
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 25 January 2022
Sec. Industrial Biotechnology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.824859
Ectoine, an osmotic pressure-compensated solute, is used in the food, agriculture, medicine, and cosmetics industries due to its ability to protect macromolecules. In this study, an ectoine-producing variant of Escherichia coli, ET08, was genetically constructed by introducing the ectABC gene cluster and eliminating metabolic pathways involving lysine and pyruvate. Medium optimization enhanced ectoine production from 1.87 to 10.2 g/L. Analysis of the transcriptional levels revealed that supplementation with ammonium sulfate enhanced the metabolic flux towards the biosynthesis of ectoine. Furthermore, by optimizing the copy number of ectA, ectB, and ectC, the recombinant E. coli ET11 (ectA:ectB:ectC = 1:2:1) produced 12.9 g/L ectoine in the shake flask and 53.2 g/L ectoine in a fed-batch fermenter, representing the highest ectoine titer produced by E. coli, which has great industrial prospects.
As a compatible solute, ectoine (4S-2-methyl-1,4,5,6-tetrahydro-4-pyrimidinecarboxylic acid) is commonly found in halophilic and halotolerant microorganisms and was first discovered in Ectothiorhodospira halochloris by Galinski et al. (1985). In addition to its primary function of maintaining cell osmotic balance and resisting the impact of high osmotic pressure, it also displays remarkable bioprotective properties, which have garnered increasing attention (Roychoudhury et al., 2012; Kunte et al., 2014; Hahn et al., 2017). There are many potential applications of ectoine in the food biotechnology, agriculture, skin care, and medical industries (Nakayama et al., 2000; Kentaro et al., 2005; Graf et al., 2008; Unfried et al., 2016; Boroujeni and Nayeri, 2018).
Ectoine has been synthesized from the precursor L-aspartate-β-semialdehyde (ASA) with L-2,4-diaminobutyrate transaminase (EctB), 2,4-diaminobutyrate acetyltransferase (EctA), and ectoine synthase (EctC) as catalysts (Göller et al., 1998; Calderon et al., 2004; Schwibbert et al., 2011; Li et al., 2017). These three enzymes are encoded by genes which are typically organized in the ectABC gene cluster, and may also comprise the ectD gene (Bursy et al., 2008). Similar gene clusters involved in ectoine biosynthesis were disclosed in Marinococcus halophilus (Louis and Galinski, 1997), Halobacillus dabanensis D-8T (Zhao et al., 2006), Methylomicrobium alcaliphilum 20Z (Reshetnikov et al., 2006), and Nesterenkonia halobia DSM20541 (Zhang et al., 2008).
Commercial ectoine production can be realized through the microbial fermentation of halophiles in a complex process called “bacterial milking” (Sauer and Galinski, 1998; Fallet et al., 2010). Although this method can be used to obtain ectoine on a large scale, the quantity of salt used corrodes the equipment. In addition, during the process, a higher resistance to osmotic pressure by the bacteria is required. To address the shortcomings of this process, great efforts have been made, including optimizing the process conditions and improving ectoine production performance by breeding halophilic bacteria. However, using transgenic non-halophilic bacteria for ectoine production has proven more efficient in recent years. For example, Becker et al. successfully integrated the ectABCD gene operon of Pseudomonas stutzeri A1501 into Corynebacterium glutamicum using systematic metabolic engineering, and then mutated aspartate kinase to ensure a sufficient supply of ASA. This resulted in a final overall spacetime yield of 6.7 g/L ectoine per day (Becker et al., 2013). In 2015, the ectABC gene cluster of Halomonas elongata DSM 2581 was overexpressed in E. coli K-12/BW25113. The overall production of ectoine was 25.1 g/L with aspartate and glycerol as substrates (He et al., 2015). Subsequently, an engineered strain of E. coli, ECT05, was constructed using a series of metabolic engineering strategies. The reported final titer was 25.1 g/L, and the overall yield of ectoine was 0.11 g/g of glucose (Ning et al., 2016). Recently, it was reported that ectoine production reached 65 g/L within 56 h, following transcriptional balancing of the ectoine pathway in Corynebacterium glutamicum (Gießelmann et al., 2019).
Although ectoine production can be realized by the fermentation of engineered microbial strains, low glucose conversion rate and low ectoine production efficiency are still observed. Therefore, recent reports have been primarily focused on the metabolic modification of engineered microbes. There are few studies on the nutritional optimization and fermentation regulation regarding engineered microbial strains. A study reported by Xu et al. revealed that through the regulation and optimization of the nitrogen source, the production of ε-poly-L-lysine was increased (Xu et al., 2018). The regulation of the nitrogen source was also reported to have a significant effect on ethanol and antibiotic production (Aharonowitz and Demain, 1979; Yue et al., 2012). This may also be applied to the synthesis of ectoine.
In this study, we first constructed a variant of E. coli by introducing an ectoine biosynthetic pathway while eliminating the lysine synthesis branch and by-product metabolic pathways (Figure 1). Then, the effects of the nitrogen sources and amino donors on the production of ectoine were investigated. Next, the transcription levels of the key genes responsible for ectoine and ammonium metabolism were analyzed. Lastly, the copy number of ectA, ectB, and ectC were optimized to improve the final ectoine titer. This strategy provides a commercially attractive method for the microbial production of ectoine.
