- 1School of Life Sciences, Faculty of Science, University of Technology Sydney, Sydney, NSW, Australia
- 2ARC Research Hub for Integrated Device for End-user Analysis at Low-levels (IDEAL), Faculty of Science, University of Technology Sydney, Sydney, NSW, Australia
- 3School of Women’s and Children’s Health, Faculty of Medicine and Health, University of New South Wales, South Wales, NSW, Australia
Ovarian Cancer
Ovarian cancer (OC) is a disease that most often affects post-menopausal women who present abdominal discomfort and bloating over a few months prior to detection. The majority of patients are diagnosed at advanced stages of the disease as the early stages are commonly asymptomatic (Jayson et al., 2014). According to Global Cancer Statistics 2020, OC is the seventh most common cancer in women worldwide accounting for around 314,000 new cases (3.4% of all new cancer cases in females) annually (Sung et al., 2021). In 2021 it established itself as the ninth most commonly diagnosed cancer amongst Australian women with 1720 new cases and 1,042 reported deaths (www.canceraustralia.gov.au). Prognosis is significantly determined by the stage of diagnosis where survival for stages I, II, III and IV is 73–92%, 45–55%, 21% and <6%, respectively1, underscoring the need for better early detection of ovarian cancer.
There are four main types of OC: epithelial, Germ cell, sex chord and stromal, with each having different epidemiological statistics, and with epithelial ovarian cancer (EOC) accounting for approximately 90% of all cases (Smith et al., 2006). In EOC there are histological subtypes, though most patients have high-grade serous ovarian cancer, a disease characterised by p53 gene abnormalities (Köbel, 2010). Some high grade serous ovarian cancers are caused by deleterious mutations of BRCA1 and BRCA2 genes, while others arise from a combination of somatic mutations (Network, 2011). The most common and effective management of the disease is a combination of surgery with chemotherapy. Surgical removal of cancer mass almost always occurs after diagnosis however, this is not always feasible when the cancer is very advanced. Furthermore, although surgery has proven effective in early stages of the disease, most patients diagnosed at advanced stages will go on to develop many iterations of recurrent disease (Jayson et al., 2014).
Cancer Antigen-125 (CA-125) and trans-vaginal imaging are currently routinely used as part of ovarian cancer diagnosis. Blood level of CA-125 is the most widely used serum biomarker, but it lacks the sensitivity or specificity to be used alone as a screening test (Jayson et al., 2014). It is also not useful for early diagnosis as CA-125 expression levels are too low for accurate detection and there are also several other conditions including endometriosis (Nisenblat et al., 2016), gall bladder (Wang, 2014) and liver cancer (Devarbhavi et al., 2002) where CA-125 levels are elevated, leading to false positive results. Likewise, it is often difficult to detect small early-stage tumors with trans-vaginal imaging. Hence, diagnosis often involves invasive techniques like laporascopy and tissue biopsies. It is therefore clear that early diagnosis of ovarian cancer requires improved screening tests that can be performed easily and inexpensively, as well as achieving high sensitivity and specificity.
New Advances in the Screening of Ovarian Cancer
Several different approaches have been proposed to combat the shortcomings of current techniques. Studies focused on smaller molecules, such as circulating miRNA and cell-free DNA (cf-DNA) have gained significant attention in recent times (Jørgensen, 2013; Ling et al., 2013; Peng and Croce, 2016). These molecules have emerged as promising cancer biomarkers suited for improved non/minimally invasive diagnostic, prognostic and therapeutic applications. Multiple miRNAs associated with ovarian cancer, such as the Let-7 family miRNAs, together with a variety of miRNA assays including RT-qPCR, microarrays and RNA sequencing have been reported as potential screening techniques (Banno et al., 2014; Deb et al., 2018; Aziz et al., 2020). As of 2021 there were over 250 miRNAs associated with ovarian cancers on the online database miRCancer, and depending on the cellular context, their up or down regulation was shown to be involved in disease progression (Xie et al., 2013). Despite demonstrating good analytical performance, a lack of reproducibility and the high costs associated with miRNA profiling mean their development remains a work in progress [for in-depth discussion of these challenges, see reviews (Tiberio et al., 2015; Witwer, 2015)]. In response to these issues, the research focus has expanded to include alternate approaches like the use of exosomes (EX) which are gaining increased interest in recent literature.
