- School of Biological Sciences, University of Edinburgh, Edinburgh, United Kingdom
Fed-batch cultures of Chinese Hamster Ovary cells have been used to produce high quantities of biotherapeutics, particularly monoclonal antibodies. However, a growing number of next-generation biotherapeutics, such as bi-specific antibodies and fusion proteins, are difficult to express using standard fed-batch processes. Decoupling cell growth and biotherapeutic production is becoming an increasingly desired strategy for the biomanufacturing industry, especially for difficult-to-express products. Cells are grown to a high cell density in the absence of recombinant protein production (the growth phase), then expression of the recombinant protein is induced and cell proliferation halted (the production phase), usually by combining an inducible gene expression system with a proliferation control strategy. Separating the growth and production phases allows cell resources to be more efficiently directed toward either growth or production, improving growth characteristics and enhancing the production of difficult to express proteins. However, current mammalian cell proliferation control methods rely on temperature shifts and chemical agents, which interact with many non-proliferation pathways, leading to variable impacts on product quality and culture viability. Synthetic biology offers an alternative approach by strategically targeting proliferation pathways to arrest cell growth but have largely remained unused in industrial bioproduction. Due to recent developments in microbial decoupling systems and advances in available mammalian cell engineering tools, we propose that the synthetic biology approach to decoupling growth and production needs revisiting.
Introduction
Industrial-scale production of many biotherapeutics relies on the culture of Chinese Hamster Ovary (CHO) cells. In fed-batch cultures, nutrients are added to the culture to prevent nutrient limitation, prolong culture duration, and maximize product titer. Once culture viability drops below a certain threshold, the culture is harvested and the product is purified. Advances in cell engineering, expression vector design, and process development have allowed monoclonal antibody (mAb) titers to reach 10 g/L. However, high expression of recombinant genes places a large metabolic burden on the cell, which can result in the downregulation of recombinant gene expression. To avoid unstable cell lines entering the manufacturing stage, stability studies (which remain a significant bottleneck in cell line development timelines) are performed to identify cell lines that maintain high gene expression over the culture period. Additionally, a growing range of next-generation biotherapeutics (such as bi-specific antibodies, fusion proteins, and toxic proteins) are being produced as a result of recent developments in protein engineering. These are often difficult to express in CHO cells at desired quantities. For example, production of hyperactive human DNase I is particularly challenging with standard culture processes, because protein expression negatively impacts cell growth, viability, and stability (Lam et al., 2017).
An increasingly attractive method of production is to decouple the growth and production phases of the culture process, usually by combining an inducible gene expression system with a proliferation control strategy (Misaghi et al., 2014; Lam et al., 2017; Poulain et al., 2017). Under such a system, cells are initially grown up to high cell density in the absence of recombinant protein production (the growth phase). Once the desired cell density is reached, expression of the recombinant protein is induced and cell proliferation is halted (the production phase). Proliferation is usually arrested in the G1 phase of the cell cycle, which is associated with larger cells and increased ribosomal protein S6 levels (Carvalhal et al., 2003; Bi et al., 2004). Separating the growth and production phases allows cell resources to be more efficiently utilized in each phase, improving growth characteristics and enhancing production stability (Misaghi et al., 2014). It also avoids expression of cytotoxic recombinant proteins during the growth phrase, reducing the negative impact on cell growth and viability (Lam et al., 2017). Inducing global downregulation of translation (apart from the product gene) during the production phase can increase product purity to aid downstream processing (Bojar et al., 2019).
However, the benefits of decoupling growth and production in mammalian cells are challenged by suboptimal proliferation control strategies. Most commonly, cells are arrested by shifting the culture temperature from 37°C to 30–35°C, or through the addition of chemical agents [such as sodium butyrate (Hong et al., 2014)] to the culture. Both strategies successfully arrest cell growth, usually in the G1 phase, while improving specific productivity and reducing nutrient uptake and waste production (Marchant et al., 2008). However, both strategies also impact other non-proliferation pathways. Transcriptomic studies have shown that temperature shifts cause wide-scale changes in gene expression in the cell (Bedoya-López et al., 2016) and can impact certain product quality attributes (e.g., glycosylation profile; Nam et al., 2008). In addition to negatively impacting product quality (Hong et al., 2014), chemical agents such as sodium butyrate also trigger apoptotic pathways (Kim and Lee, 2000). Model-based approaches have been developed to reduce the number of experiments required to determine the optimum timing for the temperature shift, to maximize product titer (Paul et al., 2019). However, decoupling growth and production usually requires extensive and time-consuming process development work to maximize titer, while avoiding negative product quality consequences (Xu et al., 2019). Lack of a suitable targeted proliferation control approach hinders rapid development of a decoupled production process.
Recently, there have been significant advances in decoupling growth and production in microbial systems using synthetic biology approaches (Izard et al., 2015; Li et al., 2016; Durante-Rodríguez et al., 2018; Stargardt et al., 2020). However, it has been over 20 years since the overexpression of cyclin-dependent kinase inhibitors (p21Cip1, p27Kip1, and p53) was first used to decouple growth and production in mammalian cells (Fussenegger et al., 1997). Implementation of this strategy was limited by a lack of appropriate inducible gene expression systems and its weak impact on mammalian cell proliferation (Weber and Fussenegger, 2007). Since then, there have been significant advances in synthetic biology tool development, but their application in decoupling growth and production in mammalian cells has largely remained unexplored (Guha et al., 2017; Kallunki et al., 2019; Trauth et al., 2019). Here, we discuss the benefits of a targeted approach to proliferation control and discuss how recently developed tools in mammalian synthetic biology provide a promising solution for decoupling growth and production.
