first focused on the effect of the residues contributing to generate 3′HE or 6,3′diHE as minor or major products, respectively. If the residues stabilize the binding mode of (S)-equol for 3′-hydroxylation, any possible mutations on the residues might reduce the hydroxylation activity at C3′ at (S)-equol (via losing hydrogen bonding), while maintaining hydroxylation activity at C6. Because we did not want to lose the robust (S)-equol hydroxylation activity of EcHpaB, the residues allowing hydrogen bonds or hydrophobic interactions with (S)-equol in both binding modes were excluded from the mutation candidates. An interaction between O1 of 6HE and Y301 was the sole difference between the two binding modes. However, mutation of Y301 might deteriorate the activity of EcHpaB on 6HE because Y301 seems to stabilize the binding of 6HE via hydrogen bonding.
Instead, we paid attention to T292 potentially forming a hydrogen bond with 6-OH when 6HE was docked. T292 appeared to contribute more stability to the binding of 6HE than (S)-equol due to hydroxylation at C3’ (Supplementary Figure S1). In order to confirm our predictions, the T292A mutant was constructed and evaluated by using whole-cell assay. The mutant showed no detectable production of 3′HE compared to WT, but yielded 6HE as the major product (ca. 98%) and significantly diminished the production of 6,3′diHE (Figure 5), suggesting that the binding of (S)-equol or 6HE to wild-type EcHpaB for hydroxylation at C3′ is likely to be stabilized by possible interactions with T292. Especially, hydrogen bonds between 6-OH of 6HE and T292-OH could explain the diminished production of 6,3′diHE by EcHpaB T292A.
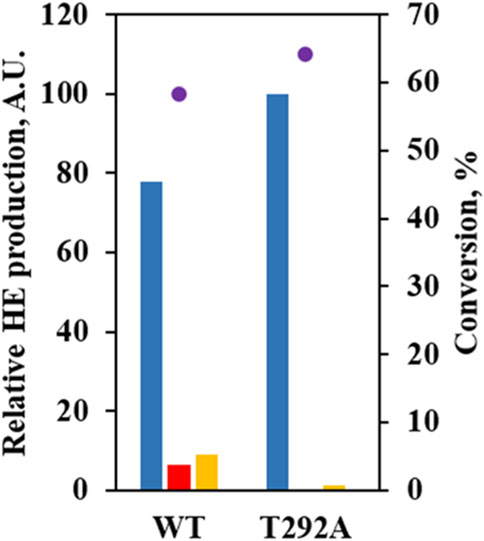
FIGURE 5. Mutational study to investigate the role of T292 in (S)-equol hydroxylations. Relative abundances of 6HE (blue bar), 3′HE (red bar), and 6,3′diHE (yellow bar) were shown with (S)-equol conversion at 4 h (purple dot) for EcHpaBC-WT and T292A. The 6HE production level was arbitrarily set to 100, and relative concentrations of the other HEs were estimated with peak areas on gas chromatogram.
One-Pot Synthesis of Ortho-hydroxylated (S)-Equols From Daidzein
In previous studies, we suggested that an equol-producing recombinant E. coli strain that converted daidzein into (S)-equol would be a useful biocatalytic platform to prepare various equol analogs because the whole-cell biotransformation system of the transformant, tDDDT tolerated aerobic production of equol from daidzein (or 5-hydroxyequol from genistein) and displayed a high yield (g/L titer) and productivity (Lee P.-G. et al., 2016; Lee et al., 2017; Lee et al., 2018). It would be fascinating to synthesize novel HEs rarely found in the human body, but with unknown new biological roles, from common soy isoflavone daidzein. The previous development of the whole-cell catalysts and plasmids drove us to investigate the possibility of a combined use of equol and HE producing strains developed in this study in one-pot reaction. Then, one E. coli strain could perform the series of reduction steps of daidzein to (S)-equol, and the other strain could subsequently oxidize (S)-equol into HEs. The concept has proven to be valid in combinatorial whole-cell biotransformation exploiting tDDDT and the transformant with tEcHpaBC-T292A strains as biocatalysts (Figure 6). All the initial amount of daidzein (ca. 1 mM) was consumed in 4 h, converted into (S)-equol rapidly, and then finally 0.95 mM of 6HE (ca. 244 mg/L) was synthesized in a highly regioselective manner. During the whole-cell biotransformation, no significant intermediates or byproducts were detected. This interesting finding shows that EcHpaBC monooxygenase selectively reacts with (S)-equol rather than other isoflavone intermediates including daidzein, dihydrodaidzein, and tetrahydrodaidzein, which helped in maintaining the regioselectivity of EcHpaBC-T292A in the microbial cells. This is the first study of one-pot synthesis involving oxidation and reduction of isoflavones using two individual whole cells. Because isoflavone derivatives including equol freely pass through the cell membrane, the two whole-cell compartments can perform simultaneous oxidoreductions of isoflavone with high efficiency (Lee et al., 2017). As a result, it becomes an efficient biocatalytic platform to prepare diverse equol derivatives including hydroxyequols or other chemically modified equols if active equol-catalyzing enzymes are provided. In addition, this platform is an economical choice for producing invaluable hydroxyequol derivatives, for example, the price of non-derivatized (S)-equol (>97%, in Sigma, St. Louis, MO, US at 01/10/2022) is $329/25 mg which is about 4.5 fold higher than daidzein (>98%, in Sigma at 01/10/2022, $73.5/25 mg).
