- 1School of Life Sciences, Shandong Normal University, Jinan, China
- 2Biotechnology Research Center, Shandong Academy of Agricultural Sciences, Shandong Provincial Key Laboratory of Genetic Improvement, Ecology and Physiology of Crops, Jinan, China
- 3Key Laboratory of Aquatic-Ecology and Aquaculture of Tianjin, College of Fishery, Tianjin Agricultural University, Tianjin, China
Serine/threonine kinases (STKs) play important roles in prokaryotic cellular functions such as growth, differentiation, and secondary metabolism. When the external environment changes, prokaryotes rely on signal transduction systems, including STKs that quickly sense these changes and alter gene expression to induce the appropriate metabolic changes. In this study, we examined the roles of the STK genes spkD and spkG in fatty acid biosynthesis in the unicellular cyanobacterium Synechocystis sp. PCC6803, using targeted gene knockout. The linoleic acid (C18: 2), γ-linolenic acid (C18: 3n6), α-linolenic acid (C18: 3n3), and stearidonic acid (C18: 4) levels were significantly lower in spkD and spkG gene knockout mutants than in the wild type at a culture temperature of 30°C and a light intensity of 40 μmol⋅m–2⋅s–1. The expression levels of fatty acid desaturases and STK genes differed between the spkD and spkG gene knockout mutants. These observations suggest that spkD and spkG may directly or indirectly affect the fatty acid composition in Synechocystis sp. PCC6803 by regulating the expression of fatty acid desaturases genes. Therefore, the STK genes spkD and spkG play important roles in polyunsaturated fatty acid biosynthesis in Synechocystis sp. PCC6803. These findings could facilitate the development of cyanobacteria germplasm resources that yield high levels of fatty acids. In addition, they provide a theoretical basis for the genetic engineering of cyanobacteria with improved yields of secondary metabolites and increased economic benefits.
Introduction
Polyunsaturated fatty acids (PUFAs) are fatty acids with multiple unsaturated bonds (any number of carbon atoms). Long chain PUFAs has multiple unsaturated bonds and more than 18 carbon atoms in the fatty acid chain. PUFAs are divided into multiple categories based on the position where the unsaturated bond starts relative to the methyl end of the fatty acid carbon chain, using the ω (omega) numbering system. ω-3 and ω-6 PUFAs play important roles in various organisms (Mayra et al., 2020). In addition to having important physiological functions, these PUFAs, including linoleic acid (C18: 2), α-linolenic acid (C18: 3n3), γ-linolenic acid (C18: 3n6), stearidonic acid (C18: 4), arachidonic acid (C20: 4n6), eicosapentaenoic acid (C20: 5), and docosahexaenoic acid (C22: 6), have high nutritional and medicinal value. PUFAs are critical to the function and structure of the nervous system, especially in children. Thus, the demand for PUFAs is increasing. The large-scale industrialization of PUFAs would provide a solution to the PUFA supply and demand (Jan et al., 2010).
PUFAs have traditionally been derived from oils obtained from deep-sea fish and shellfish (Pauly et al., 2005; Hibbeln et al., 2006). However, factors such as seasonal restrictions, increased environmental pollution, overfishing, and residual fishy odor during the purification of PUFAs from fish oil have limited the production of PUFAs (Drexler et al., 2003). Cyanobacteria have been genetically engineered to produce PUFAs that do not have the shortcomings of traditional PUFAs sources (Gong and Miao, 2019; Somayeh et al., 2019). These prokaryotes use sunlight and carbon dioxide to produce a variety of valuable metabolites. Cyanobacteria have beneficial characteristics that make them ideal for large-scale cultivation, including a rapid growth rate and high metabolic yield (Jan et al., 2010). The single-cell cyanobacterium Synechocystis sp. PCC6803, a facultative autotrophic organism that uses light energy, is considered a model organism to study PUFA biosynthesis (Guedes et al., 2011; Sara et al., 2015).
Serine/threonine kinases (STKs) play important roles in the growth, differentiation, and secondary metabolism of prokaryotic cells (Rajagopal et al., 2003). When the external environment changes, prokaryotes such as cyanobacteria rely on signal transduction systems that include STKs to quickly perceive changes, and through the precise regulation of gene expression, alter their metabolic processes accordingly. The mechanism employed by STKs in cyanobacteria is similar to that in eukaryotes. After receiving the signal, STKs become activated through phosphorylation of their serine and threonine residues, and cooperate with other signal molecules. For example, when exposed to ultraviolet radiation, osmotic stress, heat stress, salt stress, or cold stress, STK proteins are activated and phosphorylate their downstream molecules, ultimately allowing these external signals to be transmitted to the nucleus (Zhang et al., 2007). The precise regulation of gene expression (Hasegawa et al., 2000) leads to changes in metabolite levels, allowing the cyanobacterium to withstand various types of adversity (Liu et al., 2015; Piyoosh et al., 2019).
Synechocystis is the first cyanobacterium whose entire genome has been sequenced. Based on its genome sequence, Synechocystis contains seven STK genes, including spkA (sll1574), spkB (slr1697), spkC (slr0599), spkD (sll0776), spkE (slr1443), spkF (slr1225), and spkG (slr0152) (Zhou et al., 2011). spkA and spkB of Synechocystis sp. PCC6803 contribute to cell movement, spkE encodes a protein lacking kinase activity, spkC, spkF, and spkK have been found to be involved in the phosphorylation of the small molecular chaperone protein GroES (Zorina et al., 2011) and PknD of Anabaena sp. PCC7120 is involved in regulating nitrogen metabolism (Zhang and Libs, 1998; Zhou et al., 2015). Therefore, we first constructed spkD and spkG knockout mutant strains, and identified a potential connection between these genes and PUFAs.
The main function of STKs is signal transduction. Several STK genes are involved in cell growth and cell survival, but their roles in unsaturated fatty acid biosynthesis is remaining unknown. We previously performed qRT-PCR of STK genes in Synechocystis sp. PCC6803 at 2, 4, and 6 days of culture, finding that spkD and spkG were expressed at significantly higher levels in strains with high levels of PUFAs compared to the wild type when grown at a temperature of 30°C and a light intensity of 40 μmol⋅m–2⋅s–1. In addition, there have been related reports showing that spkG plays an important role in STKs (Liang et al., 2011), and we obtained two knockout mutants, spkG and spkD. These findings suggest that spkD and spkG play important roles in unsaturated fatty acid biosynthesis in Synechocystis sp. PCC6803.
Most studies of the unsaturated fatty acid biosynthesis pathway conducted in Synechocystis to date have aimed to identify gene functions through analyses of mutants with metabolic deficiencies. However, our knowledge of the genes that function in Synechocystis fatty acid metabolism is incomplete. STKs have a variety of physiological functions in the cyanobacteria Synechocystis sp. PCC6803 and Anabaena sp. PCC7120, such as biological movement, osmotic pressure regulation, and cell survival. For example, the STK genes spkA and spkB are thought to function in cell movement (Kamei et al., 2001); spkH is thought to help maintain osmotic pressure in the cell (Kalyanee et al., 2004); and spkD affects cell survival (Laurent et al., 2008). However, the roles of other STK genes are still being explored, and the roles of STKs in PUFA biosynthesis in cyanobacteria are currently unknown.
Through the screening of STKs genes and the construction of plasmids, we obtained two mutants: spkD and spkG. Preliminary experimental results showed that PUFA accumulation in spkD and spkG deletion mutants was significantly lower than that in the wild type. Therefore, we carried out subsequent experiments using these mutants.
In this study, we further studied the role of spkD and spkG in PUFA biosynthesis in Synechocystis sp. PCC6803. We constructed spkD and spkG deletion mutant strains of Synechocystis sp. PCC6803 (spkD- and spkG-) and measured the expression levels of fatty acid desaturases and STK genes in the mutants at various points throughout the lifecycle. Fatty acid desaturases catalyze the dehydrogenation of carrier-bound fatty acids to form double bonds in the fatty acid chain. Fatty acid desaturases play an important role in the metabolism of fatty acids and in the maintenance of the correct structure and biological function of the membrane in the organism. Therefore, fatty acid desaturases were analyzed in our experiment. The contents of four unsaturated fatty acids were lower in the spkD and spkG deletion mutants than in the wild type, whereas the expression levels of fatty acid desaturases genes were higher in the mutants, indicating that spkD and spkG play important roles in PUFA biosynthesis in Synechocystis sp. PCC6803. This study lays the foundation for further investigating the effects of other STK genes on fatty acid biosynthesis.