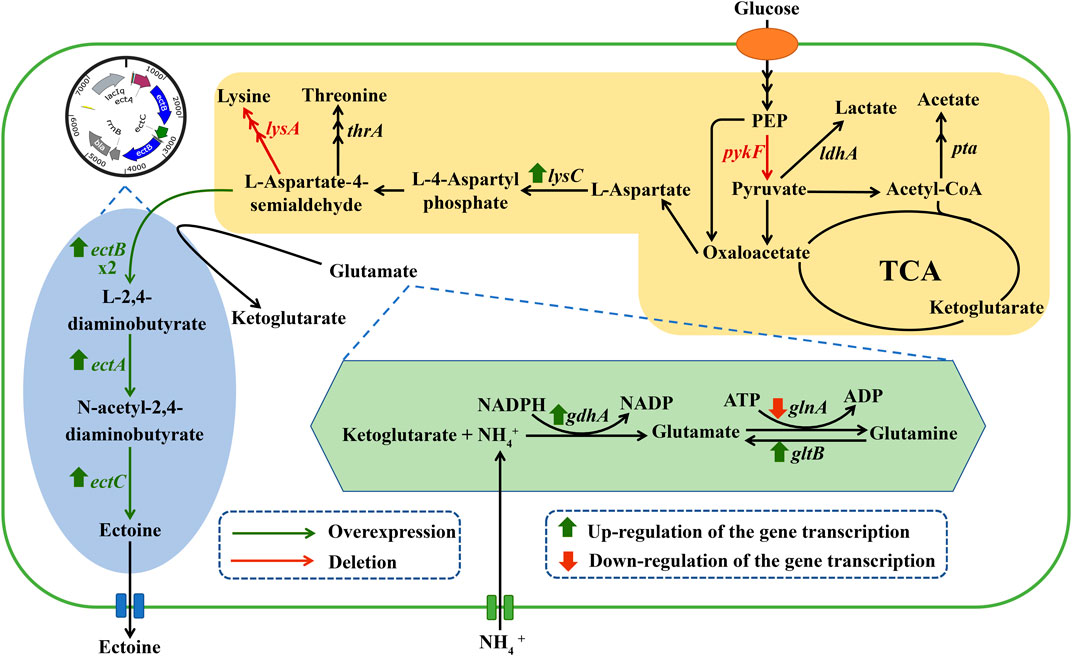
FIGURE 1. Related metabolic pathways for the synthesis of ectoine in engineered E. coli. The genes marked in green indicate the key genes of biosynthetic ectoine introduced into the engineered E. coli. The genes marked in red indicate deletion of the corresponding gene. Abbreviations: PEP, phosphoenolpyruvate.
The strains and plasmids constructed and used in this study are listed in Table 1. All restriction digest enzymes, DNA polymerase and DNA ligase were obtained from Takara Bio Inc. (Dalian, China). Oligonucleotides were synthesized by GENEWIZ Bio Inc. (Suzhou, China). Ectoine was purchased from Sigma-Aldrich (Shanghai, China).
The ectABC (MW414529) operon was amplified from the genome of Halomonas venusta ZH using the primers ET01-F and ET01-R. The PCR products were purified and then ligated into the linearized vector pTrc99a by in-fusion cloning with restriction sites of Kpn I and BamH I, forming plasmid pTrc-ectABC. For the construction of the plasmid pET-ectABC, the ectABC was amplified by PCR with ET02-F and ET02-R using the genomic DNA of H. venusta ZH as a template. The PCR products were purified and then inserted into pET-28a by in-fusion cloning with restriction sites of BamH I and Xho I.
The genes (ectA, ectB, ectC) were amplified from the genome of Halomonas venusta ZH respectively. These gene fragments were fused with corresponding trc promoter using overlap-extension PCR to form trcpro-ectA, trcpro-ectB, and trcpro-ectC. The fragment trcpro-ectA, trcpro-ectB, and trcpro-ectC were assembled with linearized plasmid pTrc-ectABC (BamH I/Sal I) using ClonExpress Entry One Step Cloning Kit to from plasmid pTETA, pTETB, and pTETC, respectively. A similar method can be used to construct a plasmid pTETATA, pTETBTB, pTETCTC, and pTETBTC.
Deletion of the lysA, thrA, pykF, ldhA, and pta genes were carried out using the Red recombination method (Datsenko and Wanner, 2000). The primers used for gene cloning and chromosomal manipulation are listed in Supplementary Table S1.
The initial fermentation medium contained (per liter) 5 g glucose, 20 g yeast extract, 0.5 g NaCl, 2.5 g KH2PO4, 7.5 g K2HPO4, 1 g MgSO4·7 H2O. Seed cultures were carried out in 500 ml Erlenmeyer flasks containing 50 ml LB medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) at 37°C and 200 rpm for 8 h. The seed culture (5 ml; 10% v/v) was inoculated into a 500 ml baffled shake flask containing 50 ml initial fermentation medium and cultivated at 37°C and 200 rpm for 48 h. Both media were supplemented with 100 mg/L ampicillin. As the inducer, 0.1 mmol/L IPTG was added at 4 h. 20 g/L of glucose was supplied by adding feeding solution (500 g/L glucose) every 12 h during the fermentation process.
To optimize the types of organic nitrogen sources, five organic nitrogen sources (yeast extract, peptone, beef extract, soybean meal extract, corn steep liquor) were chosen for E. coli ET08 fermentation of ectoine. The concentrations of each nitrogen source were 20 g/L. Then, complex nitrogen sources were optimized by mixing various concentrations of yeast extract (10 g/L, 15 g/L, 20 g/L) up with ammonium chloride (152 mM) and sodium nitrate (152 mM) respectively. The medium without inorganic nitrogen addition was used as control check. To optimize the exogenous amino donor, the effects of various concentrations of ammonium chloride (0, 38, 76, 152, 228 mM), ammonium sulfate (0, 19, 38, 76, 114 mM), and sodium glutamate (0, 38, 76, 152, 228 mM) were investigated in shake-flask tests.
Fed-batch fermentation of E. coli ET11 was carried out in a 7.5 L bioreactor (New Brunswick BioFlo/CelliGen 120, New Brunswick, Germany) containing 3.5 L medium. The fed-batch fermentation medium contained (per liter) 15 g glucose, 20 g yeast extract, 0.5 g NaCl, 2.5 g KH2PO4, 7.5 g K2HPO4, 1 g MgSO4·7 H2O, 10 g ammonium sulfate. The primary seed culture was prepared by transferring colonies from LB agar plate into a 50 ml shake flask containing 10 ml LB medium and culturing at 37°C and 200 rpm for 10 h. Then, the second seed culture was obtained by transferring 2 ml primary seed culture into a 1 L baled shake flask containing 200 ml LB medium and culturing under the same conditions. When the optical density at 600 nm (OD600) reached about 5, the second seed culture was inoculated into fermentation medium using a 10% (v/v) inoculum size. The pH was automatically controlled at 7.0 by adding NH4OH (25%, v/v), and the temperature was maintained at 37°C. The dissolved oxygen (DO) was maintained at 40% during 0–24 h by variation of the stirrer speed and the aeration rate. Subsequently, the DO was maintained at 20% from 24 to 48 h. The fed-batch culture was implemented in the early stage of fermentation (0–18 h), while the glucose concentration was maintained below 1 g/L by adding feeding solution (600 g/L glucose) at a designed rate in the middle-to-late period (24 h-end). As the inducer, 0.1 mmol/L IPTG was added when OD600 reached 0.4.
