
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bioeng. Biotechnol. , 11 October 2021
Sec. Industrial Biotechnology
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.765396
This article is part of the Research Topic Engineering Yeast to Produce Plant Natural Products View all 11 articles
Non-conventional yeasts have attracted a growing interest on account of their excellent characteristics. In recent years, the emerging of CRISPR/Cas technology has improved the efficiency and accuracy of genome editing. Utilizing the advantages of CRISPR/Cas in bioengineering of non-conventional yeasts, quite a few advancements have been made. Due to the diversity in their genetic background, the ways for building a functional CRISPR/Cas system of various species non-conventional yeasts were also species-specific. Herein, we have summarized the different strategies for optimizing CRISPR/Cas systems in different non-conventional yeasts and their biotechnological applications in the construction of cell factories. In addition, we have proposed some potential directions for broadening and improving the application of CRISPR/Cas technology in non-conventional yeasts.
Non-conventional yeasts have been considered as potential eukaryotic chassis for scientific research and industrial application. Owing to their outstanding natural characteristics. These advantageous attributes include thermotolerance, utilization of extensive carbon sources, and the capacity to produce high-titer proteins, lipids, or other commercial metabolites. For instance, Scheffersomyces stipitis is continually used in ethanol fermentation via lignocellulosic feedstock due to its inherent xylose metabolism (Agbogbo and Coward-Kelly, 2008). Methylotrophic yeasts like Ogataea polymorpha, Pichia pastoris and Ogataea thermomethanolica, possess an efficient ability to secrete heterologous protein and glycosylate, and were utilized commercially for producing a variety of proteins (Gellissen, 2000; Hartner and Glieder, 2006; Bredell et al., 2018). Oleaginous yeasts Rhodotorula toruloides and Yarrowia lipolytica, are capable of storing large amounts of cellular lipids from low-cost carbon sources (Castañeda et al., 2018; Chattopadhyay et al., 2021). The heat resistance of Kluyveromyces marxianus allows it to ferment at higher temperature, thus decreasing the probability of contamination (Marcišauskas et al., 2019). Furthermore, Kluyveromyces lactis finds extensive usage in lactose metabolism to secrete proteins (Spohner et al., 2016).
Genetic engineering is fundamental to study gene functions and control the expression of genes for producing specific compounds or otherwise regulating the gene expression when these yeasts are employed for scientific research or industrial applications. The efficient genome editing approaches and the corresponding tools are critical for rapid genome and metabolic engineering. The traditional gene manipulation tools such as Cre-loxP system can improve the genome editing efficiency to some extent, but these systems can only modify single locus in one step, and the marker recycling is time-consuming and left many scars in genome, which is not conducive to genomic stability (Xie et al., 2014). Moreover, the natural homologous recombination (HR) depends on a DNA break which occurs accidentally at the target locus (Raschmanová et al., 2018). In the last few decades, several new, better, and more accurate genetic tools have been developed to improve the efficiency of genome editing, such as zinc-finger nucleases (ZFNs) (Doyon et al., 2011) and transcription activator-like effector nucleases (TALENS) (Li et al., 2011). The targeting of specific DNA sequences by ZNFs and TALENs depends on the protein-DNA interaction, and then the DNA break is introduced by FokI. However, the construction of specific DNA binding proteins is still a laborious and time-consuming task.
Recently, the CRISPR/Cas system has revolutionized genome editing technology due to its efficiency, accuracy, and convenience (Hsu et al., 2014). This system essentially comprises a DNA endonuclease (e.g. Cas9 or Cpf1) which can bind to the target DNA sequence by the guidance of sgRNA, and then generating DNA double-strand breaks (DSB), and later the repair mechanism including HR, non-homologous end-joining (NHEJ), or microhomology-mediated end joining (MMEJ) is activated (Zetsche et al., 2015; Burstein et al., 2017) (Figure 1A). The CRISPR/Cas system relies on the DNA-RNA recognition for inducing precise DNA cleavage, which is marker-free and capable of simultaneous multi-loci editing, thus greatly accelerates the genome editing.
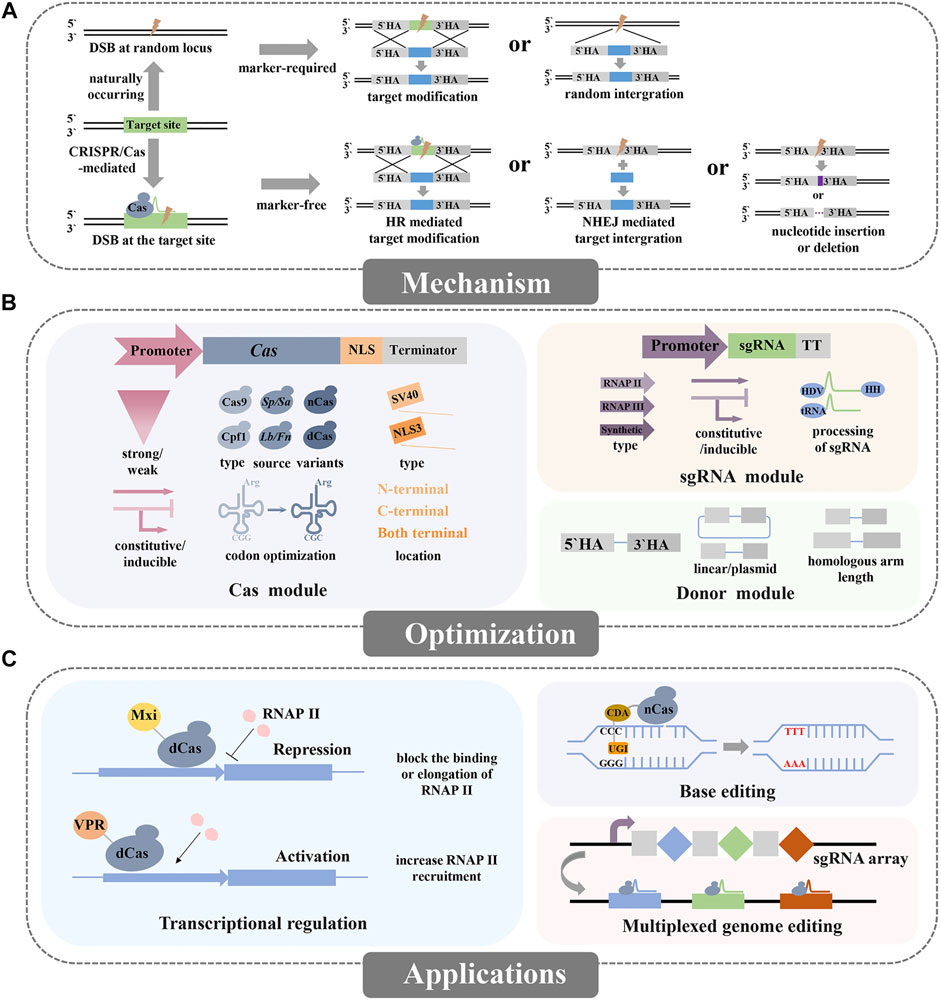
FIGURE 1. Overview of the mechanism, optimizations, and applications for CRISPR/Cas mediated genome editing in non-conventional yeasts. (A) Genome editing via naturally occurring and CRISPR/Cas-mediated DNA repair mechanisms. (B) The optimizations of the CRISPR/Cas system include the Cas module, sgRNA module, and donor module. (C) The applications of the CRISPR/Cas system mainly include the basic applications (knockin or knockout) and advanced applications (repression, activation, base editing or multiplexed genome editing).