Results
Inactivation of spkD and spkG in Synechocystis sp. PCC6803
We constructed spkD and spkG deletion plasmids (Figures 1A,B) by replacing these genes with a kanamycin resistance cassette and spkD and spkG were separated into 0.9 and 1.0 kb fragments, respectively. These plasmids were used to transform into Synechocystis sp. PCC6803 to obtain the spkD Kanr locus and spkG Kanr locus mutants, here after referred to as spkD-and spkG-, respectively. To verify that spkD/spkG were successfully knocked out, we conducted PCR analysis using primer pair 1 (spkD-F and spkD-R) and primer pair 2 (spkG-F and spkG-R; Table 1). Whereas primer pairs 1 and 2 amplified 1.9 kb fragments from wild type genomic DNA, they amplified 3.3 kb fragments from the mutant genomic DNA. All amplification products were of the predicted size (Figures 1C,D). PCR is confirmed that all copies of spkD/spkG in the Synechocystis sp. 6,803 genome were replaced by the deletion cassette. Thus, we successfully knocked out the spkD and spkG genes in Synechocystis, and obtained two deletion mutant strains.
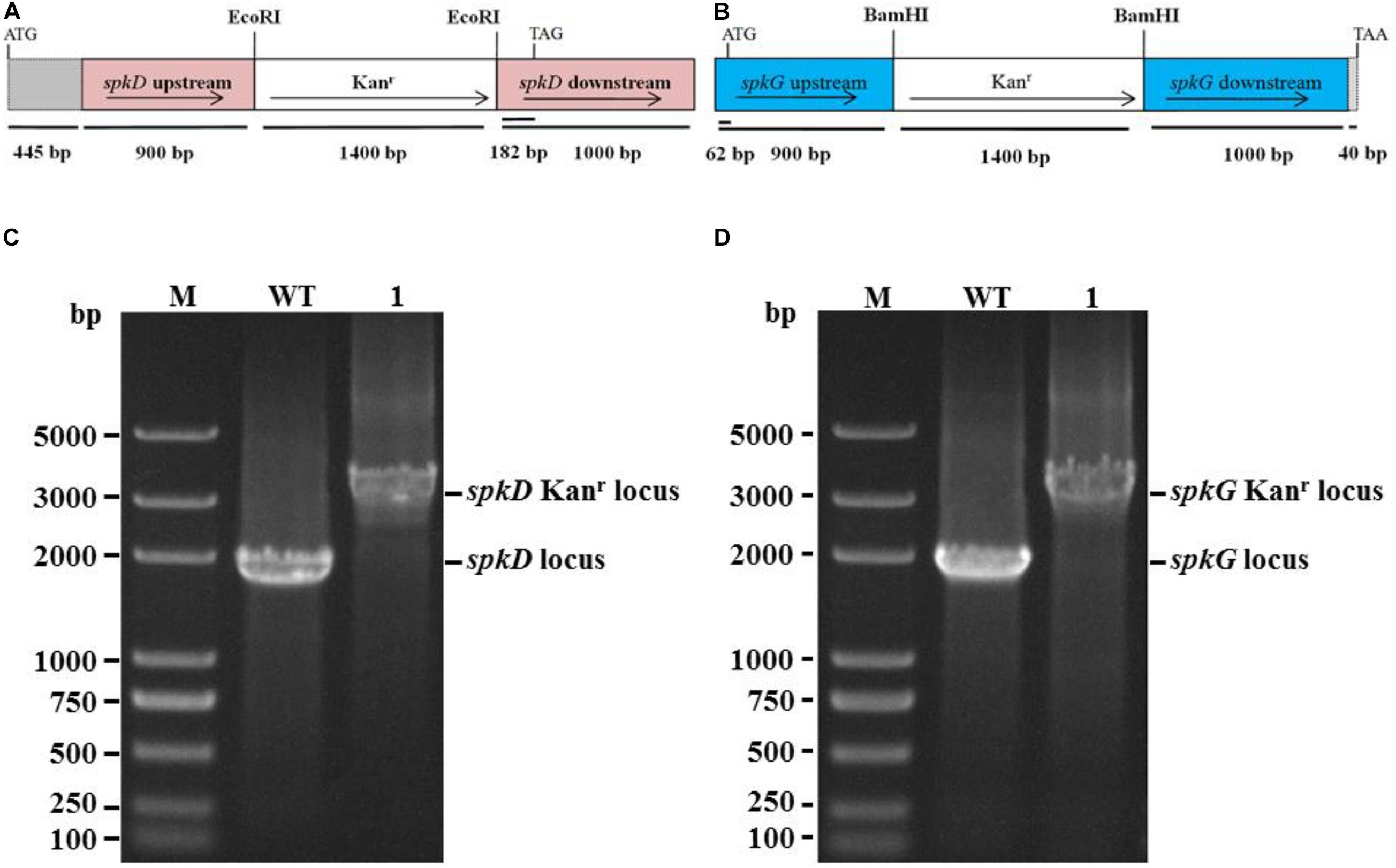
Figure 1. Construction of Synechocystis spkD and spkG gene knockout vectors and PCR detection of mutant strains. (A) Diagram of the spkD gene-directed knockout vector. The gray region (445 bp) shows the unamplified area in the ORF of spkD; (B) Diagram of the spkG gene-directed knockout vector. The gray region (40 bp) shows the unamplified area in the ORF of spkG; (C) PCR detection of spkD gene-directed knockout, M: Trans 2K plus DNA Marker, WT: spkD gene in wild type Synechocystis, Lane 1: spkD gene knockout mutant; (D) PCR detection of spkG gene knockout, M: Trans 2K plus DNA Marker, WT: spkG gene in wild type Synechocystis; Lane 1: spkG gene knockout mutant.
Growth Characteristics of Wild Type and Mutant Strains
To determine whether the knockout of spkD and spkG genes would affect the normal growth of Synechocystis, we incubated the cultures for 10 days under normal light intensity (40 μmol⋅m–2⋅s–1) and measured the OD730 value once daily. Based on these OD730 values, we generated growth curves of the wild type and two mutant strains. The growth patterns of all three Synechocystis strains were roughly similar (Figure 2A). The OD730 values of the knockout mutants were slightly higher than that of the wild type under the same conditions; the final OD730 value of the spkG knockout mutant was the highest. Nevertheless, the overall differences among the three strains were small, indicating that the algal cell density and growth rates were approximately the same.
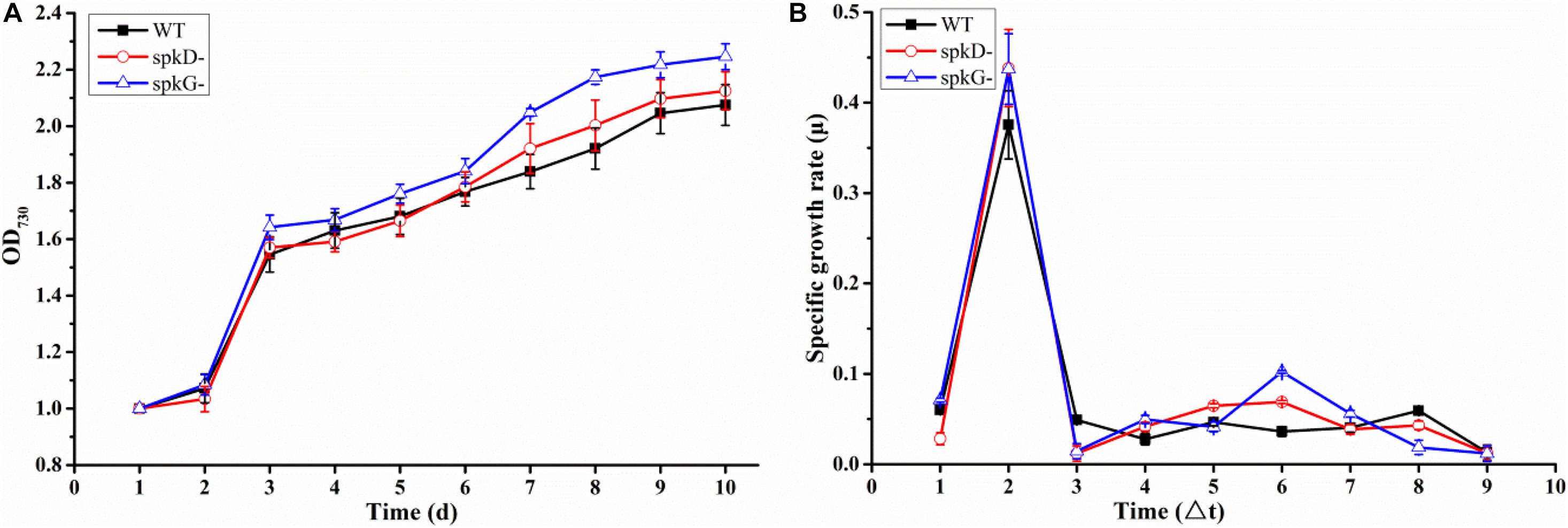
Figure 2. Growth curves and specific growth rates of mutant and wild type cultures under normal light conditions (A) Growth curves of wild type Synechocystis sp. PCC6803 and two knockout mutants; (B) Specific growth rates of wild type Synechocystis sp. PCC6803 and two knockout mutants. WT represents wild type; spkD represents spkD knockout mutant; and spkG represents spkG knockout mutant. The experiment was carried out under a normal light intensity of 40 μmol⋅m–2⋅s–1. △t is the length of the time interval in days. Values are means ± SD (bars) of three independent experiments conducted on different days. The absence of a bar indicates that the SD falls within the symbol.
