- 1Glycomics and Glycan Bioengineering Research Center (GGBRC), College of Food Science and Technology, Nanjing Agricultural University, Nanjing, China
- 2Protein Analysis Group, Department of Pharmacy, University of Copenhagen, Copenhagen, Denmark
- 3Natural and Medical Sciences Institute (NMI), University of Tubingen, Reutlingen, Germany
Peptide-N4-(N-acetyl-β-glucosaminyl) asparagine amidases (PNGases, N-glycanases, EC 3.5.1.52) are indispensable tools in releasing N-glycans from glycoproteins. So far, only a limited number of PNGase candidates are available for the structural analysis of glycoproteins and their glycan moieties. Herein, a panel of 13 novel PNGase H+ candidates (the suffix H+ refers to the acidic pH optimum of these acidobacterial PNGases) was tested in their recombinant form for their deglycosylation performance. One candidate (originating from the bacterial species Dyella japonica) showed superior properties both in solution-phase and immobilized on amino-, epoxy- and nitrilotriacetate resins when compared to currently acidic available PNGases. The high expression yield compared to a previously described PNGase H+, broad substrate specificity, and good storage stability of this novel N-glycanase makes it a valuable tool for the analysis of protein glycosylation.
Introduction
The study of protein N-glycosylation is an active area of biotechnological research, given that all antibodies, and many biotherapeutics as well as extracellular and cell surface proteins bear these post-translational modifications (Overington et al., 2006; Saldova et al., 2017). The detailed analysis of therapeutic glycoproteins is of great importance, as the quality and effectiveness of these drugs strongly depend on the consistency of their glycan moieties (Berkowitz et al., 2012; Duivelshof et al., 2019). Peptide-N4-(N-acetyl-β-glucosaminyl) asparagine amidases (PNGases) enzymatically release asparagine-linked glycan moieties from glycoproteins and have been described as valuable tools in the analysis of glycotherapeutics (Wang and Voglmeir, 2014). A subset of these PNGases used in bioanalytical applications (namely PNGase A and PNGase At) originate from plants and show a peculiar promiscuity toward a broad range of eukaryotic glycoproteins, including those from plants and insects (Takahashi, 1977; Altmann et al., 1998; Yan et al., 2018). Furthermore, the low pH optimum of these enzymes is of particular interest for analytical methods which require acidic conditions, such as rapid N-glycans derivatization methods (Du et al., 2015) or hydrogen-deuterium exchange mass spectrometry (HDX-MS) of glycoproteins (Jensen et al., 2016).
One limitation of PNGase A (and based on the close homology, also PNGase At) is that this enzyme itself is a glycoprotein extracted from almonds (Takahashi, 1977), thus it can undergo auto-deglycosylation, thereby contaminate analyzed samples with endogenous PNGase A glycan structures (Altmann et al., 1998). Moreover, there are currently no reports describing the successful recombinant expression of PNGase A in prokaryotic systems, and therefore limiting the use of engineered fusion tags for the facilitated purification and immobilization of this enzyme. Interestingly, our research group described recently a PNGase A homologue from Acidobacteria instead of plants (Wang et al., 2014). This phylum of soil bacteria was virtually unknown until 2009 (Ward et al., 2009). The discovered PNGase originated from Terriglobus roseus and was recombinantly expressed in E. coli, and due to its surprisingly low pH optimum of pH 2.5 designated as “PNGase H+.” Although the enzymatic activity of PNGase H+ from Terriglobus roseus could be successfully used for conventional N-glycan analysis protocols (Wang et al., 2017; Du et al., 2018), its use in a wider range of analytical workflows was hampered by difficulties in optimizing expression and purification of the enzyme at larger scales. With the rapid progress and the availability of whole-genome sequences in recent years, >70 acidobacterial PNGase H+ homologs can be currently found using BLAST (Altschul et al., 1990) in March 2020. This pool of candidate genes, therefore, allows the evaluation of alternative PNGase candidate genes for their biotechnological potential. In this Brief Research Report, we studied the activity and stability of 12 unstudied acidobacterial PNGase H+ candidates and we compared them to the previously described PNGase from Terriglobus roseus, both in solution and immobilized on various solid supports.
Materials and Methods
Materials
Epoxy- and amino activated methacrylate beads were kindly donated by Purolite (Zhejiang, China). Ni-NTA (nickel-ion nitrilotriacetic acid) superflow resin was obtained from Qiagen (Shanghai, China). WST-1 (sodium 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolate) was purchased from Sangon Biotech Company (Shanghai, China). Horseradish peroxidase (HRP) was obtained from Duly Biotech Ltd. (Nanjing, China). Human IgG was purchased from ProteinTech (Nanjing, China). Lactoferrin was obtained from Wako Pure Chemical Industries (Nanjing, China). PNGase F was provided by Qlyco Ltd. (Nanjing, China). All other standard chemicals and buffer reagents were of the highest grade available.