Strategic Proliferation Control: Towards a Targeted Approach
A targeted approach to cell proliferation control could hold the key to arresting cell proliferation at the G1 phase, the phase associated with larger cells and increased ribosomal protein S6 levels, while maintaining product quality. Progression through the cell cycle is governed by the regulated activation, degradation, and synthesis of a series of cell-cycle regulators. During G1, cells make a decision whether to irreversibly commit to a new round of cell division or remain in a non-proliferative G0 state (the restriction point). Progression past the restriction point is governed by the retinoblastoma (RB) protein, an inhibitor of G1/S gene transcription (Figure 1). Cyclin D-CDK4/6 and Cyclin E-CDK2 complexes phosphorylate RB, enabling the expression of G1/S genes. Cyclin-dependent kinase inhibitors (CKIs) regulate Cyclin-CDK activity (Alberts et al., 2002; Harper and Brooks, 2004).
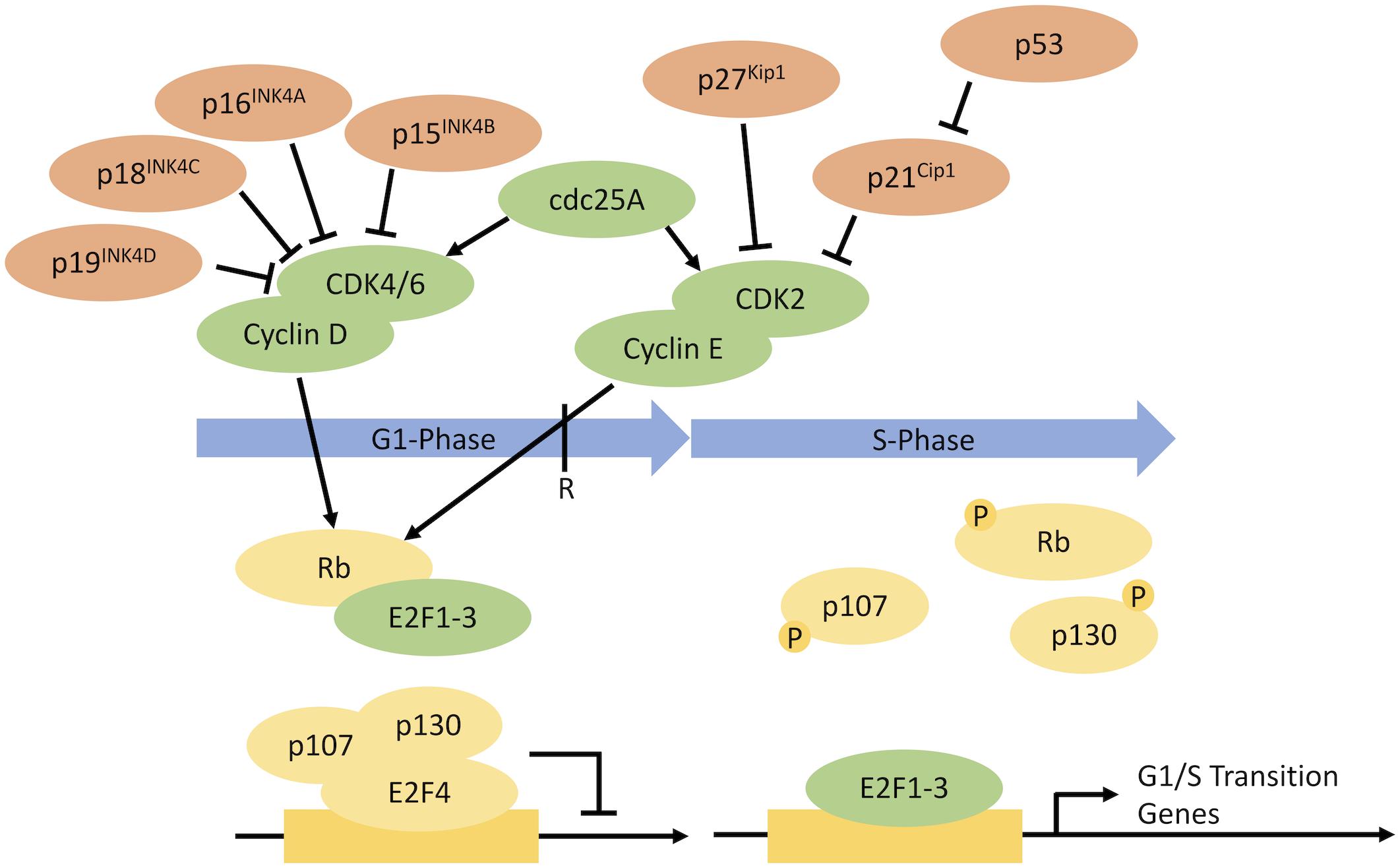
Figure 1. Regulation of the progression through G1 phase into S phase (Sunley and Butler, 2010). The progression through G1 into S phase is governed by a series of cell-cycle regulators, with some regulators promoting progression and others inhibiting progression. CKIs for potential overexpression are shown in red. Potential targets for gene knockdown/protein degradation are shown in green.
