
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 07 February 2022
Sec. Synthetic Biology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.805429
This article is part of the Research Topic Genomic Strategies for Efficient Microbial Cell Factories View all 5 articles
 Hai-Yun Gao1†
Hai-Yun Gao1† Huan Zhao1†
Huan Zhao1† Tian-Yuan Hu2
Tian-Yuan Hu2 Zhou-Qian Jiang1
Zhou-Qian Jiang1 Meng Xia1
Meng Xia1 Yi-Feng Zhang3
Yi-Feng Zhang3 Yun Lu1
Yun Lu1 Yuan Liu1
Yuan Liu1 Yan Yin4
Yan Yin4 Xiao-Chao Chen1
Xiao-Chao Chen1 Yun-Feng Luo1
Yun-Feng Luo1 Jia-Wei Zhou5
Jia-Wei Zhou5 Jia-Dian Wang1
Jia-Dian Wang1 Jie Gao1
Jie Gao1 Wei Gao1,3*
Wei Gao1,3* Lu-Qi Huang6*
Lu-Qi Huang6*Friedelin, the most rearranged pentacyclic triterpene, also exhibits remarkable pharmacological and anti-insect activities. In particular, celastrol with friedelin as the skeleton, which is derived from the medicinal plant Tripterygium wilfordii, is a promising drug due to its anticancer and antiobesity activities. Although a previous study achieved friedelin production using engineered Saccharomyces cerevisiae, strains capable of producing high-level friedelin have not been stably engineered. In this study, a combined strategy was employed with integration of endogenous pathway genes into the genome and knockout of inhibiting genes by CRISPR/Cas9 technology, which successfully engineered multiple strains. After introducing an efficient TwOSC1T502E, all strains with genetic integration (tHMG1, ERG1, ERG20, ERG9, POS5, or UPC2.1) showed a 3.0∼6.8-fold increase in friedelin production compared with strain BY4741. Through further double knockout of inhibiting genes, only strains GD1 and GD3 produced higher yields. Moreover, strains GQ1 and GQ3 with quadruple mutants (bts1; rox1; ypl062w; yjl064w) displayed similar increases. Finally, the dominant strain GQ1 with TwOSC1T502E was cultured in an optimized medium in shake flasks, and the final yield of friedelin reached 63.91 ± 2.45 mg/L, which was approximately 65-fold higher than that of the wild-type strain BY4741 and 229% higher than that in ordinary SD-His-Ura medium. It was the highest titer for friedelin production to date. Our work provides a good example for triterpenoid production in microbial cell factories and lays a solid foundation for the mining, pathway analysis, and efficient production of valuable triterpenoids with friedelin as the skeleton.
Friedelin is a typical friedelane-type pentacyclic triterpenoid extracted from cork and stem bark of various plants, such as Quercus suber (Cristina et al., 2006), Calophyllum pinetorum (Bello et al., 2008), and Drypetes tessmanniana (Kuete et al., 2010). In recent years, a considerable number of studies have reported that friedelin exhibits remarkable pharmacological activities, such as anti-inflammatory, hypolipidaemic, and antidiabetic activities (Antonisamy et al., 2011; Duraipandiyan et al., 2016; Sunil et al., 2021). In particular, friedelin is the critical precursor of celastrol, which is isolated from the medicinal plant Tripterygium wilfordii and represents one of the most promising drugs used in anticancer and antiobesity studies (Zhou et al., 2019). Thus, friedelin and its derivatives provide potential resources for the development of new drugs or dietary supplements. Additionally, friedelin also plays an important role in agriculture. For example, friedelin shows excellent natural anti-insect activity for plant protection (Baskar et al., 2014), and Dong et al. found that friedelin could regulate soil microbial community dynamics as an allelochemical (Dong et al., 2014).
However, the supply of friedelin from plant sources is insufficient, while chemical methods are often complex, include extreme reaction conditions, and produce toxic chemicals; thus, it is urgent to develop an environmentally friendly and cost-effective method of securing the friedelin supply (Moses et al., 2013; Guo et al., 2020). Synthetic biology and metabolic engineering have provided promising and green approaches to reconstruct microorganisms to yield these natural high-value products. Sharing the common upstream biosynthetic pathway with plant triterpenes, Saccharomyces cerevisiae could provide the essential precursor 2,3-oxidosqualene, which is cyclized into diverse triterpene skeletons by oxidosqualene cyclases (OSCs) (Thimmappa et al., 2014; Rahimi et al., 2019). In S. cerevisiae, the compatibility with most membrane proteins and feasibility of genetic engineering make it a perfect chassis for the production of numerous triterpenes (Carsanba et al., 2021), such as ginsenosides Rh2 (Wang et al., 2019), α-amyrin (Yu et al., 2020), and oleanolic acid (Zhao et al., 2018).
Friedelin is the most highly rearranged pentacyclic triterpene in plants, (Han et al., 2019; Zhou et al., 2019; Lu et al., 2021), the cyclization mechanism of which can be seen in Figure 1. In our previous study, friedelin production in engineered yeast was achieved by combining CRISPR/Cas9 technology with plasmid overexpression of genes, and the yield of friedelin was 37.07 mg/L. However, when strain ZH1, stored in the refrigerator at −80°C, was fermented and tested under the same operating conditions, the yield of friedelin was only comparable to that of the wild-type strain BY4741, which was likely caused by the instability of the plasmid (Zhang et al., 1996; Flagfeldt et al., 2009). In the process of strain preservation and activation, the characteristics of the engineered strain were lost due to the loss of plasmids or partial functional fragments, which led to a sharp decline in the production of friedelin. Hansen et al. obtained a haploid strain containing seven deletions, and it showed friedelin production of only 0.5 mg/L (Hansen et al., 2020). Obviously, the low yield of friedelin and instability of the strains limit its further application.
