
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 09 September 2022
Sec. Biomaterials
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.1002291
This article is part of the Research Topic Synthesis, Functionalization, and Clinical Translation of Pharmaceutical Biomaterials, Volume II View all 8 articles
Human dental pulp stem cells (hDPSCs) have been a focus of pulp regeneration research because of their excellent odontogenic potential and availability. Applying the odontoblastic differentiation of hDPSCs to tooth regeneration has been challenging. Metformin-based carbon nanodots (MCDs) were synthesized and characterized to investigate their effects in vitro on odontoblastic hDPSC differentiation and the underlying mechanism. MCDs were synthesized by a hydrothermal treatment method and characterized using transmission electron microscopy (TEM), Fourier transform infrared spectroscopy, and X-ray photoelectron spectroscopy. The biocompatibility and fluorescence properties of the MCDs in Dulbecco’s modified Eagle’s medium high-glucose culture medium and the in vitro odontogenic potential and related mechanism of the bioactive nanomaterial was explored. TEM images showed that MCDs were spherical in shape with a size of approximately 5.9 nm. MCDs showed biological safety in cell viability, apoptosis, and fluorescence labelling ability at a concentration up to 200 μg/ml in vitro. The presence of MCDs facilitated high-efficiency odontogenic differentiation of hDPSCs by promoting odontogenic gene and protein expression. Moreover, MCDs promoted odontoblastic hDPSC differentiation via autophagy. MCDs are capable of activating autophagy and enhancing the odontogenic differentiation of hDPSCs by upregulating odontoblast gene marker (DMP1, DSPP, RUNX2, and SP7) and protein (DSPP and DMP1) expression.
Dental pulp regeneration is an ideal solution for destructive diseases of the dental pulp, such as pulpitis and pulp necrosis. Human dental pulp stem cells (hDPSCs) are prime candidates for such treatment, given their multipotent differentiation potential and lineage specification into odontoblasts (Yang et al., 2012; Mozaffari et al., 2019). To achieve functional pulp regeneration, hDPSCs need to be differentiated into different types of cells, such as odontoblasts, fibroblasts, and vascular endothelial cells (Sui et al., 2019; Ahmed et al., 2020). Studies have focused on exploring new biomaterials to manipulate hDPSC differentiation, particularly in promoting odontogenic differentiation, which is crucial for dentin regeneration, as dentin is a specific hard tissue of great significance in increasing the mechanical resistance of the root and building the dental pulp defensive system (Ahmed et al., 2020). However, applying bioactive materials to orchestrate hDPSC differentiation into odontoblasts, which specifically generate dentin, remains a challenge.
Recently, carbon dots (CDs), defined as nanoparticles <10 nm in size, which are easily surface-functionalized, have demonstrated the ability to promote mesenchymal stem cell differentiation (Shao et al., 2017; Wang et al., 2019a; Han et al., 2019; Meng et al., 2019; Zhang et al., 2020). CDs synthesized from medications hold great promise for biological application, as they not only show excellent biocompatibility, water solubility, and fluorescence but also may maintain and even enhance the original medication effect (Liu et al., 2017; Wang et al., 2019a; Han et al., 2019; Meng et al., 2019). For example, CDs prepared from zinc gluconate, magnesium gluconate, adenosine, and aspirin as raw materials all promote osteogenic differentiation of bone marrow mesenchymal stem cells, in the absence of any external osteo inductive factors (Wang et al., 2019b; Han et al., 2019; Meng et al., 2019). However, limited data are available on the effect of such CDs on hDPSCs.
As a first-line medication for type 2 diabetes, metformin exhibits pleiotropic effects on a variety of tissues (Lv and Guo, 2020). Qin’s team has reported metformin’s potential odontogenic effect by enhancing hDPSCs differentiation via the AMPK signal pathway, and designed novel calcium phosphate cement with metformin-loaded chitosan for odontogenic differentiation of hDPSCs (Qin et al., 2018a; Qin et al., 2018b). Resultantly, we propose that metformin-based CDs (MCDs) could be effective and safe odontoinductive agents for deriving odontoblasts from hDPSCs. Herein, we aimed to first synthesize MCDs and then employ them to investigate the effects of MCDs on the biocompatibility and odontoblastic differentiation of hDPSCs in vitro (Figure 1).
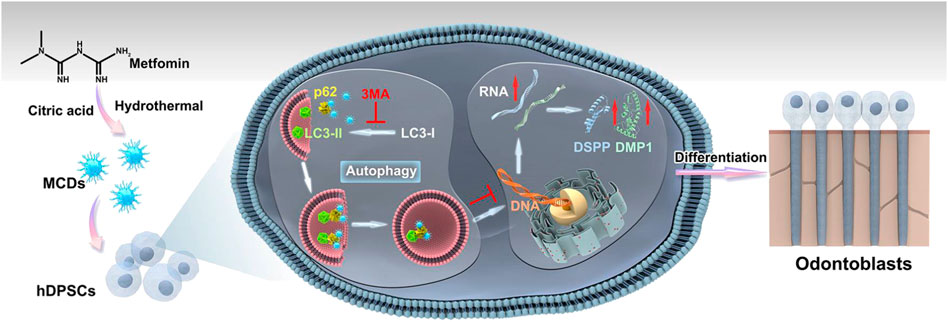
FIGURE 1. Experimental design—analytical workflow. Metformin-based carbon nanodots (MCDs) were synthesized by a hydrothermal treatment method and characterized using transmission electron microscopy (TEM), Fourier transform infrared spectroscopy, and X-ray photoelectron spectroscopy. Moreover, MCDs are capable of activating autophagy and enhancing the odontogenic differentiation of human dental pulp stem cells (hDPSCs) by upregulating odontoblast gene marker (DMP1, DSPP, RUNX2, and SP7) and protein (DSPP and DMP1) expression.
