
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bioeng. Biotechnol. , 16 November 2020
Sec. Biomaterials
Volume 8 - 2020 | https://doi.org/10.3389/fbioe.2020.598450
Membrane proteins (MPs) are essential to many organisms’ major functions. They are notorious for being difficult to isolate and study, and mimicking native conditions for studies in vitro has proved to be a challenge. Lipid nanodiscs are among the most promising platforms for MP reconstitution, but they contain a relatively labile lipid bilayer and their use requires previous protein solubilization in detergent. These limitations have led to the testing of copolymers in new types of nanodisc platforms. Polymer-encased nanodiscs and polymer nanodiscs support functional MPs and address some of the limitations present in other MP reconstitution platforms. In this review, we provide a summary of recent developments in the use of polymers in nanodiscs.
MPs are essential for cell homeostasis, signaling, transport, and energy production. They play critical roles in normal function and human disease, and as such they are important drug targets (Overington et al., 2006). MPs have an integral role in the pathophysiology of diseases, such as cystic fibrosis, hypertension, diabetes, cerebrovascular accidents, cardiac infarcts, and neurodegenerative diseases, to name a few (Lee and White, 2009; Orellana et al., 2014; Beyder and Farrugia, 2016; Kumar et al., 2016; Schmidt et al., 2016; Zaucha et al., 2020). At least 50% of current druggable targets are classified as MPs, and about half of all known small molecule drugs bind to them (Russell and Eggleston, 2000; Overington et al., 2006). Therefore, there is high priority in understanding MPs because of their importance to improve therapeutics for many medical conditions. Yet, progress in the MP field has been slow in comparison to that of other biomolecules because of difficulties in overexpression, membrane extraction, and purification, which frequently vary even for proteins within the same family (Seddon et al., 2004; Hardy et al., 2018). Maintaining an environment conducive to MP function and stability for evaluation in vitro is still a major challenge.
Studies of proteins in their native environment are critical to understanding their function and regulation in organisms and cells, but details on their structures and mechanisms at the atomic and molecular level are better investigated on purified proteins under well-controlled conditions. In the case of MPs, a complete understanding requires working with purified proteins reconstituted in model membranes that are robust enough to support research in an abiotic environment for extended periods of time. And yet, MPs are notoriously difficult to isolate and study due to the hurdle of discovering native-membrane-mimicking environments that cater to their stability while also support their functions. Ideally, a purified MP of interest should be incorporated in a membrane in which it can operate as it would in the native cell membrane, but in a solubilized and stable state where it can be subjected to analyses of its structure, function, and regulation.
In most cases, studies of purified MPs include expression through recombinant methods followed by extraction from membranes using detergents to produce the proteins solubilized in detergent micelles (Figure 1; Seddon et al., 2004; Hardy et al., 2018; Stroud et al., 2018). Once solubilized, purification can proceed using methodologies typically employed to purify soluble proteins, but in the continuous presence of detergent to keep the MP in a soluble state. The purification methodologies frequently include liquid chromatography based on fusion tags added to the protein (e.g., His tag, FLAG tag) and/or on the intrinsic properties of the protein (e.g., size-exclusion chromatography, ion-exchange chromatography) (Seddon et al., 2004; Morrison et al., 2016). While detergents are the most common agents used for MP extraction, finding the best detergent can be time consuming (Le Maire et al., 2000). Even if the best detergent is found, decreased MP stability and alterations in MP structure and function are possible (Leitz et al., 2006; Brewer et al., 2011; Hagn et al., 2013; Denisov and Sligar, 2016; Viegas et al., 2016; Zoghbi et al., 2016). Therefore, it is desirable to reconstitute the MPs into a lipid-bilayer-like environment. The simplest solution is adding lipids to the detergent to form detergent-lipid mixed micelles (Seddon et al., 2004; Skrzypek et al., 2018). However, heterogeneities make these micelles suboptimal for biophysical studies (Zoghbi et al., 2017). Drawbacks on traditional methods have prompted investigations of new methodologies for the extraction, isolation, and reconstitution of MPs that are aimed at overcoming logistical hurdles. There are several platforms available for MP reconstitution, including planar lipid bilayers, liposomes, and nanodiscs (Skrzypek et al., 2018). Here we focus on the latter, which constitute an MP platform of excellent properties that offers great potential.