We generated a standard curve of Synechocystis density and OD730 (y = 32,798x + 6,249, R2 = 0.9896). Using this growth curve, the density of Synechocystis under different OD values could be determined. We conducted a statistical analysis of the growth rates of the two mutant strains and the wild type under normal light conditions (Figure 2B). The growth rates of all three cyanobacterial strains peaked on the second day of culture, began to increase slightly on the fourth day, and reached a second small peak on the sixth day. However, after this stage of culture, the growth rate began to decline until it reached the lowest value and became stable. Overall, there was no significant difference in growth rates between the mutant and wild type strains.
Effects of Knocking Out spkD and spkG Under Normal Light Conditions on STK Gene Expression in Synechocystis
To examine the effect of spkD and spkG gene knockout on the expression of other STK genes, we conducted follow-up experiments investigating differences in the expression levels of these related genes in wild type and mutant Synechocystis sp. PCC6803 under normal light conditions. The STK-related genes were generally expressed at lower levels in the spkD knockout mutant than in the wild type (Figure 3). Except for spkA transcript levels at 24 h of light treatment, the differences in gene expression patterns between the mutant and wild type strains were substantial, with a maximum difference in expression observed at approximately 6 h of light treatment, especially in the case of spkE. Knockout of spkD had the greatest effect on spkE expression, pointing to a possible association between these genes. In addition, the STK-related genes were expressed at higher levels in the mutant than in the wild type Synechocystis sp. PCC6803 before 6 h of light treatment, suggesting that spkD might be involved in regulating the expression of some inhibitors during the early stages of STK biosynthesis.
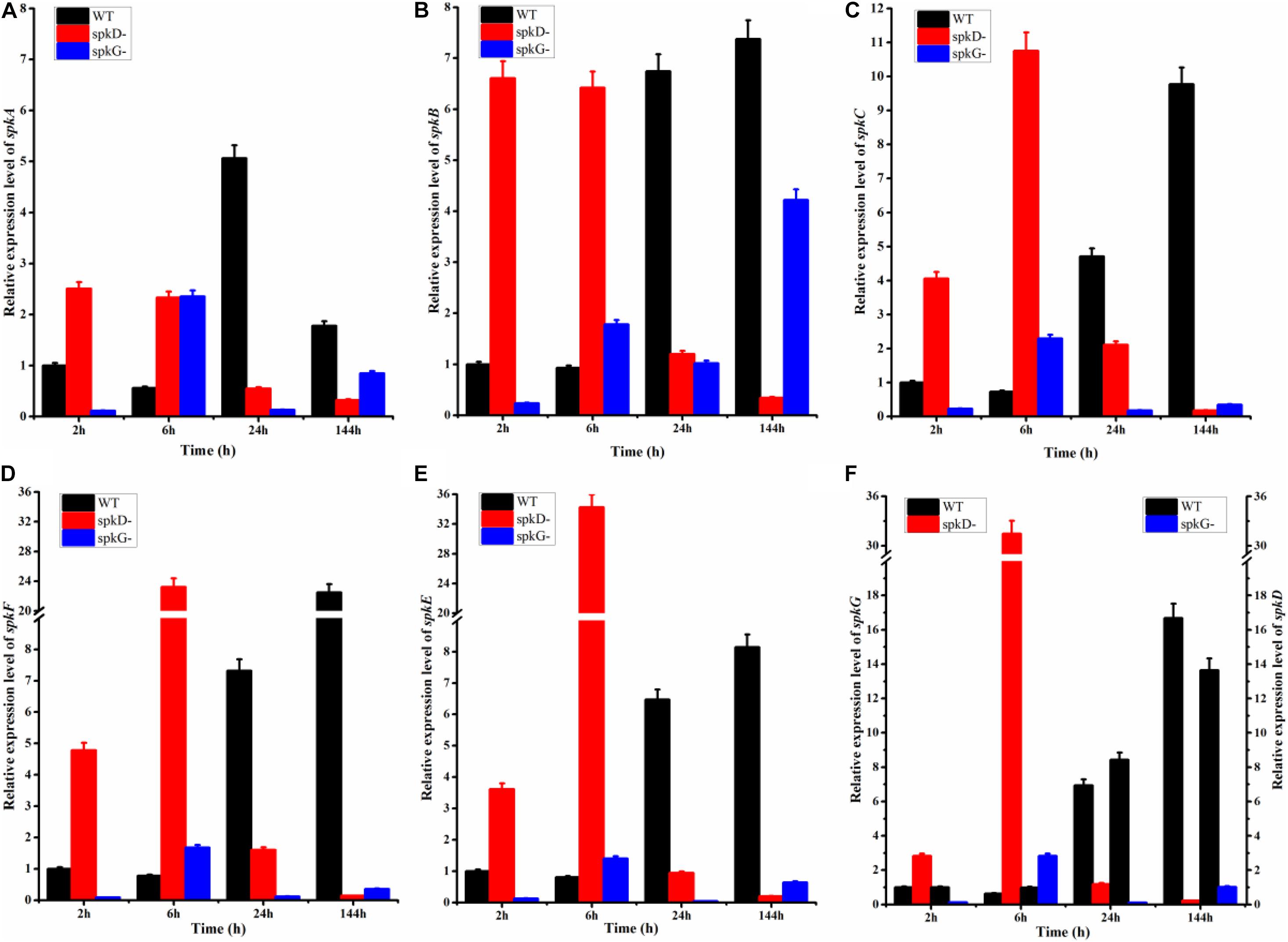
Figure 3. Changes in serine/threonine kinase (STK) gene expression in wild type and mutant strains detected after different periods of exposure to normal light. WT represents wild type Synechocystis sp. PCC6803; spkD- represents the spkD knockout mutant; spkG- represents the spkG knockout mutant. The experiment was carried out under a normal light intensity of 40 μmol⋅m–2⋅s–1. (A–E) show the relative expression levels of spkA, spkB, spkC, spkF, and spkE, respectively, in the wild type and two mutant strains. (F) Relative expression levels of spkG in the wild type and spkD-. The data correspond to the left vertical axis. The black bars represent the wild type. Synechocystis sp. PCC6803, and the red bars represent the mutation that knocked out spkD. The right vertical axis shows the relative expression levels of spkD in the wild type and spkG-, and the black bar represents the wild type. The blue bar represents the mutation that knocked out spkG. Values are means ± SD (bars) of three independent experiments conducted on different days. The absence of a bar indicates that the SD falls within the symbol.
We also examined the expression of STK-related genes in the spkG knockout mutant under the same conditions. Under normal light conditions, the difference in spkA and spkB gene expression in the spkG knockout mutant vs. the wild type was greatest at 24 h, and the expression of spkC, spkD, spkE, and spkF increased overtime (Figure 3). The difference in gene expression also gradually increased over time. Since the knockout of spkG strongly affected spkC, spkD, spkE, and spkF expression, we speculate that spkG is associated with the expression of these genes in Synechocystis sp. PCC6803.