The mechanism by which temperature shifts induce cell-cycle arrest is thought to involve membrane-dependent activation of the ataxia telangiectasia mutated and Rad3-related kinase (ATR)–p53–p21 pathway (Roobol et al., 2011). However, as previously mentioned, temperature shifts also cause wide-scale changes in gene expression in the cell (Bedoya-López et al., 2016) which can negatively impact product quality attributes. By contrast, targeted approaches to proliferation control should have far fewer off-target effects. Indeed, targeted inhibition of CDK4/6 activity using a highly selective cancer therapeutic molecule (PD 0332991) (Fry et al., 2004) increased specific mAb productivity and prolonged cell culture, while having minimal impact on product quality (Du et al., 2015). In addition, by contrast with temperature shift, targeted CDK4/6 inhibition did not lead to changes in expression of genes involved in the glycosylation pathway (Du et al., 2015; Bedoya-López et al., 2016). Thus, targeted inhibition approaches may avoid the negative impact from off-target effects of current cell proliferation control strategies for decoupling growth and production in mammalian cells. However, the high cost and physiological activity of cancer drugs limit their potential to be applied for such purposes in industrial bioproduction (Weber and Fussenegger, 2007). Synthetic biology strategies, which selectively target the activity of cell-cycle regulators through cell engineering approaches, could provide a less expensive and more industrially viable solution.
Previous synthetic biology strategies have focused on the overexpression of cyclin-dependent kinase inhibitors (CKIs), with p21Cip1, p53, and P27Kip1 the most studied (Fussenegger et al., 1997; Mazur et al., 1998; Carvalhal et al., 2003; Bi et al., 2004). The original approach controlled the expression of the CKI and product gene, on a dicistronic vector, with a doxycycline-inducible gene expression system (Fussenegger et al., 1997). Upon addition of doxycycline, cells were arrested in G1 and product expression was switched on, decoupling the growth and production phases of the culture process (Table 1A). Since then, multiple approaches have been tested, using a variety of inducible promoters and gene combinations (Mazur et al., 1998; Carvalhal et al., 2003; Bi et al., 2004). The overexpression of CKIs caused increases in specific productivity, but the degree of specific productivity increase was variable (likely due to the inherent variability in transfection methods available at the time and the expression systems used). Overexpression of p21Cip1 led to a fourfold increase in specific productivity in a stable mAb-producing CHO cell line but not in a stable secreted embryonic alkaline phosphatase (SEAP)-producing CHO cell line (Mazur et al., 1998; Bi et al., 2004). However, the implementation of CKI overexpression for cell proliferation control has been limited by its weak impact on proliferation. Combining overexpression of CKIs with a temperature shift showed that the temperature shift had the dominant impact on cell proliferation (Kaufmann et al., 2001). This strongly suggests that novel synthetic biology approaches are required for the manipulation of proliferation pathways.
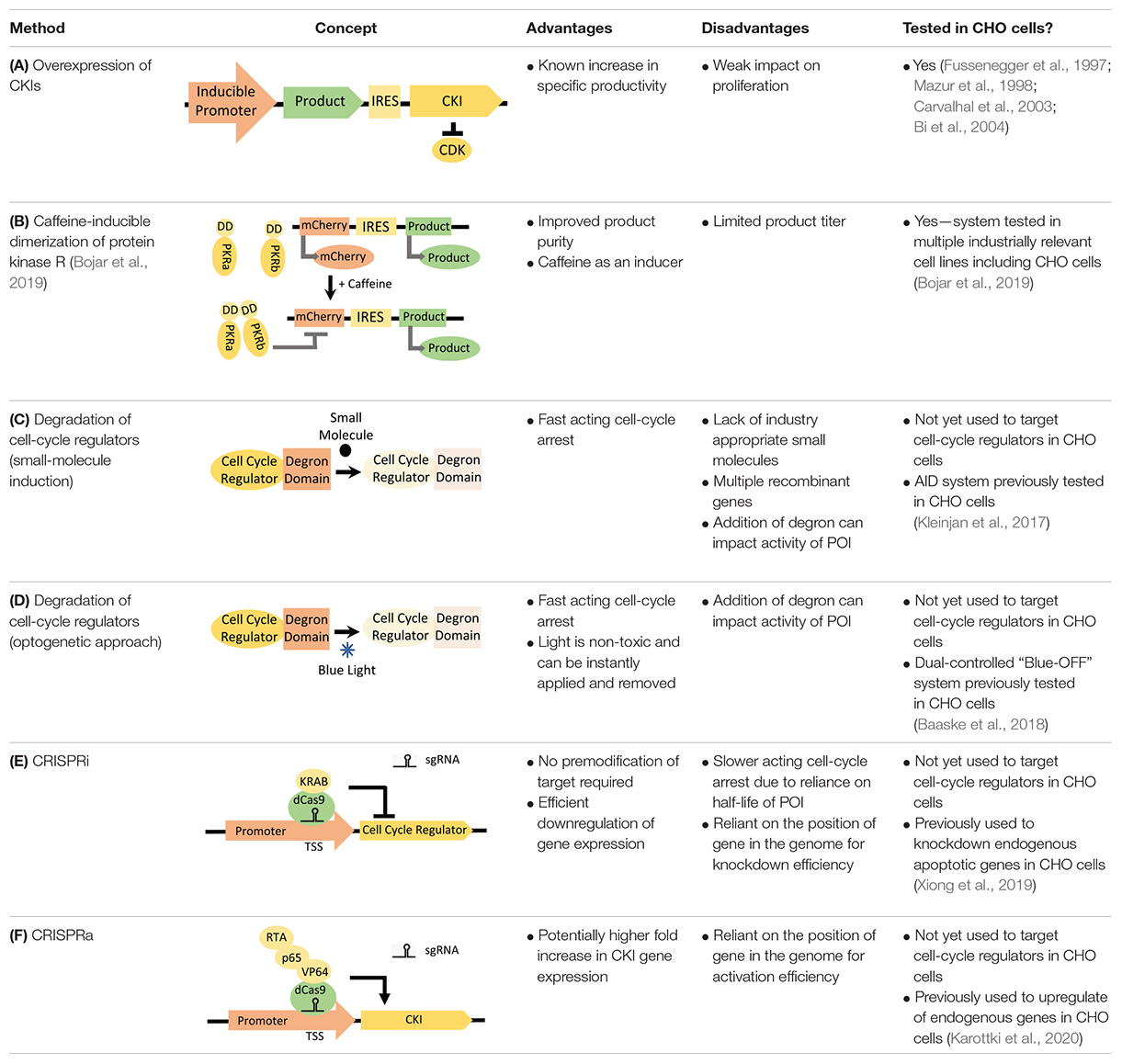
Table 1. Mammalian synthetic biology tools which could be used for decoupling growth and production.
