- 1State Key Laboratory of Microbial Technology, Shandong University, Qingdao, China
- 2Center for Gene and Immunotherapy, The Second Hospital of Shandong University, Jinan, China
- 3State Key Laboratory of Microbial Metabolism, Joint International Research Laboratory of Metabolic and Developmental Sciences, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
Pseudomonas putida KT2440 is a promising chassis of industrial biotechnology due to its metabolic versatility. Butane-2,3-diol (2,3-BDO) is a precursor of numerous value-added chemicals. It is also a microbial metabolite which widely exists in various habiting environments of P. putida KT2440. It was reported that P. putida KT2440 is able to use 2,3-BDO as a sole carbon source for growth. There are three stereoisomeric forms of 2,3-BDO: (2R,3R)-2,3-BDO, meso-2,3-BDO and (2S,3S)-2,3-BDO. However, whether P. putida KT2440 can utilize three stereoisomeric forms of 2,3-BDO has not been elucidated. Here, we revealed the genomic and enzymic basis of P. putida KT2440 for dehydrogenation of different stereoisomers of 2,3-BDO into acetoin, which will be channeled to central mechanism via acetoin dehydrogenase enzyme system. (2R,3R)-2,3-BDO dehydrogenase (PP0552) was detailedly characterized and identified to participate in (2R,3R)-2,3-BDO and meso-2,3-BDO dehydrogenation. Two quinoprotein alcohol dehydrogenases, PedE (PP2674) and PedH (PP2679), were confirmed to be responsible for (2S,3S)-2,3-BDO dehydrogenation. The function redundancy and inverse regulation of PedH and PedE by lanthanide availability provides a mechanism for the adaption of P. putida KT2440 to variable environmental conditions. Elucidation of the mechanism of 2,3-BDO catabolism in P. putida KT2440 would provide new insights for bioproduction of 2,3-BDO-derived chemicals based on this robust chassis.
Introduction
Pseudomonas putida is known for its rapid growth, low nutrient demand, and adaption to various physicochemical stresses (Nikel and de Lorenzo, 2018). P. putida KT2440, the first Gram-negative soil bacterium which was certified as generally recognized as safe (i.e., GRAS) (Nelson, 2002; Nelson et al., 2002), has attracted substantial attention as a new workhorse for bioindustries (Nikel and de Lorenzo, 2018; Weimer et al., 2020). Based on the toolbox for genetic engineering of P. putida KT2440 (Poblete-Castro et al., 2020; Sun et al., 2020; Weimer et al., 2020; Zhou et al., 2020), it has been engineered for heterologous production of diverse value-added products (Nikel and de Lorenzo, 2013; Johnson and Beckham, 2015; Dvořák and de Lorenzo, 2018; Sánchez-Pascuala et al., 2019; Bator et al., 2020; Bentley et al., 2020; Tiso et al., 2020) and environmental remediation (Franden et al., 2018). In addition, the intrinsically high metabolic diversity gives P. putida KT2440 a wide substrate spectrum for chemical production (Nelson et al., 2002; Wu et al., 2011). Elucidation of the diverse biological processes in P. putida KT2440 would incorporate new substrates for bioproduction (Li et al., 2019; Salvachúa et al., 2020; Wada et al., 2021).
Butane-2,3-diol (2,3-BDO) is a physiological metabolic product excreted by many microorganisms such as Bacillus subtilis, B. amyloliquefaciens, Klebsiella oxytoca, K. pneumoniae, Enterobacter cloacae and Serratia marcescens (Ryu et al., 2003; Xu et al., 2014). Some economical 2,3-BDO fermentation processes using various inexpensive carbohydrate raw materials have been established (Liu et al., 2011; Lian et al., 2014; Xu et al., 2014; Chu et al., 2015; Ge et al., 2016; Yang et al., 2017). However, development of efficient derivative processes for 2,3-BDO is still a prerequisite for value-added utilization of biotechnologically produced 2,3-BDO. P. putida KT2440 can metabolize 2,3-BDO as the sole carbon source for growth. Recently, a bioconversion process for mevalonate production from 2,3-BDO has been established by using recombinant P. putida KT2440 (Yang et al., 2020).
2,3-BDO is a chiral compound with two chiral carbon atoms and it has three stereoisomeric forms, (2R,3R)-2,3-BDO, meso-2,3-BDO, and (2S,3S)-2,3-BDO. The anabolism mechanism of the three stereoisomeric forms of 2,3-BDO has been detailedly elucidated. Three key enzymes participate in 2,3-BDO biosynthesis. α-Acetolactate synthase catalyzes two molecules of pyruvate into α-acetolactate, which is then converted to (3R)-acetoin [(3R)-AC] by α-acetolactate decarboxylase (Celińska and Grajek, 2009; Ji et al., 2011). (3R)-AC is reduced to meso-2,3-BDO and (2R,3R)-2,3-BDO by meso-2,3-BDO dehydrogenase (meso-BDH, belonging to short-chain dehydrogenase/reductase SDR family) and (2R,3R)-2,3-BDO dehydrogenase (R,R-BDH, belonging to zinc-containing medium-chain dehydrogenase/reductase MDR family), respectively (Celińska and Grajek, 2009; Ji et al., 2011; Xu et al., 2014; Zhang et al., 2014; Yang et al., 2017). In addition, diacetyl, which is produced from α-acetolactate by non-enzymatic oxidative decarboxylation, can be reduced to (2S,3S)-2,3-BDO by meso-BDH (Xiao and Xu., 2007; Ge et al., 2016). Existence of multiple stereospecific dehydrogenases in 2,3-BDO producing bacteria results in the presence of three stereoisomeric forms in natural habitats (Yang et al., 2017). Whether P. putida KT2440 can use three stereoisomers of 2,3-BDO has not been clarified.
In this study, P. putida KT2440 was identified to be able to use three 2,3-BDO stereoisomers for growth. The first step of 2,3-BDO catabolism is its dehydrogenation to AC and three key enzymes catalyzing different stereoisomers of 2,3-BDO were identified in P. putida KT2440. Briefly, R,R-BDH catalyzes the dehydrogenation of (2R,3R)-2,3-BDO and meso-2,3-BDO. Two quinoprotein alcohol dehydrogenases are critical for (2S,3S)-2,3-BDO dehydrogenation. In addition, the lanthanide-responsive switch of the two quinoprotein alcohol dehydrogenases was identified to lead to growth of P. putida KT2440 in (2S,3S)-2,3-BDO depending on the presence or absence of lanthanides.
Materials and Methods
Bacteria and Culture Conditions
Bacterial strains and plasmids used in this work are listed in Supplementary Table S1. P. putida KT2440 and its derivatives were cultured in MSM (Ma et al., 2007) with 2 g L−1 different substances at 200 rpm and 30°C. Lanthanide concentration-dependent growth of P. putida KT2440 was monitored using a Bioscreen microbiology reader (Bioscreen C Labsystems, Helsinki, Finland). E. coli DH5α and BL21 (DE3) were cultured in Luria-Bertani (LB) medium at 180 rpm and 37°C. Antibiotics were added to the medium when necessary, at the following concentrations: kanamycin at 50 μg mL−1 and ampicillin at 100 μg mL−1.
Reverse Transcription-Polymerase Chain Reaction
RT-PCR experiments were conducted as described previously (Zhang et al., 2018). Total RNA was isolated from P. putida KT2440 cells grown to mid-log phase in MSM supplemented with different carbon sources using Qiagen RNeasy total RNA Kit. DNA contamination was eliminated by RNase-free DNase I treatment. cDNA was generated using Superscript II RT Kit. Samples were incubated at 25°C for 10 min and 42°C for 30 min, then heated at 85°C for 5 min. RT-PCR was performed with Taq DNA Polymerase (Transgen, China) using corresponding oligonucleotides (Supplementary Table S2). The genomic DNA and total RNA of P. putida KT2440 were used as positive and negative controls, respectively.