Effects of Knocking Out spkD and spkG on the Expression of Fatty Acid Desaturases Genes in Synechocystis
Fatty acid desaturases is a key enzyme in the PUFA biosynthesis pathway, catalyzing the formation of double bonds at specific locations on the fatty acid chain. There are four types of fatty acid desaturasess in cyanobacteria: delta 6 fatty acid desaturases, delta 9 fatty acid desaturases, delta 12 fatty acid desaturases and delta 15 fatty acid desaturases. Referred to, respectively, as d6D, d9D, d12D, and d15D, these fatty acid desaturases differ depending on where they form double bonds in fatty acids.
Using qRT-PCR, we compared the expression patterns of the four fatty acid desaturases genes in the wild type and mutant strains at different time points under normal light conditions. The expression of the fatty acid desaturases related genes tended to decrease over time in both mutant strains, but to increase in the wild type. These genes were expressed at higher levels in the spkD knockout mutant than in the spkG knockout mutant. Under normal light conditions, desaturases are expressed at higher levels in the spkD deletion mutants than in the wild type during the early stages of growth, with peak expression at 6 h in the mutant. Because the expression level of the wild type was low in the early stages of culture, the difference between the expression level of the wild type and that of the two mutant strains reached a maximum at 6 h. However, this difference decreased over time, and the genes were ultimately expressed at higher levels in the wild type (Figure 4). These results indicate that spkD and spkG are strongly associated with the expression of fatty acid desaturases genes in Synechocystis sp. PCC6803 (Figure 4).
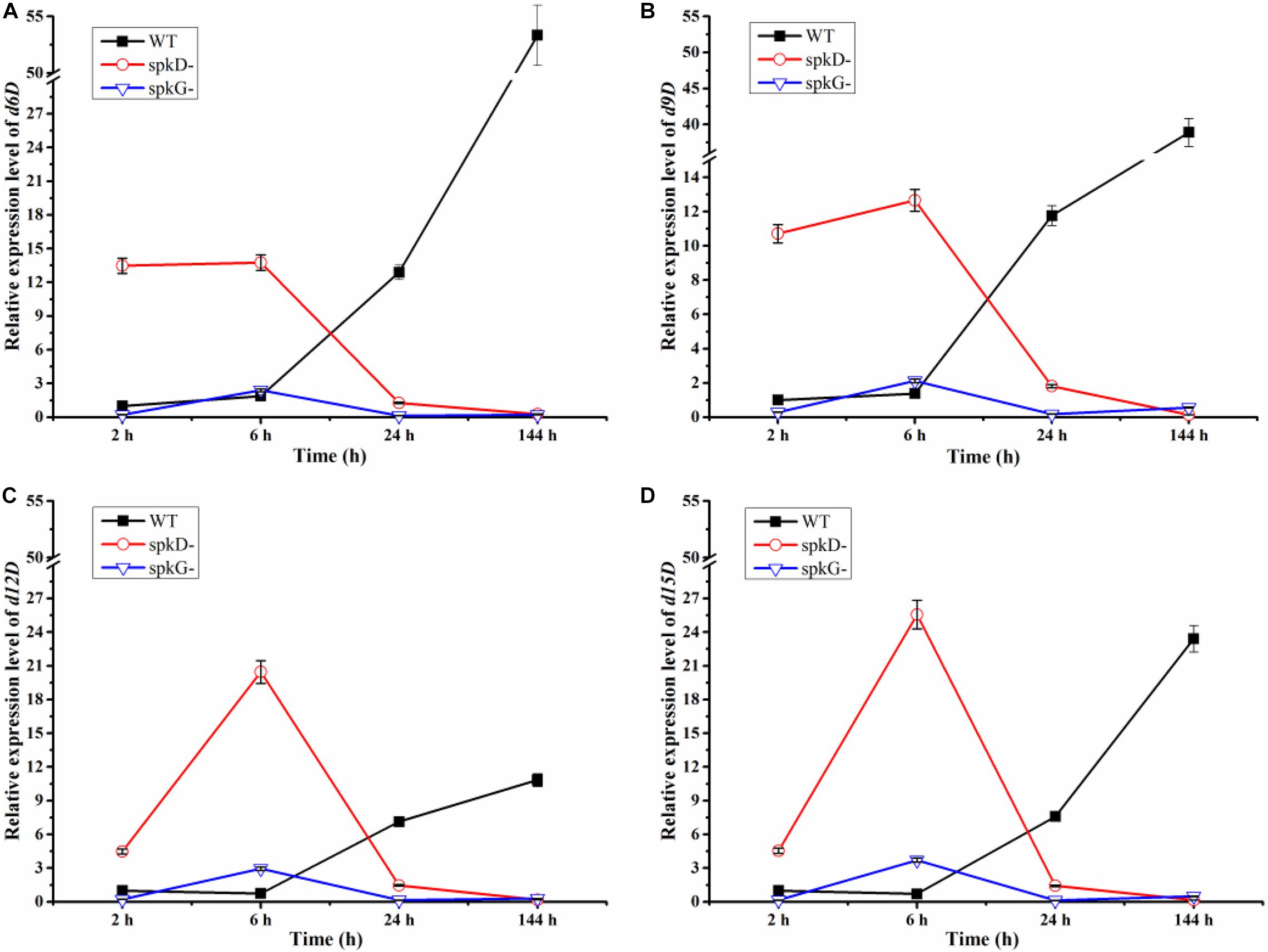
Figure 4. Changes in fatty acid desaturases gene expression in the wild type, spkD and spkG knockout mutants under normal light conditions. WT represents wild type Synechocystis sp. PCC6803; spkD- represents the spkD knockout mutant; spkG- represents the spkG knockout mutant. The experiment was carried out under a normal light intensity of 40 μmol⋅m–2⋅s–1. (A–D show the relative expression levels of the d6D, d9D, d12D, and d15D fatty acid desaturases genes, respectively, in wild type Synechocystis sp. PCC6803 and the two knockout mutants) Values are means ± SD (bars) of three independent experiments conducted on different days. The absence of a bar indicates that the SD falls within the symbol.
In addition, we compared the expression levels of the four fatty acid desaturases genes in the two mutant strains. In the spkG knockout mutant, the expression of the four fatty acid desaturases genes was maintained at relatively low levels. The expression patterns of all four genes in the spkD knockout mutants were opposite to those of the wild type. These results indicate that knockout of spkG affects the expression of fatty acid desaturases genes and that this gene has a greater influence on fatty acid desaturases gene expression than spkD.
We speculate that, after 6 h of light exposure, the expression level of fatty acid enzymes of the mutant strains transiently increased compared with the wild type. After 24 h of light treatment, the expression level of these genes decreased in the mutants, to levels that were lower than in wild-type Synechocystis sp. PCC6803. After 144 h of light exposure, the knockout of spkD and spkG in the mutant strain further inhibited the expression of fatty acid enzyme genes, leading to further reductions in their expression in the mutants. Thus, deletion of spkD and spkG decreased the expression of the four genes (encoding d6D, d9D, d12D, and d15D).
Changes in Fatty Acid Contents in the spkD and spkG Knockout Mutants
To characterize differences in fatty acid content between the mutant and wild type, we analyzed the fatty acid contents of Synechocystis sp. PCC6803 cultured at 30°C and a light intensity of 40 μmol⋅m–2⋅s–1 by gas chromatography. There was little difference in total fatty acid content in the wild type and mutant strains (Figure 5). However, the contents of C18: 2, C18: 3n6, C18: 3n3, and C18: 4 were slightly lower in the mutants than in the wild type. The contents of other fatty acids were lower in the wild type than in the mutants, and the contents of these fatty acids in the two mutants were similar. The C18: 3n3 and C18: 4 contents were significantly lower in the spkG than in the spkD knockout mutant.
