
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 14 February 2022
Sec. Industrial Biotechnology
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.802031
This article is part of the Research Topic Applied ‘Model’ Organisms in Industrial Biotechnology View all 10 articles
 Sunday Bulus Peter
Sunday Bulus Peter Zhina Qiao
Zhina Qiao Hero Nmeri Godspower
Hero Nmeri Godspower Samaila Boyi Ajeje
Samaila Boyi Ajeje Meijuan Xu
Meijuan Xu Xian Zhang
Xian Zhang Taowei Yang
Taowei Yang Zhiming Rao*
Zhiming Rao*Lactic acid bacteria represent a worthwhile organism within the microbial consortium for the food sector, health, and biotechnological applications. They tend to offer high stability to environmental conditions, with an indicated increase in product yield, alongside their moderate antimicrobial activity. Lack of endotoxins and inclusion bodies, extracellular secretion, and surface display with other unique properties, are all winning attributes of these Gram-positive lactic acid bacteria, of which, Pediococcus is progressively becoming an attractive and promising host, as the next-generation probiotic comparable with other well-known model systems. Here, we presented the biotechnological developments in Pediococcal bacteriocin expression system, contemporary variegated models of Pediococcus and lactic acid bacteria strains as microbial cell factory, most recent applications as possible live delivery vector for use as therapeutics, as well as upsurging challenges and future perspective. With the radical introduction of artificial intelligence and neural network in Synthetic Biology, the microbial usage of lactic acid bacteria as an alternative eco-friendly strain, with safe use properties compared with the already known conventional strains is expected to see an increase in various food and biotechnological applications in years to come as it offers better hope of safety, accuracy, and higher efficiency.
The idea that bacteria are useful can be difficult to understand. Today, however, some are known as friendly, good, or healthy bacteria that are widely used in foods, beverages, and supplements that help promote a healthy digestive tract and immune system. Currently, research continued to support these findings and suggest even more benefits.
Among these bacteria, is lactic acid bacteria (LAB), made up of different sets of Gram-positive, non-pathogenic bacteria that are diverse and taxonomically heterogeneous (George et al., 2018). Generally, they are catalase negative and occur as cocci or rod shaped, although pseudocatalase can be seen in rare cases. The genera Pediococcus, Streptococcus, and certain species of Lactobacillus carry out homolactic fermentation. As observed in Lactobacillus species, Pediococcus can also create a number of enzymes that releases unique food flavor compounds. Heterolactic fermentation on the other hand is performed by bacteria of the genus Leuconostoc and some Lactobacillus species (González-Centeno et al., 2017). The Lactobacillus and Pediococcus genera are found within the family Lactobacillaceae, made up of 80 identified species and 15 subspecies that are uniquely different on the basis of the outcome of genotyping. So far, the current classification is based on 16S rRNA (ribosomal RNA) and 23S rRNA sequence comparative analysis, for phylogenetic relationships. This catalogs and sequences reveal that the evolutionary lineage is within the clostridial class for Pediococcus, with a low G + C ratio less than 50% compared with other related Lactobacillus species, ranging from 84% to 87%. Their cell wall is made up of the amino acid sequence Lys-d-Asp, a peptidoglycan or diaminopimelic acid in some cases (Raccach, 2014). The common sources of these genera are plants, cheese, and processed meats, with industrial relevance as starter culture.
The health-promoting and disease-preventing function of LAB in immune modulation, gut epithelial barrier integrity maintenance, antioxidative capacity, with their important safe record of application in food processing, has warranted the generally recognized as safe (GRAS) status for consumption. In accordance with the Qualified Presumption of Safety (QPS) recommendation, Lactobacillus, Leuconostoc, Bifidobacterium, Streptococcus, and Pediococcus genera have been considered safe based on taxonomy and appropriate identification at species level (Ricci et al., 2018). In order to properly convey the expected health benefits, the incorporation of beneficial organisms, prebiotics, or symbiotics into food products is important for the gastrointestinal tract (GIT) microbiota and of great technological concern. The right dosage of probiotic, therefore, is required during transit, as it passes through the GIT. In protecting probiotics, enteric coating, microencapsulation, and prebiotic usage are directed toward stimulating the proliferation and activity of the required bacteria, by initiating low-redox potential environment and other factors that address their sensitivity to metabolites formed in the course of growth or as co-adjuvant with other starter culture (Davani-Davari et al., 2019). The generation of off-flavors in food are posed by some metabolites; however, mixed culture with inducer strains, might result in growth-enhancing metabolite production or oxygen content reduction by adjuvant starter culture (Kongo and Malcata, 2016). Withal, the molecular processes attributed to these probiotic bacteria in exerting their health-enhancing benefits is relatively unknown; therefore, precise molecular processes of probiotics action are required.
As the gastrointestinal tract remains the pathway that allows the passage of food and removal of solid waste from the body, it is also understood that the gut microflora that occupies it plays a unique function in various facet of preserving human health, including promoting nutrient digestion and absorption, defending against pathogens, and modulating the immune system. A variety of disorders, like inflammatory bowel disease, asthma, obesity, cardiovascular and autoimmune conditions, are associated with gut microbiota imbalance (Bull and Plummer, 2014). Recently, with the growing understanding of the human microbiome, to prevent and cure some health conditions, probiotics have received considerable attention in bioengineering to support human health. Helped by the extended creation of tools and methods, probiotics are engineered as live biotherapeutics for studying and prevention of many diseases (Mazhar et al., 2020). By stabilizing the gastrointestinal tract, it might offer health-promoting benefits.
Genomic proves for adaptations to the GIT via cell-surface anchoring proteins associated with the intestinal barrier causes these bacteria to induce an increase in polysaccharides diversity that are directed toward degradation and the expression of host genes innate immunity. Analogy with completely sequenced LAB genomes shows that the primary impetus of evolution within these genomes is lateral gene transfer. Genomic analysis to a great extent has unravel the core mechanisms by which “Probiogenome” encodes key functions that regulates the growth, survival, signaling, fermentative mechanism, and relatively, the ultimate potential of probiotic activities within multifaceted microbial and host biota (Goh and Klaenhammer, 2015). The evolutionary distance between distant phyla might have an effect on the array of gene availability or its absence in a given set of genomes. Therefore, engineering, in order to maintain a favorable balance in the intestinal microbiota, is essential for a better health status.
Despite LAB therapeutic prowess, these microbes could also be employed to manufacture substances that are currently unavailable, owing to a number of favorable characteristics (Bosma et al., 2017), resulting in the introduction of novel products and microbially produced compounds with the capability of replacing existing products. This might allow for an alternative and cost-effective manufacturing methods in sustainable bulk biochemical production (Bosma et al., 2013; Budzianowski and Postawa, 2016; Budzianowski, 2017), with applications in food, medicine, and pharmaceutical industries. LAB has been utilized in the production of low-calorie polyols including high value products like mannitol and sorbitol produced naturally by several heterofermentative and homofermentative LAB as well (Bozell and Petersen, 2010). Fructose is employed as a carbon source as well as an electron acceptor in certain LAB, where it is transformed to mannitol by mannitol dehydrogenase (Saha and Racine, 2011). Mannitol phosphate dehydrogenase and lactate dehydrogenase (LDH) enzymes were inactivated in Lactobacillus plantarum for sorbitol synthesis, and a sorbitol dehydrogenase gene was overexpressed, allowing more fructose 6-phosphate to be converted to sorbitol 6-phosphate (Nissen et al., 2005; Ladero et al., 2007). In addition, there has also been interest in producing xylitol from LAB, a low-calorie sugar alcohol with five carbons that is not formed naturally by LAB. In a Lactococcus lactis strain modified with xylose reductase from Pichia stiptis and a xylose transporter from Lactobacillus brevis, and cultured in a glucose-limited fed-batch cultivation method with high xylose concentrations, xylitol synthesis was observed (Nyyssölä et al., 2005). Lactate may be converted to 1,2-propanediol (1,2-PDO) by Lactobacillus buchneri, Lactobacillus brevis, and Lactobacillus fermentum, a polyol commonly utilized in anti-freeze fluids, polyester resins, non-ionic detergents, coolants, and as an addition in cosmetics, nutritional goods, medicines, and colors (Bennett and San, 2001; Oude Elferink et al., 2001; Bosma et al., 2017).
Lactic acid as a globally integrated bio-product, in the manufacture of biodegradable polymers, replacement of oil-based plastics and building-block molecule in the chemical industry (Singhvi et al., 2019), is of great importance, especially in the usage of LAB as new field of application in medicine. New possibilities are posed by genetically modified LAB (Figure 1), which could establish a strong trend in few years.