Novel Synthetic Biology Methods for Proliferation Control
Since the original, cell engineering-based proliferation control studies (Fussenegger et al., 1997; Mazur et al., 1998; Carvalhal et al., 2003; Bi et al., 2004), multiple synthetic biology tools have emerged, including inducible gene expression systems (Kallunki et al., 2019), degradation systems (Trauth et al., 2019), and gene knockout/repression systems (Guha et al., 2017). However, the application of such tools in decoupling growth and production in mammalian cells has largely remained unexplored. There have been significant advances in synthetic biology approaches to decoupling growth and production in microbial systems, based on mRNA interferases (Suzuki et al., 2005), control of host RNA polymerase activity (Izard et al., 2015; Stargardt et al., 2020), and regulation of DNA replication machinery (Li et al., 2016). Recently, a bacteriophage-derived Escherichia coli RNA polymerase inhibitor was used to arrest cell growth by inhibiting σ-factor 70-driven host gene expression (Stargardt et al., 2020). Although mammalian proliferation control pathways are more complex, similar approaches could be used to target the mammalian cell cycle. The potential of synthetic biology in decoupling growth and production in mammalian cells has been partly realized in “purity by design” cell lines. Activation of a caffeine-inducible mammalian protein kinase R switches off non-product protein translation, while a viral IRES sequence protects recombinant protein production (Bojar et al., 2019). However, titer is limited because translation from IRES sequences does not have access to the full ribosomal machinery (Table 1B; Bojar et al., 2019). Although this method could be incredibly valuable for products where the presence of host cell proteins may be particularly harmful to product quality, it is inherently limited when improving product titer is a priority, as is common need for difficult-to-express proteins.
Degradation systems target endogenous cellular proteins and could be used to rapidly knock down cell-cycle regulator activity (Trauth et al., 2019). A degron, added to the N- or C-terminus of a protein of interest (POI), allows targeted and rapid protein degradation upon addition of a small-molecule inducer (Table 1C). An example of such a system is the Auxin-Inducible Degron (AID) system. AID-mediated degradation of RAD21-mAC rapidly arrested growth of human cells (Natsume et al., 2016). However, fusion of the endogenous POI with the degron tag requires considerable effort and, with the AID system, implementation of the strategy requires the expression of additional heterologous proteins (TIR1). Moreover, a degron-tagged cell-cycle regulator could negatively impact cell proliferation during the growth phase if protein activity is reduced. However, a degron-tagged topoisomerase IIα did not impact proliferation rate or culture viability, despite the enzyme’s essential role in the cell cycle (Farr et al., 2014). Imperative for the application of degron systems in industrial bioproduction is that the small-molecule inducer is non-toxic, physiologically inactive, and inexpensive (Weber and Fussenegger, 2007). However, metabolic bi-products of auxin have displayed toxicity in mammalian cell lines (Kim et al., 2004, 2010). Optogenetic approaches could overcome such limitations, since light is non-toxic and can be instantly applied and removed during culture. For example, the dual-controlled “Blue-OFF” system combines a KRAB-EL222 light-inducible repressor with a B-LID (blue light-inducible degradation domain) protein degradation module, to rapidly downregulate target protein levels in response to blue light (Baaske et al., 2018). By combining such systems with recently developed light-inducible gene expression tools (Yamada et al., 2020), blue light could be used to trigger both cell-cycle regulator degradation (to halt proliferation) and expression of the product gene (to start the production phase).