The genome-editing system mediated by CRISPR/Cas has already been widely employed in the genetic engineering of non-conventional yeasts and promoted the biotechnological development of these yeasts. Considering the increasing attention of non-conventional yeasts as the chassis for synthetic biology, we systematically summarized the optimization strategies for highly efficient adopting CRISPR/Cas systems in non-conventional yeasts and highlighted the advanced applications of this technology on functional genomics and constructing non-conventional yeast cell factories. Based on current achievements and challenges, we presented our perspectives on building more efficient and adaptable CRISPR/Cas derived system, which would provide new insights in further study CRISPR/Cas technology in non-conventional yeasts.
CRISPR/Cas system comprises two main components, namely Cas protein and sgRNA. To ensure efficient genome editing in non-conventional yeasts, species-specific optimizations of these two components are essentially required (Figure 1B).
Sometimes the efficient expression of sgRNA comes out to be a challenge in non-conventional yeasts due to the lack of suitable promoters. The promoters used for sgRNA expression should have suitable strength and the sgRNAs require nuclear localization and both ends trimming. In non-conventional yeasts, the transcription of functional sgRNA is usually achieved by four types of promoters: 1) The RNA polymerase II (RNAP II) dependent promoters; 2) The RNA polymerase III (RNAP III) dependent promoters; 3) Synthetic hybrid promoters; 4) T7 polymerase dependent artificial promoter.
The RNAP II dependent promoters are usually responsible for producing mRNAs, so the ribozymes executing cleavage sequences are generally flanked on both ends of sgRNAs for modification and maturation. Hammer head ribozyme (HHR) and hepatitis delta virus (HDV) ribozyme are the commonly used elements, which have been employed in building CRISPR/Cas9 system in S. cerevisiae (Gao and Zhao, 2013), P. pastoris (Weninger et al., 2016), O. thermomethanolica (Phithakrotchanakoon et al., 2018), and Y. lipolytica (Gao et al., 2016). However, the genome editing efficiency was lower in comparison to RNAP III dependent promoters based CRISPR/Cas system. This may be presumably due to the extension of the sgRNA variable region from 20 bp to 69 bp because of the homologous arm of the 5′ hammerhead ribozyme (Gao and Zhao, 2013), another reason may be the sgRNAs transcribed by these promoters were exported from the nucleus into the cytosol for translation. Hence RNAP II dependent promoters have rarely been used in sgRNA expression. Interestingly, in the CRISPR/Cpf1 system of Y. lipolytica, the editing efficiency reached 93.3% ± 11.5% where sgRNA expression was controlled by RNAP II dependent promoter TEFin without ribozymes (Yang et al., 2020), which may be caused by the inherent capacity of Cpf1, thereby enabling to produce mature sgRNA from pre-sgRNA array (Fonfara et al., 2016).
RNAP III dependent promoters such as SNR52, RPR1 have been considered as the most suitable candidates for the expression of sgRNA, because the RNAs transcribed by them remain in the nucleus. By far, the SNR52 promoter is the most commonly employed promoter for sgRNAs expression, which allowed for gene editing in various non-conventional yeasts, including S. stipites, K. marxianus, K. lactis, C. albicans and O. polymorpha. The RPR1 promoter was also employed in P. kudriavzevii and I. orientalis. In Table 1, we summarized the great and broad effect of these type of promoters in genetic engineering of non-conventional yeasts.
The RNAP III binding sites of some RNAP III dependent promoters are located within their mature transcript, which may add additional nucleotides to the sgRNA, thereby preventing the maturity and release of sgRNA. The sgRNAs expressed by these promoters are thus fused with tRNA, then the tRNA is isolated through its internal maturation mechanism. In P. pastoris, the orthogonal tRNA-sgRNA cassettes were expressed by the tRNA promoter that enabled multiplexed genome integration of three genes (gnt1, mns1, and mnn2) involved in glycosylation (Dalvie et al., 2020). In O. polymorpha, an improved system with tRNALeu-sgRNA fusion was constructed to enhance the sgRNA expression. The efficiencies of indel mutations were significantly improved to 17–71% in comparison to native RNAP III dependent promoter OpSNR6 which resulted in less than 1% gene disruption (Gao et al., 2021). In I. orientalis, a series of native and synthetic promoters used for sgRNA expression were evaluated, and the synthetic RPR1’-tRNALeu promoter was identified as the most effective promoter; the efficiency of single, double, and triple gene disruption was recorded as 97, 90, and 46.7% (Tran et al., 2019), respectively. A similar study was up taken in Y. lipolytica where synthetic promoters based on the RNAP III dependent promoters and tRNAGly were employed for the expression of sgRNA. The editing efficiency of PEX10 reached up to 92% by the synthetic promoter SCR1’-tRNAGly, which enabled more than a 2-fold increase over the native SNR52 promoter (Schwartz et al., 2016). In K. marxianus, three synthetic RNAP III dependent promoters, including RPR1-tRNAGly, SCR1-tRNAGly, and SNR52-tRNAGly promoter, were applied to optimize the expression of sgRNA. The highest editing efficiency observed in this case was 66% which was achieved by the RPR1-tRNAGly promoter (Löbs et al., 2017). Similarly, in the CRISPR/Cpf1 system, synthetic SCR1’-tRNAGly promoter and native the RNAP III dependent promoter 5sRNA were tested for their potential to enhance sgRNA expression in Y. lipolytica. The highest efficiency was achieved in the case of SCR1′-tRNAGly promoter; the editing efficiency of CAN1 via indels mutation was found to be as high as 86.6% ± 5.7% (Yang et al., 2020).