EXs are extracellular vesicles, ranging between 40 and 160 nm in size (Ruivo et al., 2017), released from the phospholipid membrane of the cell. These lipid bilayer-bound vehicles carry nucleic acids, proteins, and lipids to neighbouring or distant cells that influence a multitude of processes (Schey et al., 2015). Since EXs mimic the profile of the host cell from which they originate, it is not surprising that EXs released from cancer cells have been shown to aid in tumor progression including cellular proliferation, angiogenesis, migration, invasion, metastasis, and drug resistance (Tai et al., 2018). These properties make them suitable targets not only for novel therapeutics but also as specific markers for use in cancer diagnostics, prognosis, and even as chemotherapy drug-delivery systems. A recent review by Huda et al. (2021) highlighted several clinical applications and preclinical advancement outlining the importance of exosomes for use in targeted drug delivery, biomarker study, and vaccine development (Huda et al., 2021). A recent study by Zhao et al. (2016) reported the development of a microfluidic chip to isolate EXs expressing certain ovarian cancer biomarkers including CA-125 (Zhao et al., 2016). In addition, use of specific antibodies resulted in a greater number of exosomes being captured in patient samples, compared to healthy controls (Zhao et al., 2016). Although this study highlighted how exosomal CA-125 may be used to distinguish between healthy controls and cancer patients (Zhao et al., 2016), the associated drawbacks of CA-125 as a promising ovarian cancer biomarker for early detection remain a challenge.
It is now well established that the proteomic profile of EXs can act as a useful tool or biomarker for diagnosing various types of cancer (Nuzhat et al., 2017; Sharma et al., 2017; Kok and Yu, 2020; Croft et al., 2021). For example, a study by Klein-Scory et al. (2014) illustrated that EXs secreted from pancreatic cancer cells had a distinctive proteomic profile allowing for possible use as an early stage biomarker (Klein-Scory et al., 2014). Thus, it is seemingly reasonable to exploit the proteomic profile of exosomes from ovarian cancer cells to identify novel biomarkers. One such example is the Chloride Intracellular Ion Channels (CLIC) family of proteins which has been shown to be elevated in patients with ovarian cancer (Kobayashi et al., 2012).
Chloride Intracellular Ion Channels as Biomarkers for Ovarian Cancer
The CLIC family in vertebrates consists of six evolutionarily conserved protein members (Littler et al., 2010). These proteins are unusual, existing in cells as both soluble and membrane bound proteins, where they demonstrate “moonlighting” activity, exhibiting two independent functions. In the membrane bound form, CLIC proteins act as ion channels (Valenzuela et al., 2013) while in the soluble form, they act as oxidoreductase enzymes, which are most likely involved in cell protective functions like detoxification and/or anti-oxidative roles (Al Khamici et al., 2015). Several studies have also shown CLIC proteins to have important extracellular activities (see review (Argenzio and Moolenaar, 2016)), with CLIC1, CLIC3 and CLIC4 found on endometrium/placenta cell exosomes (Zhao et al., 2016). CLICs are also known to be up-regulated in various types of cancers where they are primarily involved in cancer metastasis via exosomal and anti-apoptotic activities (GururajaRao et al., 2020). In a recent bioinformatics study of patient survival as a function of the relative mRNA levels, across several human cancers, the data clearly indicates both low or high CLIC expression levels, influence patient survival (GururajaRao et al., 2020). For example, high CLIC1 and CLIC3 mRNA expression were associated with advanced stages of liver cancer (Huang et al., 2021) while high CLIC4 mRNA expression correlates with poor patient survival in EOC (Singha et al., 2018). However, to better understand the therapeutic potential of these proteins, it is important to further deduce details regarding their tissue expression and release into blood.
Studies have shown two members of the CLIC protein family, CLIC1 and CLIC4, to be significantly upregulated in EOC patients compared to healthy controls (Tang et al., 2012; Tang et al., 2013; Ye et al., 2015; Singha et al., 2018; Yu et al., 2018), and shed into the blood of ovarian cancer patients (Tang et al., 2012; Tang et al., 2013). At the tissue level, combining CLIC1 and CLIC4 staining with CA-125, resulted in better sensitivity, compared to using CA-125 alone, hence a panel consisting of CLIC proteins and CA-125 may outperform individual biomarkers (Singha et al., 2018). Expression levels of CLIC1 and CLIC4 have also been implicated in patient survival, with elevated CLIC4 expression a negative indicator of patient survival in ovarian cancer (Singha et al., 2018). Knockdown of either CLIC4 or CLIC1 resulted in slower growth of ovarian cancer cells suggesting a role of CLICs in cell proliferation and cell migration (Singha et al., 2018). Furthermore, consistent with other reports, these proteins have also been shown to play important roles in EOC progression across multiple EOC subtypes (Ye et al., 2015; Singha et al., 2018; Yu et al., 2018). Similarly, CLIC3 levels have been shown to be elevated in 90% of ovarian cancer patients, where upregulation of CLIC3 in breast cancer tissue was associated with increased cancer cell invasiveness (Hernandez-Fernaud et al., 2017). The fact that exosome associated CLICs display cancer-specific signatures for EOCs, allows for the coupling of CLIC expression with emerging technologies capable of detecting and profiling exosomes, leading to simple, non-invasive, early and specific detection of ovarian cancer.