At the forefront of synthetic biology improvements has been the development of CRISPR/Cas9 as a tool for genetic manipulation (Boettcher and McManus, 2015). The sgRNA-guided Cas9 endonuclease generates double-stranded breaks in target-gene sequences, which are repaired by the error-prone non-homologous end-joining pathway, generating a variety of mutations (termed CRISPRn). CRISPR/Cas9 methods have already been extensively used for enhancing recombinant gene expression in CHO cell bioproduction (Dangi et al., 2018). A fusion of a catalytically inactive Cas9 (dCas9) and the Krüppel-associated box (KRAB) repression domain, targeted to a transcriptional start site (TSS), can be used to silence expression of a target gene (termed CRISPR interference or CRISPRi) (Table 1D). An inducible version of CRISPRi is created by controlling dCas9 or sgRNA expression with an inducible gene expression system. The application of CRISPRi for decoupling growth and production has already been realized in bacterial cells, where knockdown of DNA replication machinery or nucleotide synthesis was shown to cause cell arrest (Li et al., 2016). Using RNAi to knock down cyclin E1 and E2 expression, cell proliferation was arrested in the G1 phase in human hepatocellular carcinoma cell lines (Geng et al., 2018). This could be repeated with CRISPRi, a method that gives more consistent and robust gene knockdown than RNAi, while exhibiting less off-target effects (Boettcher and McManus, 2015). Alternatively, dCas9 can be used for the upregulation of target-gene expression (termed CRISPR activation or CRISPRa). The VPR CRISPRa approach requires the direct fusion of dCas9 with transcriptional activators (VP64, p65, and Rta), targeted to the TSS of endogenous genes with sgRNAs (Table 1E; Chavez et al., 2015). CRISPRa could be used to arrest proliferation by upregulating endogenous CKI gene expression (Table 1F). Although the degree of target-gene upregulation is highly loci dependent, CRISPRa allows for much higher fold increases in target-gene expression than what is common for standard inducible promoter systems (Chavez et al., 2015). This could overcome the problem of weak cell-cycle arrest in previous CKI-overexpression studies.
Selection of Suitable Targets
Identification of cell-cycle targets for the purposes of cancer treatment has shown that the impact of knocking down cell-cycle regulators is often cell line specific. For example, Cdk2 knockdown had no significant impact on cell proliferation in certain mammalian cell lines, whereas in other studies, Cdk2 knockdown induced cell-cycle arrest (Tetsu and McCormick, 2003; Liu et al., 2020). Potential targets for knockdown or overexpression are outlined in Figure 1. Selection of certain cell-cycle regulators for gene knockdown/protein degradation may be limited by their interaction with apoptotic pathways. To minimize the risk of inducing apoptosis after knockdown of cell-cycle regulators, proliferation control strategies could be combined with overexpression or knockout of key apoptotic genes (Kim and Lee, 2000, 2002). Reducing apoptosis could also help maintain high cell densities during the production phase for a longer time period, thus improving product titer.
Transcriptional inhibition approaches, such as CRISPRi, are inherently slower acting than degradation methods, due to a reliance on the inherent half-life of the protein, meaning proteins with longer half-lives take longer to remove. Cell-cycle regulators with short half-lives, such as D-type cyclins (half-life of 30 min) (Donjerkovic and Scott, 2000), are likely to provide more suitable targets for transcription-based regulation. Challenges for implementing this strategy will also arise from the genetic plasticity that CHO cells exhibit, giving rise to modifications in gene copy number, changes in transcriptional activity, and substantial chromosome rearrangements (Vcelar et al., 2018). A mutation in the proliferation control pathway could lead to cells escaping arrest. Additionally, the gene knockdown strategy would have to arrest the majority of cells in the culture. The efficiency of CRISPRi-mediated knockdown is known to be largely dependent on the genomic context of the target gene, meaning that high knockdown efficiencies of cell-cycle regulators may not easily be achieved. If these challenges are not addressed, populations of fast-growing cells may outcompete the arrested cells, limiting culture duration and product titer. Targeting multiple proliferation pathways is a strategy that is common in cancer therapies and could reduce the number of cells escaping cell-cycle arrest. To improve CRISPRi efficiency, cell-cycle regulators could be targeted more easily by knocking out the endogenous gene and expressing a recombinant version of the gene at a more easily targeted locus, using a targeted integration system.
Discussion
With the increasing demands on biomanufacturing facilities to produce a wider range of protein biotherapeutics at higher titers and in a shorter period of time, decoupling growth and production provides a viable alternative to traditional constitutive expression strategies. Moving the product expression window away from the growth phase mitigates the negative impact that cytotoxic proteins have on cell growth, viability, and stability. Additionally, there is increasing evidence that recombinant protein expression places a substantial metabolic burden on the cell, particularly on its secretory capacity, resulting in decreased cell line stability and downregulation of recombinant gene expression (Jones et al., 2005; Ong et al., 2019; Poulain et al., 2019). For example, removing the expression of a non-essential recombinant gene substantially improved the growth characteristics of a CHO cell line (Kallehauge et al., 2017). This suggests that decoupling growth and production could have wider application in the production of biotherapeutics, beyond just those that are cytotoxic.
The application of decoupling strategies in bioproduction has been limited by the variable impact of temperature shifts and chemical agents on product titer, culture viability, and product quality. This is largely due to the interaction of these strategies with many other cellular pathways that are not related to cell proliferation. Although industrially inviable due to their cost, the use of highly selective cancer drugs for proliferation control has yielded promising results for targeted approaches (Du et al., 2015). Synthetic biology offers a potentially more industrially relevant approach, but its use in proliferation control has remained largely unexplored. Here, we have discussed synthetic biology tools that have emerged over the last 20 years that could be used to arrest cell proliferation. Synthetic biology approaches to decoupling growth and production in microbial systems have been developed, but a mammalian cell engineering approach remains largely unexplored. The implementation of such strategies in mammalian cells has considerable advantages over conventional approaches to proliferation control, allowing the selective targeting of cell-cycle regulators to induce cell-cycle arrest (with the associated increase in specific productivity), while avoiding negative impacts on product quality.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
JD and MD developed the presented idea. JD wrote the manuscript in consultation with SR and MD. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. (2002). An Overview of the Cell Cycle. Molecular Biology of the Cell. 4th Edn. New York: Garland Science.
