
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bioeng. Biotechnol. , 15 October 2020
Sec. Industrial Biotechnology
Volume 8 - 2020 | https://doi.org/10.3389/fbioe.2020.594081
This article is part of the Research Topic Engineering Yeast to Produce Plant Natural Products View all 11 articles
Cocoa butter is extracted from cocoa beans, and it is mainly used as the raw material for the production of chocolate and cosmetics. Increased demands and insufficient cocoa plants led to a shortage of cocoa butter supply, and there is therefore much interesting in finding an alternative cocoa butter supply. However, the most valuable component of cocoa butter is rarely available in other vegetable oils. Saccharomyces cerevisiae is an important industrial host for production of chemicals, enzyme and pharmaceuticals. Advances in synthetical biology and metabolic engineering had enabled high-level of triacylglycerols (TAG) production in yeast, which provided possible solutions for cocoa butter equivalents (CBEs) production. Diverse engineering strategies focused on the fatty acid-producing pathway had been applied in S. cerevisiae, and the key enzymes determining the TAG structure were considered as the main engineering targets. Recent development in phytomics and multi-omics technologies provided clues to identify potential targeted enzymes, which are responsible for CBE production. In this review, we have summarized recent progress in identification of the key plant enzymes for CBE production, and discussed recent and future metabolic engineering and synthetic biology strategies for increased CBE production in S. cerevisiae.
Cocoa butter (CB) is mainly extracted from cocoa beans of cocoa tree (Theobroma cacao), and is usually used as food flavor and cosmetics additive (Jahurul et al., 2013). With the economic development, global demands of chocolate and other CB-based products increase. As the main sources of CB, the cocoa tree can only grow in the tropical area with limited planting area, and the large-scale cocoa farming would occupy the rain forest space and thus threat the global food supplies (Clough et al., 2009). Moreover, the pest damage and infectious disease of cocoa tree might reduce CB yields (Drenth and Guest, 2016). Therefore, CB supply is quite limited, and an alternative CB supply is of interest.
CB is mainly composed of three different kinds of triacylglycerols (TAG), including 1,3-dipalmitoyl-2-oleoyl-glycerol (POP, C16:0–C18:1–C16:0), 1-palmitoyl-3-stearoyl-2-oleoyl-glycerol (POS, C16:0–C18:1–C18:0), and 1,3-distearoyl-2-oleoyl-glycerol (SOS, C18:0–C18:1–C18:0) (Lipp and Anklam, 1998; Vieira et al., 2015). SOS is the key CB flavor composition, and it is the most valuable composition in CB (Jahurul et al., 2013). Cocoa butter equivalents (CBEs) are lipids that have similar physicochemical properties as CB, and vegetable oils are often used as CBEs (Lipp and Anklam, 1998; Sonwai et al., 2014). Though the POP and POS contents are high in some vegetable oils, such as coconut oil and palm oil, the SOS content in vegetable oils is low (Lipp and Anklam, 1998). The properties of these vegetable oils-derived CBEs normally are different with CB, since they have lower melting temperature and the mouth feeling is different with CB (de Silva Souza and Block, 2018). Shea butter and a few other tropical butters are ideal CBEs (Zeng et al., 2020), but their supply is limited due to their limited distribution in the tropical area.
Yeasts are attractive choices for industrial-scale microbial oleochemical production (Zhou et al., 2016b; Spagnuolo et al., 2019). The accumulation of TAGs is a way for carbon storage in yeast, and the main TAGs of yeast are composed of C16 and C18 fatty acids (Klug and Daum, 2014). Many yeasts are generally recognized as safe (GRAS) species and can be used in the food and cosmetic industry, enabling yeasts as potential CBE production hosts. Several different wild-type yeast species had been used for CBE production (Wei et al., 2017b), and Saccharomyces cerevisiae is the most widely used microbial cell factories. However, no SOS was detected in the lipidome of S. cerevisiae BY4741 (Ejsing et al., 2009), and the SOS content of S. cerevisiae CEN.PK113-7D was low (Ejsing et al., 2009). Therefore, metabolic engineering and other synthetic biology strategies need to be applied to increase CBE production of S. cerevisiae, especially the SOS production. Metabolic engineering of S. cerevisiae for CBE or SOS production requires understanding the lipid biosynthetic pathway of S. cerevisiae and plants.
In S. cerevisiae, glucose is converted into Glycerol-3-phosphate (G-3-P) through glycolysis, which is the precursor of TAG backbone. Part of G-3-P is further converted into pyruvate, which is used to synthesize acetyl-CoA by pyruvate dehydrogenase complex (PDHC) in mitochondria (Krivoruchko et al., 2015). Most acetyl-CoA generated in mitochondria is consumed in the tricarboxylic acid (TCA) cycle. In the cytosol, the pyruvate dehydrogenase (PDH) bypass converts pyruvate to acetyl-CoA via three steps catalyzed by pyruvate decarboxylase (PDC), acetaldehyde dehydrogenase and two acetyl-CoA synthetase (ACS1 and ACS2) (Figure 1A; Zhang et al., 2018).