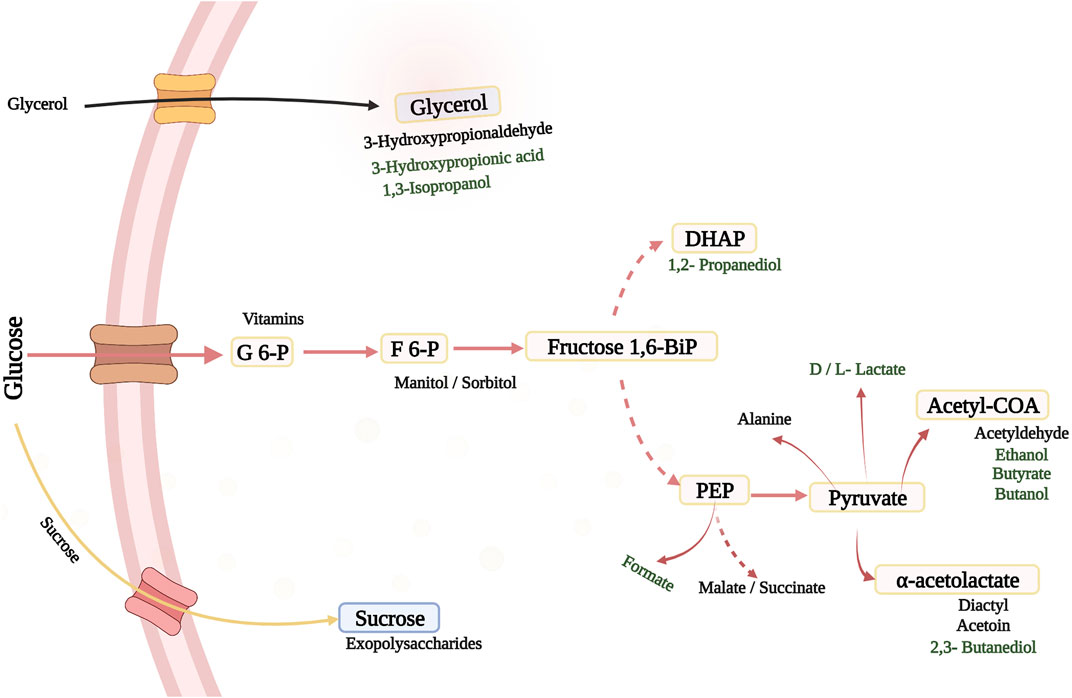
FIGURE 1. Possible Products in the Natural Metabolic Pathways of LAB and Pediococci under Various Conditions. The dashed arrows indicate different enzymatic steps. Black: taste, texture, medicinal ingredients and health uses; Green: chemicals and fuels. F 6-P, fructose 6-phosphate; DHAP, dihydroxyacetone phosphate; Fructose 1,6BiP, fructose 1,6 bisphosphate; PEP, phosphoenolpyruvate; G 6-P, Glucose-6-Phosphate.
Plasmids are broadly found in many LAB sources, as a self-replicating material and have gained much attention due to their proximity with a lot of essential functions and industrial importance (O’Connor et al., 2007). Pediococcus pentosaceus and Pediococcus acidilactici of Pediococcus species harbor various plasmids, from 1.82 to 190 kb size that encodes a variety of traits. Some plasmids from Pediococcus includes pEOC01, pMD136, pRS4 (A cryptic plasmid), pUCL287 (A cryptic plasmid), and pSMB 74. All plasmids mentioned, with the exception of pRS4, exhibit theta replicating mechanism.
Plasmids enable direct distribution of multiple copies of recombinant genes and the degree of expression is decided by promoter properties. Inducible promoters are more preferred as they allow efficient control over the expression of recombinant protein than constitutive promoters (Table 1). In the expression of genome-encoded proteins, integration of genome offers lower copy number of gene. However, genome integration brings various support-like stability, acceptability for various species, and void of surrounding effects (Zhu et al., 2015). Introducing classical methods are not often correlated with probiotic effectiveness and safe use. Nevertheless, the use of genome-based approaches for characterization of promising probiotic strains like LAB will help validate the role of these species and might stand as a useful instrument in the regulatory scenario for LAB, allowing them to acquire new attributes and increase in their beneficial characteristics. For human applications, genetic modifications may be used, if antibiotic resistance is eliminated and these bacteria are altered in their genetic code. This could be accomplished by genetic integration or chromosomal deletion, use of food-grade vectors, and clustered regularly interspaced short palindromic repeats (CRISPR) technology (Bravo and Landete, 2017; Castro-López et al., 2021) (Figure 2A).
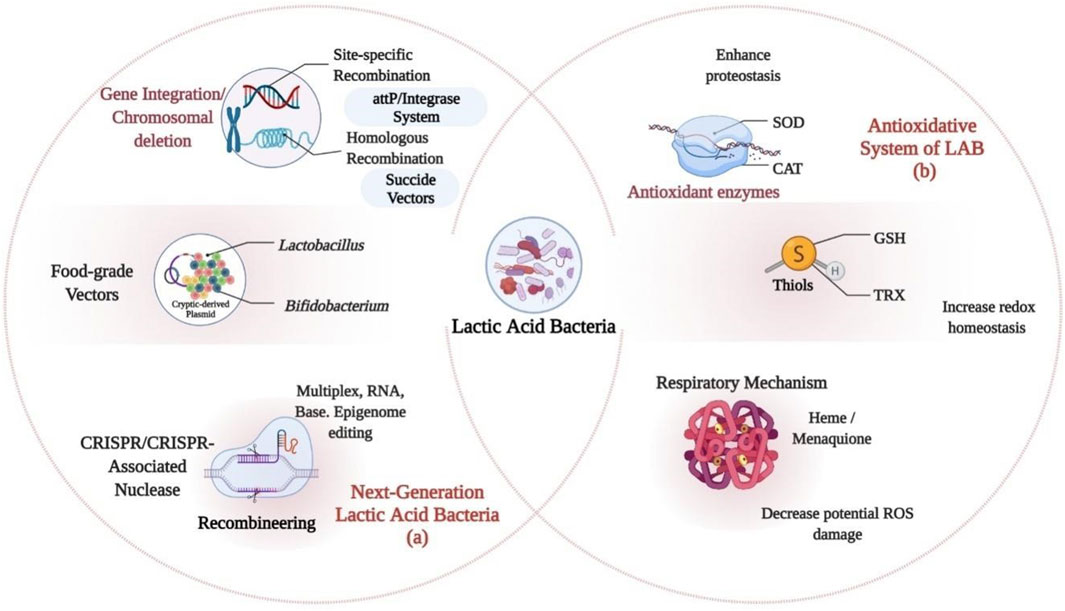
FIGURE 2. Genomic Approaches and Antioxidative System of Lactic Acid Bacteria (LAB): (A) Genome-based approaches to the characterization of promising probiotic strains. (B) Antioxidative system of LAB; Thiols, Antioxidant enzymes and Respiratory repertoire which allows them to withstand various environmental stresses that generates highly reactive species or through the NOX (NADH oxidase) like H2O2. ROS, Reactive oxygen species; SOD, Superoxide dismutase; GSH, Glutathione; TRX, Thioredoxin-thioredoxin reductase system; CAT, catalase.
These will provide a broad range of opportunities for commercially important Lactobacillus and Bifidobacteria to be engineered by genetically modifying and improving their therapeutic potential for vaccination or host immune response modulation. Gene editing tools such as transcription activator-like effector nucleases (TALEN), zinc-finger nucleases (ZFNs), CRISPR, and various approaches to omics, and systems biology could allow better comprehension and restructuring of metabolic pathways. These methods will strengthen the awareness of researchers toward the gut microbiome and provide novel routes for probiotic bacteria to be studied (Yadav et al., 2018). CRISPR, as a revolutionary genome editing technology has provided a key bioengineering tool for investigating different diseases with great sensitivity, speed, precision, and a comprehension of biological processes (Yadav et al., 2021). However, an extensive strategy to diversifying the genome engineering toolbox is to discover new CRISPR systems. Recently, developments in detection of specific CRISPR systems and the restructuring of the Cas9 protein for extended functionalities have been made possible. This could improve and expand the efficiency of the Cas9 nuclease.
Novel techniques have been introduced recently that seems to overcome the time-consuming and relatively inefficient procedures of insertion by classical chromosomal integration techniques. Recombineering allows precision genome engineering, while CRISPR Cas-based engineering helps in putrefying selection for both recombineering and homologous recombination (Table 2; Figure 2) (Doudna and Charpentier, 2014; Ortiz-Velez and Britton, 2017), which help circumvent the need for mutation screening.
In LAB and starter cultures of industrial concern, this technology grasps a better assurance for application. Its DNA-encoded, RNA-mediated, and DNA-targeting protective roles offer sequence-specific targeting of DNA (Rath et al., 2015), by producing alteration at the excision site using different pathways, that results in the initiation of double-stranded DNA breaks. These technologies will continue to remodel total bacterial exploitation and the editing of eukaryotic genomes. In CRISPR-based technologies for probiotic organisms. Application of (ss)DNA recombineering merged with CRISPR-Cas selection for gene editing of bacteria has an immense value in the production of industrial bacteria of the next century (Barrangou and van Pijkeren, 2016). CRISPR–Cas systems are extensively found in most LAB with eight subtypes and can be present in bacterial genomes alone or in multiple occurrences. These helps enhance the efficacy of fermentation, allows phage resistance or provide basic perception into starter cultures. Problems, particularly discharge and the release of antibiotic resistance markers, can arise from their engineering. Nevertheless, this may be alleviated by biocontainment of genetically modified LAB in a closed system, use of other selection markers and homologous DNA (Plavec and Berlec, 2020). However, unlike (Ds)DNA recombineering, in which the selection of positive mutants is marker dependent, which allows excision of the selection marker, leaving a scar on the loxP site of the modification locus, in (ss)DNA recombineering, it is uniformly avoided.