However, finding a suitable RNAP III dependent promoter in some hosts is still a challenge. In a recent study, an artificially constructed promoter based on the T7 expression system was successfully used for sgRNA expression in some yeasts. An optimized T7 polymerase mutant (P266L) fused with an SV40 nuclear localization sequence (NLS) was developed to ensure a functional T7 promoter for the expression of sgRNA. This system was widely applied in Y. lipolytica, K. lactis, and S. cerevisiae and its editing efficiency was more than 60% (Morse et al., 2018). The optimized T7 expression system from bacteria provides an alternative tool for hosts with no suitable promoters for sgRNA expression. Overall, these innovative approaches of sgRNA expression have great potential for enhancing genome editing in non-conventional yeasts.
Nuclear localization, codon optimization, and promoter screening (strong/weak or constitutive/inducible) were often adapted for optimizing the expression of Cas protein. In eukaryotic chassis, the Cas protein should be localized in the nucleus to generate DSB, the most common strategy being the fusion of an NLS with the Cas to achieve nuclear targeting. The most typical strategy involves the fusion of an SV40 NLS to the N- or C-terminal of Cas (Schwartz et al., 2016; Yang et al., 2020), or even both C- and N-terminal (Cao et al., 2017; Tran et al., 2019; Hou et al., 2020). It has been reported that in R. toruloides, an SV40 NLS fused to the C-terminal of Cas9 is insufficient to achieve its nuclear targeting. Therefore, NLS3 (an endogenous NLS) was appended to the C-terminus of the Cas9 to ensure its import to the nucleus (Schultz et al., 2019).
Codon optimization of the Cas gene can affect the functionality of the CRISPR/Cas system, however, in terms of the efficiency of genome editing, it does not seem necessary in some non-conventional yeasts. The Homo sapiens codon-optimized Cas9 has been used for genetic editing in P. pastoris (Weninger et al., 2016) and Y. lipolytica (Gao et al., 2016). ScCas9 (S. cerevisiae codon-optimized) has been used in K. lactis (Horwitz et al., 2015) and C. glabrata (Enkler et al., 2016). Both C. glabrata and S. cerevisiae codon-optimized sequences of Cas9 enabled the genetic editing in C. glabrata (Enkler et al., 2016) and both H. sapiens and Y. lipolytica codon-optimized sequences are functional in Y. lipolytica (Gao et al., 2016; Schwartz et al., 2016). In contrast, the codon optimization of Cas9 seriously influenced the efficiency of editing in P. pastoris (Weninger et al., 2016) and must be required for the non-canonical codon assignment yeast C. albicans (Vyas et al., 2015).
In non-conventional yeasts, the promoters used for Cas expression commonly focus on strong and constitutive promoters, such as PTEFinor PTEF in Y. lipolytica (Holkenbrink et al., 2018; Bae et al., 2020), PENO1 in C. albicans (Vyas et al., 2015), PFBA1 in K. lactis (Horwitz et al., 2015), and PScTEF1 in C. glabrata (Enkler et al., 2016). In O. thermomethanolica, the expression of Cas9 was controlled by an inducible promoter PAOX. The high expression of Cas9, when triggered by methanol, has a detrimental influence on its fitness. This negative effect was ameliorated when the expression of Cas9 was induced by glycerol instead (Schwartz et al., 2017a). However, the inducible promoter tested in P. pastoris was not successful (Schwartz et al., 2018). In C. glabrata, the choice of the promoter may influence the type of mutation, a single base pair or larger insertions were observed when different promoters were used to express Cas9 (Enkler et al., 2016). In Y. lipolytica, different promoters were screened for nCas9-pmCDA1-UGI expression, and the highest efficiency was achieved by TFFin promoter (Ramesh et al., 2020). These findings show that an appropriate level of Cas9 expression is beneficial to the strain’s resilience and genome editing efficiency. As a result, species-specific improvements of the CRISPR system are required for it to function properly.
Except for the expression of sgRNA and Cas, the targeted gene loci and the sequence of sgRNA also have a significant impact on the genome-editing efficiency. In addition, if the DSB must be repaired by HR, a donor DNA must be given. The type (linear or plasmid) and homologous arm length of donor DNA also impact HR effectiveness.
Besides the exploitation of the basic function of the CRISPR/Cas system like gene deletion or integration, some advanced CRISPR/Cas technologies have been applied in non-conventional yeasts such as the regulation of transcription, base editing and homology-independent gene integration (Figure 1C).
Metabolic engineering and functional genomics both benefit from targeted gene transcriptional regulation. CRISPR activation (CRISPRa) and CRISPR interference (CRISPRi) have been constructed to enhance or weaken the expression of the target gene in non-conventional yeasts. These technologies are based on inactive Cas (dCas), which still preserving the capacity to target and bind to particular DNA sequences.
CRISPRi is a simple but useful tool that can down-regulate the expression level of the target gene. By binding to region of the promoter, dCas could sterically block the binding or elongation of the RNA polymerase, resulting in transcription repression. The effect of repression can be further strengthened by fusing a repressor domain like Mxi1 to dCas. For example, a CRISPRi system based on dCas9 was developed in P. Pastoris, and through this system, a more precise sequence of PAOX1 was obtained which can control the transcription and translation of AOX1 (Hou et al., 2020). In Y. lipolytica, the CRISPRi system was also developed to repress NHEJ. By multiplexed targeting to ku70 and ku80, the HR efficiency, in this case, is as high as 90% compared to the ku70 deficient strain (Schwartz et al., 2017a). In K. marxianus, a multiple CRISPRi system was developed for redirecting carbon flux of the central metabolic pathway towards ethyl acetate production, causing an improved ethyl acetate titer by 3.8-fold (Löbs et al., 2018). Alternatively, the selection of sgRNA has a great influence on the repression efficiency of CRISPRi. Zhang et al. designed a multiplexed sgRNA targeting strategy in Y. lipolytica. Through simultaneous targeting to gfp gene with three different sgRNAs, the repression efficiency reached 92% and 85% with dCas9 and dCpf1 respectively. Furthermore, the efficient repression of three target genes (vioA, vioB, and vioE) in protodeoxy-violaceinic acid (PVA) synthetic pathway was also realized in one step by this strategy, the content of PVA was reduced by 61% and 75% with dCpf1 and dCas9 separately compared with their corresponding control strains (Zhang et al., 2018).