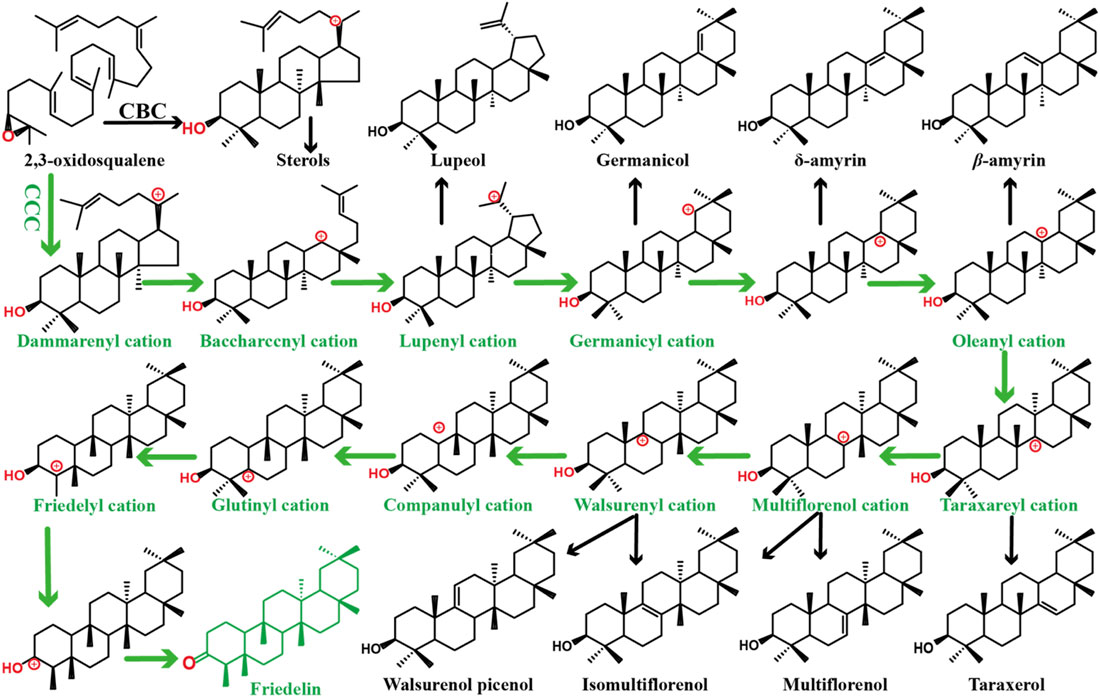
FIGURE 1. Mechanism for the cyclization of 2,3-oxidosqualene into the most rearranged pentacyclic friedelin and diverse triterpenoid skeletons. The green arrows indicate rearrangement steps including 2,3-oxidosqualene protonation, cyclization, several rearrangements, and deprotonation. The black arrows represent the formation of other triterpenoid skeletons. C–C–C, chair–chair–chair; C–B–C, chair–boat–chair.
In this study, we constructed a stable and high-yielding engineered strain, GQ1, by integrating multiple endogenous genes into yeast chromosomes and regulating genes of competing pathways. These endogenous genes include truncated 3-hydroxyl-3-methylglutaryl-CoA reductase (tHMG1), farnesyl diphosphate synthase (ERG20), squalene synthase (ERG9), and 2,3-oxidosqualene synthase (ERG1), which are involved in the mevalonate (MVA) pathway, as well as an uptake control transcriptional regulator (UPC2.1) and POS5 gene (encoded a NADH kinase), which participate in the auxiliary pathway. Overexpression of these genes represented an effective strategy for increasing the metabolic flow of terpenes according to the literature (Dai et al., 2012; Paddon et al., 2013; Dai et al., 2014; Paramasivan and Mutturi, 2017; Li et al., 2020; Sun et al., 2021). At the same time, to further increase the metabolic flux of 2,3-oxidosqualene, the genes BTS1 (geranylgeranyl diphosphate synthase), YPL062w, YJL064w, and ROX1 (repressor of hypoxia) on the competing pathways were knocked out by the CRISPR/Cas9 system (Figure 2). Then, the catalytic efficiency of friedelin synthase (FRS) from different species was evaluated, and TwOSC1T502E from T. wilfordii showed the highest efficiency in the production of friedelin. Finally, TwOSC1T502E was transformed into the dominant strain GQ1 and cultured in an optimized medium in shake flasks. The final yield of friedelin was 63.91 ± 2.45 mg/L, which was approximately 65-fold higher than that of the wild-type strain BY4741 and 229% higher than that in ordinary SD-His-Ura medium. To the best of our knowledge, the 63.91 ± 2.45 mg/L yield of friedelin produced in a shake flask represented the highest titer to date. Our work sets a good example for triterpenoid production in microbial cell factories and lays a solid foundation for the mining of valuable triterpenoids with friedelin as the precursor skeleton and the analysis of biosynthesis pathways and efficient synthesis.
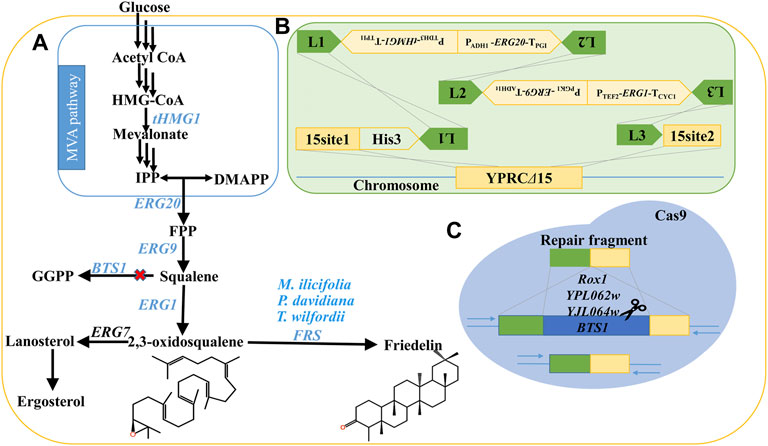
FIGURE 2. Strategies employed to enhance the production of friedelin in S. cerevisiae. (A) The biosynthesis pathway of friedelin in S. cerevisiae. (B) Key genes (tHMG1, ERG20, ERG9, ERG1) of the MVA pathway were integrated into the genome of BY4741 to increase the metabolic flux of precursor 2,3-oxidosqualene. (C) Knock out genes affecting the MVA pathway by CRISPR/Cas9 technology, including BTS1, ROX1, YPL062w and YJL064w. The red X represents blocking. IPP: isopentenyl diphosphate; FPP: farnesyl diphosphate; GGPP: (E,E,E)-geranylgeranyl diphosphate; FRS: friedelin synthase.
The initial strain used in this study was BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), which was cultivated in YPD (1% yeast extract (OXOID, England), 2% peptone (OXOID), and 2% glucose) medium. Recombinant yeast strains were selected on synthetic dropout (SD) medium (Fun-Genome Company, China) at 30°C. E. coli Trans1-T1 (TransGen Biotech, Beijing, China) was used for plasmid construction, maintenance and amplification, and the bacteria were grown at 37°C in Luria-Bertani (LB) medium (5 g/L yeast extract, 10 g/L tryptone, and 10 g/L NaCl; 20 g/L agar was added for solid medium) with 100 μg/ml ampicillin or 50 μg/ml kanamycin if necessary.
To integrate multiple transcription units simultaneously, a modularized two-step (M2S) technique was performed (Li et al., 2016), and the recombinant strain GH1 was taken as an example. The plasmids used for gene overexpression were stored in our laboratory, and detailed plasmid information is shown in Supplementary Table S1.
