
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 11 January 2022
Sec. Nanobiotechnology
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.808933
This article is part of the Research Topic Nanotechnology for Innovative Cancer Diagnosis and Therapy View all 6 articles
Exosomes are small nanoscale vesicles with a double-layered lipid membrane structure secreted by cells, and almost all types of cells can secrete exosomes. Exosomes carry a variety of biologically active contents such as nucleic acids and proteins, and play an important role not only in intercellular information exchange and signal transduction, but also in various pathophysiological processes in the human body. Surface-enhanced Raman Spectroscopy (SERS) uses light to interact with nanostructured materials such as gold and silver to produce a strong surface plasmon resonance effect, which can significantly enhance the Raman signal of molecules adsorbed on the surface of nanostructures to obtain a rich fingerprint of the sample itself or Raman probe molecules with ultra-sensitivity. The unique advantages of SERS, such as non-invasive and high sensitivity, good selectivity, fast analysis speed, and low water interference, make it a promising technology for life science and clinical testing applications. In this paper, we briefly introduce exosomes and the current main detection methods. We also describe the basic principles of SERS and the progress of the application of unlabeled and labeled SERS in exosome detection. This paper also summarizes the value of SERS-based exosome assays for early tumor diagnosis.
The exosome was originally isolated from sheep reticulocytes as a small vesicle with the characteristics of the reticulocyte cytosol (Trams et al., 1981; Pan et al., 1985; Grasso et al., 1990). Researchers only considered it to be a “waste product” of erythrocyte maturation, and as exosomes have been studied more closely, it has been confirmed that exosomes are extracellular vesicles (EVs) secreted by the cell in the form of cytosol (Han et al., 2021b; Clark et al., 2021; Isaac et al., 2021). Exosomes are multi-vesicular endosomes that originate from the endocytic system and are formed through four processes: outgrowth, invagination, multivesicular body formation and secretion, and are spherical, flat or cup-shaped vesicles with a diameter of 30–150 nm (Spilak et al., 2021; Staubach et al., 2021; Sun et al., 2021). In recent years, it has been found that exosomes are distributed in human body fluids, including blood, urine, saliva, thoracoabdominal fluid, cerebrospinal fluid, bile, milk, tears and semen (Yu et al., 2021a; Zhang et al., 2021b; Lin et al., 2021; Mohammadi et al., 2021; Richter and Lehr, 2021). In addition, exosomes of different tissue sources carry different nucleic acids, proteins and lipids (Teng and Fussenegger, 2020; Shehzad et al., 2021; Wu et al., 2021). Exosomes can transmit information and function between cells through the nucleic acids, proteins, and lipids they carry (Xu et al., 2020; Zhu et al., 2020; Azparren-Angulo et al., 2021). Moreover, exosomes play an important role in tumorigenesis and progression, invasive metastasis and immune escape (Hu et al., 2020; Morrissey and Yan, 2020; Thietart and Rautou, 2020; Yan and Jiang, 2020). It can be used not only as a biomarker for tumor diagnosis, but also as a target for tumor therapy (Ariston Gabriel et al., 2020; Dai et al., 2020; Zhou et al., 2020).
As a non-destructive, non-contact and repeatable rapid detection technology, Raman spectroscopy can provide fingerprint information of molecular structure vibration and the peaks are narrow and not easy to overlap. It has significant advantages in the detection of biochemical substances. However, Raman spectroscopy has a cross section of only 10–30 cm2/molecule and is susceptible to fluorescence interference (Prieto et al., 2016), which limits the wide application of Raman spectroscopy. In the 1970s, it was discovered that when pyridine was adsorbed on a rough silver surface, a high-intensity Raman spectrum signal could be obtained (Leitch et al., 2012). The Raman effect of Raman spectroscopy is derived from molecular vibrations and rotations, which enables the non-invasive analysis of the composition and structure of substances (Markina et al., 2021; Tanwar et al., 2021; Naqvi et al., 2022). However, conventional Raman spectroscopy is extremely weak, susceptible to fluorescence interference, and has low detection sensitivity, which limits its application. Surface-enhanced Raman spectroscopy (SERS) is a method that uses light to interact with nanostructured materials such as gold and silver to produce a strong surface plasmon resonance effect, which can significantly enhance the Raman signal of molecules adsorbed on the surface of nanostructures to obtain ultra-sensitive fingerprints of the sample itself or Raman probe molecules (Zhang et al., 2021a; Berry et al., 2021; Liu et al., 2021b). SERS has become a promising technique for bio-detection (Wang et al., 2021a; Wang et al., 2021b) by compensating for the shortcomings of conventional Raman spectroscopy with local enhancement of up to 1014 in the plasma gap and sharp protrusions. In addition, SERS has a number of advantages. It is highly sensitive and can be used for single molecule detection and resistant to photobleaching and photodegradation. Besides, it is suitable for long-term monitoring (Zhao et al., 2021a; Lopez-Lorente, 2021). SERS technology is widely used in various biomedical fields, such as nucleic acid, protein, human cells, human tissues and human body fluids (Calderon et al., 2021; Ma et al., 2021; Xi and Liang, 2021), and has important applications in the diagnosis of tumors (Guerrini et al., 2021; Shen et al., 2021; Sun et al., 2021). Nowdays, it is believed that physical enhancement mechanism and chemical enhancement mechanism are two explanation mechanisms of SERS enhancement. Among them, physical enhancement is also called electromagnetic enhancement. The interaction of Ag nanostructures will cause the collective oscillation of free electrons on the metal surface. When the frequency of incident light matches the natural oscillation frequency of free electrons in the metal, localized surface plasmon excitation Element Resonance (LSPR) will occur, which leads to the enhancement of the incident light field. Studies have shown that the enhancement factor of electromagnetic field enhancement can reach 1011 (Ding et al., 2017). Chemical enhancement is a charge transfer enhancement (Tong et al., 2011; Zhan et al., 2019), which requires the analyte to be directly adsorbed or bonded to the rough metal surface through chemical bonds, and there is a charge transfer with the metal surface. When the energy of the incident photon and electrons are on the metal substrate and the adsorbate When the transfer energy difference between them is equal, resonance will occur, which will increase the effective polarizability of the system and increase the Raman signal. Studies have shown that the chemical enhancement factor is 101–103 (Gruenke et al., 2016). It is generally believed that the SERS effect is the result of the above-mentioned two enhancement mechanisms. At present, a large number of researches on the selection of SERS metal substrates show that Au and Ag nanostructures have good stability, repeatability and easy preparation, It has become the first choice of researchers for SERS substrates, and the anisotropic and sharp morphology nanoparticles have a better SERS enhancement effect than symmetrical nanoparticles (Abdulhalim, 2014).
In recent years, attempts have been made to use the ultra-high-sensitivity SERS technique for exosome detection in an attempt to develop a new technique for efficient and nondestructive liquid biopsy (Chen et al., 2021b; Guerrini et al., 2021). Strategies for exosomes detection based on SERS technology can be divided into two types. One is the unlabeled SERS technique that directly acquires the Raman signal of the exosome itself without the introduction of a Raman probe (Yan et al., 2019; Shin et al., 2020b). The advantage of this method is that it is simple and fast and allows a more direct analysis of the exosome composition. However, the signals of various biomolecules may overlap with each other, resulting in partial loss of diagnostic signal. The other is the labeled SERS technique, which uses Raman probe molecules to label exosomes and indirectly detects the presence and concentration of exosomes by detecting the Raman intensity of the labeled molecules (Shin et al., 2020a; Dong et al., 2020; Fan et al., 2021). In this review, we summarized the application and research of SERS technology in the detection of exosomes, and discussed the advantages and disadvantages of SERS compared with other exosome detection technologies. In addition, we also summarized the research on the detection of exosomes based on SERS in the early diagnosis of tumors, and proposed new insights for future research directions.
Under physiological and pathological conditions, exosomes can be actively secreted extracellularly by various cells including immune cells, stem cells and tumor cells, and can transport a large number of biomolecules from the parent cell to other cells, which is closely related to the pathogenesis of tumors (Claridge et al., 2021; Oey et al., 2021; Yamada, 2021). Because of their small size, exosomes are able to avoid phagocytosis by mononuclear macrophages and cross the vascular wall to the extracellular matrix, they are widely present in various body fluids including blood, saliva, urine, cerebrospinal fluid, and thoracoabdominal fluid (Clark et al., 2021; Leggio et al., 2021; Mao et al., 2021; Prieto-Vila et al., 2021). In addition, exosomes are relatively biologically stable and can be found in tissues and cells due to their phospholipid bilayer structure (Chen et al., 2021a; Hu et al., 2021). Information transfer between exosomes and target cells is achieved through 3 main pathways (Figure 1). Exosomes can enter the recipient cell through receptor-ligand interactions (Isaac et al., 2021; Staubach et al., 2021). Besides, exosomes can also be endocytosed by the recipient cells through phagocytosis (Chen et al., 2021c; Hur and Lee, 2021). Exosomes have an important role in tumor development and can serve as potential diagnostic markers and therapeutic targets for many tumors. In the tumor microenvironment, non-coding RNAs can be distributed in cancer cells, stromal cells and immune cells through small extracellular vesicles (sEVs) and participate in intercellular communication. Sev-nc RNA can be used as a good diagnostic and prognostic fluid marker for tumors (Reese and Dhayat, 2021; Shin et al., 2021; Tang et al., 2021). Exosomal-derived long noncoding RNA (lncRNA) TMZ-associated lnc RNA in GBM recurrence (lnc-TALC) modulates the glioblastoma (GBM) microenvironment and reduces the sensitivity of GBM to temozolomide chemotherapy, targeting exosomal-lnc-TALC may overcome TMZ resistance in GBM (Li et al., 2021b). Lnc RNA colorectal neoplasia differentially expressed (lnc-CRNDE) is highly expressed in tumor-associated macrophages (TAM) of gastric cancer (GC) patients, and CRNDE can be transferred from M2-polarized macrophages to GC cells via exosomal form and further promote GC cell proliferation and inhibit cisplatin resistance in GC cells (Xin et al., 2021).
