- 1Department of Agricultural Chemistry and Food Science Technology, Division of Applied Life Science (BK21 Plus Program), Institute of Agriculture & Life Science (IALS), Gyeongsang National University, Jinju, South Korea
- 2Department of Life Sciences, CHRIST (Deemed to be University), Bengaluru, India
- 3Applied and Industrial Biotechnology Laboratory (AIBL), Department of Life Sciences, CHRIST (Deemed to be University), Bengaluru, India
Strains of Clostridium genus are used for production of various value-added products including fuels and chemicals. Development of any commercially viable production process requires a combination of both strain and fermentation process development strategies. The strain development in Clostridium sp. could be achieved by random mutagenesis, and targeted gene alteration methods. However, strain improvement in Clostridium sp. by targeted gene alteration method was challenging due to the lack of efficient tools for genome and transcriptome engineering in this organism. Recently, various synthetic biology tools have been developed to facilitate the strain engineering of solventogenic Clostridium. In this review, we consolidated the recent advancements in toolbox development for genome and transcriptome engineering in solventogenic Clostridium. Here we reviewed the genome-engineering tools employing mobile group II intron, pyrE alleles exchange, and CRISPR/Cas9 with their application for strain development of Clostridium sp. Next, transcriptome engineering tools such as untranslated region (UTR) engineering and synthetic sRNA techniques were also discussed in context of Clostridium strain engineering. Application of any of these discussed techniques will facilitate the metabolic engineering of clostridia for development of improved strains with respect to requisite functional attributes. This might lead to the development of an economically viable butanol production process with improved titer, yield and productivity.
Introduction
Strain improvement for production of fuels or any biobased industrial product could be achieved by employing any of the following two strategies: (i) heterologous expression of metabolic pathway genes in a non-native producers, and (ii) improvement of native producers (Arora et al., 2019; Banerjee et al., 2019; Choi et al., 2019). However, achieving titer values in heterologous host matching to those being produced by native organisms, it requires a significant effort with high chances of failure. Therefore, the strategy of improving native strains with necessary genes of the desired pathway and cofactor regeneration capability is preferred (Park et al., 2018; Rhie et al., 2019). However, this strategy of strain improvement in Clostridium sp. has been limited by the availability of appropriate genome engineering tools.
Clostridium genus comprises many industrially important strains for biorefinery applications such as cellulosic and hemicellulosic biomass degradation, carbon fixation, advanced biofuel and platform chemical production and as anti-cancer therapeutics (Jang et al., 2012; Malaviya et al., 2012; Liu J. et al., 2015; Jones et al., 2016; Staedtke et al., 2016; Noh et al., 2018; Woo et al., 2018; Xin et al., 2018; Strecker et al., 2019a). The full potential of Clostridium genus for biorefinery applications could only be realized by advancement in the synthetic biology toolkits for strain improvement. During the last decade, tremendous progresses have been made in the development of genome engineering toolkit for strain engineering of Clostridium species. Development of genetic tools in Clostridium have been well reviewed by various research groups (Pyne et al., 2014; Liu Y. J. et al., 2015; Minton et al., 2016; Moon et al., 2016; Joseph et al., 2018; Kuehne et al., 2019; McAllister and Sorg, 2019; Wen et al., 2019b, c). Most of these reports are focused on couple of tools with an explanation in depth.
In this work, we have reviewed overall recent toolbox for genome and transcriptome engineering in solventogenic Clostridium, which could be used to develop improved clostridia strains, for production of sustainable and commercially viable industrial scale products. Brief features of the synthetic toolbox are summarized in Table 1. Consolidated information in this review dealing with strain improvement tools for Clostridium will aid the scientific and industrial sector to select the appropriate tools for strain improvement.
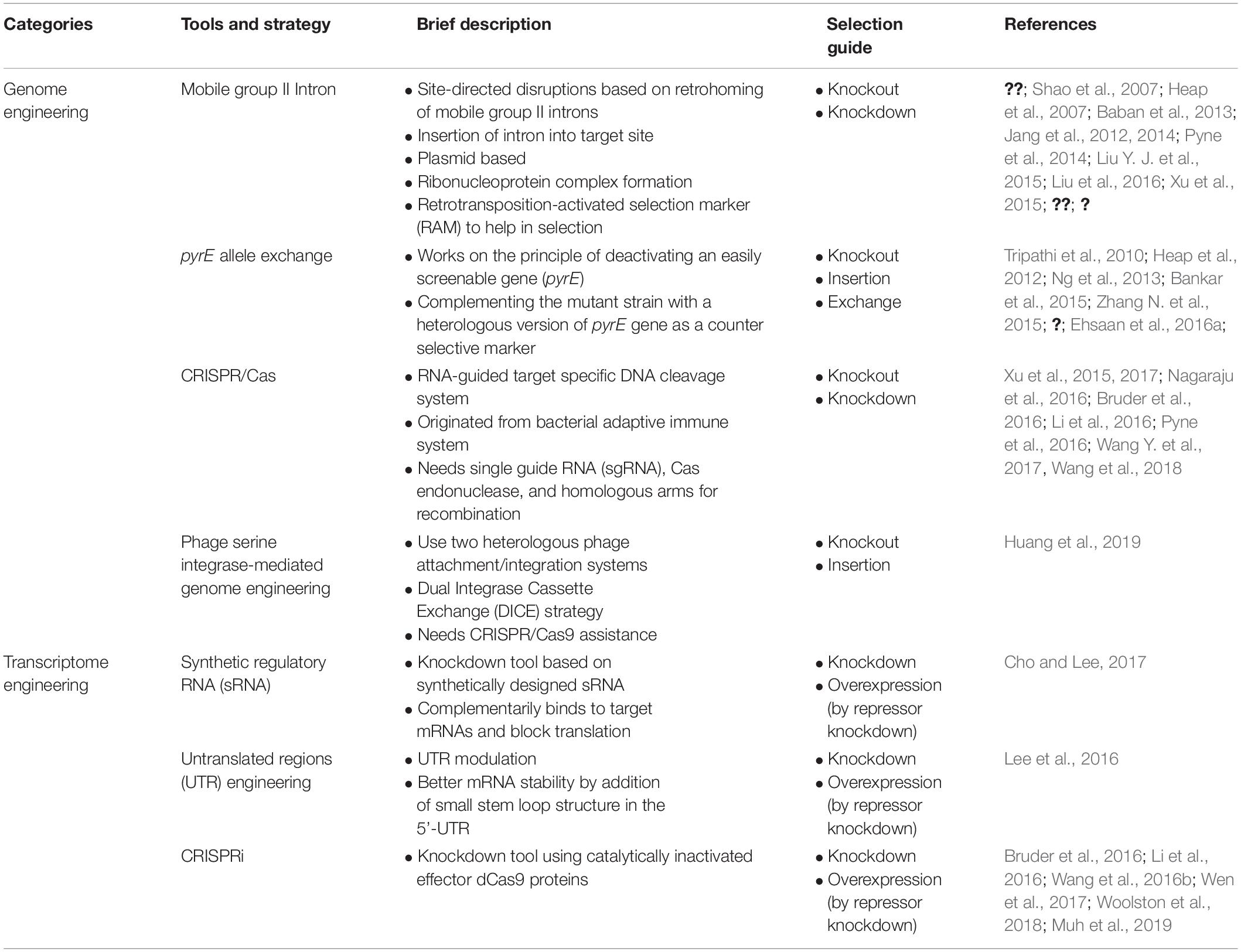
Table 1. Summary of synthetic biology tools and strategies applied for genome and transcriptome engineering of solventogenic Clostridium.