Construction of the Recombinant Plasmids
The genes encoding the putative PNGase homologs Ac (from Acidobacterium capsulatum, UniProt ID C1F2K4) and Ka (from Kutzneria albida, UniProt ID W5W1E9) were amplified from bacterial DNA and cloned into a pET30a expression vector as described for the PNGase Tr (from Terriglobus roseus, UniProt ID I3ZL27) previously (Wang et al., 2014). The other 10 candidate genes were synthesized in Escherichia coli K12 codon preference and ligated on the pET30a vector by Genscript Ltd. (Nanjing, China) and designated as PNGases Le (from Lysobacter enzymogenes, UniProt ID A0A0S2DLB6), Sp (from Streptomyces populi, UniProt ID A0A2I0SFR1), Sa (from Streptomyces albulus, UniProt ID A0A059W230), Ss (from Solimicrobium silvestre, UniProt ID A0A2S9GXY8), Sb (Silvibacterium bohemicum), Ab (from Acidobacteria bacterium, GenBank ID PYX41174), Ea (from Edaphobacter aggregans, GenBank ID WP_035357016), Xt (Xanthomonass theicola, GenBank ID PPT80182), Ft (from Frateuria terrea, UniProt ID A0A1H6SY22), and Dj (from Dyella japonica, UniProt ID A0A075JY55).
Expression, Purification, and Immobilization of Recombinant PNGases
The expression and Ni-NTA affinity purification of the 13 PNGase candidate genes were performed as previously described for the Terriglobus roseus homolog (Wang et al., 2014). The enzymes’ activities were evaluated using both a gel-based deglycosylation assay (section “Gel-Based Deglycosylation Assay”) and a microplate-based activity test (section “Microplate-Based Activity Assay”). The most active PNGase (Dj) was selected for detailed biochemical characterization and for immobilization studies.
For the immobilization using amino-functionalized methacrylate resins (ECR8305F and ECR8310F) or the epoxy-activated methacrylate resins (ECR8214F and ECR8205F), 500 mg of each resin was equilibrated in immobilization buffer (0.5 M NaCl, 50 mM sodium phosphate buffer, pH 8.0). Then, 4 mL of each purified enzyme solution was mixed with the resins and incubated at 4°C for 18 h with orbital shaking. Then, the mixture was filtered and washed with an immobilization buffer. For the immobilization of PNGase Dj on Ni-NTA resin, 500 mg aliquots were removed after the washing step of the affinity purification. The immobilization efficiency was determined by measuring the PNGase activity of either the purified enzyme solutions (20 μL), the supernatants of the immobilization reactions (20 μL), or the supporting resins (50 mg). In all cases, the immobilization yield (Ψ) was calculated as follows:
where ABSinitial is the determined absorbance values at 582 nm using the microplate-based activity assay of the purified PNGase solutions offered to the resins, while ABSsupernatant is the absorbance values of the remaining PNGase proteins that remain in the supernatant after the immobilization time.
Gel-Based Deglycosylation Assay
The enzymatic activity of the PNGase candidates was evaluated using a gel-based deglycosylation of horseradish peroxidase (HRP) previously described (Wang et al., 2014). A 20 μL assay mixture consisting of 5 μL of purified PNGase enzyme solution, 1 μL of HRP substrate (10 μg/μL, denatured at 95°C for 10 min), 6 μL of sodium phosphate-citrate buffer (1 M, pH 2.0) and 8 μL of water was incubated at 37°C for 16 h. For the deglycosylation experiments of Lactoferrin or human IgG, 5 μg of the dried glycoproteins were either incubated with 20 μL the PNGase Dj reaction mixture or with 20 μL of a PNGase F reaction mixture (consisting of 0.1 μg of PNGase F and 150 mM sodium phosphate buffer (pH 7.5) in water). Then the assay mixtures were mixed with 5 μL of Laemmli denaturation buffer (fivefold concentrated) and incubated at 95°C for 10 min. These samples were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and protein bands visualized by Coomassie Brilliant Blue R-250 staining.
Microplate-Based Activity Assay
This rapid colorimetric activity test relies on the reduction of the yellowish tetrazolium dye WST-1 into a blue formazan dye by the reducing end of N-glycans, which were enzymatically released from glycoprotein substrates (Wang et al., 2019). Typically, reaction mixtures of 20 μL contained 5 μL of purified PNGase enzyme solution, 5 μL of HRP substrate (20 μg/μL, denatured at 95°C for 10 min), 6 μL of sodium phosphate-citrate buffer (1 M, pH 2.0), and 4 μL of distilled water. For negative controls purified PNGase protein solutions were replaced with water. Samples were incubated at 37°C for 3 h. Then the reaction was quenched by adding 20 μL of 2.5 M trichloroacetic acid, samples centrifuged at 12,000 g for 30 min, and 10 μL of the supernatants transferred into a 384-well microplate. After adding 5 μL of NaOH solution (4 M) and 10 μL of aqueous WST-1 solution (1.7 mM), the mixture was incubated at 50°C for 1 h and subsequently analyzed at a wavelength of 584 nm using a microplate reader (Thermo Multiscan FC, Shanghai, China).
