
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 25 May 2020
Sec. Nanobiotechnology
Volume 8 - 2020 | https://doi.org/10.3389/fbioe.2020.00421
This article is part of the Research Topic Biointerfacing 2D Nanomaterials and Engineered Heterostructures View all 7 articles
 Valentina Palmieri1,2*†
Valentina Palmieri1,2*† Lorena Di Pietro3†
Lorena Di Pietro3† Giordano Perini1,2
Giordano Perini1,2 Marta Barba2,3
Marta Barba2,3 Ornella Parolini2,3
Ornella Parolini2,3 Marco De Spirito1,2
Marco De Spirito1,2 Wanda Lattanzi2,3
Wanda Lattanzi2,3 Massimiliano Papi1,2*
Massimiliano Papi1,2*With recent advances in nanotechnology, graphene nanomaterials are being translated to applications in the fields of biosensing, medicine, and diagnostics, with unprecedented power. Graphene is a carbon allotrope derived from graphite exfoliation made of an extremely thin honeycomb of sp2 hybridized carbons. In comparison with the bulk materials, graphene and its water-soluble derivative graphene oxide have a smaller size suitable for diagnostic platform miniaturization as well as high surface area and consequently loading of a large number of biological probes. In this work, we propose a nanotechnological method for concentrating total RNA solution and/or enriching small RNA molecules. To this aim, we exploited the unique trapping effects of GO nanoflakes in the presence of divalent cations (i.e., calcium and magnesium) that make it flocculate and precipitate, forming complex meshes that are positively charged. Here, we demonstrated that GO traps can concentrate nucleic acids in the presence of divalent cations and that small RNAs can be selectively released from GO-magnesium traps. GO nano-concentrators will allow better analytical performance with samples available in small amounts and will increase the sensitivity of sequencing platforms by short RNA selection.
With advances in next-generation sequencing, the human transcriptome has been deciphered and the regulatory roles of a large fraction of small RNAs suggest that they might represent poorly invasive disease biomarkers in diagnostics (Masud et al., 2019). Liquid biopsy, as an example, investigates cell-free nucleic acids in the peripheral blood of patients and is considered one of the most advanced non-invasive tool for early diagnosis, staging, and prognosis (Finotti et al., 2018). The analysis of circulating free RNAs has been mostly focused on microRNA (miRNA) patterns as epigenetic signatures associated with the neoplastic transformation but also many other diseases (Burgos et al., 2014; Finotti et al., 2018). However, the clinical significance of miRNAs might not be clearly demonstrated without an accurate optimization and standardization of the protocols used for their isolation and assessment (El-Khoury et al., 2016). In order to quantify miRNA levels in plasma or in other patient fluids, several types of nanotechnologies have been proposed (Finotti et al., 2018). However, cell-free nucleic acid measurement approaches suffer from important drawbacks, such as the instability of small RNAs and their low concentration (e.g., femtomolar to picomolar levels) and complicate tagging and isolation procedures (El-Khoury et al., 2016; Finotti et al., 2018). Even if new methods based on miniaturization and nanomaterials have been exploited to improve current detection limits (Chen et al., 2018) the development of a sensitive and reliable assay for miRNA enrichment is still lacking.
In recent years, the carbon allotrope graphene has become extremely popular in the biomedical field, due to its unique interactions with cells and organic compounds (Dhinakaran et al., 2020). Graphene is a bi-dimensional material made of a honeycomb of carbon atoms and available in sizes ranging from few nanometers (graphene quantum dots) to large flakes of several microns in lateral size. Among the many graphene derivatives, the soluble oxidized form graphene oxide (GO) is the most studied for biomedical applications, as it interacts with cells, proteins, and bacteria in unique ways (Dreyer et al., 2014; Palmieri et al., 2018a, b).
Both GO and its reduced by-product rGO have a high affinity for single-stranded nucleic acids (ssNAs) via hydrogen bonds and/or π-π interactions, especially for short ssNAs (i.e., <20 nucleotides in length), given the slower diffusivity of longer ssNAs (Liu et al., 2016; Yan et al., 2017). This observation led to the development of GO-based methods for NA extraction from complex media (Hashemi et al., 2014). However, some restrictions in the use of GO and its derivatives are still limiting their widespread use.
Graphene oxide binding to nucleic acids is based on the interaction with backbone phosphate, and this could lead to indiscriminate reactions with either double-stranded nucleic acids (dsNAs) or ssNAs, and to reduced sensitivity in distinguishing the dsNAs to the ssNAs. For this reason, rGO has been preferred, given its direct interaction with nucleotide residues exposed in ssNAs like miRNAs (Yan et al., 2017). On the other hand, rGO, compared with GO, is difficult to handle because it precipitates in complex biological fluids. Recent studies indeed proposed to functionalize rGO with magnetic beads (Yan et al., 2017) or gold nanoparticles (Bao et al., 2019) to avoid this limitation. rGO samples can be obtained from GO using several alternative reduction protocols, which causes high variability in oxygen content, stability, and size (Dreyer et al., 2014; Palmieri et al., 2019) and could lead to poor reproducibility of results from different laboratories. Finally, there is a lack of a standardized method for efficient desorption of adsorbed NAs from both GO and rGO surface (Liu et al., 2016). This step is crucial for the subsequent analysis of the isolated RNA samples.
In this work, we propose a nanotechnological method for enriching total RNA or selectively small RNA molecules (<100 nucleotides in length as miRNAs) in solution. To this aim, we developed a protocol based on the unique trapping effects of GO nanoflakes in the presence of divalent cations. Indeed, cations like calcium and magnesium quickly adsorb on GO surface. After adsorption, GO flocculates and precipitates forming complex structures with size and stability largely dependent on the type and concentration of the ion involved (Wu et al., 2013; Yang et al., 2016). This has been also named the “trapping effect of GO,” when it occurred in the presence of bacteria that remain encapsulated and insulated in large GO traps (Ji et al., 2016; Palmieri et al., 2017a, b; De Maio et al., 2019). Here, we demonstrate that the trapping occurs also with nucleic acids and that can be reversed selectively at high pH to selectively release and concentrate small RNAs. Conversely, large RNAs remain on GO surface, especially in the presence of Mg2+, which is known to form RNA clamps in cells (Petrov et al., 2011). This method is simple and specific and does not involve any complex preparation process, multistep probe functionalization, or tagging. GO nano-concentrators of small RNAs will give the opportunity for refining the analytical performance of existing platforms by reducing the saturation of purification columns by large RNA species (including the large amount of mRNAs and rRNAs), increasing the sensitivity of sequencing platforms and will allow short RNA selection even in samples available in small amounts such as cerebrospinal fluids (Kopkova et al., 2018) and tears (Pieragostino et al., 2015).
