
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 28 April 2020
Sec. Biomaterials
Volume 8 - 2020 | https://doi.org/10.3389/fbioe.2020.00374
This article is part of the Research Topic 3D Printing for Implantable Medical Devices: From Surgical Reconstruction to Tissue/Organ Regeneration View all 13 articles
 Daniela F. Duarte Campos1*
Daniela F. Duarte Campos1* Christopher D. Lindsay1
Christopher D. Lindsay1 Julien G. Roth2
Julien G. Roth2 Bauer L. LeSavage3
Bauer L. LeSavage3 Alexis J. Seymour3
Alexis J. Seymour3 Brad A. Krajina1
Brad A. Krajina1 Ricardo Ribeiro4
Ricardo Ribeiro4 Pedro F. Costa4
Pedro F. Costa4 Andreas Blaeser5
Andreas Blaeser5 Sarah C. Heilshorn1*
Sarah C. Heilshorn1*Human tissues, both in health and disease, are exquisitely organized into complex three-dimensional architectures that inform tissue function. In biomedical research, specifically in drug discovery and personalized medicine, novel human-based three-dimensional (3D) models are needed to provide information with higher predictive value compared to state-of-the-art two-dimensional (2D) preclinical models. However, current in vitro models remain inadequate to recapitulate the complex and heterogenous architectures that underlie biology. Therefore, it would be beneficial to develop novel models that could capture both the 3D heterogeneity of tissue (e.g., through 3D bioprinting) and integrate vascularization that is necessary for tissue viability (e.g., through culture in tissue-on-chips). In this proof-of-concept study, we use elastin-like protein (ELP) engineered hydrogels as bioinks for constructing such tissue models, which can be directly dispensed onto endothelialized on-chip platforms. We show that this bioprinting process is compatible with both single cell suspensions of neural progenitor cells (NPCs) and spheroid aggregates of breast cancer cells. After bioprinting, both cell types remain viable in incubation for up to 14 days. These results demonstrate a first step toward combining ELP engineered hydrogels with 3D bioprinting technologies and on-chip platforms comprising vascular-like channels for establishing functional tissue models.
Three-dimensional (3D) cell culture systems that model the microenvironment of tissues and organs are expected to yield results with higher predictive value in drug discovery, preclinical testing, and personalized medicine (Langhans, 2018). It is well-accepted that 3D culture systems that mimic key factors of native extracellular matrix (ECM) are more representative of the in vivo microenvironment than comparative two-dimensional (2D) cultures (Petersen et al., 1992; Ravi et al., 2015). For example, 3D cancer models have shown more physiologically relevant outcomes in migration and invasion assays compared to 2D models (Katt et al., 2016). However, existing 3D models remain inadequate to recapitulate the complex and heterogenous architectures present in vivo. In particular, vascularization is typically absent from many 3D tissue models (Zhang et al., 2016). Vascular tissue interfaces are particularly important in in vitro models of the neural stem cell niche (Tavazoie et al., 2008), blood-brain-barrier (Brown et al., 2015), and in vitro models of cancer metastasis (Carey et al., 2013; Curtin et al., 2018).
Microfluidic and on-chip technologies are experimental models that can include dynamic vascular-like channels (Cochrane et al., 2019). In a recent study, a low permeability microfluidic platform was developed for screening pharmaceuticals that target neurodegenerative diseases (Bang et al., 2017). Although such platforms have shown vascular permeability comparable to reported in vivo studies, they fail to recapitulate the 3D architecture of the native tissue, as cells are cultured on 2D polydimethylsiloxane (PDMS) substrates. In vitro models of the neural stem cell niche commonly use random co-culture mixtures or transwell inserts that do not mimic the spatial proximity and geometry of the cross-talk between neural progenitor cells (NPCs) and endothelial cells (Shen et al., 2004). Similar culture systems have been reported in cancer research (Sontheimer-Phelps et al., 2019). Here, we hypothesized that conventional microfluidic devices could be combined with 3D bioprinting technology to fabricate ex vivo tissue mimics with on-chip vascular-like networks.
3D bioprinting technologies are key biomanufacturing methods used to create 3D constructs by sequential deposition of cell-laden bioink layers (Murphy and Atala, 2014; Leberfinger et al., 2019). Several recent examples have demonstrated the promise of 3D bioprinting to create in vitro models of human tissues and disease. For example, microextrusion bioprinting was used to generate expansion lattices for neural research (Gu et al., 2018; Lindsay et al., 2019), whereas microextrusion and laser-based bioprinting were used to construct 3D co-culture models of interacting cancer and endothelial cells (Phamduy et al., 2015; Zhou et al., 2016). Despite these exciting advances, the biomaterials commonly used as bioinks, such as alginate and gelatin methacrylate, poorly capture the biochemical complexity and biodegradability of the native ECM.
Previous studies have identified bioink stiffness as a key element for directing cell morphology and differentiation in 3D cultures after bioprinting (Blaeser et al., 2015; Duarte Campos et al., 2015). Cells encapsulated within polymeric 3D microenvironments also require matrix remodeling to spread, migrate, and proliferate. Unfortunately, a trade-off frequently exists between printability and biological outcome when designing bioinks (Duarte Campos et al., 2016). In general, increasing the bioink stiffness can also improve printing precision, whereas cell spreading and differentiation are often improved by decreasing the bioink stiffness. For this reason, proteolytically degradable hydrogels, such as elastin-like protein (ELP) hydrogels, have been successfully engineered to control encapsulated cell phenotype and stemness (Madl et al., 2017). ELP hydrogels are a family of recombinant engineered-protein materials that contain elastin-like repeat units alternating with modular and customizable bioactive domains (Straley and Heilshorn, 2009). The initial stiffness of ELP hydrogels can be tuned by variation of the final concentration of ELP or variation of the crosslinker concentration. For example, in previous work, ELP hydrogel stiffness was varied between 0.5 and 50 kPa in 3–10 wt% ELP hydrogels (Madl et al., 2017). Cell-laden ELP hydrogels were shown to be stable for at least 2 weeks. These materials are proteolytically degradable by collagenases, elastases, and other proteases, resulting in local remodeling of the matrix and enabling cell proliferation over 2 weeks (Chung et al., 2012a; Madl et al., 2017).
In this study, we explore the feasibility of ELP hydrogels with the fibronectin-derived, cell-adhesive RGD amino acid sequence (ELP-RGD) as bioinks for engineering 3D in vitro models with on-chip vascular-like channels (Figure 1). Bioink printability, single-cell and cell-spheroid viability after bioprinting, as well as proof-of-concept bioprinting of a neural tissue-on-chip, were assessed using ELP-RGD hydrogels. Analysis of neural progenitor cell and cancer spheroid survival after bioprinting showed encouraging results after 7 days of culture. Prolonged cultures up to 14 days showed that NPCs spread and cancer spheroids continued growing at a comparable rate as non-bioprinted controls. Preliminary analysis of the endothelialized channels demonstrated distribution of endothelial cells along the entire lumen of the channel.
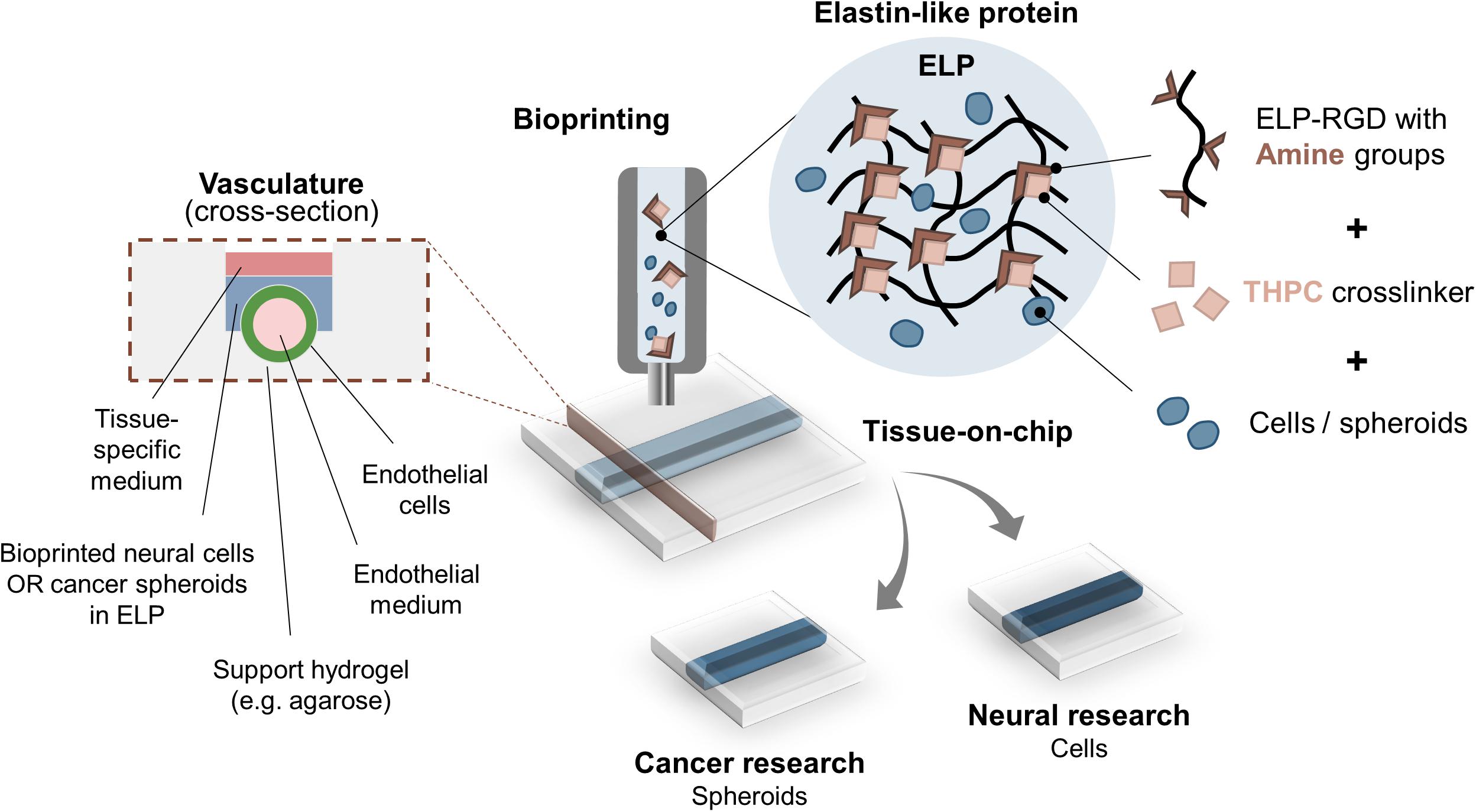
Figure 1. Bioprinting ELP-RGD bioinks with encapsulated cells or spheroids as tissue-on-chip platforms. ELP-RGD bioinks with pendant primary amine groups crosslink in the presence of tetrakis(hydroxymethyl)phosphonium chloride (THPC), which is a tetra-functional, amine-reactive, small molecule crosslinker. Pre-mixed ELP-RGD/THPC bioinks with cells or spheroids are loaded into the bioprinting cartridge and bioprinted directly onto microfluidic chips that enable fabrication of endothelialized vascular-like channels. Two distinct types of culture media are used: endothelial medium is perfused in the lumen of the vascular channel, while neural- or cancer-specific medium is added to the upper culture chamber.