As the fermentation in shake flasks reached 36 h, the production was closed to maximum production. At this timepoint, the transcriptional levels of the key genes in ammonium metabolic pathways and ectoine synthesis were determined. Total RNA was isolated using RNAiso Plus (9108Q, TaKaRa Biotechnology Company, China). The synthesis of cDNA was then performed with PrimeScriptTM II 1st Strand cDNA Synthesis Kit (6210A, TaKaRa Biotechnology Company, China), using the total RNA as template. Real-time PCR (StepOnePlusTM Real-Time PCR System, Applied Biosystems, United States) was carried out with SYBR® Premix Ex TaqTM (RR420Q, TaKaRa Biotechnology Company, China) as fluorochrome, with 16S rDNA as an endogenous control gene. The primers used for qPCR were designed according to the E. coli genome sequence and were summarized in Supplementary Table S2. The amplification program consisted of one cycle at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s, 60°C for 30 s. All reactions were repeated three folds. Data from RT-qPCR were treated with the 2−ΔΔCt method for relative quantification (Livak and Schmittgen, 2000). To present the results in a better way, the formula 2−ΔΔCt was multiplied by 1. Therefore, the comparative expression level of each gene under the control was always 1 (Liu et al., 2017).
Cell growth was monitored by measuring the absorbance at 600 nm (OD600). Dry cell weight (DCW) was calculated based on the calibration curve (OD600 = 2.6131DCW–6.4197). The concentration of extracellular ectoine was determined via high-performance liquid chromatography (HPLC) using TSKgel ODS-80Ts column (4.6 × 250 mm, Tosoh, Tokyo) with an acetonitrile/water mixture (2:98 v/v) at a flow rate of 0.5 ml/min as the mobile phase. Ectoine was monitored by a UV detector at a wavelength of 210 nm. The residual sugar in the fermentation broth was measured using an SBA-40C biological analyzer (Shandong Academy of Sciences, China). Organic acid was determined by HPLC, using an Aminex HPX-87H column (Bio-Rad, Hercules, United States) kept at 65°C, with 5 mM sulfuric acid as the mobile phase at a low rate of 0.5 ml/min (Dong et al., 2021).
To achieve the heterologous synthesis of ectoine, two different host cells, E. coli MG1655 and E. coli BL21 (DE3), were chosen based on their genetic background and the maturity of the genetic tools. The constructed plasmid pTrc-ectABC was transferred into E. coli BL21 (DE3) and E. coli MG1655, respectively to construct the recombinant strains ET00 and ET01 (Table 1). The extracellular ectoine titer reached 0.52 g/L after 48 h cultivation using ET01 (Figure 2). However, the production of ectoine by ET00 was not detected (Figure 2) despite similar expression levels of ectABC in ET00 and ET01 (Supplementary Figure S1). To further improve the expression level of ectABC, the plasmid pET-ectABC was transferred into E. coli BL21 (DE3), to generate strains ET02. As expected, the expression of ectABC increased significantly, but ectoine was still not detected. Accordingly, E. coli MG1655 was a better chassis for the production of ectoine. The recombinant strain ET01 was chosen for further genetic manipulation.
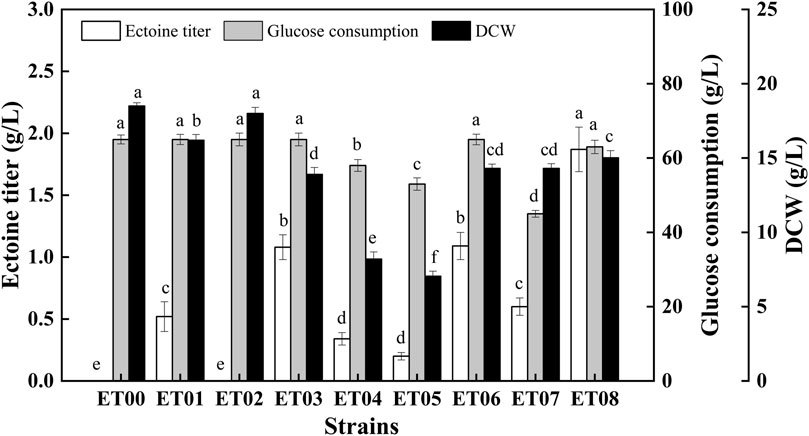
FIGURE 2. Comparison of ectoine titer, glucose consumption, and DCW in recombinant E. coli strains. Values represent the mean ± SD. Statistical analysis was performed by Duncan’s test (p < 0.05). Different lowercase letters indicate significant differences.
To reduce L-aspartate-β-semialdehyde shunting via branch metabolism, the genes lysA (encoding diaminopimelate decarboxylase) and thrA (encoding aspartate kinase/homoserine dehydrogenase) in E. coli ET01 were knocked out individually and collectively, generating the recombinant strains E. coli ET03, E. coli ET04, and E. coli ET05, respectively. As shown in Figure 2, compared to E. coli ET01, the ectoine titer of E. coli ET03 (1.08 g/L) was increased 2.08-fold, however the biomass (14 g/L) decreased with the same glucose consumption (65 g/L). When thrA was deleted, the ectoine titer of E. coli ET04 was decreased to 0.34 g/L, while glucose consumption and cell growth showed a similar trend, declining to 58 g/L and 8.2 g/L, respectively. Similarly, as lysA and thrA were knocked out collectively, the ectoine titer of E. coli ET05 decreased (0.22 g/L) with 53 g/L of glucose consumption and 7.1 g/L of DCW. The results indicated that the deletion of thrA significantly inhibited the growth of E. coli ET01, which was consistent with previous reports (Ning et al., 2016). The deletion of thrA might increase the metabolic burden of cells (Fang et al., 2014), which further affected the growth of the strain and synthesis of ectoine.