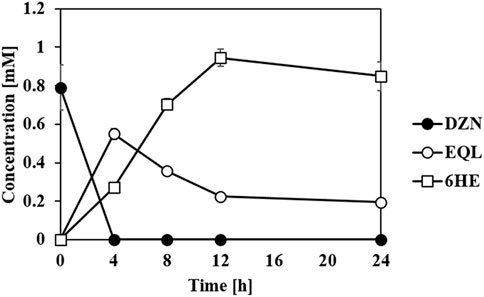
FIGURE 6. Whole-cell biotransformation of daidzein (DZN) into (S)-equol (EQL) and subsequently to 6HE. The tDDDT strain (previously developed by Lee et al. (2016)) and tEcHpaBC-T292A strain set up in one-pot reaction catalyzed daidzein (initial concentration: 1 mM) into equol (reduction step), and then subsequently into 6HE (oxidation step) in a highly regioselective manner. All the data were recorded using mean values with standard deviations for triplicates.
Estrogenic Efficacies of (S)-Equol and Hydroxyequol Derivatives
With the aid of tDDDT whole-cell biotransformation, the other HEs including 3′HE and 5HE were synthesized through the conversion of corresponding isoflavones and purified to evaluate their estrogenic efficacies. The yeast-two-hybrid (Y2H) systems between ERs and SRC1 were exploited and agonistic activities of the estrogenic compounds synthesized throughout this study were measured (Supplementary Figure S3 and Figure 7). In these systems, EC50s of 17β-estradiol (E2) were about 0.2 nM for two ER isotypes with a slight potency preference on ERβ. Genistein and (S)-equol had about 3-orders of magnitude weaker estrogenic activity both on ERα and ERβ than E2 but had SERM activity with the preference to ERβ with the β/α ratio of 1.25 and 1.31, respectively. 3′ and 6HE showed much lower activities than the other examined compounds having their EC50s with concentrations in the range of 10–5∼10–4 M and had weaker SERM efficacies than (S)-equol displaying β/α ratios of 1.15 and 1.00, respectively, indicating that 6HE is a slightly ERα preferring SERM compared to E2. 5HE did not show any response with concentrations in the range of 10–8∼10–3 M. However, a previous report demonstrated that 5HE had a higher binding affinity to ERα than ERβ, suggesting that 5HE is an ERα-selective antagonist (Lee et al., 2017). In general, all tested HEs did not show prominent estrogenic activity, which suggests that equol loses its estrogenic activity through metabolism in the liver (Rufer et al., 2006).
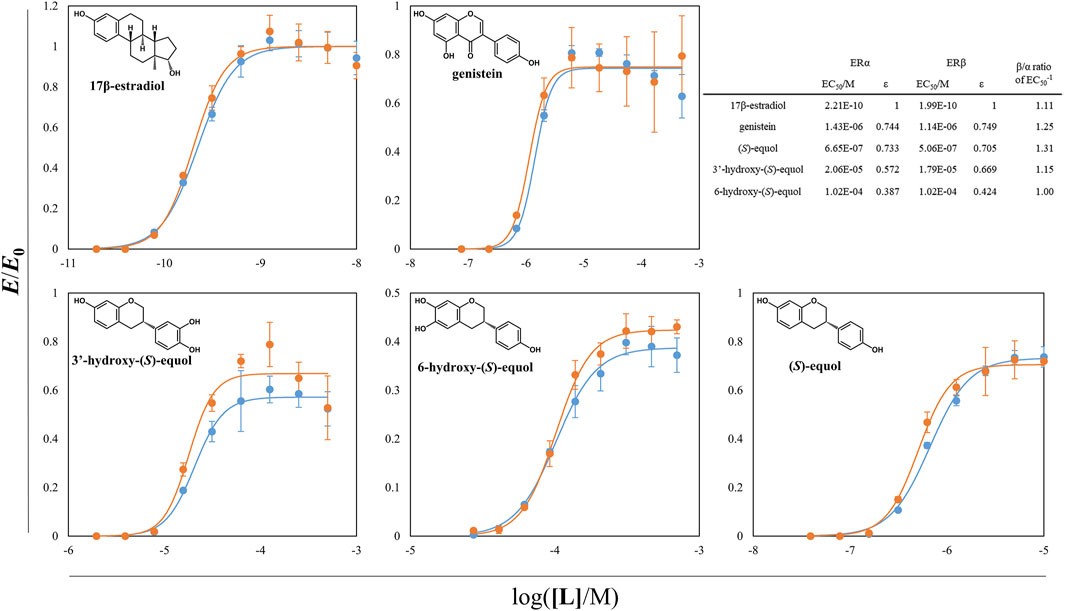
FIGURE 7. ER-SRC1 agonistic Y2H assay of equol derivatives and the other estrogenic compounds. ß-galactosidase activity of yeast L40 cells harboring ERα (or ERβ) LBD hybrid and tSRC1 hybrid protein was assessed. The normalized responses of ERα-SRC1 and ERβ-SRC1 interaction were expressed as blue and orange circles, respectively. The lines with two different colors were the regressed values. The inward table is the summary of EC50s, εs, and β/α ratios of each compounds. All the data were recorded using mean values with standard deviations for triplicates.