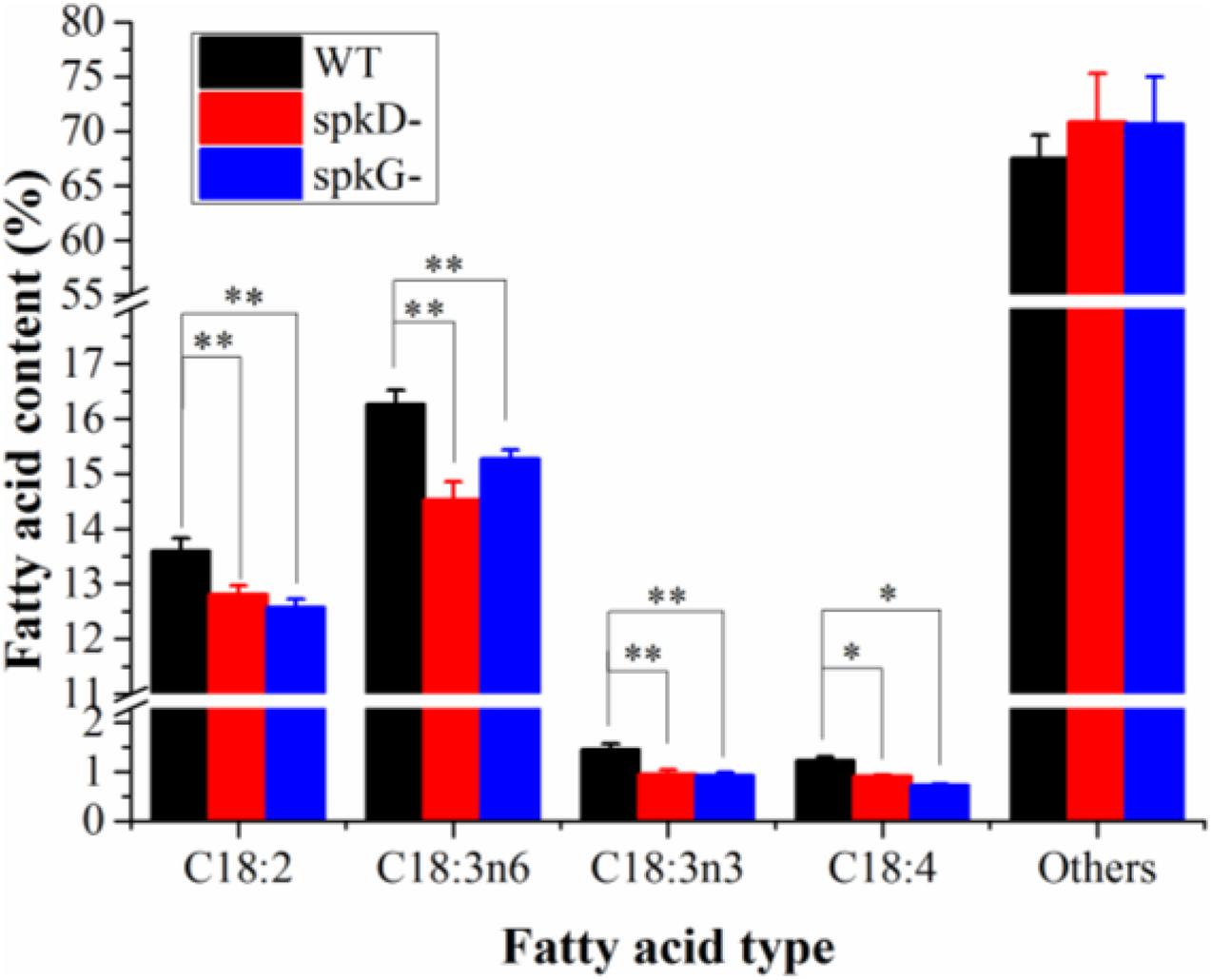
Figure 5. Fatty acid composition and content in spkD and spkG knockout mutants. Data are the mean values of three experiments; the cultivation temperature was 30°C and the light intensity was 40 μmol⋅m–2⋅s–1; WT: Synechocystis sp. PCC6803; spkD-: spkD knockout mutant strain; spkG-: spkG knockout mutant strain. Values are means ± SD (bars) of three independent experiments conducted on different days. The absence of a bar indicates that the SD falls within the symbol. Asterisks mark statistically (t-test) significant differences from the WT value (∗∗p < 0.01; ∗p < 0.05).
Discussion
Genetic engineering to modify metabolic pathways represents an efficient, convenient method for increasing the yields and types of unsaturated fatty acids produced by an organism. The fatty acid biosynthetic pathways in plants and cyanobacteria have been well studied. Synechocystis sp. PCC6803 is a single-cell cyanobacterium, making it an ideal choice for studying fatty acid biosynthetic pathways whose potential as a tool for the commercial production of biofuels and fatty acids has been explored (Ruiz-López et al., 2012). For example, the delta 6 and delta 15 fatty acid desaturases genes were transferred into Synechocystis sp. PCC6803 to produce unsaturated fatty acids (Chen et al., 2014).
Cyanobacteria are the earliest photosynthetic autotrophic organisms. During the long evolutionary process, cyanobacteria developed an efficient adaptive system to cope with the constant changes in the external environment. For example, a complex signal transduction network system in Synechocystis sp. PCC6803 allows it to adapt to a variety of environments (Lei et al., 2014). The prokaryotic cyanobacteria are dominated by a binary signal transduction system. In 1991, eukaryotic protein kinases including STKs were also identified in the prokaryotic organism Myxococcus and shown to play major roles in prokaryotic cells (Zorina et al., 2014).
STKs contain multiple genes, and two mutant strains, spkD and spkG, were successfully obtained by knockout. The genes knocked out in the spkD and spkG strains have been well characterized, but the association between these genes and fatty acids remains to be explored. Based on this, we studied these two genes.
Here, to verify the roles of STKs in unsaturated fatty acid biosynthesis in Synechocystis sp. PCC6803, we knocked out these genes in this cyanobacterium and obtained gene knockout mutants. Gas chromatography analysis revealed that the contents of C18: 2, C18: 3n6, C18: 3n3, and C18: 4 were significantly lower in the two mutant strains than in the wild type. The C18: 3n3 and C18: 4 contents were significantly lower in the spkG than in the spkD knockout mutant, whereas the contents of other unsaturated fatty acids such as C18: 2 and C18: 3n6 in the two mutants were similar.
To verify this finding, we extracted RNA from the cultures and performed qRT-PCR to analyze fatty acid-related gene expression levels. Fatty acid desaturases genes were expressed at higher levels in the two mutant strains vs. the wild type, and the expression levels of these genes in the spkD and spkG knockout mutants significantly differed at different time points and under different light intensities. These findings suggest that spkD and spkG play different roles in unsaturated fatty acid biosynthesis.
We also analyzed the changes in fatty acid desaturases gene expression in the mutants and wild type at different time points under normal light intensity. The expression levels of these genes in the wild type and mutant strains were significantly different. Under normal light conditions (40 μmol⋅m–2⋅s–1), the fatty acid desaturases genes were expressed at higher levels in the spkD- mutant than in the spkG- mutant prior to 144 h of cultivation. Unlike the wild type, these genes were expressed at their highest levels in the mutants at 6 h of light treatment, with the most obvious differences detected in the spkD knockout mutant. After the 6 h time point, the expression levels of these genes in the wild type continued to increase. These results indicate that knockout of spkD and spkG indeed affects the relative fatty acid desaturases contents of Synechocystis sp. PCC6803, indirectly demonstrating that the two genes are closely related to fatty acid biosynthesis.
Subsequently, we detected the changes in expression levels of other STK genes and established that spkD and spkG influence the expression patterns of these genes as well, as the knockout of these genes significantly altered the expression levels of other STK genes. Our findings suggest that in Synechocystis sp. PCC6803, spkD is most closely associated with spkE expression and spkG is somewhat associated with spkC, spkD, spkE, and spkF expression.
In addition, with the extension of light time, in contrast to wild type Synechocystis, the expression of other STKs genes in the spkD knockout mutants initially increased, before decreasing. Perhaps spkD is related to the pre-expression phase of these genes, a process that might involve some inhibitory factors or other genes. Compared to the spkD knockout mutant, other STK genes were significantly down regulated in the spkG knockout mutant, indicating that spkG is more closely associated with the expression of other STK genes than is spkD. The expression levels of the STK genes of these two knockout mutants peaked after approximately 6 h of light exposure. The importance of this time point will be the subject of subsequent experiments.
As we had expected, in the subsequent determination of fatty acid content, we also noticed that several fatty acids were significantly higher in the wild type than in the two knockout mutant strains. This difference has to do with fatty acid desaturases and coincides with the results of STKs obtained previously, further illustrating that spkD and spkG are associated with fatty acid metabolism and are worth further examination.