ELP-RGD hydrogels were synthesized as previously described (Madl et al., 2017; LeSavage et al., 2018). Briefly, ELP was cloned into pET15b plasmids and expressed in BL21(DE3)pLysS Escherichia coli. Bacterial cultures containing the plasmids grew to an OD600 of 0.8 before inducing ELP expression with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After culture, bacteria were harvested, resuspended and lysed by repetitive freeze-thaw cycles. ELP was next purified by inverse temperature cycling, dialyzed, and lyophilized to a solid state. ELP-RGD hydrogels were prepared by first dissolving the lyophilized ELP in phosphate-buffered saline (PBS) to a concentration of 3.75, 5.0, or 6.75 wt%. These ELP solutions were then mixed with a diluted solution of the amine-reactive crosslinker, tetrakis(hydroxymethyl)phosphonium chloride (THPC), in a 4:1 volumetric ratio (ELP solution:THPC solution), resulting in final ELP-RGD hydrogel concentrations of 3, 4, or 5 wt%, respectively. For each hydrogel condition, the THPC solution was initially diluted in PBS such that the hydrogels had a final crosslinking ratio of 0.5:1 (THPC reactive sites:primary amines on ELP). For cell experiments, all components used to prepare the ELP-RGD hydrogels and bioinks were sterile-filtered prior to culture using a 0.22 μm syringe filter.
For bioprinting experiments using the chip design with sacrificial gel-made channels, 5 wt% gelatin was used to encapsulate human umbilical vein endothelial cells (HUVECs) that endothelialized the channel. Stocks of 5 wt% gelatin were prepared by dissolving gelatin powder (Sigma) in deionized water at 37°C, sterile filtering with a 0.22 μm syringe filter, aliquoting in 1 ml tubes, and storing at −20°C. Prior to bioprinting experiments, frozen stocks were thawed at 37°C, mixed with HUVECs, and extruded directly inside the chip with a syringe coupled to an 18G needle. Agarose hydrogels were used to support the formation of gelatin channels. A stock solution of 3 wt% agarose (Bio-Rad) was prepared by dissolving agarose powder in deionized water, followed by autoclave sterilization at 121°C.
For printability experiments using the microextruder, Pluronic F127 (Sigma) was used as a supporting gel in a reservoir. 26 wt% Pluronic gel was prepared by dissolving it in deionized water at 4°C. Molds containing 26 wt% Pluronic were cast prior to ELP-RGD printability testing.
Matrigel was used as non-bioprinted control for encapsulation of cells and spheroids, and for the expansion of cancer spheroids prior to bioprinting experiments. Stocks of Matrigel (#354277, Corning) were stored frozen at −20°C and thawed at 4°C prior to use.
Printability of ELP-RGD bioinks was tested by drop-on-demand (DoD) and microextrusion bioprinting. DoD experiments were performed using a hand-held bioprinter (DropGun, BlackDrop Biodrucker GmbH, Aachen, Germany), consisting of a hand-held bioink reservoir connected to an air compressor, mounted with a 300 μm micro-valve, and regulated by a controller unit (Duarte Campos et al., 2019). Average drop diameter and weight of ELP-RGD bioinks at concentrations of 3, 4, and 5 wt%, and deionized water (as liquid control) were analyzed. First, the hand-held bioprinter, ELP-RGD hydrogel precursors, and THPC solutions were placed on ice for 15 min prior to the printability experiments in order to slow down the crosslinking speed of ELP-RGD bioinks. ELP-RGD hydrogels at concentrations of 3.75, 5.0, and 6.75 wt% were mixed with a diluted THPC solution in 4:1 volumetric ratio using manual pipetting, and immediately transferred to the bioprinter reservoir. Each printing experiment was performed using a single ejection valve or needle, without use of coaxial needles. These formulations have previously reported to have crosslinking times that vary between 5 and 30 min (Chung et al., 2012b). The loaded hand-held bioprinter was fixed at a 1 cm distance from the printing substrate (glass slide). All materials were dispensed with a valve opening time (gating time) of 450 μs, and at defined pressures of 0.25, 0.5, 0.75, 1.0, or 1.5 bar. Images of the printed drops (n = 10 for each material and variable) were recorded immediately after printing, and drop diameter was measured with ImageJ (National Institutes of Health, NIH). For assessing drop weight, 100 drops of each testing material were collected in 500 μl tubes and weighed.
Qualitative printability tests were performed with 3 wt% ELP-RGD bioink by DoD in circular and S shapes (by hand). Additional printability tests by microextrusion were performed using a 3D-bioprinter (Biobot1, Allevi, Philadelphia, PA, United States) mounted with a flat-tip 27G needle. 3 wt% ELP-RGD bioink was pre-mixed with 5 μl green food color, added to one printer head, and extruded in a spiral shape in a Pluronic print bath. Images were taken of both DoD and microextruded specimens.
Two distinct chips were custom-designed using an online platform (Biofabics Toolbox)1, and manufactured (Biofabics, Porto, Portugal): (1) chip with ready-made channel and (2) chip with sacrificial gel-made channel.
Chip 1 was designed with tool number 4 of the Biofabics Toolbox (Figure 2A), consisting of a 96-well plate layout with 6.5 mm well diameter, 3.5 mm well height, 11.5 mm channel length, 1 mm channel diameter, 1 mm base thickness, and a square chamber with 6.5 mm width/length. A column of two device units was selected, and all remaining parameters were kept at zero. Each chamber was perfused with a 0.8 mm diameter molding line, which was removed after bioprinting of ELP bioinks.
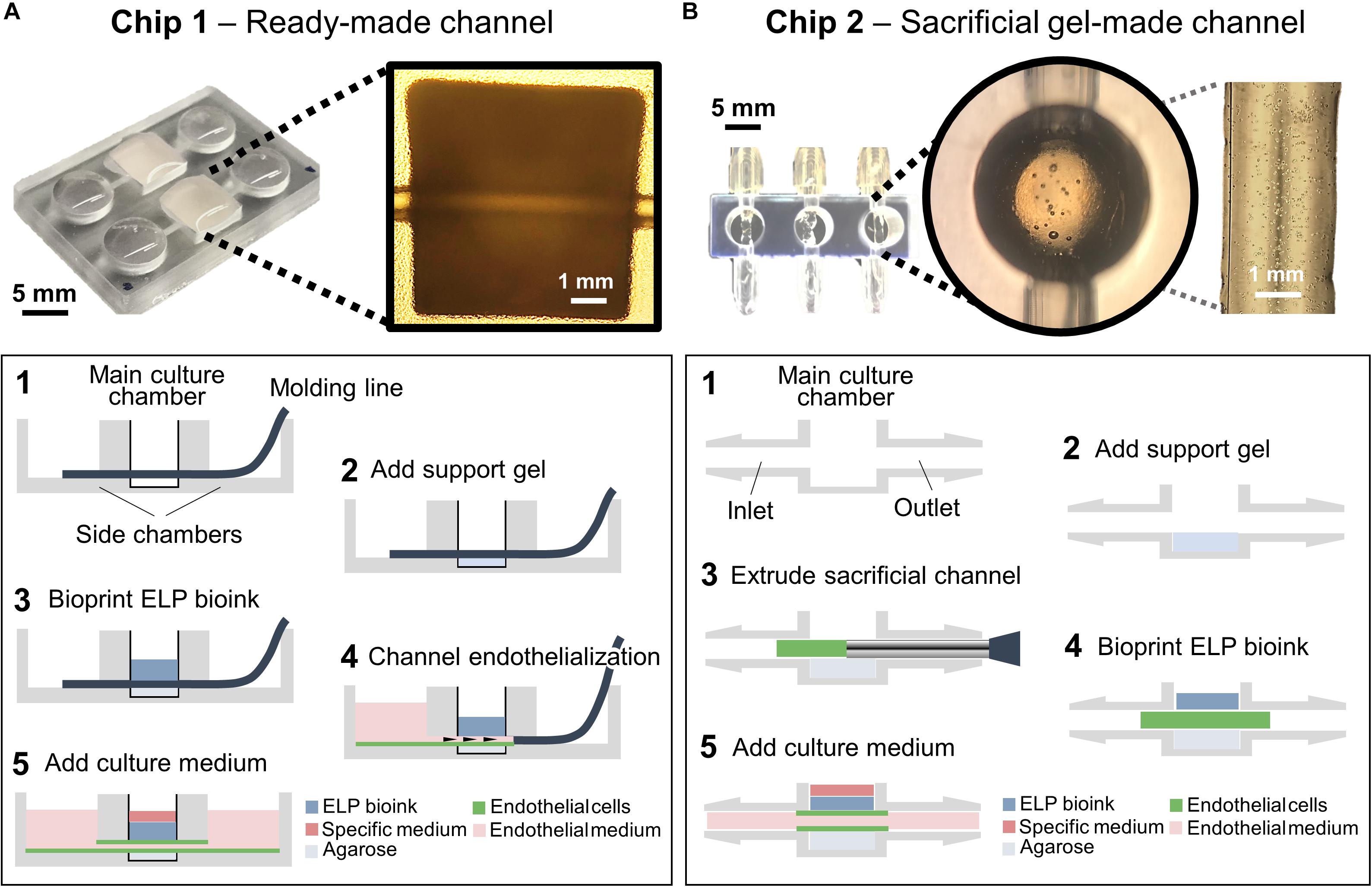
Figure 2. Chip designs used for the biofabrication of printed tissue frameworks with on-chip vascular-like channels. Chips designs with (A) ready-made channels, and (B) sacrificial gel-made channels were evaluated. Sequence of events (1–5) indicates the necessary working steps to fabricate both types of tissue models. Sacrificial gel-made channels can be used to incorporate endothelial cells that will form monolayers on the channel surface after removing the sacrificial gel. The inset photograph in (A) shows the channel formation at step 5. The inset photograph in (B) shows the presence of endothelial cells inside the sacrificial gel at step 3.