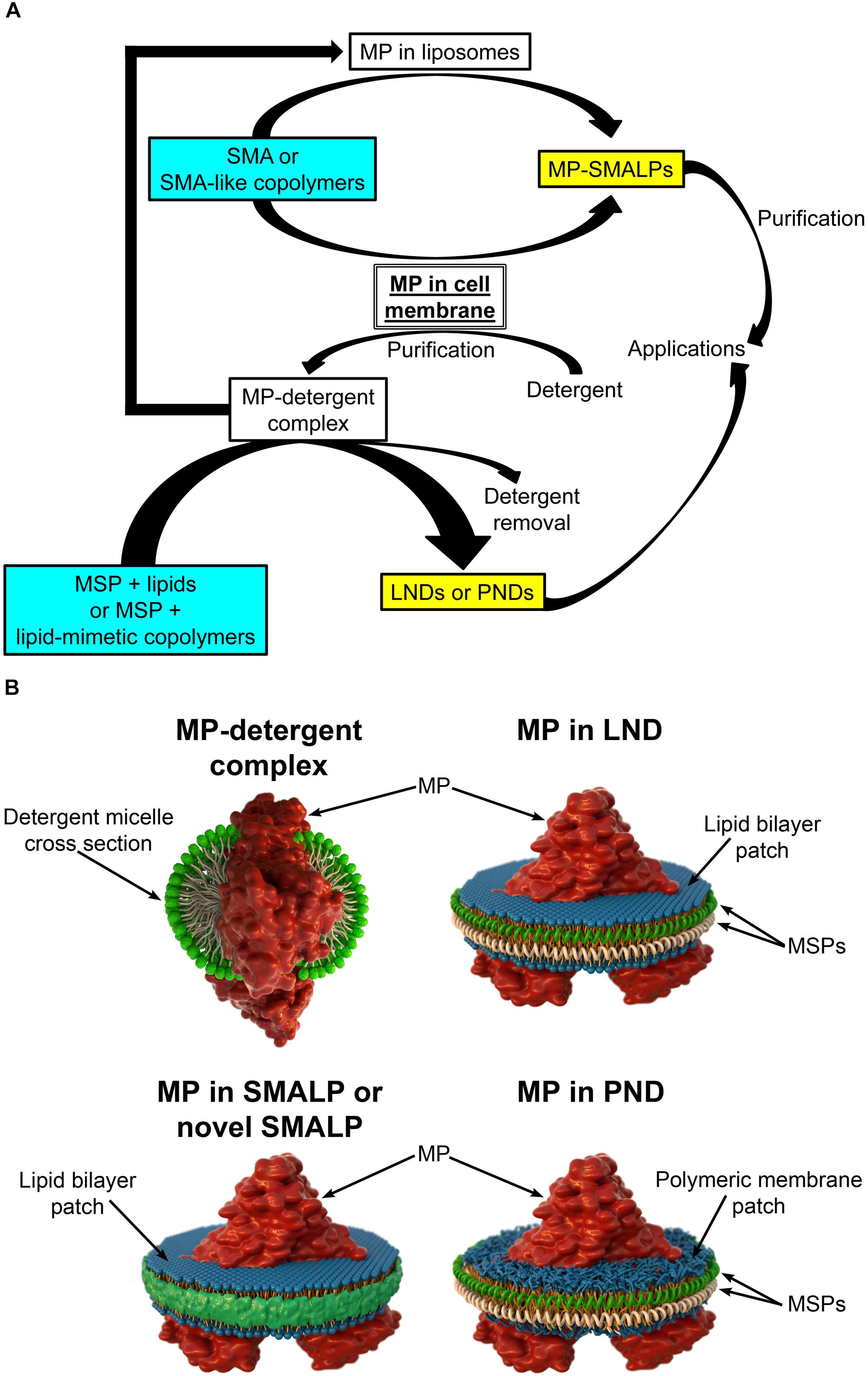
Figure 1. Different types of nanodiscs. (A) Schematic representation of the steps involved in the production of different types of membrane protein (MP)-loaded nanodiscs. Note that styrene maleic acid (SMA) and SMA-like copolymers can yield SMA lipid particles (SMALPs or novel SMALPs) in a single solubilization/reconstitution step without the need for detergents if cell membranes are the starting material. The soluble MPs reconstituted in SMALPs can then be purified in the absence of detergent. In contrast, production of lipid (LNDs) and polymer (PNDs) nanodiscs requires MPs solubilized in detergent. (B) Schematic representation of a MP in a detergent micelle and in different types of nanodiscs.
Lipid nanodiscs (LNDs) are composed of a disc-shaped phospholipid bilayer surrounded by a two-molecule belt of a membrane scaffold protein (MSP) (Figure 1B), a protein derived from apolipoprotein A1, a major component of serum high-density lipoprotein complexes (Denisov and Sligar, 2016). Usually, these nanostructures are referred to as “nanodiscs.” However, here we use the term LNDs to differentiate them from other types of nanodiscs discussed below. LNDs contain a lipid bilayer that provides MPs with stability and access from both sides of the membrane. MP-loaded LNDs are nanometer-scale structures that can be produced in a monodisperse solution and are therefore suitable for most biochemical and biophysical assays, including optical and NMR spectroscopies, as well as cryo-electron microscopy (Denisov and Sligar, 2016, 2017; Thonghin et al., 2018). Although LNDs have emerged as an excellent MP platform for many applications, reconstitution into LNDs requires the use of detergents for extraction of the MP from the membrane (Figure 1A). Thus, composition of the lipid bilayer in the nanodiscs is not the same as that of the native lipid bilayer but instead that of the composition selected during LNDs preparation, potentially affecting protein structure, activity, and regulation. Furthermore, even though LNDs are still not broadly used in biotechnology and biomedical applications, the labile nature of lipid bilayers and poor chemical versatility could restrict those applications. Due to these limitations, there is a continuous interest in new developments, which has resulted in new types of nanodiscs based on the use of polymers. These can have advantages over LNDs for some applications and can replace detergents for the extraction of MPs from membranes and for protein purification.