Metformin and citric acid were purchased from Shanghai Aladdin Chemicals (Shanghai, China). KCl, HCl, NaOH, Na2SO4, MgSO4, CaSO4, Fe2(SO4)3, CuSO4, ZnSO4, and AlCl3 were purchased from Beijing Chemical Reagent Co. (Beijing, China). Dulbecco’s modified Eagle’s medium high glucose (H-DMEM) and antibiotics (100 U/ml penicillin and 100 U/ml streptomycin) were purchased from Hyclone (Logan, UT, United States). Fetal bovine serum (FBS), phosphate-buffered saline (PBS), and TRIzol reagent were purchased from (Gibco, Waltham, MA, United States). Crystal violet staining reagent was purchased from Solarbio (Beijing, China). The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was purchased from Sigma‒Aldrich (St Louis, MO, United States). An Annexin V APC apoptosis analysis kit was purchased from Sungene Biotech (Tianjin, China). All antibodies, except DMP1 and DSPP, were purchased from Proteintech Group Laboratory (Wuhan, China). DMP1 and DSPP antibodies were purchased from ImmunoWay Biotechnology Co. (Plano, TX, United States) and Santa Cruz Biotechnology (Dallas, TX, United States), respectively. CD44-PE, CD90-PE, CD45-FITC, and CD11b-FITC antibodies were purchased from BioLegend (San Diego, CA, United States). 3-Methyladenine (3-MA) was purchased from Topscience (Shanghai, China). Enhanced chemiluminescence, the Hieff First-Strand cDNA Synthesis Super Mix for the RT-qPCR kit, and the Hieff qPCR SYBR Green Master Mix kit were purchased from Yeasen Biotech (Shanghai, China).
MCDs were synthesized by a one-pot hydrothermal process, as previously described (Liu et al., 2017). Briefly, 1 mmol each of metformin and CA were dissolved in 10 ml of deionized water. The mixture was then transferred to a poly (tetrafluoroethylene) (Teflon)-lined autoclave (25 ml) and heated to 200°C for 4 h. The product was cooled naturally to room temperature. Large impurities in the product were removed with a 0.22 μm filter, and the dark brown solution obtained was transferred to a 500–1,000 Da dialysis bag against deionized water for 24 h to remove excess precursors and by-products. MCD powder was collected after freeze-drying.
Transmission electron microscopy (TEM) was performed using a JEM-2100F microscope (JEOL, Tokyo, Japan). Photoluminescence (PL) spectra were recorded on an RF-5301 PC spectrophotometer (Shimadzu Corp, Kyoto, Japan). Ultraviolet-visual light (UV-vis) absorption spectra were measured on a Shimadzu 3100 UV-Vis spectrophotometer. Fourier transform infrared (FT-IR) spectra were recorded on a Nicolet AVATAR 360 FTIR spectrophotometer (Thermo Fisher Scientific; Waltham, MA, United States). X-ray photoelectron spectroscopy (XPS) analysis was performed on an ESCALAB 250 spectrometer (Thermo Fisher Scientific) with a monochromatic Al Kα excitation source (1,486.6 eV).
hDPSCs were isolated, cultured, and characterized as previously described (Gronthos et al., 2002). The procedure was approved by the Ethics Committee of the Hospital of Stomatology, Jilin University. Cells were isolated from the third molar of healthy individuals (18–25 years old) undergoing tooth extraction for orthodontic treatment. The isolated cells were maintained in H-DMEM containing 20% FBS and antibiotics, at 37°C in a humidified 5% CO2 atmosphere. When the primary cell culture reached 80% confluence, the cells were trypsinized and subcultured in H-DMEM with antibiotics and 10% FBS. The cells from the second passage were washed and resuspended in PBS and incubated with anti-human antibodies, including CD44-PE, CD90-FITC, CD45-PE, and CD11b-FITC antibodies, for 30 min at 4°C, in the dark, to examine their surface antigen expression. The cell samples were then evaluated by flow cytometry (FACS calibur; Becton Dickinson Biosciences, Franklin Lakes, CA, United States). hDPSCs from the third to fifth passages were used for subsequent experiments. Supplementary Figures S1A,B present microscope images of hDPSCs after 10 days of primary passage and in the third passage, respectively. Consistent with other mesenchymal stem cell populations, hDPSCs expressed high levels of mesenchymal markers and low levels of hematopoietic markers (Supplementary Figure S1C) (Cui et al., 2018).
MTT assays were used to detect the cytotoxic effects of MCDs. First, 200-μl aliquots with a density of 2×103 cells/mL were placed in each well of a 96-well plate and were then treated with MCD concentrations of 50, 100, 200, 400, and 800 μg/ml, followed by incubation for 1, 3, 7, 10, and 14 days at 37°C in a humidified 5% CO2 atmosphere. The medium was replaced with 180 μl PBS and 20 μl of stock MTT (5 mg/ml), after which the cells were incubated for another 4 h. Then, the medium was removed, and the dark blue formazan crystals that had formed were dissolved in DMSO. Absorbance was measured at 490 nm using a microplate reader in the shaking mode (RT-6000; Lei Du Life Science and Technology Co., Shenzhen, China). Untreated cells were used as a control to determine the relative cell viability.
Apoptosis analysis was performed using an Annexin V APC apoptosis analysis kit. Briefly, hDPSCs were seeded onto 6-well plates at a seeding density of 3×105 cells/well. Cells (1×105) from the 50, 100, 200, 400, and 800 μg/ml MCD-treated groups were collected at 7 days, washed with PBS, and resuspended in 100 μl of binding buffer. Annexin V APC (5 μl) and propidium iodide solution (5 μl) were added to the cell solution for 15 min at room temperature, followed by flow cytometry analysis within 1 h.
hDPSCs were seeded onto clean coverslips in a 6-well plate at a density of 1×105 cells/well for 24 h. Subsequently, the cells were treated with 50, 100, 200, 400, and 800 μg/ml MCD in medium for another 24 h and washed three times with PBS. Finally, the cell imaging under ultraviolet excitation wavelengths were observed under an inverted fluorescence microscope (Olympus, Tokyo, Japan). Supplementary Figures S2 shows the bright blue colour within the cytoplasm when hDPSCs were incubated with MCDs, indicating that MCDs might be applied to track the dynamic changes of cells through fluorescence.