Chip 2 was designed with tool number 5 of the Biofabics Toolbox (Figure 2B), consisting of a 96-well plate layout with a round chamber with 8 mm chamber height, 5 mm chamber diameter, 2 mm channel diameter, 2 mm channel to chamber bottom distance, and 1 mm base thickness. A column of three device units was selected, and all remaining parameters were kept at zero. The inlet/outlet fittings of each chip were fastened to silicone tubes, which were coupled to syringes as inlets (fresh medium supply, three inlets), or outlets (culture medium waste, three outlets).
Murine NPCs from micro-dissected dentate gyrus of adult female mice (C57Bl/6) were kindly provided by Prof. Theo Palmer (Stanford Neurosurgery) (Babu et al., 2007). All animal work followed protocols reviewed and approved by the Stanford Administrative Panel on Laboratory Animal Care. NPCs were cultured and expanded in 2D, and encapsulated in 3D ELP-RGD hydrogels for bioprinting experiments, following previously established protocols (Madl et al., 2017; LeSavage et al., 2018). After isolation, NPCs were expanded in maintenance medium [Neurobasal-A (Thermo Fisher Scientific), 2% B27 supplement (Gibco), 2 mM GlutaMAX (Life Technologies), 20 ng/ml fibroblast growth factor 2 (FGF-2), and 20 ng/ml epidermal growth factor (EGF) (PeproTech)] on Poly-L-ornithine and laminin coated tissue culture plastic (Madl et al., 2016). For cell passaging and encapsulation in ELP-RGD bioinks, NPCs were trypsinized, pelleted, resuspended, and counted. For expansion, NPCs were plated at 1 × 104 cells/cm2, and, for encapsulation, cells were mixed with hydrogel components to achieve a final density of 1.5 × 107 cells/ml. For bioprinting experiments, NPCs were encapsulated in 3 wt% ELP-RGD bioinks, pre-mixed with THPC, as described above, and loaded into the printing cartridge. After bioprinting, specimens were incubated at room temperature for 15 min, followed by incubation at 37°C for 30 min prior to adding culture medium. Culture medium was replaced every 2 days during cell expansion and every day after bioprinting.
Human induced pluripotent stem cells (hiPSCs, line: 8343.2) were differentiated into cortical neural progenitors following a previously established protocol (Shi et al., 2012). Briefly, hiPSCs were maintained in Essential 8 (E8) medium (Gibco) on Matrigel-coated tissue culture plastic and differentiated in N3 medium consisting of 50% DMEM/F-12, 50% Neurobasal A, 1% N-2 supplement, 2% B-27 supplement, 2 mM GlutaMAX, 1% MEM Non-Essential Amino Acid Solution (NEAA), and 2.5 μg/ml human recombinant insulin (all Thermo Fisher Scientific). N3 medium was further supplemented with 5 μM transforming growth factor beta (TGF-beta) receptor inhibitor [SB-431542 (Tocris)] and 100 nM activin receptorlike kinase 2 (ALK2) and ALK3 inhibitor [LDN-193189 (Stemgent)] for the first 11 days of culture. At day 12, hiPSCs were dissociated with Cell Dissociation Solution (Sigma) and plated on pre-coated plates with 50 μg/ml Poly-D-Lysine (Sigma) and 5 μg/ml Laminin (Roche). Next, hiPSC-derived NPCs were further cultured in N3 medium without SB-431542 or LDN-193189 until day 16. Prior to bioprinting experiments, hiPSC-NPCs were dissociated and encapsulated in 3 wt% ELP-RGD bioinks pre-mixed with THPC at a density of 1.5 × 107 cells/ml. After bioprinting, specimens were incubated at room temperature for 15 min, followed by incubation at 37°C for 30 min prior to adding culture medium. Culture medium was replaced daily before and after bioprinting.
HUVECs were purchased from PromoCell. HUVECs were expanded in EGM-2 growth medium (Lonza) containing 0.04% hydrocortisone, 0.4% human FGF-2, 0.1% vascular endothelial growth factor (VEGF), 0.1% recombinant analog of insulin-like growth factor (R3-IGF-1), 0.1% ascorbic acid, 0.1% human EGF, 0.1% gentamicin sulfate-amphotericin (GA-1000), 0.1% heparin, and 2% fetal bovine serum (FBS). For cell passaging and encapsulation in gelatin, HUVECs were trypsinized, pelleted, resuspended, and counted. HUVECs were plated at 1 × 104 cells/cm2 for expansion. For encapsulation, cells were mixed with 5 wt% gelatin to achieve a final density of 1 × 107 cells/ml. Culture medium was replaced every 2 days during cell expansion and every day after bioprinting.
Human premalignant breast epithelial cells (MCF10ATs) were kindly provided by Jan Liphardt (Stanford Bioengineering) and expanded, following a previously published protocol (Leung and Brugge, 2012). Briefly, MCF10ATs were expanded in DMEM/F-12 medium (Gibco) supplemented with 100 μg/ml EGF (Gibco), 1 mg/ml hydrocortisone (Sigma), 1 mg/ml cholera toxin (Sigma), 10 mg/ml insulin (Sigma), and 100x penicillin/streptomycin (Gibco). For cell passaging and encapsulation in Matrigel, MCF10ATs were trypsinized, pelleted, resuspended, and counted. For expansion, MCF10ATs were plated at 1 × 104 cells/cm2, and for encapsulation, cells were mixed with Matrigel to achieve a final density of 5 × 105 cells/ml. After 7 days culture in Matrigel, MCF10AT spheroids with approximately 50 μm diameter, containing dozens of cells, were harvested for bioprinting experiments. Briefly, MCF10AT-laden Matrigel cultures were incubated in 5 mM EDTA in PBS for 45 min on ice. Dissociated Matrigel, containing MCF10AT spheroids, was pelleted and resuspended in basal medium. MCF10AT spheroids were encapsulated at a density of 1 × 107 spheroids/ml in 3 wt% ELP-RGD bioinks pre-mixed with THPC, as described above, and loaded into the printing cartridge. After bioprinting, specimens were incubated at room temperature for 15 min, followed by incubation at 37°C for 30 min prior to adding culture medium. Culture medium was replaced every 2 days during expansion and daily after bioprinting.
Before bioprinting experiments, chips were washed thrice in 70% ethanol and sterile PBS. Cell and spheroid-laden 3 wt% ELP-RGD bioinks were prepared as described above and bioprinted by DoD. For chip 1, bioprinting of bioinks was performed prior to channel endothelialization. First, each main culture chamber was filled with 100 μl bioink. After crosslinking, HUVECs suspended in culture medium were added to one of the supporting chambers, and the molding line was carefully removed, allowing the HUVEC suspension to fill the channel by capillary action. Subsequently, chips were flipped and incubated bottom-up at 37°C for 4 h to allow for cell seeding on the upper segment of the channel, followed by upright incubation at 37°C for 5 days. Culture media for both cell types was replaced daily.
Bioprinting into chip 2 was performed after channel formation. First, sterile 18G needles were inserted in the chips through the side tube connectors. Twenty microliter 3 wt% agarose was cast in the bottom of each main culture chamber to support the formation of a HUVEC-laden gelatin channel. HUVEC-laden 5 wt% gelatin was prepared as described above and extruded via syringe inside the culture chamber at room temperature. After gelation of the gelatin channel, 50 μl cell-laden ELP-RGD bioinks were bioprinted by DoD into each chamber and allowed to crosslink for 15 min at room temperature. Next, chips were flipped and incubated bottom up at 37°C for 4 h. At this step, gelatin channels melted and HUVECs sedimented by gravity on the upper segment of the channel. Chips were once again flipped and incubated upright for 5 days. Syringes with fresh EGM-2 medium for HUVEC culture were connected to one side of the chamber, and empty syringes were connected to the opposite side to collect media waste. Cell-specific culture medium was added to the upper part of the main chamber, and both culture media were replaced daily.
Three, seven, and fourteen days after cell and spheroid encapsulation in hydrogels and bioprinting experiments, specimens were removed from the incubator, washed once with PBS, and stained with Live/Dead viability staining (Thermo Fisher Scientific). Staining conditions were optimized for cells encapsulated in Matrigel and ELP-RGD. For Matrigel samples, stock solutions were diluted to 0.5 μg/ml calcein and 2 μl/ml ethidium homodimer-1 (EthD-1) in PBS, and incubated for 30 min at 37°C. Cells in 3 wt% ELP-RGD were incubated in solutions of 1 μl/ml calcein and 2 μl/ml EthD-1 diluted in PBS for 30 min at 37°C. After incubation, samples were washed once with PBS and imaged using a Leica SPE confocal microscope. Percent (%) viability was calculated based on the number of counted live and dead cells (Equation 1). Three independent biological replicates were used for each time point and variable (N = 3).
Percent (%) viability of cells and spheroids.
For immunocytochemistry, samples were fixed with 4% paraformaldehyde in PBS for 30 min at 37°C. Samples were permeabilized with 0.25% Triton X-100 in PBS (PBST) for 1 h at room temperature. The samples were then blocked with 5% bovine serum albumin (BSA) and 5% goat serum in PBST for 3 h at room temperature. Primary antibodies against sex determining region Y-box 2 (Sox2) (1:400, rabbit, Cell Signaling Technology, 23064S) and/or cluster of differentiation 31 (CD31) (1:200, mouse, PECAM-1, Sigma, P8590) were diluted in PBST containing 2.5% goat serum, added to samples, and incubated overnight at 4°C. The samples were then washed thrice with PBST, incubated with secondary antibodies [AlexaFluor488 (goat anti-rabbit, 1:500); AlexaFluor647 (goat anti-mouse, 1:500); and/or tetramethylrhodamine (TRITC) phalloidin 532 (F-actin, 1:500)], and counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) overnight at 4°C. Samples were washed thrice with PBST and imaged using a Leica SPE confocal microscope. Three independent biological replicates were used for each time point and variable (N = 3).