Expression and Purification of R,R-BDH
The gene encoding R,R-BDH was amplified from genomic DNA of P. putida KT2440 using primers pp0552-F/pp0552-R (Supplementary Table S2). The PCR product was cloned into the pETDuet-1 plasmid to obtain plasmid pETDuet-pp0552. E. coli BL21(DE3) containing pETDuet-pp0552 was grown to an optical density at 600 nm (OD600nm) of 0.6–0.8 in LB medium with 100 μg mL−1 ampicillin at 37°C, 180 rpm and induced at 16°C with 1 mM isopropyl-D-1-thiogalactopyranoside (IPTG). The cells were harvested, washed twice, and resuspended in buffer A (pH 7.4, 20 mM sodium phosphate and 500 mM sodium chloride), then lysed by sonication in an ice bath after the addition of 1 mM phenylmethanesulphonyl fluoride (PMSF) and 10% glycerol (vol/vol). The cell lysate was centrifuged at 12,000 × g for 40 min at 4°C to remove cell debris. The supernatant was loaded onto a HisTrap HP column (5 mL) and eluted with buffer B (pH 7.4, 20 mM sodium phosphate, 500 mM sodium chloride, and 500 mM imidazole). The quality of purified protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 12.5% polyacrylamide gels and the concentration was determined by the Bradford protein assay Kit.
Determination of the Native Molecular Weight of R,R-BDH
The native molecular weight of R,R-BDH was estimated by using a gel filtration column (Superdex 200 10/300 GL; GE Healthcare, United States) as described previously (Zhang et al., 2019) with the eluent buffer (pH 7.2, 50 mM sodium phosphate and 150 mM sodium chloride) at a flow rate of 0.5 mL min−1. Thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), and ribonuclease A (13.7 kDa) were used as standard proteins.
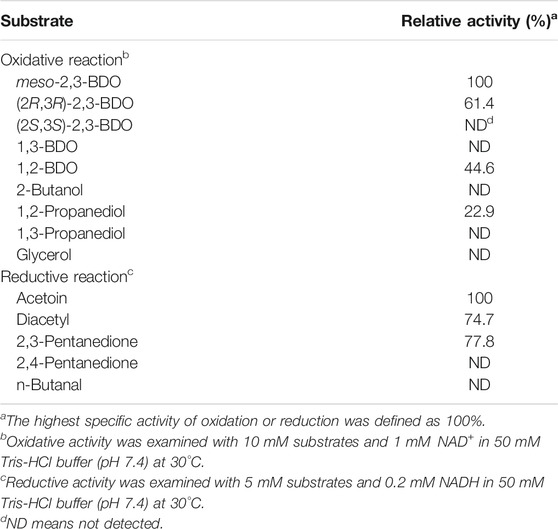
Enzymatic Assays of R,R-BDH
The activity of R,R-BDH was assayed by measuring the change in absorbance at 340 nm at 30°C. For assay of the oxidative activity, the reaction solution contained 10 mM substrates and 1 mM NAD(P)+ in Tris-HCl buffer (pH 7.4, 50 mM). For assay of the reductive activity, the reaction solution contained 5 mM substrates and 0.2 mM NAD(P)H. One unit of enzyme activity was defined as the amount of enzyme that consumed or produced 1 μmol of NAD(P)H per min.
To determine the reductive product of the R,R-BDH, the 1 ml reaction solution containing 1 mM AC or diacetyl, 1 mM NADH, 10 U purified R,R-BDH in Tris-HCl (pH 7.4, 50 mM) was incubated at 30°C and 180 rpm for 1 h. The solutions were centrifuged at 13,000 × g for 10 min and the supernatant was used for detection. The concentrations of 2,3-BDO and AC were analyzed by a gas chromatography system (Varian 3800, Varian) equipped with a flame ionization detector and a 30-m SPB-5 capillary column (0.32-mm inner diameter, 0.25-μm film thickness; Supelco, Bellefonte, PA) using nitrogen as the carrier gas. The injector and detector temperatures were 280°C while the column oven temperature was maintained at 40°C for 3 min and then raised to 240°C at a rate of 20°C min−1 (Ma et al., 2009; Xu et al., 2014). The stereo configuration of 2,3-BDO produced from AC or diacetyl was detected by a gas chromatography system (Agilent GC6820) equipped with a flame ionization detector and a fused silica capillary column (Supelco Beta DEX TM 120, inside diameter, 0.25 mm; length, 30 m) using nitrogen as the carrier gas. The injector and detector temperatures were 280°C while the column oven temperature was maintained at 40°C for 3 min, raised to 80°C at a rate of 1.5°C min−1, then increased to 86°C at a rate of 0.5°C min−1 and finally raised to 200°C at a rate of 30°C min−1 (Xiao et al., 2010).
Construction of P. putida KT2440 Mutants
To construct the Δpp0552 mutant strain of P. putida KT2440, the upstream and downstream homologous arms of the pp0552 gene were amplified using the primers pp0552-up-F/R and pp0552-down-F/R, respectively (Supplementary Table S2). Homologous arms were fused together via recombinant PCR and amplified with the primers pp0552-up-F and pp0552-down-R. The product was inserted into a mobilizable plasmid pK18mobsacB (Schäfer et al., 1994), which does not replicate in P. putida, to construct pK18mobsacB-Δpp0552. Then the plasmid was transformed into P. putida KT2440 by electroporation and integrated into the chromosome. The correct single-crossover mutants were selected on LB plate supplemented with 50 μg mL−1 kanamycin. The double-crossover allelic exchange mutant strains were screened on LB plates containing 10% (wt/vol) sucrose. The pedH and pedE mutants of P. putida KT2440 were constructed by using the same procedure.
Results
Utilization of Three Stereoisomers of 2,3-BDO by P. putida KT2440
Rhizosphere is a natural habitat of different microorganism. Some rhizobacteria such as Aerobacter spp., Bacillus spp., and Serratia spp., can produce three stereoisomers of 2,3-BDO as the volatile compounds to trigger induced systemic resistance and promote plant growth (Ryu et al., 2003; Han et al., 2006; Taghavi et al., 2010). P. putida KT2440 is a widely studied rhizosphere-dwelling microorganism. Growth of P. putida KT2440 in minimal salt medium (MSM) supplemented with different stereoisomers of 2,3-BDO was firstly studied. As shown in Figure 1, P. putida KT2440 grew robustly in all three 2,3-BDO stereoisomers with the consumption of each substrate.
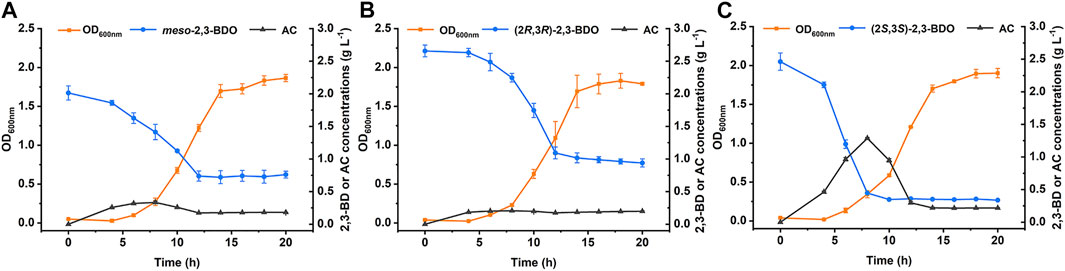
FIGURE 1. Growth of P. putida KT2440 in three stereoisomers of 2,3-BDO. The biomass, 2,3-BDO consumption, and AC production were measured in MSM supplemented with 2 g L−1 different stereoisomers of 2,3-BDO as the sole carbon source. (A)meso-2,3-BDO. (B) (2R,3R)-2,3-BDO. (C) (2S,3S)-2,3-BDO. Error bars indicate deviation from the means (n = 3).
Comparative Genomics of 2,3-BDO Utilization Operon in P. putida KT2440
Recently, a 2,3-BDO utilization operon was identified in P. aeruginosa PAO1 (Liu et al., 2018). Comparative genomics analysis indicated that PP0555, PP0554, and PP0553 exhibit strikingly high homology to AcoA, AcoB, and AcoC in AC dehydrogenase enzyme system (AoDH ES) of P. aeruginosa PAO1. R,R-BDH and (2S,3S)-2,3-BDO dehydrogenase (S,S-BDH) encoded in 2,3-BDO utilization operon of P. aeruginosa PAO1 catalyze three 2,3-BDO stereoisomers into AC and then AC is cleaved by AoDH ES into acetyl-CoA and acetaldehyde. As shown in Figure 2A, a putative R,R-BDH (PP0552) is also annotated in the genome of P. putida KT2440. Its protein sequence exhibits high homology to R,R-BDHs of P. aeruginosa PAO1 (86.2% sequence identity), Bacillus cereus YUF-4 (44.9% sequence identity), Paenibacillus polymyxa ATCC 12321 (41.8% sequence identity), P. polymyxa ZJ-9 (38.2% sequence identity), Rhodococcus erythropolis WZ010 (38.0% sequence identity), and Saccharomyces cerevisiae S288C (31.7% sequence identity) (Supplementary Figure S1).