In addition to ectoine, acetate, as a main organic acid, was observed in the broth (Dong et al., 2021). Metabolic pathway analysis showed that acetate and lactate are derived from pyruvate (Figure 1). Since the production of organic acids distributes the carbon flux and can significantly inhibit cell growth by reducing the pH level, this may have affected the final titer. As such, to further increase the final quantity of ectoine, recombinant strains E. coli ET06, E. coli ET07, and E. coli ET08, were constructed by deleting genes of pta, ldhA, and pykF in E. coli ET03, respectively. which are responsible for acetate, lactate, and pyruvate synthesis, respectively (Figure 1). When pta was deleted, the ectoine production and biomass of E. coli ET06 were similar to those of E. coli ET03 but with an extended lag phase. Although the deletion of pta significantly reduced the titer of acetate from 6.52 g/L to 1.81 g/L in the broth, the titers of lactate, and pyruvate increased significantly (Supplementary Figure S2). Meanwhile, the ectoine titer of E. coli ET07 was decreased to 0.61 g/L with the lowest glucose consumption (45 g/L). These results indicated that the deletion of pta and ldhA had no positive impact on ectoine production. However, the ectoine titer of E. coli ET08 was increased by 73.1% (reached to 1.87 g/L) compared with that of E. coli ET03. A slight increase in the biomass (15 g/L) was observed in E. coli ET08, which indicated that the deletion of pykF can promote the growth of the strain. Organic acid analysis showed that acetate (3.25 g/L) was reduced by 50.2%, while lactate and pyruvate no longer accumulated (Supplementary Figure S2). This might explain the reason for the increase in biomass. In addition, the pykF deficient decreased the flux from phosphoenolpyruvate (PEP) to pyruvate and increased the PEP pool (Siddiquee et al., 2004). The accumulation of PEP, as a prerequisite for ectoine synthesis, might further promote the synthesis of ectoine. Based on these results, E. coli ET08 was chosen as the potential strain for further studies.
To further improve ectoine production, it is necessary to optimize the type and quantity of the nitrogen sources. As shown in Figure 3A, the yeast extract was more conducive to ectoine synthesis and bacterial growth than other organic nitrogen sources. In addition, as shown in Figure 3B, with the increase in the concentration of the yeast extract concentration in the medium, the production of ectoine by E. coli ET08 also increased, except for the titer obtained by sodium nitrate addition. The production of ectoine (7.29 g/L) was achieved with the supplementation of 20 g/L yeast extract and 152 mM ammonium chloride, which was 3.90-fold higher than that of the control (1.87 g/L). With the same yeast extract concentration, the ectoine titer with the addition of extra ammonium chloride was significantly higher than that of the blank control and sodium nitrate. The combination of the yeast extract and nitrate did not have a positive effect on the production of ectoine. Both the biomass and sugar consumption of E. coli ET08 were also decreased (Figure 3B). This result implied that ammonium salt played a more significant role in promoting the production of ectoine compared to nitrate salt. In addition, the ammonium salt was more conducive to microbial absorption than the nitrate, and may act as an exogenous amino donor by providing the NH4+ ions involved in the synthesis of glutamate (Figure 1). This is beneficial as glutamate is involved in the catalytic reaction of the key enzyme EctB in the ectoine synthesis pathway, as a co-substrate, and provides an amidogen to L-aspartate-β-semialdehyde (Li et al., 2017).
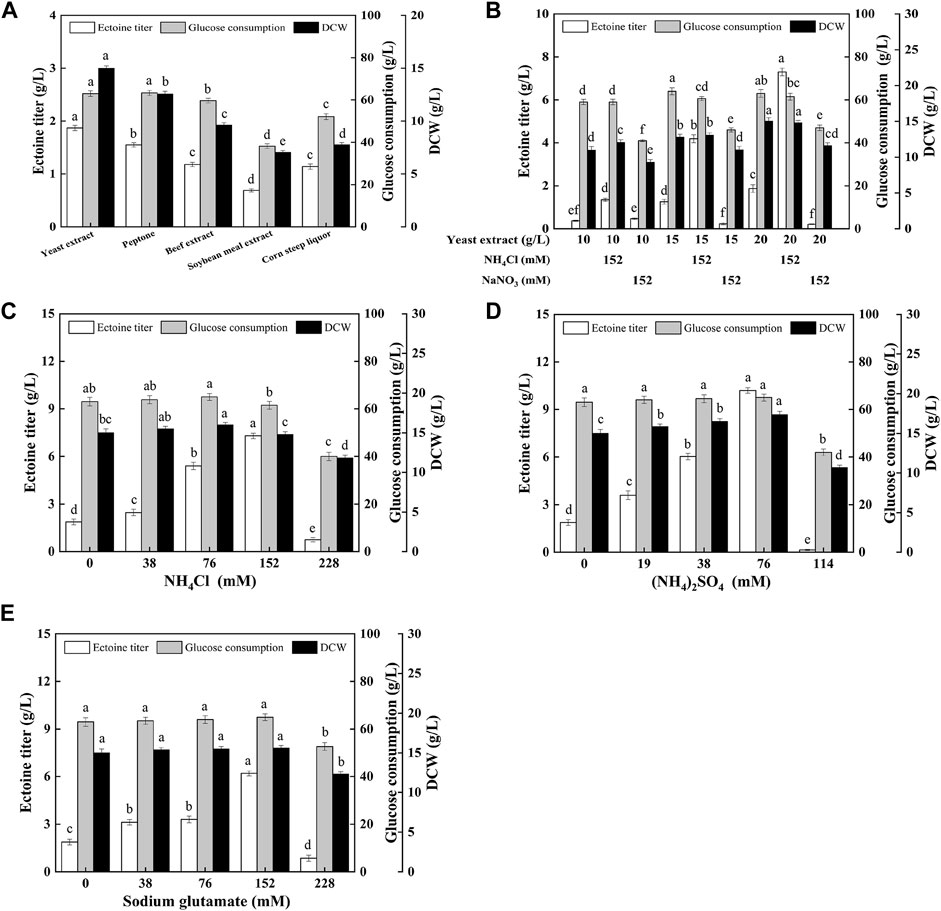
FIGURE 3. Effects of (A) different organic nitrogen sources, (B) the yeast extract mixed with inorganic nitrogen, (C) different ammonium chloride concentrations, (D) different ammonium sulfate concentrations, (E) different sodium glutamate concentrations on ectoine production, glucose consumption, and cell growth in E. coli ET08. Values represent the mean ± SD. Statistical analysis was performed by Duncan’s test (p < 0.05). Different lowercase letters indicate significant differences.