Antioxidative Effect of Hydroxyequol Derivatives
Antioxidative effects of the HEs and the prominent antioxidants were assessed through DPPH radical scavenging assay (Figure 8). Among the examined compounds, the compounds with a catechol moiety showed better radical scavenging activity even among the equol derivatives. Especially, all the HEs of interest had EC50 values in the range of 7–11 μM and had superior scavenging activities than 3′HD which is a well-known antioxidative isoflavone. 5HE and 3′HE had lower EC50 values than their original isoflavones, suggesting that the backbone of isoflavanes tends to be more potent direct antioxidants than the backbone of isoflavones.
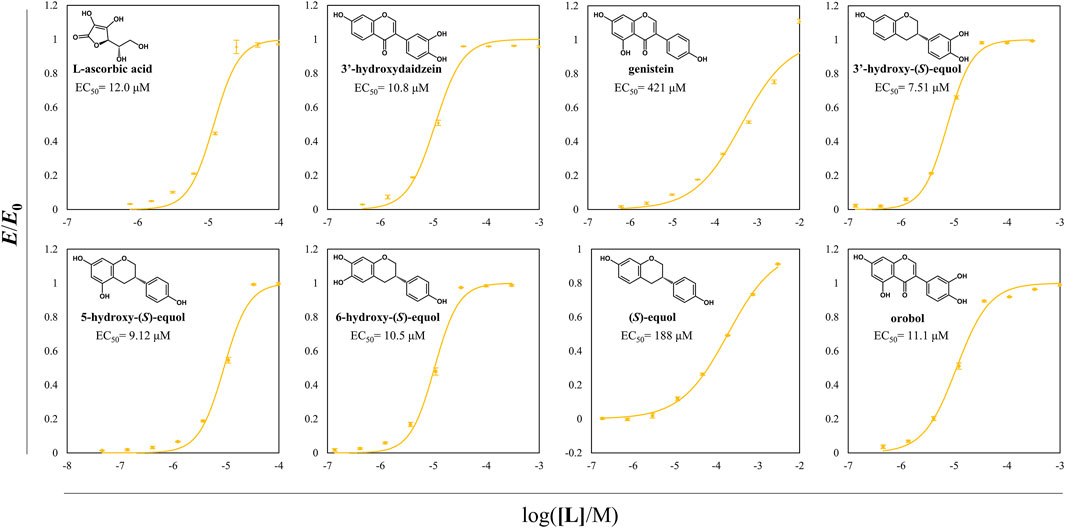
FIGURE 8. DPPH radical scavenging assay of probable direct antioxidative compounds including HEs. DPPH radical scavenging activity of direct antioxidants was assessed according to the changes in the value of OD520/OD495. EC50 of DPPH radical scavenging of each compound was written in the plots. The normalized OD ratio change values were expressed as yellow circles, and the lines of yellow color were the regressed values. All the data were recorded using mean values with standard deviations for triplicates.
Conclusion
Despite the possible presence of HEs in the human body, the lack of the synthetic methods of HE has prohibited us from pursuing further studies on their biological roles in hormonal regulation or intended nutritional effect in the human gut microbiome. In this study, we demonstrated that EcHpaBC exhibited significant catalytic activity toward (S)-equol (a dominant form of equol enantiomer in the gut), generating 6HE and 6,3′diHE as major products. To enhance the yield of regioselective synthesis of 6HE from (S)-equol or daidzein, protein engineering of EcHpaBC using the semi-rational approach, such as ligand-docking analysis and site-directed mutagenesis on the selected target residues, was carried out. Based on this approach, we achieved a 95% yield of 6-hydroxy-(S)-equol in 4 h from 1 mM daidzein with a one-pot synthesis procedure. Along with 6HE, the other HEs such as 3′HE and 5HE were synthesized from daidzein and genistein and were assessed for ER binding efficacy and antioxidant efficiency. This work provides valuable information on the construction of an efficient biocatalytic platform to prepare diverse HEs and evaluation of the synthesized HE compounds for ER-binding assays, for the first time. On the basis of our findings, other studies such as the evaluation of HEs’ antagonism and/or in vivo testing for exploring novel activities of HEs could be supported, and a facile one-pot high-titer production of another equol derivatives is anticipated.