Confocal images were analyzed with ImageJ (NIH). Images recorded with the same magnification (20× objective) were transformed into 8-bit images. Color channels of each image were split, and the threshold was manually adjusted prior to measuring percent cell area values. Total cell area was calculated as a function of the total image area of 2.9 × 105 μm2. Three independent biological replicates were used for each time point and variable (N = 3).
Comparisons between two experimental groups were made using two-tailed Student’s t-tests (Prism 8, version 8.3.0, GraphPad Software). Comparisons between three or more experimental groups were made using one-way ANOVA with Tukey’s post hoc test. Statistical significance was considered as ∗p < 0.05, ∗∗p < 0.005, and ∗∗∗p < 0.001. Independent biological replicates and exact P-values are indicated in each Figure including statistical analyses. Experimental data are presented as means ± standard deviation.
The printability of ELP-RGD bioinks at varying concentrations was tested by DoD (Figure 3A) and microextrusion bioprinting (Figure 3B). For all tests, the ELP-RGD engineered protein was solubilized in PBS and pre-mixed with the tetra-functional crosslinker THPC at a 0.5:1 stoichiometric ratio between THPC functional groups and primary amines in the protein. DoD printability tests were performed with a hand-held bioprinter connected to an air compressor, which allowed for hand-guided dispensing of single bioink drops in circular and S shapes. Microextrusion printability tests were performed using a 3D-bioprinter, which allowed for continuous extrusion of the bioink in spiral shapes submerged in a Pluronic print-bath. The smallest diameter observed for single drops of ELP-RGD bioinks printed by DoD was 1.4 mm (5 wt% ELP-RGD at 0.25 bar, Figure 3C). By increasing the applied pressure during printing, it was possible to increase the drop size without varying bioink concentration or nozzle diameter. Drops larger than 4 mm in diameter were difficult to measure due to impaired droplet integrity wherein large drops tended to splatter into smaller drops. Nevertheless, drop size and weight continued to increase upon increasing the applied pressure, as shown in Figures 2C, 3D. Interestingly, the mass of the printed drops decreased with increasing ELP-RGD concentration and constant pressure. For example, at 0.75 bar, the 3 wt% ELP-RGD formed single drops with an average mass of 342 μg, whereas that of 4 wt% and 5 wt% ELP-RGD inks had average single drops of 311 and 257 μg, respectively.
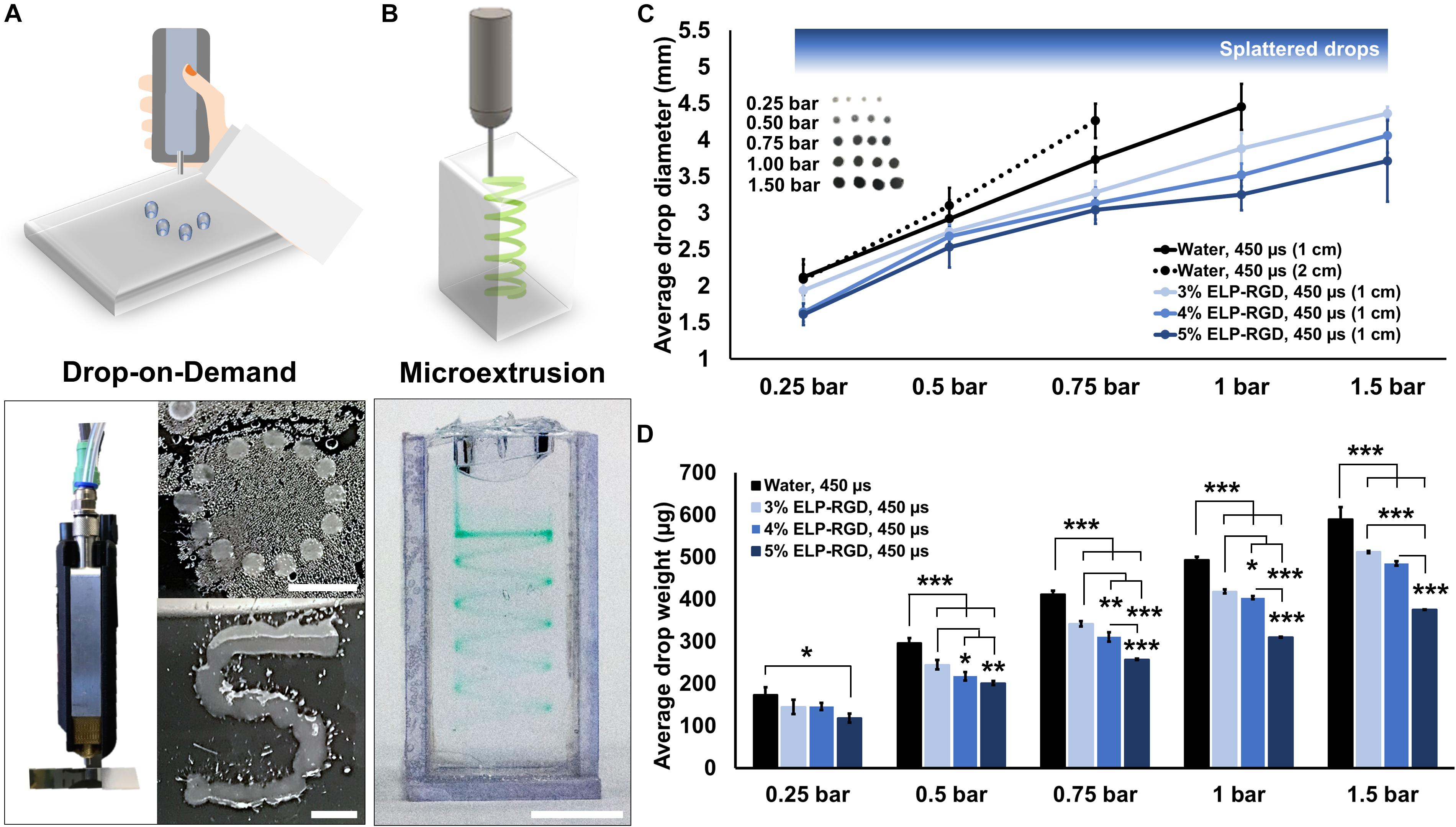
Figure 3. Printability of ELP-RGD bioinks. (A) Qualitative drop-on-demand (DoD) printability tests of 3 wt% ELP-RGD printed as single drops into circular and S shapes. Scale bars represent 5 mm. (B) Qualitative microextrusion printability test of 3 wt% ELP-RGD in a spiral shape within a Pluronic bath. Scale bar represents 5 mm. (C) Average single drop diameter of deionized water, 3, 4, and 5 wt% ELP-RGD printed by DoD at applied pressures ranging from 0.25 to 1.5 bar (n = 10). (D) Average single drop mass of deionized water, 3, 4, and 5 wt% ELP-RGD printed by DoD at applied pressures ranging from 0.25 to 1.5 bar (n = 3, 100 drops each). Valve opening time (gating time) was fixed at 450 μs for all experiments. Statistical significance marked as *p < 0.05, **p < 0.005, and ***p < 0.001 (One-way ANOVA with Tukey’s post hoc test).
Toward the long-term goal of culturing bioprinted 3D tissue models with vascular-like channels, we evaluated two different chip designs and fabrication protocols. In one method, a simple, easy-to-use chip design with ready-made channels was evaluated (Figure 2A). This chip comprised two main culture chambers, where the tissue models were bioprinted, and four supporting chambers, where cell culture medium was added. Two plastic molding lines with 0.8 mm diameters were inserted in each chip by traversing the main culture chambers and the two supporting chambers connected to them. After DoD bioprinting of the ELP-RGD layers onto the plastic wire, HUVECs suspended in medium were added to one of the sides of the chip and were subsequently seeded in the inner wall of the channel by removing the plastic line. As a second method, a chip design comprising culture chambers with tubing connectors suitable for dynamic perfusion culture was evaluated (Figure 2B). This type of chip design allowed for HUVEC seeding in the vascular-like channel within a sacrificial gelatin hydrogel. The ELP-RGD ink was bioprinted directly on top of the sacrificial gelatin to form an upper layer of printed tissue. After the last bioprinting step, the chips were placed in an incubator at 37°C allowing for further crosslinking and solidification of the ELP-RGD bioink, and simultaneous fluidization and removal of the sacrificial gelatin channel.
Several variables have been reported to impact cell viability during different bioprinting methods, including bioink viscosity, printing nozzle size, and applied printing pressure (Duarte Campos et al., 2015; Dubbin et al., 2017). Therefore, an important step toward developing a bioprinted in vitro tissue-on-chip model is to evaluate cell viability. In this study, cell and spheroid viability was analyzed after DoD bioprinting using dissociated NPCs (Figure 4A) and MCF10AT spheroids (Figure 4C). Cells and spheroids encapsulated in 3 wt% ELP-RGD bioinks and Matrigel without bioprinting were included as controls. NPC survival in 3 wt% ELP-RGD was greater than 88.9% in both experimental groups, including non-bioprinted and bioprinted samples (Figure 4B). The lowest NPC survival of about 77.4% was observed in non-bioprinted Matrigel controls after 7 days of culture. The overall MCF10AT spheroid viability rates at day three of culture were lower compared to NPC cultures (Figure 4D). The lowest MCF10AT spheroid survival rate observed was about 70.2% 3 days after bioprinting. Interestingly, upon further time in culture, bioprinted MCF10AT viability rates increased to about 88.3% at day seven of culture. This cell viability at 7-days post-printing was not statistically significantly different than the 7-day viability without printing in either ELP-RGD (82.3%) or Matrigel (93.3%).