Diagnostic Approaches for the Analysis of Exosome Associated CLICs as Biomarkers for Early Detection of Ovarian Cancer
EXs have been successfully isolated from blood, urine, ascites, cerebrospinal fluid, amniotic fluid, semen, saliva, and bile (Colombo et al., 2014; Sullivan et al., 2017) thus providing an array of possibilities for their detection using several different techniques, with some examples as highlighted in Figure 1. Some of the currently available techniques that analyse EX proteins from different human body fluids, with or without EX isolation, include flow cytometry, protein microarray (EX Array), diagnostic magnetic resonance, nanoplasmonic sensing technology and microfluidics (Orozco and Lewis, 2010; Shao et al., 2012; Jørgensen, 2013; Im et al., 2014; Zhao et al., 2016; Sullivan et al., 2017). Recently, a number of studies have proposed exosomal proteins as diagnostic biomarkers of breast cancer (Rupp et al., 2011; Soung et al., 2017; Sullivan et al., 2017), where one such study showed EXs isolated using anti-CD24 and anti-EpCAM-coupled magnetic beads as potential breast cancer-specific markers (Rupp et al., 2011). An assay called “ExoScreen” available for the detection of colon cancer is based on the detection of cancer-specific circulating double-positive (CD146/CD9) EXs using photosensitive-beads, from as little as 5 µL of patient serum (Yoshioka et al., 2014). Numerous commercial kits are also available where specific EXs are isolated and analysed using microfluidic platforms for detecting different types of cancer (Contreras-Naranjo et al., 2017).
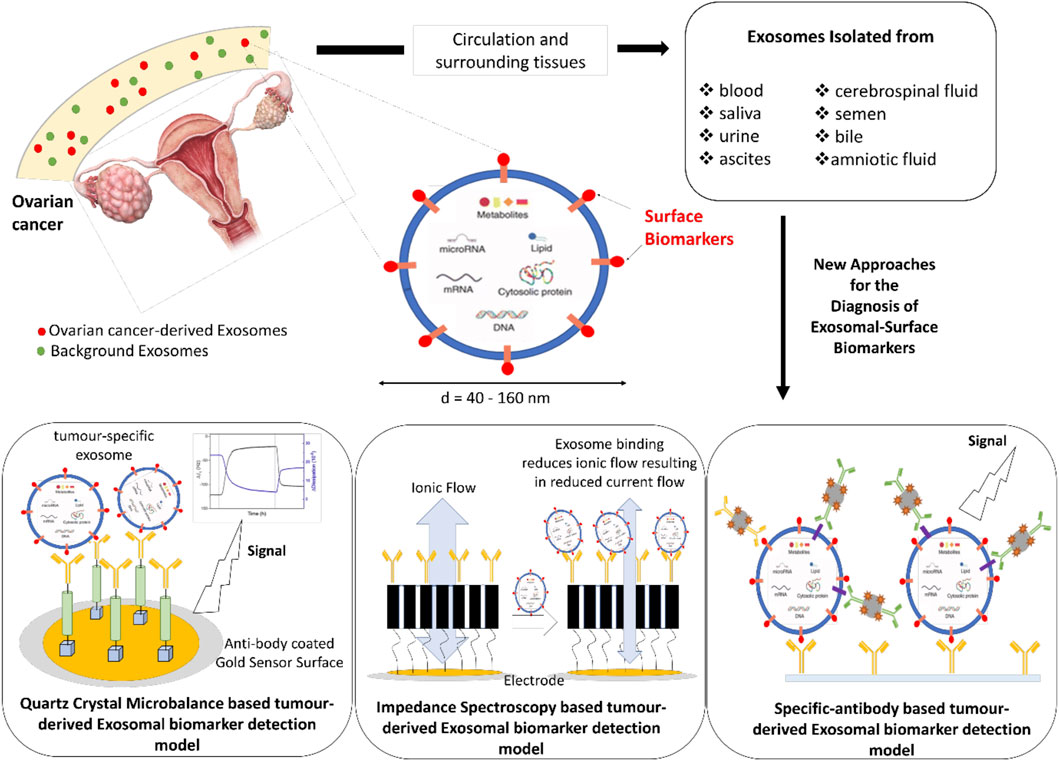
FIGURE 1. Schematic representation of new diagnostic approaches for the detection of Exosomal-Surface Biomarkers.