A previous related study indicated that sll0776 (spkD) is part of the gene cluster (sll0775, sll0776, sll0777, and sll0778). However, the intergenic region of this gene cluster could not be amplified by qRT-PCR, indicating that sll0776 was not organized in an operon with the adjacent genes, and the protein sequences encoded by the other genes had low similarity with the corresponding proteins in the database. Therefore, we chose spkD gene as the research object. spkD is not thought to be an essential gene, but may be associated with the TCA cycle. Deletion of spkD maybe affects cyanobacterial growth, and the growth can be recovered by adding specific metabolites of HCO3– or TCA cycle as auxiliary elements (Laurent et al., 2008). In our research, the STK-related genes were expressed at higher levels in the spkD deletion mutant strains in the first 6 h, and gradually decreased in the later stages. We speculated that this may be due to a lack of a bit substances related to growth (Figure 3). Another study revealed that certain cyanobacterial genes are expressed soon after exposure to light, and their expression gradually decreases or even disappears at later stages (He et al., 2001). This may also explain why spkA, spkB, spkC, spkF, and spkE are expressed at relatively high levels suddenly after exposure to light, however, the specific mechanism remains to be elucidated.
Recent studies of spkG (slr0152) also identified a relationship between spkG kinase and fatty acids. The slr0144–slr0152 gene cluster encodes a protein that was recently annotated as Ferredoxin 5 (Fd5), which appears to be a phosphoprotein. SpkG kinase is involved in the phosphorylation of Ferredoxin 5 (Fd5) (Angeleri et al., 2018). Our study confirms that spkG influences the contents of fatty acids. Furthermore, knockout of spkG (Δslr0152) will not have an obvious effect on the expression of genes that function downstream of genes in the cluster. We are planning to study other genes in the gene cluster to further explore the connection between spkG and fatty acid metabolism. As slr0151 and spkG have similar functions, we speculate that spkG kinase and slr0151 protein may have complementary functions, which would result in the loss of spkD inhibiting cyanobacterial growth to a greater extent than the loss of spkG. This would also explain why other STKs and fatty acid desaturases are altered greater in spkD deletion mutants than in spkG deletion mutants (Figures 3, 4).
In this study, we successfully constructed spkD and spkG gene knockout mutants in Synechocystis sp. PCC6803 via insertional inactivation to study the effects of these genes on fatty acid biosynthesis. Under normal light conditions, the expression levels of fatty acid desaturases genes were significantly higher in the wild type than in the two knockout mutants. Knockout of spkD and spkG also affected the expression of other STK-related genes to different extents, indicating that these genes play different roles in fatty acid biosynthesis. In addition, the contents of several major fatty acids were lower in the mutant strains than in the wild type. Our results indicate that STKs affect fatty acid biosynthesis, and spkD and spkG directly or indirectly participate in the regulation of STK gene expression. In addition, the knockout of these two genes affected fatty acid biosynthesis. However, how specific genes cooperate with each other to regulate fatty acid biosynthesis mechanisms remains to be determined. We plan to conduct more in-depth research to improve the biosynthesis of fatty acids in cyanobacteria. This study lays the foundation for further improving the fatty acid biosynthesis pathway in cyanobacteria and for the efficient production of PUFAs.
Materials and Methods
Strains and Growth Conditions
The cyanobacterium Synechocystis sp. PCC6803 was obtained from the Freshwater Algae Culture Collection of the Institute of Hydrobiology, Chinese Academy of Sciences. Synechocystis sp. PCC6803 was cultivated in BG-11 medium at 30°C (Chen et al., 2014). For solid BG-11 medium, 1.5% (w/v) Difco Bacto-agar (Becton Dickinson, Sparks, MD, United States), 0.3% (w/v) sodium thiosulfate, and 10 mM TES 2- [(2-hydroxy-1, 1-bis (hydroxylmethyl) ethyl) amino] ethanesulfonic acid pH 8.2 were added to BG-11 medium. The culture was bubbled with air under a light intensity of 40 μmol⋅m–2⋅s–1 (Teruo and Kaplan, 2003). Transformed strains were selected by adding 50 μg/mL kanamycin (Dingguo Company, Beijing, China) to both liquid and solid BG-11 medium. The mutant and wild type in the logarithmic growth phase were added to the liquid medium without kanamycin. Under a light intensity of 40 μmol⋅m–2⋅s–1, 30°C, after waiting for a period of growth (about 3–5 days), the OD730 value was uniformly adjusted to 1.0. Samples were collected and measured at the same time each day. Cell density was determined by measuring the optical density (OD) of the suspension at 730 nm (OD730) with a spectrophotometer (DU-70, Beckman Coulter, Brea, CA, United States).
Generation of the spkD and spkG Mutants
A 1.9 kb DNA fragment including the spkD coding region (sll0776, GenBank: AB046600) was amplified by PCR from genomic DNA of Synechocystis sp. PCC6803 using the primer pair spkD-F and spkD-R (Table 1). A 1.9 kb DNA fragment including the spkG (slr0152, GenBank: CP028094) coding region was amplified by PCR from genomic DNA of Synechocystis sp. PCC6803 using the primer pair spkG-F and spkG-R (Table 1). The amplified DNA fragments were cloned separately into the pClone007 simple vector (TSINGKE Biological Technology). In our experiments, the EcoRI restriction site was found at position 1,327 bp of the spkD gene, and the BamHI restriction site was found at position 866 bp of the spkG gene. We designed primers for the two gene sequences and inserted Kan fragments using enzyme digestion. The mutant was constructed by inserting a 1.2 kb kanamycin resistance cassette into the restriction site of the amplified DNA fragment. The Synechocystis sp. PCC6803 strain was transformed as described (Kamei et al., 2001). Transformants were selected on standard medium containing 50 μg/mL kanamycin. Complete segregation of the mutant was confirmed by PCR.
RNA Isolation and cDNA Synthesis
Wild type and transformant cell lines were cultured and harvested during the exponential growth phase, and total RNA was isolated from the samples using Trizol Reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instructions. First-strand cDNA was synthesized using M-MLV reverse transcriptase and modified oligo (dT) following the manufacturer’s instructions (TaKaRa, Dalian, China).
Quantitative Reverse-Transcription PCR
Quantitative reverse-transcription PCR (qRT-PCR) of STK gene expression was carried out RNA Bio-Rad iQ5 real-time PCR system. The resulting cDNA molecules were amplified by PCR using the following gene-specific primers: rnpB-F and rnpB-R to amplify the rnpB gene and 16S rRNA-F and 16S rRNA-R to amplify the 16S rRNA gene, which was used as the loading control. Primers were also designed to amplify the following STK genes (Table 1): spkA (spkB, spkC, spkD, spkG, spkF) -RT-F and spkA (spkB, spkC, spkD, spkG, spkF) -RT-R to amplify spkA (spkB, spkC, spkD, spkG, spkF); d6D (d9D, d12D, d15D) -RT-F and d6D (d9D, d12D, d15D) -RT-R to amplify d6D (d9D, d12D, d15D). Reactions were prepared following the manufacturer’s instructions, and qRT-PCR was performed using a Bio-Rad iQ5 system. Each PCR was repeated four times in a total volume of 20 μL containing 2 × SYBR Green I PCR Master Mix (TaKaRa), 100 nM of each primer, and 1 μL diluted (1:20) template cDNA. Reactions were carried out in 96-well optical-grade PCR plates and a matched optical-grade membrane (TaKaRa). The amplification program was as follows: an initial denaturation step of 1 min at 95°C; 42 cycles of 10 s at 95°C, 30 s at 60°C, and 30 s at 72°C; and then an additional cycle of 10 s at 95°C, 30 s at 58°C, and 5 min at 72°C, followed by 10 s at 95°C for melting curve analysis. The data were analyzed using Bio-Rad iQ5 software. The relative expression levels of STK genes at various developmental stages were calculated using the relative 2–Δ Δ Ct method (Livak and Schmittgen, 2001); the error bars indicate SD (n = 3). Sterile water was used as negative control instead of template in each primer set.