In general, the fusion of transcriptional activators like VP64 and VPR with dCas9 causes gene upregulation by increasing RNA polymerases recruitment. In Y. lipolytica, a dCas9-VPR fusion was used to activate β-glucosidases that allow its growth with cellobiose as the single carbon source (Schwartz et al., 2018). Truncating sgRNA could inhibit the nuclease activity of Cas protein, but not influence the targeting effect. On this basis, a Cpf1-VPR fusion with truncated sgRNA (16 bp) increased hrGFP expression by 10-fold in Y. lipolytica (Ramesh et al., 2020).
In addition to the on/off states of gene expression, controlling the gene expression at a suitable level allows for the creation of the desired phenotype. Graded gene expression strengths were obtained by altering the sgRNA target site in the promoter region using CRISPRi or CRISPRa. It resulted in a dynamic gene expression range from zero to several 10-fold improvement, allowing for fine-tuning of metabolic pathway expression and optimization of optimal phenotypes. The balance between cell growth and products biosynthesis is the major issue to be addressed in construction cell factories. CRISPR/Cas-mediated multiple genes synchronized regulation maybe a powerful tool to build a highly efficient non-conventional yeast cell factories.
Base editing is a valuable tool with a lot of potential in genetic editing. By fusing deaminase with the nCas or dCas proteins, these fusion proteins may directly create precise point mutations in genomic DNA. A Target-AID (target activation-induced cytidine deaminase) base editor based on the nCas9-pmCDA1 fusion protein was created to execute the conversion of C to T in human cells and S. cerevisiae, which was tested by inserting a nonsense mutation into the coding sequence.
In non-conventional yeasts, the Target-AID base editor was employed to realize gene disruption in Y. lipolytica (Bae et al., 2020). Similarly in K. marxianus, this base editor was also constructed to disrupt Nej1 or Dnl4 which was involved in NHEJ to enhance the proportions of HR-mediated integration (Nambu-Nishida et al., 2017). Apart from gene disruption, this base editor was further employed for situ mutagenesis, thus enabling it to obtain the desired phenotype. Recently, the general transcription factor gene SPT15 in S. cerevisiae was mutated by Target-AID base editor to enhance the stress tolerances (Liu et al., 2021). Furthermore, this strategy has also been applied in mammals (Ma et al., 2016) and plant (Li et al., 2020) but has not been widely reported in non-conventional yeasts. Though the overall usage of base editing is less in comparison to conventional CRISPR/Cas genetic editing, it has the potential to complement genetic editing tools because of its procedures being donor-free and DSB-free.
Constructing homologous arms for homology-dependent genome editing is laborious and time-consuming. In most non-conventional yeasts such as Y. lipolytica, S. stipites, R. toruloides, P. pastoris, and K. marxianus, NHEJ is the dominant repair pathway of DSB. A recently study took advantage of this inherent property and constructed a CRISPR/Cas9-mediated homology-independent gene integration tool in Y. lipolytica. The targeted gene integration rate was up to 55% by optimizing the cleavage efficiency of Cas9, manipulating repair fidelity of NHEJ, cell cycle and integration sites. By using this tool, iterative integration of canthaxanthin biosynthesis pathway including four genes (GGS1, carB, carRP and CrtW) was achieved (Cui et al., 2021). It is worth noting that integration of an 8,417 bp fragment composed of GGS1, carB, and carRP into genome by one step may still be a challenge for the HR dependent targeted genome integration, indicating this tool paves a new avenue to realize the accurate and efficient targeted genome integration in some non-conventional yeasts.
Relative to the conventional yeast S. cerevisiae, the genome annotations for non-conventional yeast are not particularly thorough. In Y. lipolytica, CRISPR/Cas9 has been applied to research functional genomics through the construction of a sgRNA library that covers the whole genome and targeting 7,845 coding sequences (CDS). A total of 1,377 CDSs were identified as necessary CDSs by employing this approach. This sgRNA library facilitates the screening of growth and non-growth related phenotypes, such as canavanine resistance (Schwartz et al., 2019). In P. pastoris, one to three nucleotides have been precisely inserted or deleted at the S215 of the methanol expression regulator Mxr1, and the S215 also has been mutated to A215 through a single base replacement. The frameshift mutation of Mxr1 resulted in almost no transcription of its target genes DAS1, DAS2 and AOX1, with AOX2 transcription, decreased by 40%. For the Mxr1S215A, the transcription of these four targeted genes was decreased by nearly 60% (Hou et al., 2020). In K. marxianus, the CRISPR/Cas9 system was developed to characterize functional genes within the ethanol and ethyl acetate biosynthesis pathway by disrupting the genes involved, thereby demonstrating that ADH7 (alcohol dehydrogenase) played a major role as an alternative pathway for the biosynthesis of ethyl acetate (Löbs et al., 2017). These researches demonstrated that CRISPR/Cas was a valuable method for determining gene function and identifying candidate genes.
The creation and application of strains often require repetitive design-build-test cycles, the CRISPR/Cas method can speed up this process because it enables marker-free gene editing. Wang et al. used CRISPR/Cas9 to engineer O. polymorpha to produce resveratrol. By targeting the rDNA site, 10 copies of resveratrol biosynthetic pathway (three genes, composed of TAL, 4CL and STS) were integrated into the genome, resulting in higher resveratrol production, which reached as high as 97.23 ± 4.84 mg/L, representing a 20-fold increase compared with single-copy integration (Wang et al., 2018). In Y. lipolytica, CRISPR/Cas9 was used by Schwartz et al. to screen integration loci that not only allow for high expression of the integrated gene but also have no detrimental impact on cell resilience after the gene has been integrated. Five of the 17 loci were found, and four genes (crtB, crtE, crtI and Ggs1) involved in the lycopene production pathway were each integrated into a separate locus enabling an 8.6 folds increase in lycopene production in comparison to the wildtype strain (Schwartz et al., 2017b). In P. kudriavzevii, the CRISPR/Cas9 was adapted to deleting the downstream competing pathway and then integrating the itaconic acid (IA) biosynthesis pathway into the genome. The production of IA achieved afterward was 1,232 mg/L in fed-batch fermentation (Sun et al., 2020). Overall, CRISPR/Cas technology is a powerful tool in bioengineering these traditional hardly genetically engineered yeasts for basic research or industrial applications (Table 1).