Graphene oxide (4 mg/ml) was purchased from GrapheneA (Spain) and diluted in MilliQ RNA and DNA free water for further experiments. GO size and thickness were evaluated by atomic force microscopy (AFM) (Nanowizard II JPK Instruments AG, Berlin, Germany) as previously reported (Bugli et al., 2018). GO hydrodynamic properties and surface charge were measured by dynamic light scattering (DLS) and zeta potential analysis by means of Zetasizer Nano ZS (Malvern, Herrenberg, Germany) as previously reported (Palmieri et al., 2019). The lateral size of samples was calculated using intensity particle size distribution peaks as reported previously (Lotya et al., 2013). Samples have been prepared using a fixed GO concentration (2 mg/ml) and a final concentration of cations in solution of 5, 1, or 0.5 mM adding CaCl2 or MgCl2. After the interaction, samples were diluted to avoid multiple scattering previous DLS measurements.
Genomic DNA (gDNA) and total RNA samples were alternatively extracted from U87 human glioblastoma cells (purchased from ATTC). Cells were maintained in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS, EuroClone), 2% penicillin–streptomycin (Sigma-Aldrich), and 2% L-glutamine (Sigma-Aldrich). gDNA was extracted using E.Z.N.A.® SQ Blood DNA Kit (Omega BioTek), according to the manufacturer’s instructions. Total RNA samples were collected by TRIzol reagent method (Invitrogen), using the standard protocol (Di Pietro et al., 2017; Barba et al., 2018). gDNA and RNA purity and concentration were assessed using a UV spectrophotometer (DU 800, Beckman Coulter).
Ten microliters of GO (at initial concentration of 2 mg/ml) have been added to 1 μl of ddH2O, CaCl2, or MgCl2. After 10 min, 9 μl of gDNA or total RNA (60 ng/μl) have been added to the solution. The final concentration of cations in solution was 5, 1, or 0.5 mM. A synthetic miRNA (cel-miR-39-3p, mature miRNA sequence: UCACCGGGUGUAAAUCAGCUUG, ThermoFisher) was added to RNA samples, to a final concentration of 30 pM, to be assessed as an exogenous control for small RNAs for qPCR analysis. In particular, the use of this exogenous miRNA allowed overcoming the possible issue deriving from the different expression levels of any endogenous cellular miRNAs among biological samples. Samples were incubated for 15 min and then centrifuged for 10 min at 14,000 × g. Nucleic acid concentration was measured in the supernatant of samples by measuring absorbance at 260 nm with Cytation 3 Take 3 reader (BioTek, United States) or fluorescence peak at 520 nm when 5′6-FAM-labeled ssNAs were used in the experiment. In particular, a 22-nucleotide-long DNA sequence (ACTGCGGCTTAGGTTAGCATTG) was randomly designed, considering a sequence with the same length and CG and AT content of a typical miRNA, and purchased by IDT Integrated DNA technologies with a 5′-56 FAM modification. The presence and integrity of total RNA in the supernatant of samples have been additionally evaluated by agarose gel electrophoresis in all the experimental conditions.
The levels of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) transcripts and of cel-miR-39-3p in samples were evaluated using reverse transcription quantitative real-time PCR (qPCR). More in detail, the same quantity of each sample was reverse transcribed using GoScript Reverse Kit (Promega) and GAPDH transcript levels were subsequently amplified with StepOne system (Applied Biosystems), using Syber Green master mix (GoTaq qPCR Master Mix Kit, Promega) and specific primer pairs (forward primer: AACTTTGGTATCGTGGAAGGA; reverse primer: GGCAGTGATGGCATGGAC) (Barba et al., 2012; Di Pietro et al., 2017). In parallel, total RNAs were also used as templates for miRNA reverse transcription, using TaqManTM Advanced miRNA cDNA Synthesis Kit (ThermoFisher), according to the manufacturer’s instructions. cel-miR-39-3p levels were amplified using TaqManTM Fast Advanced Master Mix (ThermoFisher) and a specific TaqManTM Advanced miRNA Assay (Assay ID#478293_mir, ThermoFisher).
Graphene oxide nanoflakes used in our experiments had a mean lateral size of ∼700 nm as measured from AFM imaging (Figure 1A). AFM was used to measure the average GO flakes thickness, which was ∼0.8 nm (see a representative line profile of a GO flake in Figure 1B). The size of GO flakes was confirmed by DLS, used to analyze hydrodynamic properties of samples (Figure 1C). Data represent the peak of size intensity distributions reported in Supplementary Figure S1, together with the lateral size of each sample obtained by DLS (Supplementary Table S1). It is known that GO is stable in water but rapidly aggregates in the presence of divalent cations. This effect was exploited in environmental applications envisaging water purification (Hegab and Zou, 2015) and was reported to trap biological entities such as bacteria and fungi (Palmieri et al., 2017a, b). In order to induce RNA trapping, we have chosen Ca2+ and Mg2+ ions among cations known to change GO properties in solution mainly because heavy metal cations (Cr3+, Pb2+, Cu2+, Cd2+, and Ag+) destabilize too strongly GO (Yang et al., 2016). Ca2+ and Mg2+ have similar hydration shells (Yang et al., 2016) and the same positive charge, so that a reduced repulsion between RNA phosphates and GO surface can be expected, though Mg2+ is known to be most effective in stabilizing RNA in folded structures (Draper, 2004).