Cell metabolism rewiring occurs progressively and often at low throughput, but for efficient microbial cell factories, a combinatorial metabolic engineering method based on an orthogonal trifunctional CRISPR system (CRISPRAID) with three CRISPR proteins, a nuclease-deficient CRISPR protein fused to an activation domain (CRISPRa), nuclease-deficient mutant fused to a repression domain (CRISPRi), and finally a catalytically active CRISPR protein (CRISPRd) have been proposed for an effective metabolic and regulatory network with high performance permitting the examination of all possible gRNA combinations to build optimal cell factories (Lian et al., 2017). This has contributed to a 2.5-fold increase in β-carotene and an improved presence of endoglucanase on yeast surface. The ideal genome editing method for achieving any desired modification with minimal adverse effects is still a work in progress. Nevertheless, unlike previous methods, prime editing has implemented exogenous reverse transcriptase action to “write” DNA modifications directly to genomic DNA using a programmable RNA nickase linked to a reverse transcriptase and prime editing guide RNA (Anzalone et al., 2019). Both the genomic target and the sequence may be edited using approaches. By creating a powerful, inducible, broad-spectrum vector-based mutagenesis system that amplifies mutations above baseline and exceeds the mutation efficiencies and range of commonly used in vivo and in vitro approach, a mechanism-driven approach could have advantages over current methods as it offers improved random mutagenesis in cells with control, genomic stability, improved efficiency, and wider mutation spectra. The introduction of a system for the detection of antibiotic resistance that overcome chemical mutagens, ultraviolet light, and the mutant strain XL1Red subjected to the same conditions in E. coli could allow for the progressive evolution of T7 RNA polymerase variants with the potential to initiate transcription using the T3 promoter on both bacterial and bacteriophage-mediated laboratory evolution procedures (Badran and Liu, 2015).
Biopreservation systems have gained growing attention in recent years, causing consumers to be more conscious of the danger to health triggered by chemical preservatives in foods. Interestingly, most producers of bacteriocin are part of LAB, a category found in food with a historic record of safe utilization in the dairy industry. Since bacteriocins, either refined or released by its producing strains show no health risk issues, they offer an alternative to the use of deleterious preservatives (Silva et al., 2018). The possibility of engineering and improving their biological activity could be realized when the alteration systems in bacterial cell use in processing these proteins are known. In LAB, this is a current frontier and one with a lot of possible rewards.
A proposed universal scheme for bacteriocin alongside the original scheme of classification for lactic acid bacteria bacteriocins by Klaenhammer has been put forward, by integrating the revised scheme of Cotter and establishing linear, globular, and multi-component into the lantibiotics subgroups. Antimicrobial peptides (AMPs) with its broad multifunctional tools to fight microbial infections and modulate immune response of the host enhance or restrict the entrance of cells and harmful chemicals to the infection site, henceforth, protecting cells from serious tissue damage. Pediococcus pentosaceus zy-B, produced by bacteriocin PE-ZYB1, shows heat resistance and a wide variety of antimicrobial action with increased electrical extracellular conductivity and bactericidal degradation of L. monocytogenes cell membrane integrity (Heng and Tagg 2006; Zhang Y. et al., 2020). For pediocin usage as food additive, the most desired feature is its stability in heterogenous food system. Various bacteriocins from Pediococcus contain residues of methionine, the sulfur particle of which might be oxidized, leading to bacteriocin instability. With an emphasis on Pediocin PA-1 methionine residue, substituted Met13 residue with Ala, Leu, or Ile has proven to shield peptide from the effect of oxidation with moderate influence on the action of antimicrobials, while the replacement of Asp results in obvious fall in efficiency against indicator strains. Therefore, a relevant point in transforming Pediocin PA-1 into a valuable food additive is to increase its stability by substituting methionine with a different hydrophobic residue to conserve its function (Brumfitt et al., 2002). Continuous research can bring about pediocins with enhanced stability and improved functions. However, acetic acid, which is indirectly associated with the cell center metabolism, influencing and controlling bacteriocin production, in its undissociated state was found to freely disperse and dissociate through the hydrophobic layer of the membrane (Ge et al., 2019), influencing the development, yield, and activity of bacteriocin production.
Pediocins (Bacteriocin Class II) are made by Pediococcus acidilactic and Pediococcus pentosaceus strains with GRAS status in certain food applications, characterized by exceptional antimicrobial potency, good thermostability, wide pH range, and efficacy against spoilage and pathogenic organisms. In targeting members of the same species, bacteriocins can exhibit a narrow range of activity; meanwhile, others can show a wider activity for targeting other species and genus. Variants of bacteriocins have proven more significant activity than their native counterpart. This is made possible through DNA shuffling techniques by creating chimeric gene sequences having desirable traits. Bacteriocin like Pediocin PA-1 applies its role by disrupting the cytoplasmic membrane, causing outflow of ions and small molecules (Villalobos-Delgado et al., 2019). Nisin was, until recently, the only bacteriocin to be sold as a food biopreservative. However, a bacteriocin produced by Pediococcus species has also been commercially sold under the name Alta 2341™ with an effectiveness relatively as that of Nisin against L. monocytogenes, Staphylococcus aureus, Pseudomonas, and E. coli (Deegan et al., 2006). Pediocin A, Pediocin N5p, Pediocin L, Pediocin S, Pediocin ACCEL, Pediocin ST18, Pediocin SM-1, Pediocin pK23-2, Pediocin 05–10, Bacteriocin ST44AM, and Pediocin P are among the examples of pediocins (Papagianni and Anastasiadou, 2009). Single or co-adjuvant cultures of lactic acid bacteria producing bacteriocin help regulate adventitious flora and induce cell lysis, which improves food quality and sensory attributes. Recently, the incorporation into bioactive films and coatings of bacteriocins into food surfaces and packaging of bacteriocin-producing LAB has been made possible (Silva et al., 2018). Research findings on the efficacy of bacteriocins as bio-preservatives is remarkable, but the industry is a bit reluctant to commit itself financially to the production of commercial preparations of bacteriocins due to low production rates, costly downstream processing, and the legal problems that could occur from their production.
Effect of temperature, acid, osmotic pressure, and oxidative effects are some of the environmental conditions encountered by LAB in the gastrointestinal tract and during food processing that possess a significant effect on their biological functionalities. However, LAB is empowered with adaptation mechanisms that protectively saves them from harsh environmental stresses and gene alteration (Figure 2B) (Aakko et al., 2014). Some LAB like Lactobacillus fermentum (KGPMF28, KGPMF2) can survive at 45°C for 24 h. At high temperatures, cellular activities are disrupted with increase membrane fluidity due to degradation and loss of function of proteins and nucleic acids. Condition optimization for γ-aminobutyric acid (GABA) production by P. pentosaceus MN12 has provided the basis for the formulation of a GABA-rich fermented product. The use of heat shock proteins and enzymes has recently been applied to reduce biomolecule denaturation and degradation (Chen et al., 2017; Thuy et al., 2021). Understanding LAB adaptation mechanisms at extreme temperatures is crucial for sorting out LAB species as starter cultures and probiotics.
In single-strain fermentation, the development of most products is typically poor. However, improving bacterial cocultures suited for high yield, diversification, and sustainability for fermented products might be required. Fermentation of products made from selected species has shown increased productivity, reduced volatile hexanal off-flavor-generating compounds, and created several attractive flavor compounds (Sharma et al., 2020). In assessing the complex growth of P. pentosaceus and S. cerevisiae during fermentation, the forms and contents of esters showed an increase in co-culture with P. pentosaceus. Also, the concentration of bacteriocins yield by P. pentosaceus 147 increased from 1.92 × 104 AU/ml to 5.12 × 104 AU/ml as coadjuvant with Lactobacillus plantarum LE27 (Gong and Qi, 2020). Interestingly, in our laboratory, C. glutamicum G01 in coculture with a multimutagenized L. plantarum GB01-21 yielded a high concentration of 80.5 g/L, 2.68 g/L/h productivity of γ-aminobutyric acid (GABA) from glutamic acid using glucose from cassava via a two-stage fermentation strategy (Yang T. et al., 2015). These present exceptional strains required for microbial co-adjuvant in increasing desired product yield from a glucose-based substrate for health and food.
Lactic acid bacteria can form protective biofilms that allows its survival in harsh environment. Nevertheless, surrounding factors, like additives, can serve a major function in biofilm formation (BF), pH and addition of sugar supplements, with effect on LAB metabolism, growth, lactate output, and bacteriocin production. Research using P. pentosaceus ATCC 43200 grown at pH 5.0–6.0 in MRS medium (control) or in the same amounts after inclusion of 0.5, 1.0, and 1.5% (w/w) of sucrose and inulin. A variation in these parameters existed at pH 6.0 between the control medium and the supplemented media. At exponential level, both sucrose and inulin accelerated P. pentosaceus growth (de Souza de Azevedo et al., 2017). Recently reported as a potential threonine producer, P. pentosaceus TL-3 showed a boost in threonine output via optimized medium (Lim et al., 2019). These can help ensure a more economical growth medium for prospective large-scale application. However, the addition of certain substances (e.g., honey phenolic extract) might decrease LAB biofilm formation.