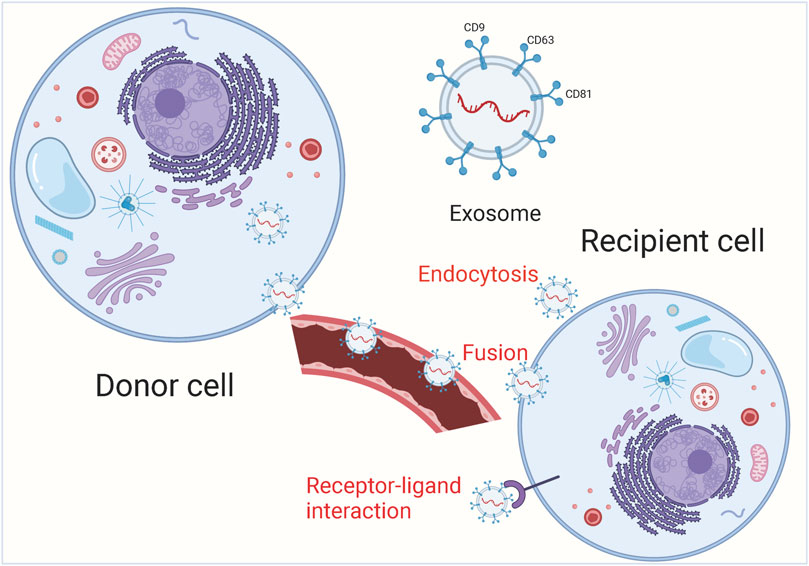
FIGURE 1. The main process of exosome release. Exosomal cargoes from the source cell can be further delivered to recipient cells via endocytosis, direct membrane fusion and receptor-ligand interaction.
Exosome-related assays currently include isolation and purification, physical characterization, and analysis for downstream functional studies. The main techniques for the isolation and purification of exosomes include ultracentrifugation, density gradient centrifugation, size exclusion chromatography, ultrafiltration, affinity assay and polymer-based co-precipitation method, while ultracentrifugation-based isolation techniques are the most used for the isolation of exosomes (Figure 2; Table 1) (Ng et al., 2021). Exosomes are detected, isolated and purified in normal or tumor cell culture medium supernatants, blood, saliva and other body fluids (Liu et al., 2021a; Yu et al., 2021c). Physical characterization techniques for exosomes include electron microscopy, dynamic light scattering, flow cytometry and nanoparticle tracking analysis, and tunable resistive pulse sensing assays (Zhao et al., 2021b; Liu et al., 2021c). Techniques for exosomal protein analysis include enzyme linked immunosorbent assay (ELISA), protein blotting and mass spectrometry (Lee et al., 2021; Xu et al., 2021). Nucleic acid analysis techniques include second-generation sequencing, gene chips, and polymerase chain reaction (PCR) (Yu et al., 2021b; Wang et al., 2021c; Zhang et al., 2021c). These analytical techniques have greatly facilitated the research process of exosomes, but there are some limitations: ELISA requires a large number of highly concentrated samples and cannot achieve rapid analysis of complex samples. Nanoparticle tracking analysis techniques have a limited range of measurement of exosome concentrations. Flow cytometry analysis of exosomes may have weak recovery signals; transmission electron microscopy, PCR and tamseq second-generation sequencing techniques are expensive, complicated and time-consuming. In conclusion, there is still no unified standard for exosome analysis and therefore, there is an urgent need to further develop new methods for exosome detection to accelerate the development of cancer body fluid biopsy.
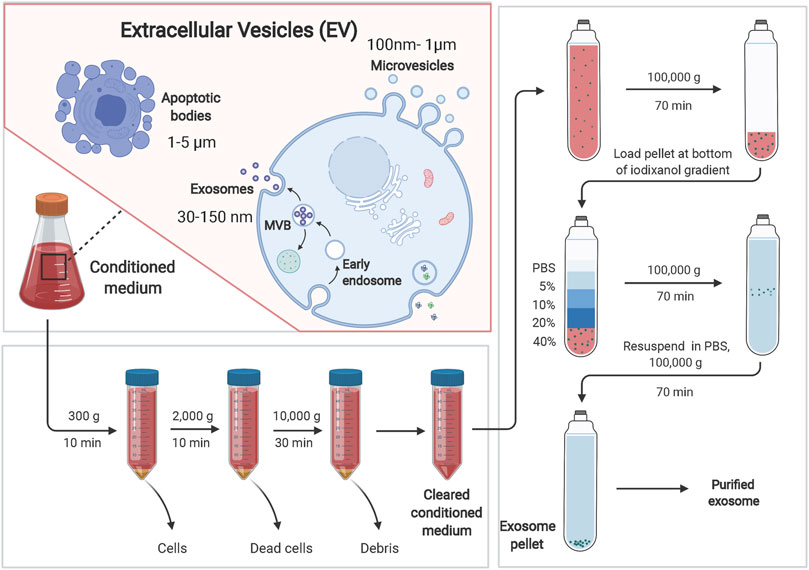
FIGURE 2. Exosomes can be separated by ultracentrifugation-based isolation techniques. Extracellular vesicles (EVs) can be divided into three main categories: microvesicles, exosomes and apoptotic bodies. Ultracentrifugation-based isolation techniques are the most used for the isolation of exosomes. Brifely, the culture supernatant was centrifuged at 300 × g for 10 min at 4°C, at 2,000 × g for 10 min, and at 10,000 × g for 30 min. After centrifugation, the supernatant was collected. The supernatant was ultra-centrifuged at 100,000 × g for 70 min to pellet the exosomes. Exosomes were washed with PBS and pelleted by ultracentrifugation for 70 min at 100,000 × g.
The unlabeled assay is a Raman spectroscopy of exosomes obtained by adsorbing the purified exosomes directly or very close to the SERS substrate surface (Soares Martins et al., 2018; Zhang et al., 2019b). The signal intensity in SERS decays exponentially as the analyte moves away from the hot spot (Liu et al., 2021b; Tahir et al., 2021). To obtain the desired exosome Raman signal, efficient SERS-enhanced substrates need to be prepared. To use unlabeled SERS for exosomes detection, several issues need to be addressed. First, since exosomes are mainly composed of non-chromophore biomolecules, it is difficult to detect their unique signal patterns. Exosomes are larger relative to SERS hotspots, and therefore it is difficult to locate exosomes in large numbers in SERS hotspots. It is important to develop a dedicated exosome detection strategy to acquire their signals sensitively. The Raman spectra of exosomes are heterogeneous and complex, and the molecular fingerprint signals also involve information from many biomolecules. In addition, since exosomes are larger than small biomolecules, identifying the relative position of exosome on the surface of metal nanostructures is challenging for signal intensity. The complex composition of the exosome in body fluids makes the analysis even more difficult. Therefore, effective methods are needed to interpret and analyze complex and heterogeneous signals.
Stremersch et al. (2016) A positively charged gold nanoparticle with enhanced Raman signal was prepared and analyzed using partial least squares discriminant analysis (PLS-DA) with multivariate curve resolution alte RNAting least squares (MCR-ALS) analysis algorithm to analyze the obtained Raman spectral data and be able to distinguish between vesicles from normal and cancer cells. The use of SERS probes decorated with Raman reporter genes and specific aptamer gold nanoparticles for targeting exosomes to capture substrates can be of great benefit, as it can capture most types of exosomes by recognizing the universal surface protein CD63. Based on this, exosomes can be detected in the blood of patients (Wang et al., 2018). The non-tagged SERS technology presents a “fingerprint” of the exosome itself and then uses multivariate statistics and computerized deep learning algorithms to dig deeper into the diagnostic information and build a discriminatory model that can eventually distinguish and trace exosomes from different sources. Currently, there are many difficulties in translating the non-labeled SERS exosome detection technology to the clinical setting, but with the expansion of the number of exosome samples and the development of related technologies, it may be applied to clinical testing in the future. In the non-label detection method, the sample to be tested is directly adsorbed on the surface of the metal nanostructure, and the SERS technology is used to directly obtain the Raman fingerprint of the biological sample itself. This method requires a combination of special nano-substrate structure and spectral analysis technology to distinguish between Spectral information of biomolecules or cells of different species and microorganisms. The advantage of non-label SERS nucleic acid detection technology is that the steps are simple and no special processing is required for samples. However, in general, this method has not yet met the sensitivity of clinical detection and specific requirements.
The SERS labeling assay is a nano-labeling immunoassay technique that combines SERS spectroscopy with the specific action of antibody antigens, which are plasma nanoparticles containing a strong Raman signal. The SERS nanomarkers are a class of plasma nanoparticles containing strong Raman signals. The SERS nanomarkers form a complex with the target exosomes through a specific reaction similar to that of antibody antigens, and the distribution and concentration of the target molecules are indirectly characterized by measuring the Raman intensity of the Raman-labeled molecules in the complex. The common structures can be mainly classified into exosome-magnetic bead-based SERS model (Figure 3) and exosome-chip-based SERS model (Figure 4).
Compared with normal cells, tumor cells secrete more exosomes and have specific biomarkers on their membrane surface, which makes tumor-derived exosomes an important cancer biomarker. Therefore, an exosome-magnetic bead-based SERS assay model can be designed to take advantage of the fact that tumor-derived exosomes contain a variety of specific proteins. This model typically uses two types of nanoparticles-magnetic nanobeads and SERS nanoprobes, and two specific antibodies are modified on the surface of both particles, which can recognize two different proteins on exosomes separately. In the presence of targeted exosomes, an immune complex can be formed between the SERS nanoprobe, the exosome and the magnetic nanoparticles. In the presence of a magnet, the immunocomplex can precipitate, so the SERS nanoprobe will detect the SERS signal in the precipitation. In contrast, in the absence of targeting exosomes, the SERS signal is barely detectable because no immune complexes can be formed and only magnetic nanoparticles are precipitated. In this way, the SERS signal can be used to discriminate the presence of target exosomes. In recent years, more and more people are discovering the potential of exosome-magnetic bead-based SERS detection models in many studies. In order to identify three different exosomes secreted by cancer cells, researchers prepared capture probes (Wang et al., 2018) by modifying three corresponding aptamers and Raman reporter molecules on the surface of gold nanoparticles. Three probes were present in the supernatant, and when a target exosome was added, it was specifically adsorbed with the corresponding magnetic beads and probes, and the complexes were aggregated to the bottom of the tube under the action of a magnet. At this point, the capture probe for this exosome in the supernatant is significantly reduced, and the corresponding Raman peak for this probe is significantly weakened. In addition, Tian et al. (2018) Similarly demonstrated that the exosome-bead-based SERS assay model has considerable potential as a convenient and highly sensitive quantitative tool for the detection of exosomes in biological samples. Pang et al. (2020) proposed a method that can capture and analyze PD-L1 exosomes directly from serum. Because tio2 shells can bind to the hydrophilic phosphate of phospholipid molecules on exosomes, they used Fe3O4@tio2 nanoparticles to enrich exosomes. PD-L1 exosomes can be quantitatively labeled using PD-L1 antibody-modified Au@Ag@ MBA-SERS labeling. Mi RNA plays an important role in the post-transcriptional regulation of gene expression and plays an essential role in the diagnosis and treatment of tumors (Li et al., 2021a; Andrikopoulou et al., 2021; Crudele et al., 2021; Konoshenko et al., 2021; Oh-Hohenhorst and Lange, 2021). The nature of the exosomal lipid bilayer avoids miRNA degradation, making the specific miRNAs in exosomes well suited as biomarkers for early cancer diagnosis and prognosis. Ma et al. (2018) established a SERS method based on double-stranded specific nuclease-assisted circular amplification for the quantitative detection of miRNAs in human blood exosomes. Pang et al. (2019) validated the feasibility of a SERS biosensor with double-stranded specific nuclease-assisted cycle amplification. This assay further amplifies the signal with nuclease assistance, allows for single base recognition with detection limits, and is suitable for clinical diagnosis without the need for multiple washes and hybridizations (Pang et al., 2019). A Raman scattering probe with a strong electromagnetic hot spot was developed by assembling gold nanoparticles in triangular pyramidal DNA, which can be used for efficient and sensitive detection of circulating tumor cells and exosomes (Zhang et al., 2019a). The MIO structure allows the capture and analysis of exosomes from the plasma of cancer patients without any labeling process. The SERS intensity at 1,087/cm of exosomes caused by P-O bonding within phosphoproteins can be used as a standard for liquid tumor biopsies. The 1,087/cm SERS peak intensity of exosomes extracted from the plasma of cancer patients is at least twice that of healthy individuals (Dong et al., 2020). Chen et al. (2021b) utilized 4-mercaptobenzoic acid-labeled silver nanoparticles attached to the outer surface of exosomes, where the silver nanoparticles can be used as SERS generators. The Raman reagent 4MBA is susceptible to specific intracellular stimuli and can exhibit different vibrational spectral features accordingly. The use of SERS spectroscopy enables the tracking of intracellular distribution of exosomes and simultaneous quantitative sensing of environmental ph (Chen et al., 2021b).