The cell samples were collected and washed twice with PBS on ice. Total RNA was extracted using TRIzol reagent, according to the manufacturer’s protocol. DNase I-treated RNA (1 µg) was used for the reverse transcription reaction with a Hieff First Strand cDNA Synthesis Super Mix for RT-qPCR. Quantitative PCR analysis was performed using the Hieff qPCR SYBR Green Master Mix kit. The expression of each investigated gene was normalized to that of the housekeeping gene β-actin, and fold-differences were calculated using the comparative Ct method. The odontogenic markers were DSPP, DMP1, RUNX2, SP7, and COL1. The following primer sequences, generated by Sangon Biotech (Shanghai, China), were used: β-actin (forward primer: 5′-TGGCACCCAGCACAATGAA-3′, reverse primer: 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′); DSPP (forward primer: 5′-GCTGGAAGCAATAACAGTACAG-3′, reverse primer: 5′-TGCTGTTGATCTGAGGTGTTAT-3′); DMP1 (forward primer: 5′-CAAAGAAGATAGCAACTCCACG-3′, reverse primer: 5′-CATCAACTGTTAATTTCCGGCT-3′); RUNX2 (forward primer: 5′-CACTGGCGCTGCAACAAGA-3′, reverse primer: 5′-CATTCCGGAGTCAGCAGAATAA-3′); SP7 (forward primer: 5′-TGGCGTCCTCCCTGCTTG-3′, reverse primer: 5-'TGCTTTGCCCAGAGTTGTTG-3′); and COL1 (forward primer: 5′-TGTTGGTCCTGCTGGCAAGAATG-3′, reverse primer: 5′-GTCACCTTGTTCGCCTGTCTCAC-3′). Data were calculated using the 2−ΔΔCT method.
The cell samples were harvested, washed with PBS on ice, and subsequently lysed in lysis buffer containing a protease inhibitor cocktail. Proteins were separated on sodium dodecyl sulfate-polyacrylamide gels and transferred to polyvinylidene fluoride membranes. The membranes were incubated overnight with primary antibodies against LC3 at 1:500, p62 at 1:1,000, Beclin1 at 1:1,000, DMP1 at 1:500, DSPP at 1:1,000, and GAPDH at 1:5,000 dilution. Subsequently, the membranes were incubated with secondary antibodies for 60 min, and blotted bands were visualized using enhanced chemiluminescence.
hDPSCs were cultured with 50 μg/ml MCD for 24, 48, or 72 h, fixed with 2% glutaraldehyde for 1 h at 4°C, then incubated in PBS overnight, and post-fixed in 1% osmium tetroxide in the same buffer for 2 h at room temperature. After dehydration with 50, 70, 90, and 100% ethanol, the samples were embedded in Epon. Ultrathin sections of cells were cut using a Leica EMUC6 ultramicrotome (Leica Biosystems, Wetzlar, Germany) and sections were settled on 200-mesh carbon-coated copper grids. After staining with 2% uranyl acetate and lead citrate, the samples were observed using an EM-14100BINC TEM (JEOL, Tokyo, Japan) at 80 kV. TEM images were taken to observe autophagic vesicles.
To evaluate whether MCDs promote odontogenic differentiation of hDPSCs via autophagy, autophagy was blocked in hDPSCs by adding 3-MA. In this experiment, hDPSCs were treated with 50 μg/ml MCDs for 0, 6, 12, 24, 48, and 72 h. Additionally, 3-MA (0.5 mM) or 25 μg/ml metformin (as control), equivalent to the amount of metformin used for synthesis of 50 μg/ml MCDs, was added to the hDPSCs for 12 h prior to the addition of MCDs. Protein levels of LC3 and p62 were detected by western blotting, with GAPDH protein as a loading control.
Data were analysed using one-way analysis of variance with post hoc Tukey’s test using GraphPad Prism software (GraphPad Software, Inc., San Diego, CA, United States). The level of statistical significance for all tests was set at p < 0.05.
TEM images illustrated that MCDs had good dispersibility and exhibited a spherical particle shape. High-resolution TEM of MCDs showed a distinct lattice structure with a lattice spacing of 0.26 nm (Figure 2A). The average particle size was approximately 5.9 nm (Figure 2B). The PL spectrum (Figure 2C) and the interpolated images revealed that an aqueous solution of the CDs exhibited strong blue fluorescence with an emission peak at 444 nm and an optimum excitation wavelength of 367 nm. The UV-vis absorption spectrum (Figure 2D) of the MCDs contained a distinct absorption peak at 332 nm corresponding to the n→π* transition of the C=O/C=N chemical bond (Feng et al., 2017). The peak at 232 nm contributed to the π→π* transition of the aromatic C=C bond (Yang et al., 2014; Liu et al., 2017).
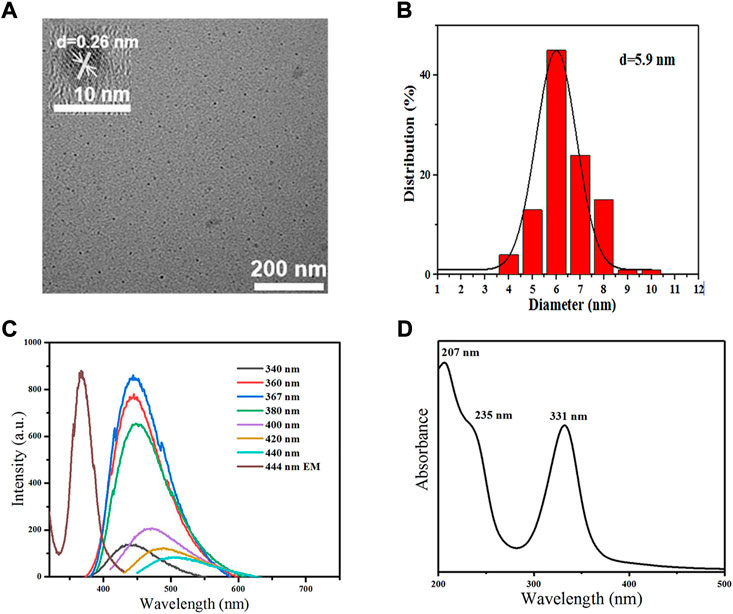
FIGURE 2. Characterization of MCDs. (A) TEM images of MCDs (scale bar: 200 nm); the inset is the HRTEM image of MCDs (scale bar: 10 nm). (B) Size distribution histogram of MCDs. (C) PL spectra of MCDs with different excitation wavelength and emission spectra of MCDs (D) UV-vis absorption of MCDs. HRTEM: high-resolution transmission electron microscopy, MCDs: metformin-based carbon nanodots, PL: photoluminescence, TEM: transmission electron microscopy, UV-vis: ultraviolet-visible.
FT-IR spectroscopy (Figure 3A) was performed to characterize the chemical groups of MCDs, and the following peaks were observed: stretching vibration of O–H at 3,300 cm−1, N–H and 3,115 cm−1, C=O at 1,680 cm−1, C=C/C=N at 1,500–1,620 cm−1, and C–N–C at 1,400 cm−1 (Feng et al., 2017; Liu et al., 2018; Yan et al., 2019), which revealed that the MCDs retained the chemical groups of metformin.