Undoubtedly, probiotics is currently becoming a great deal of focus in scientific probing and application in prevention or treatment of health conditions (Tables 3 and 4). In the pathology and development of chronic inflammation of digestive tract, the gastrointestinal microbiota is considered a critical factor, through the preservation of gut membrane integrity and regulation of the host immune system. Reduced colitis, decreased weight loss and disease activity index, and short-chain fatty acid formation are but a few unique observed features (Bian et al., 2020).
The resultant effect of exposure to highly reactive intermediates is oxidative damage to proteins, lipids, and nucleic acids. This is due to instability in the production of these intermediates and the antioxidant activity of the organism (Tan et al., 2018). Interestingly, the amount of oxidative damage increase and NAD+ decline has been linked to aging symptoms and can be at the root of a number of age-associated diseases. According to recent proof, increasing NAD+ levels might help reduce or even reverse the effects of aging, as well as retard the progression of age-related diseases. At the moment, a great deal of research has been centered on foodstuff sources, nutritional ingredients, and components that confers antioxidant result in humans and other life forms (Amaretti et al., 2013). However, the principal mechanism by which LAB relieves oxidative stress and intestinal damage is not fully comprehended. Another increasing health-related concern is the enormous increase level in heavy metal toxicants in human body. P. pentosaceus (FB145, FB181) reduced cadmium bioaccessibility by 46%, serving as an effective biosorbent to prevent cadmium harmful effect and reduce its assimilation into the human body (Le and Yang, 2019), thereby reducing toxicity to essential organs.
The capacity to degrade histamine, reduce bile salts from cholesterol, strain susceptibility to gastrointestinal diseases, adhesion to CaCO3 cells, antibiotic tolerance, and the removal of virulence genes are of importance as a unique attribute of a good probiotic strain. Lactobacillus paracasei L3C21M6 offers an increase in cholesterol and histamine-lowering potentials, absence of virulence genes, and good susceptibility to relevant antibiotics (Amaral et al., 2017; Domingos-Lopes et al., 2020). Pediococcus parvulus shows no ability to produce tyramine, histamine, or putrescine (Garai et al., 2007), suggesting that this ability may be strain specific instead of being associated to certain bacteria species. The main cause of sepsis is by lipopolysaccharide (LPS), which is linked to an increased death toll in patients. There are currently no effective therapeutic agents available to prevent patients from sepsis, which is depicted by LPS-mediated tissue damage and organ breakdown (Hu et al., 2013). Among intestinal bacteria, Escherichia coli or Salmonella spp. possesses lipopolysaccharide, which could cause inflammation of the intestines of humans. LPS, particularly its lipid A portion, is actually toxic. Using Pediococcus pentosaceus AK-23, with the capacity to bind LPS (Asami et al., 2017; Kawahara, 2021), these harmful bacteria might be managed effectively. Additionally, melanocytes, a specialized pigment-generating cell, houses pigmented particles called melanosomes, responsible for the production of melanin. Previously, a melanin synthase of melanocytes that can suppress tyrosinase, from kimchi-derived Pediococcus acidilactici PMC48, was developed and used as a medicinal component to actively fight the overproduction and accumulation of melanin that induces skin darkening and abnormalities (Sukyung et al., 2020). In anticipation, this could be of great importance as a basic material for melanin degradation in pharmaceutical products.
Probiotic candidates like Pediococcus and other well-known LAB are chosen to assess their sufficient tolerance to physiological barriers. Further research is required, however, to explain the awareness of their metabolic capacities, main adaptation characteristics, health and physiological functions. Bioengineering techniques can create characteristics that may be very rewarding, yet, they can also be detrimental, if not properly handled. Nevertheless, as the consumption pattern of minimally processed and preserved foods is growing, the use of pediocins of the food industry, as moderate antimicrobials, may provide solutions and alternatives to traditional means of preservation. Pediocin is projected to see more uses in medicinal application in the future (Bron and Kleerebezem, 2011). Understanding the mechanisms in Pediococcus and certainly other probiotic species will result to the creation of a molecular toolbox for applications in various sectors.
Probiotics are primarily delivered to humans by dairy-based products. The next likely food category appearance of this bacteria would be in non-dairy products. Methods that will provide probiotics with the requisite safety allow them to step beyond medicinal and supplemental uses into the field of food ingredients. Probiotics can be protected from environmental stresses using microcapsules and released in a regulated manner. Its absorption in the intestinal tract can also be aided by mucoadhesive polymers (Razavi et al., 2021). Nanocomposite, nano-emulsification, and nano-structuration will help open up a whole new chapter of possibilities for probiotics applications. Food with nanostructures might possess the ability to enhance flavor, texture, consistency, and increase yield in other important metabolite production. By using alginate, these can be applicable in several probiotic strains, with high survival rate besides non-encapsulated cells at low pH and moderate heat treatment. Microencapsulation techniques with gelatin, to offer a protective coat to acid-sensitive Lactobacillus and Bifidobacterium, have also been tried. Encapsulation of food additives has the potential to prolong product shelf life, enhance interaction with specific receptors (Sekhon, 2010), and function as de novo vaccines (Ravi Sankar and Dastagiri Reddy, 2010) to modulate immune responses.
Current approaches in “Probiogenomics” study have incited much interests and provided means for studying probiotic mechanisms and the identification of new strains from broad microbial sources. PROBIO and other probiotics database were put in place for these efforts. With the continuous change in sequencing and bioinformatic techniques, genomic sequence data for the species of interest could allow for improved regulation, optimization, and recognition of novel genes necessary for survival during processing and technical versatility of probiotics by allowing gene expression profiles of the strain during fermenter growth and different production steps (Ahmed, 2003; Song et al., 2021). In the future, this might help to tailor the technical properties of probiotic strains. Also, detecting quorum-sensing signal of pathogenic bacteria in GIT by release of reporter enzymes (Mao et al., 2018), generation and secretion of anti-enterococcal peptides upon sensing Enterococcus peptide pheromone with precise suppression of enterococcal growth (Borrero et al., 2015), the use of nisin-producing strains and immunity proteins in combination, to express reporter proteins in a tractable spatial manner (Kong et al., 2017), and an independent inducible degradation of marked proteins with accurate control of the abundance of specific proteins (Cameron and Collins, 2014), all these ushers probiotics and LAB to the world of synthetic engineering for therapeutic applications.
The anti-malarial medicine artemisinin took a very long time to develop; however, we could revolutionize what we can achieve with bioengineering if we can grow new cells to specification in a matter of weeks or months rather than years. Scientists are now working on a new tool that will adapt machine learning algorithms to the demands of synthetic biology in order to methodically steer progress and avoid a lengthy process (Zhang J. et al., 2020; Radivojević et al., 2020). The options are limitless. Scientists will no longer have to spend years learning about each part of a cell and what it does in order to manipulate it; instead, using a small set of training data, the algorithms will be able to predict how changes in the DNA of a cell or biochemistry will affect its behavior, then make recommendations for the next engineering phase, as well as probabilistic predictions for achieving the desired goal.
One of such development is Unified Representation (UniRep), a versatile summary of fundamental protein features that applies deep learning to unlabeled amino acid sequences, in order to distill the essential properties of a protein into a statistical representation. This tends to integrate unpredictable protein sequences into fixed-length vectors that resemble the capabilities, function, and configuration of critical proteins in the absence of structural or evolutionary histories. UniRep pushes protein computing to move straight from sequence to design by learning a novel base from scratch (Alley et al., 2019). Surprisingly, the UniRep might be used to rapidly generalize distant and invisible sections, revamp protein engineering roadmaps, or, at best, facilitate the detection of sequence variants that are not accessible to conventional procedures. Although it is limited by sampling bias in the length of the training sequence data, the size and coverage of the sequence databases and specific deep learning computer hardware has improved exponentially. UniRep-guided protein design pledges to boost the rate of manufacturing biosensors, DNA and protein binders, and develop genome-editing enzymes. Experts have also sought for decades to use handcoded heuristic algorithms to prescribe the rules of chemistry to computers. Discovering retrosynthetic routes using Monte Carlo tree search and symbolic artificial intelligence (AI) might assist in delivering high-quality findings. This scheme uses an expansion rule network to help choose the most efficient retrosynthetic steps more quickly than the usual computational search approach. Almost every published answer in organic chemistry has been trained into these deep neural networks (Segler et al., 2018). This makes the target molecules simpler precursors that facilitates the development of a suitable strategy for the synthesis of small organic compounds. This could be crucial in keeping the machine a useful chemical synthesis assistant capable of solving the most pressing problems in agriculture, health, and materials science.