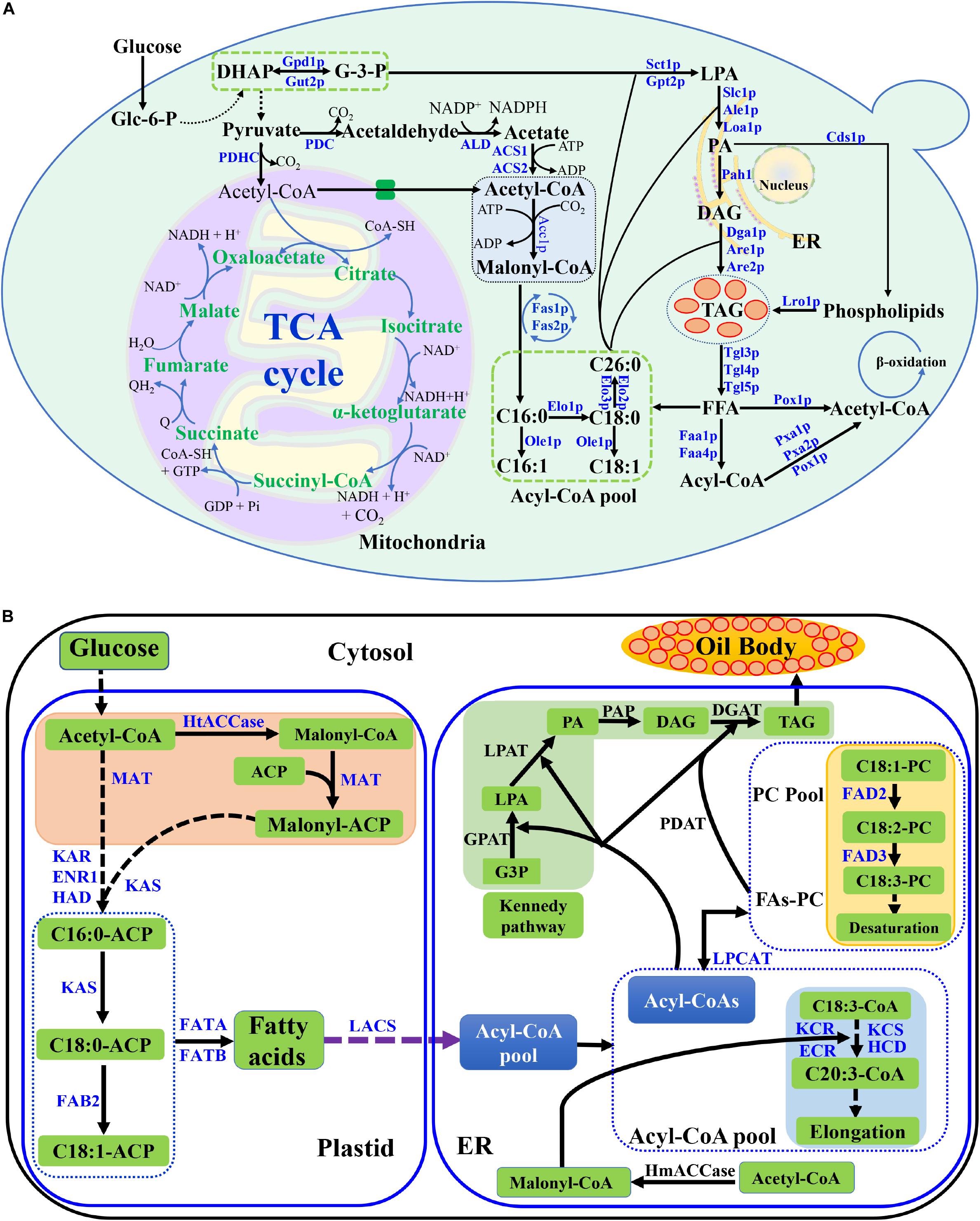
Figure 1. Lipid metabolic pathway in S. cerevisiae and lipid biosynthetic pathway in plant cell. (A) Lipid metabolic pathway in S. cerevisiae, including the biosynthesis and degradation, was described. The key enzymes were listed. Abbreviations: Glc-6-p, glucose 6 phosphate; DHAP, dihydroxyacetone phosphate; G-3-P, glyceraldehyde 3-phosphate; LPA, lysophophatidic acid; PA, phosphatidic acid; DAG, diacylglycerol; TAG, triacylglycerols; FFA, free fatty acid; TCA cycle, tricarboxylic acid cycle; PDHC, pyruvate dehydrogenase complex; PDC, pyruvate decarboxylase; ALD, acetaldehyde dehydrogenase; ACS, acetyl-CoA synthetase; ER, endoplasmic reticulum; (B) Lipid biosynthetic pathway in plant cell. Abbreviations: HtACCase, heteromeric acetyl-CoA carboxylase; ACP, acyl carrier protein; MAT, plastidial malonyl-CoA-ACP malonyltransferase; KAS, ketoacyl-ACP synthase; KAR, plastidial ketoacyl-ACP reductase; HAD, plastidial hydroxyacyl-ACP dehydrase; ENR1, plastidial enoyl-ACP reductase; FAB2, stearoyl-ACP desaturase; FATA, acyl-ACP thioesterase; FATB, acyl-ACP thioesterase; LACS, long-chain acyl-CoA synthetase; FAD2, ER oleate desaturase; FAD3, ER linoleate desaturase; KCS, β-Ketoacyl-CoA synthase; KCR, ketoacyl-CoA reductase; ECR, enoyl-CoA reductase; HmACCase, homomeric acetyl-CoA carboxylase; LPCAT, lysophosphatidylcholine acyltransferase; PDAT, phospholipid:diacylglycerol acyltransferase; GPAT, glycerol-3-phosphate acyltransferase; LPAT, lysophosphatidic acid acyltransferase; PAP, phosphatidic acid phosphatase; DGAT, acyl-CoA:diacylglycerol acyltransferase; G3P, glycerol-3-phosphate. Dashed arrows indicate multiple steps, and solid lines indicate a single step in the metabolic pathway.
In an initial step, acetyl-CoA is carboxylated by the addition of CO2 to malonyl-CoA with Acetyl-CoA carboxylase (Acc1p). Malonyl-CoA is used as building blocks, and acetyl-CoA is used as the precursor in the fatty acid biosynthesis. The fatty acid biosynthesis is catalyzed by fatty acid synthase, a multienzyme complex consisting of Fas1p and Fas2p. The fatty acid types of S. cerevisiae are determined by the genetic background, carbon source and other effects, which usually contain C16 and C18 fatty acids with one or none double bond (Wei et al., 2017b). The ACC1, FAS1, and FAS2 in wild-type yeast strains were replaced with same strong constitutive promoter in order to produce fatty acid consistently (Tang et al., 2015). The fatty acyl-CoA length is decided by the elongases of Elo1p, Elo2p, and Elo3p (Tehlivets et al., 2007). Usually, Elo1p controls medium length-chain fatty acid synthesis (C12 to C16), while Elo2p and Elo3p are responsible for long-chain fatty acid synthesis (up to C26) (Rossler et al., 2003; Wenning et al., 2017). Approximately 70–80% of the total fatty acids in yeasts are monounsaturated fatty acid in a wide range of cultivation conditions, which are synthesized from saturated fatty acyl-CoA precursors by the △9-fatty acid desaturase of Ole1p (Martin et al., 2007).