In the past several years, various electrochemical assays have been developed for the detection and characterisation of exosomes. One such development is an integrated magneto-electrochemical sensor (iMEX) where antibody-coated-magnetic beads are used for EXs capture and labelling following detection via electrochemical sensing (Jeong et al., 2016). Doldan et al. have developed an electrochemical sandwich approach using gold electrodes prefunctionalised with specific antibodies for exosome determination (Doldán, 2016). Electrical impedance technology is another well-established technique, which over the last 30 years has gained increasing interest in its applications in medicine. Impedance biosensors are usually label free and do not depend on any specific enzyme for analyte detection. Instead, impedimetric biosensors rely on unique bioreceptors or tags, which when specifically bound to the biomarker, produce changes in an electric current that lead to detection and in some cases, quantification of the biomarkers (Leva-Bueno et al., 2020). Impedance spectroscopy with tethered membrane technology has also been used for E. coli and Staphylococcus aureus detection, where specific antibodies embedded in the membrane detect the presence of bacteria in different fluid samples (Tan et al., 2011). This technology has been adapted to detect prostate (Mishra et al., 2012) and skin (Braun et al., 2017) cancer specific biomarkers and has the potential to be reliable, specific and selective, while being inexpensive and fast. Similarly, the detection of disease-related biomarkers based on acoustic sensors has created global research interest, with biosensors based on the quartz crystal microbalance (QCM) exploited for cancer diagnosis. Commercial QCM systems have been utilised for detecting highly metastatic human breast cancer cells (Atay et al., 2016; Bakhshpour et al., 2019), while an aptamer-based QCM was applied for the detection of leukemia cells (Shan et al., 2014). QCM technology with different signal enhancing approaches has been employed for the detection of several cancer biomarkers such as the carcinoembryonic antigen (CEA) for colorectal cancer (Chi et al., 2020), potential cancer biomarker poly (ADP-ribose) polymerase-1 (PARP-1) (Yang et al., 2019) and even for the detection of volatile compounds like propanol, ethyl benzene, hexanal and decane which are under investigation as non-invasive lung cancer biomarkers (AthirahAwatif Abdul Rahman et al., 2019). Recently, Suther et al. (2020) highlighted the potential of a QCM with dissipation monitoring (QCM-D) system to detect exosomal-CD63 biomarker by exploiting their surface protein profile (Suthar et al., 2020). Thus, the QCM system has emerged as a robust biosensing platform which allows the detection and quantification of a wide range of biomolecules. This, together with their high sensitivity and short detection time, makes QCM based-biosensors attractive for the early detection of various cancer types and the routine monitoring of disease progression.
Achieving early detection of ovarian cancer remains the greatest hope for improved patient survival outcomes. Therefore, investigation of these novel tools and technologies for early ovarian cancer diagnosis is a high priority.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank the Australian Research Council (ARC) Industry Transformational Research Hub Scheme (Grant IH150100028 to SV) ARC website URL: http://www.arc.gov.au/. The funder had no role in study design or data collection and analysis.
Footnotes
1UK, C.R. Cancer Research UK, 2013.
