
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 10 January 2022
Sec. Industrial Biotechnology
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.770302
This article is part of the Research Topic Advances in Protein Structure, Function, and Design View all 6 articles
In this study, a novel enzymatic approach to transform levulinic acid (LA), which can be obtained from biomass, into value-added (R)-4-aminopentanoic acid using an engineered glutamate dehydrogenase from Escherichia coli (EcGDH) was developed. Through crystal structure comparison, two residues (K116 and N348), especially residue 116, were identified to affect the substrate specificity of EcGDH. After targeted saturation mutagenesis, the mutant EcGDHK116C, which was active toward LA, was identified. Screening of the two-site combinatorial saturation mutagenesis library with EcGDHK116C as positive control, the kcat/Km of the obtained EcGDHK116Q/N348M for LA and NADPH were 42.0- and 7.9-fold higher, respectively, than that of EcGDHK116C. A molecular docking investigation was conducted to explain the catalytic activity of the mutants and stereoconfiguration of the product. Coupled with formate dehydrogenase, EcGDHK116Q/N348M was found to be able to convert 0.4 M LA by more than 97% in 11 h, generating (R)-4-aminopentanoic acid with >99% enantiomeric excess (ee). This dual-enzyme system used sustainable raw materials to synthesize (R)-4-aminopentanoic acid with high atom utilization as it utilizes cheap ammonia as the amino donor, and the inorganic carbonate is the sole by-product.
Chiral γ-amino acids have attracted increasing attention for their usefulness as building blocks in the pharmaceutical industry and peptide chemistry (Vasudev et al., 2011; Gómez et al., 2017). γ-amino acid scaffolds involved in the synthesis of the peptidomimetics increase the in vivo stability and the diversity of peptide molecules (Comegna et al., 2015). Moreover, many peptides containing γ-amino acids have biological activity (Kato et al., 1986; Pettit et al., 1987; Stratmann et al., 1994; Chen et al., 2014). (R)-4-aminopentanoic acid is a γ-amino acid with high added-value, which is an important intermediate for the synthesis of Gly-Pro-Glu-OH (GPE*) analogue—a novel class of pharmaceutical agents for the treatment of central nervous system injuries and neurodegenerative diseases including Alzheimer’s, Parkinson’s, and Huntington’s diseases (Trotter et al., 2005)—and muscarinic M4 receptor agonist (Brown et al., 2018) (Figure 1), and it can also participate in the formation of physiologically active artificial peptides (Grison et al., 2016). Therefore, the efficient synthesis of (R)-4-aminopentanoic acid has become a research hotspot.
Levulinic acid (LA) is a promising platform chemical that can be obtained from biomass (Weingarten et al., 2012; Pileidis and Titirici, 2016; Girisuta and Heeres, 2017; Habe et al., 2020; Wang et al., 2020). The synthesis of (R)-4-aminopentanoic acid by reductive amination of LA is an attractive reaction route for its sustainable characteristics. The chemical synthesis from LA to (R)-4-aminopentanoic acid suffers from poor stereoselectivity (Du et al., 2011; Wu et al., 2019; Xie et al., 2019). Compared with the chemical synthesis of (R)-4-aminopentanoic acid, enzymatic methods are environmentally friendly and highly enantioselective. (R)-selective amine transaminases can catalyze the synthesis of (R)-4-aminopentanoic acid with high optical purity (>99% ee) (Jiang et al., 2015); but the reductive amination of carbonyl compounds by transaminases requires organic amine as an amino donor and strategies for shifting the unfavorable thermodynamic equilibrium (Knaus et al., 2017). The reductive amination of LA directly by dehydrogenase to obtain the product is an ideal enzymatic reaction route for its environmentally friendly and high atom economy. Mayol et al. (2016) identified a wild-type amine dehydrogenase from Petrotoga mobilis (PmAmDH), which is capable of reductive amination of LA to (S)-4-aminopentanoic acid. Subsequently, Cai et al. (2020) engineered PmAmDH through directed evolution and obtained mutants with increased activity, thereby achieving efficient synthesis of (S)-4-aminopentanoic acid. However, there are still no relevant reports on the dehydrogenase that converts LA into (R)-4-aminopentanoic acid. Engineered amine dehydrogenases modified based on leucine and phenylalanine dehydrogenases are promising enzymes for asymmetric synthesis of (R)-chiral amines, as they can use cheap ammonia as amino donor and generate only water as byproduct (Abrahamson et al., 2012; Ye et al., 2015; Patil et al., 2018); but their substrate scopes are restricted to carbonyl compounds without a carboxyl group. Nevertheless, these findings provide motivation for engineering of glutamate dehydrogenase (GDH) for the synthesis of γ-amino acids from LA (Figure 2).
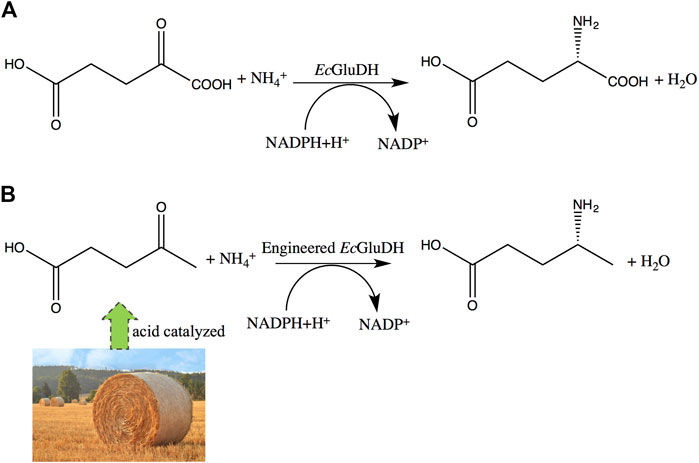
FIGURE 2. Expected changes in substrate specificity of GDH. (A) Wild-type GDH reaction. (B) engineered GDH reductive amination of LA to (R)-4-aminopentanoic acid.
In this study, a combinatorial saturation mutagenesis library on the key sites, which were determined by crystal structure analysis, of GDH from Escherichia coli (EcGDH) was constructed and screened, and the best engineered EcGDHK116Q/N348M was obtained. This engineered enzyme can use LA as substrate to efficiently synthesize (R)-4-aminopentanoic acid. Hence, this study expands the synthetic scope of amine dehydrogenases and the toolbox of enzymes involved in the synthesis of γ-amino acids.