Since ammonium chloride, as an amino donor, plays an important role in improving ectoine, production, the types, and various concentrations of amino donors were further optimized, as shown in Figures 3C–E. The maximal titer of ectoine (10.2 g/L) was obtained with the addition of ammonium sulfate, which was 1.40- and 1.65-fold higher than that obtained after the addition of ammonium chloride (7.29 g/L) and sodium glutamate (6.18 g/L), respectively. With an increase in the concentration of the amino donor, the ectoine titer showed a trend from increasing to decreasing. It was noted that the maximum titer was obtained when the concentration of NH4+ (for ammonium salt) or −NH2 (for sodium glutamate) was 152 mM, regardless of the presence of ammonium chloride (Figure 3C), ammonium sulfate (Figure 3D), or sodium glutamate (Figure 3E). The concentration of NH4+ or −NH2 had little effect on the growth of bacteria when it was less than or equal to 152 mM. However, when the concentration of NH4+ or −NH2 reached 228 mM, bacterial growth was significantly inhibited, which was viewed as an important factor for the decrease in the total glucose consumption and ectoine production. Although all these three amino donors improved ectoine biosynthesis, ammonium sulfate was the most efficient. This may be attributed to the higher transport efficiency of ammonium sulfate compared with sodium glutamate.
To exploit the mechanism of NH4+ on ectoine production, the transcription levels of the related genes were investigated by adding 76 mM ammonium sulfate. The addition increased the transcription levels of gdhA and gltB. It was observed that the transcription levels of gdhA and gltB increased by 3.36- and 2.92-fold, respectively, whereas the transcription levels of glnA decreased significantly compared with the control (Figure 4). This was expected as it is known that ammonium is mainly assimilated by glutamate dehydrogenase (GDH) encoded by gdhA, or glutamine synthetase/glutamate synthase (GS/GOGAT) encoded by glnA and gltB, respectively (Merrick and Edwards, 1995; Reitzer, 2003). As previously reported, the up-regulation of gltB and gdhA promoted the synthesis and accumulation of glutamate in the microbial strains during fermentation (Huang et al., 2011). The downregulation of glnA on the other hand, indicated the inhibition of glutamine synthesis, but the promotion of glutamate accumulation. Furthermore, glutamate, as an essential amino acid, may be directly involved in the synthesis of ectoine through transamination (Figure 1). Therefore, the addition of ammonium sulfate may enhance the synthesis of ectoine by improving glutamate synthesis.
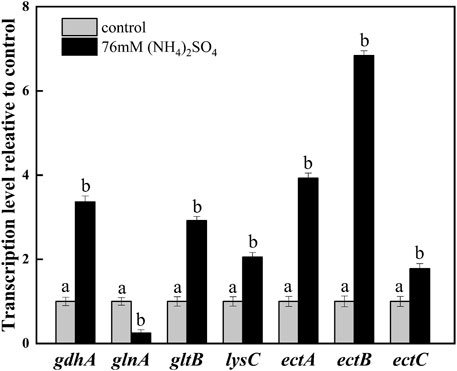
FIGURE 4. Transcription levels of the key genes in ammonium metabolic pathways and ectoine synthesis. The level of transcription was calculated relative to transcription of the control (0 mM ammonium sulfate), which were defined as 1. Values represent the mean ± SD. Different lowercase letters indicate significant differences at p < 0.05.
Aspartokinase (AK), EctA, EctB, and EctC play significant roles in ectoine synthesis. As depicted in Figure 4, the transcription level of the aspartate kinase gene (lysC) was upregulated by 2.05-fold, which demonstrated the improved synthesis ability of downstream products in the aspartate metabolic pathway. Significant increases in the transcription levels of ectABC which acted as the principal gene cluster in the synthesis of ectoine, were observed. The transcription level of ectB was up-regulated by 6.84-fold, which was higher than that of ectA (3.93-fold) and ectC (1.78-fold). This indicated that EctB was the most important enzyme among the key enzymes involved in the synthesis of ectoine. These results are consistent with that of a previous report (Hillier et al., 2020). It was speculated that the addition of the ammonium sulfate enhanced the supplementation of L-aspartate-β-semialdehyde and glutamate, providing an affluent co-substrate for the key enzyme EctB. At the same time, the strong expression of ectABC, especially ectB, promoted the flow of substrate in the ectoine synthesis pathway, improving the production of ectoine.
A previous report found that the expression of ectABC was not similar among the high-yielding strains, and that the expression ratios of ectA, ectB, and ectC were crucial for obtaining high ectoine production (Gießelmann et al., 2019). In addition, the copy number of the gene may significantly affect the protein expression level. Thus, it is necessary to study the effect of different copy numbers of ectA, ectB, and ectC on ectoine synthesis. As such, the plasmids pTETA, pTETB, pTETC, pTETATA, pTETBTB, and pTETCTC were constructed and introduced into the ET08-N strain, which is the strain ET08 without plasmid, producing ET09-ET14, respectively. The flask fermentation results showed that ectoine titers of ET09 (ectA:ectB:ectC = 2:1:1) and ET10 (ectA:ectB:ectC = 3:1:1) were 6.85 g/L and 2.39 g/L lower than that of ET08 (ectA:ectB:ectC = 1:1:1), respectively (Figure 5A). This indicated that the synthesis of ectoine was not effectively enhanced with an increase of the copy number of ectA. It was also noted that as the copy number of ectA increased, the DCW was significantly reduced (Figure 5C), revealing that the overexpression of ectA had an inhibitory effect on cell growth. Therefore, it was speculated that the overexpression of EctA is toxic to cells.
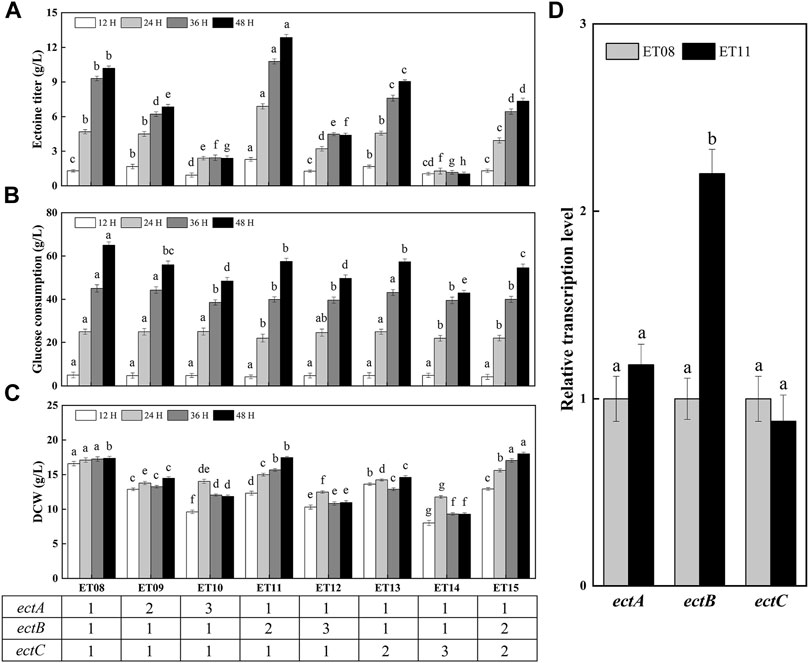
FIGURE 5. Ectoine production in metabolically engineered E. coli with different copy numbers of ectA, ectB, ectC. (A) Ectoine production; (B) Glucose consumption; (C) Cell growth; (D) Transcription levels of gene ectA, ectB, ectC. Values represent the mean ± SD. Statistical analysis was performed by Duncan’s test (p < 0.05). Different lowercase letters indicate significant differences.