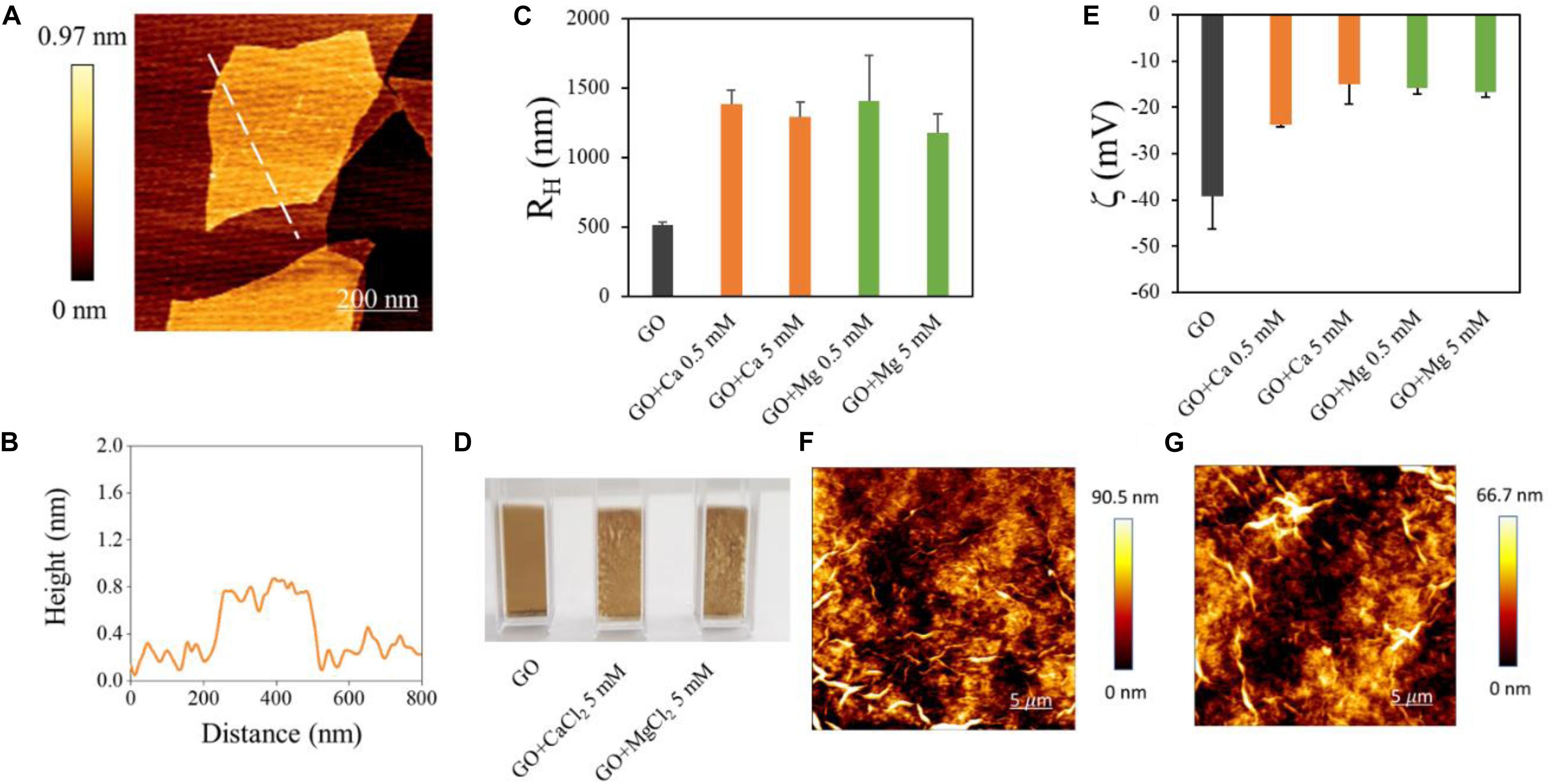
Figure 1. Characterization of GO samples used and of GO interaction with divalent cations. (A) AFM representative image of GO flake and a height profile obtained along the dashed line (B). Hydrodynamic radius of GO samples in different solutions obtained by DLS (C). Photographs of cuvettes containing GO in different solutions (D). Zeta potential of GO in different solutions (E). Representative images of GO aggregates in Ca2+ (F) or Mg2+ (G) obtained by AFM.
From a macroscopic point of view, GO and cations quickly formed a dense cloud in solution as soon as they were mixed (Figure 1D). Representative AFM images of aggregates are shown in Figure 1E. The hydrodynamic radius of GO was measured after the addition of Ca2+ or Mg2+ at two concentrations (0.5 and 5 mM). The addition of divalent cations caused an increase of RH up to ∼1400 nm from ∼500 nm as reported in Figure 1C. From a negative value of ∼−40 mV of bare GO, the surface zeta potential (Figure 1E) increased with Ca2+ and Mg2+, confirming a positive charging of GO surface in the presence of cations. GO meshes created with cations can be separated from the rest of the solution by precipitation or more quickly with centrifugation; this avoids functionalization with magnetic beads or other nanoparticles for further analysis. Representative images of GO aggregates in the presence of Ca2+ (Figure 1F) and Mg2+ (Figure 1G) obtained by AFM.
We evaluated GO interaction with single-strand and double-strand NAs. First, we measured GO affinity for dsDNA by incubating GO with gDNA at a final concentration of 30 ng/μl. gDNA concentration was measured in the supernatant fractions recovered after incubation with different GO samples and centrifugation, and compared to the control sample (gDNA with buffer; Figure 2).
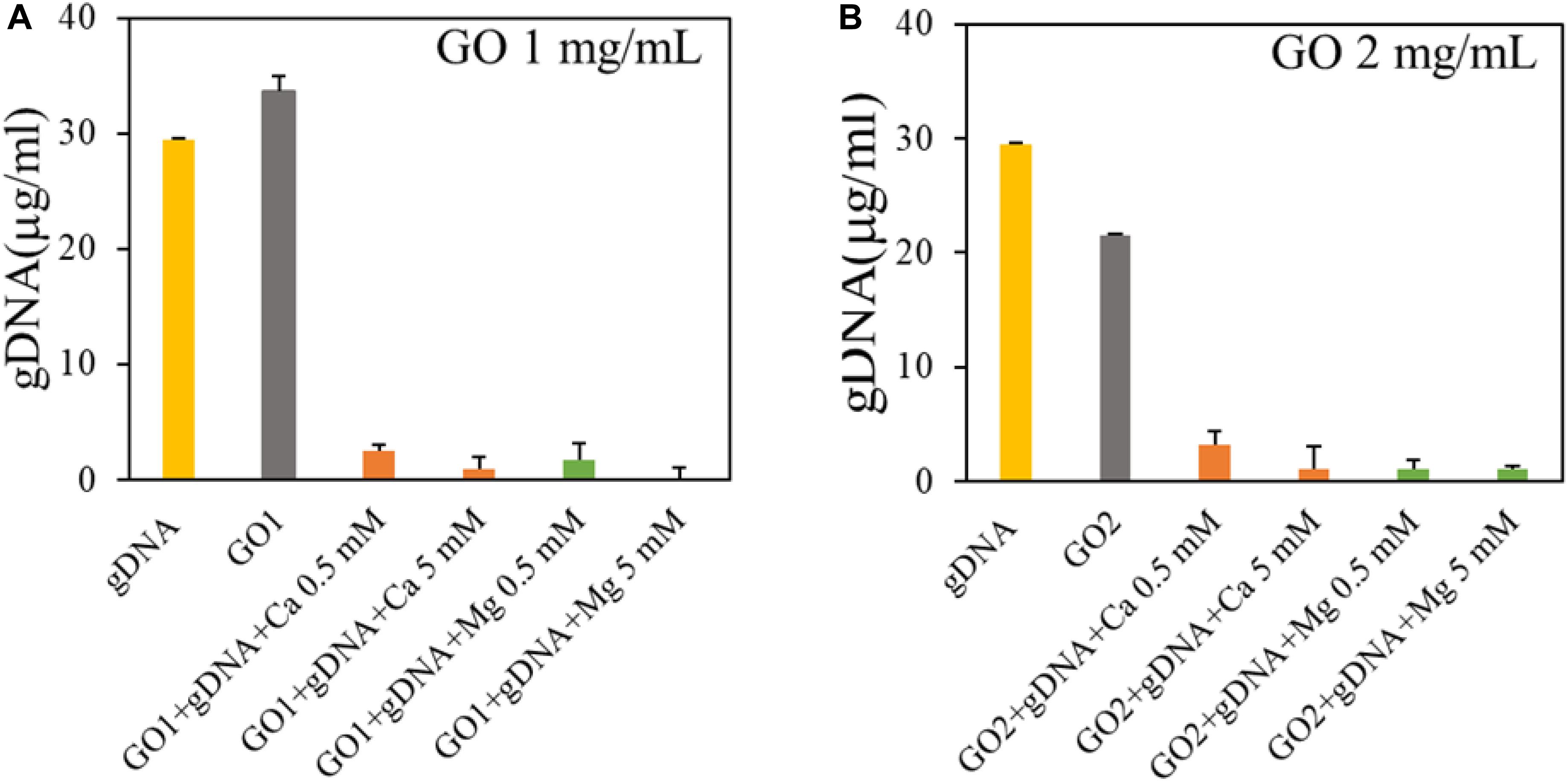
Figure 2. Analysis of binding of GO with genomic DNA in the presence of GO at 1 mg/ml (A) or GO at 2 mg/ml (B). Concentration of DNA in supernatants is reported after UV spectroscopic analysis.