Glutamate dehydrogenase from E. coli-K12 (EcGDH, GenBank no. CP047127.1) and formate dehydrogenase of Burkholderia stabilis (BsFDH, GenBank no. ACF35003.1) were synthesized by Sangon Biotech (Shanghai, China). The ClonExpress II One Step Cloning Kit were purchased from Vazyme Biotech Company, Ltd. (Nanjing, China). The primers were synthesized by Sangon Biotech (Shanghai, China). Luria-Bertani (LB) media was used for the growth of E. coli. PCR reagents were purchased from TaKaRa. The Plasmid Mini Kit I (100), BioSpin PCR Purification Kit, and DNA Gel Extraction Kit (100) were purchased from Omega. LA was supplied by Shanghai Macklin Biochemical Co., Ltd. (Shanghai, China). Racemic 4-aminopentanoic acid was purchased from Taizhou Runsun Chemical Co., Ltd. (Zhejiang, China). All other chemicals were analytical grade and commercially available.
In order to obtain better mutants, combinatorial saturation mutagenesis is selected to combine the sites confirmed by structural analysis. The primers used in this study are shown in Supplementary Table S1. The plasmid pET28a-EcGDH was used as the template. The screening volume of the clones reached 95% library coverage (Reetz et al., 2008).
Single colonies created by combinatorial saturation mutagenesis were picked into 96-well microtiter plates containing 1 ml of LB media with 50 μg/ml kanamycin using a QPix420 colony picker. In each plate, four wells were reserved as the controls, two of which are positive controls and the other two are negative controls. The plates were shaken at 37°C for 10 h, then shifted 50 μL of culture to 48-well microtiter plates containing 2 ml of LB media with 50 μg/ml kanamycin for an additional 2.5 h at 37°C, and finally induced with 0.15 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After 12 h of induction at 17°C, cells were harvested by centrifugation (12857 × g, 5 min, 4°C), resuspended in 200 μL lysis buffer (100 mM Tris-HCl buffer, 4 mg/ml lysozyme, pH 7.5), and disrupted by freezing at −80°C for 30 min and by heat shocking at 45°C for 3 min, and then incubated for 10 min at 37°C. After centrifugation (3,724 × g, 20 min, 4°C), the crude enzyme extracts were used for downstream enzymatic reactions.
Combinatorial saturation mutagenesis library was screened using an NADPH auto fluorescence assay (Abrahamson et al., 2012). The assay involved reading the well’s absorbance at two wavelengths, 340 and 600 nm. The decreased absorbance at 340 nm corresponds to the consumption of NADPH, while the 600 nm reading estimates the biomass present in the well. Differences over background in absorbance at 340 nm are normalized by the 600 nm absorbance readings. The change in absorbance at 340 nm for 1 minute was divided by absorbance at 600 nm. The mutants with higher absorbance than wild-type enzyme were selected. The reaction mixture (200 μL) contained 20 μL of crude enzyme extracts, 0.3 mM NADPH, 40 mM LA, 0.8 M NH4Cl and 0.1 M Tris-HCl buffer (pH 8.5).
The wild-type and mutants were chosen for purification. Cells were grown in LB medium at 37°C until the OD600 reached about 0.6, and then they were induced by 0.1 mM IPTG. The culture was collected by centrifugation and washed three times using normal saline. Then, cell breaking was performed with an ultrasonic cell disruption system (SCIENTZ-IID). After centrifugation at 12857 × g for 30 min at 4°C, the clarified supernatant was purified by Ni2+-NTA chromatography. The molecular mass of the purified GDH was examined by SDS-PAGE (Schägger, 2006).
The activity of EcGDH and its mutants was determined at 30°C by monitoring the absorbance change at 340 nm which was corresponding to the concentration variation of NADPH (Li et al., 2014). For reductive amination, the reaction mixture (200 μL) contained 80 mM LA, 0.2 mM NADPH, 0.8 M NH4Cl, Tris-HCl buffer (100 mM, pH 8.5), and 20 μL purified enzyme. One unit of enzyme activity was defined as the amount of enzyme catalyzing the reduction of 1 μmol substrate or oxidation of 1 μmol production per minute.
Kinetic parameters of the mutants were determined in Tris-HCl buffer (100 mM, pH 8.5) at 30°C with varied concentration of substrate (with concentration range from 80 to 1,600 mM) or NADPH (with concentration range from 0.05 to 0.8 mM). The Michaelis-Menten constants (Km and kcat) were calculated using the nonlinear curve in Origin 8.5.3.
To compare the reaction performances of EcGDHK116Q/N348M under different conditions (NADP+ concentration, pH and temperature), a series of experiments are carried out. In the experiment of the effect of NADP+ (with concentration range from 0.25 to 1 mM) on the reaction, 2 ml of reaction mixture contains 0.8 M NH4COOH, 100 mM Tris-HCl buffer (pH 8), 80 mM LA, 0.5 mg/ml purified mutant EcGDHK116Q/N348M, and 0.03 mg/ml purified BsFDH at 30°C. In the experiment of the effect of pH (pH 7, 8, and 9) on the reaction, 2 ml of reaction mixture contains 0.8 M NH4COOH, 100 mM Tris-HCl buffer, 80 mM LA, 0.5 mM NADP+, 0.5 mg/ml purified mutant EcGDHK116Q/N348M, and 0.03 mg/ml purified BsFDH at 30°C. In the experiment of the effect of temperature (30, 35, 40, and 45°C) on the reaction, 2 ml of reaction mixture contains 0.8 M NH4COOH, 100 mM Tris-HCl buffer, 80 mM LA, 1 mM NADP+, 0.5 mg/ml purified mutant EcGDHK116Q/N348M, and 0.03 mg/ml purified BsFDH. Under the optimal conditions (1 mM NADP+, pH 8 and 45°C), the mutant EcGDHK116Q/N348M was tested in 10 ml of reaction mixture, containing 3.2 M NH4COOH, 200 mM Tris-HCl buffer, 400 mM LA, 1.51 mg/ml purified mutant EcGDHK116Q/N348M, and 0.20 mg/ml purified BsFDH.