As the rate-limiting enzyme in the ectoine biosynthetic pathway, the increase in the expression level of ectB is of great significance for increasing the production of ectoine (Chen et al., 2015). Gideon Gießelmann et al. (2019) found that all high-yielding strains in the mutant library had high expression levels of EctB. As such, in this study, the copy number of ectB was increased 2-fold to generate ET11 (ectA:ectB:ectC = 1:2:1). The production of ectoine by ET11 increased to 12.9 g/L, which was 1.26-fold higher than that of ET08. As shown in Figure 5B, the glucose consumption of ET11 (58 g/L) was lower than that of strain ET08 (65 g/L). As a result, the yield of ectoine based on glucose consumption was increased by 38%. When the copy number of ectB was further increased to three copies, the DCW of ET12 (ectA:ectB:ectC = 1:3:1) was significantly decreased to 11 g/L. Glucose consumption and ectoine titers were also reduced. These results indicate that the increase in the copy number of ectB can effectively improve ectoine production and enhance the carbon flux to promote ectoine synthesis. A sufficient supply of EctB may form a strong driving force for the subsequent biosynthetic steps. However, a further increase in the copy number did not have a positive effect on the ectoine titer. It is possible that a further increase in the copy number of ectB increases the bacterial metabolic burden and causes metabolic imbalance, thereby affecting production and growth.
Similarly, when the copy number of ectC was increased, the production of ectoine by ET13 (ectA:ectB:ectC = 1:1:2) and ET14 (ectA:ectB:ectC = 1:1:3) was only 9.05 g/L and 1.04 g/L, respectively. This may be due to the lack of precursors that can be used as a substrate for EctC to be converted into the final ectoine product. Although the total output of ET13 was reduced, the specific production of ectoine by ET13 was 0.62 g/g DCW, which was 1.1-fold higher than that of ET08 (0.59 g/g DCW). Therefore, we further increased the copy number of ectC to two copies in ET11 to generate ET15 (ectA:ectB:ectC = 1:2:2). Unfortunately, the specific ectoine production of ET15 was 0.41 g/g DCW, which was significantly reduced compared with that of ET11 (0.74 g/g DCW). In conclusion, ET11 proved to be the most effective at producing ectoine. Transcription analysis showed that the transcription levels of the ectA, ectB, and ectC genes of E. coli ET11 were 1.18-, 2.20-, and 0.88-fold that of ET08, respectively (Figure 5D). This indicated that the transcription level of ectB in ET11 increased as the copy numbers increased. The optimal copy numbers of ectA, ectB, and ectC were 1, 2, and 1, respectively. The optimal copy number ratio of ectA, ectB, and ectC was found to be 1:2:1. In this case, the transcriptional balancing of ectA, ectB, and ectC might be more suitable for the biosynthesis of ectoine. These results further confirms that the ratio of EctA, EctB, and EctC, and the transcriptional balancing of ectA, ectB, and ectC in the cell, were crucial for the biosynthesis of ectoine, as observed in a previous study using C. glutamicum (Gießelmann et al., 2019).
Previously, we reported a fermentation regulation strategy based on the kinetic analysis of ectoine production in fed-batch fermentation (Dong et al., 2021). The two-stage feeding method was combined with a two-step DO control strategy to efficiently increase ectoine production. Based on this fermentation strategy, fed-batch fermentation was implemented in a 7.5 L bioreactor to assess the overall production performance of E. coli ET11 with the addition of an exogenous amino donor (Figure 6). Within 18 h of fermentation, the fast-growing strains produced 9.43 g/L of ectoine. When the duration of fermentation was extended to 48 h, 53.2 g/L of ectoine was accumulated with slow bacterial growth. Additionally, the productivity was 1.11 g/L/h. To the best of our knowledge, our work shows the highest ectoine titer and productivity of E. coli. Compared with previous studies (Table 2), whole-cell catalysis seems to have certain advantages in yield from substrate, but it requires the addition of extra glycerol and aspartate, which results in high cost of extra substrate and cell culture before the catalytic reaction (Ning et al., 2016). Although C. glutamicum ectABCopt (Gießelmann et al., 2019) achieved higher total titer, the yield was only 0.19 g/g, which was lower than our yield (0.33 g/g glucose). Thus, the metabolically engineered E. coli strain ET11 and the fermentation strategy of supplementing amino donors display a potential for application in industrial production of ectoine.
In this study, we constructed an efficient strain E. coli ET08 by metabolic engineering. By optimizing nutritional element and analyzing the transcription levels, we could conclude that ammonium sulfate, as the optimal amino donor, has a positive effect on ectoine synthesis. Furthermore, optimizing the copy number of ectA, ectB, and ectC was employed to improve ectoine titer to 12.9 g/L in the shake flask. Fed-batch fermentation of E. coli ET11 with supplementing amino donor led to an accumulation of 53.2 g/L ectoine. The yield and productivity reached 0.33 g/g glucose and 1.11 g/L/h respectively. It provides a novel strategy for the synthesis of ectoine by engineered strain in industry. This research provides the basis for an effective process for ectoine production and could also be used to produce other high value amino acid derivative.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
HZ performed the experiments and drafted the manuscript. SL, HZ, and ZSL designed this study. ZL, MZ, and YM supervised the experiments and helped to draft the manuscript. SL and HX conceived the study and reviewed the final manuscript. All authors read and approved the final manuscript.