Phosphatidic acid (PA) is a vital component in the acylglycerol lipid metabolism. PA is synthesized by two different pathways, the G-3-P pathway and the dihydroxyacetone phosphate (DHAP) pathways (Figure 1A). The conversion between G-3-P and DHAP can reversely be catalyzed by glycerol-3-phosphate dehydrogenase (Gpd1p and Gut2p). The acyl-CoA is transferred to the sn-1 position of glycerol-3-phosphate (G-3-P) to form 1-acyl-G-3-P (LPA) which is mainly catalyzed with the enzyme of acyl-CoA:glycerol-sn-3-phosphate acyl-transferase (GPAT, Sct1p, and Gpt2p) or to DHAP which is catalyzed with acyl-CoA:DHAP acyltransferase (DHAPAT). G-3-P pathway is the main pathway for lysophosphatidic Acid (LPA) formation in S. cerevisiae (Klug and Daum, 2014; Fakas, 2017). Subsequently, an acyl chain was added to the sn-2 position by lysophophatidate acyl-transferase (LPAT, Slc1p, Ale1p, or Loa1p) to yield PA. PA can either be dephosphorylated to diacylglycerol (DAG) by phosphatidic acid phosphatase (Pah1p), or converts to phospholipids by Phosphatidate cytidylyltransferase (Cds1p). Phospholipids can further convert to TAGs via the acyltransferase Lro1p. DAG is the substrate for TAG synthesis by acyl-CoA:diacylglycerol acyl-transferase (DGAT) of Dga1p (Coleman and Lee, 2004; de Kroon et al., 2013). Besides, PA can also be used for other lipids synthesis, such as phosphatidylcholine (PC), phosphatidylserine (PS), and phosphatidylinositol (PI) (de Kroon et al., 2013). TAG can be degraded to free fatty acids (FFA) with triacylglycerol lipase (Tgl3p, Tgl4p, and Tgl5p). The FFA is further converted to acetyl-CoA via Faa1p, Faa4p, Pox1p and other enzymes in peroxisome (Figure 1A).
The de novo lipid biosynthetic pathway of plant and S. cerevisiae is different (Figure 1; Bates et al., 2013; Fakas, 2017). In plant, the major biochemical reactions for TAG biosynthesis includes plastid fatty acid synthesis step, acyl editing step and TAG synthesis step, which mainly occur in plastid, mitochondria and endoplasmic reticulum (ER). The precursors for plant fatty acid synthesis are Acetyl-CoA and malonyl-ACP, which are converted to fatty acids-ACP through catalysis by ketoacyl-ACP synthase (KAS) and several other enzymes in plastid and mitochondria (Baud and Lepiniec, 2010; Bates et al., 2013). The fatty acid ACP are further converted to fatty acids via ACP thioesterase (FATA and FATB) (Baud and Lepiniec, 2010; Argout et al., 2011). The long-chain acyl-CoA synthetase catalyzes fatty acids to acyl-CoA in ER. The acyl-CoA are further elongated or desaturated with several enzymes. The final TAG products are formed via Kennedy pathway, and the main enzymes are GPAT, LPAT, PAP, and DGAT in ER (Figure 1B). The TAGs are stored in plant oil body.
The C18 content are lower than C16 content in S. cerevisiae (Ejsing et al., 2009). SOS is composed with one glycerol backbone, two steric acids (C18:0), and one oleic acid (C18:1). In order to increase CBE production in S. cerevisiae, the current efforts focused on: (1) increasing fatty acids especially C18 compositions via directing metabolic flux toward lipid synthesis; (2) overexpression of acyl-CoA transferases specially for CBE synthesis.
The Acetyl-CoA carboxylase Acc1p can convert acetyl-CoA to malonyl-CoA, and the ACC1 expression level affects fatty acid composition in S. cerevisiae. By overexpression of ACC1, FAS1, and FAS2, TAG production increased four-fold over the control strain (Runguphan and Keasling, 2014). In another study, ACC1 variant ACC1S659A S1157A (ACC1**) can abolish snf1 regulation and increased fatty acid production (Shi et al., 2014). Expression of ACC1** can increase TAG production, especially increased C18 composition and decreased C16:1 composition (Bergenholm et al., 2018). The Elo1p is responsible for the elongation of C16 and C18 fatty acids, while Elo2p and Elo3p are responsible for very long chain fatty acids biosynthesis. The overexpression of ELO1 increase C18:1 titer. However the TAG content didn’t change. Combination of engineering ACC1**, OLE1 and ELO1 significantly increases TAG content and SOS content 5.8-fold and 48-fold, respectively (Bergenholm et al., 2018).
Several yeast fatty acid production platforms for oleochemics have been established (Supplementary Table S1; Li et al., 2014; Runguphan and Keasling, 2014; Leber et al., 2015; Zhou et al., 2016a,b; Dai et al., 2018; Ferreira et al., 2018a). Strengthening fatty acid biosynthetic pathway and weakening the degradation pathway increased C18 compositions and decreased C16:1 composition, and the final FFA titer reached 10.4 g/L (Zhou et al., 2016b). Further relieving the side-pathway competition by harnessing yeast peroxisomes increased the production of fatty-acid-derived chemical of fatty alcohols, alkanes and olefins up to seven-fold (Zhou et al., 2016a). By simplifying the lipid metabolic network with the redirection of fatty acid metabolism and the reduction of feedback regulation, a strain with 129 mg⋅g DCW–1 free fatty acid production was achieved (Ferreira et al., 2018b). Moreover, by global rewiring of cellular metabolism and adaptive laboratory evolution, the Crabtree effect of S. cerevisiae can be abolished, which is help for the acetyl-CoA derived product accumulation (Dai et al., 2018). The S. cerevisiae strain was reprogrammed from alcoholic fermentation to lipogenesis, and the final FFA titer reached 33.4 g/L (Yu et al., 2018). Overexpression of ACC1∗∗, PAH1 and DGA1, and disruption of TGL3, TGL4, TGL5, ARE1, POX1, and PXA1, lead to the final TAG accumulation of 254 mg⋅g DCW–1, reaching 27.4% of the maximum theoretical yield in S. Cerevisiae, which is the highest TAG titer reported in S. cerevisiae (Ferreira et al., 2018a).