The structures EcGDHK116C and EcGDHK116Q/N348M were modeled using SWISS-MODEL (Schwede et al., 2003). The X-ray structure of EcGDH (PDB: 4BHT) was used as the structural template. The 3D structures of 2-ketoglutarate and LA were obtained by using ChemDraw Ultra. The docking study was processed in AutoDockTool. The wild-type GDH protein was defined as the receptor and 2-ketoglutarate or LA as the ligand, Lys126 was selected as the Flexible Residue, and a box containing Lys92, Ser380, Lys126, Lys116 and Asn348 was set. (The active sites were confirmed by multiple sequence alignments with 5IJZ, 4XGI and 1BGV) And EcGDHK116C or EcGDHK116Q/N348M protein was defined as the receptor and LA as the ligand, Lys126 was selected as the Flexible Residue, and a box containing Lys92, Ser380 and Lys126 was set. Afterward, the docking was automatically processed by running Autogrid and AutoDock in AutoDockTool. After AutoDock running, 10 ligand conformations were generated and their corresponding binding energies were calculated. The best docking model was screened among the 10 docking poses according to the ranks of the models and hydrogen bond formation determined by structure alignment. Both the structures and docking were visualized with PyMOL.
The conversion of LA was determined using HPLC with an Aminex HPX-87H column (300 × 7.8 mm). With the column temperature maintained at 30°C, mobile phase H2SO4 (5 mM) ran at a flow rate of 0.6 ml/min. (Cai et al., 2020). (R)-4-aminopentanoic acid in the reaction mixtures were labeled using 1-fluoro-2,4-dinitrophenyl-5-L-alanineamide (FDAA) and then analyzed on a Develosil ODS-UG-5 column (150 × 4.6 mm). A 10 μL sample of the amino acid, 8 μL of 1 M NaHCO3, and 40 μL of 1% (w/v) FDAA in acetone were mixed and heated for 1 h at 40°C. When the sample was cooled to room temperature, 8 μL of 1 N HCl and 934 ml of 40% (v/v) aqueous acetonitrile were added to the mixture, after which it was vortexed and filtered (0.22 mm) for HPLC (Hanson et al., 2008). The HPLC conditions were: mobile phase A 5% acetonitrile (0.05% trifluoroacetic acid, 1% methanol), mobile phase B 60% acetonitrile (0.05% trifluoroacetic acid, 1% methanol), linear gradient from 0% B to 100% B over 45 min at a flow rate 1 ml/min and detected at 340 nm, injection volume 20 μL (Bhushan and Bruckner, 2004; Zhang et al., 2019).
By querying the Protein Data Bank database, a large number of crystal structures of GDH were found, including 1BGV (Stillman et al., 1993), 4XGI, 5IJZ, 5GUD (Son et al., 2015) and 4BHT (this study). Unfortunately, there was no ligand in 4BHT. Therefore, the structure of 4BHT was compared with the other four crystal structures (enzyme–ligand complex) to identify the key residues that bind to the ligand. These structures were highly similar to 4BHT, and the root-mean-square deviation (RMSD) was between 0.904 and 2.442. As shown in Figure 3, lysine residues formed hydrogen bonds with the main chain carboxyl group of the ligand in four crystal structures, and additional asparagine residues of 5GUD and 4XGI formed hydrogen bonds with the main chain carboxyl group of the ligand. Therefore, two sites (lysine and asparagine residues), especially lysine residue, were found to play an important role in the binding of the carboxyl group of the ligand backbone. Subsequently, these protein sequences were aligned (Sievers et al., 2011) (Robert and Gouet, 2014), further revealing that these two sites interacted with the main chain carboxyl group are conserved within these enzymes (Figure 4). Therefore, K116 and N348 residues of EcGDH were selected as important sites for targeted mutation.
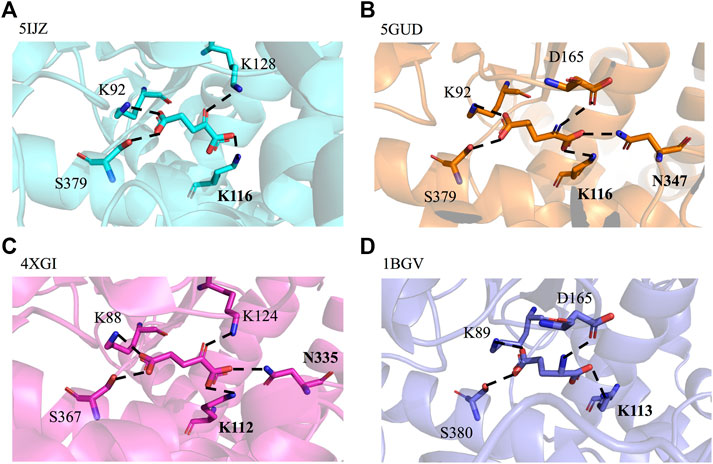
FIGURE 3. GDH active sites with bound ligand. (A) GDH from Corynebacterium glutamicum (CgGDH, PDB: 5IJZ), (B) GDH from Corynebacterium glutamicum (CgGDH, PDB: 5GUD), (C) GDH from Burkholderia thailandensis (BtGDH, PDB: 4XGI), and (D) GDH from Clostridium symbiosum (CsGDH, PDB: 1BGV). (the boldface indicates the residues that interact with the main chain carboxyl group of the ligand).
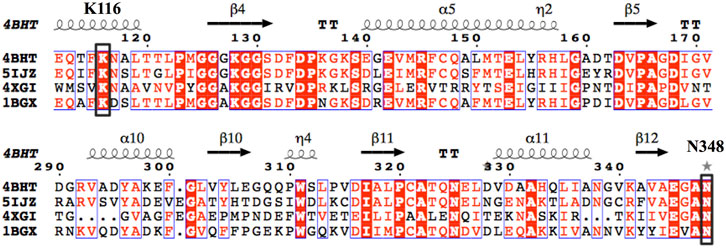
FIGURE 4. Multiple-sequence alignment of the four selected GDHs as the template. (The sequences were aligned using ClustalO and were presented using ESPript3.0. The two conserved amino acid sites K116 and N348 (numbering according to EcGDH) were indicated by black box.).