Labeled SERS has great potential for qualitative and quantitative studies of exosomes. The method of exosome isolation and enrichment can be done using SERS chips in addition to magnetic beads. Using two different proteins on the exosome and then modifying two specific antibodies on the substrate and probe respectively, an immune complex can be formed between the SERS probe, exosome and the chip in the presence of the targeted exosome, and then the excess non-targeted exosome can be rinsed for SERS detection. Lee and others invented a SERS-based sensing platform for the quantitative determination of exosomal miRNA (Lee et al., 2019). They first modified the capture probe on a homogeneous gold nanopillar substrate, and the target exosomal miRNAs were captured by the probe sequence when they were present. Since the target exosomal miRNAs have long sequences and some sequences are exposed, sequences with Raman reporter molecules are added to complementarily ligate with the exposed sequences to target both the target exosomal miRNAs and the Raman probe on the substrate. SERS detection probes were made using bimetallic SERS active nanotags, gold-silver-silver core-shell-shell nano sea cucumbers (gssnts), decorated with junctional DNA, complementary to the aptamer of the targeted exosome. In the presence of exosomes, the aptamer specifically recognizes and captures the exosomes and subsequently releases the gssnts into the supernatant. The presence of the target exosome is indicated by the detection of an attenuated SERS signal on the MB (Ning et al., 2020). A densely packed and ordered Au octahedral array was used as a sensing platform for the quantitative determination of let-7a in MCF-7 cell-derived exosomes. Where the gold octahedra in the array stand uniformly on their triangular faces, significantly improving detection sensitivity and uniformity. This model enables let-7a detection with single-base specificity in the presence of a target by detecting the SERS intensity change caused by the conversion of DNA from a hairpin structure to a double-stranded body (Kang et al., 2021). Chen et al. (2020) A novel exosome/metal nanohybrid was constructed. The heterogeneous exosomes can be internalized by lattice-protein-mediated endocytosis and then translocated to lysosomes. The metal nanoparticles in the hybrid were shown to have no effect on exosome features while serving as a surface-enhanced Raman scattering generator. This technique may improve the utility of exosome nanohybrids in therapeutics such as multifunctional drug delivery systems (Chen et al., 2020).
Protein is one of the vital biological macromolecules in living organisms, and is the main bearer of life activities in energy storage, metabolism, and cell function regulation. Abnormally expressed exosomal proteins usually appear in the early or late stages of tumors. It has important diagnostic significance in its occurrence and progress. Therefore, sensitive, accurate and specific quantitative analysis is of great significance. SERS has been proven to have great potential in high-throughput biomolecular detection, including protein Detection. The research on protein detection by SERS only involves the side chains of amino acids, and some studies have obtained signals from the two amide groups and side chains. The inte RNAl structure information of biomolecules (non-labeled method) and the exte RNAl SERS label (labeled method) are two independent methods for direct and indirect detection of biomolecules. The labeling method requires an appropriate dye label. If the label has resonance under a certain excitation line, it can provide an additional enhanced signal, so the detection sensitivity of the labeling method is higher. Instead of labeling, all the information obtained is the amino acid skeleton of the protein. The reproducibility of the spectrum is poor, the non-reproducibility of protein adsorption sites and the denaturation of protein adsorption on the metal surface will all be studied for non-labeling methods. Bring difficulties. However, the information given by non-labeled means can analyze protein structure more directly, which is of great help to protein analysis.
Nucleic acid is a biological macromolecular compound formed by nucleotide as a synthetic unit. It is the most basic genetic material of life and plays a decisive role in a series of major life phenomena such as growth, inheritance, and evolution of life. Carrying out nucleic acid molecular diagnosis is of great significance to promoting the development of human health and medical treatment. SERS has unique technical advantages in the detection and analysis of biological samples such as nucleic acids. First of all, SERS has low requirements on the shape and size of the sample to be tested, which can save complicated sample pretreatment steps. Secondly, SERS has a small sample requirement and is suitable for the detection of trace and trace samples. In addition, SERS can achieve rapid detection, generally within 30 s to complete the detection of a sample. The introduction of Raman labeling technology can achieve the purpose of strong specific detection. Moreover, because the Raman scattering of water is very weak, the Raman spectroscopy technique is very suitable for the detection of samples in the water environment. SERS can also be combined with other detection methods to increase detection sensitivity and specificity. SERS combined with PCR nucleic acid detection method can successfully realize the recombination and concentration amplification of the target sequence, so as to achieve the purpose of ultra-sensitive detection. The nucleic acid SERS detection based on enzyme digestion technology mainly relies on the excellent SERS enhancement performance of the “hot spots” between metal nanoparticles, uses the self-assembly of ss-DNA to increase the probability of hot spots formation, and uses enzyme digestion reactions to eliminate hot spots to achieve SERS signals. Change from weak to strong and then from strong to weak.
However, there is no research report comparing SERS and other traditional detection methods in the detection of exosomal-derived proteins or nucleic acids. Whether SERS has outstanding advantages in the detection of exosomal-derived proteins or nucleic acids still needs further research.
Exosomes are promising biomarkers present in body fluids and cell lines in cancers. As described above, plenty of SERS-based exosomes have been proposed in combination with specific recognition/separation/amplification techniques for qualitative/quantitative analysis of individual targets (Table 2).
Ovarian cancer is the most lethal gynecologic malignancy with a 5-years survival rate of only 47.4%, seriously endangering women’s physical and mental health (Baert et al., 2021; Wang et al., 2021d). A silver film coated on the surface of 3D nanobowls as a SERS substrate was used to detect the SERS signal of exosomes of ovarian cancer cells over time using sputtering technique (Lee et al., 2015). In contrast to silver nanoparticles prepared by reduction method, sputtering iso-expressed ovarian cancer exosomes were specifically captured. In addition, this method allows selective detection and analysis of target exosomes from exosome-rich body fluids, a technique with great potential for early disease detection.
Lung cancer has developed into one of the most common malignant tumors in the world, with the highest incidence and mortality rate (Jiang et al., 2021a; Drusbosky et al., 2021). The incidence of lung cancer has been on the rise due to factors such as smoking and environmental pollution (Koch et al., 2021; Raguraman et al., 2021). Developed a label-free, highly sensitive method for exosome classification by combining SERS technology with statistical algorithms (Park et al., 2017). They used gold nanoparticles as an enhanced substrate for SERS and used principal component analysis to analyze the spectra of exosomes secreted by lung cancer cells and normal cells separately and found a sensitivity of 95.3% and specificity of 97.3%. Shin and others used computerized deep learning to perform SERS signals of exosomes from normal and lung cancer cell lines trained and modeled after extracting features of both types of exosome spectra for analysis and prediction of SERS spectra of human plasma-derived exosomes (Shin et al., 2020a). This trial combined SERS spectral analysis of exosomes with deep learning to successfully identify lung cancer patients, even those in stage 1. The method is noninvasive, safe, sensitive, and important for the development of early liquid biopsy methods for lung cancer. Shin and others demonstrated correlation between non-small cell lung cancer (NSCLC) cell-derived exosomes and potential protein markers for cancer diagnosis by unique Raman scattering spectroscopy and principal component analysis (PCA). Based on the SERS signals of exosomes from NSCLC cells and normal cells, they extracted the Raman patterns of cancerous exosomes by PCA and clarified the specific patterns into unique peaks by quantitative analysis of the proportional mixture of cancerous and normal exosomes. Subsequently, they compared these unique peaks with the characteristic Raman bands of several exosomal protein markers. The correlation was used for the diagnosis of NSCLC by analysis (Shin et al., 2018). Pang et al. (2020) Enrichment of exosomes by binding of tio2 shells and hydrophilic phosphate heads of exosomal phospholipids was performed using Fe3O4@tio2 nanoparticles. Exosomal PD-L1 was labeled and quantified by adding anti-PD-L1 antibody-modified Au@Ag@MBA SERS tags. Based on personalized SERS signal analysis, it was possible to differentiate NSCLC patients from healthy controls and the potential efficacy of immunotherapy (Pang et al., 2020). Preparation of SERS signal reporter molecules with Au@R6G@agau nanoparticles with small nanogaps produces stable SERS signals. Covalent attachment of isolated substrates of ARANP and silica beads to the 3′- and 5′-termini of capture probes targeting exosomal miRNAs specifically recognizes exosomal miRNAs. This technique was used to detect exosomal miRNAs in the plasma of NSCLC cancer patients (Ma et al., 2018).