Our results showed that GO (1 mg/ml) does not adsorb gDNA (Figure 2A), only if the concentration of GO is doubled, 40 μg of GO adsorb ∼0.15 μg of gDNA. As expected the interaction between GO and dsDNA is not favored, since the binding between GO and nucleic acids is largely dependent on hydrogen bonds and/or π-π interactions with nucleobases and nucleobases are occluded inside the double helix in gDNA (Liu et al., 2016). Further, repulsive interactions occur between phosphate backbones and GO negative surface. With the addition of divalent salts, the gDNA is completely adsorbed on GO even at very low salt concentration (0.5 mM). This can be explained by the bridging of cations between GO and the dsDNA phosphate backbone both charged negatively (Wu et al., 2011). With Ca2+/Mg2+ in solution, the concentration of gDNA adsorbed is ∼5 times higher (0.58 μg of gDNA per 40 μg of GO) without significant differences between the cations.
We then analyzed the interaction of GO with total RNA extracted from cells in culture (U87 glioblastoma cells). First, we incubated GO (2 mg/ml) with total RNA (final concentration 30 ng/μl) added with the synthetic cel-miR-39-3p as an exogenous control for small RNA (final concentration 15 pM) (Figure 3A). As observed with gDNA, GO alone has poor affinity for total RNA, which is adsorbed only if a solution with high Ca2+ or Mg2+ concentration is added to GO prior to the incubation with RNA. Twenty micrograms of GO adsorbed 0.4 and 0.55 μg of total RNA with 5 mM Ca2+ and 5 mM Mg2+, respectively. This result might seem in contrast with reports from literature that have shown, even if in low amounts, adsorption of RNA even from bare GO (Wu et al., 2011; Yan et al., 2017). We wanted to reproduce conditions of previous studies and we used, instead of total RNA, the FAM-ssNA, as a model NA. We incubated FAM-ssNA (final concentration, 30 ng/μl) with different GO samples and then measured fluorescence from supernatants. We observed that FAM-labeled ssNA is readily adsorbed also by GO (Figure 3B). This led us to hypothesize that studying adsorption of FAM-tagged nucleic acids might be biased by the strong interaction of FAM probe with GO surface.
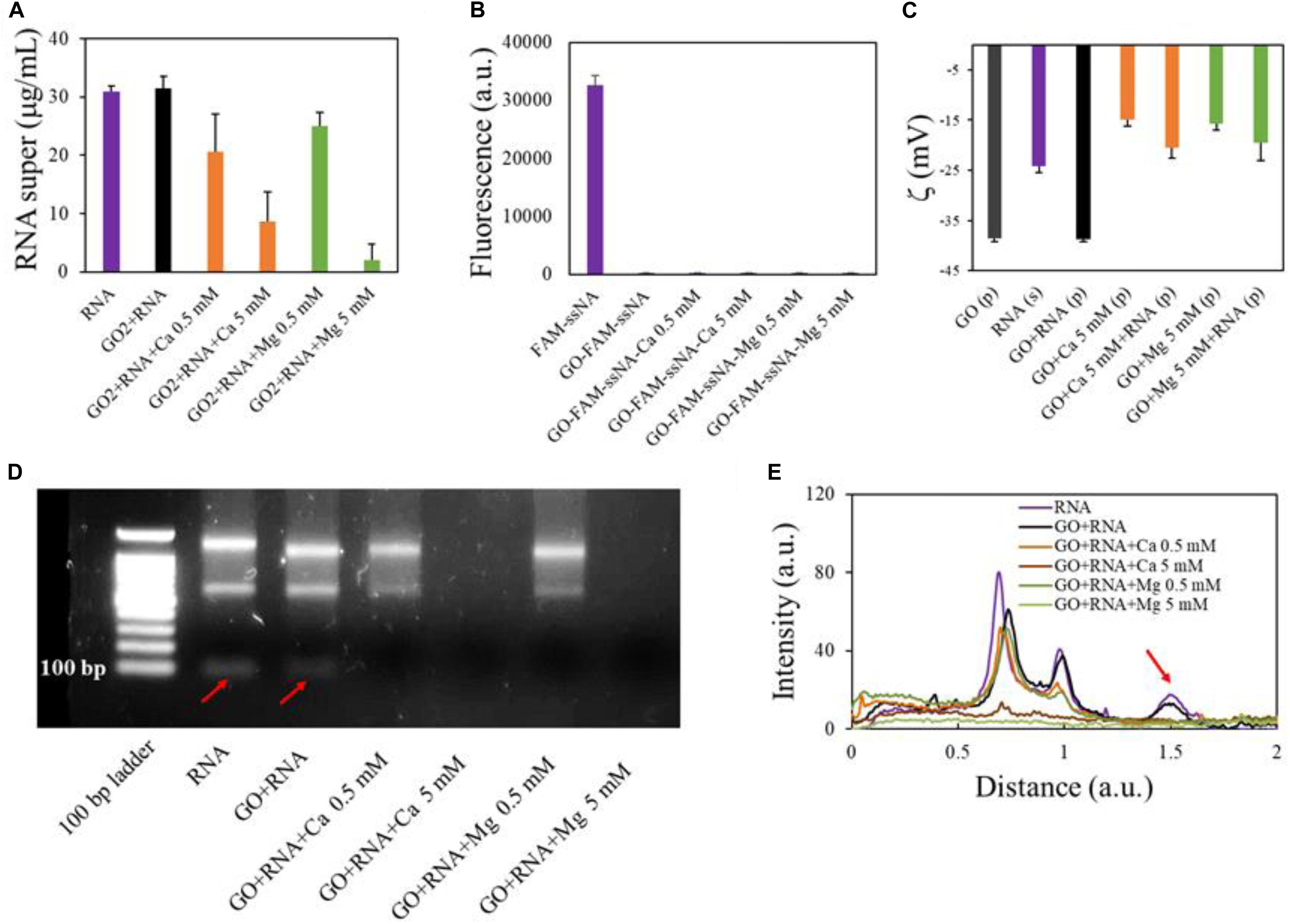
Figure 3. Analysis of GO binding with total RNA. Concentration of RNA in supernatants after interaction with GO or GO and cations measured by UV spectroscopy (A). Quantification of fluorescent FAM-ssNA in supernatants after interaction with GO (B). Zeta potential of GO samples after RNA adsorption in different conditions (C). Electrophoretic analysis of supernatants after interaction with GO (D) and line profile analysis of gels (E) with red arrow highlighting small RNA peak (corresponding to the band under 100 bases).