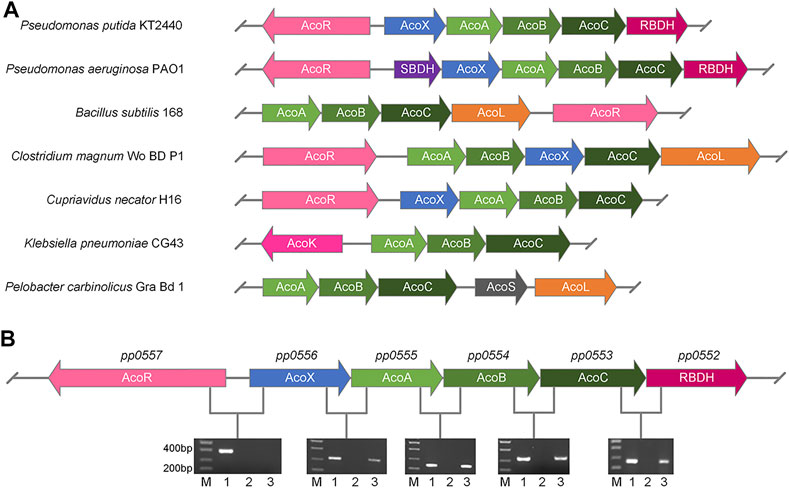
FIGURE 2. Identification of R,R-BDH in 2,3-BDO utilization operon of P. putida KT2440. (A) Gene clusters arrangement of genes for 2,3-BDO and AC catabolism in different species. (B) Co-transcription analysis of genes in 2,3-BDO utilization operon of P. putida KT2440 by RT-PCR (lane 3). The genomic DNA (lane 1) and RNA (lane 2) of P. putida KT2440 were used as positive and negative control, respectively. P. putida KT2440 were cultured in MSM supplemented with 2 g L−1 mixed 2,3-BDO as the sole carbon source.
Since pp0555, pp0554, pp0553 and pp0552 were located adjacent to acoX (pp0556) and acoR (pp0557) gene homologues of AC utilization operons in other bacteria, we hypothesized that these genes may also comprise a 2,3-BDO utilization operon in P. putida KT2440 (Figure 2A). The mRNA from cells of P. putida KT2440 grown in 2,3-BDO was extracted and reverse-transcribed into cDNA as template for reverse transcription-polymerase chain reaction (RT-PCR). As shown in Figure 2B, the five genes including pp0556, pp0555, pp0554, pp0553 and pp0552 were co-transcribed and might be regulated by AcoR.
Enzymatic Properties of R,R-BDH in P. putida KT2440
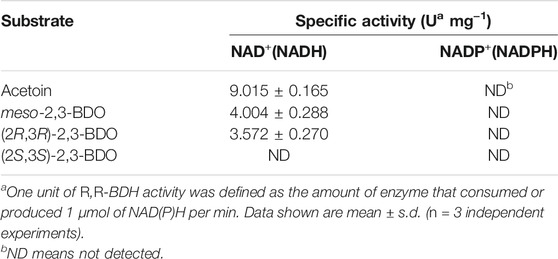
The putative R,R-BDH in P. putida KT2440 was overexpressed and then purified by using a HisTrap column (Figure 3A). Based on the mass of the R,R-BDH monomer and the results of size exclusion chromatography, this protein exists as a homotetramer (Figure 3B). For the oxidative reaction, R,R-BDH can catalyze NAD-dependent dehydrogenation of (2R,3R)-2,3-BDO and meso-2,3-BDO. For the reductive reaction, R,R-BDH can catalyze NADH-dependent reduction of diacetyl and AC (Table 1 and Table 2). It had no activity toward 1,3-butanediol, 2-butanol, 1,3-propanediol, and glycerol.
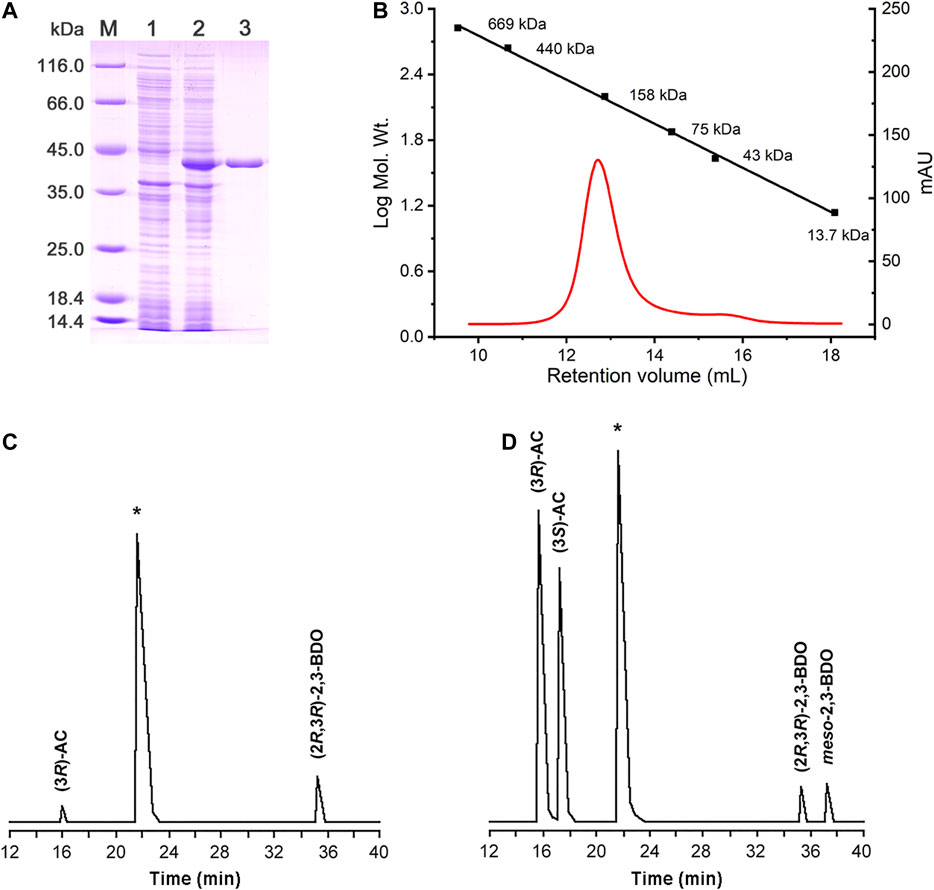
FIGURE 3. Enzymatic characterization of R,R-BDH in P. putida KT2440. (A) SDS-PAGE analysis of the purified R,R-BDH. Lane M, molecular weight markers; Lane 1, crude cell extract of E. coli BL21 (DE3) (blank control); Lane 2, crude cell extract of E. coli BL21 (DE3) harboring pETDuet-pp0552; Lane 3, purified R,R-BDH. (B) Determination of the native molecular weight of R,R-BDH by gel filtration column. GC analysis of 2,3-BDO produced by R,R-BDH catalyzed reduction of diacetyl (C) and racemic AC (D).
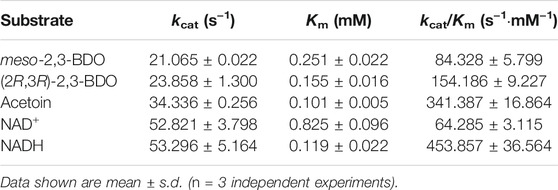
The kinetic parameters of R,R-BDH in P. putida KT2440 toward different substrates were determined (Table 3). In oxidative direction, double-reciprocal plots of the initial rates plotted against the concentrations of (2R,3R)-2,3-BDO, meso-2,3-BDO, and cofactor NAD+ yielded Km values of 0.155 ± 0.016 mM, 0.251 ± 0.022, and 0.825 ± 0.096 mM, respectively. The kcat values were estimated to be 23.858 ± 1.300 s−1, 21.065 ± 0.022 s−1, and 52.821 ± 3.798 s−1, respectively. R,R-BDH in P. putida KT2440 exhibited higher catalytic efficiency toward meso-2,3-BDO and (2R,3R)-2,3-BDO dehydrogenation than other reported R,R-BDHs (Supplementary Table S3). In reductive direction, the apparent Km values of R,R-BDH toward AC and NADH was 0.101 ± 0.005 mM and 0.119 ± 0.022 mM, respectively. The kcat values were estimated to be 34.336 ± 0.256 s−1 and 53.296 ± 5.164 s−1, respectively.