Mobile Group Ii Intron Based Gene-Knockout
Mobile group II intron technology is also known as “ClosTron” when applied in context of Clostridium genus. In this method a gene is disrupted by inserting the mobile intron into a target locus in the chromosome by a process termed as retrohoming, making this technology a convenient, efficient and specific method of gene disruption (Heap et al., 2007, 2010; Shao et al., 2007; Jang et al., 2012, 2014; Mohr et al., 2013; Liu Y. J. et al., 2015). Among various mobile group II introns, Ll.LtrB and TeI3c/4c have been extensively used for gene knockout in the solventogenic Clostridium. Ll.LtrB intron includes intron RNA domain and open reading frame (ORF) domain. Intron RNA domain contains splicing sites consisting of exon binding sites (EBS) 1, EBS 2, and δ (Figure 1A). The ORF domain contains genes encoding reverse transcriptase (RTase), maturase, and endonuclease (Figure 1A). TeI3c/4c intron has been employed to develop genome engineering tool for thermophilic Clostridium thermocellum, since the intron could be melted down at high temperatures (Mohr et al., 2013).
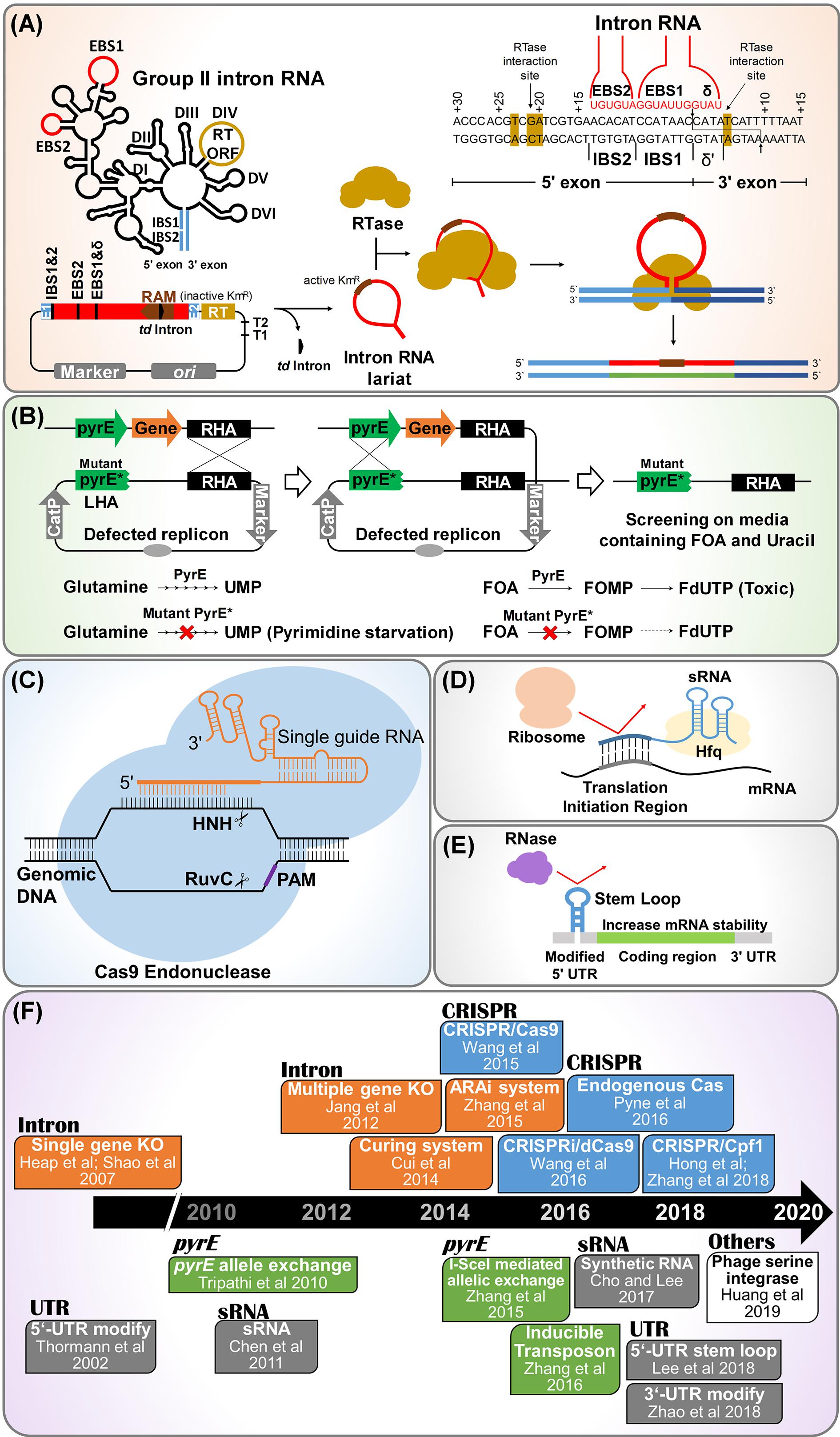
Figure 1. Synthetic biology tools developed for genome and transcriptome engineering of solventogenic Clostridium. (A) Mobile group II intron-based genome engineering. Also known as ClosTron in context of Clostridium sp. In this technology, site directed gene disruption is achieved by insertion of the mobile group II intron into the target locus of chromosome. Abbreviations: RAM, retrotranscription-activated marker (typically kanamycin resistant marker containing self-splicing group I intron, phage T4 td intron); RTase, reverse transcriptase; EBS, exon binding site; IBS, intron binding site. (B) pyrE based allele exchange technology for genome engineering. Here, pyrE encoding orotate phosphoribosyl transferase is used as counter selection marker to ensure double crossover event. The pyrE-mutant (PyrE∗) and wild type (PyrE) are resistant and sensitive to 5-fluoroorotic acid (FOA), respectively. Abbreviation: RHA, right homology arm. (C) CRISPR/Cas system for genome engineering. This needs single guide RNA containing crRNA and tracrRNA, Cas endonuclease, and homologous arm for recombination. Abbreviation: PAM, protospacer-adjacent motif. (D) Synthetic regulatory RNA (sRNA) based knockdown strategy. sRNA are having regulatory role in gene expression, mediated by chaperon Hfq. sRNA binds to complimentary mRNA sequences, prohibiting ribosome clamping at ribosome binding site located in translation initiation region. (E) 5′-UTR engineering for regulation of gene expression. The insertion of a small stem loop structure in the 5′-UTR increases the mRNA stability by blocking RNase, resulting in a high gene expression. (F) Timeline of notable events in the development of synthetic biology tools for genome and transcriptome engineering of solventogenic Clostridium.
Moreover, Ll.LtrB intron has further been modified to include a retrotransposition-activated selection marker (RAM) (Zhong et al., 2003). RAM consists of a selection marker and is inserted into the intron. A group I intron is inserted into the marker to inactivate the marker itself. Inserted group I intron is self catalytically spliced out of mRNA in an orientation dependent manner, so that a functional marker gene can only be expressed after successful chromosomal insertion occurs (Joseph et al., 2018).