Breast cancer is a malignant tumor that occurs in the epithelial cells of the breast and is highly heterogeneous in time and space, and its extremely high incidence poses a serious threat to women’s physical and mental health and life safety (Boix-Montesinos et al., 2021; Burstein et al., 2021). Zhang developed an assembling gold nanoparticles in triangular pyramid DNA (TP-Au nps) based on the principle of electrostatic attraction between negatively charged DNA tetrahedra and positively charged Au nps (Zhang et al., 2019a). This sensor with a strong electromagnetic hot spot can greatly enhance the SERS signal. The epithelial cell adhesion molecule content on human breast cancer cell MCF-7 exosomes is high, so epcam aptamers and cholesterol-modified DNA can be selected for exosome identification and enrichment when detecting exosomes. Streptavidin-modified magnetic beads react with biotin-modified epcam aptamers to capture enriched exosomes. The cholesterol-modified DNA hybridizes with TP-Au nps SERS probes and the exosomes are identified by hydrophobic interactions between the cholesterol attached to the SERS probes and the exosomal lipid membrane. This method can effectively distinguish between exosomes extracted from plasma of healthy individuals and breast cancer patients. Circulating exosomal PD-L1 can be used as a predictor of clinical efficacy of anti-PD-1/PD-L1. A template array was immobilized on a standard gold-plated glass lens and then a miniaturized affinity-based device for exosome capture was prepared by modifying antibodies on the array well plate (Kwizera et al., 2018). Gold nanorods bind to exosomes via electrostatic interactions between cetyltrimethylammonium bromide and lipid membranes on exosomes, enabling quantitative detection of several surface protein markers on exosomes derived from breast cancer cell cultures and human epidermal growth factor receptor 2 (HER2)-positive breast cancer patients. HER2-positive Exosomes from plasma of breast cancer patients have more significant levels of HER2 and epcam than those from healthy donors, suggesting the diagnostic potential of these markers for HER2 and epcam in breast cancer diagnosis. Lee et al. (2019) developed a SERS-based sensing platform for the quantitative determination of exosomal miRNA. They obtained ultrasensitive exosomal miRNAs with single nucleotide specificity in an enhanced SERS signal from a homogeneous plasma head flocked gold nanopillar substrate. The proposed SERS sensor showed extremely low detection limits without any amplification process, a wide dynamic range and good multiplex sensing capability. Using this model they could analyze the differences in miRNA expression in plasma exosomes of breast cancer patients and differentiate breast cancer subtypes based on the results (Lee et al., 2019).
Pancreatic cancer is a highly aggressive and malignant malignancy of the gastrointestinal tract, and its incidence is increasing year by year (Hou et al., 2021; Huang et al., 2021; Tao et al., 2021). SERS with polydopamine-modified immunocapture substrate and ultrathin polydopamine-encapsulated antibody-reporter gene-Ag (shell)-Au (nucleus) multilayer (PEARL) was developed. Migration inhibitory factor (MIF) antibody-based SERS immunoassay can distinguish not only between pancreatic cancer patients and healthy controls, but also between metastatic and non-metastatic tumors, as well as tumor lymph node metastasis carmicheal used the unlabeled SERS technique in combination with a principal component differential function analysis algorithm to differentiate between exosomes derived from pancreatic cancer and normal pancreatic epithelial cells with 90% discriminatory accuracy. They further applied this method to the analysis of exosomes in serum and also obtained satisfactory results, which is expected to develop a new method for early detection of pancreatic cancer (Carmicheal et al., 2019). A DSN-assisted dual SERS biosensor was developed for the detection of miRNA in exosomes and blood sample residual plasma based on Fe3O4 @Ag-dnaau@Ag@DTNB (SERS tag) affixes. Early diagnosis of patients can be made by detecting differences in SERS signals in plasma-derived exosomes and residual supernatant plasma from pancreatic ductal adenocarcinoma (PDAC), chronic pancreatitis and normal control blood samples (Pang et al., 2019). Jiang et al. (2021b) Designed an in situ platform for direct extraction of exosomal miRNA from serum samples. They synthesized locking nucleic acid-modified Au@DTNB as a SERS tag into exosomes and assembled with target miRNAs to induce hotspot SERS signals. The exosomes were then enriched by the addition of Fe3O4@tio2 nanoparticles for further SERS detection. Using this technique to detect exosomal miRNA-10b can be used to effectively identify patients with PDAC (Jiang et al., 2021b).
Melanoma is a highly aggressive malignancy originating from melanocytes, the incidence of which is increasing every year (Han et al., 2021a; Dimitriou et al., 2021; Worrede et al., 2021). Fraire et al. (2019) demonstrated that a new strategy using core-shell plasma nanoparticles as SERS substrates showed higher near-field enhancement than previous methods, which significantly increased the signal-to-noise ratio of SERS spectra. Individual vesicles of melanoma cells and erythrocytes can be distinguished using this technique (Fraire et al., 2019).
Osteosarcoma is the most common primary malignant bone tumor that occurs in the epiphysis of long bones, mainly in the distal femur, proximal tibia and humerus (Lin et al., 2017; Lu et al., 2020). Approximately 18% of patients with osteosarcoma already have micrometastases at the time of diagnosis (Ballatori and Hinds, 2016). For patients with osteosarcoma who develop metastases and recurrence, their 5-years survival rate remains at about 20% (Gill et al., 2013). Han et al. (2021c) analyzed the potential of SERS and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) for the detection of osteosarcoma. They characterized plasma-derived exosomes from osteosarcoma patients and healthy individuals using SERS and MALDI-TOF MS, and further combined the two types of spectroscopic techniques using a data fusion approach, which revealed through analysis that non-invasive liquid biopsy methods of SERS and MALDI-TOF MS fingerprint mapping of exosomes have great potential for rapid diagnosis of osteosarcoma (Han et al., 2021c).
Exosomes are found in a variety of body fluids, carry characteristic markers and play a wide range of functions (Han et al., 2021b; Cheng et al., 2021; Peng et al., 2021). The study of exosomes has great potential for new diagnostic and medical translational applications, yet no significant breakthroughs have been achieved to date. Our understanding of the molecular mechanisms of exosomes development and sorting of contents is still in its infancy, and the standardization of exosomes collection and detection techniques for clinical samples is still immature, both of which have limited the development of exosomesas clinically available diagnostic and therapeutic tools. The precise molecular mechanisms of exosomes, the innovation of isolation and purification techniques, and the multi-omics analysis of exosomes are the future directions of exosomes research.
There are still many challenges and limitations in using Raman spectroscopy to analyze biological samples. Higher sensitivity techniques such as SERS can be used when the Raman spectral signal of the sample is weak, but sample preparation plays an important role. When analyzing Raman spectra of biological samples, the complexity of the composition requires a reliable calibration model to ensure reliable and quantitative measurements. There are also challenges when using probes for in vivo analysis, where deficiencies such as background fluorescence, artifacts and weak signals need to be overcome and diagnostic models need to be constructed by multivariate statistical analysis methods before introducing Raman spectroscopy as a basis for diagnosis. Raman spectroscopy can analyze changes in diseases at the molecular level, providing objective and quantifiable information for disease diagnosis and evaluation of treatment efficacy. The use of Raman spectroscopy provides more insight into the infiltration process of tumors and the chemical information obtained allows quantitative and qualitative analysis of the disease. Interference such as fluorescence poses great difficulties in acquiring and resolving Raman spectra of biological samples, so there is still a need to develop techniques for acquiring, processing and classifying data to determine the nature of tumor tissue lesions by pre-processing the raw data, eliminating unwanted signals and enhancing Raman spectral features to achieve qualitative and quantitative analysis of the data. With the continuous improvement of Raman spectroscopy database, tissue classification methods and instrument design, it will become possible to acquire Raman spectroscopy data with higher resolution and accuracy and shorter acquisition time, thus enabling early cancer diagnosis. The reproducibility, reliability and sensitivity of SERS biological detection technology depend on the reasonable design of the SERS substrate, the proper handling of biological samples, the precise control of the detection conditions and the adequate analysis of the spectral data. In the future, efforts should be made to control its potential factors. We should prepare SERS substrates with high uniformity, high enhancement effect and clean surface, which can be modified appropriately. We can also choose appropriate Raman probe molecules to improve their biocompatibility, avoid non-specific adsorption, and to be tested In addition, when storing and pretreating biological samples, keep them in an environment close to the normal physiological state as much as possible to avoid irreversible changes. Moreover, select appropriate SERS detection conditions to obtain reliable detection information. More importantly, establish a standard SERS database of biomolecules to accurately identify spectral information. If necessary, the probe response in the detection system can be corrected, or appropriate data processing and statistical analysis methods can be selected. Future SERS technology The prospect in the field of early detection of tumor biomarkers lies in the development of an ultra-sensitive, highly specific, high-throughput and universal detection platform to meet the needs of life sciences and clinical diagnosis.
In recent years, SERS technology has been applied to the study of exosome detection, which has greatly improved the sensitivity of exosome detection, which is significant for the development of new liquid biopsy techniques based on exosomes (Lee et al., 2020; Han et al., 2021c; Das et al., 2021; Fan et al., 2021; Guerrini et al., 2021). The forms of exosome detection by SERS technology are mainly divided into non-labeled and labeled assays. Non-labeled assays provide overall biochemical information about the exosome itself and are easy, fast and inexpensive to perform (Russo et al., 2020; Jiang et al., 2021b; Guerrini et al., 2021). Marker assays, on the other hand, improve the specificity of exosome identification and can also identify and detect secreted exosomes from multiple cancer cell lines simultaneously (Blanco-Formoso and Alvarez-Puebla, 2020; Chen et al., 2020; Hao et al., 2020). Although some progress has been made in exosome detection with SERS technology, many challenges remain for further research. First, we need to continue to explore efficient SERS detection substrates to further improve the Raman signal of exosomes and enhance the detection limit. Secondly, we should continue to develop SERS detection strategies for exosomal surface proteins and internal nucleic acids to obtain high quality and reproducible Raman signals and to extract comprehensive diagnostic information. Moreover, it is important to develop efficient machine learning algorithms for deep mining of exosomal SERS signals to achieve rapid and accurate identification of exosomes from different sources. In the future, we should also explore the functionalization of exosomes and characterize the biological behavior of exosomes in combination with SERS technology. For clinical applications, we should also continue to expand clinical testing samples to further validate the feasibility and applicability of SERS-based exosome detection technology. With further development of related technologies, we believe that SERS-based exosome detection technology is expected to become a new method for nondestructive, convenient, and accurate liquid biopsy.