Experimental Methods
Chemicals. Daidzein and genistein were purchased from Sigma Aldrich, and (S)-equol was purchased from Tocris Bioscience. l-Ascorbic acid was purchased from Junsei. 2,2-Diphenyl-1-(2,4,6-trinitrophenyl)hydrazin-1-yl (DPPH) radical and 17β-estradiol (E2) were purchased from Sigma Aldrich. 3′-Hydroxydaidzein and orobol were prepared from daidzein and genistein respectively using the reported method (Lee S.-H. et al., 2016).
Microbial strains. Cloning information and used primers for the construction of EcHpaBC and MaFMO encoded vectors are listed in Supplementary Table S1. In brief, a two-component flavin-dependent monooxygenase, EcHpaBC, was newly cloned from the genomic DNA of Escherichia coli ATCC 8739, and MaFMO was cloned from the genomic DNA of Methylophaga aminisulfidivorans. The vector encoding CYP102G4 was previously constructed (Kim et al., 2018). Respective E. coli BL21 (DE3) transformants were verified for the soluble expression in SDS-PAGE (Supplementary Figure S2) and were then used as microbial strains for the whole-cell biotransformation. Three EcHpaB mutants were made by site-directed mutagenesis using the listed overlapping primer-containing intended mutations. Saccharomyces cerevisiae L40 (MATa his3-200 trp1-901 leu2-3,112 ade2 lys2-801a.m. URA3(lexAop)8- lacZ LYS2(lexAop)4-HIS3) strain was utilized as a host for yeast-two-hybrid (Y2H) assays. The bait hybrid proteins were cloned in the pBTM116 vector as fusions of LexA DNA-binding domain (DBD)-human estrogen receptor a ligand-binding domain (ERα LBD, 312–595) or LexA DBD-ERβ LBD (264–510). The prey hybrid protein was cloned in the pVP16 vector as a fusion of the VP16 activator domain (AD)-(Gly4Ser)2 linker-truncated steroid receptor coactivator 1 (tSRC1) (613–773). These sets of two vectors (pBTM116:LexA DBD-ERα LBD & pVP16:VP16 AD-(Gly4Ser)2-tSRC1 or pBTM116:LexA DBD-ERβ LBD & pVP16:VP16 AD-(Gly4Ser)2-tSRC1) were transformed in the L40 strains.
Whole-cell biotransformation (GC-MS analysis). The aforementioned E. coli transformants were grown overnight, 1 ml of which was transferred to 50 ml of LB containing appropriate antibiotics. When the optical density (OD600) of the cells reached 0.6 to 0.8, addition of 0.1 mM isopropyl-thio-β-d-galactopyranoside (IPTG) or 0.5 mM 5-aminolevulinic acid for heme formation of CYP102G4 induced the heterologous expression of monooxygenases overnight at 18°C. The monooxygenase-expressing cells were harvested via centrifugation (4°C, 4,000 rpm; 3,480 rcf for 10 min), washed with phosphate buffer saline (0.5x volume), and then centrifuged again for subsequent whole-cell catalysis.
The prepared whole cells were resuspended in the reaction solution (final OD600 = 10, total volume = 5 ml) composed of 0.1 M potassium phosphate buffer (KPB) pH 7.0, 1% (w/v) glucose, 5 mM l-ascorbic acid, and 0.2 mM (S)-equol. Whole-cell biotransformation was performed in the baffled flask shaken by 180 rpm at 30°C. In the case of consecutive oxidoreduction of daidzein into HEs, the reaction condition was slightly modified by referring to apreviously reported study (0.2 M KPB pH 8.0, 2% (w/v) glucose, 1% (v/v) glycerol, and polyvinylpyrrolidone 1% (w/v) in a 140 rpm shaking incubator) (Lee et al., 2018). The equol-producing whole-cells were also prepared following the previously reported method (Lee P.-G. et al., 2016).
Preparation of hydroxyequols. 6-Hydroxyequol (6HE) was synthesized with the aforementioned method. 5HE was synthesized with the previously reported method using genistein as the initial substrate (Lee P.-G. et al., 2016; Lee et al., 2017). 3′HE was synthesized using the same method from 3′-hydroxydaidzein which was prepared by ortho-hydroxylation of the tyrosianse derived from Bacillus megaterium with borate chelation (Lee P.-G. et al., 2016; Lee S.-H. et al., 2016). All of the isoflavonoids were purified through EA extraction, HPLC preparation (Econosil C18 prep column, 10 μm, 22 × 250 mm; Alltech, United States), and freeze-drying. The final purity of 6HE, 3′HE, and 5HE was confirmed with GC-MS or HPLC and 1H-NMR analysis.
6-Hydroxy-(S)-equol: 1H-NMR [(CD3)2SO, 400 MHz] δ 2.73 (m, 2H, H-4), 2.98 (m, 1H, H-3), 3.81 (t, J = 10.4 Hz, 1H, H-2β), 4.05 (m, 1H, H-2α), 6.19 (s, 1H, H-8), 6.44 (s, 1H, H-5), 6.71 (d, J = 8.4 Hz, 2H, H-3′), 7.09 (d, J = 8.4 Hz, 2H, H-2’), 8.26 (s, 1H, OH), 8.69 (s, 1H, OH), and 9.26 (s, 1H, OH).