This work was supported by the National Key R&D Program of China (No. 2019YFA0905203), and the National Natural Science Foundation of China (No. 22178177).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.824859/full#supplementary-material
Aharonowitz, Y., and Demain, A. L. (1979). Nitrogen Nutrition and Regulation of Cephalosporin Production in Streptomyces Clavuligerus. Can. J. Microbiol. 25, 61–67. doi:10.1139/m79-010
Becker, J., Schäfer, R., Kohlstedt, M., Harder, B. J., Borchert, N. S., Stöveken, N., et al. (2013). Systems Metabolic Engineering of Corynebacterium Glutamicum for Production of the Chemical Chaperone Ectoine. Microb. Cell Fact. 12, 110. doi:10.1186/1475-2859-12-110
Boroujeni, M. B., and Nayeri, H. (2018). Stabilization of Bovine Lactoperoxidase in the Presence of Ectoine. Food Chem. 265, 208–215. doi:10.1016/j.foodchem.2018.05.067
Bursy, J., Kuhlmann, A. U., Pittelkow, M., Hartmann, H., Jebbar, M., Pierik, A. J., et al. (2008). Synthesis and Uptake of the Compatible Solutes Ectoine and 5-hydroxyectoine by Streptomyces Coelicolor A3(2) in Response to Salt and Heat Stresses. Appl. Environ. Microbiol. 74, 7286–7296. doi:10.1128/aem.00768-08
Calderón, M. I., Vargas, C., Rojo, F., Iglesias-Guerra, F., Csonka, L. N., Ventosa, A., et al. (2004). Complex Regulation of the Synthesis of the Compatible Solute Ectoine in the Halophilic Bacterium Chromohalobacter Salexigens DSM 3043T. Microbiology 150, 3051–3063. doi:10.1099/mic.0.27122-0
Chen, J., Liu, P., Chu, X., Chen, J., Zhang, H., Rowley, D. C., et al. (2020). Metabolic Pathway Construction and Optimization of Escherichia coli for High-Level Ectoine Production. Curr. Microbiol. 77 (8), 1412–1418. doi:10.1007/s00284-020-01888-6
Chen, W., Zhang, S., Jiang, P., Yao, J., He, Y., Chen, L., et al. (2015). Design of an Ectoine-Responsive AraC Mutant and its Application in Metabolic Engineering of Ectoine Biosynthesis. Metab. Eng. 30, 149–155. doi:10.1016/j.ymben.2015.05.004
Datsenko, K. A., and Wanner, B. L. (2000). One-step Inactivation of Chromosomal Genes in Escherichia coli K-12 Using PCR Products. Proc. Natl. Acad. Sci. 97, 6640–6645. doi:10.1073/pnas.120163297
Dong, Y., Zhang, H., Wang, X., Ma, J., Lei, P., Xu, H., et al. (2021). Enhancing Ectoine Production by Recombinant Escherichia coli through Step-wise Fermentation Optimization Strategy Based on Kinetic Analysis. Bioproc. Biosyst Eng 44, 1557–1566. doi:10.1007/s00449-021-02541-7
Fallet, C., Rohe, P., and Franco-Lara, E. (2010). Process Optimization of the Integrated Synthesis and Secretion of Ectoine and Hydroxyectoine under Hyper/hypo-Osmotic Stress. Biotechnol. Bioeng. 107, 124–133. doi:10.1002/bit.22750
Fang, H., Zhang, C., Xie, X., Xu, Q., Zhou, Y., and Chen, N. (2014). Enhanced Cytidine Production by a Recombinant Escherichia coli Strain Using Genetic Manipulation Strategies. Ann. Microbiol. 64, 1203–1210. doi:10.1007/s13213-013-0760-4
Galinski, E. A., Pfeiffer, H.-P., and Truper, H. G. (1985). 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic Acid. A Novel Cyclic Amino Acid from Halophilic Phototrophic Bacteria of the Genus Ectothiorhodospira. Eur. J. Biochem. 149, 135–139. doi:10.1111/j.1432-1033.1985.tb08903.x
Gießelmann, G., Dietrich, D., Jungmann, L., Kohlstedt, M., Jeon, E. J., Yim, S. S., et al. (2019). Metabolic Engineering of Corynebacterium Glutamicum for High-Level Ectoine Production: Design, Combinatorial Assembly, and Implementation of a Transcriptionally Balanced Heterologous Ectoine Pathway. Biotechnol. J. 14, e1800417. doi:10.1002/biot.201800417
Göller, K., Ofer, A., and Galinski, E. A. (1998). Construction and Characterization of an NaCl-Sensitive Mutant of Halomonas Elongata Impaired in Ectoine Biosynthesis. FEMS Microbiol. Lett. 161, 293–300. doi:10.1111/j.1574-6968.1998.tb12960.x
Graf, R., Anzali, S., Buenger, J., Pfluecker, F., and Driller, H. (2008). The Multifunctional Role of Ectoine as a Natural Cell Protectant. Clin. Dermatol. 26, 0–333. doi:10.1016/j.clindermatol.2008.01.002
Hahn, M. B., Meyer, S., Schroter, M. A., Kunte, H. J., Solomun, T., and Sturm, H. (2017). DNA protection by Ectoine from Ionizing Radiation: Molecular Mechanisms. Phys. Chem. Chem. Phys. 19, 25717–25722. doi:10.1039/c7cp02860a
He, Y. Z., Gong, J., Yu, H. Y., Tao, Y., Zhang, S., and Dong, Z. Y. (2015). High Production of Ectoine from Aspartate and Glycerol by Use of Whole-Cell Biocatalysis in Recombinant Escherichia coli. Microb. Cell Fact. 14, 55. doi:10.1186/s12934-015-0238-0
Hillier, H. T., Altermark, B., and Leiros, I. (2020). The crystal Structure of the Tetrameric DABA-Aminotransferase EctB, a Rate-Limiting Enzyme in the Ectoine Biosynthesis Pathway. Febs J. 287, 4641–4658. doi:10.1111/febs.15265
Huang, B. Q., Qin, P. Y., Xu, Z. W., Zhu, R. Y., and Meng, Y. H. (2011). Effects of CaCl2 on Viscosity of Culture Broth, and on Activities of Enzymes Around the 2-oxoglutarate branch, in Bacillus Subtilis CGMCC 2108 Producing Poly-(γ-Glutamic Acid). Bioresour. Technol. 102, 3595–3598. doi:10.1016/j.biortech.2010.10.073
Kentaro, F., Toshihiro, Y., and Shoji, S. (2005). Ectoine Alters Subcellular Localization of Inclusions and Reduces Apoptotic Cell Death Induced by the Truncated Machado–Joseph Disease Gene Product with an Expanded Polyglutamine Stretch. Neurobiol. Dis. 20, 170–178. doi:10.1016/j.nbd.2005.02.011