Soluble expression phage-assisted continuous evolution (SE PACE) is a scheme for swiftly developing proteins with high solubility in expression, applicable in many proteins to increase their soluble expression through the rapid generation of improved variants without library cloning, or to improve time-consuming methods that allows protein-driven evolutionary methods. By using a PACE compatible splitintein pIII that functions as an AND gate to join two orthogonal positive selections, SE-PACE is able to develop proteins in the presence or lack of concurrent selection for protein action and improve the expression of a wide range of proteins, along with bacterially derived proteins (MBP), genetically modified antibodies (scFv), and eukaryotic enzymes, as well as proteins with lower stability that have developed via protracted directed evolution. In conclusion, SE-PACE may provide a method for swiftly reversing the negative consequences of large-scale mutations in order to improve the usability of advanced or previously designed proteins (Wang et al., 2018).
The introduction of phage-assisted continuous evolution (PACE) of TEV protease was an early attempt at this, which involves protease generated from approximately 2,500 generations of PACE with 20 non-silent mutations that clears human IL23 at the target peptide bond and suppresses IL23-mediated immune activation when pre-mixed with IL23 in primary mouse splenocyte cultures. The specificity profile of mutation cleavage and an additional protease showed the molecular basis for some changes and supports the creation of custom-made proteases that catalytically change or terminate target proteins for biotechnological and therapeutic applications (Packer et al., 2017). This work establishes not only the ability to change the substrate specificity of a protease at various positions but also on a suitable time scale.
Lactic acid bacteria not only offer great potential as an established organic product, but as public interest in the importance of probiotic usage in food and health-related aspect rises, researchers of this millennium are increasingly introducing classical therapies into genomic approaches. While the results for recombinant bacteria and their metabolites are relatively mixed, they are anticipated in the near future. The importance of recombinant probiotics has been reported in mouse models, but some few human clinical studies is also encouraging. However, the main problem with the development and usage of probiotics and other well-established organisms lies in the selection of the strains, the appropriate dose, and the lateral transfer of genes from recombinants to other types of bacteria. The use of CRISPR Cas9 selection with LAB recombination tools, PACE, and the continuous introduction of AI-based protocols (machine learning and deep neural network) could be useful in the production of important compounds and the ability to restore health with high efficiency and precision. In summary, Pediococcus and other related LAB species remain an effective, safe, and innovative alternative for food, health, and biotechnology applications.
SP and ZR wrote the first draft of the manuscript. ZQ, HN, SA made all the necessary corrections and carried out the final editing of the manuscript. Visualization and Graphic design was by SP. MX, XZ and TY reviewed the manuscript. ZR gave the final approval for publication.
This research was funded by the National Key Research and Development Program of China (grant number 2021YFC2100900), the National Natural Science Foundation of China (grant number 32071470), and the Science and Technology Project of Xinjiang Production and Construction Corps (grant number 2019AB009).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aakko, J., Sánchez, B., Gueimonde, M., and Salminen, S. (2014). Assessment of Stress Tolerance Acquisition in the Heat-Tolerant Derivative Strains of Bifidobacterium Animalis Subsp. Lactis BB-12 and Lactobacillus Rhamnosus GG. J. Appl. Microbiol. 117, 239–248. doi:10.1111/jam.12520
Ahmed, F. E. (2003). Genetically Modified Probiotics in Foods. Trends Biotechnol. 21, 491–497. doi:10.1016/j.tibtech.2003.09.006
Alley, E. C., Khimulya, G., Biswas, S., AlQuraishi, M., and Church, G. M. (2019). Unified Rational Protein Engineering with Sequence-Only Deep Representation Learning. Nat. Methods 16, 589333. doi:10.1038/s41592-019-0598-1
Amaral, D. M. F., Silva, L. F., Casarotti, S. N., Nascimento, L. C. S., and Penna, A. L. B. (2017). Enterococcus Faecium and Enterococcus Durans Isolated from Cheese: Survival in the Presence of Medications under Simulated Gastrointestinal Conditions and Adhesion Properties. J. Dairy Sci. 100, 933–949. doi:10.3168/jds.2016-11513
Amaretti, A., di Nunzio, M., Pompei, A., Raimondi, S., Rossi, M., and Bordoni, A. (2013). Antioxidant Properties of Potentially Probiotic Bacteria: In Vitro and In Vivo Activities. Appl. Microbiol. Biotechnol. 97, 809–817. doi:10.1007/s00253-012-4241-7
Asami, K., Kondo, A., Suda, Y., Shimoyamada, M., and Kanauchi, M. (2017). Neutralization of Lipopolysaccharide by Heat Shock Protein inPediococcus pentosaceusAK-23. J. Food Sci. 82, 1657–1663. doi:10.1111/1750-3841.13679
Badran, A. H., and Liu, D. R. (2015). Development of Potent In Vivo Mutagenesis Plasmids with Broad Mutational Spectra. Nat. Commun. 6, 8425. doi:10.1038/ncomms9425
Barrangou, R., and van Pijkeren, J.-P. (2016). Exploiting CRISPR-Cas Immune Systems for Genome Editing in Bacteria. Curr. Opin. Biotechnol. 37, 61–68. doi:10.1016/j.copbio.2015.10.003
Benbouziane, B., Ribelles, P., Aubry, C., Martin, R., Kharrat, P., Riazi, A., et al. (2013). Development of a Stress-Inducible Controlled Expression (SICE) System in Lactococcus Lactis for the Production and Delivery of Therapeutic Molecules at Mucosal Surfaces. J. Biotechnol. 168, 120–129. doi:10.1016/j.jbiotec.2013.04.019
Bennett, G. N., and San, K.-Y. (2001). Microbial Formation, Biotechnological Production and Applications of 1,2-propanediol. Appl. Microbiol. Biotechnol. 55, 1–9. doi:10.1007/s002530000476
Berlec, A., Škrlec, K., Kocjan, J., Olenic, M., and Štrukelj, B. (2018). Single Plasmid Systems for Inducible Dual Protein Expression and for CRISPR-Cas9/CRISPRi Gene Regulation in Lactic Acid Bacterium Lactococcus Lactis. Sci. Rep. 8, 1009. doi:10.1038/s41598-018-19402-1
Bermúdez-Humarán, L. G., Cortes-Perez, N. G., Lefèvre, F., Guimarães, V., Rabot, S., Alcocer-Gonzalez, J. M., et al. (2005). A Novel Mucosal Vaccine Based on Live Lactococci Expressing E7 Antigen and IL-12 Induces Systemic and Mucosal Immune Responses and Protects Mice against Human Papillomavirus Type 16-induced Tumors. J. Immunol. 175, 7297–7302. doi:10.4049/jimmunol.175.11.7297
Bermúdez-Humarán, L. G., Langella, P., Cortes-Perez, N. G., Gruss, A., Tamez-Guerra, R. S., Oliveira, S. C., et al. (2003). Intranasal Immunization with Recombinant Lactococcus Lactis Secreting Murine Interleukin-12 Enhances Antigen-specific Th1 Cytokine Production. Infect. Immun. 71, 1887–1896. doi:10.1128/IAI.71.4.1887-1896.2003
Bian, X., Yang, L., Wu, W., Lv, L., Jiang, X., Wang, Q., et al. (2020). Pediococcus Pentosaceus LI05 Alleviates DSS‐induced Colitis by Modulating Immunological Profiles, the Gut Microbiota and Short‐chain Fatty Acid Levels in a Mouse Model. Microb. Biotechnol. 13, 1228–1244. doi:10.1111/1751-7915.13583
Borrero, J., Chen, Y., Dunny, G. M., and Kaznessis, Y. N. (2015). Modified Lactic Acid Bacteria Detect and Inhibit Multiresistant Enterococci. ACS Synth. Biol. 4, 299–306. doi:10.1021/sb500090b
Bosma, E. F., Forster, J., and Nielsen, A. T. (2017). Lactobacilli and Pediococci as Versatile Cell Factories - Evaluation of Strain Properties and Genetic Tools. Biotechnol. Adv. 35, 419–442. doi:10.1016/j.biotechadv.2017.04.002
Bozell, J. J., and Petersen, G. R. (2010). Technology Development for the Production of Biobased Products from Biorefinery Carbohydrates-The US Department of Energy's "Top 10" Revisited. Green. Chem. 12, 539–554. doi:10.1039/b922014c
Bravo, D., and Landete, J. M. (2017). Genetic Engineering as a Powerful Tool to Improve Probiotic Strains. Biotechnol. Genet. Eng. Rev. 33, 173–189. doi:10.1080/02648725.2017.1408257
Bron, P. A., and Kleerebezem, M. (2011). Engineering Lactic Acid Bacteria for Increased Industrial Functionality. Bioengineered Bugs 2, 80–87. doi:10.4161/bbug.2.2.13910
Brumfitt, W., Salton, M. R. J., and Hamilton-Miller, J. M. T. (2002). Nisin, Alone and Combined with Peptidoglycan-Modulating Antibiotics: Activity against Methicillin-Resistant Staphylococcus aureus and Vancomycin-Resistant Enterococci. J. Antimicrob. Chemother. 50, 731–734. doi:10.1093/jac/dkf190
Budzianowski, W. M. (2017). High-value Low-Volume Bioproducts Coupled to Bioenergies with Potential to Enhance Business Development of Sustainable Biorefineries. Renew. Sustainable Energ. Rev. 70, 793–804. doi:10.1016/j.rser.2016.11.260
Budzianowski, W. M., and Postawa, K. (2016). Total Chain Integration of Sustainable Biorefinery Systems. Appl. Energ. 184, 1432–1446. doi:10.1016/j.apenergy.2016.06.050
Bull, M. J., and Plummer, N. T. (2014). Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. (Encinitas) 13, 17–22. Available at: https://pubmed.ncbi.nlm.nih.gov/26770121.