We reviewed the latest research and progress in the identification of exosomes by SERS in the early diagnosis of tumors. First, we discussed the current research status of SERS and the main detection methods of exosomes. Then, we summarized the exosomes detection strategies used to obtain the exosomes and unlabeled SERS signals, and classified and summarized the detection strategies according to how to capture exosomes on the SERS substrate. At the same time, we summarized the advantages and disadvantages of SERS in detecting proteins and nucleic acids. Moreover, we analyzed and summarized the research and progress of SERS identification of exosomes in the early diagnosis of different tumors, and proposed possible future research directions based on the current research status. With the further development of related technologies, we believe that the exosomes detection technology based on SERS technology is expected to become a new non-destructive, convenient and accurate liquid biopsy method for tumors.
LY made important contributions to the study conception and design. JJ conducted data analysis and interpretation. SL participated in the drafting and revision of the manuscript. All authors read, revised and approved this manuscript and agreed to be responsible for all aspects of the research to ensure the data accuracy and integrity of this work.
This work was supported by the Fundamental Research Funds for the Central Universities (No. LD202110).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the generous support by Liaoning Cancer Hospital and Institute (Shenyang) and Dalian University of Technology (Dalian).
ELISA, enzyme linked immunosorbent assay; EVs, extracellular vesicles; GC, gastric cancer; GBM, glioblastoma; HER2, human epidermal growth factor receptor 2; lncRNA, long noncoding RNA; MCR-ALS, multivariate curve resolution alte RNAting least squares; MIF, Migration inhibitory factor; NSCLC, non-small cell lung cancer; PDAC, pancreatic ductal adenocarcinoma; PCA, principal component analysis; PLS-DA, partial least squares discriminant analysis; PCR, Polymerase chain reaction; sEVs, small extracellular vesicles; SERS, Surface-enhanced Raman spectroscopy; TAM, tumor-associated macrophages.
Abdulhalim, I. (2014). Plasmonic Sensing Using Metallic Nano-Sculptured Thin Films. Small 10 (17), 3499–3514. doi:10.1002/smll.201303181
Andrikopoulou, A., Shalit, A., Zografos, E., Koutsoukos, K., Korakiti, A.-M., Liontos, M., et al. (2021). MicroRNAs as Potential Predictors of Response to CDK4/6 Inhibitor Treatment. Cancers 13 (16), 4114. doi:10.3390/cancers13164114
Ariston Gabriel, A. N., Wang, F., Jiao, Q., Yvette, U., Yang, X., Al-Ameri, S. A., et al. (2020). The Involvement of Exosomes in the Diagnosis and Treatment of Pancreatic Cancer. Mol. Cancer 19 (1), 132. doi:10.1186/s12943-020-01245-y
Azparren-Angulo, M., Royo, F., Gonzalez, E., Liebana, M., Brotons, B., Berganza, J., et al. (2021). Extracellular Vesicles in Hepatology: Physiological Role, Involvement in Pathogenesis, and Therapeutic Opportunities. Pharmacol. Ther. 218, 107683. doi:10.1016/j.pharmthera.2020.107683
Baert, T., Ferrero, A., Sehouli, J., O'Donnell, D. M., González-Martín, A., Joly, F., et al. (2021). The Systemic Treatment of Recurrent Ovarian Cancer Revisited. Ann. Oncol. 32 (6), 710–725. doi:10.1016/j.annonc.2021.02.015
Ballatori, S. E., and Hinds, P. W. (2016). Osteosarcoma: Prognosis Plateau Warrants Retinoblastoma Pathway Targeted Therapy. Sig Transduct Target. Ther. 1, 16001. doi:10.1038/sigtrans.2016.1
Berry, M. E., Kearns, H., Graham, D., and Faulds, K. (2021). Surface Enhanced Raman Scattering for the Multiplexed Detection of Pathogenic Microorganisms: towards point-of-use Applications. Analyst 146 (20), 6084–6101. doi:10.1039/d1an00865j
Blanco-Formoso, M., and Alvarez-Puebla, R. A. (2020). Cancer Diagnosis through SERS and Other Related Techniques. Int. J. Mol. Sci. 21 (6), 2253. doi:10.3390/ijms21062253
Boix-Montesinos, P., Soriano-Teruel, P. M., Armiñán, A., Orzáez, M., and Vicent, M. J. (2021). The Past, Present, and Future of Breast Cancer Models for Nanomedicine Development. Adv. Drug Deliv. Rev. 173, 306–330. doi:10.1016/j.addr.2021.03.018
Burstein, H. J., Curigliano, G., Thürlimann, B., Weber, W. P., Poortmans, P., Regan, M. M., et al. (2021). Customizing Local and Systemic Therapies for Women with Early Breast Cancer: the St. Gallen International Consensus Guidelines for Treatment of Early Breast Cancer 2021. Ann. Oncol. 32 (10), 1216–1235. doi:10.1016/j.annonc.2021.06.023
Calderon, I., Guerrini, L., and Alvarez-Puebla, R. A. (2021). Targets and Tools: Nucleic Acids for Surface-Enhanced Raman Spectroscopy. Biosensors 11 (7), 230. doi:10.3390/bios11070230
Carmicheal, J., Hayashi, C., Huang, X., Liu, L., Lu, Y., Krasnoslobodtsev, A., et al. (2019). Label-free Characterization of Exosome via Surface Enhanced Raman Spectroscopy for the Early Detection of Pancreatic Cancer. Nanomedicine: Nanotechnology, Biol. Med. 16, 88–96. doi:10.1016/j.nano.2018.11.008
Chen, H., Chengalvala, V., Hu, H., and Sun, D. (2021a). Tumor-derived Exosomes: Nanovesicles Made by Cancer Cells to Promote Cancer Metastasis. Acta Pharmaceutica Sinica B 11 (8), 2136–2149. doi:10.1016/j.apsb.2021.04.012
Chen, H., Luo, C., Yang, M., Li, J., Ma, P., and Zhang, X. (2020). Intracellular Uptake of and Sensing with SERS-Active Hybrid Exosomes: Insight into a Role of Metal Nanoparticles. Nanomedicine 15 (9), 913–926. doi:10.2217/nnm-2019-0419
Chen, H., Luo, C., and Zhang, S. (2021b). Intracellular Imaging and Concurrent pH Sensing of Cancer-Derived Exosomes Using Surface-Enhanced Raman Scattering. Anal. Bioanal. Chem. 413 (15), 4091–4101. doi:10.1007/s00216-021-03365-w
Chen, L., Qu, J., Mei, Q., Chen, X., Fang, Y., Chen, L., et al. (2021c). Small Extracellular Vesicles from Menstrual Blood-Derived Mesenchymal Stem Cells (MenSCs) as a Novel Therapeutic Impetus in Regenerative Medicine. Stem Cel Res Ther 12 (1), 433. doi:10.1186/s13287-021-02511-6
Cheng, Y.-C., Chang, Y.-A., Chen, Y.-J., Sung, H.-M., Bogeski, I., Su, H.-L., et al. (2021). The Roles of Extracellular Vesicles in Malignant Melanoma. Cells 10 (10), 2740. doi:10.3390/cells10102740
Claridge, B., Lozano, J., Poh, Q. H., and Greening, D. W. (2021). Development of Extracellular Vesicle Therapeutics: Challenges, Considerations, and Opportunities. Front. Cel Dev. Biol. 9, 734720. doi:10.3389/fcell.2021.734720
Clark, R. A., Garman, Z. G., Price, R. J., and Sheybani, N. D. (2021). Functional Intersections between Extracellular Vesicles and Oncolytic Therapies. Trends Pharmacol. Sci. 42, 883–896. doi:10.1016/j.tips.2021.09.001
Crudele, F., Bianchi, N., Astolfi, A., Grassilli, S., Brugnoli, F., Terrazzan, A., et al. (2021). The Molecular Networks of microRNAs and Their Targets in the Drug Resistance of Colon Carcinoma. Cancers 13 (17), 4355. doi:10.3390/cancers13174355
Dai, J., Su, Y., Zhong, S., Cong, L., Liu, B., Yang, J., et al. (2020). Exosomes: Key Players in Cancer and Potential Therapeutic Strategy. Sig Transduct Target. Ther. 5 (1), 145. doi:10.1038/s41392-020-00261-0
Das, G. M., Managò, S., Mangini, M., and De Luca, A. C. (2021). Biosensing Using SERS Active Gold Nanostructures. Nanomaterials 11 (10), 2679. doi:10.3390/nano11102679
Dimitriou, F., Long, G. V., and Menzies, A. M. (2021). Novel Adjuvant Options for Cutaneous Melanoma. Ann. Oncol. 32 (7), 854–865. doi:10.1016/j.annonc.2021.03.198
Ding, S.-Y., You, E.-M., Tian, Z.-Q., and Moskovits, M. (2017). Electromagnetic Theories of Surface-Enhanced Raman Spectroscopy. Chem. Soc. Rev. 46 (13), 4042–4076. doi:10.1039/c7cs00238f
Dong, S., Wang, Y., Liu, Z., Zhang, W., Yi, K., Zhang, X., et al. (2020). Beehive-Inspired Macroporous SERS Probe for Cancer Detection through Capturing and Analyzing Exosomes in Plasma. ACS Appl. Mater. Inter. 12 (4), 5136–5146. doi:10.1021/acsami.9b21333
Drusbosky, L. M., Dawar, R., Rodriguez, E., and Ikpeazu, C. V. (2021). Therapeutic Strategies in METex14 Skipping Mutated Non-small Cell Lung Cancer. J. Hematol. Oncol. 14 (1), 129. doi:10.1186/s13045-021-01138-7
Fan, C., Zhao, N., Cui, K., Chen, G., Chen, Y., Wu, W., et al. (2021). Ultrasensitive Exosome Detection by Modularized SERS Labeling for Postoperative Recurrence Surveillance. ACS Sens. 6 (9), 3234–3241. doi:10.1021/acssensors.1c00890