Analysis of surface zeta potential of GO pellets after RNA adsorption confirmed that RNA molecules were deposited on GO surface (Figure 3C) only in the presence of divalent cations. Indeed, zeta potential of GO pellets after incubation with RNA was equal to GO alone. Conversely, after incubation, pellets of GO-Ca2+-RNA and GO-Mg2+-RNA had surface charge substantially different from GO-Ca2+ and GO-Mg2+ pellets, respectively.
Electrophoresis analysis of supernatants confirmed the UV spectroscopy results (Figure 3D) and in particular displayed that smaller RNAs (<100 nucleotides in length) were better adsorbed since they were not visible in the supernatant even in samples with small cation concentrations as shown by the disappearance of the low-molecular-weight peak in the line profiles of electrophoretic gels (red arrow in Figure 3E). This is confirmed by results in literature that reported that longer RNAs bind to GO with slower diffusivity (Liu et al., 2016).
To better clarify the higher affinity of GO for smaller RNAs in the presence of divalent cations, qPCR analysis was performed on supernatant samples recovered after incubation of RNA samples with GO. Considering that higher Ct (cycle threshold) values correspond to a lower amount of transcript levels in the sample, we have compared the different samples amplifying the same volume of each supernatant. Our data showed that longer RNAs are significantly adsorbed more at high concentration of cations, as demonstrated by the increased Ct for GAPDH transcript amplification (one-way ANOVA, p < 0.0001; Figure 4A). In particular, qPCR analysis indicated that both Ca2+ and Mg2+ had the same ability to retain RNA. Instead, miRNAs started binding even if only GO is present (Figure 4B). Very interestingly, in the presence of the high concentration (5 mM) of both Ca2+ or Mg2+, GO was able to adsorb the total amount of small RNAs, since cel-miR-39 became undetectable in the supernatant fraction after incubation (one-way ANOVA, p < 0.0001; Figure 4B).
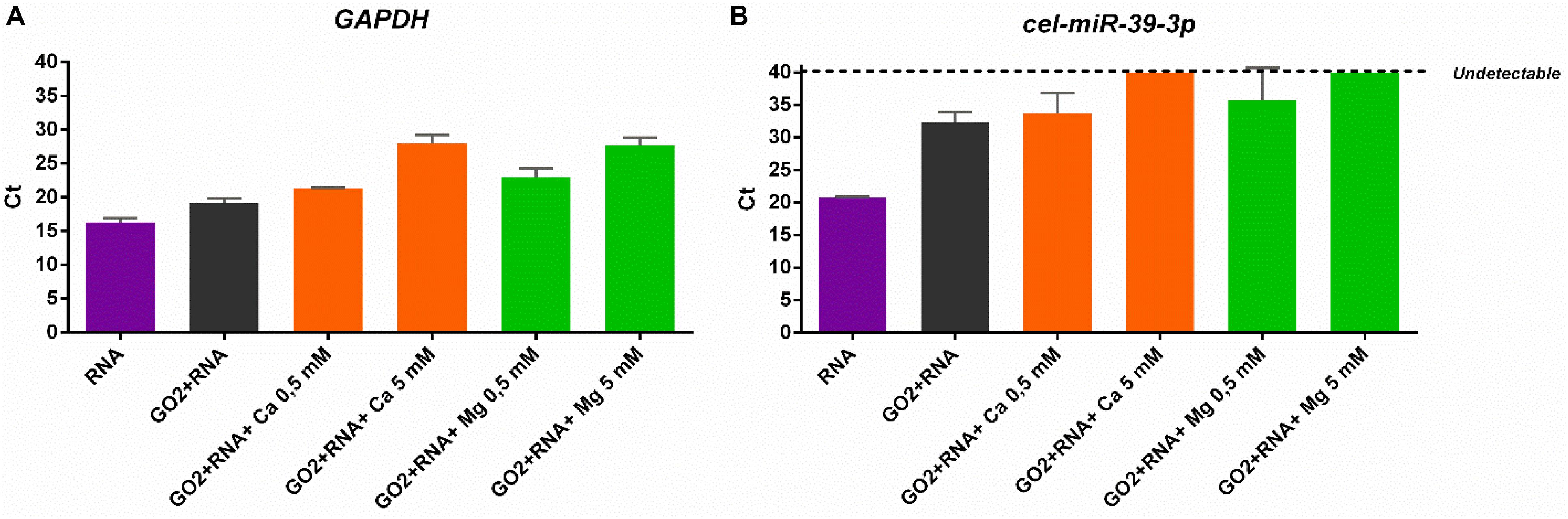
Figure 4. (A) qPCR analysis of GAPDH and (B) cel-miR-39-3p levels in supernatants after incubation with GO and different cations.
Altogether, these data demonstrate efficient trapping of total RNA in solution by GO-divalent cations with faster recognition of small RNAs.