At the first stage of the clostridia gene knockout using Ll.LtrB intron, single gene knockouts mutant, such as spo0A, pta, ack, ptb, buk, hbd, hydA and argA variants have been constructed across the Clostridium genus, including C. acetobutylicum, C. beijerinckii, C. botulinum, and C. difficile (Heap et al., 2010; Dingle et al., 2011; Jang et al., 2012; Baban et al., 2013; Honicke et al., 2014; Lawson and Rainey, 2016; Liu et al., 2016). In 2012, a new method for second gene deletion was reported which could overcome the necessity of removing the plasmid used for the first gene deletion and resulted in the construction of various C. acetobutylicum strains, including pta/buk, pta/ctfB, ptb/buk, and triple mutant pta/buk/ctfB strains (Jang et al., 2012). In this technique, two genes encoding the erythromycin and chloramphenicol resistance enzymes were used as mutant selection marker and the concept of plasmid incompatibility was employed (Jang et al., 2012). In 2014, the same group reported the fourth and fifth gene deletion process for the construction of mutants pta/buk/ctfB/adhE1 and pta/buk/ctfB/adhE1/hydA of C. acetobutylicum (Jang et al., 2014).
Curing and off-target manipulation remained one of the major limitations of mobile group II intron technology (Wen et al., 2019c). Curing efficiency of the plasmid containing mobile intron was enhanced by cloning pyrF (orotidine 5-phosphate decarboxylase) to ClosTron plasmid. The pyrF encodes essential enzyme of pyrimidine biosynthesis which can use 5-fluoroorotic acid (FOA) as a substrate and converts it to toxic compound and is widely used as counter selection marker (Sato et al., 2005; Tripathi et al., 2010; Heap et al., 2012). Once FOA gets converted to toxic compound by pyrF in the ClosTron plasmid, only cured strain could survive in the FOA added media. The cured strain can be rapidly selected by pyrF-based screening system, even on one plate (Cui et al., 2014).
Another problem with ClosTron is that it accidently affects and manipulates the off-target genome and cause unexpected genotypes and phenotypes (Heap et al., 2012). To overcome this, a highly regulated ClosTron system has been developed by inducing L-arabinose inducer (ARAi) to reduce off-target possibility (Zhang J. et al., 2015). To verify the impact of inducible ClosTron using ARAi system, pSY6-mspI (Cui et al., 2012) and pGZ-pyrF-cipC (Cui et al., 2014) were modified by introducing ARAi system in C. cellulolyticum H10 ΔpyrF strain. Surprisingly, it was found that the off-target manipulation frequency was decreased to 0 by inducible ClosTron ARAi system (Zhang J. et al., 2015).
Genome Editing Using PYRE Alleles
Recently, allele coupled exchange (ACE) method has been developed which facilitates the insertion of complex heterologous DNA of varying size into the host genome (Ng et al., 2013; Zhang N. et al., 2015; Ehsaan et al., 2016a; Minton et al., 2016). In ACE, a counter selection marker is coupled to a desired double crossover event (Figure 1B). The counter selection marker entitles the isolation of double cross over through homologous recombination. The pyrE and codA genes are the most frequently used selectable marker in ACE Technology. The gene codA encodes for the enzyme cytosine deaminase while, pyrE encodes orotate phosphoribosyl transferase, which is a key enzyme required in the de novo pathway for pyrimidine biosynthesis.
In clostridia genome editing, pyrE allele has been primarily employed. Mutant and wild type pyrE allele confers resistance and sensitivity to FOA, respectively. The advantages of pyrE allele based recombination includes: (i) rapid insertion of heterologous DNA, (ii) double crossovers which forms the stable integration, (iii) allows large insert size, and (iv) has higher efficiency as compared to simple ClosTron and random mutagenesis (Ng et al., 2013; Ehsaan et al., 2016b; Minton et al., 2016).
The pyrE cassettes consists of two arms, i.e., right homology arm (RHA) and left homology arm (LHA) with the internal region comprising of pyrE gene (Figure 1B). A plasmid is constructed with a selectable marker (antibiotic resistance gene), origin of replication and a sequence containing ∼300-bp homologous to pyrE gene and a longer sequence of ∼1,200-bp homologous to the adjacent region of 3′ end of pyrE. Once the pyrE based pseudo-suicide plasmid is delivered into Clostridium cells, single crossover is formed through homologous recombination. Subsequently, the single crossover mutant is inoculated into the media containing FOA and uracil (Heap et al., 2012). Metabolization of FOA kills the single crossover cells carrying the active pyrE gene. Inactivation of pyrE happens only if double recombination had occurred on both 1200-bp long sequence and 300-bp short sequence and the FOA does not affect the cells obtained by such double crossovers (Ng et al., 2013). The final double crossovers are formed by ACE of shorter left homology arm of 300-bp by the second single crossover, which also leads to the excision of the plasmid (Minton et al., 2016). This technology has been found to be applicable for many species of Clostridium genus (Heap et al., 2012).
Butanol yield in C. pasteurianum has been reported to be improved by application of pyrE based genome editing toolkit. For this, deletion mutations were created in three genes of C. pasteurianum: hydrogenase (hydA), redox response regulator (rex), and glycerol dehydratase (dhaBCE), using plasmid pMTL-KS01. This resulted in increased availability of NADPH in cell due to depletion of 1,3-propanediol synthesis, which eventually contributed to improved butanol production (Schwarz et al., 2017). Similarly, successful expression of cellulosomal subunits in C. acetobutylicum has also been achieved using this method (Kovacs et al., 2013). Few other Clostridium species modified using ACE technology includes C. acetobutylicum, C. sporogenes, and C. difficile (Heap et al., 2012; Ng et al., 2013; Bankar et al., 2015; Zhang J. et al., 2015; Ehsaan et al., 2016b; Minton et al., 2016; Willson et al., 2016).
CRISPR/Cas Based Clostridia Genome Engineering
Clustered regulatory interspaced short palindromic repeats (CRISPR) have been developed as one of the most advanced genetic engineering tools along with CRISPR-associated (Cas) protein (Doudna and Charpentier, 2014). As bacterial genome manipulation tool, CRISPR/Cas system needs single guide RNA (sgRNA), Cas endonuclease, and homologous arms for recombination (Jiang et al., 2013). The Streptococcus pyogenes type II CRISPR was the first CRISPR system which was exploited for genome engineering applications. Cas9 endonuclease is the basis of CRISPR based genome editing system. Cas9 recognize the protospacer adjacent motif (PAM) site (5′-NGG-3′ in S. pyogenes) and cleave at the 3′ end of the target gene (Mojica et al., 2009; Garneau et al., 2010; Jinek et al., 2012) (Figure 1C).
Various strains of Clostridium genus have been manipulated using the CRISPR/Cas9 system including C. acetobutyricum (Bruder et al., 2016; Li et al., 2016; Wasels et al., 2017), C. beijerinkii (Wang et al., 2016b), C. autoethanogenum (Nagaraju et al., 2016), C. difficile (McAllister et al., 2017; Wang et al., 2018), C. cellulolyticum (Xu et al., 2015, 2017), C. pasteurianum (Pyne et al., 2016), C. ljungdahlii (Huang et al., 2016), and C. saccharoperbutylacetonicum (Wang S. et al., 2017). However, the expression of Cas9 becomes detrimental for bacteria, including clostridia, in terms of the toxicity it causes. The mutation of 10th amino acid (aspartic acid to alanine) in Cas9 inactivates it’s RuvC-like nuclease domain resulting in formation of Cas9 nickase (Cas9n), which can cleave only single-strand of DNA (Jinek et al., 2012; Nishimasu et al., 2014; Swarts and Jinek, 2018; Li et al., 2019). Cas9n have advantage in terms of overcoming the toxicity caused by expression of Cas9. Introduction of highly regulated inducible promoter for Cas9 expression is another strategy to circumvent the associated toxicity (Wang et al., 2016a; McAllister et al., 2017; Wasels et al., 2017). Nevertheless, the CRISPR/Cas9n system is still being used for clostridia genome editing (Wang et al., 2016b; Wang S. et al., 2017; Wang Y. et al., 2017; Wang et al., 2018; McAllister et al., 2017; Wasels et al., 2017).