Cameron, D. E., and Collins, J. J. (2014). Tunable Protein Degradation in Bacteria. Nat. Biotechnol. 32, 1276–1281. doi:10.1038/nbt.3053
Castro-López, C., García, H. S., Guadalupe Martínez-Ávila, G. C., González-Córdova, A. F., Vallejo-Cordoba, B., and Hernández-Mendoza, A. (2021). Genomics-based Approaches to Identify and Predict the Health-Promoting and Safety Activities of Promising Probiotic Strains - A Probiogenomics Review. Trends Food Sci. Technology 108, 148–163. doi:10.1016/j.tifs.2020.12.017
Chahuki, F. F., Aminzadeh, S., Jafarian, V., Tabandeh, F., and Khodabandeh, M. (2019). Hyaluronic Acid Production Enhancement via Genetically Modification and Culture Medium Optimization in Lactobacillus Acidophilus. Int. J. Biol. Macromolecules 121, 870–881. doi:10.1016/j.ijbiomac.2018.10.112
Chen, M.-J., Tang, H.-Y., and Chiang, M.-L. (2017). Effects of Heat, Cold, Acid and Bile Salt Adaptations on the Stress Tolerance and Protein Expression of Kefir-Isolated Probiotic Lactobacillus Kefiranofaciens M1. Food Microbiol. 66, 20–27. doi:10.1016/j.fm.2017.03.020
Daniel, C., Repa, A., Wild, C., Pollak, A., Pot, B., Breiteneder, H., et al. (2006). Modulation of Allergic Immune Responses by Mucosal Application of Recombinant Lactic Acid Bacteria Producing the Major Birch Pollen Allergen Bet V 1. Allergy 61, 812–819. doi:10.1111/j.1398-9995.2006.01071.x
Davani-Davari, D., Negahdaripour, M., Karimzadeh, I., Seifan, M., Mohkam, M., Masoumi, S., et al. (2019). Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods 8, 92. doi:10.3390/foods8030092
de Souza de Azevedo, P. O., Converti, A., Domínguez, J. M., and de Souza Oliveira, R. P. (2017). Stimulating Effects of Sucrose and Inulin on Growth, Lactate, and Bacteriocin Productions by Pediococcus Pentosaceus. Probiotics Antimicro. Prot. 9, 466–472. doi:10.1007/s12602-017-9292-8
Deegan, L. H., Cotter, P. D., Hill, C., and Ross, P. (2006). Bacteriocins: Biological Tools for Bio-Preservation and Shelf-Life Extension. Int. Dairy J. 16, 1058–1071. doi:10.1016/j.idairyj.2005.10.026
Domingos‐Lopes, M. F. P., Stanton, C., Ross, R. P., and Silva, C. C. G. (2020). Histamine and Cholesterol Lowering Abilities of Lactic Acid Bacteria Isolated from Artisanal Pico Cheese. J. Appl. Microbiol. 129, 1428–1440. doi:10.1111/jam.14733
Doudna, J. A., and Charpentier, E. (2014). The New Frontier of Genome Engineering with CRISPR-Cas9. Science 346, 1258096. doi:10.1126/science.1258096
Dubey, V., Mishra, A. K., Ghosh, A. R., and Mandal, B. K. (2019). Probiotic Pediococcus Pentosaceus GS 4 Shields brush Border Membrane and Alleviates Liver Toxicity Imposed by Chronic Cadmium Exposure in Swiss Albino Mice. J. Appl. Microbiol. 126, 1233–1244. doi:10.1111/jam.14195
F. Bosma, E., van der Oost, J., M. de Vos, W., and van Kranenburg, R. (2013). Sustainable Production of Bio-Based Chemicals by Extremophiles. Cbiot 2, 360–379. doi:10.2174/18722083113076660028
Feng, P., Ye, Z., Han, H., Ling, Z., Zhou, T., Zhao, S., et al. (2020). Tibet Plateau Probiotic Mitigates Chromate Toxicity in Mice by Alleviating Oxidative Stress in Gut Microbiota. Commun. Biol. 3, 242. doi:10.1038/s42003-020-0968-3
Garai, G., Dueñas, M. T., Irastorza, A., and Moreno-Arribas, M. V. (2007). Biogenic Amine Production by Lactic Acid Bacteria Isolated from Cider. Lett. Appl. Microbiol. 45, 473–478. doi:10.1111/j.1472-765X.2007.02207.x
Ge, J., Kang, J., and Ping, W. (2019). Effect of Acetic Acid on Bacteriocin Production by Gram-Positive. J. Microbiol. Biotechnol. 29, 1341–1348. doi:10.4014/jmb.1905.05060
George, F., Daniel, C., Thomas, M., Singer, E., Guilbaud, A., Tessier, F. J., et al. (2018). Occurrence and Dynamism of Lactic Acid Bacteria in Distinct Ecological Niches: A Multifaceted Functional Health Perspective. Front. Microbiol. 9, 2899. doi:10.3389/fmicb.2018.02899
Glenting, J., Poulsen, L. K., Kato, K., Madsen, S. M., Frøkiær, H., Wendt, C., et al. (2007). Production of Recombinant Peanut Allergen Ara H 2 Using Lactococcus Lactis. Microb. Cel Fact. 6, 28. doi:10.1186/1475-2859-6-28
Goh, Y. J., and Klaenhammer, T. R. (2015). Genetic Mechanisms of Prebiotic Oligosaccharide Metabolism in Probiotic Microbes. Annu. Rev. Food Sci. Technol. 6, 137–156. doi:10.1146/annurev-food-022814-015706
Gong, Y., and Qi, X. (2020). A Study Revealing Volatile Aroma Produced by Pediococcus Pentosaceus in Dough Fermentation. Food Sci. Nutr. 8, 5077–5085. doi:10.1002/fsn3.1807
González-Centeno, M. R., Chira, K., and Teissedre, P.-L. (2017). Comparison between Malolactic Fermentation Container and Barrel Toasting Effects on Phenolic, Volatile, and Sensory Profiles of Red Wines. J. Agric. Food Chem. 65, 3320–3329. doi:10.1021/acs.jafc.6b05497
Guo, T., Xin, Y., Zhang, Y., Gu, X., and Kong, J. (2019). A Rapid and Versatile Tool for Genomic Engineering in Lactococcus Lactis. Microb. Cel Fact. 18, 22. doi:10.1186/s12934-019-1075-3
Hao, L., Cheng, Y., Su, W., Wang, C., Lu, Z., Jin, M., et al. (2021). Pediococcus Pentosaceus ZJUAF-4 Relieves Oxidative Stress and Restores the Gut Microbiota in Diquat-Induced Intestinal Injury. Appl. Microbiol. Biotechnol. 105, 1657–1668. doi:10.1007/s00253-021-11111-6
Heng, N. C. K., and Tagg, J. R. (2006). What's in a Name? Class Distinction for Bacteriocins. Nat. Rev. Microbiol. 4, 160. doi:10.1038/nrmicro1273-c1
Hu, L., Sun, C., Wang, S., Su, F., and Zhang, S. (2013). Lipopolysaccharide Neutralization by a Novel Peptide Derived from Phosvitin. Int. J. Biochem. Cel Biol. 45, 2622–2631. doi:10.1016/j.biocel.2013.09.002
Kalyanasundram, J., Chia, S. L., Song, A. A.-L., Raha, A. R., Young, H. A., and Yusoff, K. (2015). Surface Display of Glycosylated Tyrosinase Related Protein-2 (TRP-2) Tumour Antigen on Lactococcus Lactis. BMC Biotechnol. 15, 113. doi:10.1186/s12896-015-0231-z
Kawahara, K. (2021). Variation, Modification and Engineering of Lipid A in Endotoxin of Gram-Negative Bacteria. Int. J. Mol. Sci. 22, 2281. doi:10.3390/ijms22052281
Kong, W., Blanchard, A. E., Liao, C., and Lu, T. (2017). Engineering Robust and Tunable Spatial Structures with Synthetic Gene Circuits. Nucleic Acids Res. 45, 1005–1014. doi:10.1093/nar/gkw1045
Kongo, J. M., and Malcata, F. X. (2016). “Acidophilus Milk” in, eds. B. Caballero, P. M. Finglas, and H. Toldrá (Oxford: Academic Press), 6–14. doi:10.1016/B978-0-12-384947-2.00002-7
Ladero, V., Ramos, A., Wiersma, A., Goffin, P., Schanck, A., Kleerebezem, M., et al. (2007). High-level Production of the Low-Calorie Sugar Sorbitol by Lactobacillus Plantarum through Metabolic Engineering. Appl. Environ. Microbiol. 73, 1864–1872. doi:10.1128/AEM.02304-06