Culture Conditions
Three Synechocystis strains (wild type and mutants) were cultured under normal light conditions (40 μmol⋅m–2⋅s–1) for 10 days at 30°C. After a brief centrifugation (6,000 × g, 10 min at room temperature), the cyanobacteria were added to BG-11 medium to a cell density of 4.578 × 109 ind./L (individual/liter) and cultured under a normal light intensity of 40 μmol⋅m–2⋅s–1 for 6 days. All treatments had four replicate flasks, and cyanobacteria cultured continuously under 40 μmol⋅m–2⋅s–1 served as controls. During the light culture stage, algal growth was estimated each day based on the OD730, as measured with a spectrophotometer. A standard curve relating Synechocystis cell density to OD730 was established using serial dilutions of cyanobacterial culture. Total RNA for subsequent qRT-PCR analysis was isolated from the cyanobacteria at 0, 2, 4, 6, 24, and 144 h.
Calculation of Specific Growth Rate
The specific growth rate of the cyanobacteria was calculated using the following formula: μ = (lnNt–lnN0)/△t, where N0 is the population cell density at the beginning of the time interval, Nt is the cell density at the end of the time interval, and △t is the length of the time interval in days. The growth rate was calculated based on measurements taken from the first 2 days of the experiment onward. Data from the second day was compared with that from the first day, and the rate was calculated by comparison.
Extraction of Total Fatty Acids From Synechocystis
Synechocystis cells were collected in 1,000 mL autoclaved flasks, each containing 400 mL of sterile BG-11 medium, and grown for 10 days at a light intensity of 40 μmol⋅m–2⋅s–1 and a constant temperature of 30°C. When the culture reached OD730 = 2.2, the cyanobacterial fluid was collected by centrifugation (4,500 × g, 10 min at room temperature), and the pellet was washed with distilled water and centrifuged again (4,500 × g, 10 min at room temperature). The washing and centrifugation steps were performed three times (Chen et al., 2017). The cultured cyanobacterial cells were harvested by centrifugation (6,000 × g, 15 min at room temperature) and dried under a vacuum. Each 0.2 g sample of dried cyanobacterial powder was placed in a mortar, repeatedly ground in liquid nitrogen, extracted with 7.0 mL of methanol-chloroform (2:1, v: v), and sonicated for 10 min. After the sample was centrifuged at 6,000 × g for 15 min at room temperature, 1.5 mL of methanol-chloroform (2: 1, v: v) was added to the residue. The sample was centrifuged (6,000 × g, 10 min at room temperature) and the organic phase was retained; this step was repeated once. The organic phase extracts were combined in a separatory funnel. After adding 2.5 mL of chloroform and 3.0 mL of sodium chloride solution (1:100, v: v), the sample was mixed well and allowed to stand for layer separation. The lower layer was recovered, and 2.5 mL of chloroform was added to the original upper and middle layers and extracted again. The lower layers were combined; this step was repeated once. The combined lower layers were placed in a fat-lifting bottle, and the solvent was evaporated to constant weight with nitrogen at 50°C (Sheng et al., 2011; Ryckebosch et al., 2012). The total fatty acids and the total weight of the fat-lifting bottle were measured using an electronic scale.
Analysis of Fatty Acid Composition in Synechocystis
The extracted total fat was dissolved in 4 mL of chloroform in a 10 mL test tube with a stopper, combined with 5 mL of 0.04 M potassium hydroxide-methanol solution, and mixed well. The sample was incubated in a 60°C water bath for 60 min for saponification, with oscillation every 10 min during saponification. The saponified sample was removed from the water bath, cooled, combined with 4.0 mL hydrochloric acid-methanol (1:9) solution, and mixed well. Then 20.0 μL of 1.5 mg/mL non-adecanoic acid (C19: 0) was added to the sample as an internal standard. The sample was methylated in a water bath at 60°C for 20 min with shaking every 10 min during the methylation process (Ichihara and Fukubayashi, 2010). After cooling, the methylated sample was combined with 3.0 mL of saturated saline, followed by 1.0 mL of n-hexane, and shaken thoroughly. After letting the sample stand (at room temperature for about 30 min), the n-hexane layer was subjected to chromatographic analysis. The extracted n-hexane was dehydrated with an appropriate amount of anhydrous sodium sulfate and centrifuged at high speed (13,000 × g, 3 min) at room temperature (about 30°C). The supernatant [Synechocystis sample fatty acid methyl ester (FAME) eluent] was subjected to gas chromatography (GC) using an Elite-wax column in an ASXL instrument (Perkin-Elmer, Waltham, MA, United States) (Kenichi and Yumeto, 2010). The flame ionization detection temperature was 250°C, and the operating temperature was maintained at 220°C. The samples used in the experiment are from the same batch. The value for each sample was averaged over three experiments.
Statistical Analysis
Data are expressed as means ± SD (n = 3). The data were subjected to a t-test to determine significant differences between treatments (∗p < 0.05; ∗∗p < 0.01).
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
GC, XB, and MD conceived and designed the experiments. YC, HZ, XW, YL, and XC performed the experiments. GC contributed reagents, materials, and analysis tools. GC, YC, and XW wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Key Research and Development Program of Shandong (2019GSF107098 and 2018GSF121019), the National Key Research and Development Program of China (2018YFE0108600), the National Natural Science Foundation of China (31772857), the Science and Technology Plan Project of Tianjin (19YFZCSN00070), and the Key Laboratory of Algal Biology, Institute of Hydrobiology, Chinese Academy of Sciences (2018-006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Angeleri, M., Zorina, A., Aro, E. M., and Battchikova, N. (2018). Interplay of SpkG kinase and the Slr0151 protein in the phosphorylation of Ferredoxin 5 in Synechocystis sp. strain PCC 6803. FEBS Lett. 592, 411–421. doi: 10.1002/1873-3468.12970
Chen, G., Chen, J., He, Q. F., Zhang, Y., Peng, Z. Y., Fan, Z. X., et al. (2017). Functional expression of the Arachis hypogaea L. Acyl-ACP thioesterases AhFatA and AhFatB enhances fatty acid production in Synechocystis sp. PCC6803. Energies 10:2093. doi: 10.3390/en10122093
Chen, G., Qu, S. J., Wang, Q., Bian, F., Peng, Z. Y., Zhang, Y., et al. (2014). Transgenic expression of delta-6 and delta-15 fatty acid desaturases enhances omega-3 polyunsaturated fatty acid accumulation in Synechocystis sp. PCC6803. Biotechnol. Biofuels 7, 1–10. doi: 10.1186/1754-6834-7-32
Drexler, H., Spiekermann, P., Meyer, A., Domergue, F., Zank, T., Sperling, P., et al. (2003). Metabolic engineering of fatty acids for breeding of new oilseed crops, strategies, problems and first results. J. Plant Physiol. 16, 779–802. doi: 10.1078/0176-1617-01025
Gong, Y., and Miao, X. L. (2019). Short chain fatty acid biosynthesis in microalgae Synechococcus sp. PCC 7942. Mar. Drugs 17:255. doi: 10.3390/md17050255
Guedes, A., Amaro, H. M., Barbosa, C. R., Pereira, R. D., and Malcata, F. X. (2011). Fatty acid composition of several wild microalgae and cyanobacteria, with a focus on eicosapentaenoic, docosahexaenoic and α-linolenic acids for eventual dietary uses. Food Res. Int. 44, 2721–2729. doi: 10.1016/J.FOODRES.2011.05.020
Hasegawa, P. M., Bressan, R. A., Zhu, J. K., and Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 163–199. doi: 10.1146/ANNUREV.ARPLANT.51.1.463