Despite the numerous successful applications of CRISPR/Cas technology in different non-conventional yeasts, there are still many obstacles to overcome and CRISPR/Cas based techniques to develop in non-conventional yeasts.
In most non-conventional yeasts, NHEJ weakens their precise genome engineering. To increase the HR efficiency, the most common strategy is to knock out or inhibit NHEJ-related genes such as ku70/80, Nej1, and Dnl4, which have proven effective in Y. lipolytica (Kretzschmar et al., 2013), K. marxianus (Nambu-Nishida et al., 2017). However, many studies reported that the robustness of NHEJ-deficient cells is poor in comparison to NHEJ-proficient cells (Kretzschmar et al., 2013). Alternatively, some studies show that the HR efficiency can be enhanced by overexpressing HR-associated genes like Rad51/52 and Sae2. Expression of the codon-optimized ScRad52 demonstrated an obvious improvement in the HR efficiency of Y. lipolytica (Ji et al., 2020). The combined expression of ScRad51, ScRad52, and ScSae2 significantly improved the HR rate of O. polymorpha (Gao et al., 2021). On the contrary, ScRad51/Rad52 expressed in P. pastoris (Cai et al., 2021) and S. stipitis (Cao et al., 2018) had no distinct improvement in HR efficiency, but overexpression of endogenous Rad51 and Rad52 resulted in higher HR activity of P. pastoris (Cai et al., 2021). Presumably, the mechanism of DSB repair is mysterious and complex, the choice of proper genes and the expression strength of these genes are important to increase HR efficiency, which is essential for efficient operation of the multiple genes simultaneously in non-conventional yeasts. Alternatively, CRISPR/Cas9-mediated homology-independent targeted gene integration maybe a potential tool for precise genome engineering.
In addition to the current commonly used Cas9 and Cpf1, other Cas proteins have gradually started receiving attention. Recently, Walton et al. developed a near-PAMless SpCas9 variant (SpRY), which can target almost all PAMs (Walton et al., 2020). For Cpf1, two AsCpf1 variants were developed, which recognize TYCV and TATV PAMs, respectively (Gao et al., 2017). Cas13a previously known as C2c2 can induce precise cleavage of RNA, therefore, it can perform RNA interference without DNA damage (Cox et al., 2017). Furthermore, gene editing at the RNA level is reversible and changeable, it is a viable method for developing dynamic regulatory elements to control the level and timing of Cas13a expression. Cas14, which has only 400 to 700 amino acids, may target ssDNA and induce cleavage without requiring a specific sequence (Harrington et al., 2018). Up to now, the applications of Cas9, Cpf1 variants, Cas13a, and Cas14 are mainly focused on nucleic acid testing (Gootenberg et al., 2017; Ge et al., 2021) or mammalian (Gao et al., 2017; Walton et al., 2020) and plant cells (Abudayyeh et al., 2017). Based on their advantages, they will have promising potentials to be developed in non-conventional yeasts.
The use of the CRISPR/Cas system in conjunction with other methods or effectors has considerably increased the total capability of cellular engineering. In the realm of synthetic biology, the combination of dynamic regulation with CRISPR is highly efficient in improving biological processes. Recently, the muconic acid production in E. coli was increased by 1.3-fold through using an optogenetic CRISPRi system (Wu et al., 2020). Similar to Y. lipolytica, by the combination of a fatty acid biosensor and CRISPRi, the production of naringenin was increased by 74.8% (Lv et al., 2020). On the other hand, the target nucleotide diversification was performed by the fusion of nCas9 and error-prone DNA polymerase in E. coli (Halperin et al., 2018) and S. cerevisiae (Tou et al., 2020) respectively. Although the base editing accomplished by Cas-deaminases fusion has been widely investigated, it has only been reported in few non-conventional yeasts. The introduction of these systems into non-conventional yeasts would be a huge help in fine-tuning gene expression or global genome engineering, both of which are important techniques for building highly efficient cell factories or scientific research.
LS: writing-original draft. ZD and QW: writing-review and editing and project administration. All authors contributed to the article and approved the submitted version.