It is important to release small RNAs bound to GO surface for further PCR/sequencing analysis. Indeed, at the critical concentration of 4 μg/ml, PCR reactions are inhibited by the GO in solution (Wang et al., 2017). The desorption of NAs from GO is indeed a complex problem (Liu et al., 2016). Many studies in the literature have reported that the addition of either cDNA, random DNA, or albumin could detach RNA from GO surfaces as well as the use of solutions of isopropanol or base that could facilitate the specific desorption of miRNAs (Hashemi et al., 2014). We have tried several buffers and incubation conditions reported in the literature but without success (data not shown). This is because the bond between RNA and GO cations has indeed proven to be very robust. The desorption of RNA was successfully reached only after two washes with H2O and with incubation of 5 min with 1.5 mM Tris buffer (pH 8.8) at room temperature. These conditions were chosen based on literature data that indicate that the adsorption is favored at lower pH and low temperature (Wu et al., 2011). Further, at high pH, the degree of deprotonation of the carboxyl groups at the edges can induce more stability of GO sheets and less precipitation and aggregation (Wu et al., 2013).
In tris buffer, the RNA was successfully released from GO-Ca2+samples (both small and long RNAs; Figure 5). The quantity of released RNA increased in a concentration-dependent manner, reaching the higher desorption levels in 5 mM samples, in line with the amount of RNA adsorbed on GO (Figure 5A). Interestingly, the desorption from GO-Mg2+ with our buffer affected only smaller RNAs and was not concentration-dependent, whereas the longer RNAs remained “trapped” and there is a small release of large RNA only at high Mg2+ concentration (see line profiles in Figures 5B,C). The GO-RNA sample did not display any signal on the electrophoretic gel as visible in Figure 5A, since the RNA did not bind to GO and was totally recovered after the first centrifugation (see Figure 3C).
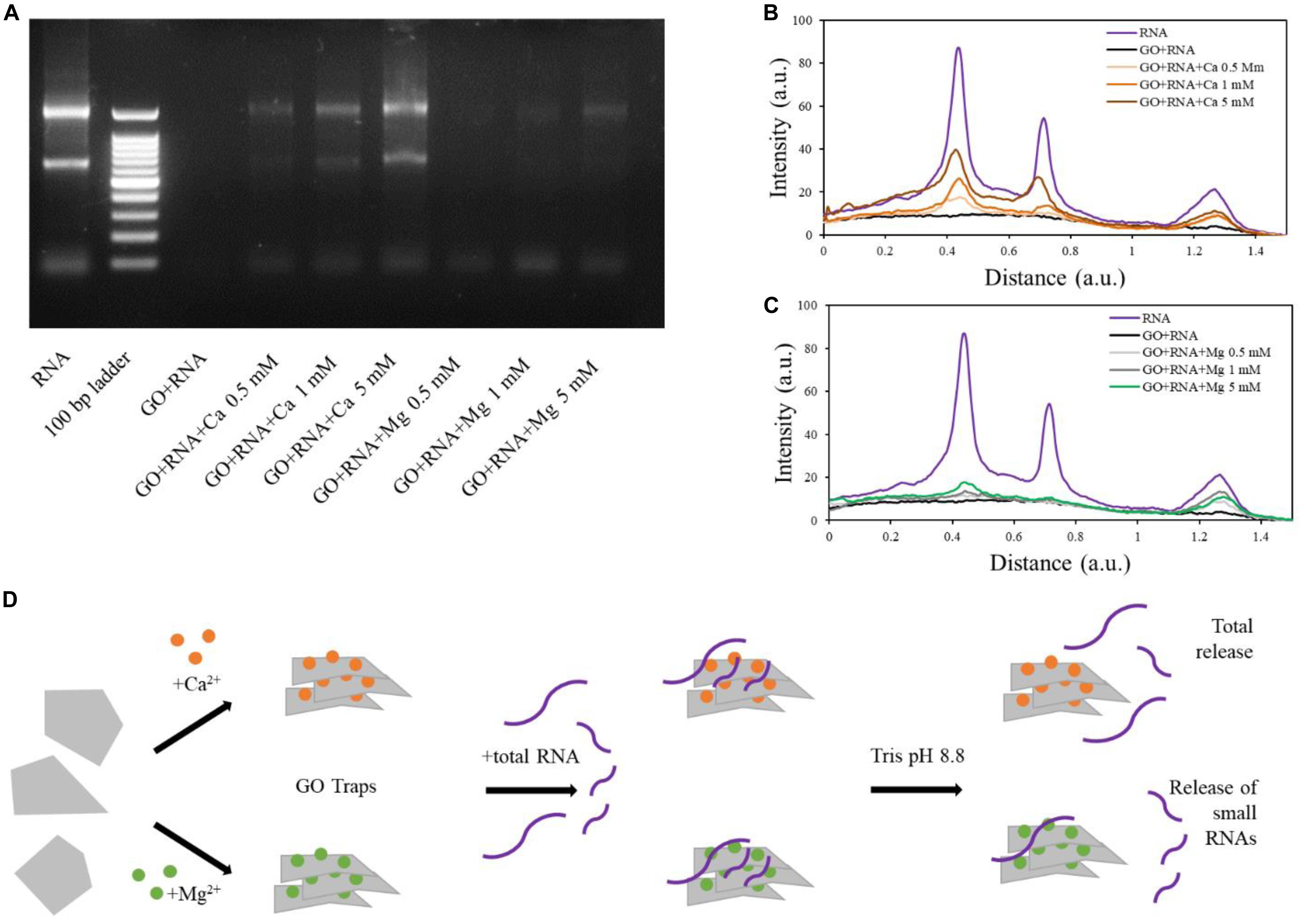
Figure 5. Electrophoretic analysis of supernatants after release from GO sheets in Tris buffer at high pH (A). Line profile analysis of electrophoresis gel and comparison of RNA and GO-RNA samples with GO-calcium (B) and GO-magnesium (C). Schematic illustration of our results (D).