The first step is to clone the promoter sequence, gene, and terminator sequence. Overexpressed gene sequences (Thmg1, ERG20, ERG9, ERG1, and POS5) were downloaded from the Saccharomyces Genome Database (SGD) (yeastgenome.org). The gene sequence of UPC2.1 was based on Hu. et al. (Hu et al., 2020) and amplified from the UPC2.1-Blunt zero plasmid. DNA sequence information of genes is listed in Supplementary Table S5. The genome of S. cerevisiae strain BY4741 was extracted as a template to amplify the open reading frame (ORF) of overexpressed genes according to the instructions of the yeast genome extraction kit (Tiagen Biochemical Technology (Beijing) Co., LTD.). According to the sequence information of genes (Ptdh3, Padh1, TPI1t, PGIt, Thmg1, and ERG20), primers with a Type IIS restriction endonuclease BsaI recognition site (GGTCTCNNNNN) were designed. The promoter Ptdh3-Padh1, terminator TPI1t-PGIt, and genes Thmg1 and ERG20 were cloned by PCR with Phusion® High-fidelity DNA Polymerase (New England Biolab, United States) using plasmids P1, T1, and BY4741 as templates. PCR products were recovered by a Gene JET Gel Extraction Kit (TransGen Biotech, Beijing, China). The Golden Gate reaction was performed on the purified DNA fragments with T4 DNA ligase under the following conditions: 25 cycles of 37°C for 3 min and 16°C for 4 min, followed by 50°C for 5 min and 80°C for 5 min, and then held at 4°C. The reaction products were transformed into E. coli Trans-T1 competent cells and cultured at 37°C, and then assembled plasmids were sequenced by Tianyibiotech (Beijing, China). The correct plasmid was stored, and overexpression module I was constructed. The results are shown in Supplementary Figure S1. Other overexpression modules were also constructed according to this method. All primers are listed in Supplementary Table S2.
The second step is to construct homologous head and tail arms for integration. Primers were designed based on the sequence information for L1, L4, pESC-His and the S. cerevisiae chromosome XVI-15 site (15 site-1: 15 site-1-F/R; His: His (15 site-1)-F/R; L1: L1 (15 site-1)-F/R; L4: L4-15 site2--F/R; 15 site2: 15 site2-F/R). The 15 site1, His, L1, L4, and 15 site2 fragments were cloned by PCR with Phusion® High-fidelity DNA Polymerase (New England Biolab, United States) using plasmids T1 and T3 and genomic DNA as templates. After the PCR products were recovered, overlap PCR was performed to construct head and tail homologous arms, and the results are shown in Supplementary Figure S1.
Finally, these overexpression modules were integrated into the yeast genome in different combinations. For genomic integration, transcription unit modules, selective markers, and integrated homologous arm modules were transformed into S. cerevisiae BY4741 cells via electroporation. Competent cells were prepared according to the following protocol: single colonies were inoculated in 4 ml liquid YPD to OD600 = 0.6–1.0 and collected via centrifugation at 8,000 × g for 1 min. The obtained cell pellet was washed twice using 1 ml precooled sterile water and then incubated in 4 ml transformation reagent (10 mM LiAc, 10 mM DTT, 0.6 M sorbitol, 10 mM pH 7.5 Tris–HCl) for 20 min at 25°C. Conditioned cells were collected by centrifugation, washed twice using 1 ml ice-cold 1 M sorbitol buffer, and then resuspended to a final volume of 100 μL in sorbitol buffer. Cells with 500 ng DNA fragments were electroporated at 3 kV, 25 μF, and 200 Ω (Bio–Rad, Hercules, CA), incubated in 1 ml sorbitol buffer for 2 h at 30°C, and then plated on selective media for 2–3 days.
The gRNA expression vector p426-SNR52p-gRNA.CAN1. Y-SUP4t (ref. no. 43803), the Cas9 expression vector p414-TEF1p-Cas9-CYC1t (43,802), and the general vectors pTY-U01 and pTY-U02 used for gene knockout were preserved in the laboratory. Specific gRNAs targeting BTS1, ROX1, YPL064w, and YJL062w were designed using an open-source tool http://yeastriction.tnw.tudelft.nl, and efficient target sequences were selected (Mans et al., 2015). All gRNA target sequences used in this study are listed in Supplementary Table S3. To obtain the single gRNAs, equal molar ratio solutions of 24 nt oligos F and R were mixed and annealed, resulting in a double-stranded insert with overhangs at both ends. Then, plasmids expressing single gRNAs were constructed by inserting the double-stranded oligos into the AarI recognition site of pTY-U01 using Golden Gate assembly (Engler et al., 2014; Lee et al., 2015). Similar to the construction of the single-gRNA plasmids, BTS1-gRNA was efficiently inserted into the plasmid pTY-U02 and resulted in a 2gRNA expression plasmid, and YJL064w-gRNA was successfully inserted into the plasmid harbouring YPL062w-gRNA (Hu et al., 2020) (Supplementary Figure S3). The details of the reaction conditions are listed, and the primers are shown in Supplementary Table S3.
To obtain strains of specific genotypes, double-stranded DNA (ds-Oligo)-mediated homologous recombination repair is required. Homologous repair fragments of ROX1, BTS1, YPL062w, and YJL064w were amplified from the genome of yeast BY4741 (300 bp upstream and downstream of the gene’s ORF) and then spliced by overlapping PCR to obtain the homologous repair fragments of four genes. The primers can be found in Supplementary Table S3. Next, the Cas9 expression vector P414-Leu-TEF1p-Cas9-CYC1t was introduced into GH1, GH2, GH3, and GH4 cells according to the directions of the Frozen-EZ Yeast Transformation II Kit (Zymo Research, Irvine, CA, United States). Four strains with the Cas9 expression plasmid were used for the following manipulations. According to different knockout purposes, a total of 500 ng of the gRNA expression plasmid was mixed with 1 μg of each corresponding homologous repair fragment and electroporated into competent cells containing the Cas9 plasmid. The plasmid information used for gRNA construction is listed in Supplementary Table S4. Electroporation conditions were the same as those mentioned above. After cultivation on synthetic drop-out medium without histidine and leucine at 30°C for 2–3 days, four mutant colonies were selected from each plate, genomic DNA was isolated for use as a template for PCR, and the products were verified by DNA sequencing. The isolated genomic DNA of strain BY4741 was used as a negative control. Then, the gRNA plasmids with URA3 labelling were removed with 5-fluoroorotic acid (Laughery et al., 2015), and Cas9 plasmids were removed as described in a previous study (Mans et al., 2015; Hu et al., 2020). The recombinant strains with desired genotypes were used in the following experiments.
MiFRS from Maytenus ilicifolia (Souza-Moreira et al., 2016) and PdFRS from Populus davidiana (Han et al., 2019) (GenBank accession numbers: KX147270.1 and KY931453.1, respectively) were synthesized by Ruibiotech (Beijing, China) and cloned into the cloning sites (BamHI/SalI) of the pESC-Leu plasmid. The sequences of FRSs were downloaded from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). And the codon-optimized plasmid pYES2-TwOSC1T502E from T. wilfordii was preserved in the laboratory (GenBank accession number: KY885467.1). These three constructed vectors were transformed into recombinant strains using the Frozen-EZ Yeast Transformation II Kit (Zymo Research, Irvine, CA, United States), and the pYES2 vector and pESC-Leu vector were transformed into yeast as a control. The transformants were screened on the corresponding solid plates and confirmed by colony PCR and DNA sequencing. Plasmid and sequence information is listed in Supplementary Tables S1, S5, respectively.