Temperature Effects and Reusability of PNGase Dj
The following parameters were measured using the microplate-based activity assay described in section “Microplate-Based Activity Assay.” The optimum temperature of PNGase Dj was determined by incubating the reaction mixtures at various temperatures ranging from 25 to 70°C. For testing the storage stability of the enzyme, the purified PNGase solution was stored for up to 60 days at 4°C, and the activity of sample aliquots tested in 7–15 day intervals. To assess the short-time thermal stability of the PNGase, enzyme samples were incubated at temperatures between 22 and 60°C for up to 24 h, and then used for activity tests. The recycling abilities of the immobilized enzyme were studied by reusing the resins in consecutive reactions. Therefore, 100 mg of enzyme carriers were incubated for 30 min at 37°C with 500 μL recycling buffer (consisting of 50 mM sodium phosphate-citrate buffer (pH 2.0), with or without 0.5% (w/v) BSA) and HRP (5 μg/μL) on a rotation mixer. After centrifugation (1 min, 2000 g), 30 μL from the supernatant was used for the microplate-based activity assay. The rest of the supernatant was discarded, and the resin washed twice with 500 μL recycling buffer. Then, the washed resin was reused again for a total of six cycles.
Analysis of N-Glycans
For the N-glycan release from mouse serum, 50 μL of the serum were incubated with either 150 μL of PNGase F reaction mixture [consisting of 50 μg of PNGase F, 0.15 % (w/v) of SDS, 0.25% (v/v) of 2-mercaptoethanol, 1.25% (w/v) of Triton X-100, and 75 mM of sodium phosphate buffer (pH 7.5) in water], or 150 μL of PNGase Dj reaction mixture [consisting of 50 μg of PNGase Dj and 470 mM of citrate/sodium phosphate buffer (pH 2.0) in water]. The deglycosylation reactions of Lactoferrin or human IgG were performed as described in section “Gel-Based Deglycosylation Assay.” After incubating the reaction mixtures for up to 16 h at 37°C, the samples were centrifuged for 5 min at 12,000 g and the supernatant subject to solid-phase extraction (Supelclean ENVI-Carb, 0.3 mL bed volume, prewashed with 1 mL of water, Supelco). After the samples were washed with 3 mL of water, the enzymatically released N-glycans were eluted from the SPE column using 3 mL of aqueous 40% acetonitrile solution containing 0.1% TFA (v/v/v). Then, the elution fractions were dried by centrifugal evaporation and subject to fluorescence derivatization using 10 μL of a 2-aminobenzamine (2-AB) labeling solution [consisting of 35 mM of 2-AB and 0.1 M of sodium cyanoborohydride in a dimethyl sulfoxide/acetic acid solution (7:3, v/v)]. After 2 h of incubation at 65°C, 5 μL of the sample was mixed with 70 μL of water and 175 μL of acetonitrile, and 40 μL of this sample solution was injected into a HILIC-UPLC system (hydrophilic interaction ultra-performance liquid chromatography, Nexera, Shimadzu) equipped with an Acquity BEH Glycan column (1.7 μm, 130Å, 2.1 × 150 mm, Waters) at an initial flow rate of 0.5 mL/min. The analytes were separated using aqueous ammonium formate (50 mM, pH 4.5) and 100% acetonitrile as mobile phases (solvent A and B, respectively). A linear gradient of 95–78% of B was applied from 0 to 6 min, and B was then decreased to 55.9% over 44.5 min. The analytes were monitored by fluorescence detection (Ex: 330 nm, Em: 420 nm) and were annotated using the GlycoBase N-glycan repository (Campbell et al., 2008). The MALDI-ToF MS analysis of underivatized HRP-derived N-glycans was performed with a crude PNGase Dj assay solution by mixing 1 μL of the sample solution and 1 μL of 6-aza-2-thiothymine (ATT) solution (consisting of 0.3% w/v in 70% v/v aqueous acetonitrile), and using a Bruker Autoflex Speed Mass Spectrometer in positive ion mode.
Results and Discussion
Evaluation of Acidobacterial PNGase Activity
Twelve putative PNGases candidate genes were selected from data from 32 publicly available acidobacterial genomes by choosing the most distinct homologs of this pool, so that the individual amino acid similarities were below 70% for each PNGase candidate. After expression, the purified enzyme solutions were subjected to the gel-based deglycosylation assay (Figure 1A) and the microplate-based activity test (Figure 1B). Both tests showed that PNGase homologs Xt, Ac, and Ka had a comparable deglycosylation activity to the previously described PNGase Tr, whereas homologs Ft and Dj exhibited increased deglycosylation of HRP relative to all other PNGases. Furthermore, the platereader assay showed that the overall deglycosylation activity of Ft and Dj was approximately fivefold higher compared to PNGase Tr. Other PNGase candidate genes showed only negligible deglycosylaton activities. Given the high activity of the PNGase Dj homology, compared to PNGase Tr, a detailed biochemical characterization and the immobilization trials focused on this enzyme.