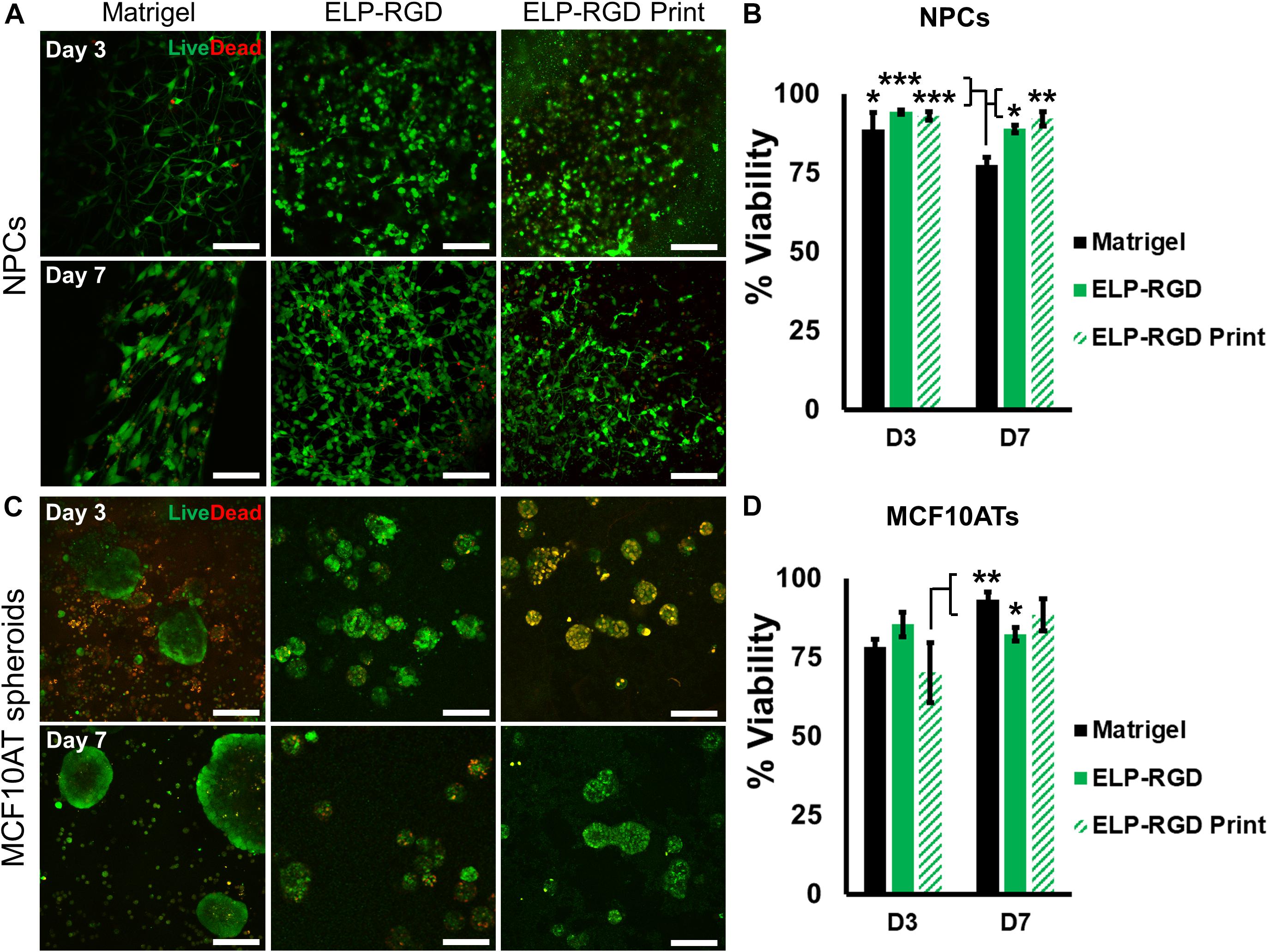
Figure 4. Viability of dissociated cells and spheroids with and without bioprinting. Live cells are stained with calcein (green) and dead cells are stained with ethidium homodimer-1 (red). (A) Live/Dead staining of murine neural progenitor cells (NPCs) on days 3 and 7 within Matrigel (not bioprinted), 3 wt% ELP-RGD (not bioprinted), and 3 wt% ELP-RGD after DoD bioprinting. Scale bars represent 100 μm. (B) Quantified viability of NPCs at days 3 and 7. Statistical significance marked as *p < 0.05, **p < 0.005, and ***p < 0.001 (N = 3, One-way ANOVA with Tukey’s post hoc test). (C) Live/Dead staining of human premalignant breast epithelial cell spheroids (MCF10ATs) in Matrigel (not bioprinted), 3 wt% ELP-RGD (not bioprinted), and 3 wt% ELP-RGD after DoD bioprinting at days 3 and 7. Scale bars represent 100 μm. (D) Quantified viability of MCF10ATs at days 3 and 7. Statistical significance marked as *p < 0.05, and **p < 0.005 (N = 3, One-way ANOVA with Tukey’s post hoc test).
Perfused and endothelialized tissue-on-chips are important components of many ex vivo tissue models. In neural research, such models may be used to mimic the neural stem cell niche to study the requirements for stem cell maintenance and activation, or in the future to recapitulate the blood-brain-barrier to study the penetration of new drugs across the barrier. In cancer research, these models may be used to investigate cancer cell migration and metastasis through blood vessels. In this work, we evaluated the endothelialization of chips with a ready-made channel design. After HUVEC seeding, culture, and fixation, entire chips were stained, sectioned, and imaged by confocal microscopy. Cross-section views with different 3D perspectives of endothelialized channels are shown in Figure 5a. A HUVEC monolayer was formed along the inner wall of the channel contained in the culture chip. Next, the endothelialized channel was imaged lengthwise to show the presence of HUVECs at different z heights (Figure 5b) and in 3D perspective (Figure 5c). A compiled view of the y-z cross-section of the channel (Figure 5d) demonstrates a uniform distribution of HUVECs along the inner wall of the channel.
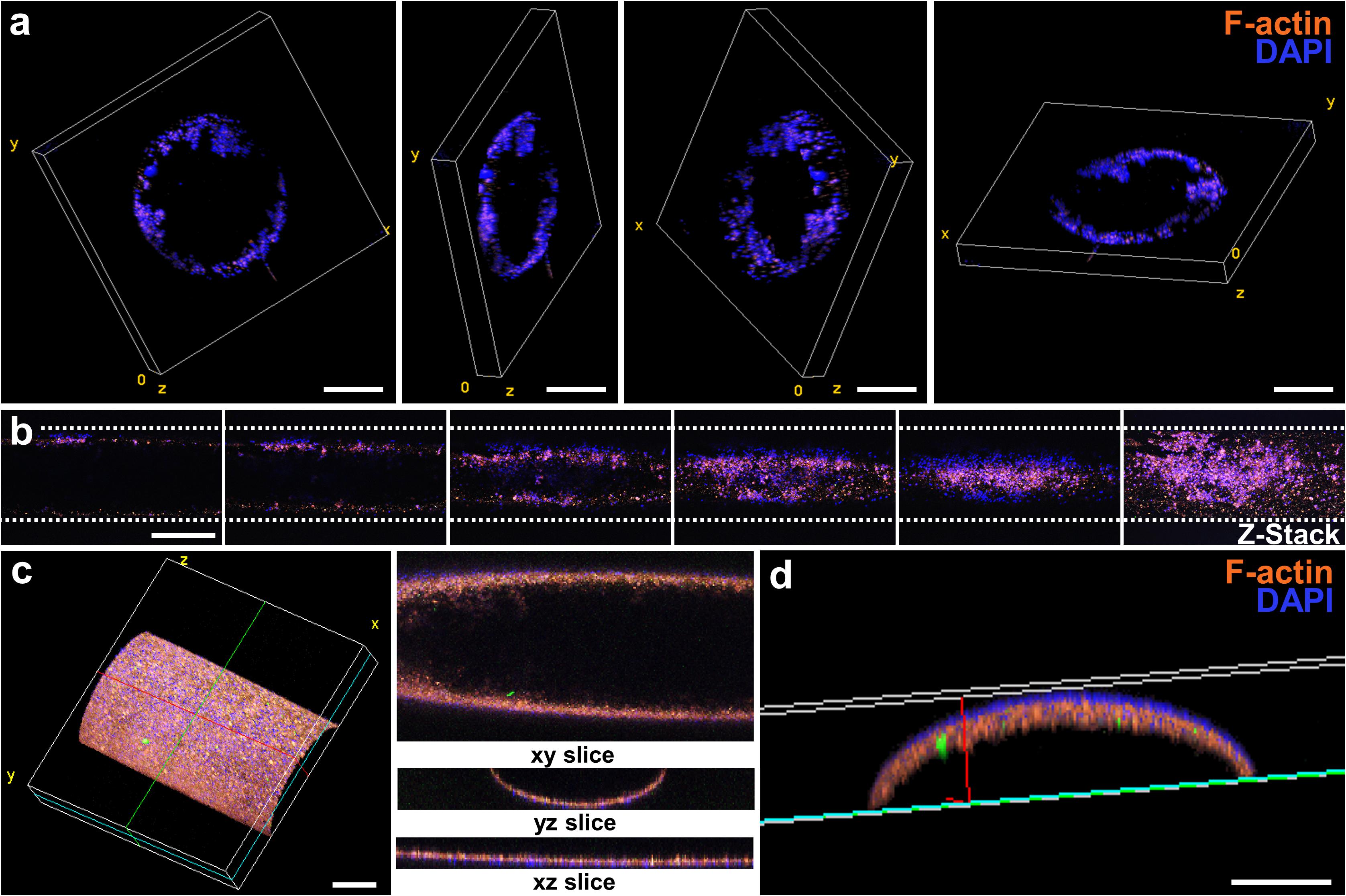
Figure 5. Endothelialization of the luminal wall of the chip. (a) Sequence of cross-sectional confocal images showing the position of HUVECs seeded within the channel. Scale bars represent 250 μm. (b) Sequence of confocal images taken lengthwise to the channel. The image at the far-right shows a z-stack projection of the sequence of images at varying z heights. Scale bar represents 500 μm. (c) 3D projection of an endothelialized channel showing the cross-section views at the xy, yz, and xz planes. Scale bar represents 250 μm. (d) Cross-section yz view of (d). Scale bar represents 100 μm.