GPAT, LPAT, and DGAT are the key enzymes in CBE/SOS production (Maraschin et al., 2019). The identification of plant SOS biosynthetic genes and their expression in S. cerevisiae can enhance SOS production (Wei et al., 2017a, 2018). There are two GPAT (Gat1p and Gat2p) and one DHAPAT in S. cerevisiae. The double deletions of both GAT1 (also known as GPT2) and GAT2 (also known as SCT1) leads to yeast lethality, showing that Gat1p and Gat2p are essential in TAG synthesis (Zheng and Zou, 2001). The acyl-specificity of Gat1p is diverse, as it can use a broad range of fatty acids as substrates; while the Gat2p displayed preference toward C16 fatty acids, suggesting that Gat1p and Gat2p can’t be used to synthesize large amount of SOS directly in S. cerevisiae (Zheng and Zou, 2001). Plants usually contains three different types of GPAT genes, which are located in the plastid, mitochondria or cytoplasm (Yang et al., 2012). Among the 10 GPAT genes of Arabidopsis thailiana, GPAT-4, -6 and -8 genes strongly preferred C16:0 and C18:1 ω-oxidized acyl-CoAs over other substrates, providing hints that these GPAT genes might be responsible for SOS production in plants (Yang et al., 2012).
LPAT (EC 2.3.1.51) is believed to have the highest substrate specificity. In S. cerevisiae, LPATs were identified to acylate LPA with a range of different acyl-CoAs, including C18:1, C22:1, and C24:0-CoA. Introducing LPAT genes from S. cerevisiae into Arabidopsis resulted in 8% to 48% increasement of very-long-chain fatty in TAGs, showing that yeast LPAT genes would not be suitable for CBE production (Zou et al., 1997). Diverse LPATs that acylate the sn-2 of LPA to form PA had been identified, and eight LPAT genes were found in cocoa genome, which can be classified into three different clusters based on amino acid identities (Argout et al., 2011). Expression of some LPAT genes can significantly increase TAG production in S. cerevisiae (Wei et al., 2017a, 2018).
As previously described, two main pathways are responsible for TAG production from DAG in S. cerevisiae. Phospholiipid:diacylglycerol acyltransferase (PDAT, EC 2.3.1.158) uses PLs as acyl donors, and it distributes in yeast and plants (Liu et al., 2012). DGAT catalyzes the last step of TAG biosynthesis from DAG and acyl-CoA. DGAT is an acyl-CoA dependent enzyme, which catalyzes the final and only committed step of the Kennedy pathway. DGAT is essential for TAG biosynthesis in seed. Overexpression of the yeast diacylglycerol acyltransferase (DGA1) can lead to TAG production increasement in the Δsnf2 disruptant of S. cerevisiae (Kamisaka et al., 2007). Two different families of DGAT1 and DGAT2 are available in yeast, plants and animals, and DGATs from different species show diversal substrate preferences (Yen et al., 2008). For example, in A. thaliana, DGAT1 displays preference to C16:0, while DGAT2 displays preference to C16:1 (Aymé et al., 2014). The Overexpression of four DGAT1 genes from Brassica napus in S. cerevisiae increased TAG biosynthesis (Greer et al., 2015). Moreover, expression of Arabidopsis DGAT gene in yeast increased 200–600 folds of yeast DGAT activity, which can lead to 3–9 folds TAG increasement, showing DGAT might be a useful target for CBE accumulation in S. cerevisiae (Bouvier-Navé et al., 2000).
Global genomic and transcriptomic analyses identified thirteen potential GPATs, eight potential LPATs and two DGATs in T. cacao (Wei et al., 2017a, 2018). The overexpression of single or multiple cocoa genes in S. cerevisiae increased CBE production. The combinational expression of GPAT, LPAT, and DGAT genes showed 134-fold production of TAG over the control strain, showing these genes from cocoa have great potential for CB production (Supplementary Table S1; Wei et al., 2018). Shea butter contains high-level SOS, and their GPATs, LPATs, and DGATs had been identified through the transcriptomics analyses and the functional heterologous expression in S. cerevisiae (Wei et al., 2019). In the future, mining more specific and efficient enzymes for CBE production would help to increase CBE composition in S. cerevisiae.
With the development of sequencing technologies, huge amounts of plant genomic or transcriptomic data had been generated (Leebens-Mack et al., 2019). The strategy to identify novel key CBE or other natural product biosynthetic genes had been extensively developed and applied (Wang et al., 2015). In general, plant samples were collected, and genome and/or transcriptome were obtained by next-generation or third generation sequencing technologies. The genome or transcriptome were assembled and annotated (Figure 2A). Plants usually harbor several enzymes for one biochemical reaction, therefore, quick identification of key enzymes for CBE production are essential. For CBE production, the potential genes involved in TAG biosynthesis can be identified based on the similarity or phylogeny (Wei et al., 2017a). Sequence similarity network, which clustered sequences based on pairwise similarity can be applied to classify sequences into subgroups. While phylogenetic analysis can give more direct insights into the relationships between newly identified genes and characterized genes. Based on phylogeny, potential CBE production genes can be identified via their neighboring characterized genes (Figure 2B). Several GPATs, LPATs, and DGATs had been identified from multi-omics data with such strategy (Wei et al., 2017a, 2018). In previous studies, cocoa GPAT, LPAT, and DGAT genes that are similar to the characterized genes or different from the known enzymes with C16 as preferred substrates were selected for experimental measurement in yeast (Wei et al., 2017a). The identified efficient/potential genes can then be used for further metabolic engineering or synthetic biology modification of S. cerevisiae. During the typical metabolic engineering cycle of Design-Build-Test-Learn (DBTL), lipid biosynthetic pathway can be redesigned and rewired (Nielsen and Keasling, 2016; Ko et al., 2020; Figure 2C). The designed pathway can be further built with advanced synthetic biology and systems biology tools to generate strains with high-level CBE production. When the TAG/CBE titer, rate and yield (TRY) of the engineered strains are high enough, they will be used for further large-scale fermentation; if the TRY are low, novel engineering strategies should be used for next round strain optimization until high TRY are obtained (Nielsen and Keasling, 2016; Ko et al., 2020).
