- 1Institute of Bioinformatics and Biotechnology, Savitribai Phule Pune University, Pune, India
- 2Abasaheb Garware College, Pune, India
A Corrigendum on
Bioengineered Platforms for Chronic Wound Infection Studies: How Can We Make Them More Human-Relevant?
by Kadam, S., Nadkarni, S., Lele, J., Sakhalkar, S., Mokashi, P., and Kaushik, K. S. (2019) Front. Bioeng. Biotechnol. 7:418. doi: 10.3389/fbioe.2019.00418
In the original article, there was a mistake in the legend for Figure 2 as published. The legend incorrectly cites a reference for Figure 2(A) as “modified from.” The figure was in fact made by the authors. The correct legend appears below.
Figure 2. (A) Typical representation of the chronic wound bed microenvironment. (B) Key features of the chronic wound bed-capillary interface. From a bioengineering standpoint, the microenvironment can be represented by a two-compartment system, where the upper compartment consists of the “infected wound bed” with host cells, matrix and microbial biofilms and the lower compartment represents the capillary interface (endothelial cells) with immune components. (C) A simplified representation of key interactions between chronic wound biofilms and other key components of the chronic wound microenvironment, which can be suitably dissected on human-relevant bioengineered platform.
Additionally, there was a mistake in Table 1 as published. The last row of the table had an incorrect placement of the figures. The corrected Table 1 appears below.
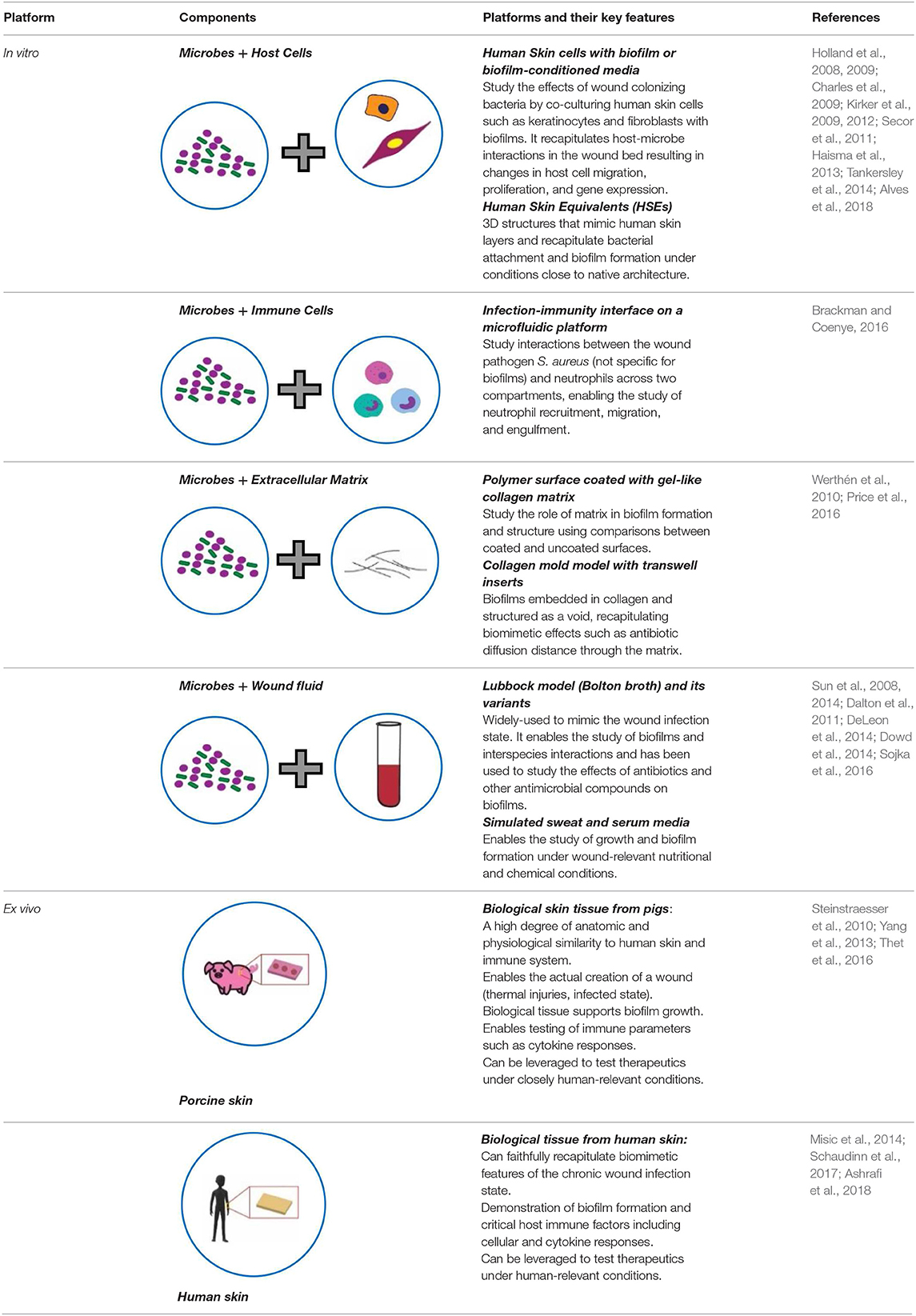
Table 1. Key features of current bioengineered platforms, in vitro and ex vivo, developed for chronic wound infection studies.