Nanoparticles based on styrene maleic acid (SMA) copolymers constitute a promising approach for MP extraction and purification in a native bilayer environment (Dorr et al., 2016; Lee et al., 2016; Stroud et al., 2018). Solubilization of membranes with SMAs results in the formation of SMA lipid particles (SMALPs), nanostructures consisting of the native lipid bilayer encircled by a belt of amphipathic SMA copolymers that replace the MSPs of LNDs (Figure 1; Morrison et al., 2016; Stroud et al., 2018). SMALPs are also referred to as native nanodiscs or Lipodisqs. SMA copolymers with various styrene-to-maleic acid ratios can be synthesized to form polymers with different hydrophobicity and solubilization properties (Morrison et al., 2016). Direct solubilization of membranes by SMAs allows for purification of MPs in the absence of detergents (Figure 1A), avoiding their detrimental effects (Stroud et al., 2018). Because MP-loaded SMALPs consist of a small patch of the native lipid bilayer with a MP, stability of the latter is expected to be higher than that of the protein solubilized in detergent, allowing for studies of structure and function in a native-like environment (Dorr et al., 2014; Lee et al., 2016; Parmar et al., 2018; Stroud et al., 2018; Routledge et al., 2020). Although transport measurements cannot be performed in SMALPs (or LNDs), both sides of the MP are accessible for straightforward measurements of activity and structural changes that result from binding of ligands and modulators.
While SMAs are promising alternatives to traditional solubilization and reconstitution methods, their use presents serious limitations. First, there is a limited pH range in which SMAs are stable and will form SMALPs, with the styrene-to-maleic acid ratio and the charge of carboxylic groups on the maleic acid units affecting the pH range in which SMALPs are soluble (Banerjee et al., 2012; Fiori et al., 2017a; Ravula et al., 2017a; Hall et al., 2018a). At lower pHs, SMAs become insoluble because of their increased hydrophobicity, making them unsuitable for forming SMALPs with MPs that function at low pH or for downstream applications that involve studies at pH <∼6.5 (Fiori et al., 2017a; Ravula et al., 2017a, 2019). Second, the size of SMALPs may be too small to contain “large” MPs. The average diameter of SMALPs is 6–9 nm, which can restrict the proteins that can be studied (Fiori et al., 2017a, 2020; Ravula et al., 2017b, 2019). Third, SMA polymers can bind divalent cations, such as Mg2+ and Ca2+, reducing the charge of the maleic acid units, which results in precipitation of the copolymers out of solution (Fiori et al., 2017a, 2020; Ravula et al., 2017a). The sensitivity to divalent cations can be restrictive to downstream analyses that require their presence, such as measurements of ATPase activities (Fiori et al., 2017a).
Recently, a method was developed to improve downstream applications of MPs solubilized with SMA (Hesketh et al., 2020). The approach takes advantage of the precipitation of the SMA with Mg2+ while maintaining the MP soluble with added amphipols or detergents (Hesketh et al., 2020). This platform exchange method results in a product that is compatible with millimolar concentrations of Mg2+, displays increased homogeneity when compared to SMALPs, and is suitable for mass spectrometry, and probably other downstream applications, such as electron microscopy (Hesketh et al., 2020). Since native membrane lipids present in the SMALPs are transferred to the new exchange platform (Hesketh et al., 2020), stability of the purified MP may be enhanced and studies on the regulation of function by native lipids are possible.
Given the difficulties in using traditional SMAs, there has been emerging research aimed at creating new SMA-like copolymers with broader buffer compatibilities that can also form SMALPs of larger sizes. The maleic acid groups of SMA are easily accessible and amenable to modification, enabling the addition of different chemical groups to create new polymers. One of the promising SMA-like copolymers that we developed to address the buffer incompatibilities of SMAs and the size limitations of SMALPs are the zwitterionic styrene maleic amides (zSMAs) (Fiori et al., 2017a, 2020). In these copolymers, the anionic carboxyl acids are replaced with zwitterionic phosphatidylcholine groups. Contrary to SMAs, zSMAs do not precipitate out of solution at low pH or in the presence of polycations (Fiori et al., 2017a). Moreover, well-defined zSMAs of varying copolymer sizes can be used to control the diameter of the resulting zSMALPs (Fiori et al., 2017a). There are other new copolymers that have been produced recently which are also stable at acidic pH, do not precipitate in the presence of millimolar concentrations of divalent cations, and/or form nanostructures larger than SMALPs. These include diisobutylene/maleic acid copolymer (DIBMA), poly(styrene-co-maleimide) (SMI), SMA-ethanolamine (SMA-EA), SMA-ethylenediamine (SMA-ED; a zwitterionic form of SMA-EA), SMAd-A (formed by dehydrating SMA-ED), styrene maleimide quaternary ammonium (SMA-QA; a positively charged SMA derivative), and SMA-SH (with thiol groups on the polar side chains) (Lindhoud et al., 2016; Oluwole et al., 2017; Ravula et al., 2017a,b, 2019; Hall et al., 2018a). The latter is unique in that fluorescent tags and dyes can be attached to it, permitting SMALP affinity purification and fluorescence detection among many potential applications (Lindhoud et al., 2016). For zSMA and other SMA-like copolymers synthesized by reversible addition-fragmentation chain transfer, or RAFT polymerization, it is simple and convenient to convert the chain-transfer moiety on the polymers to a thiol group for affinity purification or fluorescence detection.