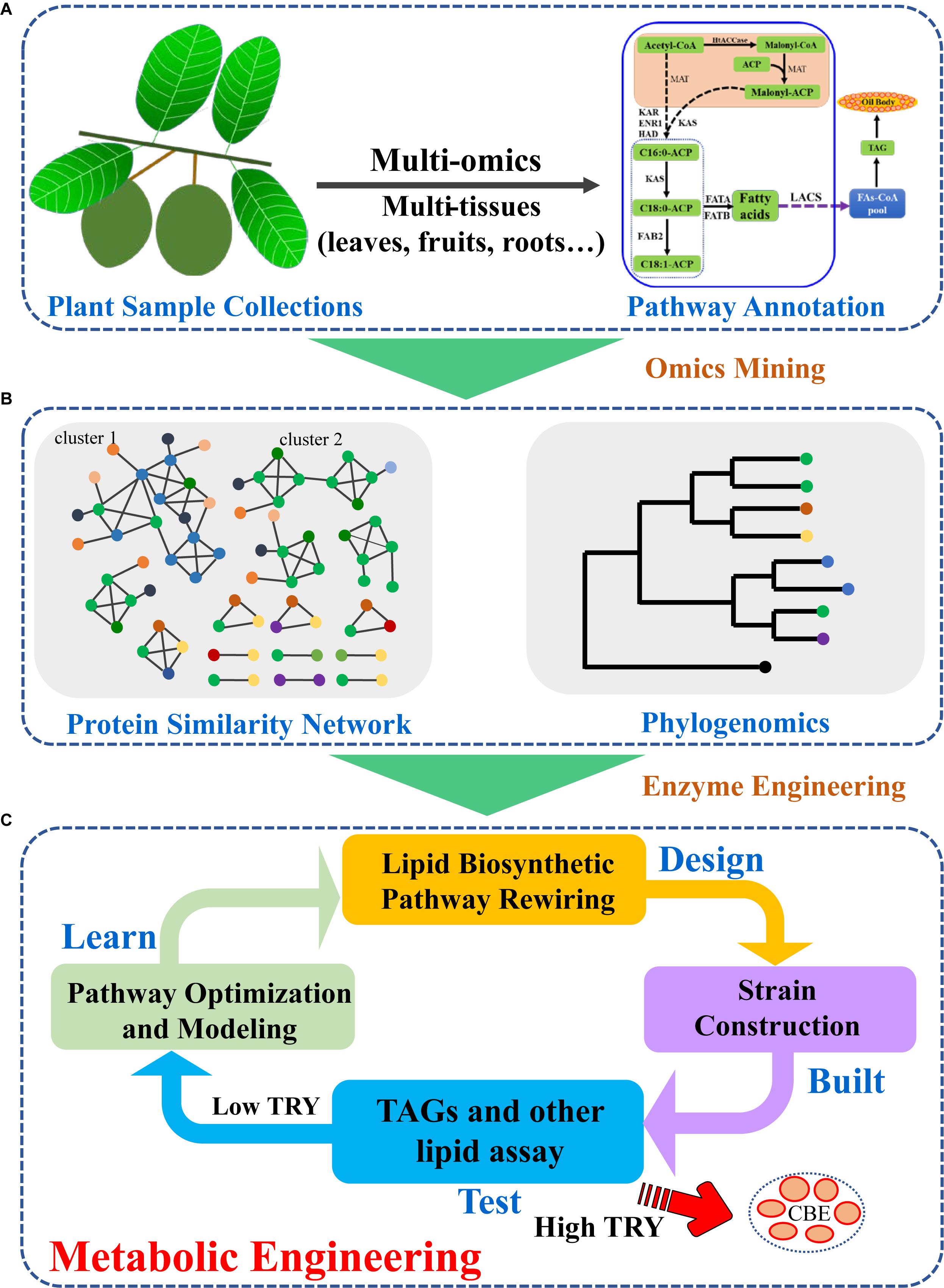
Figure 2. Strategies for plant TAG biosynthetic enzyme mining and metabolic engineering for increased CBE production. (A) Plant sample collection and multi-omics integration analyses identified potential genes in plant lipid biosynthetic pathway; (B) Phylogenetic and protein similarity network analyses led to efficient enzyme selection; (C) Classic design-build-test-learn cycle enhanced CBE production in S. cerevisiae by introduction of efficient plant lipid biosynthetic enzymes.
Based on the high-fatty acid production S. cerevisiae platform strains (Ferreira et al., 2018a; Yu et al., 2018), metabolic engineering to increase specific TAG productivity by introducing efficient enzymes of GPAT, LPAT, and DGAT would benefit CBE production. With the development of DNA sequencing technology, the whole-genome sequencing and transcriptomics of numerous plants have enabled systematic analysis of TAG and lipid production pathways in different plant species, which has provided the basis for future screening of efficient GPAT/LPAT/DGAT candidates (Cheng et al., 2018). By integrating the transcriptomics and lipidomics data, it will help to identify specific enzymes engaged in the production of targeted TAGs. Furthermore, in terms of engineering yeast, the enhancement of acetyl-CoA and malonyl-CoA pool by disruption of the PDC genes or engineering key fatty acid biosynthetic genes (ACC1, FAA1, FAA4, FAS, et al.), enhancing/balancing cofactor of NADPH supply, down-regulating completing pathways, harnessing yeast sub-organelle metabolism will increase CBE production in engineered S. cerevisiae (Shi et al., 2014; Zhou et al., 2016a,b; Dai et al., 2018; Yan and Pfleger, 2020). The TAG lipases encoded by TGL3 and TGL4 genes had been confirmed to involve in the TAG degradation (Dulermo et al., 2013). The blockage of TAG entering into the degradation pathways could also be an efficient strategy. Moreover, appropriate low-cost substrates (such as xylose and other lignocellulose components) can also be used to reduce microbial CBE production cost in the future (Hou et al., 2017). The adaptive laboratory evolution might help to increase strain adaptation to recalcitrant substrates, in order to produce high-level fatty acid-derived chemicals from recalcitrant substrates or one carbon source of CO2/CH4, which might be applied in the future CBE production (Pereira et al., 2019; Cao et al., 2020; Liu et al., 2020; Zhu et al., 2020). Considering oleaginous yeasts can produce high-level lipids naturally (Wei et al., 2017b), metabolic engineering of selected oleaginous yeasts can be an alternative choice in the future (Kamineni and Shaw, 2020; Wang et al., 2020).