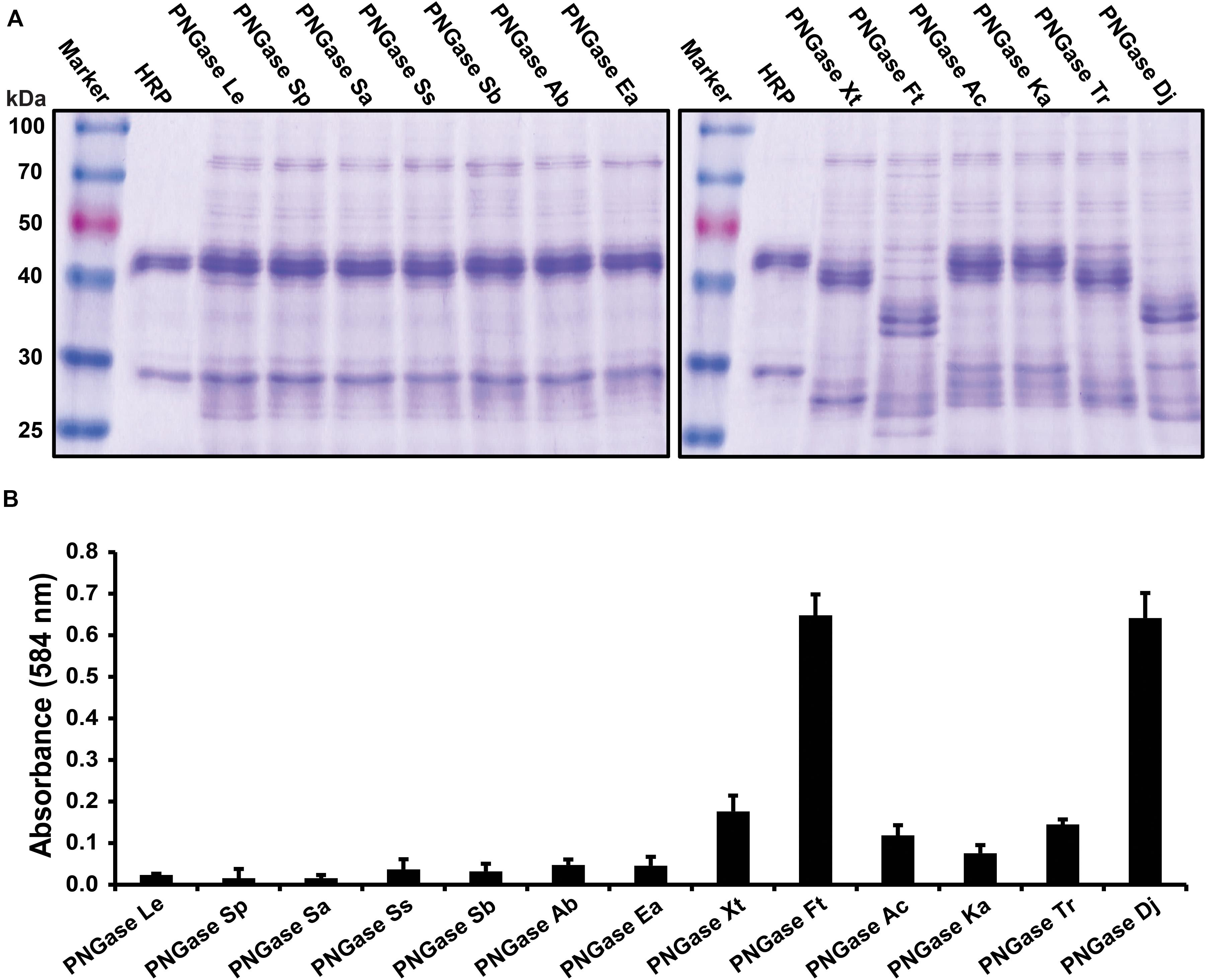
Figure 1. Activity tests of the PNGase candidate genes. (A) Gel-based deglycosylation assay to observe the molecular weight shift of the HRP glycoprotein band using SDS-PAGE. Deglycosylation of HRP is seen by a shifting of the major HRP protein band from approximately 45 kDa down to approximately 35 kDa. (B) Microplate-based activity test. The data are represented as the mean values and the error bars showing the standard deviation of three independent activity assays.
Purification and Immobilization
The purified PNGase Dj protein sample migrated as a dominant protein band with a molecular weight of approximately 65 kDa (Figure 2A). The mass of PNGase Dj was further examined using liquid chromatography and ESI mass spectrometry (LC-MS, Supplementary Figure 1), and the identity of the protein sequence was verified by LC-MS/MS analysis of peptides following digestion of PNGase Dj with Trypsin and Pepsin (Supplementary Figures 2, 3). Interestingly the MS data suggest that there is a discrepancy of 2849 Da between the expected theoretical protein mass and the measured protein mass (expected: 64417 Da, observed 61568 Da). This discrepancy can be best explained by the proteolytic cleavage of the first 28 N-terminal residues during expression (theoretical mass 61572 Da, 63 ppm deviation from the measured mass).