Fraire, J. C., Stremersch, S., Bouckaert, D., Monteyne, T., De Beer, T., Wuytens, P., et al. (2019). Improved Label-free Identification of Individual Exosome-like Vesicles with Au@Ag Nanoparticles as SERS Substrate. ACS Appl. Mater. Inter. 11 (43), 39424–39435. doi:10.1021/acsami.9b11473
Gill, J., Ahluwalia, M. K., Geller, D., and Gorlick, R. (2013). New Targets and Approaches in Osteosarcoma. Pharmacol. Ther. 137 (1), 89–99. doi:10.1016/j.pharmthera.2012.09.003
Grasso, J. A., Bruno, M., Yates, A. A., Wei, L., and Epstein, P. M. (1990). Calmodulin Dependence of Transferrin Receptor Recycling in Rat Reticulocytes. Biochem. J. 266 (1), 261–272. doi:10.1042/bj2660261
Gruenke, N. L., Cardinal, M. F., McAnally, M. O., Frontiera, R. R., Schatz, G. C., and Van Duyne, R. P. (2016). Ultrafast and Nonlinear Surface-Enhanced Raman Spectroscopy. Chem. Soc. Rev. 45 (8), 2263–2290. doi:10.1039/c5cs00763a
Guerrini, L., Garcia-Rico, E., O’Loghlen, A., Giannini, V., and Alvarez-Puebla, R. A. (2021). Surface-Enhanced Raman Scattering (SERS) Spectroscopy for Sensing and Characterization of Exosomes in Cancer Diagnosis. Cancers 13 (9), 2179. doi:10.3390/cancers13092179
Han, A., Schug, Z. T., and Aplin, A. E. (2021a). Metabolic Alterations and Therapeutic Opportunities in Rare Forms of Melanoma. Trends Cancer 7 (8), 671–681. doi:10.1016/j.trecan.2021.05.005
Han, C., Yang, J., Sun, J., and Qin, G. (2021b). Extracellular Vesicles in Cardiovascular Disease: Biological Functions and Therapeutic Implications. Pharmacol. Ther., 108025. doi:10.1016/j.pharmthera.2021.108025
Han, Z., Yi, J., Yang, Y., Li, D., Peng, C., Long, S., et al. (2021c). SERS and MALDI-TOF MS Based Plasma Exosome Profiling for Rapid Detection of Osteosarcoma. Analyst 146, 6496–6505. doi:10.1039/d1an01163d
Hao, N., Pei, Z., Liu, P., Bachman, H., Naquin, T. D., Zhang, P., et al. (2020). Acoustofluidics-Assisted Fluorescence-SERS Bimodal Biosensors. Small 16 (48), 2005179. doi:10.1002/smll.202005179
Hou, J., Li, X., and Xie, K.-P. (2021). Coupled Liquid Biopsy and Bioinformatics for Pancreatic Cancer Early Detection and Precision Prognostication. Mol. Cancer 20 (1), 34. doi:10.1186/s12943-021-01309-7
Hu, W., Liu, C., Bi, Z.-Y., Zhou, Q., Zhang, H., Li, L.-L., et al. (2020). Comprehensive Landscape of Extracellular Vesicle-Derived RNAs in Cancer Initiation, Progression, Metastasis and Cancer Immunology. Mol. Cancer 19 (1), 102. doi:10.1186/s12943-020-01199-1
Hu, Y., Wang, Y., Chen, T., Hao, Z., Cai, L., and Li, J. (2021). Exosome: Function and Application in Inflammatory Bone Diseases. Oxidative Med. Cell Longevity 2021, 1–17. doi:10.1155/2021/6324912
Huang, X., Ding, L., Liu, X., Tong, R., Ding, J., Qian, Z., et al. (2021). Regulation of Tumor Microenvironment for Pancreatic Cancer Therapy. Biomaterials 270, 120680. doi:10.1016/j.biomaterials.2021.120680
Hur, J. Y., and Lee, K. Y. (2021). Characteristics and Clinical Application of Extracellular Vesicle-Derived DNA. Cancers 13 (15), 3827. doi:10.3390/cancers13153827
Isaac, R., Reis, F. C. G., Ying, W., and Olefsky, J. M. (2021). Exosomes as Mediators of Intercellular Crosstalk in Metabolism. Cel Metab. 33 (9), 1744–1762. doi:10.1016/j.cmet.2021.08.006
Jiang, C., Zhang, N., Hu, X., and Wang, H. (2021a). Tumor-associated Exosomes Promote Lung Cancer Metastasis through Multiple Mechanisms. Mol. Cancer 20 (1), 117. doi:10.1186/s12943-021-01411-w
Jiang, S., Li, Q., Wang, C., Pang, Y., Sun, Z., and Xiao, R. (2021b). In Situ Exosomal MicroRNA Determination by Target-Triggered SERS and Fe3O4@TiO2-Based Exosome Accumulation. ACS Sens. 6 (3), 852–862. doi:10.1021/acssensors.0c01900
Kang, T., Zhu, J., Luo, X., Jia, W., Wu, P., and Cai, C. (2021). Controlled Self-Assembly of a Close-Packed Gold Octahedra Array for SERS Sensing Exosomal MicroRNAs. Anal. Chem. 93 (4), 2519–2526. doi:10.1021/acs.analchem.0c04561
Koch, A. L., Vellanki, P. J., Drezner, N., Li, X., Mishra-Kalyani, P. S., Shen, Y. L., et al. (2021). FDA Approval Summary: Osimertinib for Adjuvant Treatment of Surgically Resected Non-small Cell Lung Cancer, a Collaborative Project Orbis Review. Clin. Cancer Res. 27, 6638–6643. doi:10.1158/1078-0432.CCR-21-1034
Konoshenko, M. Y., Bryzgunova, O. E., and Laktionov, P. P. (2021). miRNAs and Androgen Deprivation Therapy for Prostate Cancer. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1876 (2), 188625. doi:10.1016/j.bbcan.2021.188625
Kwizera, E. A., O'Connor, R., Vinduska, V., Williams, M., Butch, E. R., Snyder, S. E., et al. (2018). Molecular Detection and Analysis of Exosomes Using Surface-Enhanced Raman Scattering Gold Nanorods and a Miniaturized Device. Theranostics 8 (10), 2722–2738. doi:10.7150/thno.21358
Lee, C., Carney, R. P., Hazari, S., Smith, Z. J., Knudson, A., Robertson, C. S., et al. (2015). 3D Plasmonic Nanobowl Platform for the Study of Exosomes in Solution. Nanoscale 7 (20), 9290–9297. doi:10.1039/c5nr01333j
Lee, J. U., Kim, S., and Sim, S. J. (2020). SERS-based Nanoplasmonic Exosome Analysis: Enabling Liquid Biopsy for Cancer Diagnosis and Monitoring Progression. Biochip J. 14 (3), 231–241. doi:10.1007/s13206-020-4301-5
Lee, J. U., Kim, W. H., Lee, H. S., Park, K. H., and Sim, S. J. (2019). Quantitative and Specific Detection of Exosomal miRNAs for Accurate Diagnosis of Breast Cancer Using a Surface-Enhanced Raman Scattering Sensor Based on Plasmonic Head-Flocked Gold Nanopillars. Small 15 (17), 1804968. doi:10.1002/smll.201804968
Lee, T. H., Park, E., Goh, Y.-g., Lee, H. B., Rou, W. S., and Eun, H. S. (2021). The Specific Gravity-free Method for the Isolation of Circulating Tumor KRAS Mutant DNA and Exosome in Colorectal Cancer. Micromachines 12 (8), 987. doi:10.3390/mi12080987
Leggio, L., Paternò, G., Vivarelli, S., Falzone, G. G., Giachino, C., Marchetti, B., et al. (2021). Extracellular Vesicles as Novel Diagnostic and Prognostic Biomarkers for Parkinson's Disease. Aging Dis. 12 (6), 1494–1515. doi:10.14336/AD.2021.0527
Leitch, J. J., Collins, J., Friedrich, A. K., Stimming, U., Dutcher, J. R., and Lipkowski, J. (2012). Infrared Studies of the Potential Controlled Adsorption of Sodium Dodecyl Sulfate at the Au(111) Electrode Surface. Langmuir 28 (5), 2455–2464. doi:10.1021/la204451s
Li, B., Cao, Y., Sun, M., and Feng, H. (2021a). Expression, Regulation, and Function of Exosome-Derived miRNAs in Cancer Progression and Therapy. FASEB J. 35 (10), e21916. doi:10.1096/fj.202100294RR
Li, Z., Meng, X., Wu, P., Zha, C., Han, B., Li, L., et al. (2021b). Glioblastoma Cell-Derived lncRNA-Containing Exosomes Induce Microglia to Produce Complement C5, Promoting Chemotherapy Resistance. Cancer Immunol. Res. 9, 1383–1399. doi:10.1158/2326-6066.CIR-21-0258
Lin, J., Wang, Z., Huang, J., Tang, S., Saiding, Q., Zhu, Q., et al. (2021). Microenvironment-Protected Exosome-Hydrogel for Facilitating Endometrial Regeneration, Fertility Restoration, and Live Birth of Offspring. Small 17 (11), 2007235. doi:10.1002/smll.202007235
Lin, Y.-H., Jewell, B. E., Gingold, J., Lu, L., Zhao, R., Wang, L. L., et al. (2017). Osteosarcoma: Molecular Pathogenesis and iPSC Modeling. Trends Mol. Med. 23 (8), 737–755. doi:10.1016/j.molmed.2017.06.004
Liu, X., Gao, X., Yang, L., Zhao, Y., and Li, F. (2021a). Metal-Organic Framework-Functionalized Paper-Based Electrochemical Biosensor for Ultrasensitive Exosome Assay. Anal. Chem. 93 (34), 11792–11799. doi:10.1021/acs.analchem.1c02286
Liu, X., Guo, J., Li, Y., Wang, B., Yang, S., Chen, W., et al. (2021b). SERS Substrate Fabrication for Biochemical Sensing: towards point-of-care Diagnostics. J. Mater. Chem. B 9 (40), 8378–8388. doi:10.1039/d1tb01299a
Liu, X., Zong, Z., Xing, M., Liu, X., Li, J., and Liu, D. (2021c). pH-Mediated Clustering of Exosomes: Breaking through the Size Limit of Exosome Analysis in Conventional Flow Cytometry. Nano Lett. 21, 8817–8823. doi:10.1021/acs.nanolett.1c03211
López-Lorente, Á. I. (2021). Recent Developments on Gold Nanostructures for Surface Enhanced Raman Spectroscopy: Particle Shape, Substrates and Analytical Applications. A Review. Analytica Chim. Acta 1168, 338474. doi:10.1016/j.aca.2021.338474