This work was supported by Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-CXRC-002), Hundreds of Talents Program of the Chinese Academy of Sciences (Y0J51009) and the National Natural Science Foundation of China (32071423).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., et al. (2017). RNA Targeting with CRISPR-Cas13. Nature 550, 280–284. doi:10.1038/nature24049
Agbogbo, F. K., and Coward-Kelly, G. (2008). Cellulosic Ethanol Production Using the Naturally Occurring Xylose-Fermenting Yeast, Pichia Stipitis. Biotechnol. Lett. 30, 1515–1524. doi:10.1007/s10529-008-9728-z
Bae, S. J., Park, B. G., Kim, B. G., and Hahn, J. S. (2020). Multiplex Gene Disruption by Targeted Base Editing ofYarrowia lipolyticaGenome Using Cytidine Deaminase Combined with the CRISPR/Cas9 System. Biotechnol. J. 15, 1900238. doi:10.1002/biot.201900238
Bao, Z., Xiao, H., Liang, J., Zhang, L., Xiong, X., Sun, N., et al. (2015). Homology-integrated CRISPR-Cas (HI-CRISPR) System for One-step Multigene Disruption in Saccharomyces cerevisiae. ACS Synth. Biol. 4, 585–594. doi:10.1021/sb500255k
Bredell, H., Smith, J. J., Görgens, J. F., and van Zyl, W. H. (2018). Expression of Unique Chimeric Human Papilloma Virus Type 16 (HPV-16) L1-L2 Proteins inPichia pastorisandHansenula Polymorpha. Yeast 35, 519–529. doi:10.1002/yea.3318
Burstein, D., Harrington, L. B., Strutt, S. C., Probst, A. J., Anantharaman, K., Thomas, B. C., et al. (2017). New CRISPR-Cas Systems from Uncultivated Microbes. Nature 542, 237–241. doi:10.1038/nature21059
Cai, P., Duan, X., Wu, X., Gao, L., Ye, M., and Zhou, Y. J. (2021). Recombination Machinery Engineering Facilitates Metabolic Engineering of the Industrial Yeast Pichia pastoris. Nucleic Acids Res. 49, 7791–7805. doi:10.1093/nar/gkab535
Cao, M., Gao, M., Lopez-Garcia, C. L., Wu, Y., Seetharam, A. S., Severin, A. J., et al. (2017). Centromeric DNA Facilitates Nonconventional Yeast Genetic Engineering. ACS Synth. Biol. 6, 1545–1553. doi:10.1021/acssynbio.7b00046
Cao, M., Gao, M., Ploessl, D., Song, C., and Shao, Z. (2018). CRISPR-mediated Genome Editing and Gene Repression in Scheffersomyces Stipitis. Biotechnol. J. 13, 1700598. doi:10.1002/biot.201700598
Castañeda, M. T., Nuñez, S., Garelli, F., Voget, C., and De Battista, H. (2018). Comprehensive Analysis of a Metabolic Model for Lipid Production in Rhodosporidium Toruloides. J. Biotechnol. 280, 11–18. doi:10.1016/j.jbiotec.2018.05.010
Chattopadhyay, A., Mitra, M., and Maiti, M. K. (2021). Recent Advances in Lipid Metabolic Engineering of Oleaginous Yeasts. Biotechnol. Adv., 107722. doi:10.1016/j.biotechadv.2021.107722
Cox, D. B. T., Gootenberg, J. S., Abudayyeh, O. O., Franklin, B., Kellner, M. J., Joung, J., et al. (2017). RNA Editing with CRISPR-Cas13. Science 358, 1019–1027. doi:10.1126/science.aaq0180
Cui, Z., Zheng, H., Zhang, J., Jiang, Z., Zhu, Z., Liu, X., et al. (2021). A CRISPR/Cas9-mediated, Homology-independent Tool Developed for Targeted Genome Integration in Yarrowia Lipolytica. Appl. Environ. Microbiol. 87, e02666–02620. doi:10.1128/AEM.02666-20
Dalvie, N. C., Leal, J., Whittaker, C. A., Yang, Y., Brady, J. R., Love, K. R., et al. (2020). Host-informed Expression of CRISPR Guide RNA for Genomic Engineering in Komagataella Phaffii. ACS Synth. Biol. 9, 26–35. doi:10.1021/acssynbio.9b00372
Doyon, Y., Vo, T. D., Mendel, M. C., Greenberg, S. G., Wang, J., Xia, D. F., et al. (2011). Enhancing zinc-finger-nuclease Activity with Improved Obligate Heterodimeric Architectures. Nat. Methods 8, 74–79. doi:10.1038/nmeth.1539
Enkler, L., Richer, D., Marchand, A. L., Ferrandon, D., and Jossinet, F. (2016). Genome Engineering in the Yeast Pathogen Candida Glabrata Using the CRISPR-Cas9 System. Sci. Rep. 6, 35766. doi:10.1038/srep35766
Fonfara, I., Richter, H., Bratovič, M., Le Rhun, A., and Charpentier, E. (2016). The CRISPR-Associated DNA-Cleaving Enzyme Cpf1 Also Processes Precursor CRISPR RNA. Nature 532, 517–521. doi:10.1038/nature17945
Gao, D., Smith, S., Spagnuolo, M., Rodriguez, G., and Blenner, M. (2018). Dual CRISPR‐Cas9 Cleavage Mediated Gene Excision and Targeted Integration in Yarrowia Lipolytica. Biotechnol. J. 13, 1700590. doi:10.1002/biot.201700590
Gao, J., Gao, N., Zhai, X., and Zhou, Y. J. (2021). Recombination Machinery Engineering for Precise Genome Editing in Methylotrophic Yeast Ogataea Polymorpha. iScience 24, 102168. doi:10.1016/j.isci.2021.102168
Gao, L., Cox, D. B. T., Yan, W. X., Manteiga, J. C., Schneider, M. W., Yamano, T., et al. (2017). Engineered Cpf1 Variants with Altered PAM Specificities. Nat. Biotechnol. 35, 789–792. doi:10.1038/nbt.3900
Gao, S., Tong, Y., Wen, Z., Zhu, L., Ge, M., Chen, D., et al. (2016). Multiplex Gene Editing of the Yarrowia Lipolytica Genome Using the CRISPR-Cas9 System. J. Ind. Microbiol. Biotechnol. 43, 1085–1093. doi:10.1007/s10295-016-1789-8
Gao, Y., and Zhao, Y. (2014). Self‐processing of Ribozyme‐flanked RNAs into Guide RNAs In Vitro and In Vivo for CRISPR‐mediated Genome Editing. J. Integr. Plant Biol. 56, 343–349. doi:10.1111/jipb.12152
Ge, X., Meng, T., Tan, X., Wei, Y., Tao, Z., Yang, Z., et al. (2021). Cas14a1-mediated Nucleic Acid Detectifon Platform for Pathogens. Biosens. Bioelectron. 189, 113350. doi:10.1016/j.bios.2021.113350