The effects of pH and temperature on the enzyme activity and stability of R,R-BDH were examined. The optimal pH for oxidative activity of R,R-BDH toward (2R,3R)-2,3-BDO (Supplementary Figure S2A) and meso-2,3-BDO (Supplementary Figure S2B) is 9.0, while for reductive activity toward AC is 8.0 (Supplementary Figure S2C). The enzyme is more stable under the pH range of 6.0–8.0 in citrate-Na2HPO4 buffer (Supplementary Figure S2D). The optimum temperature for R,R-BDH is about 70°C (Supplementary Figure S3A) and it is stable under 50°C (Supplementary Figure S3B). The effect of metal ions on the activity of R,R-BDH was tested by adding different metal salts (final concentration of 1 mM) to the assay buffer. As shown in Supplementary Figure S4, Fe2+, Mn2+, Co2+, Ca2+, Ba2+, Mg2+, and K+ have positively effects on R,R-BDH activity, while Zn2+, Cd2+, Cu2+, Ni2+, and EDTA have negative effects.
2,3-BDHs catalyze the stereoselective interconversion between diacetyl, 2,3-BDO and AC. As shown in Figure 3C, diacetyl was reduced into (3R)-AC and (2R,3R)-2,3-BDO by R,R-BDH, whereas racemic AC was reduced to (2R,3R)-2,3-BDO and meso-2,3-BDO (Figure 3D). These results indicated that R,R-BDH catalyzes the R-specific oxidation-reduction of hydroxyl and keto groups in its substrates and (2R,3R)-2,3-BDO and meso-2,3-BDO might be converted to (3R)-AC and (3S)-AC, respectively.
Functions of R,R-BDH in (2R,3R)-2,3-BDO and meso-2,3-BDO Utilization of P. putida KT2440
RT-PCR fragment of pp0552 was obtained from cells of P. putida KT2400 cultured with (2R,3R)-2,3-BDO, meso-2,3-BDO and (2S,3S)-2,3-BDO (Supplementary Figure S5A), indicating the transcription of pp0552 during 2,3-BDO utilization. To identify the role of R,R-BDH in 2,3-BDO utilization, its encoding gene pp0552 was knocked out in P. putida KT2440. P. putida KT2440 (Δpp0552) was cultivated in MSM supplemented with 2 g L−1meso-2,3-BDO, (2R,3R)-2,3-BDO, (2S,3S)-2,3-BDO, racemic AC or pyruvate. As shown in Figure 4, the disruption of pp0552 had no effect on the ability of P. putida KT2440 (Δpp0552) to grow in racemic AC and pyruvate but impaired its ability to utilize meso-2,3-BDO and (2R,3R)-2,3-BDO. This result indicated that R,R-BDH plays indispensable roles in meso-2,3-BDO and (2R,3R)-2,3-BDO catabolism, definitely through converting these two stereoisomers into AC. P. putida KT2440 (Δpp0552) could still utilize (2S,3S)-2,3-BDO, suggesting the presence of other proteins responding for (2S,3S)-2,3-BDO catabolism.
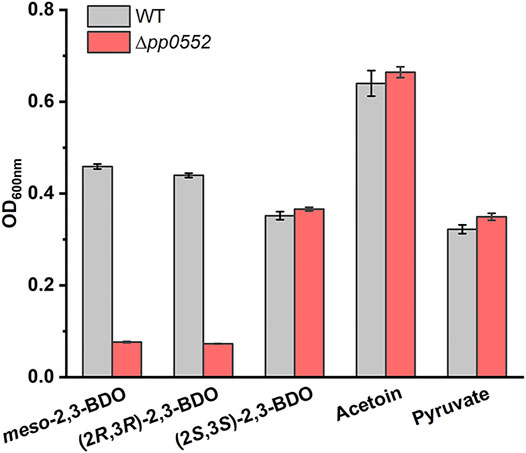
FIGURE 4. Identification of the physiological function of R,R-BDH in 2,3-BDO catabolism. P. putida KT2440 and P. putida KT2440 (Δpp0552) were cultured in MSM with 2 g L−1meso-2,3-BDO, (2R,3R)-2,3-BDO, (2S,3S)-2,3-BDO, racemic AC, or pyruvate as the sole carbon source at 30°C for 30 h by using a Bioscreen microbiology reader. Error bars indicate deviation from the means (n = 3).
Quinoprotein Ethanol Dehydrogenases Involved in (2S,3S)-2,3-BDO Utilization of P. putida KT2440
Two pyrroloquinoline quinone (PQQ)-dependent ethanol dehydrogenases (PQQ-EDHs), PP2674 (PedE) and PP2679 (PedH), were annotated in the genome of P. putida KT2440. PedH and PedE exhibit enzyme activity toward a range of substrates, including 2,3-BDO (Takeda et al., 2013; Wehrmann et al., 2017). In addition, transcription of pedE and pedH was also observed during (2R,3R)-2,3-BDO, meso-2,3-BDO and (2S,3S)-2,3-BDO utilization of P. putida KT2440 (Supplementary Figure S5B and Supplementary Figure S5C). This makes us wonder whether these two PQQ-EDHs also participate in 2,3-BDO catabolism of P. putida KT2440. Individual or combinatorial mutant strains of PedE, PedH, and R,R-BDH were constructed. As shown in Figure 5A and Figure 5B, knock out of pedE and/or pedH did not impair the growth of P. putida KT2440 in meso-2,3-BDO and (2R,3R)-2,3-BDO. P. putida KT2440 (ΔpedE) displayed no growth in MSM with (2S,3S)-2,3-BDO as sole carbon source, while P. putida KT2440 (ΔpedH) still grew normally under the same condition (Figure 5C).
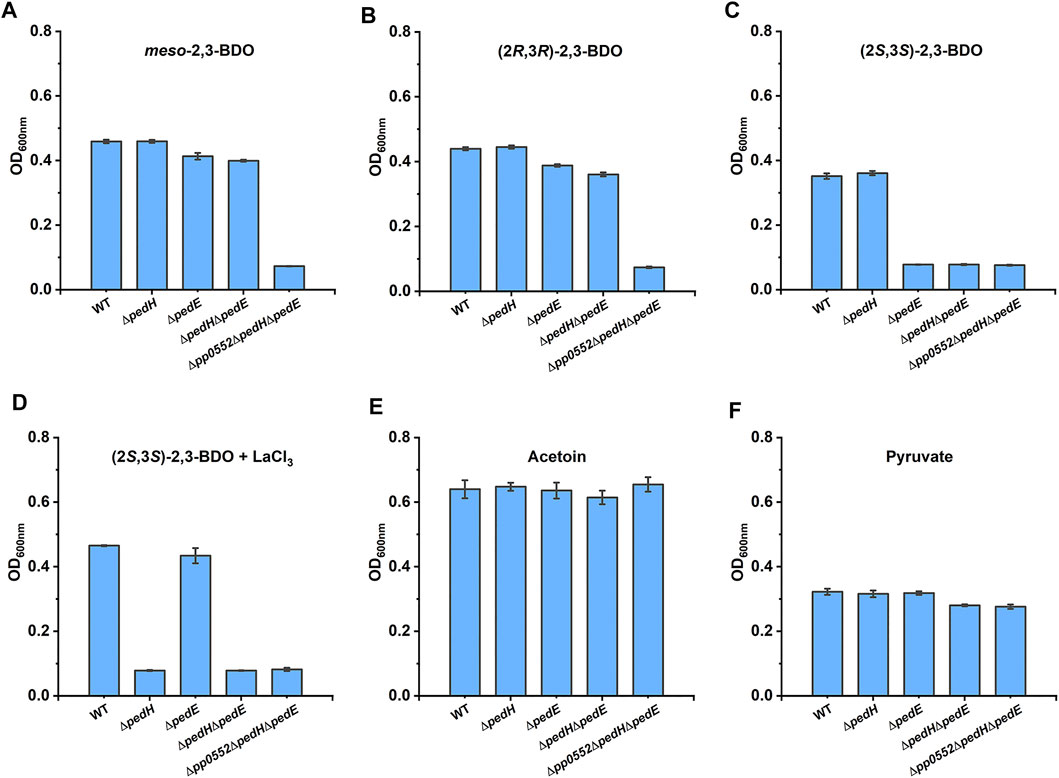
FIGURE 5. Identification of the physiological function of PedE and PedH in 2,3-BDO catabolism. Individual and combinatorial deletion mutants of P. putida KT2440 were cultured in MSM supplemented with 2 g L−1meso-2,3-BDO (A), (2R,3R)-2,3-BDO (B), (2S,3S)-2,3-BDO (C), (2S,3S)-2,3-BDO with 20 μM LaCl3(D), racemic AC (E) and pyruvate (F) as the sole carbon source at 30°C for 30 h by using a Bioscreen microbiology reader. Error bars indicate deviation from the means (n = 3).