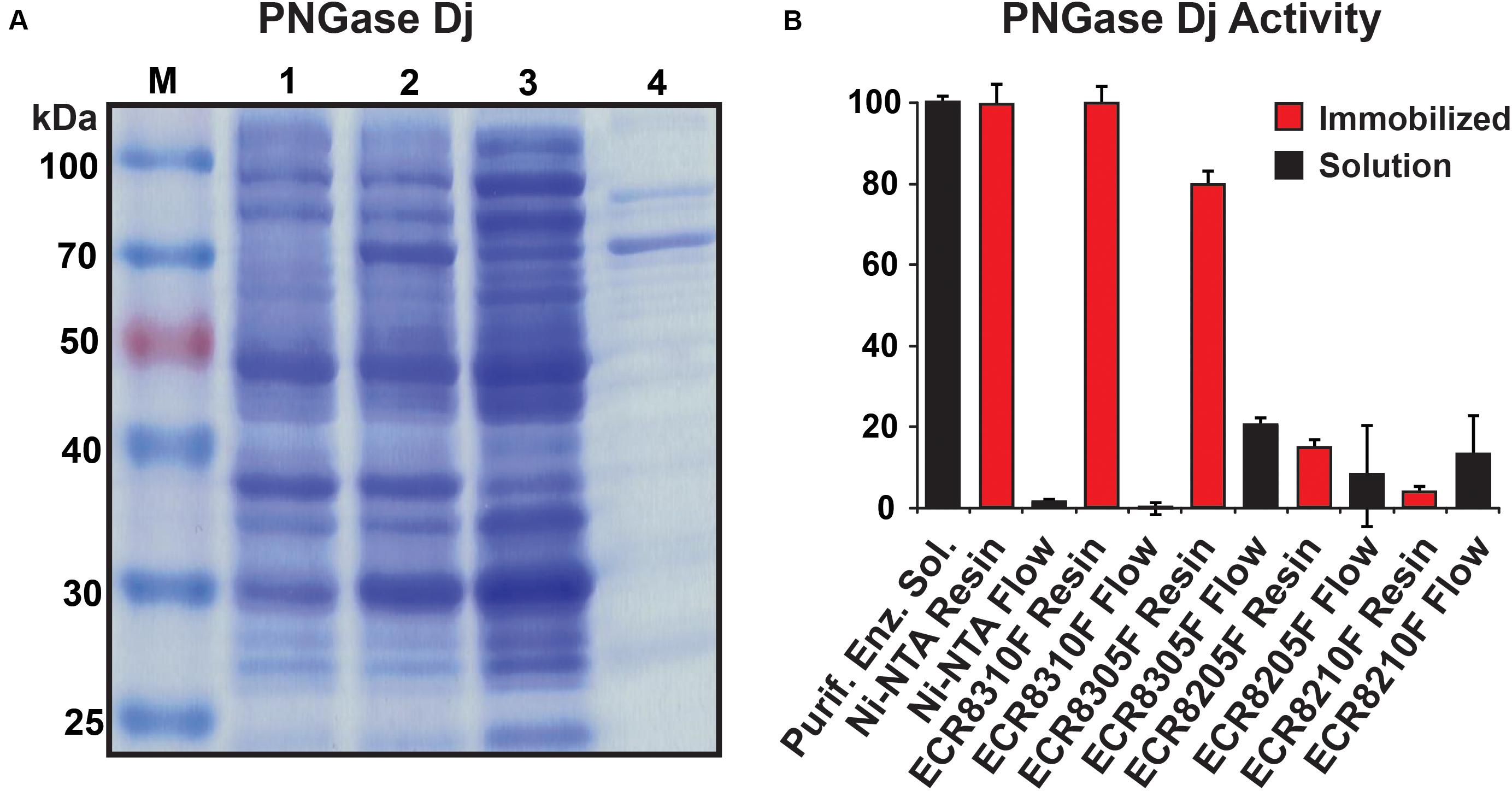
Figure 2. (A) SDS-PAGE analysis of recombinant PNGase Dj; M: protein size marker, lane 1: cells before induction, lane 2: cells after induction, lane 3: supernatant from cell lysate, lane 4: purified enzyme. (B) PNGase Dj activity in immobilized (red) and soluble form (black). The data are represented as the mean values and the error bars showing the standard deviation of three independent activity assays.
PNGase Dj was chosen for immobilization, and different enzyme carrier resins (ECRs) with different immobilization chemistries were evaluated by measuring the immobilization yield (Ψ). The Ψ values for all five tested enzyme carriers were determined to be ΨNi–NTA = 99%, ΨECR8310F = 100%, ΨECR8305F = 80%, ΨECR8205F = 92%, and ΨECR8310F = 87%. Despite these relatively high immobilization yields, only the Ni-NTA and the amino carrier resins ECR8305F and ECR8310F showed high enzymatic deglycosylation activities in the range between 75 and 95% of the enzyme in-solution activity (Figure 2B). Conversely, and despite the successful enzyme immobilization, the epoxy resins ECR8214F and ECR8205F displayed lower activities of 13 and 4% in comparison to in-solution enzyme activity. We presume that the unspecific nucleophilic reaction of the epoxy groups on the resins with lysine, histidine, cysteine, and tyrosine residues in PNGase Dj may be the reason for the inactivation of the enzyme. By using the glutaraldehyde-activated amino resins ECR8305F and ECR8310F, which specifically target lysine residues for the formation of Schiff bonds, the covalent immobilization of PNGase Dj could be achieved with high yield. Due to the simplicity of the non-covalent immobilization on Ni-NTA resins, the use of this carrier was the most promising for experimental setups where no covalent attachment of the PNGases is required.