As SMAs, SMA-like copolymers can be used to extract and reconstitute MPs directly from cell membranes in a single-step (Figure 1A; Lee et al., 2016; Stroud et al., 2018). This creates a clear advantage over LNDs in that detergents are not necessary, and the composition of the lipid bilayer that surrounds the reconstituted MP is expected to be less drastically different from that in the native membranes (Lee et al., 2016; Stroud et al., 2018). The latter is important when studies are aimed to reproduce native-like conditions as closely as possible. In addition, purified MPs reconstituted in liposomes can be re-solubilized to form traditional or novel SMALPs where the lipid composition of the bilayer can be controlled through the composition of the liposomes (Figure 1A; Ravula et al., 2017b; Fiori et al., 2020).
In the case of zSMAs, our copolymers series was produced by RAFT polymerization (Fiori et al., 2017a, 2020). Compared to those produced by other methods, RAFT polymerization yields polymers of more uniform size (lower polydispersity index), which seems to be an advantage for solubilization/reconstitution (Smith et al., 2017; Fiori et al., 2020). Although a gradient increase of styrene repeating units at the end of individual copolymer molecules is possible during any polymerization process, it can be minimized in RAFT polymerization by precisely controlling the conversion of styrene in the styrene-maleic anhydride random copolymers (Fiori et al., 2017a, 2020; Smith et al., 2017). This is important because a terminal polystyrene homopolymer segment could reduce solubilization efficiency (Hall et al., 2018b).
Using RAFT polymerization, we produced zSMA copolymers of varying sizes and styrene-to-maleic amide molar ratios (Fiori et al., 2020). Testing of these copolymers showed that they are effective agents to solubilize different MPs directly from membranes or from proteoliposomes containing purified MPs (Fiori et al., 2017a, 2020). The resulting nanoparticles have hydrodynamic diameters of 10–30 nm, depending on the polymer chain size (Fiori et al., 2017a). Extensive studies of solubilization into zSMALPs under varied experimental conditions indicate that the solubilization/reconstitution by zSMAs is better than that by the equivalent SMAs, and is very robust in the sense that it occurs with similar efficiency under a variety of conditions that include differences in buffer composition, pH and temperature (Fiori et al., 2020). This has the important practical implication that establishing solubilization/reconstitution protocols will not necessitate extensive trials.
Unexpectedly, we found that a zSMA with a 2:1 styrene:maleic amide molar ratio was better than a zSMA with a 1:1 ratio at thermal stabilization of the ATP-binding cassette protein MsbA, but the ATPase activity of MsbA was higher with the 1:1 zSMA (Fiori et al., 2020). Although the underlying reasons for the differences are unclear, the results point to our limited understanding of the interactions of the SMA-like copolymers with the lipid bilayer and/or the reconstituted MPs. Regardless, we can appreciate the objective advantages of the new SMA-like copolymers in the context of studies at low pH or in the presence of divalent cations, as well as the greater control on nanodisc size and stability.
As mentioned above, even though the use of LNDs and SMALP-like nanoparticles in biotechnology and biomedical applications is so far very limited, it is obvious that the fluid and labile characteristics of the lipid bilayer in both types of nanodiscs will constitute a limitation for stability and long-term storage without disturbances in the nanodiscs structure. Although studies of nanodiscs stability are very scarce, we found that LNDs start to aggregate within a week, even at 4°C, and that they aggregate much faster at higher temperatures or after freeze/thaw cycles (Fiori et al., 2017b).
Synthetic polymeric membranes seek to retain the soft properties of the native bilayers combined with the stability and chemical flexibility of polymers. Because natural membranes are intricate and complex, the custom design and synthesis of polymers allow for more control and versatility of the membrane properties and to tailor these properties to specific research, biotechnology, and biomedical applications. Like natural lipid bilayers, synthetic membranes can incorporate MPs as well as hydrophobic and amphipathic compounds (Goers et al., 2018; Yorulmaz Avsar et al., 2019). Given that lipids can have specific effects on MPs (Alvis et al., 2003; Opekarova and Tanner, 2003), it is notable that endogenous lipids can be inserted, if necessary, and function normally within these synthetic membranes (Seneviratne et al., 2018).
MPs, such as aquaporin and ATP synthase have been incorporated into polydimethylsiloxane (PDMS)-based triblock copolymer membranes, which are viscous and fluid at room temperature (Meier et al., 2000; Kumar et al., 2007; Kowal et al., 2014; Itel et al., 2015). To address the lack of stability of these fluid membranes, we have developed polybutadiene (PBD)- and polystyrene (PS)-based block copolymers, both of which support successful functional reconstitution of MPs while displaying remarkable versatility and stability (Hua et al., 2011; Kuang et al., 2014a,b; Fiori et al., 2017b). Thus, through varying the composition and structure of the block copolymers to change flexibility and fluidity of the self-assembled polymeric membranes, synthetic amphiphilic block copolymers are emerging as new tools for the reconstitution of MPs with preservation of their structural integrity and function.