Baaske, J., Gonschorek, P., Engesser, R., Dominguez-Monedero, A., Raute, K., Fischbach, P., et al. (2018). Dual-controlled optogenetic system for the rapid down-regulation of protein levels in mammalian cells. Sci. Rep. 8:15024. doi: 10.1038/s41598-018-32929-7
Bedoya-López, A., Estrada, K., Sanchez-Flores, A., Ramírez, O. T., Altamirano, C., Segovia, L., et al. (2016). Effect of Temperature Downshift on the Transcriptomic Responses of Chinese Hamster Ovary Cells Using Recombinant Human Tissue Plasminogen Activator Production Culture. PLoS One 11:e0151529. doi: 10.1371/journal.pone.0151529
Bi, J.-X., Shuttleworth, J., and Al-Rubeai, M. (2004). Uncoupling of cell growth and proliferation results in enhancement of productivity in p21CIP1-arrested CHO cells. Biotechnol. Bioeng. 85, 741–749. doi: 10.1002/bit.20025
Boettcher, M., and McManus, M. T. (2015). Choosing the Right Tool for the Job: rNAi. Mol. Cell. 58, 575–585. doi: 10.1016/j.molcel.2015.04.028
Bojar, D., Fuhrer, T., and Fussenegger, M. (2019). Purity by design: reducing impurities in bioproduction by stimulus-controlled global translational downregulation of non-product proteins. Metabol. Eng. 52, 110–123. doi: 10.1016/j.ymben.2018.11.007
Carvalhal, A. V., Marcelino, I., and Carrondo, M. J. T. (2003). Metabolic changes during cell growth inhibition by p27 overexpression. Appl. Microbiol. Biotechnol. 63, 164–173. doi: 10.1007/s00253-003-1385-5
Chavez, A., Scheiman, J., Vora, S., Pruitt, B. W., Tuttle, M., Iyer, E. P. R., et al. (2015). Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328. doi: 10.1038/nmeth.3312
Dangi, A. K., Sinha, R., Dwivedi, S., Gupta, S. K., and Shukla, P. (2018). Cell Line Techniques and Gene Editing Tools for Antibody Production: a Review. Front. Pharmacol. 9:630. doi: 10.3389/fphar.2018.00630
Donjerkovic, D., and Scott, D. W. (2000). Regulation of the G1 phase of the mammalian cell cycle. Cell Res. 10, 1–16. doi: 10.1038/sj.cr.7290031
Du, Z., Treiber, D., McCarter, J. D., Fomina-Yadlin, D., Saleem, R. A., McCoy, R. E., et al. (2015). Use of a small molecule cell cycle inhibitor to control cell growth and improve specific productivity and product quality of recombinant proteins in CHO cell cultures. Biotechnol. Bioeng. 112, 141–155. doi: 10.1002/bit.25332
Durante-Rodríguez, G., de Lorenzo, V., and Nikel, P. I. (2018). A Post-translational Metabolic Switch Enables Complete Decoupling of Bacterial Growth from Biopolymer Production in Engineered Escherichia coli. ACS Synth. Biol. 7, 2686–2697. doi: 10.1021/acssynbio.8b00345
Farr, C. J., Antoniou-Kourounioti, M., Mimmack, M. L., Volkov, A., and Porter, A. C. G. (2014). The α isoform of topoisomerase II is required for hypercompaction of mitotic chromosomes in human cells. Nucleic Acids Res. 42, 4414–4426. doi: 10.1093/nar/gku076
Fry, D. W., Harvey, P. J., Keller, P. R., Elliott, W. L., Meade, M., Trachet, E., et al. (2004). Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 3, 1427–1438.