To develop tissue-on-chips suitable for use as ex vivo tissue models, it is important to evaluate cell and spheroid morphological changes during culture. NPCs encapsulated in bioprinted and non-bioprinted matrices showed morphological changes from rounded at day 1 to elongated at day 14 (Figure 6A). In contrast, MCF10AT spheroids did not show morphological alterations throughout the culture period (Figure 6B). F-actin staining of NPCs encapsulated in each matrix, both with or without bioprinting, showed a significant increase (∗p < 0.05) in total cell area from days 1 to 14, suggesting increased cell spreading and elongation (Figure 6C). No statistically significant differences in NPC cell-spreading were observed between the various matrices, with or without bioprinting. Similarly, MCF10AT spheroid size was not significantly different within the different matrices. In addition, total cell area of MCF10AT spheroids encapsulated in bioprinted and non-bioprinted matrices did not significantly change between days 1 and 14 of culture (Figure 6D).
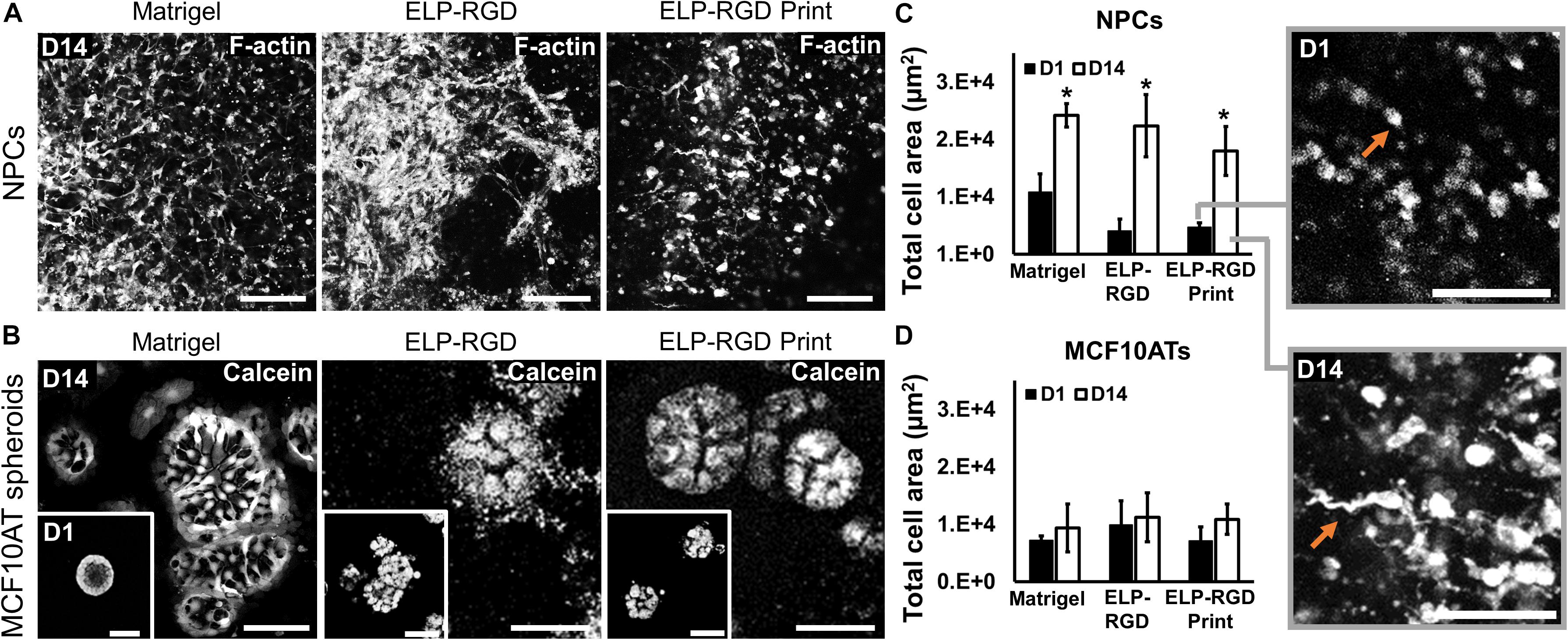
Figure 6. Morphology of dissociated NPCs and MCF10AT spheroids after 14 days of culture within 3D hydrogels. (A) Confocal images of NPCs cultured for 14 days in Matrigel, 3 wt% ELP-RGD without bioprinting, and 3 wt% ELP-RGD after DoD bioprinting. Scale bars represent 100 μm. (B) Confocal images of MCF10AT spheroids cultured for 14 days in Matrigel, 3 wt% ELP-RGD without bioprinting, and 3 wt% ELP-RGD after DoD bioprinting. Scale bars represent 50 μm. Insets show MCF10AT spheroids at day 1 of culture in the respective material. Scale bars represent 100 μm. (C) Total cell area of stained NPCs in confocal images taken at days 1 and 14 of culture. Statistical significance between days 1 and 14 of each material marked as *p < 0.05 (N = 3, two-tailed t-test, unpaired). Insets are magnified views of NPCs cultured in 3 wt% ELP-RGD after bioprinting, showing round cells at day 1 and spread cells at day 14 (orange arrows). Scale bars represent 100 μm. (D) Total cell area of stained MCF10AT spheroids in confocal images taken at days 1 and 14 of culture. One-way ANOVA with Tukey’s post hoc test did not show statistical significance between groups.
After assessing bioink printability, cell viability after bioprinting, and cell morphological changes during in vitro culture, we developed and analyzed a proof-of-concept ex vivo model of the neural stem cell niche that included both HUVECs and hiPSC-derived NPCs. For this experiment, the chip with the ready-made channel was used. hiPSC-NPCs were encapsulated in 3 wt% ELP-RGD bioinks and bioprinted by DoD over the main chamber of the chip. After bioprinting of hiPSC-NPCs, HUVECs suspended in medium were added to one side chamber of the chip, and the perfusion line was removed to induce HUVEC seeding within the vascular-like channel. Confocal images of the tissue chips after 5 days of culture showed the presence of hiPSC-NPCs (Sox-2-positive, a marker of neural progenitor cell pluripotency) within the printed ELP-RGD hydrogel and the presence of HUVECs (CD31-positive, also known as platelet endothelial cell adhesion molecule (PECAM-1), a common endothelial marker) within the vascular-like channel (Figure 7).
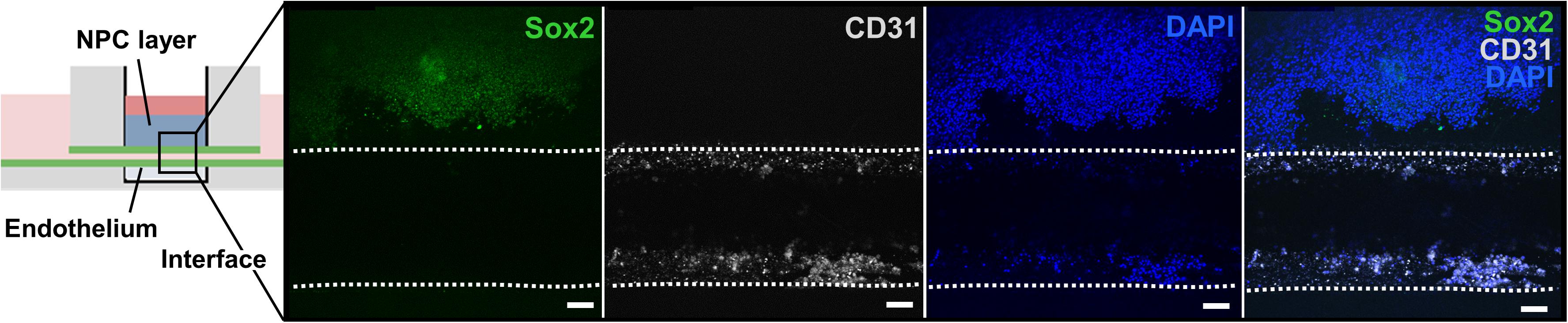
Figure 7. Bioprinted tissue-on-chip with vascular-like channel. Schematic of the tissue-on-chip design. Confocal images of hiPSC-NPCs encapsulated within 3 wt% ELP-RGD bioprinted on top of a channel seeded with HUVECs and cultured for 5 days. hiPSC-NPCs were visualized with Sox2 immunostaining, HUVECs were visualized with CD31 immunostaining, and all cells were visualized by nuclear staining with DAPI. Scale bars represent 50 μm.
The goal of this study was to explore the feasibility of using ELP-RGD hydrogels as bioinks to fabricate 3D tissue models on microfluidic chips with vascular-like channels. Following a previously established protocol (LeSavage et al., 2018), we synthesized ELP-RGD hydrogels and tested their printability as bioinks. First, the printability by DoD was tested using a hand-held bioprinting device. DoD bioprinting was chosen for this study, because it has been previously reported as a more cell-friendly and freeform method compared to, for example, microextrusion bioprinting (Blaeser et al., 2017). Drop size and weight strongly varied depending on the concentration of ELP-RGD bioink (3, 4, or 5 wt%), applied printing pressure, and distance to the printing substrate. This outcome is in accordance with previous studies, which used other hydrogels with similar rheological properties, such as polysaccharides (Blaeser et al., 2015; Duarte Campos et al., 2019). Comparable ELP-based bioinks were tested in previous works using microextrusion bioprinting (Salinas-Fernández et al., 2019). In this previous work, the geometric printability outcomes highly depended on the concentration and amino acid sequence of the ELP bioink. In our work, qualitative printability experiments by microextrusion showed well-defined 3D shapes with complex and freeform geometry (spirals, Figure 2B).
Survival of neural cells and cancer spheroids encapsulated in 3 wt% ELP-RGD was assessed after DoD bioprinting. We found that the average neural cell survival (88.9%) was in accordance with other studies that used the same bioprinting method in combination with other bioinks and cell types (Forget et al., 2017; Kreimendahl et al., 2017; Ng et al., 2017). Contrastingly, average cancer spheroid survival after bioprinting (70.2%) was markedly lower compared to neural cell survival. This result may be due to the larger diameter of cellular MCF10AT spheroids (approximately 50 μm) compared to dissociated, single NPCs (about 10 μm). It is well-known that increased fluid stresses within the printing nozzle can negatively affect cell viability (Aguado et al., 2012; Blaeser et al., 2015; Foster et al., 2017; Lindsay et al., 2019). Nozzle shape, orifice diameter, bioink viscosity, cell density in the bioink, and cell size are parameters that will influence fluid stress, thus making cell aggregates more susceptible to damage (Cidonio et al., 2019). Despite this, cell aggregates with large diameters, including cell clusters like spheroids, may be helpful for the fabrication of ex vivo tissue models (Li and Kumacheva, 2018). Therefore, future work should further evaluate the possible correlation between cell aggregate diameter and low viability post printing. Despite this decrease in cell viability immediately post-printing, by day 7 the printed cultures had recovered and displayed viability and sizes similar to those that had not been exposed to printing. Importantly, day 7 viability and day 14 spheroid size were statistically similar for cultures grown in Matrigel or bioprinted in ELP-RGD. Together, these data suggest that this engineered bioink hydrogel can support the long-term culture of these cancer spheroids after direct bioprinting on a microfluidic chip.