The positive transformants were then cultured in flasks (100 ml) containing 30 ml of SC-His-Ura and SC-His-Leu medium containing 2% glucose and incubated with shaking at 200 rpm and 30°C for 2 d. Then, the cells were collected and induced in 30 ml of SC-Ura-His medium with 2% galactose in place of glucose and further cultured at 200 rpm and 30°C for 72 h. The dominant strain harbouring pYES2-TwOSC1T502E was cultured in medium containing 5% glucose, 1% yeast extract, 3% peptone, 0.8% KH2PO4, and 0.6% MgSO4·7H2O. After culturing at 30°C and 220 rpm for 2 d, the cells were collected and induced in medium containing 5% galactose, 1% yeast extract, 3% peptone, 0.8% KH2PO4, and 0.6% MgSO4·7H2O at 30°C and 200 rpm for 72 h. Finally, 10 ml yeast cells were collected and boiled for 15 min with 10 ml 20% KOH and 50% EtOH, the supernatant was extracted with 15 ml hexane, and ultrasound was performed for 30 min, with the process repeated twice. All extracts were combined and then evaporated by rotary evaporation.
Next, the extracts were dissolved in 1 ml hexane for analysis by gas chromatography and mass spectrometry (GC–MS). For analysis, a 1 μL sample was injected into an Agilent 7000 gas chromatograph in a nonshunt manner using a DB-5 ms capillary column (injector temperature, 305°C) with a DB-5 ms (30 m × 250 μm, film thickness 0.1 μm) capillary column. One microlitre of the concentrated organic phase was then injected at a He flow rate of 1 ml/min with a temperature program of 1 min at 50°C, followed by a gradient from 50 to 260°C at 50°C/min and then to 305°C at 20°C/min, with a 15 min hold. The ion trap temperature was 230°C. The electron energy was 70 eV. Spectra were recorded in the range of 10–550 m/z. Standard chemicals were purchased from YuanYe Biotech (Shanghai, China).
When the functional MiFRS gene in expression plasmid was introduced into the wild-type strain BY4741, the product friedelin reached only 0.39 mg/L (Figure 3A). To raise the yield of friedelin in yeast, a strategy enriching supply of the precursor, 2,3-oxidosqualene, was first considered. In this study, the key genes including tHMG1, ERG20, ERG9, and ERG1 were integrated into the genome of S. cerevisiae BY4741 under the strong promoters PTDH3, PADH1, PPGK1, and PTEF2, leading to the initial recombinant strain GH1. To test the effects of the genomic integrated strains, the MiFRS gene was introduced in plasmid and the friedelin yield was determined by GC-MS. The strain GH1 harbouring MiFRS could produce 4.7-fold friedelin with the control BY4741 (Figure 3A). Some previous cases have proved increased terpene production after these genes’ overexpression, Yu et al. and Wang et al. both overexpressed the tHMG1, ERG20, ERG9, and ERG1 genes simultaneously in yeast, thereby significantly increasing the production of α-amyrin and ginsenoside Rh2 (Wang et al., 2019; Yu et al., 2020), which further demonstrated the feasibility of our research design. POS5, which encodes an NADH kinase, can provide more NADPH to serve as electron donor for HMGR and squalene synthase. Various increased yields were achieved with different gene combinations. Thus, an alternative recombinant strain GH2 was generated after replacement of ERG20 with POS5, and yielded 1.35 mg/L of friedelin, which was equivalent to GH1 (Figure 3A). Li et al. verified that the co-expression of tHMG1 and POS5 could form a push-pull strategy, thus improving the yield of squalene (Li et al., 2020). In S. cerevisiae, overexpression of tHMG1 has commonly been used to increase the production of many terpenoids, such as artemisinin (Paddon et al., 2013), ginsenosides (Dai et al., 2014), and β-carotene (Yan et al., 2012). Multi-copy overexpression of tHMG1 benefits the production of different terpenoids (Yu et al., 2020), UPC2.1 enables S. cerevisiae to take up external sterols during aerobic cultivation. Therefore, an extra overexpression module with tHMG1 and UPC2.1 was constructed in the strains GH1 and GH2. However, the resulting strains GH3 and GH4 showed no more increase compared with strains GH1 and GH2. In brief, the recombinant strains GH1∼GH4 harbouring MiFRS improved the production of friedelin to varying degrees compared with the wild-type strain BY4741, which was GH1>GH2>GH4>GH3 (Figure 3A). The initial recombinant strains GH1 and GH2 had obvious advantages over GH3 and GH4 in friedelin yield improvement through genomic integration. Moreover, to obtain better expression level of the genes, the multiple overexpression cassettes were integrated into the YPRCΔ15 site of BY4741, because the YPRCΔ15 site was reported to be one of the integration sites with high β-galactosidase activity, and simultaneous integration of multiple genes was achieved at this site (Flagfeldt et al., 2009; Li et al., 2016). The schematic diagrams for overexpression module construction correspond to A, B, C, and D in Supplementary Figure S1. Genotypes of each strain are listed in Table 1.
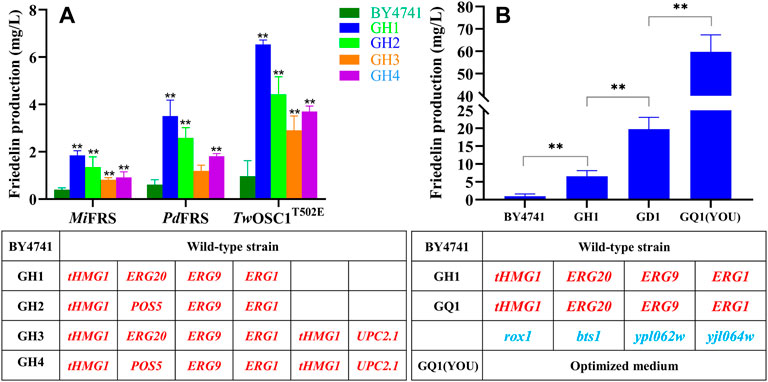
FIGURE 3. Comparison of the yield of FRSs from different plant sources and the transformation process of the dominant strain GQ1. (A) The comparison chart of the yield of friedelin obtained by introducing three FRSs into the recombinant strain GH1, GH2, GH3, GH4, respectively. The asterisk shows a statistical significance between engineered strains and wild-type strain BY4741. (B) Diagram of the transformation process of the dominant strain GQ1, (YOU) represents the use of optimized medium for cultivation. The asterisk shows a statistical significance between engineered strains (*p < 0.05, **p < 0.01). In the table below, uppercase red indicates overexpressed genes, and the lowercase blue indicates knocked out genes.
In the strategy of constructing engineered strains of yeast, the catalytic efficiency of enzymes is another key to realizing high-yield friedelin in addition to a sufficient precursor supply. To date, several FRSs from only four plant species have been functionally characterized (Zhou et al., 2021), among which MiFRS (Souza-Moreira et al., 2016) and PdFRS (Han et al., 2019) were reported to be monofunctional, and others were multifunctional, with friedelin as the major product. Multifunctional TwOSC1T502E proved to significantly increase friedelin production in our previous study, and monofunctional MiFRS and PdFRS were selected and transformed into strains BY4741, GH1, GH2, GH3, and GH4, respectively. The catalytic activity of different FRSs was evaluated in shake flask experiments. Notably, all strains harbouring TwOSC1T502E displayed a higher yield of friedelin than those with MiFRS or PdFRS (Figure 3A). Hence, TwOSC1T502E was finally confirmed as the most efficient FRS and introduced into the recombinant strains for yield evaluation.