Gellissen, G. (2000). Heterologous Protein Production in Methylotrophic Yeasts. Appl. Microbiol. Biotechnol. 54, 741–750. doi:10.1007/s002530000464
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., et al. (2017). Nucleic Acid Detection with CRISPR-Cas13a/C2c2. Science 356, 438–442. doi:10.1126/science.aam9321
Halperin, S. O., Tou, C. J., Wong, E. B., Modavi, C., Schaffer, D. V., and Dueber, J. E. (2018). CRISPR-guided DNA Polymerases Enable Diversification of All Nucleotides in a Tunable Window. Nature 560, 248–252. doi:10.1038/s41586-018-0384-8
Harrington, L. B., Burstein, D., Chen, J. S., Paez-Espino, D., Ma, E., Witte, I. P., et al. (2018). Programmed DNA Destruction by Miniature CRISPR-Cas14 Enzymes. Science 362, 839–842. doi:10.1126/science.aav4294
Hartner, F. S., and Glieder, A. (2006). Regulation of Methanol Utilisation Pathway Genes in Yeasts. Microb. Cel Fact. 5, 39. doi:10.1186/1475-2859-5-39
Holkenbrink, C., Dam, M. I., Kildegaard, K. R., Beder, J., Dahlin, J., Doménech Belda, D., et al. (2018). EasyCloneYALI: CRISPR/Cas9-based Synthetic Toolbox for Engineering of the Yeast Yarrowia Lipolytica. Biotechnol. J. 13, 1700543. doi:10.1002/biot.201700543
Horwitz, A. A., Walter, J. M., Schubert, M. G., Kung, S. H., Hawkins, K., Platt, D. M., et al. (2015). Efficient Multiplexed Integration of Synergistic Alleles and Metabolic Pathways in Yeasts via CRISPR-Cas. Cel Syst. 1, 88–96. doi:10.1016/j.cels.2015.02.001
Hou, C., Yang, Y., Xing, Y., Zhan, C., Liu, G., Liu, X., et al. (2020). Targeted Editing of Transcriptional Activator MXR1 on the Pichia pastoris Genome Using CRISPR/Cas9 Technology. Yeast 37, 305–312. doi:10.1002/yea.3462
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157, 1262–1278. doi:10.1016/j.cell.2014.05.010
Ji, Q., Mai, J., Ding, Y., Wei, Y., Ledesma-Amaro, R., and Ji, X.-J. (2020). Improving the Homologous Recombination Efficiency of Yarrowia Lipolytica by Grafting Heterologous Component from Saccharomyces cerevisiae. Metab. Eng. Commun. 11, e00152. doi:10.1016/j.mec.2020.e00152
Jiao, X., Zhang, Y., Liu, X., Zhang, Q., Zhang, S., and Zhao, Z. K. (2019). Developing a CRISPR/Cas9 System for Genome Editing in the Basidiomycetous Yeast Rhodosporidium Toruloides. Biotechnol. J. 14, 1900036. doi:10.1002/biot.201900036
Kretzschmar, A., Otto, C., Holz, M., Werner, S., Hübner, L., and Barth, G. (2013). Increased Homologous Integration Frequency in Yarrowia Lipolytica Strains Defective in Non-homologous End-Joining. Curr. Genet. 59, 63–72. doi:10.1007/s00294-013-0389-7
Li, C., Zhang, R., Meng, X., Chen, S., Zong, Y., Lu, C., et al. (2020). Targeted, Random Mutagenesis of Plant Genes with Dual Cytosine and Adenine Base Editors. Nat. Biotechnol. 38, 875–882. doi:10.1038/s41587-019-0393-7
Li, T., Huang, S., Zhao, X., Wright, D. A., Carpenter, S., Spalding, M. H., et al. (2011). Modularly Assembled Designer TAL Effector Nucleases for Targeted Gene Knockout and Gene Replacement in Eukaryotes. Nucleic Acids Res. 39, 6315–6325. doi:10.1093/nar/gkr188
Liu, Q., Shi, X., Song, L., Liu, H., Zhou, X., Wang, Q., et al. (2019). CRISPR-Cas9-mediated Genomic Multiloci Integration in Pichia pastoris. Microb. Cel Fact. 18, 144. doi:10.1186/s12934-019-1194-x
Liu, Y., Lin, Y., Guo, Y., Wu, F., zhang, Y., Qi, X., et al. (2021). Stress Tolerance Enhancement via SPT15 Base Editing in Saccharomyces cerevisiae. Biotechnol. Biofuels 14, 155. doi:10.1186/s13068-021-02005-w
Löbs, A.-K., Engel, R., Schwartz, C., Flores, A., and Wheeldon, I. (2017). CRISPR-Cas9-enabled Genetic Disruptions for Understanding Ethanol and Ethyl Acetate Biosynthesis in Kluyveromyces Marxianus. Biotechnol. Biofuels 10, 164. doi:10.1186/s13068-017-0854-5
Löbs, A.-K., Schwartz, C., Thorwall, S., and Wheeldon, I. (2018). Highly Multiplexed CRISPRi Repression of Respiratory Functions Enhances Mitochondrial Localized Ethyl Acetate Biosynthesis in Kluyveromyces Marxianus. ACS Synth. Biol. 7, 2647–2655. doi:10.1021/acssynbio.8b00331
Lv, Y., Gu, Y., Xu, J., Zhou, J., and Xu, P. (2020). Coupling Metabolic Addiction with Negative Autoregulation to Improve Strain Stability and Pathway Yield. Metab. Eng. 61, 79–88. doi:10.1016/j.ymben.2020.05.005
Ma, Y., Zhang, J., Yin, W., Zhang, Z., Song, Y., and Chang, X. (2016). Targeted AID-Mediated Mutagenesis (TAM) Enables Efficient Genomic Diversification in Mammalian Cells. Nat. Methods 13, 1029–1035. doi:10.1038/nmeth.4027
Marcišauskas, S., Ji, B., and Nielsen, J. (2019). Reconstruction and Analysis of a Kluyveromyces Marxianus Genome-Scale Metabolic Model. BMC Bioinformatics 20, 551. doi:10.1186/s12859-019-3134-5
Morse, N. J., Wagner, J. M., Reed, K. B., Gopal, M. R., Lauffer, L. H., and Alper, H. S. (2018). T7 Polymerase Expression of Guide RNAs In Vivo Allows Exportable CRISPR-Cas9 Editing in Multiple Yeast Hosts. ACS Synth. Biol. 7, 1075–1084. doi:10.1021/acssynbio.7b00461
Nambu-Nishida, Y., Nishida, K., Hasunuma, T., and Kondo, A. (2017). Development of a Comprehensive Set of Tools for Genome Engineering in a Cold- and Thermo-Tolerant Kluyveromyces Marxianus Yeast Strain. Sci. Rep. 7, 8993. doi:10.1038/s41598-017-08356-5
Phithakrotchanakoon, C., Puseenam, A., Wongwisansri, S., Eurwilaichitr, L., Ingsriswang, S., Tanapongpipat, S., et al. (2018). CRISPR-Cas9 Enabled Targeted Mutagenesis in the Thermotolerant Methylotrophic Yeast Ogataea Thermomethanolica. FEMS Microbiol. Lett. 365. doi:10.1093/femsle/fny105