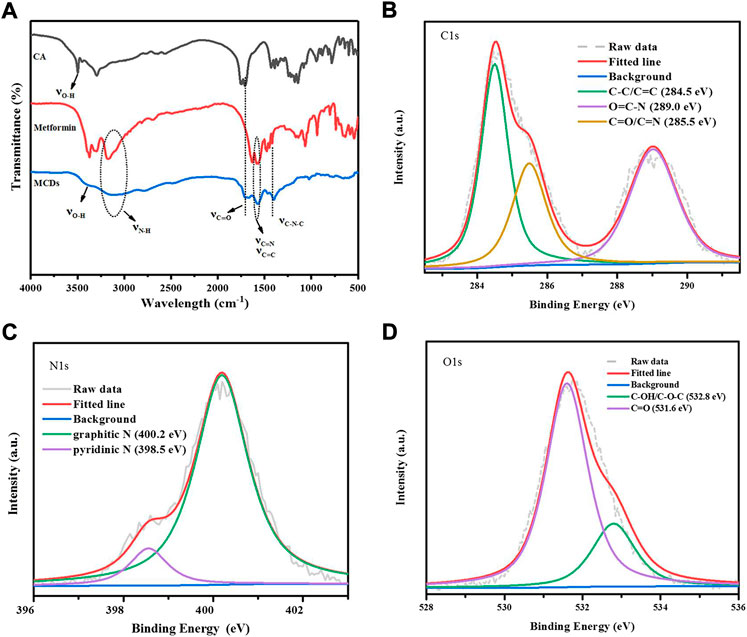
FIGURE 3. (A) FT-IR spectra of CA, MH, and MCDs, respectively. XPS analysis of MCDs. High resolution C1s (B), N1s (C), and O1s (D) peaks of MCDs. FT-IR: Fourier transform infrared, MCDs: metformin-based carbon nanodots, XPS: X-ray photoelectron spectroscopy.
XPS (Supplementary Figure S3A) was employed to further investigate the chemical structures of the MCDs. The C1s spectra (Figure 3B) showed three peaks of C–C/C=C (284.5 eV), O=C–N (289.0 eV), and C=O/C=N (285.5 eV). The N1s spectra (Figure 3C) showed two peaks: graphitic N (398.5 eV) and pyridinic N (400.2 eV). The O1s spectra (Figure 3D) appeared as two peaks of C–OH/C–O–C at 532.8 eV and C=O at 531.6 eV (Narayanan et al., 2006; D'Souza et al., 1997; Bae et al., 2018; Li et al., 2011), consistent with the FT-IR results. The above data indicated that the MCDs were rich in functional groups.
The photostability of the MCDs was also evaluated before the biological experiments. MCDs showed excellent fluorescence stability in different metal ion solutions (Supplementary Figure S3B). The PL intensity remained nearly unchanged in the pH range of 5–14 (Supplementary Figure S3C) and in high-concentration KCl solution (Supplementary Figure S3D), indicating that MCDs were applicable in the bio-microenvironment.
As shown in Figure 4A, MCDs at concentrations of 50, 100, and 200 μg/ml significantly promoted cell proliferation at 3, 7, and 10 days, while concentrations of 400 and 800 μg/ml MCDs inhibited cell proliferation at 10 and 14 days. Moreover, we determined whether the same concentration could induce apoptotic cell death using Annexin VAPC assessment by flow cytometry (Figure 4B). The group of MCDs concentrations up to 400 μg/ml showed no significant increase in apoptosis observed at 7 days, as compared with the control group. However, MCDs at concentrations of 800 μg/ml showed an increase in early apoptosis. Cumulatively, these results demonstrated that the effects of MCDs on hDPSC viability and apoptosis were dose-dependent. MCD concentrations of 50, 100, and 200 μg/ml exhibited minimal cytotoxicity in hDPSCs.

FIGURE 4. Biocompatibility of MCDs. (A) The relative viability of hDPSCs after incubation with MCDs at various concentrations for 1, 3, 7, 10, and 14 days. (B) Apoptotic ratio of hDPSCs after incubation with MCDs at various concentrations for 7 days were assessed by Annexin V-APC assay and measured by fluorescence-activated cell sorting analysis. hDPSCs: human dental pulp stem cells, MCDs: metformin-based carbon nanodots. MCDs promote the odontogenic differentiation of hDPSCs.
The gene expression levels of odontoblastic markers, such as dentin matrix protein 1 (DMP1), dentin sialo phosphoprotein (DSPP), runt-related transcription factor 2 (RUNX2), specificity protein 7 (SP7), and type 1 collagen (COL1) were measured to investigate the effects of MCDs on the odontoblastic differentiation of hDPSCs at 7 and 14 days (Figures 5A,B). The expression levels of DSPP, DMP1, and SP7 were significantly increased (p < 0.05) in all MCD-treated groups at 7 and 14 days, indicating that MCDs promoted the differentiation of hDPSCs into odontoblasts. The expression of RUNX2 increased significantly (p < 0.05) at 7 days and did not increase further after 14 days. The results clearly demonstrated that MCDs significantly enhanced the expression of odontogenic genes, thus preliminarily proving that MCDs can promote the odontogenic differentiation of hDPSCs.