Interestingly, the GC–MS results showed that in the extractions of yeast expressing PdFRS and MiFRS, not only friedelin but also β-amyrin and α-amyrin were detected (Supplementary Figure S2). From the perspective of the complex cyclization mechanism of friedelin (Figure 1), the production of by-products β-amyrin and α-amyrin is also reasonable. It was possible that in the functional identification of these two FRS experiments, the amount of 2,3-oxidoxysqualene provided by the chassis strain they used was insufficient, resulting in the by-products not being detected.
Next, to further improve friedelin production, we downregulated the metabolic flux towards its competing pathways. BTS1 is the first enzyme in the branching pathway for diterpenoid biosynthesis that catalyses the condensation of farnesyl diphosphate (FPP) and isopentenyl diphosphate (IPP) to (E,E,E)-geranylgeranyl diphosphate (GGPP), which would decrease the 2,3-oxidosqualene supply for triterpene production due to its high consumption of FPP and IPP (Jiang et al., 1995; Tokuhiro et al., 2009). ROX1 is a transcriptional regulator that represses many genes involved in the MVA pathway and ergosterol synthesis, and ROX1 deletion has been proven to increase the expression of multiple ERG genes, such as ERG9 and ERG1 (Henry et al., 2002; Kwast et al., 2002; Jorda and Puig, 2020). Here, two gRNA expression plasmids were constructed to target BTS1 and ROX1 (shown in Supplementary Figure S3) and transformed into strains GH1∼GH4, resulting in the double-locus knockout strains GD1∼GD4. Moreover, some studies have reported that simultaneous deletion of genes YPL062w and YJL064w would increase metabolic flow in the MVA pathway, thus increasing the yield of terpenoids (Giaever et al., 2002; Ozaydin et al., 2013; Chen et al., 2016; Zhou et al., 2019; Hu et al., 2020). Therefore, based on GD1∼GD4, YPL062w, and YJL064w were then knocked out using the same method as above. Finally, the genomically modified strains GQ1∼GQ4 were obtained with the CRISPR/Cas9 approach.
After successful genome editing of multiple genes that would influence friedelin production, the yield of friedelin from the wild-type strain and 12 recombinant strains were evaluated through fermentation in shake flasks. With the introduction of efficient TwOSC1T502E, the product friedelin was detected by GC–MS. Statistically significant differences were analysed using the independent samples t-test by SPSS software. As shown in Figure 3A, through genetic manipulation, the overexpression strains presented similarly increased production of friedelin when harbouring different FRSs. This result confirmed that the overexpression modules containing tHMG1, ERG20, ERG9, and ERG1 were critical for high production of friedelin in S. cerevisiae. The highest yield was 6.53 ± 0.13 mg/L in the GH1 strain, which was approximately 7-fold higher than that in the wild-type strain BY4741. Many studies have shown that introducing UPC2.1 into engineered yeast can increase the yield of terpenes or ergosterol (Ro et al., 2006; Dai et al., 2012; Paddon et al., 2013; Paramasivan and Mutturi, 2017; Sun et al., 2021). However, after integration of UPC2.1 and another copy of tHMG1 in the background of GH1 and GH2, the resulting strains GH3 and GH4 harbouring FRSs did not show increased harvest (Figure 3A), which indicated that more overexpression modules were not always better. Similar results about UPC2.1 integration were reported in some studies, and more overexpression cassettes introduced by multiple genetic manipulations would increase the metabolic burden on the strains (Hu et al., 2020). Hu et al. failed to improve the GGPP dephosphorylated derivative (E,E,E)-geranylgeraniol (GGOH) production, when UPC2.1 was integrated into the YPL062w deletion site in the strain BY-HZ13. Ro et al. found only a modest effect on amorphadiene production when overexpressed in the EPY208 background, which was not significantly improved (Ro et al., 2006). Another possible explanation was that the precursor flux is being channelled to branching pathways. But the ergosterol levels were measured in our study, and were consistent with friedelin production, which did not support the explanation above. The mechanism of UPC2.1 were illustrated deeply in future work.
Interestingly, strain GH1 with TwOSC1T502E increased friedelin production by 1.85-fold through integration with ERG20 compared to strain GH2 with POS5 overexpression in addition to the three common genes (tHMG1, ERG9 and ERG1). This phenomenon suggested that ERG20 was superior to POS5 for friedelin yield. As Gao reported in 2018, overexpression of POS5 gene in strain GW6 with ERG20/NADH-HMGr/PPDS-ATR1 expression cassette could enhance NADPH supply and produced protopanaxadiol 3∼4.55 fold higher under different sugar fermentations (Gao et al., 2018). Whether overexpression of ERG20 and POS5 exhibit synergistic effect needs further research.
Furthermore, when the ROX1 and BTS1 genes were knocked out simultaneously, the resulting double-mutant (rox1; bts1) strains GD1∼GD4 showed different trends of friedelin production (Figure 4). The friedelin yield of strains GD1 and GD3 increased approximately 1.4∼2.9-fold compared with that of strains GH1 (Figure 4A) and GH3 (Figure 4C), while the yield dramatically decreased in strains GD2 (Figure 4B) and GD4 (Figure 4D) when TwOSC1T502E was introduced. Moreover, strain GD4 failed to survive. The growth curve demonstrated that all recombinant strains harbouring TwOSC1T502E had higher OD600 values than BY4741 and had similar growth cycles except strain GH4. The growth remained steady from 96 to 144 h. It is worth mentioning that strain GH4 displayed slower growth than BY4741 and other recombinant strains, and GH4 had no growth after induction (Supplementary Figure S19). The reasonable hypothesis was that the overexpression of cassettes caused a considerable burden to cell growth, and further knock-out led to a loss of activity due to growth pressure. Previous literatures reported some engineered strains exhibited extended cycles (Hu et al., 2020; Yu et al., 2020). This result indicated that the recombinant strains except GH4 had better growth state and the non-extended growth cycle would not consume excessive cost. When YPL062w and YJL064w were knocked out, the quadruple mutant (rox1; bts1; ypl062w; yjl064w) showed a similar effect, leading to a great increase in friedelin production in the as-constructed strains GQ1 and GQ3, a decrease in GQ2 and even death to GQ4. Notably, among the tested strains, the quadruple mutant GQ1 harbouring TwOSC1T502E had the highest friedelin production level (19.75 ± 2.78 mg/L) in the shake flask experiments, which was approximately 20-fold higher than that of the wild-type strain BY4741 (Figure 4A). These findings suggested that the POS5 gene in strains GD2 and GD4 may interact negatively with these knockout genes under different genetic backgrounds. Meanwhile, ERG20 had a synergetic effect with the quadruple mutant genes on friedelin production. In conclusion, the deletion of ROX1 and BTS1 had significant effects on friedelin production for strains GH1 and GH3. The recombinant strain GQ1 harbouring TwOSC1T502E was determined to be the dominant strain for a high yield of friedelin.