Le, B., and Yang, S. H. (2019). Biosorption of Cadmium by Potential Probiotic Pediococcus Pentosaceus Using In Vitro Digestion Model. Biotechnol. Appl. Biochem. 66, 673–680. doi:10.1002/bab.1783
Leenay, R. T., Vento, J. M., Shah, M., Martino, M. E., Leulier, F., and Beisel, C. L. (2019). Genome Editing with CRISPR-Cas9 in Lactobacillus Plantarum Revealed that Editing Outcomes Can Vary across Strains and between Methods. Biotechnol. J. 14, 1700583. doi:10.1002/biot.201700583
Lemay, M.-L., Tremblay, D. M., and Moineau, S. (2017). Genome Engineering of Virulent Lactococcal Phages Using CRISPR-Cas9. ACS Synth. Biol. 6, 1351–1358. doi:10.1021/acssynbio.6b00388
Li, H.-S., Piao, D.-C., Jiang, T., Bok, J.-D., Cho, C.-S., Lee, Y.-S., et al. (2015). Recombinant Interleukin 6 with M Cell-Targeting Moiety Produced in Lactococcus Lactis IL1403 as a Potent Mucosal Adjuvant for Peroral Immunization. Vaccine 33, 1959–1967. doi:10.1016/j.vaccine.2015.02.061
Lian, J., HamediRad, M., Hu, S., and Zhao, H. (2017). Combinatorial Metabolic Engineering Using an Orthogonal Tri-functional CRISPR System. Nat. Commun. 8, 1688. doi:10.1038/s41467-017-01695-x
Lim, Y. H., Foo, H. L., Loh, T. C., Mohamad, R., Abdul Rahim, R., and Idrus, Z. (2019). Optimized Medium via Statistical Approach Enhanced Threonine Production by Pediococcus Pentosaceus TL-3 Isolated from Malaysian Food. Microb. Cel Fact. 18, 125. doi:10.1186/s12934-019-1173-2
Linares, D. M., Alvarez-Sieiro, P., del Rio, B., Ladero, V., Redruello, B., Martin, M. C., et al. (2015). Implementation of the Agmatine-Controlled Expression System for Inducible Gene Expression in Lactococcus Lactis. Microb. Cel Fact. 14, 208. doi:10.1186/s12934-015-0399-x
Ma, Y., Liu, J., Hou, J., Dong, Y., Lu, Y., Jin, L., et al. (2014). Oral Administration of Recombinant Lactococcus Lactis Expressing HSP65 and Tandemly Repeated P277 Reduces the Incidence of Type I Diabetes in Non-obese Diabetic Mice. PLoS One 9, e105701. doi:10.1371/journal.pone.0105701
Madsen, S. M., Arnau, J., Vrang, A., Givskov, M., and Israelsen, H. (1999). Molecular Characterization of the pH-Inducible and Growth Phase-dependent Promoter P170 of Lactococcus Lactis. Mol. Microbiol. 32, 75–87. doi:10.1046/j.1365-2958.1999.01326.x
Mao, N., Cubillos-Ruiz, A., Cameron, D. E., and Collins, J. J. (2018). Probiotic Strains Detect and Suppress Cholera in Mice. Sci. Transl. Med. 10, eaao2586. doi:10.1126/scitranslmed.aao2586
Mazhar, S. F., Afzal, M., Almatroudi, A., Munir, S., Ashfaq, U. A., Rasool, M., et al. (2020). The Prospects for the Therapeutic Implications of Genetically Engineered Probiotics. J. Food Qual. 2020, 9676452. doi:10.1155/2020/9676452
Mierau, I., and Kleerebezem, M. (2005). 10 Years of the Nisin-Controlled Gene Expression System (NICE) in Lactococcus Lactis. Appl. Microbiol. Biotechnol. 68, 705–717. doi:10.1007/s00253-005-0107-6
Miyoshi, A., Jamet, E., Commissaire, J., Renault, P., Langella, P., and Azevedo, V. (2004). A Xylose-Inducible Expression System for Lactococcus Lactis. FEMS Microbiol. Lett. 239, 205–212. doi:10.1016/j.femsle.2004.08.018
Mu, D., Montalbán-López, M., Masuda, Y., and Kuipers, O. P. (2013). Zirex: a Novel Zinc-Regulated Expression System for Lactococcus Lactis. Appl. Environ. Microbiol. 79, 4503–4508. doi:10.1128/AEM.00866-13
Neu, T., and Henrich, B. (2003). New Thermosensitive Delivery Vector and its Use to Enable Nisin-Controlled Gene Expression in Lactobacillus Gasseri. Appl. Environ. Microbiol. 69, 1377–1382. doi:10.1128/AEM.69.3.1377-1382.2003
Nissen, L., Pérez-Martínez, G., and Yebra, M. J. (2005). Sorbitol Synthesis by an Engineered Lactobacillus Casei Strain Expressing a Sorbitol-6-Phosphate Dehydrogenase Gene within the Lactose Operon. FEMS Microbiol. Lett. 249, 177–183. doi:10.1016/j.femsle.2005.06.010
Nyyssölä, A., Pihlajaniemi, A., Palva, A., von Weymarn, N., and Leisola, M. (2005). Production of Xylitol from D-Xylose by Recombinant Lactococcus Lactis. J. Biotechnol. 118, 55–66. doi:10.1016/j.jbiotec.2005.03.014
O’Connor, E. B., O’Sullivan, O., Stanton, C., Danielsen, M., Simpson, P. J., Callanan, M. J., et al. (2007). pEOC01: A Plasmid from Pediococcus Acidilactici Which Encodes an Identical Streptomycin Resistance (aadE) Gene to that Found in Campylobacter Jejuni. Plasmid 58, 115–126. doi:10.1016/j.plasmid.2007.02.002
Oh, J.-H., and van Pijkeren, J.-P. (2014). CRISPR-Cas9-assisted Recombineering in Lactobacillus Reuteri. Nucleic Acids Res. 42, e131. doi:10.1093/nar/gku623
Ohkouchi, K., Kawamoto, S., Tatsugawa, K., Yoshikawa, N., Takaoka, Y., Miyauchi, S., et al. (2012). Prophylactic Effect of Lactobacillus Oral Vaccine Expressing a Japanese Cedar Pollen Allergen. J. Biosci. Bioeng. 113, 536–541. doi:10.1016/j.jbiosc.2011.11.025
Ortiz-Velez, L., and Britton, R. (2017). Genetic Tools for the Enhancement of Probiotic Properties. Microbiol. Spectr. 5, 1. doi:10.1128/microbiolspec.BAD-0018-2016
Oude Elferink, S. J., Krooneman, J., Gottschal, J. C., Spoelstra, S. F., Faber, F., and Driehuis, F. (2001). Anaerobic Conversion of Lactic Acid to Acetic Acid and 1, 2-propanediol by Lactobacillus Buchneri. Appl. Environ. Microbiol. 67, 125–132. doi:10.1128/AEM.67.1.125-132.2001
Packer, M. S., Rees, H. A., and Liu, D. R. (2017). Phage-assisted Continuous Evolution of Proteases with Altered Substrate Specificity. Nat. Commun. 8, 956. doi:10.1038/s41467-017-01055-9
Papagianni, M., and Anastasiadou, S. (2009). Pediocins: The Bacteriocins of Pediococci. Sources, Production, Properties and Applications. Microb. Cel Fact 8, 3. doi:10.1186/1475-2859-8-3
Pinto, J. P. C., Zeyniyev, A., Karsens, H., Trip, H., Lolkema, J. S., Kuipers, O. P., et al. (2011). pSEUDO, a Genetic Integration Standard for Lactococcus Lactis. Appl. Environ. Microbiol. 77, 6687–6690. doi:10.1128/AEM.05196-11
Plavec, T. V., and Berlec, A. (2020). Safety Aspects of Genetically Modified Lactic Acid Bacteria. Microorganisms 8, 297. doi:10.3390/microorganisms8020297
Raccach, M. (2014). “Pediococcus” in, eds. C. A. Batt, and S. E. Tortorello. (Oxford: Academic Press), 1–5. doi:10.1016/B978-0-12-384730-0.00247-0
Radivojević, T., Costello, Z., Workman, K., and Garcia Martin, H. (2020). A Machine Learning Automated Recommendation Tool for Synthetic Biology. Nat. Commun. 11, 4879. doi:10.1038/s41467-020-18008-4
Rath, D., Amlinger, L., Rath, A., and Lundgren, M. (2015). The CRISPR-Cas Immune System: Biology, Mechanisms and Applications. Biochimie 117, 119–128. doi:10.1016/j.biochi.2015.03.025
Ravi Sankar, V., and Dastagiri Reddy, Y. (2010). Nanocochleate - a New Approch in Lipid Drug Delivery. Int. J. Pharm. Pharm. Sci. 2, 220–223.