Moreover, modified CRISPR systems like CRISPR interference (CRISPRi) and dCas9 has also been developed to knockdown of the essential genes required for host survival (Jinek et al., 2012; Qi et al., 2013; Peters et al., 2016; Zheng et al., 2019). The dCas9 has two silenced catalytic domains (D10A and H840A; RuvC-like and HNH domains, respectively) which remains bound and block the target DNA instead of cleavage. CRISPRi/dCas9 system has also been applied to develop several mutant strains of Clostridium sp. (Bruder et al., 2016; Wang et al., 2016a, b; Wen et al., 2017; Woolston et al., 2018; Muh et al., 2019).
Similar to Cas9, the Cpf1 from Acidaminococcus sp. is another protein that is used for PAM recognition in CRISPR based system. While Cas9 recognizes G-rich PAM site, the PAM recognition site for Cpf1 is T-rich (5′-TTTN-3′) (Swarts and Jinek, 2018) making it best suited for application in AT-rich organisms like Clostridium sp. (Zetsche et al., 2015; Yamano et al., 2016). Single CRISPR/Cpf1 system plasmid can make multiple mutants in a single application (Zetsche et al., 2017; Hong et al., 2018; Zhang et al., 2018a). CRISPR/Cpf1 system has been applied in C. ljungdahlii, C. difficile, and C. beijerinckii (Hong et al., 2018; Zhang et al., 2018a; Zhao et al., 2019).
Additionally, endogenous CRISPR systems have been developed in C. pasteurianum and C. tyrobutyricum to overcome the toxic effect associated with Cas9 and Cpf1 endonucleases (Pyne et al., 2016; Zhang et al., 2018b). The endogenous CRISPR system uses endonuclease encoded by the genome and can contain multiple pre-crRNAs under one promoter, facilitating multiple genome modification using a single plasmid (Luo et al., 2014; Makarova et al., 2015; Pyne et al., 2016; Zhang et al., 2018b).
Synthetic sRNA and Untranslated Region Engineering as Potential Domains for Clostridium Strain Improvement
Prokaryotic small RNAs (sRNA) are short strands of ribonucleotides (about 50–500 nucleotides) which have a regulatory role in maintaining the cellular processes (Gottesman, 2004). Based on the existence of natural sRNA, synthetic small RNAs are produced to alter the gene expression of the organisms. Many such naturally occurring sRNAs have been detected and analyzed in Clostridium sp. (Chen et al., 2011), which leads to the development of synthetic sRNA (Na et al., 2013).
The sRNA mediated gene expression usually results in repression of the gene which complements the sRNA nucleotide sequence, mediated by a protein called Hfq (De Lay et al., 2013). Hfq is the chaperone mediated protein which stabilizes the sRNA-mRNA binding. The translation process is prevented by sRNA binding to ribosome binding site (RBS) or by masking the access to the start codon (Na et al., 2013; Yoo et al., 2013). Recently, Cho and Lee (2017) have reported the development of synthetic small regulatory RNA (sRNA) system for controlled gene expression in C. acetobutylicum, consisting of a target recognition site, MicC scaffold, and an RNA chaperon Hfq (Figure 1D). In this study, C. acetobutylicum Hfq was found to be ineffective in binding with Escherichia coli MicC scaffold-based synthetic sRNA, however Hfq from E. coli itself resulted in much enhanced knockdown efficiency. This E. coli MicC-Hfq sRNA system was used to knockdown adhE1 gene expression resulting in 40% reduction in butanol production. Further, this synthetic sRNA system was used to knockdown the pta gene expression in PJC4BK strain, resulting in PJC4BK (pPta-HfqEco) strain with improvement of butanol titer from 14.9 to 16.9 g/l (Cho and Lee, 2017).
Untranslated regions (UTRs) are non-coding regions in the mRNA helps to regulate the gene expression. UTRs are present on both the ends of the mRNA (5′-UTR and 3′-UTR) (Figure 1E). There are sufficient reports to confirm that the 5′-UTR in C. acetobutylicum has the regulatory effect on the secondary structure of enzyme adhE1, which is involved in solvent production (Thormann et al., 2002; Scotcher et al., 2003). Lee et al. (2016) has recently found that the presence of a single stranded short 5′-UTR in the solventogenic C. acetobutylicum leads to decreased gene expression (Figure 1E). The insertion of a small stem loop structure in the 5′-UTR was found to increase the mRNA stability and gene expression by 4.6 folds, without any modification in the promoter or RBS (Lee et al., 2016). On the other hand, the 3′-UTR mostly harbors the terminator sequence for transcription process in mRNA (Richard and Manley, 2009). sRNA sequence containing the codons that regulates the post transcriptional and translation machinery is also attached to 3′-UTR. Most importantly 3′-UTR confer stability to the mRNA (Zhao et al., 2018). Although, there are very limited studies related to 3′-UTR regions in Clostridium, the presence of transcripts with long 3′-UTR is confirmed in Clostridium (Ralston and Papoutsakis, 2018). Although several RNAseq studies were reported in the Clostridium, only few studies show the data related to regulation of mRNA based on 5′- and 3′-UTRs, leaderless transcripts and non-coding RNA (Soutourina et al., 2013; Wilson et al., 2013; Sedlar et al., 2018). Further research in RNAseq and proteomics will explore the complex regulations that control mRNA stability and degradation, which will be more useful to construction synthetic toolkit.
In conclusion, many Clostridium sp. have potential to be utilized at industrial scale to produce value added chemicals, including butanol as fossil fuel substitute. Up to date, their true potential was underexploited due to challenges in strain improvement and unavailability of genome and transcriptome editing tools for this genus. Nevertheless, during the last decade, synthetic biology toolkits for Clostridium sp. have been expanded rapidly (Figure 1F). Furthermore, a recent advancement, such as phage serine integrase mediated site-specific genome engineering technique for C. ljungdahlii could be extended to other Clostridium species (Huang et al., 2019). The synthetic biology techniques that have been applied in other microorganisms may also be adopted to solventogenic clostridia in the near future: CRISPR associated site-specific insertion of transposons and base editing techniques (Ronda et al., 2015; Zhang et al., 2016; Lim and Choi, 2019; Strecker et al., 2019b). Utilization of improved clostridia strains could be a starting point for development of an industrial scale, commercially viable bio-based fuel and chemical production using Clostridium sp. using a consolidated bioprocessing concept (Wen et al., 2019a). Furthermore, these synthetic biology tools could be applied to another biotechnology fields such as degradation of plastics, such as polyethylene terephthalate and polyethylene.
Author Contributions
Y-SJ and AM conceived the project. All authors analyzed the literature, compiled data, planned content, wrote the manuscript, read, and approved the final manuscript.