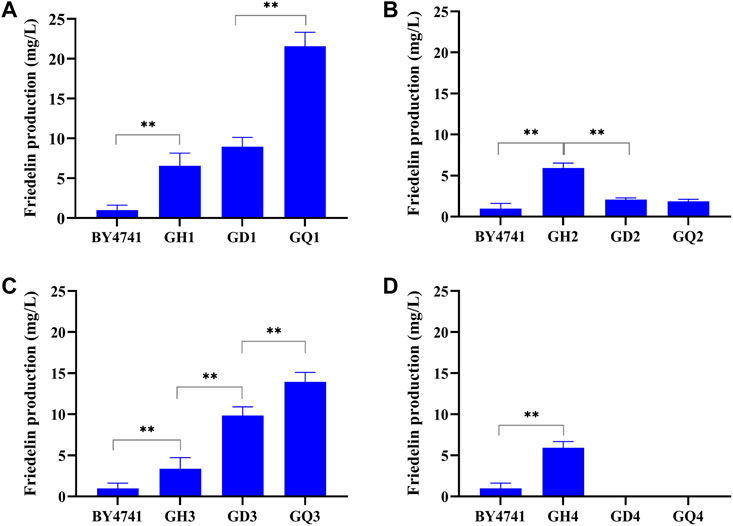
FIGURE 4. Comparison of the yield of various recombinant strains obtained by overexpressing different gene modules and knocking out different genes. BY4741 was the original strain. GHn stands for different overexpression modules, GDn stands for knockout of two genes on the basis of overexpression, and GQn stands for knockout of 4 genes on the basis of overexpression, n=1, 2, 3, 4. (A) Friedelin yield of strains GH1, GD1, GQ1. (B) Friedelin yield of strains GH2, GD2, GQ2. (C) Friedelin yield of strains GH3, GD3, GQ3. (D) Friedelin yield of strain GH4. Error bars represented standard deviations of biological quintuples.
From the above study, we screened the dominant strain GQ1 and the highly efficient FRS TwOSC1T502E. After transforming TwOSC1T502E into strain GQ1, medium optimization was used to cultivate the dominant strain. Several studies have shown that medium optimization is an effective approach to improving diterpenoid and triterpenoid production (Song et al., 2017; Zhou et al., 2019; Hu et al., 2020). Therefore, we used a medium containing 5% glucose, 1% yeast extract, 3% peptone, 0.8% KH2 PO4, and 0.6% MgSO4·7H2O to culture the friedelin high-yield strain GQ1. As shown in Figure 3B, the optimized YPD medium led to a significant increase in friedelin production. The highest yield of friedelin in strain GQ1 under shake-flask conditions was 63.91 ± 2.45 mg/L, which was approximately 65-fold higher than that of the wild-type strain BY4741 and 229% higher than that in ordinary SD-His-Ura medium. Due to the changes in metabolic flux, recombinant strains have different growth conditions and nutritional requirements compared to the original strains. The optimized YPD medium with rich nutrition can precisely meet the high-density growth of engineered bacteria, thereby increasing the yield of friedelin. These results demonstrated that the optimized YPD medium could indeed significantly increase the production of friedelin. To the best of our knowledge, the 63.91 ± 2.45 mg/L yield of friedelin produced in a shake flask was the highest titer to date.
In this study, a high-yielding friedelin engineered strain was constructed by comprehensive engineering strategies. First, multiple endogenous genes were simultaneously integrated into yeast chromosomes based on the M2S technique, and the yield of friedelin in recombinant strain GH1 was approximately 7-fold higher than that in the wild-type strain BY4741. Then, a transcriptional regulator and three genes that impact the triterpenoid synthesis pathway and the genes of the competitive pathway were knocked out by CRISPR/Cas9 technology, and the yield of friedelin in the quadruple mutant GQ1 (rox1; bts1; ypl062w; yjl064w) was approximately 20-fold higher than that in the original strain. Next, the screened high-efficiency FRS TwOSC1T502E was introduced into the dominant strain GQ1 and cultured in optimized medium in shake flasks. The final production of friedelin was 63.91 ± 2.45 mg/L, which was approximately 65-fold higher than that of the wild-type strain and 229% higher than that in ordinary SD-His-Ura medium. To the best of our knowledge, the 63.91 ± 2.45 mg/L yield of friedelin based on shake flask production was the highest titer in heterologous production reported thus far. The engineered yeast strains constructed in this work not only provide new ideas for enhancing friedelin production but can also be used as chassis strains for the mining, pathway analysis, and efficient production of valuable triterpenoids with friedelin as the skeleton.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
H-YG, HZ, WG, and L-QH conceived and designed the study. H-YG and HZ performed the experiments, drafted the manuscript. Z-QJ, MX, Y-FZ, YLu, YLi, YY, X-CC, Y-FL, J-DW, and JG participated in data analysis, discussion, and revision of the manuscript. T-YH, J-WZ, and Y-FZ provided guidance on experimental techniques. L-QH, WG, and HZ supervised the whole research and revised the manuscript. All the authors read and approved the final manuscript.