References
Al Khamici, H., Brown, L. J., Hossain, K. R., Hudson, A. L., Sinclair-Burton, A. A., Ng, J. P., et al. (2015). Members of the Chloride Intracellular Ion Channel Protein Family Demonstrate Glutaredoxin-like Enzymatic Activity. PLOS ONE 10 (1), e115699. doi:10.1371/journal.pone.0115699
Argenzio, E., and Moolenaar, W. H. (2016). Emerging Biological Roles of Cl− Intracellular Channel Proteins. J. Cel Sci 129 (22), 4165–4174. doi:10.1242/jcs.189795
Atay, S., Pişkin, K., Yılmaz, F., Çakır, C., Yavuz, H., and Denizli, A. (2016). Quartz crystal Microbalance Based Biosensors for Detecting Highly Metastatic Breast Cancer Cells via Their Transferrin Receptors. Anal. Methods 8, 153–161. doi:10.1039/c5ay02898a
Athirah Awatif Abdul Rahman, N., Hadi Ma'Radzi, A., and Zakaria, A. (2019). Fabrication of Quartz crystal Microbalance with Pegylated Lipopolymer for Detection of Non-invasive Lung Cancer Biomarker. Mater. Today Proc. 7 (2), 632–637. doi:10.1016/j.matpr.2018.12.054
Aziz, N. B., Mahmudunnabi, R. G., Umer, M., Sharma, S., Rashid, M. A., Alhamhoom, Y., et al. (2020). MicroRNAs in Ovarian Cancer and Recent Advances in the Development of microRNA-Based Biosensors. Analyst 145 (6), 2038–2057. doi:10.1039/c9an02263e
Bakhshpour, M., Piskin, A. K., Yavuz, H., and Denizli, A. (2019). Quartz crystal Microbalance Biosensor for Label-free MDA MB 231 Cancer Cell Detection via Notch-4 Receptor. Talanta 204, 840–845. doi:10.1016/j.talanta.2019.06.060
Banno, K., Yanokura, M., Iida, M., Adachi, M., Nakamura, K., Nogami, Y., et al. (2014). Application of microRNA in Diagnosis and Treatment of Ovarian Cancer. Biomed. Res. Int. 2014, 232817. doi:10.1155/2014/232817
Braun, R. P., Mangana, J., Goldinger, S., French, L., Dummer, R., and Marghoob, A. A. (2017). Electrical Impedance Spectroscopy in Skin Cancer Diagnosis. Dermatol. Clin. 35 (4), 489–493. doi:10.1016/j.det.2017.06.009
Chi, L., Xu, C., Li, S., Wang, X., Tang, D., and Xue, F. (2020). In Situ amplified QCM Immunoassay for Carcinoembryonic Antigen with Colorectal Cancer Using Horseradish Peroxidase Nanospheres and Enzymatic Biocatalytic Precipitation. Analyst 145 (18), 6111–6118. doi:10.1039/d0an01399d
Colombo, M., Raposo, G., and Théry, C. (2014). Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cel Dev. Biol. 30, 255–289. doi:10.1146/annurev-cellbio-101512-122326
Contreras-Naranjo, J. C., Wu, H.-J., and Ugaz, V. M. (2017). Microfluidics for Exosome Isolation and Analysis: Enabling Liquid Biopsy for Personalized Medicine. Lab. Chip 17 (21), 3558–3577. doi:10.1039/c7lc00592j
Croft, P. K., Sharma, S., Godbole, N., Rice, G. E., and Salomon, C. (2021). Ovarian-Cancer-Associated Extracellular Vesicles: Microenvironmental Regulation and Potential Clinical Applications. Cells 10 (9). doi:10.3390/cells10092272
Deb, B., Uddin, A., and Chakraborty, S. (2018). miRNAs and Ovarian Cancer: An Overview. J. Cel Physiol 233, 3846–3854. doi:10.1002/jcp.26095
Devarbhavi, H., Kaese, D., Williams, A. W., Rakela, J., Klee, G. G., and Kamath, P. S. (2002). Cancer Antigen 125 in Patients with Chronic Liver Disease. Mayo Clinic Proc. 77 (6), 538–541. doi:10.4065/77.6.538
Doldán, X. (2016). Electrochemical Sandwich Immunosensor for Determination of Exosomes Based on Surface Marker-Mediated Signal Amplification. Anal. Chem. 88 (21), 10466–10473.