Lu, K.-H., Lu, E. W.-H., Lin, C.-W., Yang, J.-S., and Yang, S.-F. (2020). New Insights into Molecular and Cellular Mechanisms of Zoledronate in Human Osteosarcoma. Pharmacol. Ther. 214, 107611. doi:10.1016/j.pharmthera.2020.107611
Ma, D., Huang, C., Zheng, J., Tang, J., Li, J., Yang, J., et al. (2018). Quantitative Detection of Exosomal microRNA Extracted from Human Blood Based on Surface-Enhanced Raman Scattering. Biosens. Bioelectron. 101, 167–173. doi:10.1016/j.bios.2017.08.062
Ma, J., Yan, M., Feng, G., Ying, Y., Chen, G., Shao, Y., et al. (2021). An Overview on Molecular Imprinted Polymers Combined with Surface-Enhanced Raman Spectroscopy Chemical Sensors toward Analytical Applications. Talanta 225, 122031. doi:10.1016/j.talanta.2020.122031
Mao, W., Wang, K., Wu, Z., Xu, B., and Chen, M. (2021). Current Status of Research on Exosomes in General, and for the Diagnosis and Treatment of Kidney Cancer in Particular. J. Exp. Clin. Cancer Res. 40 (1), 305. doi:10.1186/s13046-021-02114-2
Markina, N. E., Cialla-May, D., and Markin, A. V. (2021). Cyclodextrin-assisted Surface-Enhanced Raman Spectroscopy: a Critical Review. Anal. Bioanal. Chem. doi:10.1007/s00216-021-03704-x
Mohammadi, M., Zargartalebi, H., Salahandish, R., Aburashed, R., Wey Yong, K., and Sanati-Nezhad, A. (2021). Emerging Technologies and Commercial Products in Exosome-Based Cancer Diagnosis and Prognosis. Biosens. Bioelectron. 183, 113176. doi:10.1016/j.bios.2021.113176
Morrissey, S. M., and Yan, J. (2020). Exosomal PD-L1: Roles in Tumor Progression and Immunotherapy. Trends Cancer 6 (7), 550–558. doi:10.1016/j.trecan.2020.03.002
Naqvi, S. M. Z. A., Zhang, Y., Ahmed, S., Abdulraheem, M. I., Hu, J., Tahir, M. N., et al. (2022). Applied Surface Enhanced Raman Spectroscopy in Plant Hormones Detection, Annexation of Advanced Technologies: A Review. Talanta 236, 122823. doi:10.1016/j.talanta.2021.122823
Ng, C. T., Azwar, S., Yip, W. K., Zahari Sham, S. Y., Faisal Jabar, M., Sahak, N. H., et al. (2021). Isolation and Identification of Long Non-coding RNAs in Exosomes Derived from the Serum of Colorectal Carcinoma Patients. Biology 10 (9), 918. doi:10.3390/biology10090918
Ning, C.-F., Wang, L., Tian, Y.-F., Yin, B.-C., and Ye, B.-C. (2020). Multiple and Sensitive SERS Detection of Cancer-Related Exosomes Based on Gold-Silver Bimetallic Nanotrepangs. Analyst 145 (7), 2795–2804. doi:10.1039/c9an02180a
Oey, O., Ghaffari, M., Li, J. J., and Hosseini-Beheshti, E. (2021). Application of Extracellular Vesicles in the Diagnosis and Treatment of Prostate Cancer: Implications for Clinical Practice. Crit. Rev. Oncology/Hematology 167, 103495. doi:10.1016/j.critrevonc.2021.103495
Oh-Hohenhorst, S. J., and Lange, T. (2021). Role of Metastasis-Related microRNAs in Prostate Cancer Progression and Treatment. Cancers 13 (17), 4492. doi:10.3390/cancers13174492
Pan, B. T., Teng, K., Wu, C., Adam, M., and Johnstone, R. M. (1985). Electron Microscopic Evidence for Externalization of the Transferrin Receptor in Vesicular Form in Sheep Reticulocytes. J. Cel Biol 101 (3), 942–948. doi:10.1083/jcb.101.3.942
Pang, Y., Shi, J., Yang, X., Wang, C., Sun, Z., and Xiao, R. (2020). Personalized Detection of Circling Exosomal PD-L1 Based on Fe3O4@TiO2 Isolation and SERS Immunoassay. Biosens. Bioelectron. 148, 111800. doi:10.1016/j.bios.2019.111800
Pang, Y., Wang, C., Lu, L., Wang, C., Sun, Z., and Xiao, R. (2019). Dual-SERS Biosensor for One-step Detection of microRNAs in Exosome and Residual Plasma of Blood Samples for Diagnosing Pancreatic Cancer. Biosens. Bioelectron. 130, 204–213. doi:10.1016/j.bios.2019.01.039
Park, J., Hwang, M., Choi, B., Jeong, H., Jung, J.-h., Kim, H. K., et al. (2017). Exosome Classification by Pattern Analysis of Surface-Enhanced Raman Spectroscopy Data for Lung Cancer Diagnosis. Anal. Chem. 89 (12), 6695–6701. doi:10.1021/acs.analchem.7b00911
Peng, Q., Chiu, P. K.-F., Wong, C. Y.-P., Cheng, C. K.-L., Teoh, J. Y.-C., and Ng, C.-F. (2021). Identification of piRNA Targets in Urinary Extracellular Vesicles for the Diagnosis of Prostate Cancer. Diagnostics 11 (10), 1828. doi:10.3390/diagnostics11101828
Prieto, F., Su, Z., Leitch, J. J., Rueda, M., and Lipkowski, J. (2016). Quantitative Subtractively Normalized Interfacial Fourier Transform Infrared Reflection Spectroscopy Study of the Adsorption of Adenine on Au(111) Electrodes. Langmuir 32 (16), 3827–3835. doi:10.1021/acs.langmuir.6b00635
Prieto-Vila, M., Yoshioka, Y., and Ochiya, T. (2021). Biological Functions Driven by mRNAs Carried by Extracellular Vesicles in Cancer. Front. Cel Dev. Biol. 9, 620498. doi:10.3389/fcell.2021.620498
Raguraman, R., Srivastava, A., Munshi, A., and Ramesh, R. (2021). Therapeutic Approaches Targeting Molecular Signaling Pathways Common to Diabetes, Lung Diseases and Cancer. Adv. Drug Deliv. Rev. 178, 113918. doi:10.1016/j.addr.2021.113918
Reese, M., and Dhayat, S. A. (2021). Small Extracellular Vesicle Non-coding RNAs in Pancreatic Cancer: Molecular Mechanisms and Clinical Implications. J. Hematol. Oncol. 14 (1), 141. doi:10.1186/s13045-021-01149-4
Richter, R., and Lehr, C.-M. (2021). Extracellular Vesicles as Novel Assay Tools to Study Cellular Interactions of Anti-infective Compounds - A Perspective. Adv. Drug Deliv. Rev. 173, 492–503. doi:10.1016/j.addr.2021.04.010
Russo, M., Tirinato, L., Scionti, F., Coluccio, M. L., Perozziello, G., Riillo, C., et al. (2020). Raman Spectroscopic Stratification of Multiple Myeloma Patients Based on Exosome Profiling. ACS Omega 5 (47), 30436–30443. doi:10.1021/acsomega.0c03813
Shehzad, A., Islam, S. U., Shahzad, R., Khan, S., and Lee, Y. S. (2021). Extracellular Vesicles in Cancer Diagnostics and Therapeutics. Pharmacol. Ther. 223, 107806. doi:10.1016/j.pharmthera.2021.107806
Shen, Y., Yue, J., Xu, W., and Xu, S. (2021). Recent Progress of Surface-Enhanced Raman Spectroscopy for Subcellular Compartment Analysis. Theranostics 11 (10), 4872–4893. doi:10.7150/thno.56409
Shin, H., Jeong, H., Park, J., Hong, S., and Choi, Y. (2018). Correlation between Cancerous Exosomes and Protein Markers Based on Surface-Enhanced Raman Spectroscopy (SERS) and Principal Component Analysis (PCA). ACS Sens. 3 (12), 2637–2643. doi:10.1021/acssensors.8b01047
Shin, H., Oh, S., Hong, S., Kang, M., Kang, D., Ji, Y.-g., et al. (2020a). Early-Stage Lung Cancer Diagnosis by Deep Learning-Based Spectroscopic Analysis of Circulating Exosomes. ACS Nano 14 (5), 5435–5444. doi:10.1021/acsnano.9b09119
Shin, H., Seo, D., and Choi, Y. (2020b). Extracellular Vesicle Identification Using Label-free Surface-Enhanced Raman Spectroscopy: Detection and Signal Analysis Strategies. Molecules 25 (21), 5209. doi:10.3390/molecules25215209
Shin, S., Park, Y. H., Jung, S.-H., Jang, S.-H., Kim, M. Y., Lee, J. Y., et al. (2021). Urinary Exosome microRNA Signatures as a Noninvasive Prognostic Biomarker for Prostate Cancer. NPJ Genom. Med. 6 (1), 45. doi:10.1038/s41525-021-00212-w
Soares Martins, T., Catita, J., Martins Rosa, I., A. B. da Cruz e Silva, O., and Henriques, A. G. (2018). Exosome Isolation from Distinct Biofluids Using Precipitation and Column-Based Approaches. PLoS One 13 (6), e0198820. doi:10.1371/journal.pone.0198820
Špilak, A., Brachner, A., Kegler, U., Neuhaus, W., and Noehammer, C. (2021). Implications and Pitfalls for Cancer Diagnostics Exploiting Extracellular Vesicles. Adv. Drug Deliv. Rev. 175, 113819. doi:10.1016/j.addr.2021.05.029
Staubach, S., Bauer, F. N., Tertel, T., Börger, V., Stambouli, O., Salzig, D., et al. (2021). Scaled Preparation of Extracellular Vesicles from Conditioned media. Adv. Drug Deliv. Rev. 177, 113940. doi:10.1016/j.addr.2021.113940
Stremersch, S., Marro, M., Pinchasik, B.-E., Baatsen, P., Hendrix, A., De Smedt, S. C., et al. (2016). Identification of Individual Exosome-like Vesicles by Surface Enhanced Raman Spectroscopy. Small 12 (24), 3292–3301. doi:10.1002/smll.201600393
Sun, Z., Yang, J., Li, H., Wang, C., Fletcher, C., Li, J., et al. (2021). Progress in the Research of Nanomaterial-Based Exosome Bioanalysis and Exosome-Based Nanomaterials Tumor Therapy. Biomaterials 274, 120873. doi:10.1016/j.biomaterials.2021.120873
Tahir, M. A., Dina, N. E., Cheng, H., Valev, V. K., and Zhang, L. (2021). Surface-enhanced Raman Spectroscopy for Bioanalysis and Diagnosis. Nanoscale 13 (27), 11593–11634. doi:10.1039/d1nr00708d