Synthetic polymeric membranes have been used in various platform types for reconstitution of MPs, including polymer nanodiscs (PNDs) (Meier et al., 2000; Kumar et al., 2007; Hua et al., 2011; Kuang et al., 2014b; Kowal et al., 2015; Fiori et al., 2017b). PNDs constitute a new platform developed to address the limitation in stability and versatility of LNDs and SMALPs. PNDs do not contain biological lipids, but the membrane patch rather consists entirely of synthetic amphiphilic block copolymers (Figure 1). The only PNDs reported so far are our discoidal amphiphilic block polymer membrane patches encased within MSPs (Fiori et al., 2017b). In these PNDs the hydrophobic core of the lipid bilayer was replaced with hydrogenated polybutadiene, and the hydrophilic lipid headgroup moieties were replaced with the positively charged poly vinyl methylpyridine. These PNDs are easily produced and, as LNDs, assemble spontaneously under the appropriate conditions, forming uniform nanodiscs similar to LNDs in size when the same type of MSP was used (Fiori et al., 2017b). The ATPase activity of the ATP-binding cassette pump MsbA reconstituted in these PNDs was several folds higher than that of the MP solubilized in detergents, and equivalent to that of MsbA in LNDs (Fiori et al., 2017b). The PNDs showed much improved stability vis-à-vis LNDs, including lack of aggregation when stored at temperatures between 4 and 37°C, and following a freeze/thaw cycle (Fiori et al., 2017b). These studies indicate that PNDs can support functional MPs and showcase their remarkable stability.
Since some of the different types of MP platforms described in this review are fairly new, in this section we give information about the availability of the nanodisc components. The information presented is not exhaustive, but aimed at providing a starting point for interested potential users since new copolymers are becoming available rapidly, traditional components, such as MSPs and lipids are available from several sources, and new copolymers are generally available from their developers before they are commercially available. A large number of detergents and phospholipids, as well as cholesterol and other lipids, are available from Avanti Polar Lipids1, Millipore Sigma2, Larodan3, Anatrace4, and Cube Biotech5, among many sources. Purified MSPs are available from sources, such as Millipore Sigma and Cube Biotech, and plasmids for the expression and purification of MSPs in-house are available from Addgene6. SMAs and some SMA-like copolymers are available from Millipore Sigma, Orbiscope7, Anatrace, Avanti Polar Lipids, Cube Biotech, and Polyscope8. Lipid mimetic copolymers are not commercially available yet, but as with other new copolymers not yet in the market, they can be obtained on limited bases from the laboratories that developed them. The SMALP Network9 is a good source to learn about new copolymers as they become available.
The use of copolymers is a promising addition to MP research and applications. Two major advantages of copolymers over biomolecules, such as MSPs and lipids are their stability and amenability to engineering modifications tailored to specific applications. New developments in amphiphilic copolymers, such as SMA-like polymers offer the promise of direct, simultaneous solubilization of MPs in the absence of detergent, and reconstitution in the native membrane lipids (Ravula et al., 2019). Contrary to SMAs, they enjoy size control of the resulting SMALPs and compatibility with a variety of buffers (Fiori et al., 2017a; Ravula et al., 2019). The development of new SMA-like polymers with improved properties has accelerated during the last few years, but testing of the copolymers and characterization of the resulting nanostructures still lag behind.
Whereas, LNDs tend to aggregate under conditions, such as physiological mammalian temperature needed for many experiments as well as freeze/thaw cycles experienced during transport/storage/use, PNDs do not, rendering them conducive to functioning in abiotic environments for long periods of time under broader experimental conditions (Fiori et al., 2017b). PNDs’ high chemical versatility and membrane stability may potentially lead to exciting developments. It is interesting that a variety of MPs can be functionally reconstituted in synthetic polymeric membranes, including the water channel aquaporin, potassium channels, the light-driven proton pump proteorhodopsin, a bacterial reaction center, an ATP synthase, the G-protein coupled receptor protein rhodopsin, and the ATP-binding cassette transporter MsbA (Kumar et al., 2007; Hua et al., 2011; Kowal et al., 2014; Kuang et al., 2014b; Itel et al., 2015; Fiori et al., 2017b). Even more remarkably, proteorhodopsin displays a “normal” photocycle at the structural and functional level when reconstituted in “frozen” polysterene-based membranes of glass transition temperatures >70°C, which display superior bulk-state stability when compared to other synthetic membranes (Kuang et al., 2014a,b).
A representation and a comparative summary of the different types of nanodiscs described here are presented in Figure 1B and Table 1, respectively. Despite the recent developments, more studies on the basic characterization of the new copolymers and their applications are needed. In this context, the production of PNDs with highly stable polysterene-based membranes, and the possibility of engineering fully polymeric nanodiscs combining new SMA-like copolymers and synthetic polymeric membranes seem promising avenues to explore.
AC, EM, MF, HL, and GA wrote the manuscript. All authors approved the final manuscript.