Enzyme Characteristics, Temperature Stability, and Reusability
The biochemical parameters of PNGase Dj were generally comparable to the previously studied Tr homolog: Dj showed the highest activity in the pH range <3.5 with an optimum at pH 2 (Supplementary Figure 4), a temperature optimum of 55°C (Supplementary Figure 5), and its independence of metal ions (Figure 3A). The analysis of the released N-glycans from HRP using HILIC-UPLC (Supplementary Figure 6A) and MALDI-ToF mass spectrometry (Supplementary Figure 6B) showed that PNGase Dj has a comparable substrate specificity with previously described acidic PNGase homologs (Wuhrer et al., 2005; Wang et al., 2014). PNGase Dj was then used to deglycosylate bovine lactoferrin and human IgG, resulting in the complete deglycosylation of these mammalian glycoproteins within 6 h (Supplementary Figure 7). The HILIC-UPLC analysis showed a comparable N-glycan distribution as described in the literature (Reusch et al., 2015; Zhang et al., 2019), confirming that N-glycans bearing α1,6-fucosylation can be also released by this enzyme (Supplementary Figure 8). The effect of the acidic reaction conditions on acid-labile sialic acid moieties was also evaluated by releasing highly sialylated mouse serum glycoproteins using PNGase Dj, and comparing the resulting N-glycans with samples treated with PNGase F at neutral pH (pH 7.5, Supplementary Figure 9). The results showed that even after 16 h of incubation, the loss of sialylation affected mainly tetrasialylated N-glycan species, whereas the sialylation of mono-, di- and tri-sialylated N-glycans was less affected. Given that these experiments were performed at 37°C, we presume that lowering the reaction temperature would allow us to reduce the loss of sialic acids further. The recombinant PNGase was able to maintain full activity for at least 24 h of incubation at temperatures up to 55°C, but almost completely lost activities when incubated for 24 h at 60°C (Figure 3B). The reusability of the PNGase Dj enzyme immobilized on various resins showed that the Ni-NTA carrier could be reused for six cycles without loss of activity, whereas the amino resins ECR8305F and ECR8310F showed activity losses between 10 and 25% per cycle (Figure 3C). Furthermore, no PNGase activities were detected in the acidic reaction buffer, indicating the stable immobilization of PNGase Dj on the Ni-NTA resin (Supplementary Figure 10). No improvements could be achieved when 0.5% BSA (w/v) was added to the reaction mixtures. The storage stability of PNGase Dj was defined as relative activity between the first and successive reactions; No activity losses of the enzyme were observed during the test period of 60 days when stored at 4°C (Figure 3D).
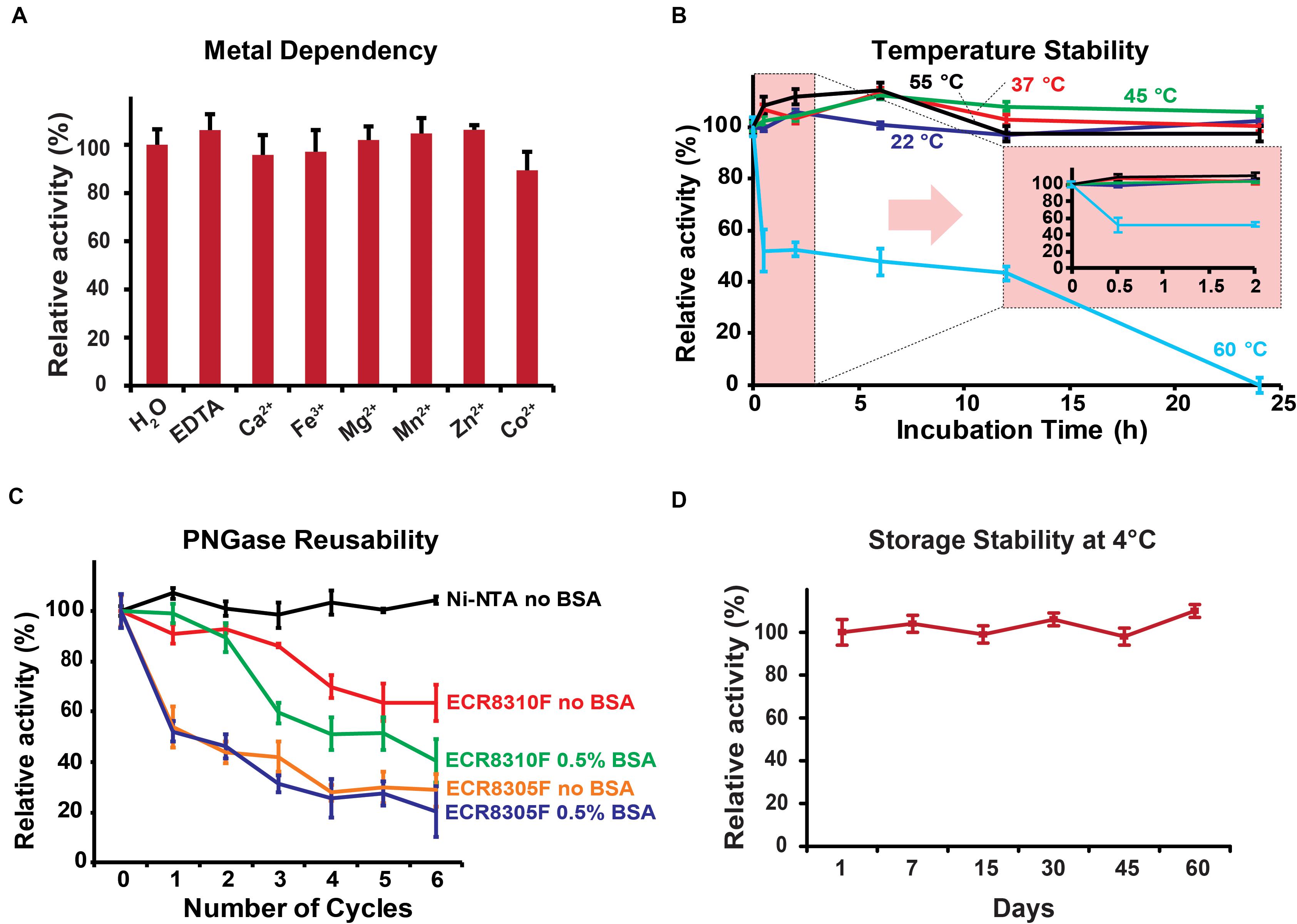
Figure 3. (A) Metal ion dependency of PNGase Dj. (B) Thermal stability of PNGase Dj at various temperatures. (C) Reusability of PNGase Dj using various resins. (D) Long-time storage stability of PNGase Dj at 4°C. The data are represented as the mean values and the error bars showing the standard deviation of three independent activity assays.
Conclusion