5-Hydroxy-(S)-equol: 1H-NMR [(CD3)2SO, 400 MHz] δ 2.47 (dd, J = 16.1, 10.9 Hz, 1H, H-4β), 2.73 (ddd, J = 16.1, 5.5, 1.4 Hz, 1H, H-4α), 2.95 (m, 1H, H-3), 3.83 (t, J = 10.4 Hz, 1H, H-2β), 4.10 (dq, J = 10.4, 1.6 Hz, 1H, H-2α), 5.70 (d, J = 2.3 Hz, 1H, H-6), 5.90 (d, J = 2.3 Hz, 1H, H-8), 6.72 (d, J = 8.5 Hz, 2H, H-3′), 7.11 (d, J = 8.5 Hz, 2H, H-2’), 8.93 (s, 1H, OH), 9.17 (s, 1H, OH), and 9.25 (s, 1H, OH).
3′-Hydroxy-(S)-equol: 1H-NMR [(CD3)2SO, 400 MHz] δ 2.78 (m, 2H, H-4), 2.94 (m, 1H, H-3), 3.87 (t, J = 10.5 Hz, 1H, H-2β), 4.15 (m, 1H, H-2α), 6.19 (d, J = 2.4 Hz, 1H, H-8), 6.29 (dd, J = 8.2, 2.4 Hz, 1H, H-6), 6.56 (dd, J = 8.2, 2.2 Hz, 1H, H-6′), 6.67 (d, J = 2.3 Hz, 1H, H-2′), 6.69 (d, J = 8.2 Hz, 1H, H-5′), 6.87 (d, J = 8.2 Hz, 1H, H-5), 8.78 (s, 2H, OH), and 9.14 (s, 1H, OH).
Ligand-docking simulation. AutoDock Vina 1.1.2 (Trott and Olson, 2010) predicted docking modes of (S)-equol or 6HE in the reported EcHpaB crystal structure (PDB code: 6QYI) (Deng et al., 2020), which were visualized by using Chimera 1.15.
Bioinformatic analyses. MetaQuery, an open-source web server for quantitative analysis of a given gene in the human gut microbiome, was utilized to calculate the abundance and prevalence of HpaB (from E. coli) and daidzein reductase (from Slakia isoflavoniconvertens, reference) in a human gut microbiome database (Nayfach et al., 2015). The minimum percent identity/maximum E-value/query alignment coverage/target alignment coverage were set to 30%/1e-5/70%/70% (web default values), respectively.
Yeast-two-hybrid assay. The transformed L40 strains were grown in synthetic leucine and tryptophan drop-out (SD L−W-) media. In 96-deep well plates, each well with the cultural medium of 500 μL SD L−W- contained initial OD600 = 0.1 of L40 preculture, 2% (v/v) DMSO, and specific amount of isoflavonoids or 17β-estradiol (E2). These plates were incubated at 30 °C with shaking of 1,000 rpm for overnight (∼20 h). After the incubation, the cultures were centrifuged, and the supernatants were discarded. In each well, 500 μL Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 1 mM MgSO4; pH 7.0) with 1 mM dithiothreitol (DTT) was filled and resuspended. 50 μL of each suspension was taken for OD600 measurement before chloroform treatment. (This value was not used for the measurement of the Miller unit.) Chloroform 50 μL was added to each 450 μL of the suspension, and the solution mixture was shaked for 1 min at 1,000 rpm to disrupt the cell membrane. The disrupted suspensions were kept still at room temperature for 2 min, and 50 μL aqueous aliquots were taken. In polystyrene flat-bottomed 96-well plate, each well was charged with 100 μL of Z-buffer with 1 mM DTT, 50 μL aqueous aliquot, and 50 μL of Z-buffer with 1 mM DTT and 0.4 mg/ml ortho-nitrophenyl-β-galactoside (ONPG). Initially, OD600 of all the wells was measured, and subsequently, OD420 of the same wells was measured for 30 min with a minute interval. The Miller units (dOD420/dt/OD600) of all the wells were calculated and normalized by using the value of the positive control (10 nM E2). The data were triplicated, and the points of average values were regressed using the following formula (Buchwald, 2020).
where E/E0 is the normalized Miller unit and ε is the maximum response relative to E2 (ε(E2) = 1). [L] is the concentration of ligands and n is the Hill’s coefficient. EC50 is the value of effective concentration at E/E0 = 0.5ε.