This work was supported in part by the NSF grants DMR-1623241, CBET-1623240, and DMR-1810767, the Cancer Prevention and Research Institute of Texas (CPRIT) award RP180827, and the Medical Student Summer Research Program Scholarships to AC and EM from the TTUHSC School of Medicine.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alvis, S. J., Williamson, I. M., East, J. M., and Lee, A. G. (2003). Interactions of anionic phospholipids and phosphatidylethanolamine with the potassium channel KcsA. Biophys. J. 85, 3828–3838. doi: 10.1016/s0006-3495(03)74797-3
Banerjee, S., Sen, K., Pal, T. K., and Guha, S. K. (2012). Poly(styrene-co-maleic acid)-based pH-sensitive liposomes mediate cytosolic delivery of drugs for enhanced cancer chemotherapy. Int. J. Pharm. 436, 786–797. doi: 10.1016/j.ijpharm.2012.07.059
Beyder, A., and Farrugia, G. (2016). Ion channelopathies in functional GI disorders. Am. J. Physiol. Gastrointest Liver Physiol. 311, G581–G586.
Brewer, K. D., Li, W., Horne, B. E., and Rizo, J. (2011). Reluctance to membrane binding enables accessibility of the synaptobrevin SNARE motif for SNARE complex formation. Proc. Natl. Acad. Sci. U.S.A. 108, 12723–12728. doi: 10.1073/pnas.1105128108
Denisov, I. G., and Sligar, S. G. (2016). Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 23, 481–486. doi: 10.1038/nsmb.3195
Denisov, I. G., and Sligar, S. G. (2017). Nanodiscs in membrane biochemistry and biophysics. Chem. Rev. 117, 4669–4713. doi: 10.1021/acs.chemrev.6b00690
Dorr, J. M., Koorengevel, M. C., Schafer, M., Prokofyev, A. V., Scheidelaar, S., van der Cruijsen, E. A., et al. (2014). Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc. Natl. Acad. Sci. U.S.A. 111, 18607–18612. doi: 10.1073/pnas.1416205112
Dorr, J. M., Scheidelaar, S., Koorengevel, M. C., Dominguez, J. J., Schafer, M., van Walree, C. A., et al. (2016). The styrene-maleic acid copolymer: a versatile tool in membrane research. Eur. Biophys. J. 45, 3–21. doi: 10.1007/s00249-015-1093-y
Fiori, M. C., Jiang, Y., Altenberg, G. A., and Liang, H. (2017a). Polymer-encased nanodiscs with improved buffer compatibility. Sci. Rep. 7:7432.
Fiori, M. C., Jiang, Y., Zheng, W., Anzaldua, M., Borgnia, M. J., Altenberg, G. A., et al. (2017b). Polymer nanodiscs: discoidal amphiphilic block copolymer membranes as a new platform for membrane proteins. Sci. Rep. 7:15227.
Fiori, M. C., Zheng, W., Kamilar, E., Simiyu, G., Altenberg, G. A., and Liang, H. (2020). Extraction and reconstitution of membrane proteins into lipid nanodiscs encased by zwitterionic styrene-maleic amide copolymers. Sci. Rep. 10:9940.
Goers, R., Thoma, J., Ritzmann, N., Di Silvestro, A., Alter, C., Gunkel-Grabole, G., et al. (2018). Optimized reconstitution of membrane proteins into synthetic membranes. Commun. Chem. 1:35.
Hagn, F., Etzkorn, M., Raschle, T., and Wagner, G. (2013). Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc. 135, 1919–1925. doi: 10.1021/ja310901f
Hall, S. C. L., Tognoloni, C., Charlton, J., Bragginton, E. C., Rothnie, A. J., Sridhar, P., et al. (2018a). An acid-compatible co-polymer for the solubilization of membranes, and proteins into lipid bilayer-containing nanoparticles. Nanoscale 10, 10609–10619. doi: 10.1039/c8nr01322e
Hall, S. C. L., Tognoloni, C., Price, G. J., Klumperman, B., Edler, K. J., Dafforn, T. R., et al. (2018b). Influence of Poly(styrene- co-maleic acid) copolymer structure on the properties and self-assembly of SMALP nanodiscs. Biomacromolecules 19, 761–772. doi: 10.1021/acs.biomac.7b01539
Hardy, D., Desuzinges Mandon, E., Rothnie, A. J., and Jawhari, A. (2018). The yin and yang of solubilization and stabilization of wild-type and full-length membrane protein. Methods 147, 118–125. doi: 10.1016/j.ymeth.2018.02.017
Hesketh, S. J., Klebl, D. P., Higgins, A. J., Thomsen, M., Pickles, I. B., Sobott, F., et al. (2020). Styrene maleic-acid lipid particles (SMALPs) into detergent or amphipols: an exchange protocol for membrane protein characterisation. Biochim. Biophys. Acta Biomembr. 1862:183192. doi: 10.1016/j.bbamem.2020.183192
Hua, D. B., Kuang, L. J., and Liang, H. J. (2011). Self-directed reconstitution of proteorhodopsin with amphiphilic block copolymers induces the formation of hierarchically ordered proteopolymer membrane arrays. J. Am. Chem. Soc. 133, 2354–2357. doi: 10.1021/ja109796x
Itel, F., Najer, A., Palivan, C. G., and Meier, W. (2015). Dynamics of membrane proteins within synthetic polymer membranes with large hydrophobic mismatch. Nano Lett. 15, 3871–3878. doi: 10.1021/acs.nanolett.5b00699
Kowal, J., Wu, D. L., Mikhalevich, V., Palivan, C. G., and Meier, W. (2015). Hybrid polymer-lipid films as platforms for directed membrane protein insertion. Langmuir 31, 4868–4877. doi: 10.1021/acs.langmuir.5b00388