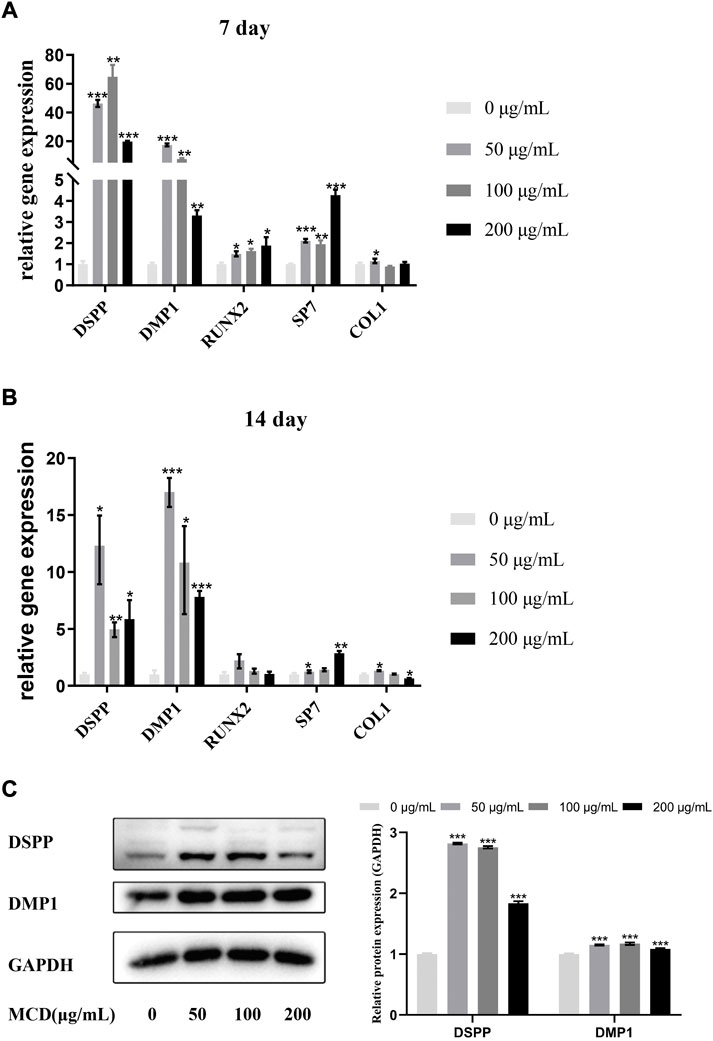
FIGURE 5. The effects of MCD treatment on the odontoblastic differentiation of hDPSCs for 7 and 14 days. (A,B) The messenger RNA expressions of DSPP, DMP1, Runx2, SP7, and Col1 were analyzed using real-time reverse transcription polymerase chain reaction. These data represent three independent experiments and are expressed as the mean ± SD. (*) p < 0.05, (**) p < 0.01, and (***) p < 0.001 vs. control. (C) DMP1 protein and DSPP protein expression after 14 days exposure to MCDs. GAPDH protein expression served as a loading control. hDPSCs: human dental pulp stem cells, MCD: metformin-based carbon nanodot.
To determine the effects of MCDs on odontogenic differentiation further, we detected specific odontogenic proteins, including DSPP and DMP1, at 14 days (Figure 5C). The expression of DSPP and DMP1 in the MCD-treated group was significantly higher than that in the control group, which was coordinated with the odontogenic marker gene expression at day 14. Notably, 50 μg/ml MCDs were the most effective in promoting DSPP and DMP1 expression, while the expression in the 200 μg/ml MCD group was significantly lower than that in the other MCD groups. These results reinforce the hypothesis that MCDs promote odontogenic differentiation by upregulating the expression of odontogenic-specific markers. Given the odontogenic gene expression pattern at day 14, a lower concentration of MCDs at 50 μg/ml, was the most effective in promoting odontogenic differentiation.
To determine the mechanism by which MCDs promote odontogenic differentiation, we used TEM and western blotting to detect the autophagy-related structures and protein expression of hDPSCs. Compared to that in the controls, TEM showed typical autophagosomes in hDPSCs when the cells were cocultured with 50 μg/ml MCDs for 24 h, which increased consistently until 72 h (Figure 6A). The western blotting results showed that the expression of p62 and LC3Ⅱ was significantly increased at 12 h and peaked at 24 h (Figure 6B). Subsequently, the expression of p62 decreased. Combined with the formation of autophagosomes detected by TEM, the results indicated that MCDs were effective bioactive nanoparticles for activating autophagy in hDPSCs.
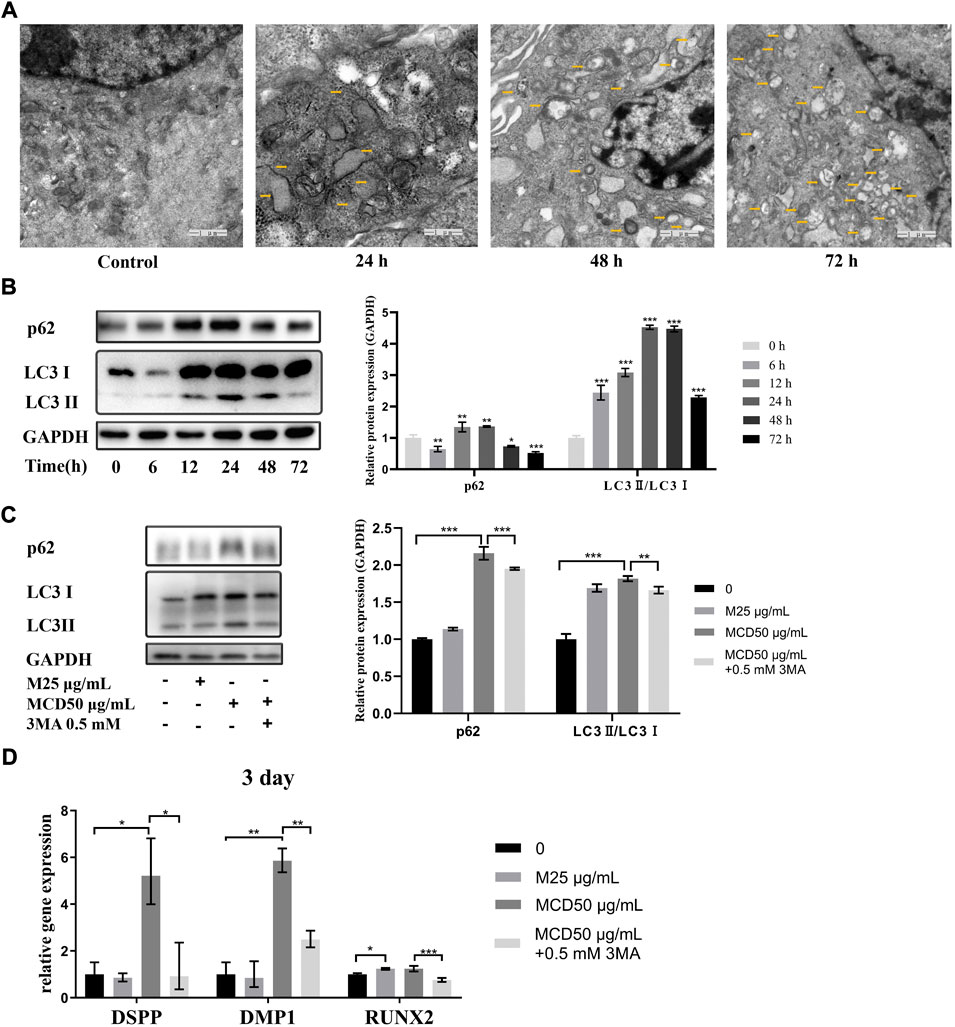
FIGURE 6. MCDs promote odontogenic differentiation of hDPSCs via autophagy. (A) TEM images of autophagic vesicles (yellow arrows) in 0, 24, 48, and 72 h (scale bar: 1 μm). (B) The hDPSCs were treated with 50 μg/ml MCDs for 0, 6, 12, 24, 48, and 72 h, and protein levels of LC3 and p62 were detected. (C) hDPSCs were treated with 3-MA (0.5 mM) for 12 h prior to addition of 50 μg/ml MCDs for 3 days. The protein levels of LC3 and p62 were detected. GAPDH protein expression served as a loading control. (D) hDPSCs were treated with 3-MA (0.5 mM) for 12 h prior to addition of 50 μg/ml MCDs for 3 days. The messenger RNA expression of DSPP, DMP1, Runx2 was analyzed using real-time reverse transcription polymerase chain reaction. These data represent three independent experiments and are expressed as the mean ± SD. (*) p < 0.05, (**) p < 0.01, and (***) p < 0.001 vs. control. hDPSCs: human dental pulp stem cells, MCDs: metformin-based carbon nanodots, 3-MA: 3-Methyladenine.