With recent advances in nanotechnology, nanomaterials are being translated to applications in the fields of biosensing, medicine, and diagnostics, with unprecedented power (Chen et al., 2018). In this paper, we exploit the unique surface features of the bidimensional GO and its trapping ability in the presence of calcium/magnesium ions to create a method to selectively concentrate total RNA/small RNAs in solution. The main advantage of using GO compared to other nanoparticles for nucleic acid-related application is its surface-to-volume ratio. One-dimensional nanomaterials indeed possess high surface-to-volume ratio, which provides large active surface area for the interactions with nucleic acids, and this strongly favors the adsorption of molecules and ultimately leads to better sensitivity (Varghese et al., 2015). Graphene derivatives have also exceptional thermal, optical, and electronic properties that allow easy building of sensor devices. We demonstrated the feasibility of applying GO-based nanotechnologies for a highly sensitive separation of RNA species based on size. In particular, we demonstrated that GO can enrich small RNAs from complex solutions of RNA and how to easily detach ssNAs from GO surface. We indeed used a two-step protocol based on GO trapping of molecules, mediated by divalent cations (calcium and magnesium) and without specific probes. GO is first decorated with divalent ions and then incubated with total RNA. When in solution with RNA, GO adsorbs nucleic acid molecules with high efficiency thanks to the bridges between phosphate backbone and negative charges of GO surface created with calcium or magnesium ions. GO trapping can completely remove RNA from the solution and concentrate it. After nano-concentration, small RNAs can be selectively released from surface by desorption induced at high pH in the presence of Mg2+ ions. Conversely, with calcium, all RNA species are released (Figure 5D). Indeed, magnesium special geometric and energetic relationship with the phosphates of RNA makes tighter packing with large RNA compared to calcium ions. We hypothesize that these results can be explained by a double effect occurring in solution. On one hand, at high cation concentrations (5 mM) that are above the critical coagulation concentration of GO, the large RNA competes for binding to GO surfaces with GO itself, and large RNA is released. On the other hand, the Mg2+ ability to make bidentate RNA clamps with large RNAs markedly influenced released studies. Indeed, it has been demonstrated that magnesium, due to its size and charge density, binds more intimately than calcium to the oxyanions of RNA, so that magnesium clamps are made stable by electrostatic interactions, charge transfer, polarization, and exchange interactions (Petrov et al., 2011). This “physiological” role of magnesium is also reflected in our binding results and makes it a selective absorbent of large RNA species. In addition, GO trapping of RNA molecules (>100 nucleotides in length) can also be exploited to improve further transcriptome sequencing that requires sufficient mRNA yield, usually obtained by either poly-A selection or depletion of ribosomal RNA (rRNA) (Choy et al., 2015; Fowler et al., 2018). rRNA indeed constitutes about 80% of the total RNA species in eukaryotic cells, while poly[A]+ mRNA constitutes only about 5%. The use of ribosomal depleted RNA has been shown to recover more information about protein-coding genes, non-coding RNAs, snRNAs, snoRNAs, and repeat elements enabling novel transcribed loci detection (Choy et al., 2015). Our GO-Mg2+ based selection represents a fast, reliable method for rRNA depletion. Our results also suggested that GO-Ca2+ complexes can instead represent a suitable method to isolate and concentrate total RNA, hence providing a tool for the simultaneous isolation and analysis of both small and large RNA species. This study can pave the way toward the development of novel methods to improve liquid biopsy performance and may contribute to developing innovative approaches for miRNA and/or total RNA detection (Fiammengo, 2017).
The datasets generated for this study are available on request to the corresponding author.
VP, LD, and MB performed experiments and contributed to manuscript writing. VP and MP designed the experiments and methodology and revised the manuscript. GP characterized the graphene samples. WL, OP, and MD contributed to manuscript writing and revision. All authors have read and agreed to the published version of the manuscript.
The research leading to these results has received funding from AIRC under IG 2019 – ID. 23124 project – P.I. MP. Università Cattolica del Sacro Cuore contributed to the funding of this research project and its publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00421/full#supplementary-material
Bao, J., Houm, C., Zhao, Y., Geng, X., Samalo, M., Yang, H., et al. (2019). An enzyme-free sensitive electrochemical MicroRNA-16 biosensor by applying a multiple signal amplification strategy based on Au/PPy–rGO nanocomposite as a substrate. Talanta 196, 329–336. doi: 10.1016/j.talanta.2018.12.082
Barba, M., Di Pietro, L., Massimi, L., Geloso, M. C., Frassanito, P., Caldarelli, M., et al. (2018). BBS9 gene in nonsyndromic craniosynostosis: role of the primary cilium in the aberrant ossification of the suture osteogenic niche. Bone 112, 58–70. doi: 10.1016/j.bone.2019.02.004
Barba, M., Pirozzi, F., Saulnier, N., Vitali, T., Natale, M. T., Logroscino, G., et al. (2012). Lim mineralization protein 3 induces the osteogenic differentiation of human amniotic fluid stromal cells through kruppel-like factor-4 downregulation and further bone-specific gene expression. BioMed Res. Int. 2012:813894 doi: 10.1155/2012/813894
Bugli, F., Cacaci, M., Palmieri, V., Di Santo, R., Torelli, R., Ciasca, G., et al. (2018). Curcumin-loaded graphene oxide flakes as an effective antibacterial system against methicillin-resistant Staphylococcus aureus. Interface Focus 8:20170059. doi: 10.1098/rsfs.2017.0059
Burgos, K., Malenica, I., Metpally, R., Courtright, A., Rakela, B., Beach, T., et al. (2014). Profiles of extracellular MiRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One 9:e94839. doi: 10.1371/journal.pone.0094839
Chen, Y.-X., Huang, K.-J., and Niu, K.-X. (2018). Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for MicroRNA detection-a review. Biosens. Bioelectron. 99, 612–624. doi: 10.1016/j.bios.2017.08.036
Choy, J., Priscilla, Y. H., Boon, L. S., Bertin, N., and Fullwood, M. J. (2015). A resource of ribosomal RNA-Depleted RNA-Seq data from different normal adult and fetal human tissues. Sci. Data 2, 1–7. doi: 10.1038/sdata.2015.63
De Maio, F., Palmieri, V., Salustri, A., Perini, G., Sanguinetti, M., De Spirito, M., et al. (2019). Graphene oxide prevents mycobacteria entry in macrophages through extracellular entrapment. Nanoscale Adv. 1, 1421–1431. doi: 10.1039/c8na00413g
Dhinakaran, V., Lavanya, M., Karthikeyan, V., Ravichandran, M., and Vijayakumar, M. D. (2020). Review on exploration of graphene in diverse applications and its future horizon. Mater. Today Proc. doi: 10.1016/j.matpr.2019.12.369
Di Pietro, L., Baranzini, M., Berardinelli, M. G., Lattanzi, W., Monforte, M., Tasca, G., et al. (2017). Potential therapeutic targets for ALS: MIR206, MIR208b and MIR499 are modulated during disease progression in the skeletal muscle of patients. Sci. Rep. 7, 1–11. doi: 10.1038/s41598-017-10161-z
Dreyer, D. R., Todd, A. D., and Bielawski, C. W. (2014). Harnessing the chemistry of graphene oxide. Chem. Soc. Rev. 43, 5288–5301.