Kowal, J. L., Kowal, J. K., Wu, D., Stahlberg, H., Palivan, C. G., and Meier, W. P. (2014). Functional surface engineering by nucleotide-modulated potassium channel insertion into polymer membranes attached to solid supports. Biomaterials 35, 7286–7294. doi: 10.1016/j.biomaterials.2014.05.043
Kuang, L. J., Fernandes, D. A., O’Halloran, M., Zheng, W., Jiang, Y. J., Ladizhansky, V., et al. (2014a). “Frozen” block copolymer nanomembranes with light-driven proton pumping performance. ACS Nano 8, 537–545. doi: 10.1021/nn4059852
Kuang, L. J., Olson, T. L., Lin, S., Flores, M., Jiang, Y. J., Zheng, W., et al. (2014b). Interface for light-driven electron transfer by photosynthetic complexes across block copolymer membranes. J. Phys. Chem. Lett. 5, 787–791. doi: 10.1021/jz402766y
Kumar, M., Grzelakowski, M., Zilles, J., Clark, M., and Meier, W. (2007). Highly permeable polymeric membranes based on the incorporation of the functional water channel protein Aquaporin Z. Proc. Natl. Acad. Sci. U.S.A. 104, 20719–20724. doi: 10.1073/pnas.0708762104
Kumar, P., Kumar, D., Jha, S. K., Jha, N. K., and Ambasta, R. K. (2016). Ion channels in neurological disorders. Adv. Protein Chem. Struct. Biol. 103, 97–136. doi: 10.1016/bs.apcsb.2015.10.006
Le Maire, M., Champeil, P., and Moller, J. V. (2000). Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta 1508, 86–111. doi: 10.1016/s0304-4157(00)00010-1
Lee, J. R., and White, T. W. (2009). Connexin-26 mutations in deafness and skin disease. Expert. Rev. Mol. Med. 11:e35.
Lee, S. C., Knowles, T. J., Postis, V. L., Jamshad, M., Parslow, R. A., Lin, Y. P., et al. (2016). A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 11, 1149–1162. doi: 10.1038/nprot.2016.070
Leitz, A. J., Bayburt, T. H., Barnakov, A. N., Springer, B. A., and Sligar, S. G. (2006). Functional reconstitution of Beta2-adrenergic receptors utilizing self-assembling Nanodisc technology. Biotechniques 40, 601–602. doi: 10.2144/000112169
Lindhoud, S., Carvalho, V., Pronk, J. W., and Aubin-Tam, M. E. (2016). SMA-SH: Modified styrene-maleic acid copolymer for functionalization of lipid nanodiscs. Biomacromolecules 17, 1516–1522. doi: 10.1021/acs.biomac.6b00140
Meier, W., Nardin, C., and Winterhalter, M. (2000). Reconstitution of channel proteins in (polymerized) ABA triblock copolymer membranes. Angew. Chem. Int. Edn. 39, 4599–4602. doi: 10.1002/1521-3773(20001215)39:24<4599::aid-anie4599>3.0.co;2-y
Morrison, K. A., Akram, A., Mathews, A., Khan, Z. A., Patel, J. H., Zhou, C., et al. (2016). Membrane protein extraction and purification using styrene-maleic acid (SMA) copolymer: effect of variations in polymer structure. Biochem. J. 473, 4349–4360. doi: 10.1042/bcj20160723
Oluwole, A. O., Danielczak, B., Meister, A., Babalola, J. O., Vargas, C., and Keller, S. (2017). Solubilization of membrane proteins into functional lipid-bilayer nanodiscs using a diisobutylene/maleic acid copolymer. Angew. Chem. Int. Edn. 56, 1919–1924. doi: 10.1002/anie.201610778
Opekarova, M., and Tanner, W. (2003). Specific lipid requirements of membrane proteins - a putative bottleneck in heterologous expression. Biochim. Biophys. Acta Biomembr. 1610, 11–22. doi: 10.1016/s0005-2736(02)00708-3
Orellana, J. A., Avendano, B. C., and Montero, T. D. (2014). Role of connexins and pannexins in ischemic stroke. Curr. Med. Chem. 21, 2165–2182. doi: 10.2174/0929867321666131228191714
Overington, J. P., Al-Lazikani, B., and Hopkins, A. L. (2006). How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996. doi: 10.1038/nrd2199
Parmar, M., Rawson, S., Scarff, C. A., Goldman, A., Dafforn, T. R., Muench, S. P., et al. (2018). Using a SMALP platform to determine a sub-nm single particle cryo-EM membrane protein structure. Biochim. Biophys. Acta Biomembr. 1860, 378–383. doi: 10.1016/j.bbamem.2017.10.005
Ravula, T., Hardin, N. Z., Ramadugu, S. K., and Ramamoorthy, A. (2017a). pH tunable, and divalent metal ion tolerant polymer lipid nanodiscs. Langmuir 33, 10655–10662. doi: 10.1021/acs.langmuir.7b02887
Ravula, T., Ramadugu, S. K., Di Mauro, G., and Ramamoorthy, A. (2017b). Bioinspired, size-tunable self-assembly of polymer-lipid bilayer nanodiscs. Angew. Chem. Int. Edn. 56, 11466–11470. doi: 10.1002/anie.201705569
Ravula, T., Hardin, N. Z., and Ramamoorthy, A. (2019). Polymer nanodiscs: Advantages and limitations. Chem. Phys. Lipids 219, 45–49. doi: 10.1016/j.chemphyslip.2019.01.010
Routledge, S. J., Jamshad, M., Little, H. A., Lin, Y. P., Simms, J., Thakker, A., et al. (2020). Ligand-induced conformational changes in a SMALP-encapsulated GPCR. Biochim. Biophys. Acta Biomembr. 1862:183235. doi: 10.1016/j.bbamem.2020.183235
Russell, R. B., and Eggleston, D. S. (2000). New roles for structure in biology and drug discovery. Nat. Struct. Biol. 7(Suppl.), 928–930.