We investigated the relationship between MCD-induced autophagy and odontoblastic differentiation. As shown in Figures 6C,D, 50 μg/ml MCDs successfully increased the expression of p62 and LC3Ⅱ and promoted the expression of odontoblastic differentiation markers. When the hDPSCs were pretreated with 3-MA for 12 h, accompanied by a significant decrease in odontogenic gene expression, the expression of p62 and LC3Ⅱ also decreased after 72 h, by which time there was no difference from the control groups. The blockade of MCD-induced autophagy led to a reduction in odontogenic gene expression. The results clearly demonstrated that MCDs promoted odontogenic differentiation of hDPSCs through autophagy. After blocking autophagy, the ability of odontogenic differentiation was inhibited, thus proving that MCDs enhance odontogenic differentiation through autophagy.
Bioactive nanomaterials have been widely proposed for regulating hDPSC differentiation into odontoblasts for pulp regeneration. However, as a novel nanomaterial, CDs were first discovered to promote odontoblastic differentiation via autophagy. Through preliminary experiments, we found that CDs could not be synthesized from metformin alone; hence, we introduced citric acid as a regular carbon source. Then, we synthesized MCDs, which had good dispersibility and exhibited a spherical particle shape of 5.9 nm in size, excitation dependence, and rich functional groups. MCDs also showed biological safety in terms of cell viability, apoptosis, and fluorescence labelling ability at a concentration up to 200 μg/ml in vitro. Importantly, the presence of MCDs facilitated high-efficiency odontogenic differentiation of hDPSCs by promoting the expression of odontogenic genes and proteins. Notably, we further revealed that MCDs promoted odontoblastic differentiation of hDPSCs via autophagy.
The MCDs exhibited typical CD properties, similar to those of other functional CDs, given its rich functional groups. For example, its hydroxyl and amino groups can promote water solubility, and its imine bonds allow easy surface modification. Moreover, the MCDs retained nitrogen-hydrogen bonds in the metformin structure. Regarding its fluorescence properties, we proved its cell labelling, as shown in Supplementary Figure S3.
Good biocompatibility is a prerequisite for the application of nanomaterials in cell function regulation. Therefore, the effect of MCDs on cell viability was assessed using an MTT assay. We found that concentrations of MCD up to 200 μg/ml did not affect cell viability. Moreover, we employed an apoptotic cell death assay to verify this, because Annexin V-APC assessment is more sensitive than MTT assays and can identify apoptosis in the same cell population by flow cytometry. We found that MCDs under 400 μg/ml did not affect apoptosis, and 800 μg/ml showed significantly increased apoptosis. We can conclude that MCDs concentrations under 200 μg/ml exhibited good biocompatibility in hDPSCs.
To investigate the effect of MCD on odontogenic differentiation, we investigated odontogenic biomarkers. DMP1 has been reported to be present during differentiation of hDPSCs and to regulate the expression of DSPP during the early differentiation of odontoblasts (Narayanan et al., 2006). DSPP is also important for specifying odontoblastic differentiation (D'Souza et al., 1997). SP7 (osterix) is required for the proliferation and differentiation of odontoblasts (Bae et al., 2018). RUNX2 is an essential transcription factor that enhances early differentiation of odontoblasts and inhibits odontoblast terminal differentiation (Li et al., 2011; Li et al., 2021; Li et al., 2022), which increases RUNX2 expression (p < 0.05) at 7 days. COL1 is a key protein in dentin formation and repair (Ni et al., 2018). The gene expression levels of DSPP, DMP1, COL1, and SP7 increased significantly at 7 days, indicating that MCDs promoted the differentiation of hDPSCs into odontoblasts. The expression of RUNX2 increased at 7 days and showed no significant change at 14 days, which may indicate that hDPSCs differentiated into odontoblasts at 14 days. The gene and protein expressions of DSPP and DMP1 increased significantly after 14 days, indicating that the differentiated cells maintained secretory activity and attained dentin function. COL1 expression increased at 7 days but declined at 14 days under MCD 200 μg/ml treatment. We speculated that at day 14, the 200 μg/ml MCDs may have affected cell activity. Thus, we chose the minimum effective concentration of 50 μg/ml to explore the mechanism by which MCD affected cell differentiation.
Autophagy refers to a relatively conservative evolutionary process in which eukaryotic double-membrane vesicles wrap the cell contents and then transport them to lysosomes for degradation (Klionsky et al., 2016). In 2013, Yang et al. (2013) demonstrated the developmental appearance of autophagy during odontogenesis in mouse lower molars, which indicated that autophagy may play an important role in odontogenesis. An increasing number of nanomaterials serve as autophagy modulators (Zhang et al., 2021). The amount of LC3Ⅱ is a common marker for monitoring autophagosome formation, and decreased expression of p62, an autophagy cargo receptor, is often associated with increased autophagic flux (Klionsky et al., 2016). The autophagic flux, which defines the amount of autophagic degradation, indicates autophagic activity. In our study, western blotting results showed that autophagy was activated after the cells were cocultured with MCDs for 12 h, and autophagic flux was enhanced after 24 h. Moreover, blockade of MCD-induced autophagy led to a reduction in odontogenic gene expression. Our findings revealed that MCDs are autophagy activators during the process of odontogenic differentiation. Similar to our research, Schisandrin C, deferoxamine, and midkine have been shown to promote odontogenic differentiation of hDPSCs by enhancing autophagy (Wang et al., 2017; Takanche et al., 2018; Park et al., 2020).