Kunte, H. J., Lentzen, G., and Galinski, E. A. (2014). Industrial Production of the Cell Protectant Ectoine: protection Mechanisms, Processes, and Products. Curr. Biotechnol. 3, 10–25. doi:10.2174/22115501113026660037
Li, Y. J., Wei, H. B., Wang, T., Xu, Q. Y., Zhang, C. L., Fan, X. G., et al. (2017). Current Status on Metabolic Engineering for the Production of L-Aspartate Family Amino Acids and Derivatives. Bioresour. Technol. 245, 1588–1602. doi:10.1016/j.biortech.2017.05.145
Liu, X. L., Zhu, P., Jiang, R. F., Wu, L. T., Feng, X. H., Li, S., et al. (2017). Enhancement of Welan Gum Production in Sphingomonas Sp. HT-1 via Heterologous Expression of Vitreoscilla Hemoglobin Gene. Carbohydr. Polym. 156, 135–142. doi:10.1016/j.carbpol.2016.08.081
Livak, K. J., and Schmittgen, T. D. (2000). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-△△Ct Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Louis, P., and Galinski, E. A. (1997). Characterization of Genes for the Biosynthesis of the Compatible Solute Ectoine from Marinococcus Halophilus and Osmoregulated Expression in Escherichia coli. Microbiology 143 (Pt 4), 1141–1149. doi:10.1099/00221287-143-4-1141
Merrick, M. J., and Edwards, R. A. (1995). Nitrogen Control in Bacteria. Microbiol. Rev. 59, 604–622. doi:10.1128/mr.59.4.604-622.1995
Nakayama, H., Yoshida, K., Ono, H., Murooka, Y., and Shinmyo, A. (2000). Ectoine, the Compatible Solute of Halomonas Elongata, Confers Hyperosmotic Tolerance in Cultured Tobacco Cells. Plant Physiol. 122, 1239–1247. doi:10.1104/pp.122.4.1239
Ning, Y. K., Wu, X. J., Zhang, C. L., Xu, Q. Y., Chen, N., and Xie, X. X. (2016). Pathway Construction and Metabolic Engineering for Fermentative Production of Ectoine in Escherichia coli. Metab. Eng. 36, 10–18. doi:10.1016/j.ymben.2016.02.013
Reitzer, L. (2003). Nitrogen Assimilation and Global Regulation in Escherichia coli. Annu. Rev. Microbiol. 57, 155. doi:10.1146/annurev.micro.57.030502.090820
Reshetnikov, A. S., Khmelenina, V. N., and Trotsenko, Y. A. (2006). Characterization of the Ectoine Biosynthesis Genes of Haloalkalotolerant Obligate Methanotroph “Methylomicrobium Alcaliphilum 20Z”. Arch. Microbiol. 184, 286–297. doi:10.1007/s00203-005-0042-z
Roychoudhury, A., Haussinger, D., and Oesterhelt, F. (2012). Effect of the Compatible Solute Ectoine on the Stability of the Membrane Proteins. Protein Pept. Lett. 19, 791–794. doi:10.2174/092986612801619570
Sauer, T., and Galinski, E. A. (1998). Bacterial Milking: a Novel Bioprocess for Production of Compatible Solutes. Biotechnol. Bioeng. 57, 306–313. doi:10.1002/(sici)1097-0290(19980205)57:3<306:aid-bit7>3.0.co;2-l
Schwibbert, K., Marin-Sanguino, A., Bagyan, I., Heidrich, G., Lentzen, G., Seitz, H., et al. (2011). A Blueprint of Ectoine Metabolism from the Genome of the Industrial Producer Halomonas Elongata DSM 2581T. Environ. Microbiol. 13, 1973–1994. doi:10.1111/j.1462-2920.2010.02336.x
Siddiquee, K. A., Arauzo-Bravo, M. J., and Shimizu, K. (2004). Metabolic Flux Analysis of pykF Gene Knockout Escherichia coli Based on C-13-Labeling Experiments Together with Measurements of Enzyme Activities and Intracellular Metabolite Concentrations. Appl. Microbiol. Biot. 63, 407–417. doi:10.1007/s00253-003-1357-9
Unfried, K., Kramer, U., Sydlik, U., Autengruber, A., Bilstein, A., Stolz, S., et al. (2016). Reduction of Neutrophilic Lung Inflammation by Inhalation of the Compatible Solute Ectoine: a Randomized Trial with Elderly Individuals. Int. J. Chronic Obstruct. Pulm. Dis. 11, 2573–2583. doi:10.2147/copd.s115061
Xu, D. L., Yao, H. Q., Cao, C. H., Xu, Z. X., Li, S., Xu, Z., et al. (2018). Enhancement of Epsilon-Poly-L-Lysine Production by Overexpressing the Ammonium Transporter Gene in Streptomyces Albulus PD-1. Bioproc. Biosyst. Eng. 41, 1337–1345. doi:10.1007/s00449-018-1961-9
Yue, G. J., Yu, J. L., Zhang, X., and Tan, T. W. (2012). The Influence of Nitrogen Sources on Ethanol Production by Yeast from Concentrated Sweet Sorghum Juice. Biomass Bioenerg. 39, 48–52. doi:10.1016/j.biombioe.2010.08.041
Zhang, B., Bao, X., Wang, L., and Yang, S. S. (2008). Cloning and Characterization of the Gene Cluster for Biosynthesis of Ectoine from Nesterenkonia Halobia DSM 20541. J. Microbiol. 46, 309–318. doi:10.1007/s12275-008-0001-x
Keywords: ectoine, Escherichia coli, metabolic engineering, medium optimization, amino donor
Citation: Zhang H, Liang Z, Zhao M, Ma Y, Luo Z, Li S and Xu H (2022) Metabolic Engineering of Escherichia coli for Ectoine Production With a Fermentation Strategy of Supplementing the Amino Donor. Front. Bioeng. Biotechnol. 10:824859. doi: 10.3389/fbioe.2022.824859
Received: 29 November 2021; Accepted: 04 January 2022;
Published: 25 January 2022.
Edited by:
Zhi-Qiang Liu, Zhejiang University of Technology, ChinaReviewed by:
Tobias Klein, Hochschule Kaiserslautern University of Applied Sciences, GermanyCopyright © 2022 Zhang, Liang, Zhao, Ma, Luo, Li and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sha Li, bGlzaGFAbmp0ZWNoLmVkdS5jbg==; Hong Xu, eHVoQG5qdGVjaC5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.