He, Q. F., Dolganov, N., Bjrkman, O., and Grossman, A. R. (2001). The high light-inducible polypeptides in Synechocystis PCC6803. Expression and function in high light. J. Korean Soc. Appl. Bi. 276, 304–314. doi: 10.1074/jbc.M008686200
Hibbeln, J. R., Nieminen, L. R., Blasbalg, T. L., Riggs, J. A., and Lands, W. E. (2006). Healthy intakes of n-3 and n-6 fatty acids, estimations considering worldwide diversity. Am. J. Clin. Nutr. 83, 1483S–1493S. doi: 10.1186/1476-511X-5-14
Ichihara, K., and Fukubayashi, Y. (2010). Preparation of fatty acid methyl esters for gas-liquid chromatography. J. Lipid Res. 51, 635–640. doi: 10.1194/jlr.D001065
Jan, P. S., Michael, H., Manuela, S., and Andreas, H. (2010). Significance of long-chain polyunsaturated fatty acids (PUFAs) for the development and behaviour of children. Eur. J. Pediatr. 169, 149–164. doi: 10.1007/s00431-009-1035-8
Kalyanee, P., Maria, A. S., Yu, K., Satoh, S., Satoshi, T., Dmitry, A. L., et al. (2004). Five histidine kinases perceive osmotic stress and regulate distinct sets of genes in Synechocystis. J. Biol. Chem. 279, 53078–53086. doi: 10.1074/jbc.M410162200
Kamei, A., Yuasa, T., Orikawa, K., Geng, X. X., and Ikeuchi, M. (2001). A eukaryotic type protein kinase, SpkA, is required for normal motility of the unicellular Cyanobacterium Synechocystis sp. strain PCC6803. J. Bacteriol. 183, 1505–1510. doi: 10.1128/JB.183.5.1505-1510.2001
Kenichi, I., and Yumeto, F. (2010). Preparation of fatty acid methyl esters for gas-liquid chromatography. J. Lipid Res. 51, 635–640. doi: 10.1194/jlr.D001065
Laurent, S., Jang, J. C., Janicki, A., Zhang, C. C., and Be′du, S. (2008). Inactivation of spkD, encoding a Ser/Thr kinase, affects the pool of the TCA cycle metabolites in Synechocystis sp. strain PCC 6803. Microbiology 154, 2161–2167. doi: 10.1099/MIC.0.2007/016196-0
Lei, H. J., Chen, G., Wang, Y. L., Ding, Q. L., and Wei, D. (2014). Sll0528, a site-2-protease, is critically involved in cold, salt and hyperosmotic stress acclimation of Cyanobacterium Synechocystis sp. PCC6803. Int. J. Mol. Sci. 15, 22678–22693. doi: 10.3390/IJMS151222678
Liang, C. W., Zhang, X. W., Chi, X. Y., Guan, X. Y., Li, Y. X., Qin, S., et al. (2011). Serine/Threonine protein kinase spkG is a candidate for high salt resistance in the unicellular Cyanobacterium Synechocystis sp. PCC 6803. PLoS One 6:e18718. doi: 10.1371/journal.pone.0018718
Liu, Z. X., Li, H. C., Wei, Y. P., Chu, W. Y., Chong, Y. L., Long, X. H., et al. (2015). Signal transduction pathways in Synechocystis sp. PCC6803 and biotechnological implications under abiotic stress. Crit. Rev. Biotechnol. 35, 269–280. doi: 10.3109/07388551.2013.838662
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods 25, 402–408. doi: 10.1006/METH.2001.1262
Mayra, M. A., Belen, T. R., Zhen, T., Owen, M. V., Mario, M. M., Shaheen, A., et al. (2020). Importance of the role of ω-3 and ω-6 polyunsaturated fatty acids in the progression of brain cancer. Brain Sci. 10:381. doi: 10.3390/brainsci10060381
Pauly, D., Watsn, R., and Alder, J. (2005). Global trends in world fisheries, impacts on marine ecosystems and food security. Philos. Trans. R. Soc. B 360, 5–12. doi: 10.1098/rstb.2004.1574
Piyoosh, K. B., Kumar, J., and Venkatesh, C. (2019). Proteomic De-regulation in Cyanobacteria in response to abiotic stresses. Front. Microbiol. 10:1315. doi: 10.3389/fmicb.2019.01315
Rajagopal, L., Clancy, A., and Rubens, C. E. (2003). A eukaryotic type Serine/Threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J. Biol. Chem. 278, 14429–14441. doi: 10.1074/JBC.M212747200
Ruiz-López, N., Sayanova, O., Napier, J. A., and Haslam, R. P. (2012). Metabolic engineering of the omega-3 long chain polyunsaturated fatty acid biosynthetic pathway into transgenic plants. J. Exp. Bot. 63, 2397–2410. doi: 10.1093/JXB/ERR454
Ryckebosch, E., Muylaert, K., and Foubert, I. (2012). Optimization of an analytical procedure for extraction of lipids from microalgae. J. Am. Oil Chem. Soc. 89, 189–198. doi: 10.1007/S11746-011-1903-Z
Sara, P. C., Miguel, A. R., Raveender, V., Lai, Y. S., Bruce, E. R., and Roberto, P. (2015). Effects of light intensity and carbon dioxide on lipids and fatty acids produced by Synechocystis sp. PCC6803 during continuous flow. Algal Res. 12, 10–16. doi: 10.1016/J.ALGAL.2015.07.018
Sheng, J., Vannela, R., and Rittmann, B. E. (2011). Evaluation of methods to extract and quantify lipids from Synechocystis PCC 6803. Bioresour. Technol. 102, 1697–1703. doi: 10.1016/J.BIORTECH.2010.08.007
Somayeh, G. F., Anithachristy, S. A., Behnam, T., Chen, H., Lu, J., and Sitther, V. (2019). Augmenting Fremyella diplosiphon cellular lipid content and unsaturated fatty acid methyl esters via sterol desaturase gene overexpression. Appl. Biochem. Biotech. 189, 1127–1140. doi: 10.1007/s12010-019-03055-5
Teruo, O., and Kaplan, A. (2003). Inorganic carbon acquisition systems in cyanobacteria. Photosynth. Res. 77, 105–115. doi: 10.1023/A:1025865500026
Zhang, C. C., and Libs, L. (1998). Cloning and characterisation of the pknD gene encoding an eukaryotic-type protein kinase in the Cyanobacterium Anabaena sp. PCC7120. Mol. Gen. Genet. 258, 26–33. doi: 10.1007/s004380050703
Zhang, X. W., Zhao, F. Q., Guan, X. Y., Yang, Y., Liang, C. W., and Qin, S. (2007). Genome wide survey of putative Serine/Threonine protein kinases in cyanobacteria. BMC Genom. 8:395. doi: 10.1186/1471-2164-8-395
Zhou, P., Wong, D., Li, W., Xie, J., and Av-Gay, Y. (2015). Phosphorylation of Mycobacterium tuberculosis protein tyrosine kinase A PtkA by Ser/Thr protein kinases. Biochem. Biophys. Res. Commun. 467, 421–426. doi: 10.1016/j.bbrc.2015.09.124
Zhou, W. G., Li, Y., Min, M., Hu, B., Chen, P., and Ruan, R. (2011). Local bioprospecting for high-lipid producing microalgal strains to be grown on concentrated municipal wastewater for biofuel production. Bioresour. Technol. 102, 6909–6919. doi: 10.1016/j.biortech.2011.04.038
Zorina, A., Stepanchenko, N., Novikova, G. V., Sinetova, M., Panichkin, V. B., Moshkov, I. E., et al. (2011). Eukaryotic-like. Ser/Thr protein kinases SpkC/F/K are involved in phosphorylation of GroES in the Cyanobacterium Synechocystis. DNA Res. 18, 137–151. doi: 10.1093/dnares/dsr006
Zorina, A. A., Bedbenov, V. S., Novikova, G. V., Panichkin, V. B., and Los, D. A. (2014). Involvement of serine/threonine protein kinases in the cold stress response in the Cyanobacterium Synechocystis sp. PCC6803: functional characterization of SpkE protein kinase. Mol. Biol. 48, 390–398. doi: 10.1134/S0026893314030212
Keywords: microalgae, serine/threonine kinase system, polyunsaturated fatty acids, biosynthesis, Synechocystis sp. PCC6803
Citation: Chen G, Cao Y, Zhong H, Wang X, Li Y, Cui X, Lu X, Bi X and Dai M (2021) Serine/threonine Kinases Play Important Roles in Regulating Polyunsaturated Fatty Acid Biosynthesis in Synechocystis sp. PCC6803. Front. Bioeng. Biotechnol. 9:618969. doi: 10.3389/fbioe.2021.618969
Received: 19 October 2020; Accepted: 04 January 2021;
Published: 21 January 2021.
Edited by:
Chengwei Liang, Qingdao University of Science and Technology, ChinaReviewed by:
Corinne Cassier-Chauvat, UMR9198 Institut de Biologie Intégrative de la Cellule (I2BC), FranceXiaojin Song, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), China
Copyright © 2021 Chen, Cao, Zhong, Wang, Li, Cui, Lu, Bi and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangdong Bi, eWw4MDExMjNAYWxpeXVuLmNvbQ==; Meixue Dai, ZGFpbWVpeHVlQHNkbnUuZWR1LmNvbQ==
 Gao Chen
Gao Chen Yuelei Cao
Yuelei Cao Huairong Zhong2
Huairong Zhong2 Meixue Dai
Meixue Dai