PedE depends on Ca2+ as a cofactor (Takeda et al., 2013), while PedH exhibits enzyme activity only in the presence of La3+ (Wehrmann et al., 2017). Thus, 20 μM La3+ was added in MSM with (2S,3S)-2,3-BDO as the sole carbon source, which resulted in robust growth of P. putida KT2440 (ΔpedE) (Figure 5D). Double-knockout strain P. putida KT2440 (ΔpedEΔpedH) was completely unable to utilize (2S,3S)-2,3-BDO both in the presence and absence of La3+ (Figures 5C,D) and there might be no other alcohol dehydrogenase catalyzing dehydrogenation of (2S,3S)-2,3-BDO in the strain. PedE and PedH function redundantly in (2S,3S)-2,3-BDO catabolism, but work under different conditions. The triple-knockout strain P. putida KT2440 (Δpp0552ΔpedEΔpedH) grew normally in AC (Figure 5E) and pyruvate (Figure 5F), while lost growth ability in either stereoisomer of 2,3-BDO (Figures 5A–D). Thus, P. putida KT2440 uses R,R-BDH, PedH, and PedE to utilize different 2,3-BDO stereoisomers.
Lanthanide Concentration-dependent Growth of Derivative Strains of P. putida KT2440 in (2S,3S)-2,3-BDO
The transcription of pedE and pedH is tightly controlled by a lanthanide-mediated switch in P. putida KT2440 (Wehrmann et al., 2017; Wehrmann et al., 2018). In the presence of La3+, pedE transcription is activated while pedH is repressed. Thus, the expression of PedH and PedE will be inversely regulated in response to various lanthanide concentrations and the lanthanide-mediated switch also influences the PQQ-EDH dependent growth of P. putida KT2440. Growth of derivative strains of P. putida KT2440 in (2S,3S)-2,3-BDO was assayed in the presence of various concentration of La3+.
As shown in Figure 6B, the growth of P. putida KT2440 (ΔpedE) in (2S,3S)-2,3-BDO was activated by increasing La3+ concentrations. In contrast, La3+ concentrations-dependent growth inhibition of P. putida KT2440 (ΔpedH) was also detected (Figure 6C). P. putida KT2440 (ΔpedH) did not grow in (2S,3S)-2,3-BDO when La3+ concentration was higher than 100 nM. Importantly, the growth of wild type P. putida KT2440 was not affected whether exogenous La3+ was existent or not (Figure 6A). P. putida KT2440 may be conferred a strong survivability by the redundancy of PedH and PedE expressed under different conditions.
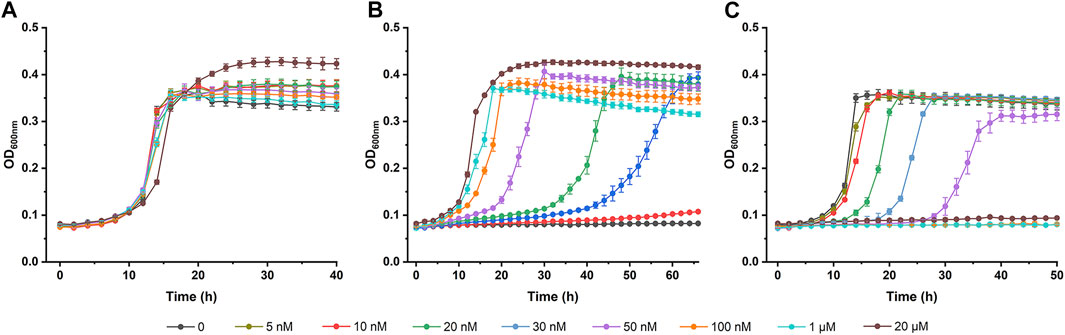
FIGURE 6. Lanthanide concentration-dependent growth of derivative strains of P. putida KT2440 in (2S,3S)-2,3-BDO. P. putida KT2440 wild-type (A), ΔpedE(B) and ΔpedH(C) mutant strains were cultured in MSM supplemented with 2 g L−1 (2S,3S)-2,3-BDO with various concentrations of LaCl3 at 30°C by using a Bioscreen microbiology reader. Error bars indicate deviation from the means (n = 3).
Discussion
2,3-BDO is widely distributed in various environments (Ryu et al., 2003; Yi et al., 2016). As a Voges-Proskauer reaction negative strain lacking the ability to produce 2,3-BDO, P. putida KT2440 can use all three stereoisomers of 2,3-BDO for its growth. The ability to efficiently use three stereoisomers of 2,3-BDO may be important for P. putida to survive and acquire nutrients in habitats like rhizosphere. Dehydrogenation to AC is the first step of 2,3-BDO catabolism and then AC will be cleaved into acetaldehyde and acetyl-CoA by AoDH ES (Xiao and Xu., 2007; Liu et al., 2018). In this work, we identified the dehydrogenation mechanism of three stereoisomers of 2,3-BDO: (2R,3R)-2,3-BDO, meso-2,3-BDO, and (2S,3S)-2,3-BDO in P. putida KT2440 (Figure 7). R,R-BDH catalyzes the dehydrogenation of (2R,3R)-2,3-BDO and meso-2,3-BDO with nearly identical kcat values, 23.858 ± 1.300 s−1 and 21.065 ± 0.022 s−1. Similar growth and consumption of substate were observed when P. putida KT2440 was cultured with (2R,3R)-2,3-BDO and meso-2,3-BDO (Figures 1A,B). PedE and PedH catalyze the irreversible dehydrogenation of (2S,3S)-2,3-BDO into (3S)-AC. Quickly consumption of (2S,3S)-2,3-BDO and temporary accumulation of AC can be detected during the (2S,3S)-2,3-BDO utilization of P. putida KT2440 (Figure 1C).
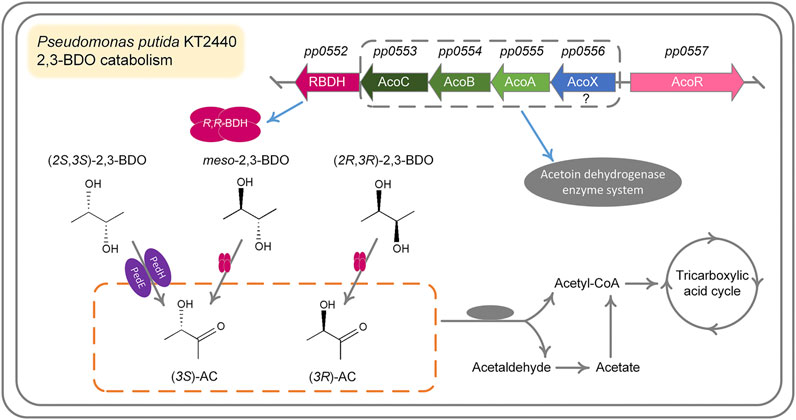
FIGURE 7. Proposed mechanism for 2,3-BDO catabolism in P. putida KT2440. meso-2,3-BDO and (2R,3R)-2,3-BDO are dehydrogenized by R,R-BDH to (3S)-AC and (3R)-AC, respectively. Dehydrogenation of (2S, 3S)-2,3-BDO to (3S)-AC is catalyzed by two PQQ-EDHs, PedH and PedE encoded by pp2679 and pp2674, respectively. (3S)-AC and (3R)-AC are subsequently cleaved by AoDH ES to acetyl-CoA and acetaldehyde for further metabolism.