Biosynthesis of CBE using engineered S. cerevisiae is one promising way to satisfy growing CB demand. Efficient genes of GPATs, LPATs, and DGATs encoding for SOS production from oil crops can be screened using both computational and experimental approaches. The expression of these lipid biosynthetic genes in S. cerevisiae chasis with high-level C18:0- and C18:1-production ability would strongly increase the CBE production. Metabolic engineering and rewiring have enabled turning S. cerevisiae from alcoholic fermentation to lipogenesis, and further systems biology, synthetic biology, evolutionary engineering and other advanced systems metabolic engineering strategies might further increase CBE production in S. cerevisiae.
JN and YW conceived the study. MW, BJ, and YW drafted the manuscript. JN revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (No. 31800079).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.594081/full#supplementary-material
Argout, X., Salse, J., Aury, J.-M., Guiltinan, M. J., Droc, G., Gouzy, J., et al. (2011). The genome of Theobroma cacao. Nat. Genet. 43, 101–108. doi: 10.1038/ng.736
Aymé, L., Baud, S., Dubreucq, B., Joffre, F., and Chardot, T. (2014). Function and localization of the Arabidopsis thaliana diacylglycerol acyltransferase DGAT2 expressed in yeast. PLoS One 9:e92237. doi: 10.1371/journal.pone.0092237
Bates, P. D., Stymne, S., and Ohlrogge, J. (2013). Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 16, 358–364. doi: 10.1016/j.pbi.2013.02.015
Baud, S., and Lepiniec, L. (2010). Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 49, 235–249. doi: 10.1016/j.plipres.2010.01.001
Bergenholm, D., Gossing, M., Wei, Y., Siewers, V., and Nielsen, J. (2018). Modulation of saturation and chain length of fatty acids in Saccharomyces cerevisiae for production of cocoa butter-like lipids. Biotechnol. Bioeng. 115, 932–942. doi: 10.1002/bit.26518
Bouvier-Navé, P., Benveniste, P., Oelkers, P., Sturley, S. L., and Schaller, H. (2000). Expression in yeast and tobacco of plant cDNAs encoding acyl CoA:diacylglycerol acyltransferase. Eur. J. Biochem. 267, 85–96. doi: 10.1046/j.1432-1327.2000.00961.x
Cao, M., Tran, V. G., and Zhao, H. (2020). Unlocking nature’s biosynthetic potential by directed genome evolution. Curr. Opin. Biotechnol. 66, 95–104. doi: 10.1016/j.copbio.2020.06.012
Cheng, S., Melkonian, M., Smith, S. A., Brockington, S., Archibald, J. M., Delaux, P. M., et al. (2018). 10KP: a phylodiverse genome sequencing plan. Gigascience 7, 1–9. doi: 10.1093/gigascience/giy013
Clough, Y., Faust, H., and Tscharntke, T. (2009). Cacao boom and bust: sustainability of agroforests and opportunities for biodiversity conservation. Conserv. Lett. 2, 197–205. doi: 10.1111/j.1755-263X.2009.00072.x
Coleman, R. A., and Lee, D. P. (2004). Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43, 134–176. doi: 10.1016/S0163-7827(03)00051-1
Dai, Z., Huang, M., Chen, Y., Siewers, V., and Nielsen, J. (2018). Global rewiring of cellular metabolism renders Saccharomyces cerevisiae crabtree negative. Nat. Commun. 9, 3059–3059. doi: 10.1038/s41467-018-05409-9
de Kroon, A. I., Rijken, P. J., and De Smet, C. H. (2013). Checks and balances in membrane phospholipid class and acyl chain homeostasis, the yeast perspective. Prog. Lipid Res. 52, 374–394. doi: 10.1016/j.plipres.2013.04.006
de Silva Souza, C., and Block, J. M. (2018). Impact of the addition of cocoa butter equivalent on the volatile compounds profile of dark chocolate. J. Food Sci. Technol. 55, 767–775. doi: 10.1007/s13197-017-2989-6
Drenth, A., and Guest, D. I. (2016). Fungal and oomycete diseases of tropical tree fruit crops. Annu. Rev. Phytopathol. 54, 373–395. doi: 10.1146/annurev-phyto-080615-095944
Dulermo, T., Tréton, B., Beopoulos, A., Kabran Gnankon, A. P., Haddouche, R., and Nicaud, J.-M. (2013). Characterization of the two intracellular lipases of Y. lipolytica encoded by TGL3 and TGL4 genes: new insights into the role of intracellular lipases and lipid body organisation. Biochim. Biophys. Acta 1831, 1486–1495. doi: 10.1016/j.bbalip.2013.07.001
Ejsing, C. S., Sampaio, J. L., Surendranath, V., Duchoslav, E., Ekroos, K., Klemm, R. W., et al. (2009). Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 106, 2136–2141. doi: 10.1073/pnas.0811700106
Fakas, S. (2017). Lipid biosynthesis in yeasts: a comparison of the lipid biosynthetic pathway between the model nonoleaginous yeast Saccharomyces cerevisiae and the model oleaginous yeast Yarrowia lipolytica. Eng. Life Sci. 17, 292–302. doi: 10.1002/elsc.201600040
Ferreira, R., Teixeira, P. G., Gossing, M., David, F., Siewers, V., and Nielsen, J. (2018a). Metabolic engineering of Saccharomyces cerevisiae for overproduction of triacylglycerols. Metab. Eng. Commun. 6, 22–27. doi: 10.1016/j.meteno.2018.01.002
Ferreira, R., Teixeira, P. G., Siewers, V., and Nielsen, J. (2018b). Redirection of lipid flux toward phospholipids in yeast increases fatty acid turnover and secretion. Proc. Natl. Acad. Sci. U.S.A. 115, 1262–1267. doi: 10.1073/pnas.1715282115