Tang, X.-H., Guo, T., Gao, X.-Y., Wu, X.-L., Xing, X.-F., Ji, J.-F., et al. (2021). Exosome-derived Noncoding RNAs in Gastric Cancer: Functions and Clinical Applications. Mol. Cancer 20 (1), 99. doi:10.1186/s12943-021-01396-6
Tanwar, S., Paidi, S. K., Prasad, R., Pandey, R., and Barman, I. (2021). Advancing Raman Spectroscopy from Research to Clinic: Translational Potential and Challenges. Spectrochimica Acta A: Mol. Biomol. Spectrosc. 260, 119957. doi:10.1016/j.saa.2021.119957
Tao, J., Yang, G., Zhou, W., Qiu, J., Chen, G., Luo, W., et al. (2021). Targeting Hypoxic Tumor Microenvironment in Pancreatic Cancer. J. Hematol. Oncol. 14 (1), 14. doi:10.1186/s13045-020-01030-w
Teng, F., and Fussenegger, M. (2020). Shedding Light on Extracellular Vesicle Biogenesis and Bioengineering. Adv. Sci. 8 (1), 2003505. doi:10.1002/advs.202003505
Thietart, S., and Rautou, P.-E. (2020). Extracellular Vesicles as Biomarkers in Liver Diseases: A Clinician's point of View. J. Hepatol. 73 (6), 1507–1525. doi:10.1016/j.jhep.2020.07.014
Tian, Y.-F., Ning, C.-F., He, F., Yin, B.-C., and Ye, B.-C. (2018). Highly Sensitive Detection of Exosomes by SERS Using Gold nanostar@Raman Reporter@nanoshell Structures Modified with a Bivalent Cholesterol-Labeled DNA Anchor. Analyst 143 (20), 4915–4922. doi:10.1039/c8an01041b
Tong, L., Zhu, T., and Liu, Z. (2011). Approaching the Electromagnetic Mechanism of Surface-Enhanced Raman Scattering: from Self-Assembled Arrays to Individual Gold Nanoparticles. Chem. Soc. Rev. 40 (3), 1296–1304. doi:10.1039/c001054p
Trams, E. G., Lauter, C. J., Norman Salem, J., and Heine, U. (1981). Exfoliation of Membrane Ecto-Enzymes in the Form of Micro-vesicles. Biochim. Biophys. Acta (Bba) - Biomembranes 645 (1), 63–70. doi:10.1016/0005-2736(81)90512-5
Wang, H.-L., You, E.-M., Panneerselvam, R., Ding, S.-Y., and Tian, Z.-Q. (2021b). Advances of Surface-Enhanced Raman and IR Spectroscopies: from Nano/microstructures to Macro-Optical Design. Light Sci. Appl. 10 (1), 161. doi:10.1038/s41377-021-00599-2
Wang, H., Liu, Y., Rao, G., Wang, Y., Du, X., Hu, A., et al. (2021a). Coupling Enhancement Mechanisms, Materials, and Strategies for Surface-Enhanced Raman Scattering Devices. Analyst 146 (16), 5008–5032. doi:10.1039/d1an00624j
Wang, Q., Liu, J., Zeng, J., Yang, Z., Ran, F., Wu, L., et al. (2021c). Determination of miRNA Derived from Exosomes of Prostate Cancer via Toehold-Aided Cyclic Amplification Combined with HRP Enzyme Catalysis and Magnetic Nanoparticles. Anal. Biochem. 630, 114336. doi:10.1016/j.ab.2021.114336
Wang, Z., Meng, F., and Zhong, Z. (2021d). Emerging Targeted Drug Delivery Strategies toward Ovarian Cancer. Adv. Drug Deliv. Rev. 178, 113969. doi:10.1016/j.addr.2021.113969
Wang, Z., Zong, S., Wang, Y., Li, N., Li, L., Lu, J., et al. (2018). Screening and Multiple Detection of Cancer Exosomes Using an SERS-Based Method. Nanoscale 10 (19), 9053–9062. doi:10.1039/c7nr09162a
Worrede, A., Douglass, S. M., and Weeraratna, A. T. (2021). The Dark Side of Daylight: Photoaging and the Tumor Microenvironment in Melanoma Progression. J. Clin. Invest. 131 (6), e143763. doi:10.1172/JCI143763
Wu, P., Zhang, B., Ocansey, D. K. W., Xu, W., and Qian, H. (2021). Extracellular Vesicles: A Bright star of Nanomedicine. Biomaterials 269, 120467. doi:10.1016/j.biomaterials.2020.120467
Xi, X., and Liang, C. (2021). Perspective of Future SERS Clinical Application Based on Current Status of Raman Spectroscopy Clinical Trials. Front. Chem. 9, 665841. doi:10.3389/fchem.2021.665841
Xin, L., Zhou, L. Q., Liu, C., Zeng, F., Yuan, Y. W., Zhou, Q., et al. (2021). Transfer of LncRNA CRNDE in TAM-Derived Exosomes Is Linked with Cisplatin Resistance in Gastric Cancer. EMBO Rep. 22, e52124. doi:10.15252/embr.202052124
Xu, H., Zheng, L., Zhou, Y., and Ye, B.-C. (2021). An Artificial Enzyme cascade Amplification Strategy for Highly Sensitive and Specific Detection of Breast Cancer-Derived Exosomes. Analyst 146 (18), 5542–5549. doi:10.1039/d1an01071a
Xu, Z., Zeng, S., Gong, Z., and Yan, Y. (2020). Exosome-based Immunotherapy: a Promising Approach for Cancer Treatment. Mol. Cancer 19 (1), 160. doi:10.1186/s12943-020-01278-3
Yamada, Y. (2021). Nucleic Acid Drugs-Current Status, Issues, and Expectations for Exosomes. Cancers 13 (19), 5002. doi:10.3390/cancers13195002
Yan, W., and Jiang, S. (2020). Immune Cell-Derived Exosomes in the Cancer-Immunity Cycle. Trends Cancer 6 (6), 506–517. doi:10.1016/j.trecan.2020.02.013
Yan, Z., Dutta, S., Liu, Z., Yu, X., Mesgarzadeh, N., Ji, F., et al. (2019). A Label-free Platform for Identification of Exosomes from Different Sources. ACS Sens. 4 (2), 488–497. doi:10.1021/acssensors.8b01564
Yu, W., Hurley, J., Roberts, D., Chakrabortty, S. K., Enderle, D., Noerholm, M., et al. (2021a). Exosome-based Liquid Biopsies in Cancer: Opportunities and Challenges. Ann. Oncol. 32 (4), 466–477. doi:10.1016/j.annonc.2021.01.074
Yu, Y., Guo, Q., Jiang, W., Zhang, H., and Cai, C. (2021b). Dual-Aptamer-Assisted and Logic Gate for Cyclic Enzymatic Signal Amplification Electrochemical Detection of Tumor-Derived Small Extracellular Vesicles. Anal. Chem. 93 (32), 11298–11304. doi:10.1021/acs.analchem.1c02489
Yu, Z., Lin, S., Xia, F., Liu, Y., Zhang, D., Wang, F., et al. (2021c). ExoSD Chips for High-Purity Immunomagnetic Separation and High-Sensitivity Detection of Gastric Cancer Cell-Derived Exosomes. Biosens. Bioelectron. 194, 113594. doi:10.1016/j.bios.2021.113594
Zhan, C., Chen, X.-J., Huang, Y.-F., Wu, D.-Y., and Tian, Z.-Q. (2019). Plasmon-Mediated Chemical Reactions on Nanostructures Unveiled by Surface-Enhanced Raman Spectroscopy. Acc. Chem. Res. 52 (10), 2784–2792. doi:10.1021/acs.accounts.9b00280
Zhang, D., Liang, P., Chen, W., Tang, Z., Li, C., Xiao, K., et al. (2021a). Rapid Field Trace Detection of Pesticide Residue in Food Based on Surface-Enhanced Raman Spectroscopy. Microchim Acta 188 (11), 370. doi:10.1007/s00604-021-05025-3
Zhang, T., Ma, S., Lv, J., Wang, X., Afewerky, H. K., Li, H., et al. (2021b). The Emerging Role of Exosomes in Alzheimer's Disease. Ageing Res. Rev. 68, 101321. doi:10.1016/j.arr.2021.101321
Zhang, W., Tian, Z., Yang, S., Rich, J., Zhao, S., Klingeborn, M., et al. (2021c). Electrochemical Micro-aptasensors for Exosome Detection Based on Hybridization Chain Reaction Amplification. Microsyst Nanoeng 7, 63. doi:10.1038/s41378-021-00293-8
Zhang, X., Liu, C., Pei, Y., Song, W., and Zhang, S. (2019a). Preparation of a Novel Raman Probe and its Application in the Detection of Circulating Tumor Cells and Exosomes. ACS Appl. Mater. Inter. 11 (32), 28671–28680. doi:10.1021/acsami.9b09465
Zhang, X., Zhang, X., Luo, C., Liu, Z., Chen, Y., Dong, S., et al. (2019b). Volume-Enhanced Raman Scattering Detection of Viruses. Small 15 (11), 1805516. doi:10.1002/smll.201805516
Zhao, J., Wang, Z., Lan, J., Khan, I., Ye, X., Wan, J., et al. (2021a). Recent Advances and Perspectives in Photo-Induced Enhanced Raman Spectroscopy. Nanoscale 13 (19), 8707–8721. doi:10.1039/d1nr01255j
Zhao, W., Zhang, L., Ye, Y., Li, Y., Luan, X., Liu, J., et al. (2021b). Microsphere Mediated Exosome Isolation and Ultra-sensitive Detection on a Dielectrophoresis Integrated Microfluidic Device. Analyst 146 (19), 5962–5972. doi:10.1039/d1an01061a
Zhou, B., Xu, K., Zheng, X., Chen, T., Wang, J., Song, Y., et al. (2020). Application of Exosomes as Liquid Biopsy in Clinical Diagnosis. Sig Transduct Target. Ther. 5 (1), 144. doi:10.1038/s41392-020-00258-9
Keywords: surface-enhanced Raman spectroscopy (SERS), exosome, early cancer diagnosis, nanoparticles, cancer
Citation: Yang L, Jia J and Li S (2022) Advances in the Application of Exosomes Identification Using Surface-Enhanced Raman Spectroscopy for the Early Detection of Cancers. Front. Bioeng. Biotechnol. 9:808933. doi: 10.3389/fbioe.2021.808933
Received: 04 November 2021; Accepted: 17 December 2021;
Published: 11 January 2022.
Edited by:
Tingting Zhao, Anhui Medical University, ChinaReviewed by:
Kar Wey Yong, University of Alberta, CanadaCopyright © 2022 Yang, Jia and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingyuan Jia, amlhamluZ3l1YW5AZGx1dC5lZHUuY24=; Shenglong Li, bGlzaGVuZ2xvbmdAY2FuY2VyaG9zcC1sbi1jbXUuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.