To investigate the effect of K116 on substrate specificity, this residue was mutated into the other 19 amino acids to determine the specific activity toward LA. As shown in Supplementary Table S2, seven mutants (A, C, M, Q, R, S, and T) were found to be active to LA, of which EcGDHK116C had the highest activity (10.1 mU/mg). To obtain mutants with higher activity toward LA, a combinatorial saturation mutagenesis library of residues K116 and N348 was constructed, and mutant EcGDHK116C was used as positive control for screening. To shorten the screening process, NNK degenerate codons were not selected for the saturation mutagenesis. Instead, based on 20 plasmids with different codons at site 116, residue 348 was mutated by four degenerate codons of AHN, TKB, CAT, and CCA encoding all 20 amino acids without introducing redundancy; in each case, 60 transformants were screened for 95% library coverage (Patrick and Firth, 2005; Tang et al., 2012). Thus, the screening amount of transformants could be downscaled from 3,066 to 1,200. (Supplementary Figure S1) Finally, the best mutant EcGDHK116Q/N348M was obtained, and its specific activity (108.6 mU/mg) was determined to be 10.8-times higher than that of EcGDHK116C.
To further explore the role of the EcGDH mutants in the acceptance of LA, the wild-type and mutants (K116C, K116Q/N348M) structure models were used for docking analysis with LA. Figure 5A shows the docking results for wild-type EcGDH and 2-ketoglutarate. The main chain carboxyl group of the ligand was found to interact with K116 and N348, and the carbonyl group of the ligand formed a hydrogen bond with the key catalytic residue K126 (with a distance of 2.7 Å). As shown in Figure 5B, when the ligand was changed from 2-ketoglutarate to LA, the carboxyl group of LA interacted with K116, K126, and N348, which caused catalytically unfavorable poses. With K126 occupied by the carboxyl group, the carbonyl group of LA cannot interact with K126, a residue that is responsible for the formation of an imine intermediate based on the reported mechanism of GDH from the natural substrate (Stillman et al., 1993; Son et al., 2015). This may explain why wild-type EcGDH has no catalytic activity for LA. As shown in Figures 5C,D, after the K116 was modified, the LA showed similar pose to that of 2-ketoglutarate in EcGDH, with a distance of 2.8 Å between the substrate carbonyl-O atom and K126 owing to a hydrogen bond. Therefore, EcGDHK116C and EcGDHK116Q/N348M have catalytic activity toward LA.
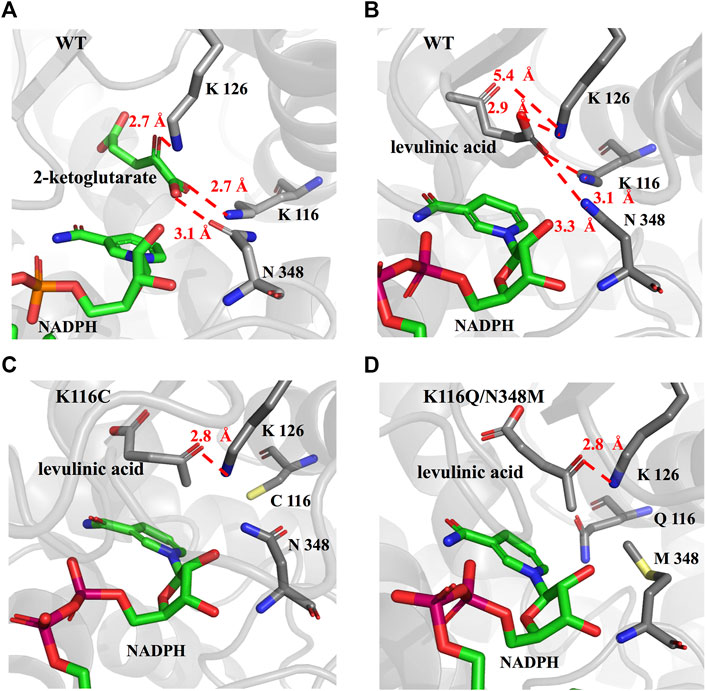
FIGURE 5. Enzyme-substrate binding pose analysis. (A) Enzyme-substrate binding pose for wild type EcGDH with 2-ketoglutarate. (B) Enzyme-substrate binding pose for wild type EcGDH with LA. (C) Enzyme-substrate binding pose for EcGDHK116C with LA. (D) Enzyme-substrate binding pose for EcGDHK116Q/N348M with LA. The distances (in angstrom) are represented by the red dashed lines.
As shown in Figure 6, the HPLC analysis of the reaction solution catalyzed by EcGDHK116Q/N348M revealed that the reaction product was (R)-4-aminopentanoic acid, which is different from the product configuration catalyzed by the natural amine dehydrogenase (Mayol et al., 2016). To clarify the reasons for the configuration of this specific product, the docking results and catalytic mechanism of GDH were analyzed. As shown in Figure 7A, two residues (K92 and S380 of the wild-type or EcGDHK116Q/N348M) formed hydrogen bones with the γ-carboxyl group of 2-ketoglutarate or the carboxyl group of LA, and the substrate carbonyl group was stabilized by the side chain of K126 before a hydride was supplied by the NADPH to the carbonyl carbon atom (Tomita et al., 2017). Moreover, the coenzyme NADPH attacked the 2-ketoglutarate from the re face (Figure 7B). However, when the substrate changed from 2-ketoglutarate to LA, the NADPH attacked the LA from the si face, which determines the stereochemistry of the product (R)-4-aminopentanoic acid (Figure 7C). These observations are consistent with the HPLC analysis of the reaction solution. The natural amine dehydrogenase was obtained from the NCBI database using the protein sequence of (2R,4S)-2,4-diaminopentanoate dehydrogenase from Clostridium sticklandii (2,4-DAPDH, EC 1.4.1.12), which can catalyze (2R)-2-amino-4-oxopentanoate to (2R,4S)-2,4-diaminopentanoate, as a template (Fukuyama et al., 2014; Mayol et al., 2016). From the perspective of the catalytic mechanism, 2,4-DAPDH and EcGDH (EC 1.4.1.4) have opposite coenzyme offensive surfaces, which results in different product stereoselectivities (Xue et al., 2018).
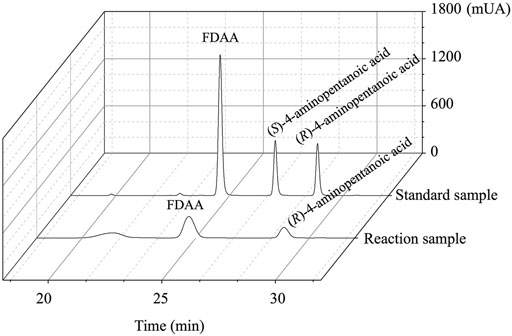
FIGURE 6. The HPLC spectrum from FDAA derivatization of the product synthesized by EcGDHK116Q/N348M and commercial racemic 4-aminopentanoic acid.