Funding
This work was supported by a grant from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea (NRF-2019R1A4A1029125). This work was further supported by Cooperative Research Program for Agricultural Science and Technology Development (Project No. PJ01492601), Rural Development Administration, Republic of Korea. AM was supported by a grant from Centre for Research Projects, CHRIST (Deemed to be University), Bangalore (MRPDSC-1829).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Arora, R., Behera, S., Sharma, N. K., and Kumar, S. (2019). Evaluating the pathway for co-fermentation of glucose and xylose for enhanced bioethanol production using flux balance analysis. Biotechnol. Bioproc. Eng. 24, 924–933. doi: 10.1007/s12257-019-0026-5
Baban, S. T., Kuehne, S. A., Barketi-Klai, A., Cartman, S. T., Kelly, M. L., Hardie, K. R., et al. (2013). The role of flagella in Clostridium difficile pathogenesis: comparison between a non-epidemic and an epidemic strain. PLoS One 8:e73026. doi: 10.1371/journal.pone.0073026
Banerjee, S., Mishra, G., and Roy, A. (2019). Metabolic engineering of bacteria for renewable bioethanol production from cellulosic biomass. Biotechnol. Bioproc. Eng. 24, 713–733. doi: 10.1007/s12257-019-0134-2
Bankar, S. B., Jurgens, G., Survase, S. A., Ojamo, H., and Granström, T. (2015). Genetic engineering of Clostridium acetobutylicum to enhance isopropanol-butanol-ethanol production with an integrated DNA-technology approach. Renew. Energy 83, 1076–1083. doi: 10.1016/j.renene.2015.05.052
Bruder, M. R., Pyne, M. E., Moo-Young, M., Chung, D. A., and Chou, C. P. (2016). Extending CRISPR-Cas9 technology from genome editing to transcriptional engineering in the genus Clostridium. Appl. Environ. Microbiol. 82, 6109–6119. doi: 10.1128/aem.02128-16
Chen, Y., Caruso, L., McClane, B., Fisher, D., and Gupta, P. (2007). Disruption of a toxin gene by introduction of a foreign gene into the chromosome of Clostridium perfringens using targetron-induced mutagenesis. Plasmid 58, 182–189. doi: 10.1016/j.plasmid.2007.04.002
Chen, Y., Indurthi, D. C., Jones, S. W., and Papoutsakis, E. T. (2011). Small RNAs in the genus Clostridium. mBio 2:e340-10.
Chen, Y., McClane, B. A., Fisher, D. J., Rood, J. I., and Gupta, P. (2005). Construction of an alpha toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl. Environ. Microbiol. 71, 7542–7547. doi: 10.1128/AEM.71.11.7542-7547.2005
Cho, C., and Lee, S. Y. (2017). Efficient gene knockdown in Clostridium acetobutylicum by synthetic small regulatory RNAs. Biotechnol. Bioeng. 114, 374–383. doi: 10.1002/bit.26077
Choi, Y. Y., Hong, M.-E., Chang, W. S., and Sim, S. J. (2019). Autotrophic biodiesel production from the thermotolerant microalga Chlorella sorokiniana by enhancing the carbon availability with temperature adjustment. Biotechnol. Bioproc. Eng. 24, 223–231. doi: 10.1007/s12257-018-0375-5
Croux, C., Nguyen, N. P., Lee, J., Raynaud, C., Saint-Prix, F., Gonzalez-Pajuelo, M., et al. (2016). Construction of a restriction-less, marker-less mutant useful for functional genomic and metabolic engineering of the biofuel producer Clostridium acetobutylicum. Biotechnol. Biofuels 9:23. doi: 10.1186/s13068-016-0432-2
Cui, G. Z., Hong, W., Zhang, J., Li, W. L., Feng, Y., Liu, Y. J., et al. (2012). Targeted gene engineering in Clostridium cellulolyticum H10 without methylation. J. Microbiol. Methods 89, 201–208. doi: 10.1016/j.mimet.2012.02.015
Cui, G. Z., Zhang, J., Hong, W., Xu, C., Feng, Y., Cui, Q., et al. (2014). Improvement of ClosTron for successive gene disruption in Clostridium cellulolyticum using a pyrF-based screening system. Appl. Microbiol. Biotechnol. 98, 313–323. doi: 10.1007/s00253-013-5330-y
De Lay, N., Schu, D. J., and Gottesman, S. (2013). Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 288, 7996–8003. doi: 10.1074/jbc.r112.441386
Dingle, T. C., Mulvey, G. L., and Armstrong, G. D. (2011). Mutagenic analysis of the Clostridium difficile flagellar proteins, FliC and FliD, and their contribution to virulence in hamsters. Infect. Immun. 79, 4061–4067. doi: 10.1128/iai.05305-11
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096.
Ehsaan, M., Kuehne, S. A., and Minton, N. P. (2016a). “Clostridium difficile genome editing using pyrE alleles,” in Clostridium Difficile, eds A. P. Roberts and P. Mullany, (Berlin: Springer), 35–52. doi: 10.1007/978-1-4939-6361-4_4
Ehsaan, M., Kuit, W., Zhang, Y., Cartman, S. T., Heap, J. T., Winzer, K., et al. (2016b). Mutant generation by allelic exchange and genome resequencing of the biobutanol organism Clostridium acetobutylicum ATCC 824. Biotechnol. Biofuels 9:4.
Garneau, J. E., Dupuis, M. E., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. doi: 10.1038/nature09523
Gottesman, S. (2004). The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58, 303–328. doi: 10.1146/annurev.micro.58.030603.123841
Heap, J. T., Ehsaan, M., Cooksley, C. M., Ng, Y. K., Cartman, S. T., Winzer, K., et al. (2012). Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucleic Acids Res. 40:e59. doi: 10.1093/nar/gkr1321
Heap, J. T., Kuehne, S. A., Ehsaan, M., Cartman, S. T., Cooksley, C. M., Scott, J. C., et al. (2010). The ClosTron: mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods 80, 49–55. doi: 10.1016/j.mimet.2009.10.018
Heap, J. T., Pennington, O. J., Cartman, S. T., Carter, G. P., and Minton, N. P. (2007). The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70, 452–464. doi: 10.1016/j.mimet.2007.05.021
Hong, W., Zhang, J., Cui, G., Wang, L., and Wang, Y. (2018). Multiplexed CRISPR-Cpf1-mediated genome editing in Clostridium difficile toward the understanding of pathogenesis of C. difficile infection. ACS Synth. Biol. 7, 1588–1600. doi: 10.1021/acssynbio.8b00087
Honicke, D., Lutke-Eversloh, T., Liu, Z., Lehmann, D., Liebl, W., and Ehrenreich, A. (2014). Chemostat cultivation and transcriptional analyses of Clostridium acetobutylicum mutants with defects in the acid and acetone biosynthetic pathways. Appl. Microbiol. Biotechnol. 98, 9777–9794. doi: 10.1007/s00253-014-6040-9
Huang, H., Chai, C., Li, N., Rowe, P., Minton, N. P., Yang, S., et al. (2016). CRISPR/Cas9-Based efficient genome editing in Clostridium ljungdahlii, an autotrophic gas-fermenting bacterium. ACS Synth. Biol. 5, 1355–1361. doi: 10.1021/acssynbio.6b00044
Huang, H., Chai, C., Yang, S., Jiang, W., and Gu, Y. (2019). Phage serine integrase-mediated genome engineering for efficient expression of chemical biosynthetic pathway in gas-fermenting Clostridium ljungdahlii. Metab. Eng 52, 293–302. doi: 10.1016/j.ymben.2019.01.005
Jang, Y.-S., Im, J. A., Choi, S. Y., Lee, J. I., and Lee, S. Y. (2014). Metabolic engineering of Clostridium acetobutylicum for butyric acid production with high butyric acid selectivity. Metab. Eng. 23, 165–174. doi: 10.1016/j.ymben.2014.03.004
Jang, Y. S., Lee, J. Y., Lee, J., Park, J. H., Im, J. A., Eom, M. H., et al. (2012). Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum. mBio 3:e00314-12.