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
References
Alves, P. M., Al-Badi, E., Withycombe, C., Jones, P. M., Purdy, K. J., and Maddocks, S. E. (2018). Interaction between Staphylococcus aureus and Pseudomonas aeruginosa is beneficial for colonisation and pathogenicity in a mixed biofilm. Pathog Dis. 76:fty003. doi: 10.1093/femspd/fty003
Ashrafi, M., Novak-Frazer, L., Bates, M., Baguneid, M., Alonso-Rasgado, T., Xia, G., et al. (2018). Validation of biofilm formation on human skin wound models and demonstration of clinically translatable bacteria-specific volatile signatures. Sci. Rep. 8:9431. doi: 10.1038/s41598-018-27504-z
Brackman, G., and Coenye, T. (2016). In vitro and in vivo biofilm wound models and their application. Adv. Exp. Med. Biol. 15–32. doi: 10.1007/5584_2015_5002
Charles, C. A., Ricotti, C. A., Davis, S. C., Mertz, P. M., and Kirsner, R. S. (2009). Use of tissue-engineered skin to study in vitro biofilm development. Dermatol. Surg. 35, 1334–1341. doi: 10.1111/j.1524-4725.2009.01238.x
Dalton, T., Dowd, S. E., Wolcott, R. D., Sun, Y., Watters, C., Griswold, J. A., et al. (2011). An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS ONE 6:e27317. doi: 10.1371/journal.pone.0027317
DeLeon, S., Clinton, A., Fowler, H., Everett, J., Horswill, A. R., and Rumbaugh, K. P. (2014). Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 82, 4718–4728. doi: 10.1128/IAI.02198-14
Dowd, S. E., Sun, Y., Smith, E., Kennedy, J. P., Jones, C. E., and Wolcott, R. (2014). Effects of biofilm treatments on the multi-species Lubbock chronic wound biofilm model. J. Wound Care 18, 508–512. doi: 10.12968/jowc.2009.18.12.45608
Haisma, E. M., Rietveld, M. H., Breij, A., Van Dissel, J. T., El Ghalbzouri, A., and Nibbering, P. H. (2013). Inflammatory and antimicrobial responses to methicillin-resistant Staphylococcus aureus in an in vitro wound infection model. PLoS ONE 8:e82800. doi: 10.1371/journal.pone.0082800
Holland, D. B., Bojar, R. A., Farrar, M. D., and Holland, K. T. (2009). Differential innate immune responses of a living skin equivalent model colonized by Staphylococcus epidermidis or Staphylococcus aureus. FEMS Microbiol. Lett. 290, 149–155. doi: 10.1111/j.1574-6968.2008.01402.x
Holland, D. B., Bojar, R. A., Jeremy, A. H. T., Ingham, E., and Holland, K. T. (2008). Microbial colonization of an in vitro model of a tissue engineered human skin equivalent–a novel approach. FEMS Microbiol. Lett. 279, 110–115. doi: 10.1111/j.1574-6968.2007.01021.x
Kirker, K. R., James, G. A., Fleckman, P., Olerud, J. E., and Stewart, P. S. (2012). Differential effects of planktonic and biofilm MRSA on human fibroblasts. Wound Repair Regen. 20, 253–261. doi: 10.1111/j.1524-475X.2012.00769.x
Kirker, K. R., Secor, P. R., James, G. A., Fleckman, P., Olerud, J. E., and Stewart, P. S. (2009). Loss of viability and induction of apoptosis in human keratinocytes exposed to Staphylococcus aureus biofilms in vitro. Wound Repair Regen. 17, 690–699. doi: 10.1111/j.1524-475X.2009.00523.x
Misic, A. M., Gardner, S. E., and Grice, E. A. (2014). The wound microbiome: modern approaches to examining the role of microorganisms in impaired chronic wound healing. Adv. Wound Care 3, 502–510. doi: 10.1089/wound.2012.0397