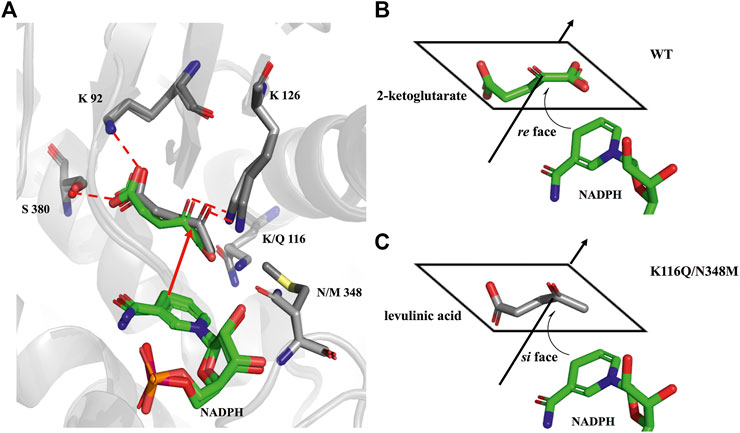
FIGURE 7. Comparison of the docking results. (A) Structural alignment analysis of docking pose for 2-ketoglutarate in the catalytic pocket of wild-type and LA in the catalytic pocket of EcGDHK116Q/N348M. (B) schematic representation of NADPH attacking 2-ketoglutarate. (C) schematic representation of NADPH attacking LA.
To compare the catalytic efficiency of the EcGDHK116C and the best mutant EcGDHK116Q/N348M, kinetic parameters for LA and NADPH were determined (Table 1). The kcat/Km of EcGDHK116Q/N348M for LA and NADPH were 42.0- and 7.9-fold higher, respectively, than that of EcGDHK116C. And EcGDHK116Q/N348M had a higher affinity and kcat for LA than EcGDHK116C. This may be because N348 has a negative effect on the binding of the enzyme and LA, thereby reducing the probability of LA to form the correct pose in the pocket. However, EcGDHK116C had higher coenzyme affinity than the EcGDHK116Q/N348M. According to the crystal structure of 5ijz, N347 (corresponding to N348 in EcGDH) can form hydrogen bonds with the coenzyme and stabilize it (Son et al., 2015). Therefore, the mutation of EcGDHK116Q/N348M at site 348 makes its affinity for NADPH weaker than that of EcGDHK116C. The Km and kcat of EcGDHK116Q/N348M to LA were very different from those of the wild-type GDH for 2-ketoglutarate (Sharkey and Engel, 2009), which indicates that the two-point mutations at residues 116 and 348 weaken the enzyme-substrate interaction, leading to a decrease in kcat and affinity.
The substrate specificity of the wild-type and mutants (K116C, and K116Q/N348M) were profiled by activity assays over a group of structurally diverse carbonyl compounds. The mutants displayed no activity toward fatty ketones (2-butanone, 2-pentanone, and 4-methyl-2-pentanone), aromatic ketones (acetophenone, phenylacetone, and phenylbutanone) and alcohol ketones (4-hydroxy-2-butanone, and 5-hydroxy-2-pentanone). Therefore, γ-carbonyl acid and its derivatives, which are more similar to the natural substrate 2-ketoglutarate, were used as substrates in this analysis. As shown in Table 2, the activity of EcGDHK116Q/N348M on natural substrates was lower than that of the wild-type, but the substrate spectrum of EcGDHK116Q/N348M was expanded compared with that of the wild-type and EcGDHK116C. Since the activity of EcGDHK116Q/N348M on non-natural substrates was generally low, further evolution of this engineered enzyme is required to improve its activity.
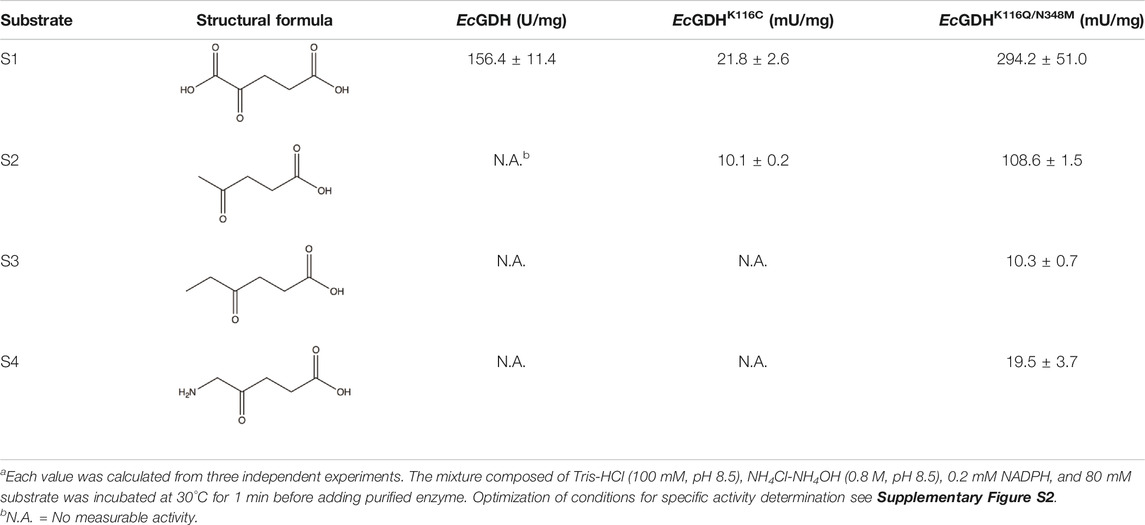
TABLE 2. Activity of wild type, EcGDHK116C and EcGDHK116Q/N348M towards various γ-carbonyl acid and its derivativesa.
Subsequently, the reductive amination reactions of LA by EcGDHK116Q/N348M coupled with BsFDH for regeneration of NADPH were performed under different conditions (different NADP+ concentration, pH and temperature). As shown in Figures 8A,C, as the concentration of the coenzyme and the temperature increased, the conversion efficiency also increased, and the reaction showed the best conversion efficiency at pH 8 (Figure 8B). Under optimal conditions (1 mM NADP+, pH 8 and 45°C), EcGDHK116Q/N348M converted 0.4 M LA to (R)-4-aminopentanoic acid (>99% ee) by more than 97% in 11 h with the addition of 1.51 mg/ml EcGDH and 0.20 mg/ml BsFDH (Figure 8D). Comparisons with other studies (Table 3) revealed that the approach for (R)-4-aminopentanoic acid production in this study had significant advantages in reaction conditions, conversion efficiency and product optical purity.