Fussenegger, M., Mazur, X., and Bailey, J. E. (1997). A novel cytostatic process enhances the productivity of Chinese hamster ovary cells. Biotechnol. Bioeng. 55, 927–939. doi: 10.1002/(SICI)1097-0290(19970920)55:6<927::AID-BIT10>3.0.CO;2-4
Geng, Y., Michowski, W., Chick, J. M., Wang, Y. E., Jecrois, M. E., Sweeney, K. E., et al. (2018). Kinase-independent function of E-type cyclins in liver cancer. PNAS 115, 1015–1020. doi: 10.1073/pnas.1711477115
Guha, T. K., Wai, A., and Hausner, G. (2017). Programmable Genome Editing Tools and their Regulation for Efficient Genome Engineering. Comput. Struct. Biotechnol. J. 15, 146–160. doi: 10.1016/j.csbj.2016.12.006
Harper, J. V., and Brooks, G. (2004). The mammalian cell cycle: an overview. Methods Mol Biol. 296, 113–154. doi: 10.1385/1-59259-857-9:113
Hong, J. K., Lee, S. M., Kim, K.-Y., and Lee, G. M. (2014). Effect of sodium butyrate on the assembly, charge variants, and galactosylation of antibody produced in recombinant Chinese hamster ovary cells. Appl. Microbiol. Biotechnol. 98, 5417–5425. doi: 10.1007/s00253-014-5596-8
Izard, J., Gomez Balderas, C. D., Ropers, D., Lacour, S., Song, X., Yang, Y., et al. (2015). A synthetic growth switch based on controlled expression of RNA polymerase. Mol. Syst. Biology. 11:840. doi: 10.15252/msb.20156382
Jones, J., Nivitchanyong, T., Giblin, C., Ciccarone, V., Judd, D., Gorfien, S., et al. (2005). Optimization of tetracycline-responsive recombinant protein production and effect on cell growth and ER stress in mammalian cells. Biotechnol. Bioeng. 91, 722–732. doi: 10.1002/bit.20566
Kallehauge, T. B., Li, S., Pedersen, L. E., Ha, T. K., Ley, D., Andersen, M. R., et al. (2017). Ribosome profiling-guided depletion of an mRNA increases cell growth rate and protein secretion. Sci. Rep. 7, 1–12. doi: 10.1038/srep40388
Kallunki, T., Barisic, M., Jäättelä, M., and Liu, B. (2019). How to Choose the Right Inducible Gene Expression System for Mammalian Studies? Cells 8:796. doi: 10.3390/cells8080796
Karottki, K. J. L. C., Hefzi, H., Xiong, K., Shamie, I., Hansen, A. H., Li, S., et al. (2020). Awakening dormant glycosyltransferases in CHO cells with CRISPRa. Biotechnol. Bioeng. 117, 593–598. doi: 10.1002/bit.27199
Kaufmann, H., Mazur, X., Marone, R., Bailey, J. E., and Fussenegger, M. (2001). Comparative analysis of two controlled proliferation strategies regarding product quality, influence on tetracycline-regulated gene expression, and productivity. Biotechnol. Bioeng. 72, 592–602.
Kim, D.-S., Jeon, S.-E., and Park, K.-C. (2004). Oxidation of indole-3-acetic acid by horseradish peroxidase induces apoptosis in G361 human melanoma cells. Cell. Signal. 16, 81–88. doi: 10.1016/S0898-6568(03)00091-3
Kim, N. S., and Lee, G. M. (2000). Overexpression of bcl-2 inhibits sodium butyrate-induced apoptosis in Chinese hamster ovary cells resulting in enhanced humanized antibody production. Biotechnol. Bioeng. 71, 184–193. doi: 10.1002/1097-0290(2000)71:3<184::AID-BIT1008>3.0.CO;2-W
Kim, N. S., and Lee, G. M. (2002). Inhibition of sodium butyrate-induced apoptosis in recombinant Chinese hamster ovary cells by constitutively expressing antisense RNA of caspase-3. Biotechnol. Bioeng. 78, 217–228. doi: 10.1002/bit.10191
Kim, S. Y., Ryu, J. S., Li, H., Park, W.-J., Yun, H.-Y., Baek, K. J., et al. (2010). UVB-Activated Indole-3-Acetic Acid Induces Apoptosis of PC-3 Prostate Cancer Cells. Anticancer Res. 30, 4607–4612.
Kleinjan, D. A., Wardrope, C., Nga Sou, S., and Rosser, S. J. (2017). Drug-tunable multidimensional synthetic gene control using inducible degron-tagged dCas9 effectors. Nat. Commun. 8:1191doi: 10.1038/s41467-017-01222-y
Lam, C., Santell, L., Wilson, B., Yim, M., Louie, S., Tang, D., et al. (2017). Taming hyperactive hDNase I: stable inducible expression of a hyperactive salt- and actin-resistant variant of human deoxyribonuclease I in CHO cells. Biotechnol. Progr. 33, 523–533. doi: 10.1002/btpr.2439
Li, S., Jendresen, C. B., Grünberger, A., Ronda, C., Jensen, S. I., Noack, S., et al. (2016). Enhanced protein and biochemical production using CRISPRi-based growth switches. Metab. Eng. 38, 274–284. doi: 10.1016/j.ymben.2016.09.003
Liu, H., Li, Z., Huo, S., Wei, Q., and Ge, L. (2020). Induction of G0/G1 phase arrest and apoptosis by CRISPR/Cas9-mediated knockout of CDK2 in A375 melanocytes. Mol. Clin. Oncol. 12, 9–14. doi: 10.3892/mco.2019.1952
Marchant, R. J., Al-Fageeh, M. B., Underhill, M. F., Racher, A. J., and Smales, C. M. (2008). Metabolic Rates, Growth Phase, and mRNA Levels Influence Cell-Specific Antibody Production Levels from In Vitro-Cultured Mammalian Cells at Sub-Physiological Temperatures. Mol. Biotechnol. 39, 69–77. doi: 10.1007/s12033-008-9032-0
Mazur, X., Fussenegger, M., Renner, W. A., and Bailey, J. E. (1998). Higher Productivity of Growth-Arrested Chinese Hamster Ovary Cells Expressing the Cyclin-Dependent Kinase Inhibitor p27. Biotechnol. Progr. 14, 705–713. doi: 10.1021/bp980062h