Price, B. L., Lovering, A. M., Bowling, F. L., and Dobson, C. B. (2016). Development of a novel collagen wound model to simulate the activity and distribution of antimicrobials in soft tissue during diabetic foot infection. Antimicrob. Agents Chemother. 60, 6880–6889. doi: 10.1128/AAC.01064-16
Schaudinn, C., Dittmann, C., Jurisch, J., Laue, M., Günday-Türeli, N., Blume-Peytavi, U., et al. (2017). Development, standardization and testing of a bacterial wound infection model based on ex vivo human skin. PLoS ONE 12:e0186946. doi: 10.1371/journal.pone.0186946
Secor, P. R., James, G. A., Fleckman, P., Olerud, J. E., McInnerney, K., and Stewart, P. S. (2011). Staphylococcus aureus biofilm and planktonic cultures differentially impact gene expression, mapk phosphorylation, and cytokine production in human keratinocytes. BMC Microbiol. 11:143. doi: 10.1186/1471-2180-11-143
Sojka, M., Valachova, I., Bucekova, M., and Majtan, J. (2016). Antibiofilm efficacy of honey and bee-derived defensin-1 on multispecies wound biofilm. J. Med. Microbiol. 65, 337–344. doi: 10.1099/jmm.0.000227
Steinstraesser, L., Sorkin, M., Niederbichler, A. D., Becerikli, M., Stupka, J., Daigeler, A., et al. (2010). A novel human skin chamber model to study wound infection ex vivo. Arch Dermatol Res. 302, 357–365. doi: 10.1007/s00403-009-1009-8
Sun, Y., Dowd, S. E., Smith, E., Rhoads, D. D., and Wolcott, R. D. (2008). In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen. 16, 805–813. doi: 10.1111/j.1524-475X.2008.00434.x
Sun, Y., Smith, E., Wolcott, R., and Dowd, S. E. (2014). Propagation of anaerobic bacteria within an aerobic multi-species chronic wound biofilm model. J. Wound Care 18, 426–431. doi: 10.12968/jowc.2009.18.10.44604
Tankersley, A., Frank, M. B., Bebak, M., and Brennan, R. (2014). Early effects of Staphylococcus aureus biofilm secreted products on inflammatory responses of human epithelial keratinocytes. J. Inflamm. 11:17. doi: 10.1186/1476-9255-11-17
Thet, N. T., Alves, D. R., Bean, J. E., Booth, S., Nzakizwanayo, J., Young, A. E. R., et al. (2016). Prototype development of the intelligent hydrogel wound dressing and its efficacy in the detection of model pathogenic wound biofilms. ACS Appl. Mater. Interfaces 8, 14909–14919. doi: 10.1021/acsami.5b07372
Werthén, M., Henriksson, L., Jensen, P. Ø., Sternberg, C., Givskov, M., and Bjarnsholt, T. (2010). An in vitro model of bacterial infections in wounds and other soft tissues. APMIS 118, 156–164. doi: 10.1111/j.1600-0463.2009.02580.x
Keywords: chronic wounds, wound infection, wound models, biofilms, bioengineered platforms, in vitro, ex vivo
Citation: Kadam S, Nadkarni S, Lele J, Sakhalkar S, Mokashi P and Kaushik KS (2020) Corrigendum: Bioengineered Platforms for Chronic Wound Infection Studies: How Can We Make Them More Human-Relevant? Front. Bioeng. Biotechnol. 7:449. doi: 10.3389/fbioe.2019.00449
Received: 16 December 2019; Accepted: 17 December 2019;
Published: 04 February 2020.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2020 Kadam, Nadkarni, Lele, Sakhalkar, Mokashi and Kaushik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karishma Surendra Kaushik, a2FyaXNobWFza2F1c2hpa0BnbWFpbC5jb20=
†These authors have contributed equally to this work
 Snehal Kadam
Snehal Kadam Shivani Nadkarni
Shivani Nadkarni Janhavi Lele
Janhavi Lele Savani Sakhalkar
Savani Sakhalkar Pratiksha Mokashi
Pratiksha Mokashi Karishma Surendra Kaushik
Karishma Surendra Kaushik