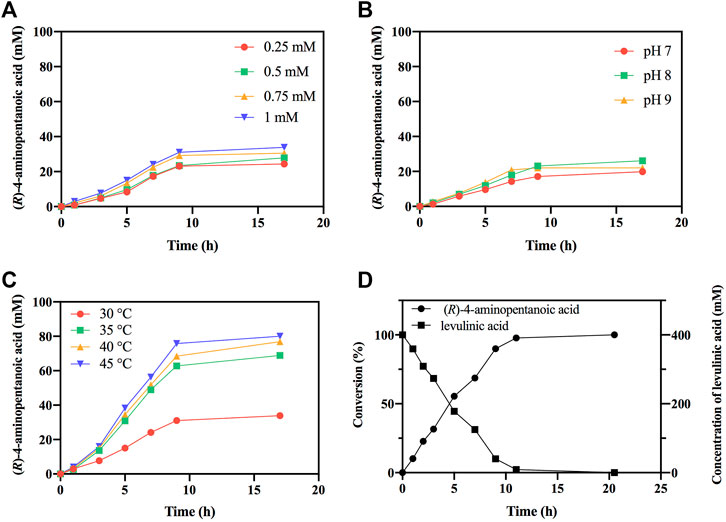
FIGURE 8. Optimization of reaction conditions. (A) The effect of NADP+ concentration on reductive amination reaction by EcGDHK116Q/N348M. (B) The effect of pH on reductive amination reaction by EcGDHK116Q/N348M. (C) The effect of temperature on reductive amination reaction by EcGDHK116Q/N348M. (D) Time courses of reductive amination of LA by EcGDHK116Q/N348M under the optimal conditions. Conversions determined by HPLC analysis.
The previous research in engineered amine dehydrogenases gained from leucine dehydrogenase or phenylalanine dehydrogenase (Abrahamson et al., 2012; Abrahamson et al., 2013; Ye et al., 2015; Chen et al., 2018) greatly solves the problem of the lack of natural amine dehydrogenase (Mayol et al., 2016; Bommarius, 2019). However, the substrate specificity of the engineered amine dehydrogenase is limited to the synthesis of aliphatic and aromatic chiral amines. (R)-selective amine transaminases can catalyze the synthesis of (R)-4-aminopentanoic acid with high optical purity; however, the amination catalysed by (R)-selective amine transaminases, using (R)-1-methylbenzylamine as an amine donor, requires removal of the co-product acetophenone to shift the unfavorable thermodynamic equilibrium. In this study, the engineered GDH provides a new way for the synthesis of γ-amino acids and enriches the toolbox of amine dehydrogenases. Coupled with BsFDH, EcGDHK116Q/N348M converted 0.4 M LA to (R)-4-aminopentanoic acid by more than 97% in 11 h with excellent stereoselectivity (>99% ee). Moreover, the reaction system can use cheap ammonia as amino donor and generate only inorganic carbonate as byproduct, and the substrate LA in the reaction is one of the top 12 carbohydrate-derived compounds listed by Department of Energy United States that can be obtained from the lignocellulosic biomass (Weingarten et al., 2012; Pileidis and Titirici, 2016; Girisuta and Heeres, 2017; Habe et al., 2020; Wang et al., 2020); the product (R)-4-aminopentanoic acid is an important intermediate for the synthesis of psychotropic drugs (Trotter et al., 2005) and muscarinic M4 receptor agonist (Brown et al., 2018), and it can also participate in the formation of physiologically active artificial peptides (Grison et al., 2016). Taken together, these results indicated that this pathway can sustainably synthesize high value-added (R)-4-aminopentanoic acid.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
FZ: Conceptualization, Methodology, Investigation, Data curation, Writing-original draft. XM: Supervision, Validation, Writing-review and editing, Funding acquisition. YX: Investigation, Data curation. YN: Resources, Writing-review and editing, Funding acquisition, Project administration.
This work was financially supported by grants from the National Key Research and Development Program of China (No. 2021YFC2100100), The National Natural Science Foundation of China (NSFC) (Nos 21336009 and 21176103), The Postgraduate Research and Practice Innovation Program of Jiangsu Province (No. KYCX19_1831), The 111 Project (No. 111-2-06), the High-end Foreign Experts Recruitment Program (No. G20190010083), The National Program for Support of Top-notch Young Professionals, The Fundamental Research Funds for the Central Universities (No. JUSRP51504), The Program for the Key Laboratory of Enzymes of Suqian (No. M201803), and the National First-Class Discipline Program of Light Industry Technology and Engineering (No. LITE 2018-09).
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Editage (www.editage.cn) for English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.770302/full#supplementary-material
Abrahamson, M. J., Vázquez-Figueroa, E., Woodall, N. B., Moore, J. C., and Bommarius, A. S. (2012). Development of an Amine Dehydrogenase for Synthesis of Chiral Amines. Angew. Chem. Int. Ed. 51, 3969–3972. doi:10.1002/anie.201107813
Abrahamson, M. J., Wong, J. W., and Bommarius, A. S. (2013). The Evolution of an Amine Dehydrogenase Biocatalyst for the Asymmetric Production of Chiral Amines. Adv. Synth. Catal. 355, 1780–1786. doi:10.1002/adsc.201201030
Bhushan, R., and Bruckner, H. (2004). Marfey?s Reagent for Chiral Amino Acid Analysis: A Review. Amino Acids 27, 231–247. doi:10.1007/s00726-004-0118-0
Bommarius, A. S. (2019). Amine Dehydrogenases Occur in Nature. Nat. Catal. 2, 288–289. doi:10.1038/s41929-019-0270-2
Brown, G. A., Congreve, M. S., Pickworth, M., Rackham, M., and Tehan, B. G. (2018). Muscarinic Agonists. U.S. Patent No US 20180228791 A1 U.S. Patent and Trademark Office Washington, DC.