El-Khoury, V., Pierson, S., Kaoma, T., Bernardin, F., and Berchem, G. (2016). Assessing cellular and circulating MiRNA recovery: the impact of the RNA isolation method and the quantity of input material. Sci. Rep. 6:19529. doi: 10.1038/srep19529
Fiammengo, R. (2017). Can nanotechnology improve cancer diagnosis through MiRNA detection? Biomark. Med. 11, 69–86. doi: 10.2217/bmm-2016-0195
Finotti, A., Allegretti, M., Gasparello, J., Giacomini, P., Spandidos, D. A., Spoto, G., et al. (2018). Liquid biopsy and PCR-Free ultrasensitive detection systems in oncology. Int. J. Oncol. 53, 1395–1434. doi: 10.3892/ijo.2018.4516
Fowler, E. K., Mohorianu, I., Smith, D. T., Dalmay, T., and Chapman, T. (2018). Small RNA populations revealed by blocking RRNA fragments in Drosophila melanogaster reproductive tissues. PLoS One 13:e0191966. doi: 10.1371/journal.pone.0191966
Hashemi, E., Akhavan, O., Shamsara, M., Valimehra, S., and Rahighi, R. (2014). DNA and RNA extractions from eukaryotic and prokaryotic cells by graphene nanoplatelets. RSC Adv. 4, 60720–60728.
Hegab, H. M., and Zou, L. (2015). Graphene oxide-assisted membranes: fabrication and potential applications in desalination and water purification. J. Membr. Sci. 484, 95–106.
Ji, H., Sun, H., and Qu, X. (2016). Antibacterial applications of graphene-based nanomaterials: recent achievements and challenges. Adv. Drug Deliv. Rev. 105, 176–189. doi: 10.1016/j.addr.2016.04.009
Kopkova, A., Sana, J., Fadrus, P., Machackova, T., Vecera, M., Vybihal, V., et al. (2018). MicroRNA isolation and quantification in cerebrospinal fluid: a comparative methodical study. PLoS One 13:e0208580. doi: 10.1371/journal.pone.0208580
Liu, B., Salgado, S., Maheshwari, V., and Liu, J. (2016). DNA adsorbed on graphene and graphene oxide: fundamental interactions, desorption and applications. Curr. Opin. Coll. Interface Sci. 26, 41–49.
Lotya, M., Rakovich, A., Donegan, J. F., and Coleman, J. N. (2013). Measuring the lateral size of liquid-exfoliated nanosheets with dynamic light scattering. Nanotechnology 24:265703. doi: 10.1088/0957-4484/24/26/265703
Masud, M. K., Umer, M., Hossain, S. A., Yamauchi, Y., Nguyen, N.-T., Shiddiky, M. J. A., et al. (2019). Nanoarchitecture frameworks for electrochemical mirna detection. Trends Biochem. Sci. 44, 433–452. doi: 10.1016/j.tibs.2018.11.012
Palmieri, V., Barba, M., Di Pietro, L., Conti, C., De Spirito, M., Lattanzi, W., et al. (2018a). Graphene oxide induced osteogenesis quantification by in-situ 2D-fluorescence spectroscopy. Int. J. Mol. Sci. 19:3336. doi: 10.3390/ijms19113336
Palmieri, V., Barba, M., Di Pietro, L., Gentilini, S., Braidotti, M. C., Ciancico, C., et al. (2018b). Reduction and shaping of graphene-oxide by laser-printing for controlled bone tissue regeneration and bacterial killing. 2D Materials 5:aa9ca7.
Palmieri, V., Boon, L. S., Bertin, N., Massimi, L., and Geloso, M. C. (2019). Biocompatible: N -acetyl cysteine reduces graphene oxide and persists at the surface as a green radical scavenger. Chem. Commun. 55, 4186–4189. doi: 10.1039/C9CC00429G
Palmieri, V., Francesca, B., Lauriola, M. C., Cacaci, M., Torelli, R., Ciasca, G., et al. (2017a). Bacteria meet graphene: modulation of graphene oxide nanosheet interaction with human pathogens for effective antimicrobial therapy. ACS Biomater. Sci. Eng. 3, 619–627.
Palmieri, V., Lauriola, M. C., Ciasca, G., Conti, C., De Spirito, M., Papil, M., et al. (2017b). The graphene oxide contradictory effects against human pathogens. Nanotechnology 28:152001. doi: 10.1088/1361-6528/aa6150
Petrov, A. S., Bowman, J. C., Harvey, S. C., and Williams, L. D. (2011). Bidentate RNA–magnesium clamps: on the origin of the special role of magnesium in RNA Folding. RNA 17, 291–297. doi: 10.1261/rna.2390311
Pieragostino, D., D’Alessandro, M., di Ioia, M., Di Ilio, C., Sacchetta, P., Del Boccio, P., et al. (2015). Unraveling the molecular repertoire of tears as a source of biomarkers: beyond ocular diseases. Proteomics Clin. Appl. 9, 169–186. doi: 10.1002/prca.201400084
Varghese, S. S., Varghese, S. H., Swaminathan, S., Singh, K. K., and Mittal, V. (2015). Two-dimensional materials for sensing: graphene and beyond. Electronics 4, 651–687.
Wang, Y., Wang, F., Wang, H., and Song, M. (2017). Graphene oxide enhances the specificity of the polymerase chain reaction by modifying primer-template matching. Sci. Rep. 7, 1–10. doi: 10.1038/s41598-017-16836-x
Wu, L., Liu, L., Gao, B., Muñoz-Carpena, R., Zhang, M., Chen, H., et al. (2013). Aggregation kinetics of graphene oxides in aqueous solutions: experiments, mechanisms, and modeling. Langmuir 29, 15174–15181. doi: 10.1021/la404134x
Wu, M., Kempaiah, R., Huang, P. J., Maheshwari, V., and Liu, J. (2011). Adsorption and desorption of DNA on graphene oxide studied by fluorescently labeled oligonucleotides. Langmuir 27, 2731–2738. doi: 10.1021/la1037926
Yan, H., Xu, Y., Lu, Y., and Xing, W. (2017). Reduced graphene oxide-based solid-phase extraction for the enrichment and detection of MicroRNA. Anal. Chem. 89, 10137–10140. doi: 10.1021/acs.analchem.7b03138
Keywords: graphene, nucleic acid, miRNA, diagnostics, nanotechnology
Citation: Palmieri V, Di Pietro L, Perini G, Barba M, Parolini O, De Spirito M, Lattanzi W and Papi M (2020) Graphene Oxide Nano-Concentrators Selectively Modulate RNA Trapping According to Metal Cations in Solution. Front. Bioeng. Biotechnol. 8:421. doi: 10.3389/fbioe.2020.00421
Received: 12 February 2020; Accepted: 14 April 2020;
Published: 25 May 2020.
Edited by:
Alessandro Martucci, University of Padua, ItalyReviewed by:
Serguei Savilov, Lomonosov Moscow State University, RussiaCopyright © 2020 Palmieri, Di Pietro, Perini, Barba, Parolini, De Spirito, Lattanzi and Papi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Valentina Palmieri, dmFsZW50aW5hLnBhbG1pZXJpQHVuaWNhdHQuaXQ=; Massimiliano Papi, TWFzc2ltaWxpYW5vLnBhcGlAdW5pY2F0dC5pdA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.