Gururaja Rao, S., Patel, N. J., and Singh, H. (2020). Intracellular Chloride Channels: Novel Biomarkers in Diseases. Front. Physiol. 11, 96. doi:10.3389/fphys.2020.00096
Hernandez-Fernaud, J. R., Ruengeler, E., Casazza, A., Neilson, L. J., Pulleine, E., Santi, A., et al. (2017). Secreted CLIC3 Drives Cancer Progression through its Glutathione-dependent Oxidoreductase Activity. Nat. Commun. 8 (1), 14206. doi:10.1038/ncomms14206
Huang, J. J., Lin, J., Chen, X., and Zhu, W. (2021). Identification of Chloride Intracellular Channels as Prognostic Factors Correlated with Immune Infiltration in Hepatocellular Carcinoma Using Bioinformatics Analysis. Medicine (Baltimore) 100 (45). doi:10.1097/md.0000000000027739
Huda, M. N., Nafiujjaman, M., Deaguero, I. G., Okonkwo, J., Hill, M. L., Kim, T., et al. (2021). Potential Use of Exosomes as Diagnostic Biomarkers and in Targeted Drug Delivery: Progress in Clinical and Preclinical Applications. ACS Biomater. Sci. Eng. 7 (6), 2106–2149. doi:10.1021/acsbiomaterials.1c00217
Im, H., Shao, H., Park, Y. I., Peterson, V. M., Castro, C. M., Weissleder, R., et al. (2014). Label-free Detection and Molecular Profiling of Exosomes with a Nano-Plasmonic Sensor. Nat. Biotechnol. 32 (5), 490–495. doi:10.1038/nbt.2886
Jayson, G. C., Kohn, E. C., Kitchener, H. C., and Ledermann, J. A. (2014). Ovarian Cancer. The Lancet 384 (9951), 1376–1388. doi:10.1016/s0140-6736(13)62146-7
Jeong, S., Park, J., Pathania, D., Castro, C. M., Weissleder, R., and Lee, H. (2016). Integrated Magneto-Electrochemical Sensor for Exosome Analysis. ACS Nano 10 (2), 1802–1809. doi:10.1021/acsnano.5b07584
Jørgensen, M. (2013). Extracellular Vesicle (EV) Array: Microarray Capturing of Exosomes and Other Extracellular Vesicles for Multiplexed Phenotyping. J. Extracell Vesicles 2.
Klein-Scory, S., Tehrani, M. M., Eilert-Micus, C., Adamczyk, K. A., Wojtalewicz, N., Schnölzer, M., et al. (2014). New Insights in the Composition of Extracellular Vesicles from Pancreatic Cancer Cells: Implications for Biomarkers and Functions. Proteome Sci. 12 (1), 50. doi:10.1186/s12953-014-0050-5
Kobayashi, E., Ueda, Y., Matsuzaki, S., Yokoyama, T., Kimura, T., Yoshino, K., et al. (2012). Biomarkers for Screening, Diagnosis, and Monitoring of Ovarian Cancer. Cancer Epidemiol. Biomarkers Prev. 21 (11), 1902–1912. doi:10.1158/1055-9965.epi-12-0646
Köbel, M. (2010). The Biological and Clinical Value of P53 Expression in Pelvic High-Grade Serous Carcinomas. J. Pathol. 222 (2), 191–198.
Kok, V. C., and Yu, C.-C. (2020). Cancer-Derived Exosomes: Their Role in Cancer Biology and Biomarker Development. Ijn Vol. 15, 8019–8036. doi:10.2147/ijn.s272378
Leva-Bueno, J., Peyman, S. A., and Millner, P. A. (2020). A Review on Impedimetric Immunosensors for Pathogen and Biomarker Detection. Med. Microbiol. Immunol. 209 (3), 343–362. doi:10.1007/s00430-020-00668-0
Ling, H., Fabbri, M., and Calin, G. A. (2013). MicroRNAs and Other Non-coding RNAs as Targets for Anticancer Drug Development. Nat. Rev. Drug Discov. 12 (11), 847–865. doi:10.1038/nrd4140
Littler, D. R., Harrop, S. J., Goodchild, S. C., Phang, J. M., Mynott, A. V., Jiang, L., et al. (2010). The enigma of the CLIC Proteins: Ion Channels, Redox Proteins, Enzymes, Scaffolding Proteins? FEBS Lett. 584 (10), 2093–2101. doi:10.1016/j.febslet.2010.01.027
Mishra, V., Bouayad, H., Schned, A., Heaney, J., and Halter, R. J. (2012). Electrical Impedance Spectroscopy for Prostate Cancer Diagnosis. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2012, 3258–3261. doi:10.1109/EMBC.2012.6346660
Network, T. C. G. A. R. (2011). Integrated Genomic Analyses of Ovarian Carcinoma. Nature 474 (7353), 609–615. doi:10.1038/nature10166