This work was supported by the National Key R&D Program of China (2020YFA0908000), the National Natural Science Foundation of China (81773830, 81973418), the Key Project at central government level: The ability establishment of sustainable use for valuable Chinese medicine resources (2060302-1806-03), and National Program for Special Support of Eminent Professionals.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past co-authorship with one of the authors LH.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.805429/full#supplementary-material
Antonisamy, P., Duraipandiyan, V., and Ignacimuthu, S. (2011). Anti-inflammatory, Analgesic and Antipyretic Effects of Friedelin Isolated from Azima Tetracantha Lam. In Mouse and Rat Models. J. Pharm. Pharmacol. 63, 1070–1077. doi:10.1111/j.2042-7158.2011.01300.x
Baskar, K., Duraipandiyan, V., and Ignacimuthu, S. (2014). Bioefficacy of the Triterpenoid Friedelin against Helicoverpa Armigera (Hub.) and Spodoptera Litura (Fab.) (Lepidoptera: Noctuidae). Pest Manag. Sci. 70, 1877–1883. doi:10.1002/ps.3742
Bello, A. A., Osmany, C.-R., Cárdenas, P. J., Lisa, P. A., and Luca, R. (2008). Constituents of the Cuban Endemic Species Calophyllum Pinetorum. J. Nat. Prod. 71, 1283–1286. doi:10.1021/np800079c
Carsanba, E., Pintado, M., and Oliveira, C. (2021). Fermentation Strategies for Production of Pharmaceutical Terpenoids in Engineered Yeast. Pharmaceuticals 14, 295. doi:10.3390/ph14040295
Chen, Y., Xiao, W., Wang, Y., Liu, H., Li, X., and Yuan, Y. (2016). Lycopene Overproduction in Saccharomyces cerevisiae through Combining Pathway Engineering with Host Engineering. Microb. Cel Fact 15, 113. doi:10.1186/s12934-016-0509-4
Cristina, M., João, M. C. M., Nagla, M., María, B., Rafael, M.-D., and Azucena, G.-C. (2006). Biovalorization of Friedelane Triterpenes Derived from Cork Processing Industry Byproducts. J. Agric. Food Chem. 54, 3566–71. doi:10.1021/jf0531151
Dai, Z., Liu, Y., Huang, L., and Zhang, X. (2012). Production of Miltiradiene by Metabolically Engineered Saccharomyces Cerevisiae. Biotechnol. Bioeng. 109, 2845–2853. doi:10.1002/bit.24547
Dai, Z., Wang, B., Liu, Y., Shi, M., Wang, D., Zhang, X., et al. (2014). Producing Aglycons of Ginsenosides in Bakers' Yeast. Sci. Rep. 4, 3698. doi:10.1038/srep03698
Dong, H.-Y., Kong, C.-H., Wang, P., and Huang, Q.-L. (2014). Temporal Variation of Soil Friedelin and Microbial Community under Different Land Uses in a Long-Term Agroecosystem. Soil Biol. Biochem. 69, 275–281. doi:10.1016/j.soilbio.2013.11.016
Duraipandiyan, V., Al-Dhabi, N. A., Stephen Irudayaraj, S., and Sunil, C. (2016). Hypolipidemic Activity of Friedelin Isolated from Azima Tetracantha in Hyperlipidemic Rats. Rev. Bras. de Farmacogn. 26, 89–93. doi:10.1016/j.bjp.2015.07.025
Engler, C., Youles, M., Gruetzner, R., Ehnert, T.-M., Werner, S., Jones, J. D. G., et al. (2014). A golden Gate Modular Cloning Toolbox for Plants. ACS Synth. Biol. 3, 839–843. doi:10.1021/sb4001504
Flagfeldt, D. B., Siewers, V., Huang, L., and Nielsen, J. (2009). Characterization of Chromosomal Integration Sites for Heterologous Gene Expression inSaccharomyces Cerevisiae. Yeast 26, 545–551. doi:10.1002/yea.1705
Gao, X., Caiyin, Q., Zhao, F., Wu, Y., and Lu, W. (2018). Engineering Saccharomyces cerevisiae for Enhanced Production of Protopanaxadiol with Cofermentation of Glucose and Xylose. J. Agric. Food Chem. 66, 12009–12016. doi:10.1021/acs.jafc.8b04916
Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Véronneau, S., et al. (2002). Functional Profiling of the Saccharomyces cerevisiae Genome. Nature 418, 387–391. doi:10.1038/nature00935
Guo, H., Wang, H., and Huo, Y.-X. (2020). Engineering Critical Enzymes and Pathways for Improved Triterpenoid Biosynthesis in Yeast. ACS Synth. Biol. 9, 2214–2227. doi:10.1021/acssynbio.0c00124
Han, J. Y., Ahn, C.-H., Adhikari, P. B., Kondeti, S., and Choi, Y. E. (2019). Functional Characterization of an Oxidosqualene Cyclase (PdFRS) Encoding a Monofunctional Friedelin Synthase in Populus Davidiana. Planta 249, 95–111. doi:10.1007/s00425-018-2985-8
Hansen, N. L., Miettinen, K., Zhao, Y., Ignea, C., Andreadelli, A., Raadam, M. H., et al. (2020). Integrating Pathway Elucidation with Yeast Engineering to Produce Polpunonic Acid the Precursor of the Anti-obesity Agent Celastrol. Microb. Cel. Fact 19, 15. doi:10.1186/s12934-020-1284-9
Henry, K. W., Nickels, J. T., and Edlind, T. D. (2002). ROX1 and ERG Regulation in Saccharomyces cerevisiae : Implications for Antifungal Susceptibility. Eukaryot. Cel. 1, 1041–1044. doi:10.1128/EC.1.6.1041-1044.2002
Hu, T., Zhou, J., Tong, Y., Su, P., Li, X., Liu, Y., et al. (2020). Engineering Chimeric Diterpene Synthases and Isoprenoid Biosynthetic Pathways Enables High-Level Production of Miltiradiene in Yeast. Metab. Eng. 60, 87–96. doi:10.1016/j.ymben.2020.03.011
Jiang, Y., Proteau, P., Poulter, D., and Ferro-Novick, S. (1995). BTS1 Encodes a Geranylgeranyl Diphosphate Synthase in Saccharomyces cerevisiae. J. Biol. Chem. 270, 21793–21799. doi:10.1074/jbc.270.37.21793
Jordá, T., and Puig, S. (2020). Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae. Genes 11, 795. doi:10.3390/genes11070795
Kuete, V., Dongfack, M. D. J., Mbaveng, A. T., Lallemand, M. C., Van-Dufat, H. T., Wansi, J.-D., et al. (2010). Antimicrobial Activity of the Methanolic Extract and Compounds from the Stem Bark of Drypetes Tessmanniana. Chin. J. Integr. Med. 16, 337–343. doi:10.1007/s11655-010-0527-8
Kwast, K. E., Lai, L.-C., Menda, N., James, D. T., Aref, S., and Burke, P. V. (2002). Genomic Analyses of Anaerobically Induced Genes in Saccharomyces cerevisiae : Functional Roles of Rox1 and Other Factors in Mediating the Anoxic Response. J. Bacteriol. 184, 250–265. doi:10.1128/JB.184.1.250-265.2002
Laughery, M. F., Hunter, T., Brown, A., Hoopes, J., Ostbye, T., Shumaker, T., et al. (2015). New Vectors for Simple and Streamlined CRISPR-Cas9 Genome Editing inSaccharomyces Cerevisiae. Yeast 32, 711–720. doi:10.1002/yea.3098
Lee, M. E., Deloache, W. C., Cervantes, B., and Dueber, J. E. (2015). A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth. Biol. 4, 975–986. doi:10.1021/sb500366v