The research fields of glycomics and glycoproteomics are developing rapidly. Therefore, the discovery of robust and highly active PNGases is key to further improve these technologies. The evaluation of the herein presented PNGase homolog from Dyella japonica in solution and on various solid supports provides a solid foundation for future applications of these enzymes. The establishment of robust and user-friendly protocols using immobilized PNGase Dj for various analytical applications including hydrogen-deuterium exchange mass spectrometry (HDX-MS) of highly glycosylated proteins, or the proteomic identification of glycoproteins after deglycosylation is the current endeavor of our research groups.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
R-RG, GC, H-HY, MG, and Y-MD performed the experiments. R-RG, GC, AZ, KR, LL, and JV analyzed the data. R-RG, GC, KR, LL, and JV wrote the manuscript. LL and JV conceived the study. AZ, KR, LL, and JV coordinated the project. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in parts by the National Natural Science Foundation of China (Grant Nos. 31471703, 31671854, 31871793, and 31871754 to JV and LL), the 100 Foreign Talents Plan (Grant No. JSB2014012 to JV), and the Independent Research Foundation Denmark (Sapere Aude Grant-DFF-4184-00537A to KR).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00741/full#supplementary-material
References
Altmann, F., Paschinger, K., Dalik, T., and Vorauer, K. (1998). Characterisation of peptide-N4-(N-Acetyl-B-Glucosaminyl) asparagine amidase a and Its N-Glycans. Eur. J. Biochem. 252, 118–123.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Berkowitz, S. A., Engen, J. R., Mazzeo, J. R., and Jones, G. B. (2012). Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nat. Rev. Drug Discov. 11, 527–540. doi: 10.1038/nrd3746
Campbell, M. P., Royle, L., Radcliffe, C. M., Dwek, R. A., and Rudd, P. M. (2008). Glycobase and autogu: tools for Hplc-based glycan analysis. Bioinformatics 24, 1214–1216. doi: 10.1093/bioinformatics/btn090
Du, Y. M., Xia, T., Gu, X. Q., Wang, T., Ma, H. Y., Voglmeir, J., et al. (2015). Rapid sample preparation methodology for plant N-Glycan analysis using acid-stable Pngase H+. J. Agric. Food Chem. 63, 10550–10555. doi: 10.1021/acs.jafc.5b03633
Du, Y. M., Zheng, S. L., Liu, L., Voglmeir, J., and Yedid, G. (2018). Analysis of N-Glycans from Raphanus sativus cultivars using PNGase H+. J. Vis. Exp. 25:57979. doi: 10.3791/57979
Duivelshof, B. L., Jiskoot, W., Beck, A., Veuthey, J. L., Guillarme, D., and D’Atri, V. (2019). Glycosylation of biosimilars: recent advances in analytical characterization and clinical implications. Anal. Chim. Acta 1089, 1–18. doi: 10.1016/j.aca.2019.08.044
Jensen, P. F., Comamala, G., Trelle, M. B., Madsen, J. B., Jorgensen, T. J., and Rand, K. D. (2016). Removal of N-Linked glycosylations at acidic pH by PNGase a facilitates hydrogen/deuterium exchange mass spectrometry analysis of N-linked glycoproteins. Anal. Chem. 88, 12479–12488. doi: 10.1021/acs.analchem.6b03951
Overington, J. P., Al-Lazikani, B., and Hopkins, A. L. (2006). How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996. doi: 10.1038/nrd2199
Reusch, D., Haberger, M., Maier, B., Maier, M., Kloseck, R., Zimmermann, B., et al. (2015). Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles–Part 1: separation-based methods. mAbs 7, 167–179. doi: 10.4161/19420862.2014.986000