BDHs in Voges-Proskauer reaction positive strains can be divided into MDR family and SDR family (Radoš et al., 2016; Liu et al., 2018). R,R-BDH belonging to MDR family is critical for (2R,3R)-2,3-BDO and meso-2,3-BDO generation and reutilization, while meso-BDH belonging to the SDR family is responsible for (2S,3S)-2,3-BDO and meso-2,3-BDO production and reutilization. The genes encoding R,R-BDH and S,S-BDH, which is only active on (2S,3S)-2,3-BDO and specifically involved in (2S,3S)-2,3-BDO metabolism, are also present in Voges-Proskauer reaction negative P. aeruginosa PAO1 and belong to an operon (Liu et al., 2018). Although there is no meso-BDH and S,S-BDH encoding genes in P. putida KT2440, this strain can still catabolize (2S,3S)-2,3-BDO through two PQQ-EDHs, PedE and PedH.
PedE and PedH exhibited enzyme activity toward many linear and aromatic primary and secondary alcohols, including 2,3-BDO (Takeda et al., 2013; Wehrmann et al., 2017). It was recently reported that mutant strains with either single-knockout or double-knockout of PedE and PedH could still grow on the plate with 2,3-BDO as sole carbon source (Wehrmann et al., 2017). This phenomenon is presumably because that mixed 2,3-BDO of three stereoisomers was used to culture those mutant strains. Although the catabolism of (2S,3S)-2,3-BDO was abolished due to the deletion of PedE and PedH, R,R-BDH could still catalyze the dehydrogenation of (2R,3R)-2,3-BDO and meso-2,3-BDO in the mixture and support the growth. In addition, we demonstrated that PedE and PedH are specifically involved in (2S,3S)-2,3-BDO metabolism. In spite of the presence of PedE and PedH, P. putida KT2440 (Δpp0552) could not grow in meso-2,3-BDO and (2R,3R)-2,3-BDO. Thus, when using 2,3-BDO as the substrate, PQQ-EDHs may be stereospecific and only active on (2S,3S)-2,3-BDO.
The growth of P. putida KT2440 (ΔpedE) in (2S,3S)-2,3-BDO was exclusively stimulated by increased concentrations of lanthanum. In contrast, the growth of P. putida KT2440 (ΔpedH) in (2S,3S)-2,3-BDO was inhibited by lanthanum in a concentration-dependent manner. These results were due to the inverse regulation of PedE and PedH by lanthanide availability, an interesting phenomenon called as lanthanide-mediated switch in several methylotrophic organisms and P. putida KT2440 (Farhan Ul Haque et al., 2015; Chu et al., 2016; Vu et al., 2016; Wehrmann et al., 2017; Wehrmann et al., 2018). Since low concentrations of lanthanum could still inhibit the growth of P. putida KT2440 (ΔpedH) in (2S,3S)-2,3-BDO at the presence of Ca2+, the lanthanide-dependent PedH seems to be the preferred PQQ-EDH when both metals are simultaneously available (Chu and Lidstrom, 2016; Vu et al., 2016; Wehrmann et al., 2017). In addition, the functional redundancy and inverse regulation of PQQ-EDHs provides a mechanism through which P. putida KT2440 can optimize its growth according to the availability of lanthanide under variable environmental conditions (Markert, 1987; Aubert et al., 2002).
P. putida KT2440 can colonize rhizospheres of different plants and induce changes in root exudation of plants like Arabidopsis (Matilla et al., 2010). Interestingly, plant root exudates also regulate expression of genes for 2,3-BDO catabolism and synthesis of some antifungal compounds in rhizosphere Pseudomonas (Mavrodi et al., 2021). As an extracellular metabolite, 2,3-BDO can act as a signaling molecule influencing the action of various organisms in symbiotic habitat of 2,3-BDO producing strains. For example, 2,3-BDO can be a kairomone or pheromone attracting insects and influencing insect propagation (Rochat et al., 2000; Moore et al., 2002). P. putida KT2440 can use endogenous chemical signals to coordinate the expression of various genes (Espinosa-Urgel and Ramos, 2004). Whether 2,3-BDO acts as a signaling molecule of P. putida KT2440 like the situation in insects needs further research. In addition, diverse mechanisms for dehydrogenation of three 2,3-BDO stereoisomers in P. putida KT2440 were ascertained in this work, whether and how the three isomers exhibit different signaling functions also deserve intensive investigation.
In summary, we proved here that P. putida KT2440 is able to utilize all three stereoisomers of 2,3-BDO for growth and revealed the dehydrogenation mechanisms of these stereoisomers. Catabolism of 2,3-BDO generates two molecules of acetyl-CoA without carbon loss (Yang et al., 2020) and thus 2,3-BDO is an ideal substrate for the production of chemicals using acetyl-CoA as the precursor. Chemical production of 2,3-BDO based on nonrenewable resources often results in a mixture of three stereoisomeric forms. Many microorganisms have been used to efficiently produce 2,3-BDO but most of them also produce a mixture of two or three stereoisomers of 2,3-BDO. The results reported in this work thus also provide a molecular basis for efficient utilization of chemical or biological produced 2,3-BDO by P. putida KT2440.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
YL, XW, ML, and LM performed experiments. YL, CL, and CM wrote the manuscript and conceived the study. CL, YL, XW, ML, LM, and WZ were involved in analysis and interpretation of experimental data. CG, CM, and PX coordinated the project.
Funding
This work was supported by the grants of National Key R&D Program of China (2019YFA0904900 and 2019YFA0904803), Natural Science Foundation of Shandong Provincial (ZR2018PC008 and ZR2020MC005), Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-005). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Xiangmei Ren from Core Facilities for Life and Environmental Sciences (State Key Laboratory of Microbial Technology, Shandong University) for assistance in gas chromatography analysis and the use of Bioscreen microbiology reader.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.728767/full#supplementary-material
References
Aubert, D., Stille, P., Probst, A., Gauthier-lafaye, F., Pourcelot, L., and Del Nero, M. (2002). Characterization and Migration of Atmospheric REE in Soils and Surface Waters. Geochim. Cosmochim. Acta 66, 3339–3350. doi:10.1016/S0016-7037(02)00913-4
Bator, I., Wittgens, A., Rosenau, F., Tiso, T., and Blank, L. M. (2020). Comparison of Three Xylose Pathways in Pseudomonas putida KT2440 for the Synthesis of Valuable Products. Front. Bioeng. Biotechnol. 7, 480. doi:10.3389/fbioe.2019.00480
Bentley, G. J., Narayanan, N., Jha, R. K., Salvachúa, D., Elmore, J. R., Peabody, G. L., et al. (2020). Engineering Glucose Metabolism for Enhanced Muconic Acid Production in Pseudomonas putida KT2440. Metab. Eng. 59, 64–75. doi:10.1016/j.ymben.2020.01.001