Shao et al. reported that citrate CDs promote osteogenic differentiation of mouse bone marrow mesenchymal cells through the reactive oxygen species (ROS)-mediated MAPK pathway (Shao et al., 2017). ROS can activate autophagy through the MAPK signalling pathway (Wang et al., 2017). Concerning the role of CDs in promoting the differentiation of stem cells, we speculate that CDs may mediate differentiation through similar or even the same signalling pathways, but further studies using citric acid and citrate CDs are required as controls to verify this. Metformin induces autophagy in various cells (Lu et al., 2021). However, in this experiment, metformin alone at 25 μg/ml did not induce autophagy in hDPSCs, and compared with the MCD group, it failed to promote odontogenic marker expression. We hypothesize that the concentration of metformin was too low to exert biological functions in hDPSCs, but this requires further investigation.
We successfully synthesized bioactive drug CDs, i.e., MCDs. These MCDs could facilitate the odontogenic differentiation of hDPSCs by upregulating the expression of odontoblast gene markers (DMP1, DSPP, RUNX2, and SP7) and proteins (DSPP, DMP1). More importantly, our findings verified that the MCDs exhibited effective odontogenic differentiation through the mechanism of autophagy. To our knowledge, no previous study has shown that CDs are capable of both activating autophagy and enhancing odontogenic differentiation of hDPSCs. Our work highlights the marked potential of these CDs in the thriving fields of regenerative endodontics and autophagy-based cell therapy.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The procedure was approved by the Ethics Committee of the Hospital of Stomatology, Jilin University.
YL proposed and designed the experiments. RL and KZ contributed to and was responsible for designing chemical experiments, performing chemical tests and analyses (Figures 1, 2; Supplementary Figure S1), and writing the article about chemical section. JL, SN, YX, XL, KZ, and YL contributed to and was responsible for designing biological experiments, performing biological tests and analyses (Figures 3–6; Supplementary Figure S2), and compolish writing biological section of this paper.
This study was supported by Natural Science Foundation from Department of Science and Technology of Jilin Province (No. 20200201409JC), Department of Finance of Jilin Province Project (#JCSZ 2021893-2), Health and Health technology innovation projects from Health and Family Planning Commission of Jilin Province (No. 2019J034), research foundation from Development and Reform Commission of Jilin Province (No. 2019C050-1), and also Bethune’s Research Project of Jilin University (No. 2018B40).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.1002291/full#supplementary-material
Ahmed, G. M., Abouauf, E. A., Abubakr, N., Dörfer, C. E., and El-Sayed, K. F. (2020). Tissue engineering approaches for enamel, dentin, and pulp regeneration: An update. Stem Cells Int. 2020, 1–15. doi:10.1155/2020/5734539
Bae, J. M., Clarke, J. C., Rashid, H., Adhami, M. D., Mccullough, K., Scott, J. S., et al. (2018). Specificity protein 7 is required for proliferation and differentiation of ameloblasts and odontoblasts. J. Bone Min. Res. 33, 1126–1140. doi:10.1002/Jbmr.3401
Cui, D., Li, H., Wan, M., Peng, Y., Xu, X., Zhou, X., et al. (2018). The origin and identification of mesenchymal stem cells in teeth: From odontogenic to non-odontogenic. Curr. Stem Cell Res. Ther. 13, 39–45. doi:10.2174/1574888x12666170913150403
D'souza, R. N., Cavender, A., Sunavala, G., Alvarez, J., Ohshima, T., Kulkarni, A. B., et al. (1997). Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (dspp) suggest distinct developmental functions in vivo. J. Bone Min. Res. 12, 2040–2049. doi:10.1359/Jbmr.1997.12.12.2040
Feng, T., Zeng, Q., Lu, S., Yan, X., Liu, J., Tao, S., et al. (2017). Color-tunable carbon dots possessing solid-state emission for full-color light-emitting diodes applications. Acs Photonics 5, 502–510. doi:10.1021/Acsphotonics.7b01010
Gronthos, S., Brahim, J., Li, W., Fisher, L. W., Cherman, N., Boyde, A., et al. (2002). Stem cell properties of human dental pulp stem cells. J. Dent. Res. 81, 531–535. doi:10.1177/154405910208100806
Han, Y., Zhang, F., Zhang, J., Shao, D., Wang, Y., Li, S., et al. (2019). Bioactive carbon dots direct the osteogenic differentiation of human bone marrow mesenchymal stem cells. Colloids Surfaces B Biointerfaces 179, 1–8. doi:10.1016/J.Colsurfb.2019.03.035
Klionsky, D. J., Abdelmohsen, K., Abe, A., Abedin, M. J., Abeliovich, H., Acevedo Arozena, A., et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. doi:10.1080/15548627.2015.11003563rd Edition
Li, J., Lai, Y., Li, M., Chen, X., Zhou, M., Wang, W., et al. (2022). Repair of infected bone defect with clindamycin-tetrahedral dna nanostructure complex-loaded 3d bioprinted hybrid scaffold. Chem. Eng. J. 435, 134855. doi:10.1016/J.Cej.2022.134855
Li, S., Kong, H., Yao, N., Yu, Q., Wang, P., Lin, Y., et al. (2011). The role of runt-related transcription factor 2 (Runx2) in the late stage of odontoblast differentiation and dentin formation. Biochem. Biophys. Res. Commun. 410, 698–704. doi:10.1016/J.Bbrc.2011.06.065
Li, S., Liu, Y., Tian, T., Zhang, T., Lin, S., Zhou, M., et al. (2021). Bioswitchable delivery of microrna by framework nucleic acids: Application to bone regeneration. Small 17, E2104359. doi:10.1002/Smll.202104359
Liu, J., Li, D., Zhang, K., Yang, M., Sun, H., and Yang, B. (2018). One-step hydrothermal synthesis of nitrogen-doped conjugated carbonized polymer dots with 31% efficient red emission for in vivo imaging. Small 14, E1703919. doi:10.1002/Smll.201703919
Liu, J., Lu, S., Tang, Q., Zhang, K., Yu, W., Sun, H., et al. (2017). One-step hydrothermal synthesis of photoluminescent carbon nanodots with selective antibacterial activity against porphyromonas gingivalis. Nanoscale 9, 7135–7142. doi:10.1039/C7nr02128c
Lu, G., Wu, Z., Shang, J., Xie, Z., Chen, C., and Zhang, C. (2021). The effects of metformin on autophagy. Biomed. Pharmacother. 137, 111286. doi:10.1016/J.Biopha.2021.111286
Lv, Z., and Guo, Y. (2020). Metformin and its benefits for various diseases. Front. Endocrinol. 11, 191. doi:10.3389/Fendo.2020.00191
Meng, Y., Yang, M., Liu, X., Yu, W., and Yang, B. (2019). Zn2+-Doped carbon dots, A good biocompatibility nanomaterial applied for bio-imaging and inducing osteoblastic differentiation in vitro. Nano 14, 1950029. doi:10.1142/S1793292019500292
Mozaffari, M. S., Emami, G., Khodadadi, H., and Baban, B. (2019). Stem cells and tooth regeneration: Prospects for personalized dentistry. Epma J. 10, 31–42. doi:10.1007/S13167-018-0156-4
Narayanan, K., Gajjeraman, S., Ramachandran, A., Hao, J., and George, A. (2006). Dentin matrix protein 1 regulates dentin sialophosphoprotein gene transcription during early odontoblast differentiation. J. Biol. Chem. 281, 19064–19071. doi:10.1074/Jbc.M600714200
Ni, S. L., Zhang, J., Liu, X., Li, X. W., Sun, Y. J., Zhang, X., et al. (2018). Effects of human bone morphogenetic protein 2 (Hbmp2) on tertiary dentin formation. Am. J. Transl. Res. 10, 2868–2876.