Jiang, W., Bikard, D., Cox, D., Zhang, F., and Marraffini, L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239. doi: 10.1038/nbt.2508
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jones, A. J., Venkataramanan, K. P., and Papoutsakis, T. (2016). Overexpression of two stress-responsive, small, non-coding RNAs, 6S and tmRNA, imparts butanol tolerance in Clostridium acetobutylicum. FEMS Microbiol. Lett. 363:fnw063. doi: 10.1093/femsle/fnw063
Joseph, R. C., Kim, N. M., and Sandoval, N. R. (2018). Recent developments of the synthetic biology toolkit for Clostridium. Front. Microbiol. 9:154. doi: 10.3389/fmicb.2018.00154
Kovacs, K., Willson, B. J., Schwarz, K., Heap, J. T., Jackson, A., Bolam, D. N., et al. (2013). Secretion and assembly of functional mini-cellulosomes from synthetic chromosomal operons in Clostridium acetobutylicum ATCC 824. Biotechnol. Biofuels 6:117. doi: 10.1186/1754-6834-6-117
Kuehne, S. A., Rood, J. I., and Lyras, D. (2019). Clostridial genetics: genetic manipulation of the pathogenic clostridia. Microbiol Spectr 7:GPP3-0040-2018.
Lawson, P. A., and Rainey, F. A. (2016). Proposal to restrict the genus Clostridium prazmowski to Clostridium butyricum and related species. Int. J. Syst. Evol. Microbiol. 66, 1009–1016. doi: 10.1099/ijsem.0.000824
Lawson, P. A., Citron, D. M., Tyrrell, K. L., and Finegold, S. M. (2016). Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prevot 1938. Anaerobe 40, 95–99. doi: 10.1016/j.anaerobe.2016.06.008
Lee, J., Jang, Y. S., Papoutsakis, E. T., and Lee, S. Y. (2016). Stable and enhanced gene expression in Clostridium acetobutylicum using synthetic untranslated regions with a stem-loop. J. Biotechnol. 230, 40–43. doi: 10.1016/j.jbiotec.2016.05.020
Li, Q., Chen, J., Minton, N. P., Zhang, Y., Wen, Z., Liu, J., et al. (2016). CRISPR-based genome editing and expression control systems in Clostridium acetobutylicum and Clostridium beijerinckii. Biotechnol. J. 11, 961–972.
Li, Q., Seys, F. M., Minton, N. P., Yang, J., Jiang, Y., Jiang, W., et al. (2019). CRISPR-Cas9(D10A) nickase-assisted base editing in the solvent producer Clostridium beijerinckii. Biotechnol. Bioeng. 116, 1475–1483. doi: 10.1002/bit.26949
Lim, H., and Choi, S. K. (2019). Programmed gRNA removal system for CRISPR-Cas9-mediated multi-round genome editing in Bacillus subtilis. Front. Microbiol. 10:1140. doi: 10.3389/fmicb.2019.01140
Liu, J., Guo, T., Wang, D., Shen, X., Liu, D., Niu, H., et al. (2016). Enhanced butanol production by increasing NADH and ATP levels in Clostridium beijerinckii NCIMB 8052 by insertional inactivation of Cbei_4110. Appl. Microbiol. Biotechnol. 100, 4985–4996. doi: 10.1007/s00253-016-7299-9
Liu, J., Sun, J., Wang, F., Yu, X., Ling, Z., Li, H., et al. (2015). Neuroprotective effects of Clostridium butyricum against vascular dementia in mice via metabolic butyrate. Biomed. Res. Int. 2015:412946.
Liu, Y. J., Zhang, J., Cui, G. Z., and Cui, Q. (2015). Current progress of targetron technology: development, improvement and application in metabolic engineering. Biotechnol. J. 10, 855–865. doi: 10.1002/biot.201400716
Luo, M. L., Mullis, A. S., Leenay, R. T., and Beisel, C. L. (2014). Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 43, 674–681. doi: 10.1093/nar/gku971
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736.
Malaviya, A., Jang, Y. S., and Lee, S. Y. (2012). Continuous butanol production with reduced byproducts formation from glycerol by a hyper producing mutant of Clostridium pasteurianum. Appl. Microbiol. Biotechnol. 93, 1485–1494. doi: 10.1007/s00253-011-3629-0
McAllister, K. N., Bouillaut, L., Kahn, J. N., Self, W. T., and Sorg, J. A. (2017). Using CRISPR-Cas9-mediated genome editing to generate C. difficile mutants defective in selenoproteins synthesis. Sci. Rep. 7:14672.
McAllister, K. N., and Sorg, J. A. (2019). CRISPR genome editing systems in the genus Clostridium: a timely advancement. J. Bacteriol. 201:e00 219–19.
Meaney, C. A., Cartman, S. T., McClure, P. J., and Minton, N. P. (2015). Optimal spore germination in Clostridium botulinum ATCC 3502 requires the presence of functional copies of SleB and YpeB, but not CwlJ. Anaerobe 34, 86–93.
Meaney, C. A., Cartman, S. T., McClure, P.J., and Minton, N. P. (2016). The role of small acid-soluble proteins (SASPs) in protection of spores of Clostridium botulinum against nitrous acid. Int. J. Food Microbiol. 216, 25–30. doi: 10.1016/j.ijfoodmicro.2015.08.024
Minton, N. P., Ehsaan, M., Humphreys, C. M., Little, G. T., Baker, J., Henstra, A. M., et al. (2016). A roadmap for gene system development in Clostridium. Anaerobe 41, 104–112. doi: 10.1016/j.anaerobe.2016.05.011
Mohr, G., Hong, W., Zhang, J., Cui, G.-Z., Yang, Y., Cui, Q., et al. (2013). A targetron system for gene targeting in thermophiles and its application in Clostridium thermocellum. PLoS ONE 8:e69032. doi: 10.1371/journal.pone.0069032
Mojica, F. J., Diez-Villasenor, C., Garcia-Martinez, J., and Almendros, C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. doi: 10.1099/mic.0.023960-0
Moon, H. G., Jang, Y. S., Cho, C., Lee, J., Binkley, R., and Lee, S. Y. (2016). One hundred years of clostridial butanol fermentation. FEMS Microbiol Lett 363:fnw001. doi: 10.1093/femsle/fnw001
Muh, U., Pannullo, A. G., Weiss, D. S., and Ellermeier, C. D. (2019). A xylose-inducible expression system and a CRISPR interference plasmid for targeted knockdown of gene expression in Clostridioides difficile. J. Bacteriol. 201, e711–e718.
Na, D., Yoo, S. M., Chung, H., Park, H., Park, J. H., and Lee, S. Y. (2013). Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 31, 170–174. doi: 10.1038/nbt.2461
Nagaraju, S., Davies, N. K., Walker, D. J., Kopke, M., and Simpson, S. D. (2016). Genome editing of Clostridium autoethanogenum using CRISPR/Cas9. Biotechnol. Biofuels 9:219.