Saldova, R., Kilcoyne, M., Stockmann, H., Millan Martin, S., Lewis, A. M., Tuite, C. M., et al. (2017). Advances in analytical methodologies to guide bioprocess engineering for Bio-therapeutics. Methods 116, 63–83. doi: 10.1016/j.ymeth.2016.11.002
Takahashi, N. (1977). Demonstration of a new amidase acting on glycopeptides. Biochem. Biophys. Res. Commun. 76, 1194–1201. doi: 10.1016/0006-291X(77)90982-2
Wang, T., Cai, Z. P., Gu, X. Q., Ma, H. Y., Du, Y. M., Huang, K., et al. (2014). Discovery and characterization of a novel extremely acidic bacterial N-Glycanase with combined advantages of PNGase F and A. Biosci. Rep. 34:e00149. doi: 10.1042/BSR20140148
Wang, T., Hu, X. C., Cai, Z. P., Voglmeir, J., and Liu, L. (2017). Qualitative and quantitative analysis of carbohydrate modification on glycoproteins from seeds of Ginkgo Biloba. J. Agric. Food Chem. 65, 7669–7679. doi: 10.1021/acs.jafc.7b01690
Wang, T., and Voglmeir, J. (2014). Pngases as valuable tools in glycoprotein analysis. Protein Pept. Lett. 21, 976–985. doi: 10.2174/0929866521666140626111237
Wang, T., Zheng, S. L., Liu, L., and Voglmeir, J. (2019). Development of a colorimetric PNGase activity assay. Carbohydr. Res. 472, 58–64. doi: 10.1016/j.carres.2018.11.007
Ward, N. L., Challacombe, J. F., Janssen, P. H., Henrissat, B., Coutinho, P. M., Wu, M., et al. (2009). Three genomes from the phylum acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75, 2046–2056. doi: 10.1128/AEM.02294-08
Wuhrer, M., Balog, C. I., Koeleman, C. A., Deelder, A. M., and Hokke, C. H. (2005). New features of site-specific horseradish peroxidase (Hrp) glycosylation uncovered by nano-Lc-Ms with repeated ion-isolation/fragmentation cycles. Biochim. Biophys. Acta 1723, 229–239. doi: 10.1016/j.bbagen.2005.02.013
Yan, S., Vanbeselaere, J., Jin, C., Blaukopf, M., Wols, F., Wilson, I. B. H., et al. (2018). Core richness of N-Glycans of caenorhabditis elegans: a case study on chemical and enzymatic release. Anal. Chem. 90, 928–935. doi: 10.1021/acs.analchem.7b03898
Keywords: acidic PNGase, N-glycans, glycoprotein deglycosylation, Dyella japonica, analytical glycoscience
Citation: Guo R-R, Comamala G, Yang H-H, Gramlich M, Du Y-M, Wang T, Zeck A, Rand KD, Liu L and Voglmeir J (2020) Discovery of Highly Active Recombinant PNGase H+ Variants Through the Rational Exploration of Unstudied Acidobacterial Genomes. Front. Bioeng. Biotechnol. 8:741. doi: 10.3389/fbioe.2020.00741
Received: 21 March 2020; Accepted: 10 June 2020;
Published: 03 July 2020.
Edited by:
Jesús Fernández Lucas, Universidad Europea de Madrid, SpainReviewed by:
Friedrich Altmann, University of Natural Resources and Life Sciences, Vienna, AustriaNicolle Hannah Packer, Macquarie University, Australia
Copyright © 2020 Guo, Comamala, Yang, Gramlich, Du, Wang, Zeck, Rand, Liu and Voglmeir. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Liu, bGljaGVuLmxpdUBuamF1LmVkdS5jbg==; Josef Voglmeir, am9zZWYudm9nbG1laXJAbmphdS5lZHUuY24=
 Rui-Rui Guo
Rui-Rui Guo Gerard Comamala2
Gerard Comamala2 Huan-Huan Yang
Huan-Huan Yang Ya-Min Du
Ya-Min Du Ting Wang
Ting Wang Kasper Dyrberg Rand
Kasper Dyrberg Rand Li Liu
Li Liu Josef Voglmeir
Josef Voglmeir