NPCs are a promising cell source for recreating a model of the neural environment. The native neural stem cell niche includes a vascular network that is critical in maintenance of NPC stemness (Tavazoie et al., 2008), with known cross-talk between endothelial cells and NPCs (Schänzer et al., 2006). In vitro differentiation toward astrocytes and neurons can be challenging in 3D, and thus, bioinks and the bioprinting process should ideally not restrict NPC differentiation potential. An important aspect of a bioink to support NPC differentiation is to enable cell-cell contact by cell-mediated matrix remodeling (Madl et al., 2019). In this work, we showed that NPCs elongated to spindle-like shapes in 3 wt% ELP-RGD bioinks after bioprinting and kept growing up to 14 days in culture. This morphology suggests that the encapsulated NPCs are able to remodel the printed matrix and make cell-cell contacts that are required for maintenance of stemness. These results are further confirmed by the continued expression of the neural stem cell marker Sox2. The use of ELP-RGD hydrogels as bioinks is an advancement in the fabrication of on-chip tissues to mimic aspects of the neural-vascular interface. Many of these previous studies have used hydrogels that allow little to no cell-driven matrix remodeling, such as agarose (Gasperini et al., 2014; Duarte Campos et al., 2016; Gu et al., 2018).
Vascularization is a frequent goal for the fabrication of engineered tissues in vitro (Zhang et al., 2016). For this reason, we used custom-designed on-chip platforms that contained vascular-like channels and that allowed for integrated 3D bioprinting. Microfluidic platforms have been used in previous studies, for example, to investigate cancer metastasis in vitro (Sontheimer-Phelps et al., 2019). However, these technologies are difficult to combine with current 3D bioprinting strategies, given that chamber and channel size is in the micrometer range. Therefore, we designed on-chip platforms consisting of culture chambers and channels in the millimeter range, which can be easily accessed with a bioprinter. Two distinct chip designs were considered for their suitability with tissue bioprinting. The first design, which was used for static culture, was simpler and easier to use, as it required fewer preparation steps. HUVECs were successfully seeded onto the inner walls of the channels in these platforms. We showed that HUVECs were distributed along the entire wall of these channels. Microvascular in vitro models containing several cell types and layers that resemble native (micro)vessels were previously reported (Jaeger et al., 2013; Cochrane et al., 2019). Although in vitro vascular-like structures are typically generated using only endothelial cells, such as HUVECs, more complex 3D models will require triple-layered vascular structures, including fibroblasts, endothelial, and smooth muscle cells (Gao et al., 2019). A limitation of our study was the use of only endothelial cells to cover the inner wall of the channels. Future experiments will consider improving the in vivo relevance of the channels by inducing additional cell types. Additionally, functional characteristics of the endothelialized layer, including permeability to nutrients, need to be assessed (Bang et al., 2017).
Finally, as proof-of-concept, a tissue-on-chip was fabricated by bioprinting hiPSC-derived NPCs using protein-engineered-bioinks onto a device with a vascular-like, endothelialized channel. Research studying the cross-talk between endothelial cells and NPCs uses random co-culture mixtures or transwell inserts (Shen et al., 2004), but these do not properly recapitulate the spatial proximity and geometry of these cells in the native stem cell niche, thus resulting in non-physiological concentration gradients of secreted, diffusible factors and inappropriate presentation of cell-surface receptors. In this study, we suggest that a combination of bioprinting and on-chip technologies can be used to realize NPC/HUVEC co-cultures with physiologically relevant geometric patterning. In our previous work, we found that NPCs would lose their Sox2 expression within 2 days in matrices that could not support stemness maintenance (Madl et al., 2017). In this experiment, we showed that both cell types remained viable over 5 days of culture, retained their proper positioning within the co-culture device, and maintained NPC stemness. Although the bioprinter used in this study is hand-held, our results provide a proof-of-concept demonstration that (i) it is feasible to bioprint ELP-RGD bioinks with precision and spatial control, (ii) these bioinks are compatible with the materials commonly used to fabricate microfluidic devices, and (iii) these bioinks are compatible with several cell types of interest. Importantly, this bioprinting technology can be easily mounted onto a stereotactic controller (Blaeser et al., 2015; Duarte Campos et al., 2019). Such automated dispensing of cell-laden hydrogels within microfluidic devices would offer higher spatial precision, greater reproducibility, and more efficient scale-up compared to manual pipetting.
Here we have successfully demonstrated that bioprinted NPCs are able to maintain their stem-like phenotype and hence remain Sox2-positive during co-culture with endothelial cells within a microfluidic device. This platform is well-suited for future cell biology studies investigating the cross-talk between ECs and NPCs that mediate stem cell maintenance and activation.
In this work, we used protein-engineered ELP-RGD hydrogels as bioinks for producing in vitro tissue-on-chip platforms. We showed that ELP-RGD bioinks are dispensable by DoD and microextrusion, and that 3D constructs can be generated. Assessment of neural progenitor cell and cancer spheroid survival after bioprinting showed encouraging results after 7 days of culture. Prolonged cultures up to 14 days showed that NPCs spread, and cancer spheroids continue growing at a comparable rate as non-bioprinted controls. Preliminary analysis of the endothelialized channels demonstrated distribution of endothelial cells along the entire lumen of the channel. The results presented here represent a first step in combining ELP engineered hydrogels with 3D bioprinting technologies and on-chip platforms for establishing vascularized in vitro tissue models. In the future, these platforms may be further developed for in vitro studies of interactions between the vascular interface and patterned, three-dimensional tissue mimics.
The datasets generated for this study are available on request to the corresponding author.
DD and SH: conceptualization. DD, CL, JR, BL, AS, and BK: data acquisition. DC: writing-original draft preparation. DD, CL, JR, BL, AS, BK, RR, PC, AB, and SH: writing and editing and final approval of manuscript. DD, SH, PC, and RR: funding acquisition.
This research was supported by the German Research Foundation (DFG-425869082 to DD) and the U.S. National Science Foundation (DMR-1808415 to SH) and the U.S. National Institutes of Health (R01-HL142718, R01-EB027171, R01-EB027666, R21-HL138042, R21-NS114549 to SH). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 798014 (to PC). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 828835 (to RR).
PC and RR were employed by the company Biofabics Lda. PC was also shareholder of the company Biofabics Lda.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aguado, B. A., Mulyasasmita, W., Su, J., Lampe, K. J., and Heilshorn, S. C. (2012). Improving viability of stem cells during syringe needle flow through the design of hydrogel cell carriers. Tissue Eng. Part A 18, 806–815. doi: 10.1089/ten.tea.2011.0391
Babu, H., Cheung, G., Kettenmann, H., Palmer, T. D., and Kempermann, G. (2007). Enriched monolayer precursor cell cultures from micro-dissected adult mouse dentate gyrus yield functional granule cell-like neurons. PLoS One 2:e388. doi: 10.1371/journal.pone.0000388
Bang, S., Lee, S.-R., Ko, J., Son, K., Tahk, D., Ahn, J., et al. (2017). A Low permeability microfluidic blood-brain barrier platform with direct contact between perfusable vascular network and astrocytes. Sci. Rep. 7:8083. doi: 10.1038/s41598-017-07416-7410
Blaeser, A., Duarte Campos, D. F., and Fischer, H. (2017). 3D bioprinting of cell-laden hydrogels for advanced tissue engineering. Curr. Opin. Biomed. Eng. 2, 58–66. doi: 10.1016/j.cobme.2017.04.003
Blaeser, A., Duarte Campos, D. F., Puster, U., Richtering, W., Stevens, M. M., and Fischer, H. (2015). Controlling shear stress in 3d bioprinting is a key factor to balance printing resolution and stem cell integrity. Adv. Healthc. Mater. 5, 326–333. doi: 10.1002/adhm.201500677
Brown, J. A., Pensabene, V., Markov, D. A., Allwardt, V., Neely, M. D., Shi, M., et al. (2015). Recreating blood-brain barrier physiology and structure on chip: a novel neurovascular microfluidic bioreactor. Biomicrofluidics 9:054124. doi: 10.1063/1.4934713
Carey, S. P., Starchenko, A., McGregor, A. L., and Reinhart-King, C. A. (2013). Leading malignant cells initiate collective epithelial cell invasion in a three-dimensional heterotypic tumor spheroid model. Clin. Exp. Metastasis 30, 615–630. doi: 10.1007/s10585-013-9565-x
Chung, C., Anderson, E., Pera, R. R., Pruitt, B. L., and Heilshorn, S. C. (2012a). Hydrogel crosslinking density regulates temporal contractility of human embryonic stem cell-derived cardiomyocytes in 3D cultures. Soft Matter 8, 10141–10148. doi: 10.1039/C2SM26082D