Greer, M. S., Truksa, M., Deng, W., Lung, S.-C., Chen, G., and Weselake, R. J. (2015). Engineering increased triacylglycerol accumulation in Saccharomyces cerevisiae using a modified type 1 plant diacylglycerol acyltransferase. Appl. Microbiol. Biotechnol. 99, 2243–2253. doi: 10.1007/s00253-014-6284-4
Hou, J., Qiu, C., Shen, Y., Li, H., and Bao, X. (2017). Engineering of Saccharomyces cerevisiae for the efficient co-utilization of glucose and xylose. FEMS Yeast Res. 17:fox034. doi: 10.1093/femsyr/fox034
Jahurul, M., Zaidul, I., Norulaini, N., Sahena, F., Jinap, S., Azmir, J., et al. (2013). Cocoa butter fats and possibilities of substitution in food products concerning cocoa varieties, alternative sources, extraction methods, composition, and characteristics. J. Food Eng. 117, 467–476. doi: 10.1016/j.jfoodeng.2012.09.024
Kamineni, A., and Shaw, J. (2020). Engineering triacylglycerol production from sugars in oleaginous yeasts. Curr. Opin. Biotechnol. 62, 239–247. doi: 10.1016/j.copbio.2019.12.022
Kamisaka, Y., Tomita, N., Kimura, K., Kainou, K., and Uemura, H. (2007). DGA1 (diacylglycerol acyltransferase gene) overexpression and leucine biosynthesis significantly increase lipid accumulation in the Deltasnf2 disruptant of Saccharomyces cerevisiae. Biochem. J. 408, 61–68. doi: 10.1042/BJ20070449
Klug, L., and Daum, G. (2014). Yeast lipid metabolism at a glance. FEMS Yeast Res. 14, 369–388. doi: 10.1111/1567-1364.12141
Ko, Y.-S., Kim, J. W., Lee, J. A., Han, T., Kim, G. B., Park, J. E., et al. (2020). Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production. Chem. Soc. Rev. 49, 4615–4636. doi: 10.1039/D0CS00155D
Krivoruchko, A., Zhang, Y., Siewers, V., Chen, Y., and Nielsen, J. (2015). Microbial acetyl-CoA metabolism and metabolic engineering. Metab. Eng. 28, 28–42. doi: 10.1016/j.ymben.2014.11.009
Leber, C., Polson, B., Fernandez-Moya, R., and Da Silva, N. A. (2015). Overproduction and secretion of free fatty acids through disrupted neutral lipid recycle in Saccharomyces cerevisiae. Metab. Eng. 28, 54–62. doi: 10.1016/j.ymben.2014.11.006
Leebens-Mack, J. H., Barker, M. S., Carpenter, E. J., Deyholos, M. K., Gitzendanner, M. A., Graham, S. W., et al. (2019). One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685. doi: 10.1038/s41586-019-1693-2
Li, X., Guo, D., Cheng, Y., Zhu, F., Deng, Z., and Liu, T. (2014). Overproduction of fatty acids in engineered Saccharomyces cerevisiae. Biotechnol. Bioeng. 111, 1841–1852. doi: 10.1002/bit.25239
Lipp, M., and Anklam, E. (1998). Review of cocoa butter and alternative fats for use in chocolate – Part A. Compositional data. Food Chem. 62, 73–97. doi: 10.1016/S0308-8146(97)00160-X
Liu, Q., Siloto, R. M. P., Lehner, R., Stone, S. J., and Weselake, R. J. (2012). Acyl-CoA:diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Prog. Lipid Res. 51, 350–377. doi: 10.1016/j.plipres.2012.06.001
Liu, Z., Wang, K., Chen, Y., Tan, T., and Nielsen, J. (2020). Third-generation biorefineries as the means to produce fuels and chemicals from CO2. Nat. Catal. 3, 274–288. doi: 10.1038/s41929-019-0421-5
Maraschin, F. D. S., Kulcheski, F. R., Segatto, A. L. A., Trenz, T. S., Barrientos-Diaz, O., Margis-Pinheiro, M., et al. (2019). Enzymes of glycerol-3-phosphate pathway in triacylglycerol synthesis in plants: function, biotechnological application and evolution. Prog. Lipid Res. 73, 46–64. doi: 10.1016/j.plipres.2018.12.001
Martin, C. E., Oh, C., and Jiang, Y. (2007). Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim. Biophys. Acta 1771, 271–285. doi: 10.1016/j.bbalip.2006.06.010
Nielsen, J., and Keasling, J. D. (2016). Engineering cellular metabolism. Cell 164, 1185–1197. doi: 10.1016/j.cell.2016.02.004
Pereira, R., Wei, Y., Mohamed, E., Radi, M., Malina, C., Herrgard, M. J., et al. (2019). Adaptive laboratory evolution of tolerance to dicarboxylic acids in Saccharomyces cerevisiae. Metab. Eng. 56, 130–141. doi: 10.1016/j.ymben.2019.09.008
Rossler, H., Rieck, C., Delong, T., Hoja, U., and Schweizer, E. (2003). Functional differentiation and selective inactivation of multiple Saccharomyces cerevisiae genes involved in very-long-chain fatty acid synthesis. Mol. Genet. Genomics 269, 290–298. doi: 10.1007/s00438-003-0836-0
Runguphan, W., and Keasling, J. D. (2014). Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 21, 103–113. doi: 10.1016/j.ymben.2013.07.003
Shi, S., Chen, Y., Siewers, V., and Nielsen, J. (2014). Improving production of malonyl coenzyme a-derived metabolites by abolishing Snf1-dependent regulation of Acc1. Mbio 5:e01130-14. doi: 10.1128/mBio.01130-14
Sonwai, S., Kaphueakngam, P., and Flood, A. E. (2014). Blending of mango kernel fat and palm oil mid-fraction to obtain cocoa butter equivalent. J. Food Sci. Technol. 51, 2357–2369. doi: 10.1007/s13197-012-0808-7
Spagnuolo, M., Yaguchi, A., and Blenner, M. (2019). Oleaginous yeast for biofuel and oleochemical production. Curr. Opin. Biotechnol. 57, 73–81. doi: 10.1016/j.copbio.2019.02.011
Tang, X., Lee, J., and Chen, W. N. (2015). Engineering the fatty acid metabolic pathway in Saccharomyces cerevisiae for advanced biofuel production. Metab. Eng. Commun. 2, 58–66. doi: 10.1016/j.meteno.2015.06.005