Ng, Y. K., Ehsaan, M., Philip, S., Collery, M. M., Janoir, C., Collignon, A., et al. (2013). Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS ONE 8:e56051. doi: 10.1371/journal.pone.0056051
Nishimasu, H., Ran, F. A., Hsu, P. D., Konermann, S., Shehata, S. I., Dohmae, N., et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949. doi: 10.1016/j.cell.2014.02.001
Noh, H. J., Woo, J. E., Lee, S. Y., and Jang, Y.-S. (2018). Metabolic engineering of Clostridium acetobutylicum for the production of butyl butyrate. Appl. Microbiol. Biotechnol. 102, 8319–8327. doi: 10.1007/s00253-018-9267-z
Park, M.-R., Kim, S.-K., and Jeong, G.-T. (2018). Biosugar production from Gracilaria verrucosa with sulfamic acid pretreatment and subsequent enzymatic hydrolysis. Biotechnol. Bioproc. Eng. 23, 302–310. doi: 10.1007/s12257-018-0090-2
Peters, J. M., Colavin, A., Shi, H., Czarny, T. L., Larson, M. H., Wong, S., et al. (2016). A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165, 1493–1506. doi: 10.1016/j.cell.2016.05.003
Pyne, M. E., Bruder, M., Moo-Young, M., Chung, D. A., and Chou, C. P. (2014). Technical guide for genetic advancement of underdeveloped and intractable Clostridium. Biotechnol. Adv. 32, 623–641. doi: 10.1016/j.biotechadv.2014.04.003
Pyne, M. E., Bruder, M. R., Moo-Young, M., Chung, D. A., and Chou, C. P. (2016). Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci. Rep. 6:25666.
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Ralston, M. T., and Papoutsakis, E. T. (2018). RNAseq-based transcriptome assembly of Clostridium acetobutylicum for functional genome annotation and discovery. AIChE J. 64, 4271–4280. doi: 10.1002/aic.16396
Rhie, M. N., Kim, H. T., Jo, S. Y., Chu, L. L., Baritugo, K.-A., Baylon, M. G., et al. (2019). Recent advances in the metabolic engineering of Klebsiella pneumoniae: a potential platform microorganism for biorefineries. Biotechnol. Bioproc. Eng. 24, 48–64. doi: 10.1007/s12257-018-0346-x
Richard, P., and Manley, J. L. (2009). Transcription termination by nuclear RNA polymerases. Genes Dev. 23, 1247–1269. doi: 10.1101/gad.1792809
Ronda, C., Maury, J., Jakociunas, T., Jacobsen, S. A., Germann, S. M., Harrison, S. J., et al. (2015). CrEdit: CRISPR mediated multi-loci gene integration in Saccharomyces cerevisiae. Microb. Cell Fact. 14:97.
Sato, T., Fukui, T., Atomi, H., and Imanaka, T. (2005). Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71, 3889–3899. doi: 10.1128/aem.71.7.3889-3899.2005
Schwarz, K. M., Grosse-Honebrink, A., Derecka, K., Rotta, C., Zhang, Y., and Minton, N. P. (2017). Towards improved butanol production through targeted genetic modification of Clostridium pasteurianum. Metab. Eng. 40, 124–137. doi: 10.1016/j.ymben.2017.01.009
Scotcher, M. C., Huang, K. X., Harrison, M. L., Rudolph, F. B., and Bennett, G. N. (2003). Sequences affecting the regulation of solvent production in Clostridium acetobutylicum. J. Ind. Microbiol. Biotechnol. 30, 414–420. doi: 10.1007/s10295-003-0057-x
Sedlar, K., Koscova, P., Vasylkivska, M., Branska, B., Kolek, J., Kupkova, K., et al. (2018). Transcription profiling of butanol producer Clostridium beijerinckii NRRL B-598 using RNA-Seq. BMC Genomics 19:415. doi: 10.1186/s12864-018-4805-8
Shao, L., Hu, S., Yang, Y., Gu, Y., Chen, J., Yang, Y., et al. (2007). Targeted gene disruption by use of a group II intron (targetron) vector in Clostridium acetobutylicum. Cell Res. 17, 963–965. doi: 10.1038/cr.2007.91
Soutourina, O. A., Monot, M., Boudry, P., Saujet, L., Pichon, C., Sismeiro, O., et al. (2013). Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 9:e1003493. doi: 10.1371/journal.pgen.1003493
Staedtke, V., Roberts, N. J., Bai, R. Y., and Zhou, S. (2016). Clostridium novyi-NT in cancer therapy. Genes Dis. 3, 144–152. doi: 10.1016/j.gendis.2016.01.003
Strecker, J., Jones, S., Koopal, B., Schmid-Burgk, J., Zetsche, B., Gao, L., et al. (2019a). Engineering of CRISPR-Cas12b for human genome editing. Nat. Commun. 10:212. doi: 10.1017/cbo9781316771440.007
Strecker, J., Ladha, A., Gardner, Z., Schmid-Burgk, J. L., Makarova, K. S., Koonin, E. V., et al. (2019b). RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53. doi: 10.1126/science.aax9181
Swarts, D. C., and Jinek, M. (2018). Cas9 versus Cas12a/Cpf1: Structure-Function Comparisons and Implications for Genome Editing. Hoboken, NJ: Wiley Interdiscip. Rev. RNA, e1481.
Thormann, K., Feustel, L., Lorenz, K., Nakotte, S., and Durre, P. (2002). Control of butanol formation in Clostridium acetobutylicum by transcriptional activation. J. Bacteriol. 184, 1966–1973. doi: 10.1128/jb.184.7.1966-1973.2002
Tripathi, S. A., Olson, D. G., Argyros, D. A., Miller, B. B., Barrett, T. F., Murphy, D. M., et al. (2010). Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl. Environ. Microbiol. 76, 6591–6599. doi: 10.1128/aem.01484-10
Wang, S., Dong, S., Wang, P., Tao, Y., and Wang, Y. (2017). Genome editing in Clostridium saccharoperbutylacetonicum N1-4 with the CRISPR-Cas9 system. Appl. Environ. Microbiol. 83:e233-17.
Wang, Y., Zhang, G., Zhao, X., and Ling, J. (2017). Genome shuffling improved the nucleosides production in Cordyceps kyushuensis. J. Biotechnol. 260, 42–47. doi: 10.1016/j.jbiotec.2017.08.021
Wang, S., Hong, W., Dong, S., Zhang, Z. T., Zhang, J., Wang, L., et al. (2018). Genome engineering of Clostridium difficile using the CRISPR-Cas9 system. Clin. Microbiol. Infect. 24, 1095–1099.
Wang, Y., Zhang, Z. T., Seo, S. O., Lynn, P., Lu, T., Jin, Y. S., et al. (2016a). Bacterial genome editing with CRISPR-Cas9: deletion, integration, single nucleotide modification, and desirable “clean” mutant selection in Clostridium beijerinckii as an example. ACS Synth. Biol. 5, 721–732. doi: 10.1021/acssynbio.6b00060
Wang, Y., Zhang, Z. T., Seo, S. O., Lynn, P., Lu, T., Jin, Y. S., et al. (2016b). Gene transcription repression in Clostridium beijerinckii using CRISPR-dCas9. Biotechnol. Bioeng. 113, 2739–2743. doi: 10.1002/bit.26020
Wasels, F., Jean-Marie, J., Collas, F., Lopez-Contreras, A. M., and Lopes Ferreira, N. (2017). A two-plasmid inducible CRISPR/Cas9 genome editing tool for Clostridium acetobutylicum. J. Microbiol. Methods 140, 5–11. doi: 10.1016/j.mimet.2017.06.010
Wen, Z., Ledesma-Amaro, R., Lin, J., Jiang, Y., and Yang, S. (2019a). Improved n-butanol production from Clostridium cellulovorans by integrated metabolic and evolutionary engineering. Appl. Environ. Microbiol 85, e2560–e2518.