Nisenblat, V., Bossuyt, P. M., Shaikh, R., Farquhar, C., Jordan, V., Scheffers, C. S., et al. (2016). Blood Biomarkers for the Non-invasive Diagnosis of Endometriosis. Cochrane Database Syst. Rev. 2016 (5), CD012179. doi:10.1002/14651858.CD012179
Nuzhat, Z., Kinhal, V., Sharma, S., Rice, G. E., Joshi, V., and Salomon, C. (2017). Tumour-derived Exosomes as a Signature of Pancreatic Cancer - Liquid Biopsies as Indicators of Tumour Progression. Oncotarget 8 (10), 17279–17291. doi:10.18632/oncotarget.13973
Orozco, A. F., and Lewis, D. E. (2010). Flow Cytometric Analysis of Circulating Microparticles in Plasma. Cytometry 77A (6), 502–514. doi:10.1002/cyto.a.20886
Peng, Y., and Croce, C. M. (2016). The Role of MicroRNAs in Human Cancer. Sig Transduct Target. Ther. 1 (1), 15004. doi:10.1038/sigtrans.2015.4
Ruivo, C. F., Adem, B., Silva, M., and Melo, S. A. (2017). The Biology of Cancer Exosomes: Insights and New Perspectives. Cancer Res. 77 (23), 6480–6488. doi:10.1158/0008-5472.can-17-0994
Rupp, A.-K., Rupp, C., Keller, S., Brase, J. C., Ehehalt, R., Fogel, M., et al. (2011). Loss of EpCAM Expression in Breast Cancer Derived Serum Exosomes: Role of Proteolytic Cleavage. Gynecol. Oncol. 122 (2), 437–446. doi:10.1016/j.ygyno.2011.04.035
Schey, K. L., Luther, J. M., and Rose, K. L. (2015). Proteomics Characterization of Exosome Cargo. Methods 87, 75–82. doi:10.1016/j.ymeth.2015.03.018
Shan, W., Pan, Y., Fang, H., Guo, M., Nie, Z., Huang, Y., et al. (2014). An Aptamer-Based Quartz crystal Microbalance Biosensor for Sensitive and Selective Detection of Leukemia Cells Using Silver-Enhanced Gold Nanoparticle Label. Talanta 126, 130–135. doi:10.1016/j.talanta.2014.03.056
Shao, H., Chung, J., Balaj, L., Charest, A., Bigner, D. D., Carter, B. S., et al. (2012). Protein Typing of Circulating Microvesicles Allows Real-Time Monitoring of Glioblastoma Therapy. Nat. Med. 18 (12), 1835–1840. doi:10.1038/nm.2994
Sharma, S., Zuñiga, F., Rice, G. E., Perrin, L. C., Hooper, J. D., and Salomon, C. (2017). Tumor-derived Exosomes in Ovarian Cancer - Liquid Biopsies for Early Detection and Real-Time Monitoring of Cancer Progression. Oncotarget 8 (61), 104687–104703. doi:10.18632/oncotarget.22191
Singha, B., Harper, S. L., Goldman, A. R., Bitler, B. G., Aird, K. M., Borowsky, M. E., et al. (2018). CLIC1 and CLIC4 Complement CA125 as a Diagnostic Biomarker Panel for All Subtypes of Epithelial Ovarian Cancer. Sci. Rep. 8 (1), 14725. doi:10.1038/s41598-018-32885-2
Smith, H. O., Berwick, M., Verschraegen, C. F., Wiggins, C., Lansing, L., Muller, C. Y., et al. (2006). Incidence and Survival Rates for Female Malignant Germ Cell Tumors. Obstet. Gynecol. 107 (5), 1075–1085. doi:10.1097/01.aog.0000216004.22588.ce
Soung, Y. H., Ford, S., Zhang, V., and Chung, J. (2017). Exosomes in Cancer Diagnostics. Cancers (Basel) 9 (1). doi:10.3390/cancers9010008
Sullivan, R., Maresh, G., Zhang, X., Salomon, C., Hooper, J., Margolin, D., et al. (2017). The Emerging Roles of Extracellular Vesicles as Communication Vehicles within the Tumor Microenvironment and beyond. Front. Endocrinol. (Lausanne) 8, 194. doi:10.3389/fendo.2017.00194
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A. Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Suthar, J., Parsons, E. S., Hoogenboom, B. W., Williams, G. R., and Guldin, S. (2020). Acoustic Immunosensing of Exosomes Using a Quartz Crystal Microbalance with Dissipation Monitoring. Anal. Chem. 92 (5), 4082–4093. doi:10.1021/acs.analchem.9b05736
Tai, Y. L., Chen, K. C., Hsieh, J. T., and Shen, T. L. (2018). Exosomes in Cancer Development and Clinical Applications. Cancer Sci. 109 (8), 2364–2374. doi:10.1111/cas.13697