Tehlivets, O., Scheuringer, K., and Kohlwein, S. D. (2007). Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta 1771, 255–270. doi: 10.1016/j.bbalip.2006.07.004
Vieira, L. R., Efraim, P., De Walle, D. V., De Clercq, N., and Dewettinck, K. (2015). Influence of Brazilian geographic region and organic agriculture on the composition and crystallization properties of cocoa butter. J. Am. Oil Chem. Soc. 92, 1579–1592. doi: 10.1007/s11746-015-2728-y
Wang, J., Ledesma-Amaro, R., Wei, Y., Ji, B., and Ji, X.-J. (2020). Metabolic engineering for increased lipid accumulation in Yarrowia lipolytica – a review. Bioresour. Technol. 313:123707. doi: 10.1016/j.biortech.2020.123707
Wang, P., Wei, Y., Fan, Y., Liu, Q., Wei, W., Yang, C., et al. (2015). Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab. Eng. 29, 97–105. doi: 10.1016/j.ymben.2015.03.003
Wei, Y., Bergenholm, D., Gossing, M., Siewers, V., and Nielsen, J. (2018). Expression of cocoa genes in Saccharomyces cerevisiae improves cocoa butter production. Microb. Cell Fact. 17:11. doi: 10.1186/s12934-018-0866-2
Wei, Y., Gossing, M., Bergenholm, D., Siewers, V., and Nielsen, J. (2017a). Increasing cocoa butter-like lipid production of Saccharomyces cerevisiae by expression of selected cocoa genes. AMB Express 7, 34–34. doi: 10.1186/s13568-017-0333-1
Wei, Y., Ji, B., Siewers, V., Xu, D., Halkier, B. A., and Nielsen, J. (2019). Identification of genes involved in shea butter biosynthesis from Vitellaria paradoxa fruits through transcriptomics and functional heterologous expression. Appl. Microbiol. Biotechnol. 103, 3727–3736. doi: 10.1007/s00253-019-09720-3
Wei, Y., Siewers, V., and Nielsen, J. (2017b). Cocoa butter-like lipid production ability of non-oleaginous and oleaginous yeasts under nitrogen-limited culture conditions. Appl. Microbiol. Biotechnol. 101, 3577–3585. doi: 10.1007/s00253-017-8126-7
Wenning, L., Yu, T., David, F., Nielsen, J., and Siewers, V. (2017). Establishing very long−chain fatty alcohol and wax ester biosynthesis in Saccharomyces cerevisiae. Biotechnol. Bioeng. 114, 1025–1035. doi: 10.1002/bit.26220
Yan, Q., and Pfleger, B. F. (2020). Revisiting metabolic engineering strategies for microbial synthesis of oleochemicals. Metab. Eng. 58, 35–46. doi: 10.1016/j.ymben.2019.04.009
Yang, W., Simpson, J. P., Libeisson, Y., Beisson, F., Pollard, M., and Ohlrogge, J. B. (2012). A land-plant-specific Glycerol-3-phosphate Acyltransferase family in Arabidopsis: substrate specificity, sn-2 preference and evolution. Plant Physiol. 160, 638–652. doi: 10.1104/pp.112.201996
Yen, C. L., Stone, S. J., Koliwad, S., Harris, C., and Farese, R. V. Jr. (2008). Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 49, 2283–2301. doi: 10.1194/jlr.R800018-JLR200
Yu, T., Zhou, Y. J., Huang, M., Liu, Q., Pereira, R., David, F., et al. (2018). Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell 174, 1549–1558.e1514. doi: 10.1016/j.cell.2018.07.013
Zeng, J., Shen, J., Wu, Y., Liu, X., Deng, Z. Y., and Li, J. (2020). Effect of adding shea butter stearin and emulsifiers on the physical properties of cocoa butter. J. Food Sci. 85, 972–979. doi: 10.1111/1750-3841.15076
Zhang, Y., Nielsen, J., and Liu, Z. (2018). Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived hydrocarbons. Biotechnol. Bioeng. 115, 2139–2147. doi: 10.1002/bit.26738
Zheng, Z., and Zou, J. (2001). The initial step of the glycerolipid pathway: identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae. J. Biol. Chem. 276, 41710–41716. doi: 10.1074/jbc.M104749200
Zhou, Y. J., Buijs, N. A., Zhu, Z., Gomez, D. O., Boonsombuti, A., Siewers, V., et al. (2016a). Harnessing yeast peroxisomes for biosynthesis of fatty-acid-derived biofuels and chemicals with relieved side-pathway competition. J. Am. Chem. Soc. 138, 15368–15377. doi: 10.1021/jacs.6b07394
Zhou, Y. J., Buijs, N. A., Zhu, Z., Qin, J., Siewers, V., and Nielsen, J. (2016b). Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat. Commun. 7:11709. doi: 10.1038/ncomms11709
Zhu, Z., Hu, Y., Teixeira, P. G., Pereira, R., Chen, Y., Siewers, V., et al. (2020). Multidimensional engineering of Saccharomyces cerevisiae for efficient synthesis of medium-chain fatty acids. Nat. Catal. 3, 64–74. doi: 10.1038/s41929-019-0409-1
Keywords: cocoa butter equivalents, Saccharomyces cerevisiae, metabolic engineering, synthetic biology, lipid biosynthesis
Citation: Wang M, Wei Y, Ji B and Nielsen J (2020) Advances in Metabolic Engineering of Saccharomyces cerevisiae for Cocoa Butter Equivalent Production. Front. Bioeng. Biotechnol. 8:594081. doi: 10.3389/fbioe.2020.594081
Received: 12 August 2020; Accepted: 21 September 2020;
Published: 15 October 2020.
Edited by:
Solange I. Mussatto, Technical University of Denmark, DenmarkReviewed by:
Isak Stephanus Pretorius, Macquarie University, AustraliaCopyright © 2020 Wang, Wei, Ji and Nielsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongjun Wei, eW9uZ2p1bndlaUB6enUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.