Chung, C., Lampe, K. J., and Heilshorn, S. C. (2012b). Tetrakis(hydroxymethyl) phosphonium chloride as a covalent cross-linking agent for cell encapsulation within protein-based hydrogels. Biomacromolecules 13, 3912–3916. doi: 10.1021/bm3015279
Cidonio, G., Glinka, M., Dawson, J. I., and Oreffo, R. O. C. (2019). The cell in the ink: improving biofabrication by printing stem cells for skeletal regenerative medicine. Biomaterials 209, 10–24. doi: 10.1016/j.biomaterials.2019.04.009
Cochrane, A., Albers, H. J., Passier, R., Mummery, C. L., van den Berg, A., Orlova, V. V., et al. (2019). Advanced in vitro models of vascular biology: human induced pluripotent stem cells and organ-on-chip technology. Adv. Drug Deliv. Rev. 140, 68–77. doi: 10.1016/j.addr.2018.06.007
Curtin, C., Nolan, J. C., Conlon, R., Deneweth, L., Gallagher, C., Tan, Y. J., et al. (2018). A physiologically relevant 3D collagen-based scaffold–neuroblastoma cell system exhibits chemosensitivity similar to orthotopic xenograft models. Acta Biomater. 70, 84–97. doi: 10.1016/J.ACTBIO.2018.02.004
Duarte Campos, D. F., Blaeser, A., Buellesbach, K., Sen, K. S., Xun, W., Tillmann, W., et al. (2016). Bioprinting Organotypic hydrogels with improved mesenchymal stem cell remodeling and mineralization properties for bone tissue engineering. Adv. Healthc. Mater. 5, 1336–1245. doi: 10.1002/adhm.201501033
Duarte Campos, D. F., Blaeser, A., Korsten, A., Neuss, S., Jäkel, J., Vogt, M., et al. (2015). The stiffness and structure of three-dimensional printed hydrogels direct the differentiation of mesenchymal stromal cells toward adipogenic and osteogenic lineages. Tissue Eng. Part A 21, 740–756. doi: 10.1089/ten.TEA.2014.0231
Duarte Campos, D. F., Zhang, S., Kreimendahl, F., Köpf, M., Fischer, H., Vogt, M., et al. (2019). Hand-held bioprinting for de novo vascular formation applicable to dental pulp regeneration. Connect. Tissue Res. 9, 1–11. doi: 10.1080/03008207.2019.1640217
Dubbin, K., Tabet, A., and Heilshorn, S. C. (2017). Quantitative criteria to benchmark new and existing bio-inks for cell compatibility. Biofabrication 9:044102. doi: 10.1088/1758-5090/aa869f
Forget, A., Blaeser, A., Miessmer, F., Köpf, M., Duarte Campos, D. F., Voelcker, N. H., et al. (2017). Mechanically tunable bioink for 3d bioprinting of human cells. Adv. Healthc. Mater. 6:1700255. doi: 10.1002/adhm.201700255
Foster, A. A., Marquardt, L. M., and Heilshorn, S. C. (2017). The diverse roles of hydrogel mechanics in injectable stem cell transplantation. Curr. Opin. Chem. Eng. 15, 15–23. doi: 10.1016/j.coche.2016.11.003
Gao, G., Kim, H., Kim, B. S., Kong, J. S., Lee, J. Y., Park, B. W., et al. (2019). Tissue-engineering of vascular grafts containing endothelium and smooth-muscle using triple-coaxial cell printing. Appl. Phys. Rev. 6:041402. doi: 10.1063/1.5099306
Gasperini, L., Mano, J. F., and Reis, R. L. (2014). Natural polymers for the microencapsulation of cells. J. R. Soc. Interface 11:20140817. doi: 10.1098/rsif.2014.0817
Gu, Q., Tomaskovic-Crook, E., Wallace, G. G., and Crook, J. M. (2018). “Engineering human neural tissue by 3D bioprinting,” in Methods in Molecular Biology, Ed. J. M. Walker (Totowa, NJ: Humana Press Inc), 129–138. doi: 10.1007/978-1-4939-7741-3_10
Jaeger, A. A., Das, C. K., Morgan, N. Y., Pursley, R. H., McQueen, P. G., Hall, M. D., et al. (2013). Microfabricated polymeric vessel mimetics for 3-D cancer cell culture. Biomaterials 34, 8301–8313. doi: 10.1016/j.biomaterials.2013.07.013
Katt, M. E., Placone, A. L., Wong, A. D., Xu, Z. S., and Searson, P. C. (2016). In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 4:12. doi: 10.3389/fbioe.2016.00012
Kreimendahl, F., Köpf, M., Thiebes, A. L., Duarte Campos, D. F., Blaeser, A., Schmitz-Rode, T., et al. (2017). Three-dimensional printing and angiogenesis: tailored Agarose-Type I collagen blends comprise three-dimensional printability and angiogenesis potential for tissue-engineered substitutes. Tissue Eng. Part C Methods 23, 604–615. doi: 10.1089/ten.tec.2017.0234
Langhans, S. A. (2018). Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 9:6. doi: 10.3389/fphar.2018.00006
Leberfinger, A. N., Dinda, S., Wu, Y., Koduru, S. V., Ozbolat, V., Ravnic, D. J., et al. (2019). Bioprinting functional tissues. Acta Biomater. 95, 32–49. doi: 10.1016/j.actbio.2019.01.009
LeSavage, B. L., Suhar, N. A., Madl, C. M., and Heilshorn, S. C. (2018). Production of elastin-like protein hydrogels for encapsulation and immunostaining of cells in 3D. J. Vis. Exp. 2018:57739. doi: 10.3791/57739
Leung, C. T., and Brugge, J. S. (2012). Outgrowth of single oncogene-expressing cells from suppressive epithelial environments. Nature 482, 410–413. doi: 10.1038/nature10826
Li, Y., and Kumacheva, E. (2018). Hydrogel microenvironments for cancer spheroid growth and drug screening. Sci. Adv. 4:aas8998. doi: 10.1126/sciadv.aas8998
Lindsay, C. D., Roth, J. G., LeSavage, B. L., and Heilshorn, S. C. (2019). Bioprinting of stem cell expansion lattices. Acta Biomater. 95, 225–235. doi: 10.1016/j.actbio.2019.05.014
Madl, C. M., Katz, L. M., and Heilshorn, S. C. (2016). Bio-orthogonally crosslinked, engineered protein hydrogels with tunable mechanics and biochemistry for cell encapsulation. Adv. Funct. Mater. 26, 3612–3620. doi: 10.1002/adfm.201505329
Madl, C. M., LeSavage, B. L., Dewi, R. E., Dinh, C. B., Stowers, R. S., Khariton, M., et al. (2017). Maintenance of neural progenitor cell stemness in 3D hydrogels requires matrix remodelling. Nat. Mater. 16, 1233–1242. doi: 10.1038/NMAT5020
Madl, C. M., LeSavage, B. L., Dewi, R. E., Lampe, K. J., and Heilshorn, S. C. (2019). Matrix remodeling enhances the differentiation capacity of neural progenitor cells in 3D Hydrogels. Adv. Sci. 6:1801716. doi: 10.1002/advs.201801716
Murphy, S. V., and Atala, A. (2014). 3D bioprinting of tissues and organs. Nat. Biotechnol. 32, 773–785. doi: 10.1038/nbt.2958
Ng, W. L., Lee, J. M., Yeong, W. Y., and Win Naing, M. (2017). Microvalve-based bioprinting-process, bio-inks and applications. Biomater. Sci. 5, 632–647. doi: 10.1039/c6bm00861e
Petersen, O. W., Ronnov-Jessen, L., Howlett, A. R., and Bissell, M. J. (1992). Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 89, 9064–9068. doi: 10.1073/pnas.89.19.9064
Phamduy, T. B., Sweat, R. S., Azimi, M. S., Burow, M. E., Murfee, W. L., and Chrisey, D. B. (2015). Printing cancer cells into intact microvascular networks: a model for investigating cancer cell dynamics during angiogenesis. Integr. Biol. 7, 1068–1078. doi: 10.1039/c5ib00151j
Ravi, M., Paramesh, V., Kaviya, S. R., Anuradha, E., and Paul Solomon, F. D. (2015). 3D cell culture systems: advantages and applications. J. Cell. Physiol. 230, 16–26. doi: 10.1002/jcp.24683
Salinas-Fernández, S., Santos, M., Alonso, M., Quintanilla, L., and Rodríguez-Cabello, J. C. (2019). Genetically engineered elastin-like recombinamers with sequence-based molecular stabilization as advanced bioinks for 3D bioprinting. Appl. Mater. Today 2019:100500. doi: 10.1016/j.apmt.2019.100500
Schänzer, A., Wachs, F.-P., Wilhelm, D., Acker, T., Cooper-Kuhn, C., Beck, H., et al. (2006). Direct stimulation of adult neural stem cells in vitro and neurogenesis in vivo by vascular endothelial growth factor. Brain Pathol. 14, 237–248. doi: 10.1111/j.1750-3639.2004.tb00060.x
Shen, Q., Goderie, S. K., Jin, L., Karanth, N., Sun, Y., Abramova, N., et al. (2004). Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science 304, 1338–1340. doi: 10.1126/science.1095505
Shi, Y., Kirwan, P., and Livesey, F. J. (2012). Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 7, 1836–1846. doi: 10.1038/nprot.2012.116
Sontheimer-Phelps, A., Hassell, B. A., and Ingber, D. E. (2019). Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 19, 65–81. doi: 10.1038/s41568-018-0104-106
Straley, K. S., and Heilshorn, S. C. (2009). Independent tuning of multiple biomaterial properties using protein engineering. Soft Matter 5, 114–124. doi: 10.1039/b808504h
Tavazoie, M., Van der Veken, L., Silva-Vargas, V., Louissaint, M., Colonna, L., Zaidi, B., et al. (2008). A specialized vascular niche for adult neural stem cells. Cell Stem Cell 3, 279–288. doi: 10.1016/j.stem.2008.07.025
Zhang, Y. S., Duchamp, M., Oklu, R., Ellisen, L. W., Langer, R., and Khademhosseini, A. (2016). Bioprinting the cancer microenvironment. ACS Biomater. Sci. Eng. 2, 1710–1721. doi: 10.1021/acsbiomaterials.6b00246
Keywords: protein engineered hydrogel, bioink, bioprinting, 3D cell culture, tissue model
Citation: Duarte Campos DF, Lindsay CD, Roth JG, LeSavage BL, Seymour AJ, Krajina BA, Ribeiro R, Costa PF, Blaeser A and Heilshorn SC (2020) Bioprinting Cell- and Spheroid-Laden Protein-Engineered Hydrogels as Tissue-on-Chip Platforms. Front. Bioeng. Biotechnol. 8:374. doi: 10.3389/fbioe.2020.00374
Received: 25 January 2020; Accepted: 06 April 2020;
Published: 28 April 2020.
Edited by:
Julien Georges Didier Barthès, Protip Medical, FranceReviewed by:
Stephanie Michelle Willerth, University of Victoria, CanadaCopyright © 2020 Duarte Campos, Lindsay, Roth, LeSavage, Seymour, Krajina, Ribeiro, Costa, Blaeser and Heilshorn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela F. Duarte Campos, ZGFuaWVsYS5jYW1wb3NAcnd0aC1hYWNoZW4uZGU=; ZGNhbXBvc0B1a2FhY2hlbi5kZQ==; Sarah C. Heilshorn, aGVpbHNob3JuQHN0YW5mb3JkLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.