Cai, R.-F., Liu, L., Chen, F.-F., Li, A., Xu, J.-H., and Zheng, G.-W. (2020). Reductive Amination of Biobased Levulinic Acid to Unnatural Chiral γ-Amino Acid Using an Engineered Amine Dehydrogenase. ACS Sustain. Chem. Eng. 8, 17054–17061. doi:10.1021/acssuschemeng.0c04647
Chen, F.-F., Zheng, G.-W., Liu, L., Li, H., Chen, Q., Li, F.-L., et al. (2018). Reshaping the Active Pocket of Amine Dehydrogenases for Asymmetric Synthesis of Bulky Aliphatic Amines. ACS Catal. 8, 2622–2628. doi:10.1021/acscatal.7b04135
Chen, Q.-Y., Liu, Y., Cai, W., and Luesch, H. (2014). Improved Total Synthesis and Biological Evaluation of Potent Apratoxin S4 Based Anticancer Agents with Differential Stability and Further Enhanced Activity. J. Med. Chem. 57, 3011–3029. doi:10.1021/jm4019965
Comegna, D., de Paola, I., Saviano, M., Del Gatto, A., and Zaccaro, L. (2015). Straightforward Entry to S-Glycosylated Fmoc-Amino Acids and Their Application to Solid Phase Synthesis of Glycopeptides and Glycopeptidomimetics. Org. Lett. 17, 640–643. doi:10.1021/ol503664t
Du, X.-L., He, L., Zhao, S., Liu, Y.-M., Cao, Y., He, H.-Y., et al. (2011). Hydrogen-Independent Reductive Transformation of Carbohydrate Biomass into γ-Valerolactone and Pyrrolidone Derivatives with Supported Gold Catalysts. Angew. Chem. Int. Ed. 50, 7815–7819. doi:10.1002/anie.201100102
Fukuyama, S., Mihara, H., Miyake, R., Ueda, M., Esaki, N., and Kurihara, T. (2014). Characterization of a Thermostable 2,4-diaminopentanoate Dehydrogenase from Fervidobacterium Nodosum Rt17-B1. J. Biosci. Bioeng. 117, 551–556. doi:10.1016/j.jbiosc.2013.11.002
Girisuta, B., and Heeres, H. J. (2017). “Levulinic Acid from Biomass: Synthesis and Applications,” in “Levulinic Acid from Biomass: Synthesis and Applications,” in Production Of Platform Chemicals from Sustainable Resources. Editors Z. Fang, R. Smith, and X. Qi (Singapore: Springer), 143–169. doi:10.1007/978-981-10-4172-3_5
Gómez, J. E., Guo, W., Gaspa, S., and Kleij, A. W. (2017). Copper-Catalyzed Synthesis of γ-Amino Acids Featuring Quaternary Stereocenters. Angew. Chem. Int. Ed. 56, 15035–15038. doi:10.1002/anie.201709511
Grison, C. M., Miles, J. A., Robin, S., Wilson, A. J., and Aitken, D. J. (2016). An α-Helix-Mimicking 12,13-Helix: Designed α/β/γ-Foldamers as Selective Inhibitors of Protein-Protein Interactions. Angew. Chem. Int. Ed. 55, 11096–11100. doi:10.1002/anie.201604517
Habe, H., Sato, Y., and Kirimura, K. (2020). Microbial and Enzymatic Conversion of Levulinic Acid, an Alternative Building Block to Fermentable Sugars from Cellulosic Biomass. Appl. Microbiol. Biotechnol. 104, 7767–7775. doi:10.1007/s00253-020-10813-7
Hanson, R. L., Davis, B. L., Goldberg, S. L., Johnston, R. M., Parker, W. L., Tully, T. P., et al. (2008). Enzymatic Preparation of a D-Amino Acid from a Racemic Amino Acid or Keto Acid. Org. Process. Res. Dev. 12, 1119–1129. doi:10.1021/op800149q
Jiang, J., Chen, X., Zhang, D., Wu, Q., and Zhu, D. (2015). Characterization of (R)-selective Amine Transaminases Identified by In Silico Motif Sequence Blast. Appl. Microbiol. Biotechnol. 99, 2613–2621. doi:10.1007/s00253-014-6056-1
Kato, Y., Fusetani, N., Matsunaga, S., Hashimoto, K., Fujita, S., and Furuya, T. (1986). Bioactive marine Metabolites. Part 16. Calyculin A. A Novel Antitumor Metabolite from the marine Sponge Discodermia Calyx. J. Am. Chem. Soc. 108, 2780–2781. doi:10.1021/ja00270a061
Knaus, T., Böhmer, W., and Mutti, F. G. (2017). Amine Dehydrogenases: Efficient Biocatalysts for the Reductive Amination of Carbonyl Compounds. Green. Chem. 19, 453–463. doi:10.1039/C6GC01987K
Li, J., Pan, J., Zhang, J., and Xu, J.-H. (2014). Stereoselective Synthesis of L-Tert-Leucine by a Newly Cloned Leucine Dehydrogenase from Exiguobacterium Sibiricum. J. Mol. Catal. B: Enzymatic 105, 11–17. doi:10.1016/j.molcatb.2014.03.010
Mayol, O., David, S., Darii, E., Debard, A., Mariage, A., Pellouin, V., et al. (2016). Asymmetric Reductive Amination by a Wild-type Amine Dehydrogenase from the Thermophilic Bacteria Petrotoga Mobilis. Catal. Sci. Technol. 6, 7421–7428. doi:10.1039/C6CY01625A
Palacios, F., Aparicio, D., Garcı́a, J., Rodrı́guez, E., and Fernández-Acebes, A. (2001). A Convenient Synthesis of Racemic and Optically Active 1-Aza-1,3-Dienes Derived from γ-amino Esters: Reduction to α,β-unsaturated and Saturated γ-amino Acid Derivatives. Tetrahedron 57, 3131–3141. doi:10.1016/S0040-4020(01)00171-5
Patil, M. D., Grogan, G., Bommarius, A., and Yun, H. (2018). Oxidoreductase-catalyzed Synthesis of Chiral Amines. ACS Catal. 8, 10985–11015. doi:10.1021/acscatal.8b02924
Patrick, W. M., and Firth, A. E. (2005). Strategies and Computational Tools for Improving Randomized Protein Libraries. Biomol. Eng. 22, 105–112. doi:10.1016/j.bioeng.2005.06.001
Pettit, G. R., Kamano, Y., Herald, C. L., Tuinman, A. A., Boettner, F. E., Kizu, H., et al. (1987). The Isolation and Structure of a Remarkable marine Animal Antineoplastic Constituent: Dolastatin 10. J. Am. Chem. Soc. 109, 6883–6885. doi:10.1021/ja00256a070