Razavi, S., Janfaza, S., Tasnim, N., Gibson, D. L., and Hoorfar, M. (2021). Microencapsulating Polymers for Probiotics Delivery Systems: Preparation, Characterization, and Applications, 120, 106882. doi:10.1016/j.foodhyd.2021.106882Food Hydrocolloids
Ricci, A., Ricci, A., Allende, A., Bolton, D., Chemaly, M., Davies, R., et al. (2018). Update of the List of QPS‐recommended Biological Agents Intentionally Added to Food or Feed as Notified to EFSA 7: Suitability of Taxonomic Units Notified to EFSA until September 2017. Efs2 16, e05131. doi:10.2903/j.efsa.2018.5131
Saha, B. C., and Racine, F. M. (2011). Biotechnological Production of Mannitol and its Applications. Appl. Microbiol. Biotechnol. 89, 879–891. doi:10.1007/s00253-010-2979-3
Segler, M. H. S., Preuss, M., and Waller, M. P. (2018). Planning Chemical Syntheses with Deep Neural Networks and Symbolic AI. Nature 555, 604–610. doi:10.1038/nature25978
Sekhon, B. S. (2010). Food Nanotechnology - an Overview. Nanotechnol. Sci. Appl. 3, 1–15. doi:10.2147/NSA.S8677
Sharma, R., Garg, P., Kumar, P., Bhatia, S. K., and Kulshrestha, S. (2020). Microbial Fermentation and its Role in Quality Improvement of Fermented Foods. Ferment 6. doi:10.3390/fermentation6040106
Silva, C. C. G., Silva, S. P. M., and Ribeiro, S. C. (2018). Application of Bacteriocins and Protective Cultures in Dairy Food Preservation. Front. Microbiol. 9, 594. doi:10.3389/fmicb.2018.00594
Singhvi, M. S., Zinjarde, S. S., and Gokhale, D. V ( (2019). Polylactic Acid: Synthesis and Biomedical Applications. J. Appl. Microbiol. 127, 1612–1626. doi:10.1111/jam.14290
Song, D., Ibrahim, S., and Hayek, S. (2021). Recent Application of Probiotics in Food and Agricultural Science. doi:10.5772/50121
Song, X., Huang, H., Xiong, Z., Ai, L., and Yang, S. (2017). CRISPR-Cas9(D10A) Nickase-Assisted Genome Editing in Lactobacillus Casei. Appl. Environ. Microbiol. 83, e01259. doi:10.1128/AEM.01259-17
Steidler, L., Hans, W., Schotte, L., Neirynck, S., Obermeier, F., Falk, W., et al. (2000). Treatment of Murine Colitis by Lactococcus Lactis Secreting Interleukin-10. Science 289, 1352–1355. doi:10.1126/science.289.5483.1352
Storaker, M. I., Kamilla, W., Zhian, S., Sigve, H. L., Daniel, S., Geir, M., et al. (2021). CRISPR Interference for Rapid Knockdown of Essential Cell Cycle Genes in Lactobacillus Plantarum. mSphere 4, e00007–19. doi:10.1128/mSphere.00007-19
Tan, B. L., Norhaizan, M. E., Liew, W.-P.-P., and Sulaiman Rahman, H. (2018). Antioxidant and Oxidative Stress: A Mutual Interplay in Age-Related Diseases. Front. Pharmacol. 9, 1162. doi:10.3389/fphar.2018.01162
Thuy, D. T. B., Nguyen, A., Khoo, K. S., Chew, K. W., Cnockaert, M., Vandamme, P., et al. (2021). Optimization of Culture Conditions for Gamma-Aminobutyric Acid Production by Newly Identified Pediococcus Pentosaceus MN12 Isolated from “Mam Nem”. A Fermented Fish Sauce. Bioengineered 12, 54–62. doi:10.1080/21655979.2020.1857626
van der Els, S., James, J. K., Kleerebezem, M., and Bron, P. A. (2021). Versatile Cas9-Driven Subpopulation Selection Toolbox for Lactococcus Lactis. Appl. Environ. Microbiol. 84, e02752–17. doi:10.1128/AEM.02752-17
van Pijkeren, J.-P., and Britton, R. A. (2012). High Efficiency Recombineering in Lactic Acid Bacteria. Nucleic Acids Res. 40, e76. doi:10.1093/nar/gks147
Villalobos-Delgado, L. H., Nevárez-Moorillon, G. V., Caro, I., Quinto, E. J., and Mateo, J. (2019). 4 - Natural Antimicrobial Agents to Improve Foods Shelf Life. Food Qual. Shelf Life 2019, 125–157. doi:10.1016/B978-0-12-817190-5.00004-5
Walker, D. C., and Klaenhammer, T. R. (1994). Isolation of a Novel IS3 Group Insertion Element and Construction of an Integration Vector for Lactobacillus Spp. J. Bacteriol. 176, 5330–5340. doi:10.1128/jb.176.17.5330-5340.1994
Wang, T., Badran, A. H., Huang, T. P., and Liu, D. R. (2018). Continuous Directed Evolution of Proteins with Improved Soluble Expression. Nat. Chem. Biol. 14, 972–980. doi:10.1038/s41589-018-0121-5
Xin, Y., Guo, T., Mu, Y., and Kong, J. (2017). Identification and Functional Analysis of Potential Prophage-Derived Recombinases for Genome Editing in Lactobacillus Casei. FEMS Microbiol. Lett. 364, 364. doi:10.1093/femsle/fnx243
Yadav, N., Narang, J., Chhillar, A. K., and Rana, J. S. (2021). CRISPR: A New Paradigm of Theranostics. Nanomedicine Nanotechnology, Biol. Med. 33, 102350. doi:10.1016/j.nano.2020.102350
Yadav, R., Kumar, V., Baweja, M., and Shukla, P. (2018). Gene Editing and Genetic Engineering Approaches for Advanced Probiotics: A Review. Crit. Rev. Food Sci. Nutr. 58, 1735–1746. doi:10.1080/10408398.2016.1274877
Yang, P., Wang, J., and Qi, Q. (2015a). Prophage Recombinases-Mediated Genome Engineering in Lactobacillus Plantarum. Microb. Cel Fact. 14, 154. doi:10.1186/s12934-015-0344-z
Yang, T., Rao, Z., Kimani, B. G., Xu, M., Zhang, X., and Yang, S.-T. (2015b). Two-step Production of Gamma-Aminobutyric Acid from Cassava Powder Using Corynebacterium Glutamicum and Lactobacillus Plantarum. J. Ind. Microbiol. Biotechnol. 42, 1157–1165. doi:10.1007/s10295-015-1645-2
Yu, D., Sawitzke, J. A., Ellis, H., and Court, D. L. (2003). Recombineering with Overlapping Single-Stranded DNA Oligonucleotides: Testing a Recombination Intermediate. Proc. Natl. Acad. Sci. U. S. A. 100, 7207–7212. doi:10.1073/pnas.1232375100
Zhang, B., Li, A., Zuo, F., Yu, R., Zeng, Z., Ma, H., et al. (2016). Recombinant Lactococcus Lactis NZ9000 Secretes a Bioactive Kisspeptin that Inhibits Proliferation and Migration of Human colon Carcinoma HT-29 Cells. Microb. Cel Fact. 15, 102. doi:10.1186/s12934-016-0506-7
Zhang, J., Petersen, S. D., Radivojevic, T., Ramirez, A., Pérez-Manríquez, A., Abeliuk, E., et al. (2020a). Combining Mechanistic and Machine Learning Models for Predictive Engineering and Optimization of Tryptophan Metabolism. Nat. Commun. 11, 4880. doi:10.1038/s41467-020-17910-1
Zhang, Y., Yang, J., Liu, Y., Wu, Y., Fang, Z., Wang, Y., et al. (2020b). A Novel Bacteriocin PE-ZYB1 Produced by Pediococcus Pentosaceus Zy-B Isolated from Intestine of Mimachlamys Nobilis: Purification, Identification and its Anti-listerial Action. LWT 118, 108760. doi:10.1016/j.lwt.2019.108760
Zhong, Y., Wu, S., Chen, F., He, M., and Lin, J. (2019). Isolation of High γ-aminobutyric Acid-Producing Lactic Acid Bacteria and Fermentation in mulberry Leaf Powders. Exp. Ther. Med. 18, 147–153. doi:10.3892/etm.2019.7557
Keywords: probiotics, Pediococcus, biotherapeutics, bacteriocin, genome editing, deep neural network
Citation: Peter SB, Qiao Z, Godspower HN, Ajeje SB, Xu M, Zhang X, Yang T and Rao Z (2022) Biotechnological Innovations and Therapeutic Application of Pediococcus and Lactic Acid Bacteria: The Next-Generation Microorganism. Front. Bioeng. Biotechnol. 9:802031. doi: 10.3389/fbioe.2021.802031
Received: 26 October 2021; Accepted: 08 December 2021;
Published: 14 February 2022.
Edited by:
Fengjie Cui, Jiangsu University, ChinaReviewed by:
Xuetuan Wei, Huazhong Agricultural University, ChinaCopyright © 2022 Peter, Qiao, Godspower, Ajeje, Xu, Zhang, Yang and Rao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiming Rao, raozhm@jiangnan.edu.cn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.