Schmidt, B. Z., Haaf, J. B., Leal, T., and Noel, S. (2016). Cystic fibrosis transmembrane conductance regulator modulators in cystic fibrosis: current perspectives. Clin. Pharmacol. 8, 127–140. doi: 10.2147/cpaa.s100759
Seddon, A. M., Curnow, P., and Booth, P. J. (2004). Membrane proteins, lipids and detergents: Not just a soap opera. Biochim. Biophys. Acta 1666, 105–117. doi: 10.1016/j.bbamem.2004.04.011
Seneviratne, R., Khan, S., Moscrop, E., Rappolt, M., Muench, S. P., Jeuken, L. J. C., et al. (2018). A reconstitution method for integral membrane proteins in hybrid lipid-polymer vesicles for enhanced functional durability. Methods 147, 142–149. doi: 10.1016/j.ymeth.2018.01.021
Skrzypek, R., Iqbal, S., and Callaghan, R. (2018). Methods of reconstitution to investigate membrane protein function. Methods 147, 126–141. doi: 10.1016/j.ymeth.2018.02.012
Smith, A. A. A., Autzen, H. E., Laursen, T., Wu, V., Yen, M., Hall, A., et al. (2017). Controlling styrene maleic acid lipid particles through RAFT. Biomacromolecules 18, 3706–3713. doi: 10.1021/acs.biomac.7b01136
Stroud, Z., Hall, S. C. L., and Dafforn, T. R. (2018). Purification of membrane proteins free from conventional detergents: SMA, new polymers, new opportunities and new insights. Methods 147, 106–117. doi: 10.1016/j.ymeth.2018.03.011
Thonghin, N., Kargas, V., Clews, J., and Ford, R. C. (2018). Cryo-electron microscopy of membrane proteins. Methods 147, 176–186. doi: 10.1016/j.ymeth.2018.04.018
Viegas, A., Viennet, T., and Etzkorn, M. (2016). The power, pitfalls and potential of the nanodisc system for NMR-based studies. Biol. Chem. 397, 1335–1354. doi: 10.1515/hsz-2016-0224
Yorulmaz Avsar, S., Kyropoulou, M., Di Leone, S., Schoenenberger, C. A., Meier, W. P., and Palivan, C. G. (2019). Biomolecules turn self-assembling amphiphilic block co-polymer platforms into biomimetic interfaces. Front. Chem. 6:645. doi: 10.3389/fchem.2018.00645
Zaucha, J., Heinzinger, M., Kulandaisamy, A., Kataka, E., Salvador, O. L., Popov, P., et al. (2020). Mutations in transmembrane proteins: diseases, evolutionary insights, prediction and comparison with globular proteins. Brief. Bioinform. 2020:bbaa132.
Zoghbi, M. E., Cooper, R. S., and Altenberg, G. A. (2016). The lipid bilayer modulates the structure and function of an ATP-binding cassette exporter. J. Biol. Chem. 291, 4453–4461. doi: 10.1074/jbc.m115.698498
Zoghbi, M. E., Mok, L., Swartz, D. J., Singh, A., Fendley, G. A., Urbatsch, I. L., et al. (2017). Substrate-induced conformational changes in the nucleotide-binding domains of lipid bilayer-associated P-glycoprotein during ATP hydrolysis. J. Biol. Chem. 292, 20412–20424. doi: 10.1074/jbc.m117.814186
Keywords: copolymer, lipid, Lipodisq, membrane-mimetic, method, native nanodisc, styrene maleic acid, synthetic membrane
Citation: Chen A, Majdinasab EJ, Fiori MC, Liang H and Altenberg GA (2020) Polymer-Encased Nanodiscs and Polymer Nanodiscs: New Platforms for Membrane Protein Research and Applications. Front. Bioeng. Biotechnol. 8:598450. doi: 10.3389/fbioe.2020.598450
Received: 24 August 2020; Accepted: 27 October 2020;
Published: 16 November 2020.
Edited by:
Emilio Isaac Alarcon, University of Ottawa, CanadaReviewed by:
Tomas Laursen, University of Copenhagen, DenmarkCopyright © 2020 Chen, Majdinasab, Fiori, Liang and Altenberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guillermo A. Altenberg, Zy5hbHRlbmJlcmdAdHR1aHNjLmVkdQ==; Hongjun Liang, aC5saWFuZ0B0dHVoc2MuZWR1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.