Pileidis, F. D., and Titirici, M.-M. (2016). Levulinic Acid Biorefineries: New Challenges for Efficient Utilization of Biomass. ChemSusChem 9, 562–582. doi:10.1002/cssc.201501405
Reetz, M. T., Kahakeaw, D., and Lohmer, R. (2008). Addressing the Numbers Problem in Directed Evolution. Chembiochem 9, 1797–1804. doi:10.1002/cbic.200800298
Robert, X., and Gouet, P. (2014). Deciphering Key Features in Protein Structures with the New ENDscript Server. Nucleic Acids Res. 42, W320–W324. doi:10.1093/nar/gku316
Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003). SWISS-MODEL: An Automated Protein Homology-Modeling Server. Nucleic Acids Res. 31, 3381–3385. doi:10.1093/nar/gkg520
Sharkey, M. A., and Engel, P. C. (2009). Modular Coenzyme Specificity: a Domain-Swopped Chimera of Glutamate Dehydrogenase. Proteins 77, 268–278. doi:10.1002/prot.22433
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, Scalable Generation of High‐quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 7, 539. doi:10.1038/msb.2011.75
Son, H. F., Kim, I.-K., and Kim, K.-J. (2015). Structural Insights into Domain Movement and Cofactor Specificity of Glutamate Dehydrogenase from Corynebacterium Glutamicum. Biochem. Biophysical Res. Commun. 459, 387–392. doi:10.1016/j.bbrc.2015.02.109
Stillman, T. J., Baker, P. J., Britton, K. L., and Rice, D. W. (1993). Conformational Flexibility in Glutamate Dehydrogenase. J. Mol. Biol. 234, 1131–1139. doi:10.1006/jmbi.1993.1665
Stratmann, K., Burgoyne, D. L., Moore, R. E., Patterson, G. M. L., and Smith, C. D. (1994). Hapalosin, a Cyanobacterial Cyclic Depsipeptide with Multidrug-Resistance Reversing Activity. J. Org. Chem. 59, 7219–7226. doi:10.1021/jo00103a011
Tang, L., Gao, H., Zhu, X., Wang, X., Zhou, M., and Jiang, R. (2012). Construction of "Small-Intelligent" Focused Mutagenesis Libraries Using Well-Designed Combinatorial Degenerate Primers. BioTechniques 52, 149–158. doi:10.2144/000113820
Tomita, T., Yin, L., Nakamura, S., Kosono, S., Kuzuyama, T., and Nishiyama, M. (2017). Crystal Structure of the 2-Iminoglutarate-Bound Complex of Glutamate Dehydrogenase fromCorynebacterium Glutamicum. FEBS Lett. 591, 1611–1622. doi:10.1002/1873-3468.12667
Trotter, N. S., Brimble, M. A., Harris, P. W. R., Callis, D. J., and Sieg, F. (2005). Synthesis and Neuroprotective Activity of Analogues of Glycyl-L-Prolyl-L-Glutamic Acid (GPE) Modified at the α-carboxylic Acid. Bioorg. Med. Chem. 13, 501–517. doi:10.1016/j.bmc.2004.10.005
Vasudev, P. G., Chatterjee, S., Shamala, N., and Balaram, P. (2011). Structural Chemistry of Peptides Containing Backbone Expanded Amino Acid Residues: Conformational Features of β, γ, and Hybrid Peptides. Chem. Rev. 111, 657–687. doi:10.1021/cr100100x
Wang, J., Cui, H., Wang, Y., Zhao, R., Xie, Y., Wang, M., et al. (2020). Efficient Catalytic Conversion of Cellulose to Levulinic Acid in the Biphasic System of Molten Salt Hydrate and Methyl Isobutyl Ketone. Green. Chem. 22, 4240–4251. doi:10.1039/D0GC00897D
Weingarten, R., Conner, W. C., and Huber, G. W. (2012). Production of Levulinic Acid from Cellulose by Hydrothermal Decomposition Combined with Aqueous Phase Dehydration with a Solid Acid Catalyst. Energy Environ. Sci. 5, 7559–7574. doi:10.1039/C2EE21593D
Wu, H., Dai, W., Saravanamurugan, S., Li, H., and Yang, S. (2019). Quasi-catalytic Approach to N-Unprotected Lactams via Transfer Hydro-Amination/cyclization of Biobased Keto Acids. ACS Sustain. Chem. Eng. 7, 10207–10213. doi:10.1021/acssuschemeng.9b00412
Xie, C., Song, J., Wu, H., Hu, Y., Liu, H., Zhang, Z., et al. (2019). Ambient Reductive Amination of Levulinic Acid to Pyrrolidones over Pt Nanocatalysts on Porous TiO2 Nanosheets. J. Am. Chem. Soc. 141, 4002–4009. doi:10.1021/jacs.8b13024
Xue, Y.-P., Cao, C.-H., and Zheng, Y.-G. (2018). Enzymatic Asymmetric Synthesis of Chiral Amino Acids. Chem. Soc. Rev. 47, 1516–1561. doi:10.1039/c7cs00253j
Ye, L. J., Toh, H. H., Yang, Y., Adams, J. P., Snajdrova, R., and Li, Z. (2015). Engineering of Amine Dehydrogenase for Asymmetric Reductive Amination of Ketone by Evolving Rhodococcus Phenylalanine Dehydrogenase. ACS Catal. 5, 1119–1122. doi:10.1021/cs501906r
Keywords: levulinic acid, reductive amination, glutamate dehydrogenase, structureguided protein engineering, (R)-4-aminopentanoic acid
Citation: Zhou F, Xu Y, Mu X and Nie Y (2022) A Sustainable Approach for Synthesizing (R)-4-Aminopentanoic Acid From Levulinic Acid Catalyzed by Structure-Guided Tailored Glutamate Dehydrogenase. Front. Bioeng. Biotechnol. 9:770302. doi: 10.3389/fbioe.2021.770302
Received: 03 September 2021; Accepted: 14 December 2021;
Published: 10 January 2022.
Edited by:
Bo Wang, Shenzhen Institutes of Advanced Technology (CAS), ChinaReviewed by:
Vytas Svedas, Lomonosov Moscow State University, RussiaCopyright © 2022 Zhou, Xu, Mu and Nie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqing Mu, eHFtdUBqaWFuZ25hbi5lZHUuY24=; Yao Nie, eW5pZUBqaWFuZ25hbi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.