Celińska, E., and Grajek, W. (2009). Biotechnological Production of 2,3-Butanediol-Current State and Prospects. Biotechnol. Adv. 27, 715–725. doi:10.1016/j.biotechadv.2009.05.002
Chu, F., Beck, D. A. C., and Lidstrom, M. E. (2016). MxaY Regulates the Lanthanide-Mediated Methanol Dehydrogenase Switch in Methylomicrobium Buryatense. PeerJ 4, e2435. doi:10.7717/peerj.2435
Chu, F., and Lidstrom, M. E. (2016). XoxF Acts as the Predominant Methanol Dehydrogenase in the Type I Methanotroph Methylomicrobium buryatense. J. Bacteriol. 198, 1317–1325. doi:10.1128/JB.00959-15
Chu, H., Xin, B., Liu, P., Wang, Y., Li, L., Liu, X., et al. (2015). Metabolic Engineering of Escherichia coli for Production of (2S,3S)-Butane-2,3-Diol from Glucose. Biotechnol. Biofuels 8, 143. doi:10.1186/s13068-015-0324-x
Dvořák, P., and de Lorenzo, V. (2018). Refactoring the Upper Sugar Metabolism of Pseudomonas putida for Co-utilization of Cellobiose, Xylose, and Glucose. Metab. Eng. 48, 94–108. doi:10.1016/j.ymben.2018.05.019
Espinosa-Urgel, M., and Ramos, J. L. (2004). Cell Density-dependent Gene Contributes to Efficient Seed Colonization by Pseudomonas putida KT2440. Appl. Environ. Microbiol. 70, 5190–5198. doi:10.1128/AEM.70.9.5190-5198.2004
Farhan Ul Haque, M., Kalidass, B., Bandow, N., Turpin, E. A., DiSpirito, A. A., and Semrau, J. D. (2015). Cerium Regulates Expression of Alternative Methanol Dehydrogenases in Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 81, 7546–7552. doi:10.1128/AEM.02542-15
Franden, M. A., Jayakody, L. N., Li, W. J., Wagner, N. J., Cleveland, N. S., Michener, W. E., et al. (2018). Engineering Pseudomonas putida KT2440 for Efficient Ethylene Glycol Utilization. Metab. Eng. 48, 197–207. doi:10.1016/j.ymben.2018.06.003
Ge, Y., Li, K., Li, L., Gao, C., Zhang, L., Ma, C., et al. (2016). Contracted but Effective: Production of Enantiopure 2,3-Butanediol by Thermophilic and GRAS Bacillus licheniformis. Green Chem. 18, 4693–4703. doi:10.1039/c6gc01023g
Han, S. H., Lee, S. J., Moon, J. H., Park, K. H., Yang, K. Y., Cho, B. H., et al. (2006). GacS-dependent Production of 2R, 3R-Butanediol by Pseudomonas chlororaphis O6 Is a Major Determinant for Eliciting Systemic Resistance against Erwinia carotovora but Not against Pseudomonas syringae pv. Tabaci in Tobacco. Mol. Plant Microbe Interact. 19, 924–930. doi:10.1094/MPMI-19-0924
Ji, X. J., Huang, H., and Ouyang, P. K. (2011). Microbial 2,3-Butanediol Production: A State-of-the-Art Review. Biotechnol. Adv. 29, 351–364. doi:10.1016/j.biotechadv.2011.01.007
Johnson, C. W., and Beckham, G. T. (2015). Aromatic Catabolic Pathway Selection for Optimal Production of Pyruvate and Lactate from Lignin. Metab. Eng. 28, 240–247. doi:10.1016/j.ymben.2015.01.005
Li, W. J., Jayakody, L. N., Franden, M. A., Wehrmann, M., Daun, T., Hauer, B., et al. (2019). Laboratory Evolution Reveals the Metabolic and Regulatory Basis of Ethylene Glycol Metabolism by Pseudomonas putida KT2440. Environ. Microbiol. 21, 3669–3682. doi:10.1111/1462-2920.14703
Lian, J., Chao, R., and Zhao, H. (2014). Metabolic Engineering of a Saccharomyces cerevisiae Strain Capable of Simultaneously Utilizing Glucose and Galactose to Produce Enantiopure (2R,3R)-Butanediol. Metab. Eng. 23, 92–99. doi:10.1016/j.ymben.2014.02.003
Liu, Q., Liu, Y., Kang, Z., Xiao, D., Gao, C., Xu, P., et al. (2018). 2,3-Butanediol Catabolism in Pseudomonas aeruginosa PAO1. Environ. Microbiol. 20, 3927–3940. doi:10.1111/1462-2920.14332
Liu, Z., Qin, J., Gao, C., Hua, D., Ma, C., Li, L., et al. (2011). Production of (2S,3S)-2,3-Butanediol and (3S)-Acetoin from Glucose Using Resting Cells of Klebsiella pneumonia and Bacillus subtilis. Bioresour. Technol. 102, 10741–10744. doi:10.1016/j.biortech.2011.08.110
Ma, C., Gao, C., Qiu, J., Hao, J., Liu, W., Wang, A., et al. (2007). Membrane-bound L- and D-Lactate Dehydrogenase Activities of a Newly Isolated Pseudomonas stutzeri Strain. Appl. Microbiol. Biotechnol. 77, 91–98. doi:10.1007/s00253-007-1132-4
Ma, C., Wang, A., Qin, J., Li, L., Ai, X., Jiang, T., et al. (2009). Enhanced 2,3-Butanediol Production by Klebsiella pneumoniae SDM. Appl. Microbiol. Biotechnol. 82, 49–57. doi:10.1007/s00253-008-1732-7
Markert, B. (1987). The Pattern of Distribution of Lanthanide Elements in Soils and Plants. Phytochemistry 26, 3167–3170. doi:10.1016/S0031-9422(00)82463-2
Matilla, M. A., Ramos, J. L., Bakker, P. A. H. M., Doornbos, R., Badri, D. V., Vivanco, J. M., et al. (2010). Pseudomonas putida KT2440 Causes Induced Systemic Resistance and Changes in Arabidopsis Root Exudation. Environ. Microbiol. Rep. 2, 381–388. doi:10.1111/j.1758-2229.2009.00091.x
Mavrodi, O. V., McWilliams, J. R., Peter, J. O., Berim, A., Hassan, K. A., Elbourne, L. D. H., et al. (2021). Root Exudates Alter the Expression of Diverse Metabolic, Transport, Regulatory, and Stress Response Genes in Rhizosphere Pseudomonas. Front. Microbiol. 12, 651282. doi:10.3389/fmicb.2021.651282
Moore, A. J., Haynes, K. F., Preziosi, R. F., and Moore, P. J. (2002). The Evolution of Interacting Phenotypes: Genetics and Evolution of Social Dominance. Am. Nat. 160, S186–S197. doi:10.1086/342899
Nelson, K. E. (2002). The Complete Genome Sequence of Pseudomonas putida KT2440 Is Finally Available. Environ. Microbiol. 4, 777–778. doi:10.1046/j.1462-2920.2002.00367.x
Nelson, K. E., Weinel, C., Paulsen, I. T., Dodson, R. J., Hilbert, H., Martins dos Santos, V. A. P., et al. (2002). Complete Genome Sequence and Comparative Analysis of the Metabolically Versatile Pseudomonas putida KT2440. Environ. Microbiol. 4, 799–808. doi:10.1046/j.1462-2920.2002.00366.x
Nikel, P. I., and de Lorenzo, V. (2013). Engineering an Anaerobic Metabolic Regime in Pseudomonas putida KT2440 for the Anoxic Biodegradation of 1,3-Dichloroprop-1-Ene. Metab. Eng. 15, 98–112. doi:10.1016/j.ymben.2012.09.006
Nikel, P. I., and de Lorenzo, V. (2018). Pseudomonas putida as a Functional Chassis for Industrial Biocatalysis: from Native Biochemistry to Trans-metabolism. Metab. Eng. 50, 142–155. doi:10.1016/j.ymben.2018.05.005
Poblete-Castro, I., Aravena-Carrasco, C., Orellana-Saez, M., Pacheco, N., Cabrera, A., and Borrero-de Acuña, J. M. (2020). Engineering the Osmotic State of Pseudomonas putida KT2440 for Efficient Cell Disruption and Downstream Processing of Poly(3-Hydroxyalkanoates). Front. Bioeng. Biotechnol. 8, 161. doi:10.3389/fbioe.2020.00161
Radoš, D., Turner, D. L., Catarino, T., Hoffart, E., Neves, A. R., Eikmanns, B. J., et al. (2016). Stereospecificity of Corynebacterium glutamicum 2,3-Butanediol Dehydrogenase and Implications for the Stereochemical Purity of Bioproduced 2,3-Butanediol. Appl. Microbiol. Biotechnol. 100, 10573–10583. doi:10.1007/s00253-016-7860-6