Wen, Z., Li, Q., Liu, J., Jin, M., and Yang, S. (2019b). Consolidated bioprocessing for butanol production of cellulolytic clostridia: development and optimization. Microb. Biotechnol. 13, 410–422. doi: 10.1111/1751-7915.13478
Wen, Z., Lu, M., Ledesma-Amaro, R., Li, Q., Jin, M., and Yang, S. (2019c). Targetron technology applicable in solventogenic clostridia: revisiting 12 years’ advances. Biotechnol. J. 15:e1900284.
Wen, Z., Minton, N. P., Zhang, Y., Li, Q., Liu, J., Jiang, Y., et al. (2017). Enhanced solvent production by metabolic engineering of a twin-clostridial consortium. Metab. Eng. 39, 38–48. doi: 10.1016/j.ymben.2016.10.013
Willson, B. J., Kovacs, K., Wilding-Steele, T., Markus, R., Winzer, K., and Minton, N. P. (2016). Production of a functional cell wall-anchored minicellulosome by recombinant Clostridium acetobutylicum ATCC 824. Biotechnol. Biofuels 9:109.
Wilson, C. M., Rodriguez, M. Jr., Johnson, C. M., Martin, S. L., Chu, T. M., Wolfinger, R. D., et al. (2013). Global transcriptome analysis of Clostridium thermocellum ATCC 27405 during growth on dilute acid pretreated Populus and switchgrass. Biotechnol. Biofuels 6:179.
Woo, J. E., Lee, S. Y., and Jang, Y.-S. (2018). Effects of nutritional enrichment on acid production from degenerated (non-solventogenic) Clostridium acetobutylicum strain M5. Appl. Biol. Chem. 61, 469–472. doi: 10.1007/s13765-018-0372-6
Woolston, B. M., Emerson, D. F., Currie, D. H., and Stephanopoulos, G. (2018). Rediverting carbon flux in Clostridium ljungdahlii using CRISPR interference (CRISPRi). Metab. Eng. 48, 243–253. doi: 10.1016/j.ymben.2018.06.006
Xin, F., Yan, W., Zhou, J., Wu, H., Dong, W., Ma, J., et al. (2018). Exploitation of novel wild type solventogenic strains for butanol production. Biotechnol. Biofuels 11:252.
Xu, T., Li, Y., He, Z., Van Nostrand, J. D., and Zhou, J. (2017). Cas9 nickase-assisted RNA repression enables stable and efficient manipulation of essential metabolic genes in Clostridium cellulolyticum. Front. Microbiol. 8:1744. doi: 10.3389/fmicb.2017.01744
Xu, T., Li, Y., Shi, Z., Hemme, C. L., Li, Y., Zhu, Y., et al. (2015). Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase. Appl. Environ. Microbiol. 81, 4423–4431. doi: 10.1128/aem.00873-15
Yamano, T., Nishimasu, H., Zetsche, B., Hirano, H., Slaymaker, I. M., Li, Y., et al. (2016). Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949–962.
Yoo, S. M., Na, D., and Lee, S. Y. (2013). Design and use of synthetic regulatory small RNAs to control gene expression in Escherichia coli. Nat. Protoc. 8, 1694–1707. doi: 10.1038/nprot.2013.105
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. doi: 10.1016/j.cell.2015.09.038
Zetsche, B., Heidenreich, M., Mohanraju, P., Fedorova, I., Kneppers, J., DeGennaro, E. M., et al. (2017). Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31–34. doi: 10.1038/nbt.3737
Zhang, J., Hong, W., Zong, W., Wang, P., and Wang, Y. (2018a). Markerless genome editing in Clostridium beijerinckii using the CRISPR-Cpf1 system. J. Biotechnol. 284, 27–30. doi: 10.1016/j.jbiotec.2018.07.040
Zhang, J., Zong, W., Hong, W., Zhang, Z. T., and Wang, Y. (2018b). Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production. Metab. Eng. 47, 49–59. doi: 10.1016/j.ymben.2018.03.007
Zhang, J., Liu, Y. J., Cui, G. Z., and Cui, Q. (2015). A novel arabinose-inducible genetic operation system developed for Clostridium cellulolyticum. Biotechnol. Biofuels 8:36. doi: 10.1186/s13068-015-0214-2
Zhang, N., Shao, L., Jiang, Y., Gu, Y., Li, Q., Liu, J., et al. (2015). I-SceI-mediated scarless gene modification via allelic exchange in Clostridium. J. Microbiol. Methods 108, 49–60. doi: 10.1016/j.mimet.2014.11.004
Zhang, Y., Xu, S., Chai, C., Yang, S., Jiang, W., Minton, N. P., et al. (2016). Development of an inducible transposon system for efficient random mutagenesis in Clostridium acetobutylicum. FEMS Microbiol. Lett. 363:fnw065. doi: 10.1093/femsle/fnw065
Zhao, J. P., Zhu, H., Guo, X. P., and Sun, Y. C. (2018). AU-rich long 3’ untranslated region regulates gene expression in bacteria. Front. Microbiol. 9:3080. doi: 10.3389/fmicb.2018.03080
Zhao, R., Liu, Y., Zhang, H., Chai, C., Wang, J., Jiang, W., et al. (2019). CRISPR-cas12a-mediated gene deletion and regulation in Clostridium ljungdahlii and its application in carbon flux redirection in synthesis gas fermentation. ACS Synth. Biol. 8, 2270–2279. doi: 10.1021/acssynbio.9b00033
Zheng, Y., Su, T., and Qi, Q. (2019). Microbial CRISPRi and CRISPRa systems for metabolic engineering. Biotechnol. Bioproc. Eng. 24, 579–591.
Keywords: Clostridium, synthetic biology, mobile intron, CRISPR, Cas, synthetic sRNA, UTR
Citation: Kwon SW, Paari KA, Malaviya A and Jang Y-S (2020) Synthetic Biology Tools for Genome and Transcriptome Engineering of Solventogenic Clostridium. Front. Bioeng. Biotechnol. 8:282. doi: 10.3389/fbioe.2020.00282
Received: 19 January 2020; Accepted: 17 March 2020;
Published: 16 April 2020.
Edited by:
Yi Wang, Auburn University, United StatesReviewed by:
Yang Gu, Shanghai Institutes for Biological Sciences (CAS), ChinaZhiqiang Wen, Nanjing University of Science and Technology, China
Copyright © 2020 Kwon, Paari, Malaviya and Jang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alok Malaviya, YWxva2t1bWFyLm1hbGF2aXlhQGNocmlzdHVuaXZlcnNpdHkuaW4=; Yu-Sin Jang, amFuZ3lzQGdudS5hYy5rcg==
†These authors have contributed equally to this work
 Seong Woo Kwon
Seong Woo Kwon Kuppusamy Alagesan Paari
Kuppusamy Alagesan Paari Alok Malaviya
Alok Malaviya Yu-Sin Jang
Yu-Sin Jang