Li, S., Ding, W., Zhang, X., Jiang, H., and Bi, C. (2016). Development of a Modularized Two-step (M2S) Chromosome Integration Technique for Integration of Multiple Transcription Units in Saccharomyces cerevisiae. Biotechnol. Biofuels 9, 232. doi:10.1186/s13068-016-0645-4
Li, T., Liu, G.-S., Zhou, W., Jiang, M., Ren, Y.-H., Tao, X.-Y., et al. (2020). Metabolic Engineering of Saccharomyces cerevisiae to Overproduce Squalene. J. Agric. Food Chem. 68, 2132–2138. doi:10.1021/acs.jafc.9b07419
Lu, Y., Liu, Y., Zhou, J., Li, D., and Gao, W. (2021). Biosynthesis, Total Synthesis, Structural Modifications, Bioactivity, and Mechanism of Action of the Quinone‐methide Triterpenoid Celastrol. Med. Res. Rev. 41, 1022–1060. doi:10.1002/med.21751
Mans, R., Van Rossum, H. M., Wijsman, M., Backx, A., Kuijpers, N. G. A., Van Den Broek, M., et al. (2015). CRISPR/Cas9: a Molecular Swiss Army Knife for Simultaneous Introduction of Multiple Genetic Modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 15, fov004. doi:10.1093/femsyr/fov004
Moses, T., Pollier, J., Thevelein, J. M., and Goossens, A. (2013). Bioengineering of Plant (Tri)terpenoids: from Metabolic Engineering of Plants to Synthetic Biology In Vivo and In Vitro. New Phytol. 200, 27–43. doi:10.1111/nph.12325
Özaydın, B., Burd, H., Lee, T. S., and Keasling, J. D. (2013). Carotenoid-based Phenotypic Screen of the Yeast Deletion Collection Reveals New Genes with Roles in Isoprenoid Production. Metab. Eng. 15, 174–183. doi:10.1016/j.ymben.2012.07.010
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., Mcphee, D., et al. (2013). High-level Semi-synthetic Production of the Potent Antimalarial Artemisinin. Nature 496, 528–532. doi:10.1038/nature12051
Paramasivan, K., and Mutturi, S. (2017). Progress in Terpene Synthesis Strategies through Engineering of Saccharomyces cerevisiae. Crit. Rev. Biotechnol. 37, 974–989. doi:10.1080/07388551.2017.1299679
Rahimi, S., Kim, J., Mijakovic, I., Jung, K.-H., Choi, G., Kim, S.-C., et al. (2019). Triterpenoid-biosynthetic UDP-Glycosyltransferases from Plants. Biotechnol. Adv. 37, 107394. doi:10.1016/j.biotechadv.2019.04.016
Ro, D.-K., Paradise, E. M., Ouellet, M., Fisher, K. J., Newman, K. L., Ndungu, J. M., et al. (2006). Production of the Antimalarial Drug Precursor Artemisinic Acid in Engineered Yeast. Nature 440, 940–943. doi:10.1038/nature04640
Song, T.-Q., Ding, M.-Z., Zhai, F., Liu, D., Liu, H., Xiao, W.-H., et al. (2017). Engineering Saccharomyces cerevisiae for Geranylgeraniol Overproduction by Combinatorial Design. Sci. Rep. 7, 14991. doi:10.1038/s41598-017-15005-4
Souza-Moreira, T. M., Alves, T. B., Pinheiro, K. A., Felippe, L. G., De Lima, G. M. A., Watanabe, T. F., et al. (2016). Friedelin Synthase from Maytenus Ilicifolia: Leucine 482 Plays an Essential Role in the Production of the Most Rearranged Pentacyclic Triterpene. Sci. Rep. 6, 36858. doi:10.1038/srep36858
Sun, Z.-J., Lian, J.-Z., Zhu, L., Jiang, Y.-Q., Li, G.-S., Xue, H.-L., et al. (2021). Combined Biosynthetic Pathway Engineering and Storage Pool Expansion for High-Level Production of Ergosterol in Industrial Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 9, 681666. doi:10.3389/fbioe.2021.681666
Sunil, C., Irudayaraj, S. S., Duraipandiyan, V., Alrashood, S. T., Alharbi, S. A., and Ignacimuthu, S. (2021). Friedelin Exhibits Antidiabetic Effect in Diabetic Rats via Modulation of Glucose Metabolism in Liver and Muscle. J. Ethnopharmacol. 268, 113659. doi:10.1016/j.jep.2020.113659
Thimmappa, R., Geisler, K., Louveau, T., O'maille, P., and Osbourn, A. (2014). Triterpene Biosynthesis in Plants. Annu. Rev. Plant Biol. 65, 225–257. doi:10.1146/annurev-arplant-050312-120229
Tokuhiro, K., Muramatsu, M., Ohto, C., Kawaguchi, T., Obata, S., Muramoto, N., et al. (2009). Overproduction of Geranylgeraniol by Metabolically Engineered Saccharomyces cerevisiae. Appl. Environ. Microbiol. 75, 5536–5543. doi:10.1128/AEM.00277-09
Wang, P., Wei, W., Ye, W., Li, X., Zhao, W., Yang, C., et al. (2019). Synthesizing Ginsenoside Rh2 in Saccharomyces cerevisiae Cell Factory at High-Efficiency. Cell Discov. 5, 5. doi:10.1038/s41421-018-0075-5
Yan, G.-l., Wen, K.-r., and Duan, C.-q. (2012). Enhancement of β-Carotene Production by Over-Expression of HMG-CoA Reductase Coupled with Addition of Ergosterol Biosynthesis Inhibitors in Recombinant Saccharomyces cerevisiae. Curr. Microbiol. 64, 159–163. doi:10.1007/s00284-011-0044-9
Yu, Y., Rasool, A., Liu, H., Lv, B., Chang, P., Song, H., et al. (2020). Engineering Saccharomyces cerevisiae for High Yield Production of α-amyrin via Synergistic Remodeling of α-amyrin Synthase and Expanding the Storage Pool. Metab. Eng. 62, 72–83. doi:10.1016/j.ymben.2020.08.010
Zhang, Z., Moo-Young, M., and Chisti, Y. (1996). Plasmid Stability in Recombinant Saccharomyces cerevisiae. Biotechnol. Adv. 14, 401–435. doi:10.1016/s0734-9750(96)00033-x
Zhao, Y., Fan, J., Wang, C., Feng, X., and Li, C. (2018). Enhancing Oleanolic Acid Production in Engineered Saccharomyces cerevisiae. Bioresour. Technol. 257, 339–343. doi:10.1016/j.biortech.2018.02.096
Zhou, J., Hu, T., Gao, L., Su, P., Zhang, Y., Zhao, Y., et al. (2019). Friedelane‐type Triterpene Cyclase in Celastrol Biosynthesis from Tripterygium Wilfordii and its Application for Triterpenes Biosynthesis in Yeast. New Phytol. 223, 722–735. doi:10.1111/nph.15809
Keywords: friedelin, triterpenes, Saccharomyces cerevisiae, genome, CRISPR/Cas9, optimized medium, engineered strain
Citation: Gao H-Y, Zhao H, Hu T-Y, Jiang Z-Q, Xia M, Zhang Y-F, Lu Y, Liu Y, Yin Y, Chen X-C, Luo Y-F, Zhou J-W, Wang J-D, Gao J, Gao W and Huang L-Q (2022) Metabolic Engineering of Saccharomyces cerevisiae for High-Level Friedelin via Genetic Manipulation. Front. Bioeng. Biotechnol. 10:805429. doi: 10.3389/fbioe.2022.805429
Received: 30 October 2021; Accepted: 17 January 2022;
Published: 07 February 2022.
Edited by:
Yun Chen, Chalmers University of Technology, SwedenReviewed by:
Yaping Yang, University of Georgia, United StatesCopyright © 2022 Gao, Zhao, Hu, Jiang, Xia, Zhang, Lu, Liu, Yin, Chen, Luo, Zhou, Wang, Gao, Gao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Gao, d2VpZ2FvQGNjbXUuZWR1LmNu; Lu-Qi Huang, aHVhbmdsdXFpMDFAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.