Park, Y. H., Lee, Y. S., Seo, Y. M., Seo, H., Park, J. S., Bae, H. S., et al. (2020). Midkine promotes odontoblast-like differentiation and tertiary dentin formation. J. Dent. Res. 99, 1082–1091. doi:10.1177/0022034520925427
Qin, W., Chen, J. Y., Guo, J., Ma, T., Weir, M. D., Guo, D., et al. (2018a). Novel calcium phosphate cement with metformin-loaded chitosan for odontogenic differentiation of human dental pulp cells. Stem Cells Int. 2018, 7173481–7173510. doi:10.1155/2018/7173481
Qin, W., Gao, X., Ma, T., Weir, M. D., Zou, J., Song, B., et al. (2018b). Metformin enhances the differentiation of dental pulp cells into odontoblasts by activating ampk signaling. J. Endod. 44, 576–584. doi:10.1016/J.Joen.2017.11.017
Shao, D., Lu, M., Xu, D., Zheng, X., Pan, Y., Song, Y., et al. (2017). Carbon dots for tracking and promoting the osteogenic differentiation of mesenchymal stem cells. Biomater. Sci. 5, 1820–1827. doi:10.1039/C7bm00358g
Sui, B., Chen, C., Kou, X., Li, B., Xuan, K., Shi, S., et al. (2019). Pulp stem cell-mediated functional pulp regeneration. J. Dent. Res. 98, 27–35. doi:10.1177/0022034518808754
Takanche, J. S., Kim, J. S., Kim, J. E., Han, S. H., and Yi, H. K. (2018). Schisandrin C enhances odontoblastic differentiation through autophagy and mitochondrial biogenesis in human dental pulp cells. Arch. Oral Biol. 88, 60–66. doi:10.1016/J.Archoralbio.2018.01.018
Wang, B., Yang, M., Liu, L., Yan, G., Yan, H., Feng, J., et al. (2019a). Osteogenic potential of Zn(2+)-passivated carbon dots for bone regeneration in vivo. Biomater. Sci. 7, 5414–5423. doi:10.1039/C9bm01181a
Wang, S., Xia, Y., Ma, T., Weir, M. D., Ren, K., Reynolds, M. A., et al. (2019b). Novel metformin-containing resin promotes odontogenic differentiation and mineral synthesis of dental pulp stem cells. Drug Deliv. Transl. Res. 9, 85–96. doi:10.1007/S13346-018-00600-3
Wang, X., Wu, T. T., Jiang, L., Rong, D., and Zhu, Y. Q. (2017). Deferoxamine-induced migration and odontoblast differentiation via ros-dependent autophagy in dental pulp stem cells. Cell. Physiol. biochem. 43, 2535–2547. doi:10.1159/000484506
Yan, F., Zu, F., Xu, J., Zhou, X., Bai, Z., Ma, C., et al. (2019). Fluorescent carbon dots for ratiometric detection of curcumin and ferric ion based on inner filter effect, cell imaging and pvdf membrane fouling research of iron flocculants in wastewater treatment. Sensors Actuators B Chem. 287, 231–240. doi:10.1016/J.Snb.2019.01.144
Yang, B., Chen, G., Li, J., Zou, Q., Xie, D., Chen, Y., et al. (2012). Tooth root regeneration using dental follicle cell sheets in combination with A dentin matrix - based scaffold. Biomaterials 33, 2449–2461. doi:10.1016/J.Biomaterials.2011.11.074
Yang, J. W., Zhu, L. X., Yuan, G. H., Chen, Y. X., Zhang, L., Zhang, L., et al. (2013). Autophagy appears during the development of the mouse lower first molar. Histochem. Cell Biol. 139, 109–118. doi:10.1007/S00418-012-1016-2
Yang, S., Sun, J., Li, X., Zhou, W., Wang, Z., He, P., et al. (2014). Large-scale fabrication of heavy doped carbon quantum dots with tunable-photoluminescence and sensitive fluorescence detection. J. Mat. Chem. A Mat. 2, 8660. doi:10.1039/C4ta00860j
Zhang, T., Tian, T., and Lin, Y. (2021). Functionalizing framework nucleic-acid-based nanostructures for biomedical application. Adv. Mat. 2021, E2107820. doi:10.1002/Adma.202107820
Keywords: autophagy, cell differentiation, carbon dots, nanomaterials, odontogenesis
Citation: Lu J, Li R, Ni S, Xie Y, Liu X, Zhang K and Li Y (2022) Metformin carbon nanodots promote odontoblastic differentiation of dental pulp stem cells by pathway of autophagy. Front. Bioeng. Biotechnol. 10:1002291. doi: 10.3389/fbioe.2022.1002291
Received: 25 July 2022; Accepted: 19 August 2022;
Published: 09 September 2022.
Edited by:
Jianxun Ding, Changchun Institute of Applied Chemistry (CAS), ChinaReviewed by:
Kunneng Liang, Sichuan University, ChinaCopyright © 2022 Lu, Li, Ni, Xie, Liu, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Li, bHlpOTlAamx1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.