Rochat, D., Meillour, P. N. L., Esteban-Duran, J. R., Malosse, C., Perthuis, B., Morin, J. P., et al. (2000). Identification of Pheromone Synergists in American palm Weevil, Rhynchophorus palmarum, and Attraction of Related Dynamis borassi. J. Chem. Ecol. 26, 155–187. doi:10.1023/A:1005497613214
Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Wei, H. X., Pare, P. W., et al. (2003). Bacterial Volatiles Promote Growth in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 4927–4932. doi:10.1073/pnas.0730845100
Salvachúa, D., Werner, A. Z., Pardo, I., Michalska, M., Black, B. A., Donohoe, B. S., et al. (2020). Outer Membrane Vesicles Catabolize Lignin-Derived Aromatic Compounds in Pseudomonas putida KT2440. Proc. Natl. Acad. Sci. USA 117, 9302–9310. doi:10.1073/pnas.1921073117
Sánchez-Pascuala, A., Fernández-Cabezón, L., de Lorenzo, V., and Nikel, P. I. (2019). Functional Implementation of a Linear Glycolysis for Sugar Catabolism in Pseudomonas putida. Metab. Eng. 54, 200–211. doi:10.1016/j.ymben.2019.04.005
Schäfer, A., Tauch, A., Jäger, W., Kalinowski, J., Thierbach, G., and Pühler, A. (1994). Small Mobilizable Multi-Purpose Cloning Vectors Derived from the Escherichia coli Plasmids pK18 and pK19: Selection of Defined Deletions in the Chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi:10.1016/0378-1119(94)90324-7
Sun, J., Lu, L. B., Liang, T. X., Yang, L. R., and Wu, J. P. (2020). CRISPR-assisted Multiplex Base Editing System in Pseudomonas putida KT2440. Front. Bioeng. Biotechnol. 8, 905. doi:10.3389/fbioe.2020.00905
Taghavi, S., van der Lelie, D., Hoffman, A., Zhang, Y. B., Walla, M. D., Vangronsveld, J., et al. (2010). Genome Sequence of the Plant Growth Promoting Endophytic Bacterium Enterobacter sp. 638. PLoS Genet. 6, e1000943. doi:10.1371/journal.pgen.1000943
Takeda, K., Matsumura, H., Ishida, T., Samejima, M., Igarashi, K., Nakamura, N., et al. (2013). The Two-step Electrochemical Oxidation of Alcohols Using a Novel Recombinant PQQ Alcohol Dehydrogenase as a Catalyst for a Bioanode. Bioelectrochemistry 94, 75–78. doi:10.1016/j.bioelechem.2013.08.001
Tiso, T., Ihling, N., Kubicki, S., Biselli, A., Schonhoff, A., Bator, I., et al. (2020). Integration of Genetic and Process Engineering for Optimized Rhamnolipid Production Using Pseudomonas putida. Front. Bioeng. Biotechnol. 8, 976. doi:10.3389/fbioe.2020.00976
Vu, H. N., Subuyuj, G. A., Vijayakumar, S., Good, N. M., Martinez-Gomez, N. C., and Skovran, E. (2016). Lanthanide-dependent Regulation of Methanol Oxidation Systems in Methylobacterium extorquens AM1 and Their Contribution to Methanol Growth. J. Bacteriol. 198, 1250–1259. doi:10.1128/JB.00937-15
Wada, A., Prates, É. T., Hirano, R., Werner, A. Z., Kamimura, N., Jacobson, D. A., et al. (2021). Characterization of Aromatic Acid/proton Symporters in Pseudomonas putida KT2440 toward Efficient Microbial Conversion of Lignin-Related Aromatics. Metab. Eng. 64, 167–179. doi:10.1016/j.ymben.2021.01.013
Wehrmann, M., Berthelot, C., Billard, P., and Klebensberger, J. (2018). The PedS2/PedR2 Two-Component System Is Crucial for the Rare Earth Element Switch in Pseudomonas putida KT2440. mSphere 3, e00376. doi:10.1128/mSphere.00376-18
Wehrmann, M., Billard, P., Martin-Meriadec, A., Zegeye, A., and Klebensberger, J. (2017). Functional Role of Lanthanides in Enzymatic Activity and Transcriptional Regulation of Pyrroloquinoline Quinone-dependent Alcohol Dehydrogenases in Pseudomonas putida KT2440. mBio 8, e00570. doi:10.1128/mBio.00570-17
Weimer, A., Kohlstedt, M., Volke, D. C., Nikel, P. I., and Wittmann, C. (2020). Industrial Biotechnology of Pseudomonas putida: Advances and Prospects. Appl. Microbiol. Biotechnol. 104, 7745–7766. doi:10.1007/s00253-020-10811-9
Wu, X., Monchy, S., Taghavi, S., Zhu, W., Ramos, J., and van der Lelie, D. (2011). Comparative Genomics and Functional Analysis of Niche-specific Adaptation in Pseudomonas putida. FEMS Microbiol. Rev. 35, 299–323. doi:10.1111/j.1574-6976.2010.00249.x
Xiao, Z., Lv, C., Gao, C., Qin, J., Ma, C., Liu, Z., et al. (2010). A Novel Whole-Cell Biocatalyst with NAD+ Regeneration for Production of Chiral Chemicals. PLoS One 5, e8860. doi:10.1371/journal.pone.0008860
Xiao, Z., and Xu, P. (2007). Acetoin Metabolism in Bacteria. Crit. Rev. Microbiol. 33, 127–140. doi:10.1080/10408410701364604
Xu, Y., Chu, H., Gao, C., Tao, F., Zhou, Z., Li, K., et al. (2014). Systematic Metabolic Engineering of Escherichia coli for High-Yield Production of Fuel Bio-chemical 2,3-Butanediol. Metab. Eng. 23, 22–33. doi:10.1016/j.ymben.2014.02.004
Yang, J., Im, Y., Kim, T. H., Lee, M. J., Cho, S., Na, J. G., et al. (2020). Engineering Pseudomonas putida KT2440 to Convert 2,3-Butanediol to Mevalonate. Enzyme Microb. Technol. 132, 109437. doi:10.1016/j.enzmictec.2019.109437
Yang, T., Rao, Z., Zhang, X., Xu, M., Xu, Z., and Yang, S. T. (2017). Metabolic Engineering Strategies for Acetoin and 2,3-Butanediol Production: Advances and Prospects. Crit. Rev. Biotechnol. 37, 990–1005. doi:10.1080/07388551.2017.1299680
Yi, H. S., Ahn, Y. R., Song, G. C., Ghim, S. Y., Lee, S., Lee, G., et al. (2016). Impact of a Bacterial Volatile 2,3-Butanediol on Bacillus subtilis Rhizosphere Robustness. Front. Microbiol. 7, 993. doi:10.3389/fmicb.2016.00993
Zhang, L., Xu, Q., Zhan, S., Li, Y., Lin, H., Sun, S., et al. (2014). A New NAD(H)-dependent meso-2,3-Butanediol Dehydrogenase from an Industrially Potential Strain Serratia marcescens H30. Appl. Microbiol. Biotechnol. 98, 1175–1184. doi:10.1007/s00253-013-4959-x
Zhang, M., Gao, C., Guo, X., Guo, S., Kang, Z., Xiao, D., et al. (2018). Increased Glutarate Production by Blocking the Glutaryl-CoA Dehydrogenation Pathway and a Catabolic Pathway Involving L-2-Hydroxyglutarate. Nat. Commun. 9, 2114. doi:10.1038/s41467-018-04513-0
Zhang, M., Kang, Z., Guo, X., Guo, S., Xiao, D., Liu, Y., et al. (2019). Regulation of Glutarate Catabolism by GntR Family Regulator CsiR and LysR Family Regulator GcdR in Pseudomonas putida KT2440. mBio 10, e01570. doi:10.1128/mBio.01570-19
Keywords: butane-2,3-diol, Pseudomonas putida KT2440, (2R,3R)-butane-2,3-diol dehydrogenase, quinoprotein alcohol dehydrogenase, lanthanide
Citation: Liu Y, Wang X, Ma L, Lü M, Zhang W, Lü C, Gao C, Xu P and Ma C (2021) Dehydrogenation Mechanism of Three Stereoisomers of Butane-2,3-Diol in Pseudomonas putida KT2440. Front. Bioeng. Biotechnol. 9:728767. doi: 10.3389/fbioe.2021.728767
Received: 22 June 2021; Accepted: 09 August 2021;
Published: 26 August 2021.
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaCopyright © 2021 Liu, Wang, Ma, Lü, Zhang, Lü, Gao, Xu and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanjuan Lü, Y2h1YW5qdWFubHZAbWFpbC5zZHUuZWR1LmNu
 Yidong Liu
Yidong Liu Xiuqing Wang1
Xiuqing Wang1 Wen Zhang
Wen Zhang Chao Gao
Chao Gao Ping Xu
Ping Xu Cuiqing Ma
Cuiqing Ma