Misaghi, S., Chang, J., and Snedecor, B. (2014). It’s time to regulate: coping with product-induced nongenetic clonal instability in CHO cell lines via regulated protein expression. Biotechnol. Progr. 30, 1432–1440. doi: 10.1002/btpr.1970
Nam, J. H., Zhang, F., Ermonval, M., Linhardt, R. J., and Sharfstein, S. T. (2008). The Effects of Culture Conditions on the Glycosylation of Secreted Human Placental Alkaline Phosphatase Produced in Chinese Hamster Ovary Cells. Biotechnol. Bioeng. 100, 1178–1192. doi: 10.1002/bit.21853
Natsume, T., Kiyomitsu, T., Saga, Y., and Kanemaki, M. T. (2016). Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep. 15, 210–218. doi: 10.1016/j.celrep.2016.03.001
Ong, E.-C., Smidt, P., and McGrew, J. T. (2019). Limiting the metabolic burden of recombinant protein expression during selection yields pools with higher expression levels. Biotechnol. Progr. 35:e2839. doi: 10.1002/btpr.2839
Paul, K., Rajamanickam, V., and Herwig, C. (2019). Model-based optimization of temperature and pH shift to increase volumetric productivity of a Chinese hamster ovary fed-batch process. J. Biosci. Bioeng. 128, 710–715. doi: 10.1016/j.jbiosc.2019.06.004
Poulain, A., Mullick, A., Massie, B., and Durocher, Y. (2019). Reducing recombinant protein expression during CHO pool selection enhances frequency of high-producing cells. J. Biotechnol. 296, 32–41. doi: 10.1016/j.jbiotec.2019.03.009
Poulain, A., Perret, S., Malenfant, F., Mullick, A., Massie, B., and Durocher, Y. (2017). Rapid protein production from stable CHO cell pools using plasmid vector and the cumate gene-switch. J. Biotechnol. 255, 16–27. doi: 10.1016/j.jbiotec.2017.06.009
Roobol, A., Roobol, J., Carden, M. J., Bastide, A., Willis, A. E., Dunn, W. B., et al. (2011). ATR (ataxia telangiectasia mutated- and Rad3-related kinase) is activated by mild hypothermia in mammalian cells and subsequently activates p53. Biochem. J. 435, 499–508. doi: 10.1042/BJ20101303
Stargardt, P., Feuchtenhofer, L., Cserjan-Puschmann, M., Striedner, G., and Mairhofer, J. (2020). Bacteriophage Inspired Growth-Decoupled Recombinant Protein Production in Escherichia coli. ACS Synth. Biol. 6, 1336–1348. doi: 10.1021/acssynbio.0c00028
Sunley, K., and Butler, M. (2010). Strategies for the enhancement of recombinant protein production from mammalian cells by growth arrest. Biotechnol. Adv. 28, 385–394. doi: 10.1016/j.biotechadv.2010.02.003
Suzuki, M., Zhang, J., Liu, M., Woychik, N. A., and Inouye, M. (2005). Single Protein Production in Living Cells Facilitated by an mRNA Interferase. Mol. Cell 18, 253–261. doi: 10.1016/j.molcel.2005.03.011
Tetsu, O., and McCormick, F. (2003). Proliferation of cancer cells despite CDK2 inhibition. Cancer Cell 3, 233–245. doi: 10.1016/s1535-6108(03)00053-9
Trauth, J., Scheffer, J., Hasenjäger, S., and Taxis, C. (2019). Synthetic Control of Protein Degradation during Cell Proliferation and Developmental Processes. ACS Omega 4, 2766–2778. doi: 10.1021/acsomega.8b03011
Vcelar, S., Jadhav, V., Melcher, M., Auer, N., Hrdina, A., Sagmeister, R., et al. (2018). Karyotype variation of CHO host cell lines over time in culture characterized by chromosome counting and chromosome painting. Biotechnol. Bioeng. 115, 165–173. doi: 10.1002/bit.26453
Weber, W., and Fussenegger, M. (2007). Inducible product gene expression technology tailored to bioprocess engineering. Curr. Opinion Biotechnol. 18, 399–410. doi: 10.1016/j.copbio.2007.09.002
Xiong, K., Marquart, K. F., Karottki, K. J. L. C., Li, S., Shamie, I., Lee, J. S., et al. (2019). Reduced apoptosis in Chinese hamster ovary cells via optimized CRISPR interference. Biotechnol. Bioeng. 116, 1813–1819.
Xu, J., Tang, P., Yongky, A., Drew, B., Borys, M. C., Liu, S., et al. (2019). Systematic development of temperature shift strategies for Chinese hamster ovary cells based on short duration cultures and kinetic modeling. mAbs 11, 191–204. doi: 10.1080/19420862.2018.1525262
Keywords: biomanufacturing, synthetic biology, decoupling production from growth, CRISPR/Cas9, CHO cell culture
Citation: Donaldson JS, Dale MP and Rosser SJ (2021) Decoupling Growth and Protein Production in CHO Cells: A Targeted Approach. Front. Bioeng. Biotechnol. 9:658325. doi: 10.3389/fbioe.2021.658325
Received: 25 January 2021; Accepted: 09 April 2021;
Published: 02 June 2021.
Edited by:
Ana Cecília Roque, New University of Lisbon, PortugalReviewed by:
Barbara Di Ventura, University of Freiburg, GermanyCleo Kontoravdi, Imperial College London, United Kingdom
Copyright © 2021 Donaldson, Dale and Rosser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James S. Donaldson, ai5zLmRvbmFsZHNvbkBzbXMuZWQuYWMudWs=
 James S. Donaldson
James S. Donaldson Matthew P. Dale
Matthew P. Dale Susan J. Rosser
Susan J. Rosser