Tan, F., Leung, P. H. M., Liu, Z.-b., Zhang, Y., Xiao, L., Ye, W., et al. (2011). A PDMS Microfluidic Impedance Immunosensor for E. coli O157:H7 and Staphylococcus aureus Detection via Antibody-Immobilized Nanoporous Membrane. Sensors Actuators B: Chem. 159 (1), 328–335. doi:10.1016/j.snb.2011.06.074
Tang, H.-Y., Beer, L. A., Chang-Wong, T., Hammond, R., Gimotty, P., Coukos, G., et al. (2012). A Xenograft Mouse Model Coupled with In-Depth Plasma Proteome Analysis Facilitates Identification of Novel Serum Biomarkers for Human Ovarian Cancer. J. Proteome Res. 11 (2), 678–691. doi:10.1021/pr200603h
Tang, H.-Y., Beer, L. A., Tanyi, J. L., Zhang, R., Liu, Q., and Speicher, D. W. (2013). Protein Isoform-specific Validation Defines Multiple Chloride Intracellular Channel and Tropomyosin Isoforms as Serological Biomarkers of Ovarian Cancer. J. Proteomics 89, 165–178. doi:10.1016/j.jprot.2013.06.016
Tiberio, P., Callari, M., Angeloni, V., Daidone, M. G., and Appierto, V. (2015). Challenges in Using Circulating miRNAs as Cancer Biomarkers. Biomed. Res. Int. 2015, 731479. doi:10.1155/2015/731479
Valenzuela, S. M., Alkhamici, H., Brown, L. J., Almond, O. C., Goodchild, S. C., Carne, S., et al. (2013). Regulation of the Membrane Insertion and Conductance Activity of the Metamorphic Chloride Intracellular Channel Protein CLIC1 by Cholesterol. PLoS ONE 8 (2), e56948. doi:10.1371/journal.pone.0056948
Wang, Y.-F. (2014). Combined Detection Tumor Markers for Diagnosis and Prognosis of Gallbladder Cancer. Wjg 20 (14), 4085–4092. doi:10.3748/wjg.v20.i14.4085
Witwer, K. W. (2015). Circulating microRNA Biomarker Studies: Pitfalls and Potential Solutions. Clin. Chem. 61 (1), 56–63. doi:10.1373/clinchem.2014.221341
Xie, B., Ding, Q., Han, H., and Wu, D. (2013). miRCancer: a microRNA-Cancer Association Database Constructed by Text Mining on Literature. Bioinformatics 29 (5), 638–644. doi:10.1093/bioinformatics/btt014
Yang, H., Li, P., Wang, D., Liu, Y., Wei, W., Zhang, Y., et al. (2019). Quartz Crystal Microbalance Detection of Poly(ADP-Ribose) Polymerase-1 Based on Gold Nanorods Signal Amplification. Anal. Chem. 91 (17), 11038–11044. doi:10.1021/acs.analchem.9b01366
Ye, Y., Yin, M., Huang, B., Wang, Y., Li, X., and Lou, G. (2015). CLIC1 a Novel Biomarker of Intraperitoneal Metastasis in Serous Epithelial Ovarian Cancer. Tumor Biol. 36 (6), 4175–4179. doi:10.1007/s13277-015-3052-8
Yoshioka, Y., Kosaka, N., Konishi, Y., Ohta, H., Okamoto, H., Sonoda, H., et al. (2014). Ultra-sensitive Liquid Biopsy of Circulating Extracellular Vesicles Using ExoScreen. Nat. Commun. 5, 3591. doi:10.1038/ncomms4591
Yu, W., Cui, R., Qu, H., Liu, C., Deng, H., and Zhang, Z. (2018). Expression and prognostic value of CLIC1 in epithelial ovarian cancer. Exp. Ther. Med. 15 (6), 4943–4949. doi:10.3892/etm.2018.6000
Keywords: ovarian cancer, CLIC proteins, exosomes, diagnostics, biomarkers
Citation: Hossain KR, Escobar Bermeo JD, Warton K and Valenzuela SM (2022) New Approaches and Biomarker Candidates for the Early Detection of Ovarian Cancer. Front. Bioeng. Biotechnol. 10:819183. doi: 10.3389/fbioe.2022.819183
Received: 21 November 2021; Accepted: 24 January 2022;
Published: 10 February 2022.
Edited by:
Dunja Aksentijevic, Queen Mary University of London, United KingdomReviewed by:
Assaf Zinger, Technion Israel Institute of Technology, IsraelManisha Sachan, Motilal Nehru National Institute of Technology Allahabad, India
Copyright © 2022 Hossain, Escobar Bermeo, Warton and Valenzuela. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: S. M. Valenzuela, U3RlbGxhLlZhbGVuenVlbGFAdXRzLmVkdS5hdQ==
†These authors have contributed equally to this work
 K. R. Hossain
K. R. Hossain J. D. Escobar Bermeo
J. D. Escobar Bermeo K. Warton
K. Warton S. M. Valenzuela
S. M. Valenzuela