DPPH radical scavenging assay. In each well of polystyrene flat-bottom 96-well plates, 200 μL of solution containing 0.1 mM 2,2-diphenyl-1-(2,4,6-trinitrophenyl)hydrazin-1-yl (DPPH) radical, 50% (v/v) MeOH, 10% (v/v) DMSO, specific amount of compounds, and 1x concentration of phosphate buffered saline (PBS) was made. The PBS was prepared with 4x concentration of its original recipe with pH 7.4 and diluted by adding 50 μL in the solution mixture. The plates were incubated at 37°C for 30 min. The OD520 and OD495 of incubated plates were measured. The radical scavenging activity was defined as (OD520/OD495)DMSO- OD520/OD495. (OD520/OD495)DMSO is the OD520/OD495 of the DMSO control without antioxidative compounds. The radical scavenging activity values were normalized with the value of the positive control (1 mM of l-ascorbic acid). The data were triplicated, and the average values were regressed using the following formula.
where E/E0 is the normalized radical scavenging activity, [L] is the concentration of compounds, and n is the Hill’s coefficient, and EC50 is the value of effective concentration at E/E0 = 0.5.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author Contributions
HS and P-GL: conceptualization, experimentation, data curation, formal analysis, validation, and writing—original draft. JK: experimentation, data curation, formal analysis and writing—review, and editing. JK and S-HL: data curation and writing—review and editing. HK, U-JL, and JK: writing—review and editing. E-JK: experimentation, data curation, and formal analysis. Byung-gee Kim: conceptualization, supervision, project administration, writing—review, and editing.
Funding
This research was supported by the Korean Medical Device Development Fund grant funded by the Korean government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health and Welfare, and the Ministry of Food and Drug Safety) (Project Number. KMDF_PR_20200901_0151) and the Korean Fund for Regenerative Medicine (KFRM) grant funded by the Korean government (the Ministry of Science and ICT and the Ministry of Health and Welfare) (21A0301L1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.830712/full#supplementary-material
References
Buchwald, P. (2020). A Single Unified Model for Fitting Simple to Complex Receptor Response Data. Sci. Rep. 10 (1), 13386. doi:10.1038/s41598-020-70220-w
Deng, Y., Faivre, B., Back, O., Lombard, M., Pecqueur, L., and Fontecave, M. (2020). Structural and Functional Characterization of 4‐Hydroxyphenylacetate 3‐Hydroxylase from Escherichia coli. Chembiochem 21 (1-2), 163–170. doi:10.1002/cbic.201900277
Dı́az, E., Ferrández, A., Prieto, M. A., and Garcı́a, J. L. (2001). Biodegradation of Aromatic Compounds by Escherichia coli. Microbiol. Mol. Biol. Rev. 65 (4), 523–569. doi:10.1128/MMBR.65.4.523-569.2001
Furuya, T., and Kino, K. (2014). Regioselective Synthesis of Piceatannol from Resveratrol: Catalysis by Two-Component Flavin-dependent Monooxygenase HpaBC in Whole Cells. Tetrahedron Lett. 55 (17), 2853–2855. doi:10.1016/j.tetlet.2014.03.076
Han, G. H., Shin, H.-J., and Kim, S. W. (2008). Optimization of Bio-Indigo Production by Recombinant E. coli Harboring Fmo Gene. Enzyme Microb. Technology 42 (7), 617–623. doi:10.1016/j.enzmictec.2008.02.004
Hashimoto, T., Nozawa, D., Mukai, K., Matsuyama, A., Kuramochi, K., and Furuya, T. (2019). Monooxygenase-catalyzed Regioselective Hydroxylation for the Synthesis of Hydroxyequols. RSC Adv. 9 (38), 21826–21830. doi:10.1039/c9ra03913a
Jackson, R. L., Greiwe, J. S., and Schwen, R. J. (2011). Emerging Evidence of the Health Benefits of S-Equol, an Estrogen Receptor β Agonist. Nutr. Rev. 69 (8), 432–448. doi:10.1111/j.1753-4887.2011.00400.x
Jones, J. A., Collins, S. M., Vernacchio, V. R., Lachance, D. M., and Koffas, M. A. G. (2016). Optimization of Naringenin Andp-Coumaric Acid Hydroxylation Using the nativeE. Colihydroxylase Complex, HpaBC. Biotechnol. Prog. 32 (1), 21–25. doi:10.1002/btpr.2185