Ramesh, A., Ong, T., Garcia, J. A., Adams, J., and Wheeldon, I. (2020). Guide RNA Engineering Enables Dual Purpose CRISPR-Cpf1 for Simultaneous Gene Editing and Gene Regulation in Yarrowia Lipolytica. ACS Synth. Biol. 9, 967–971. doi:10.1021/acssynbio.9b00498
Raschmanová, H., Weninger, A., Glieder, A., Kovar, K., and Vogl, T. (2018). Implementing CRISPR-Cas Technologies in Conventional and Non-conventional Yeasts: Current State and Future Prospects. Biotechnol. Adv. 36, 641–665. doi:10.1016/j.biotechadv.2018.01.006
Schultz, J. C., Cao, M., and Zhao, H. (2019). Development of a CRISPR/Cas9 System for High Efficiency Multiplexed Gene Deletion in Rhodosporidium Toruloides. Biotechnol. Bioeng. 116, 2103–2109. doi:10.1002/bit.27001
Schwartz, C., Cheng, J.-F., Evans, R., Schwartz, C. A., Wagner, J. M., Anglin, S., et al. (2019). Validating Genome-wide CRISPR-Cas9 Function Improves Screening in the Oleaginous Yeast Yarrowia Lipolytica. Metab. Eng. 55, 102–110. doi:10.1016/j.ymben.2019.06.007
Schwartz, C., Curtis, N., Löbs, A.-K., and Wheeldon, I. (2018). Multiplexed CRISPR Activation of Cryptic Sugar Metabolism Enables Yarrowia Lipolytica Growth on Cellobiose. Biotechnol. J. 13, 1700584. doi:10.1002/biot.201700584
Schwartz, C., Frogue, K., Ramesh, A., Misa, J., and Wheeldon, I. (2017a). CRISPRi Repression of Nonhomologous End‐joining for Enhanced Genome Engineering via Homologous Recombination in Yarrowia Lipolytica. Biotechnol. Bioeng. 114, 2896–2906. doi:10.1002/bit.26404
Schwartz, C. M., Hussain, M. S., Blenner, M., and Wheeldon, I. (2016). Synthetic RNA Polymerase III Promoters Facilitate High-Efficiency CRISPR-Cas9-Mediated Genome Editing in Yarrowia Lipolytica. ACS Synth. Biol. 5, 356–359. doi:10.1021/acssynbio.5b00162
Schwartz, C., Shabbir-Hussain, M., Frogue, K., Blenner, M., and Wheeldon, I. (2017b). Standardized Markerless Gene Integration for Pathway Engineering in Yarrowia Lipolytica. ACS Synth. Biol. 6, 402–409. doi:10.1021/acssynbio.6b00285
Spohner, S. C., Schaum, V., Quitmann, H., and Czermak, P. (2016). Kluyveromyces Lactis: an Emerging Tool in Biotechnology. J. Biotechnol. 222, 104–116. doi:10.1016/j.jbiotec.2016.02.023
Sun, W., Vila-Santa, A., Liu, N., Prozorov, T., Xie, D., Faria, N. T., et al. (2020). Metabolic Engineering of an Acid-Tolerant Yeast Strain Pichia Kudriavzevii for Itaconic Acid Production. Metab. Eng. Commun. 10, e00124. doi:10.1016/j.mec.2020.e00124
Tou, C. J., Schaffer, D. V., and Dueber, J. E. (2020). Targeted Diversification in the S. cerevisiae Genome with CRISPR-Guided DNA Polymerase I. ACS Synth. Biol. 9, 1911–1916. doi:10.1021/acssynbio.0c00149
Tran, V. G., Cao, M., Fatma, Z., Song, X., and Zhao, H. (2019). Development of a CRISPR/Cas9-based Tool for Gene Deletion in Issatchenkia Orientalis. mSphere 4, e00345–00319. doi:10.1128/mSphere.00345-19
Vyas, V. K., Barrasa, M. I., and Fink, G. R. (2015). ACandida albicansCRISPR System Permits Genetic Engineering of Essential Genes and Gene Families. Sci. Adv. 1, e1500248. doi:10.1126/sciadv.1500248
Walton, R. T., Christie, K. A., Whittaker, M. N., and Kleinstiver, B. P. (2020). Unconstrained Genome Targeting with Near-PAMless Engineered CRISPR-Cas9 Variants. Science 368, 290–296. doi:10.1126/science.aba8853
Wang, L., Deng, A., Zhang, Y., Liu, S., Liang, Y., Bai, H., et al. (2018). Efficient CRISPR-Cas9 Mediated Multiplex Genome Editing in Yeasts. Biotechnol. Biofuels 11, 277. doi:10.1186/s13068-018-1271-0
Weninger, A., Hatzl, A.-M., Schmid, C., Vogl, T., and Glieder, A. (2016). Combinatorial Optimization of CRISPR/Cas9 Expression Enables Precision Genome Engineering in the Methylotrophic Yeast Pichia pastoris. J. Biotechnol. 235, 139–149. doi:10.1016/j.jbiotec.2016.03.027
Wu, P., Chen, Y., Liu, M., Xiao, G., and Yuan, J. (2020). Engineering an Optogenetic CRISPRi Platform for Improved Chemical Production. ACS Synth. Biol. 10, 125–131. doi:10.1021/acssynbio.0c00488
Xie, W., Liu, M., Lv, X., Lu, W., Gu, J., and Yu, H. (2014). Construction of a Controllable β-carotene Biosynthetic Pathway by Decentralized Assembly Strategy inSaccharomyces Cerevisiae. Biotechnol. Bioeng. 111, 125–133. doi:10.1002/bit.25002
Yang, Z., Edwards, H., and Xu, P. (2020). CRISPR-Cas12a/Cpf1-assisted Precise, Efficient and Multiplexed Genome-Editing in Yarrowia Lipolytica. Metab. Eng. Commun. 10, e00112. doi:10.1016/j.mec.2019.e00112
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., et al. (2015). Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163, 759–771. doi:10.1016/j.cell.2015.09.038
Keywords: CRISPR/cas, cell factories, microbial production, non-conventional yeast, genome engineering
Citation: Shan L, Dai Z and Wang Q (2021) Advances and Opportunities of CRISPR/Cas Technology in Bioengineering Non-conventional Yeasts. Front. Bioeng. Biotechnol. 9:765396. doi: 10.3389/fbioe.2021.765396
Received: 27 August 2021; Accepted: 27 September 2021;
Published: 11 October 2021.
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaReviewed by:
Tian-qiong Shi, Nanjing Normal University, ChinaCopyright © 2021 Shan, Dai and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zongjie Dai, ZGFpempAdGliLmNhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.