Kim, H., Kim, J. R., Kang, H., Choi, J., Yang, H., Lee, P., et al. (2014). 7,8,4′-Trihydroxyisoflavone Attenuates DNCB-Induced Atopic Dermatitis-like Symptoms in NC/Nga Mice. PloS one 9 (8), e104938. doi:10.1371/journal.pone.0104938
Kim, J., Lee, P.-g., Jung, E.-o., and Kim, B.-G. (2018). In Vitro characterization of CYP102G4 from Streptomyces Cattleya: A Self-Sufficient P450 Naturally Producing Indigo. Biochim. Biophys. Acta (BBA) - Proteins Proteomics 1866 (1), 60–67. doi:10.1016/j.bbapap.2017.08.002
Kim, S.-H., Hisano, T., Takeda, K., Iwasaki, W., Ebihara, A., and Miki, K. (2007). Crystal Structure of the Oxygenase Component (HpaB) of the 4-hydroxyphenylacetate 3-monooxygenase from Thermus Thermophilus HB8. J. Biol. Chem. 282 (45), 33107–33117. doi:10.1074/jbc.M703440200
Lee, D. E., Lee, K. W., Byun, S., Jung, S. K., Song, N., Lim, S. H., et al. (2011a). 7,3′,4′-Trihydroxyisoflavone, a Metabolite of the Soy Isoflavone Daidzein, Suppresses Ultraviolet B-Induced Skin Cancer by Targeting Cot and MKK4. J. Biol. Chem. 286 (16), 14246–14256. doi:10.1074/jbc.M110.147348
Lee, D. E., Lee, K. W., Jung, S. K., Lee, E. J., Hwang, J. A., Lim, T.-G., et al. (2011b). 6,7,4'-trihydroxyisoflavone Inhibits HCT-116 Human colon Cancer Cell Proliferation by Targeting CDK1 and CDK2. Carcinogenesis 32 (4), 629–635. doi:10.1093/carcin/bgr008
Lee, H., Kim, B.-G., and Ahn, J.-H. (2014). Production of Bioactive Hydroxyflavones by Using Monooxygenase from Saccharothrix Espanaensis. J. Biotechnol. 176, 11–17. doi:10.1016/j.jbiotec.2014.02.002
Lee, N., Kim, E. J., and Kim, B.-G. (2012). Regioselective Hydroxylation of Trans-resveratrol via Inhibition of Tyrosinase from Streptomyces Avermitilis MA4680. ACS Chem. Biol. 7 (10), 1687–1692. doi:10.1021/cb300222b
Lee, P.-G., Kim, J., Kim, E.-J., Jung, E., Pandey, B. P., and Kim, B.-G. (2016a). P212A Mutant of Dihydrodaidzein Reductase Enhances ( S )-Equol Production and Enantioselectivity in a Recombinant Escherichia coli Whole-Cell Reaction System. Appl. Environ. Microbiol. 82 (7), 1992–2002. doi:10.1128/AEM.03584-15
Lee, P.-G., Kim, J., Kim, E.-J., Lee, S.-H., Choi, K.-Y., Kazlauskas, R. J., et al. (2017). Biosynthesis of (−)-5-Hydroxy-Equol and 5-Hydroxy-Dehydroequol from Soy Isoflavone, Genistein Using Microbial Whole Cell Bioconversion. ACS Chem. Biol. 12 (11), 2883–2890. doi:10.1021/acschembio.7b00624
Lee, P.-G., Lee, S.-H., Kim, J., Kim, E.-J., Choi, K.-Y., and Kim, B.-G. (2018). Polymeric Solvent Engineering for Gram/liter Scale Production of a Water-Insoluble Isoflavone Derivative, (S)-equol. Appl. Microbiol. Biotechnol. 102 (16), 6915–6921. doi:10.1007/s00253-018-9137-8
Lee, P. G., Lee, S. H., Hong, E. Y., Lutz, S., and Kim, B. G. (2019). Circular Permutation of a Bacterial Tyrosinase Enables Efficient Polyphenol‐specific Oxidation and Quantitative Preparation of Orobol. Biotechnol. Bioeng. 116, 19–27. doi:10.1002/bit.26795
Lee, S.-H., Baek, K., Lee, J.-E., and Kim, B.-G. (2016b). Using Tyrosinase as a Monophenol Monooxygenase: A Combined Strategy for Effective Inhibition of Melanin Formation. Biotechnol. Bioeng. 113 (4), 735–743. doi:10.1002/bit.25855
Lin, Y., and Yan, Y. (2014). Biotechnological Production of Plant-specific Hydroxylated Phenylpropanoids. Biotechnol. Bioeng. 111 (9), 1895–1899. doi:10.1002/bit.25237
Lund, T. D., Munson, D. J., Haldy, M. E., Setchell, K. D. R., Lephart, E. D., and Handa, R. J. (2004). Equol Is a Novel Anti-androgen that Inhibits Prostate Growth and Hormone Feedback1. Biol. Reprod. 70 (4), 1188–1195. doi:10.1095/biolreprod.103.023713
Marrian, G. F., and Haslewood, G. A. D. (1932). Equol, a New Inactive Phenol Isolated from the Ketohydroxyoestrin Fraction of Mares' Urine. Biochem. J. 26 (4), 1227–1232. doi:10.1042/bj0261227
Matthies, A., Loh, G., Blaut, M., and Braune, A. (2012). Daidzein and Genistein Are Converted to Equol and 5-Hydroxy-Equol by Human Intestinal Slackia Isoflavoniconvertens in Gnotobiotic Rats. J. Nutr. 142 (1), 40–46. doi:10.3945/jn.111.148247
Nayfach, S., Fischbach, M. A., and Pollard, K. S. (2015). MetaQuery: a Web Server for Rapid Annotation and Quantitative Analysis of Specific Genes in the Human Gut Microbiome. Bioinformatics 31 (20), 3368–3370. doi:10.1093/bioinformatics/btv382
Pandey, B. P., Roh, C., Choi, K. Y., Lee, N., Kim, E. J., Ko, S., et al. (2010). Regioselective Hydroxylation of Daidzein Using P450 (CYP105D7) from Streptomyces Avermitilis MA4680. Biotechnol. Bioeng. 105 (4), 697–704. doi:10.1002/bit.22582
Park, J.-S., Kim, D. H., Lee, J. K., Lee, J. Y., Kim, D. H., Kim, H. K., et al. (2010). Natural Ortho-Dihydroxyisoflavone Derivatives from Aged Korean Fermented Soybean Paste as Potent Tyrosinase and Melanin Formation Inhibitors. Bioorg. Med. Chem. Lett. 20 (3), 1162–1164. doi:10.1016/j.bmcl.2009.12.021
Park, J.-S., Park, H. Y., Kim, D. H., Kim, D. H., and Kim, H. K. (2008). ortho-dihydroxyisoflavone Derivatives from Aged Doenjang (Korean Fermented Soypaste) and its Radical Scavenging Activity. Bioorg. Med. Chem. Lett. 18 (18), 5006–5009. doi:10.1016/j.bmcl.2008.08.016
Prieto, M. A., and Garcia, J. L. (1994). Molecular Characterization of 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli. A Two-Protein Component Enzyme. J. Biol. Chem. 269 (36), 22823–22829. doi:10.1016/s0021-9258(17)31719-2
Rittle, J., and Green, M. T. (2010). Cytochrome P450 Compound I: Capture, Characterization, and C-H Bond Activation Kinetics. Science 330 (6006), 933–937. doi:10.1126/science.1193478
Rüfer, C. E., Glatt, H., and Kulling, S. E. (2006). Structural Elucidation of Hydroxylated Metabolites of the Isoflavan Equol by Gas Chromatography-Mass Spectrometry and High-Performance Liquid Chromatography-Mass Spectrometry. Drug Metab. Dispos 34 (1), 51–60. doi:10.1124/dmd.105.004929
Schwen, R. J., Nguyen, L., and Jackson, R. L. (2012). Elucidation of the Metabolic Pathway of S-Equol in Rat, Monkey and Man. Food Chem. Toxicol. 50 (6), 2074–2083. doi:10.1016/j.fct.2012.03.048
Seo, S. G., Yang, H., Shin, S. H., Min, S., Kim, Y. A., Yu, J. G., et al. (2013). A Metabolite of Daidzein, 6,7,4′-trihydroxyisoflavone, Suppresses Adipogenesis in 3T3-L1 Preadipocytes via ATP-Competitive Inhibition of PI3K. Mol. Nutr. Food Res. 57 (8), 1446–1455. doi:10.1002/mnfr.201200593
Setchell, K. D., Clerici, C., Lephart, E. D., Cole, S. J., Heenan, C., Castellani, D., et al. (2005). S-equol, a Potent Ligand for Estrogen Receptor β, Is the Exclusive Enantiomeric Form of the Soy Isoflavone Metabolite Produced by Human Intestinal Bacterial flora. Am. J. Clin. Nutr. 81 (5), 1072–1079. doi:10.1093/ajcn/81.5.1072
Trott, O., and Olson, A. J. (2010). AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 31 (2), 455–461. doi:10.1002/jcc.21334
Uchiyama, S., Ueno, T., and Suzuki, T. (2007). Identification of a Newly Isolated Equal-Producing Lactic Acid Bacterium from the Human Feces. J. Intestinal Microbiol. 21 (3), 217. doi:10.11209/jim.21.217
Keywords: hydroxy-(S)-equol, oxidoreductases, isoflavonoids, SERM (selective estrogen receptor modulator), antioxidants, one-pot synthesis, enzyme engineering
Citation: Song H, Lee P-J, Kim J, Kim J, Lee S-H, Kim H, Lee U-J, Kim JY, Kim E-J and Kim B-G (2022) Regioselective One-Pot Synthesis of Hydroxy-(S)-Equols Using Isoflavonoid Reductases and Monooxygenases and Evaluation of the Hydroxyequol Derivatives as Selective Estrogen Receptor Modulators and Antioxidants. Front. Bioeng. Biotechnol. 10:830712. doi: 10.3389/fbioe.2022.830712
Received: 07 December 2021; Accepted: 31 January 2022;
Published: 24 March 2022.
Edited by:
Didier Laurent Buisson, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Yasushi Ogasawara, Hokkaido University, JapanDirk Tischler, Ruhr University Bochum, Germany
Copyright © 2022 Song, Lee, Kim, Kim, Lee, Kim, Lee, Kim, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byung-Gee Kim, Ynl1bmdraW1Ac251LmFjLmty
†These authors have contributed equally to this work
 Hanbit Song1,2†
Hanbit Song1,2† Pyung-Gang Lee
Pyung-Gang Lee Junyeob Kim
Junyeob Kim Byung-Gee Kim
Byung-Gee Kim