- 1State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai, China
- 2Shanghai Collaborative Innovation Center for Biomanufacturing Technology, Shanghai, China
- 3Key Laboratory of Bio-based Material Engineering of China National Light Industry Council, Shanghai, China
- 4Department of Bioengineering, Rice University, Houston, TX, United States
- 5Department of Chemical and Biomolecular Engineering, Rice University, Houston, TX, United States
Hydroxy fatty acids (HFAs) are valuable compounds that are widely used in medical, cosmetic and food fields. Production of ω-HFAs via bioconversion by engineered Escherichia coli has received a lot of attention because this process is environmentally friendly. In this study, a whole-cell bio-catalysis strategy was established to synthesize medium-chain ω-HFAs based on the AlkBGT hydroxylation system from Pseudomonas putida GPo1. The effects of blocking the β-oxidation of fatty acids (FAs) and enhancing the transportation of FAs on ω-HFAs bio-production were also investigated. When fadE and fadD were deleted, the consumption of decanoic acid decreased, and the yield of ω-hydroxydecanoic acid was enhanced remarkably. Additionally, the co-expression of the FA transporter protein, FadL, played an important role in increasing the conversion rate of ω-hydroxydecanoic acid. As a result, the concentration and yield of ω-hydroxydecanoic acid in NH03(pBGT-fadL) increased to 309 mg/L and 0.86 mol/mol, respectively. This whole-cell bio-catalysis system was further applied to the biosynthesis of ω-hydroxyoctanoic acid and ω-hydroxydodecanoic acid using octanoic acid and dodecanoic acid as substrates, respectively. The concentrations of ω-hydroxyoctanoic acid and ω-hydroxydodecanoic acid reached 275.48 and 249.03 mg/L, with yields of 0.63 and 0.56 mol/mol, respectively. This study demonstrated that the overexpression of AlkBGT coupled with native FadL is an efficient strategy to synthesize medium-chain ω-HFAs from medium-chain FAs in fadE and fadD mutant E. coli strains.
Introduction
Production of bio-based chemicals has received a lot of attention due to concerns around limited non-renewable fossil fuels and the global environment. Fatty acids (FAs) from plant oils are among the most abundant resources in nature (USDA, 2018). FAs are renewable industrial chemical feedstocks, and some artificial microbial pathways have been established to synthesize high-value chemicals based on FA utilization. These reactions in the microbial pathways are mainly related to double-bond generation, double-bond oxidation, double-bond cleavage, epoxidation and functionalization of C-H bonds in alkyl chains (Biermann et al., 2011). Hydroxy fatty acids (HFAs), one kind of FA derivative, are the direct products of FA hydroxylation via C-H bond oxygenation.
HFAs are valuable chemicals due to their hydroxyl functional group and carboxyl group. HFAs have also gained great interest from various industries because of their special properties: higher reactivity, solvent miscibility, stability, and viscosity. Therefore, HFAs have wide applications and can be used as pharmaceutical intermediates and synthetic precursors (Cross et al., 1995), cosmetic ingredients, surfactants, foam improvers, emulsified agents in deodorant sticks, etc. (Koay et al., 2011), and as food bio-preservatives because of their strong antifungal activity (Cheng et al., 2010). Additionally, ω-HFAs are the ideal building blocks of green synthetic fibers for polymer materials due to the location of hydroxyl groups on the terminus of long carbon chains, which provide superior material properties (Weng and Wu, 2008; Cao and Zhang, 2013). ω-Hydroxydecanoic acid can be further derivatized to sebacic acid, which is an important precursor in the production of nylon and polyamides (PAs), primarily 4,10-PA and 5,10-PA (Bowen et al., 2016). ω-Hydroxydodecanoic acid has the potential to enable commercially relevant production of C12 α, ω-DCA, a valuable precursor of nylon-6,12 (Sugiharto et al., 2018).
Currently, considerable effort has been expended to produce HFAs by chemical synthesis; however, there are still various problems, e.g., lack of environmentally friendly solutions, complicated extraction methods, poor selectivity and harsh reaction conditions (Liu et al., 2011). Bio-catalysis is therefore regarded as a promising alternative approach because it has several advantages such as high selectivity, broad substrate spectrum, high catalytic efficiency, mufti-step reaction in a single strain, and is environmentally friendly (Manfred et al., 2011; Lin and Tao, 2017). Among all ω-HFAs produced by C-H bond oxygenases, most ω-HFAs are usually produced via cytochrome P450 monooxygenase (CYP)-based platforms. For example, these include CYP52A13 and CYP52A17 (Craft et al., 2003; Eschenfeldt et al., 2003), CYP4 (Fer et al., 2008), CYP704B2 (Li et al., 2010), CYP102A1 (Xiao et al., 2018), CYP153 (Bordeaux et al., 2012), and the CYP153AM.aq.-CPRBM3 fusion protein (Scheps et al., 2013; Ahsan et al., 2018; Joo et al., 2019). Some of these enzymes can ω-oxidize terminal methyl groups and have been applied to ω-HFA production, but there is a limitation, in that most of these enzymes are specific for chain lengths of ≥12 (Picataggio et al., 1992).
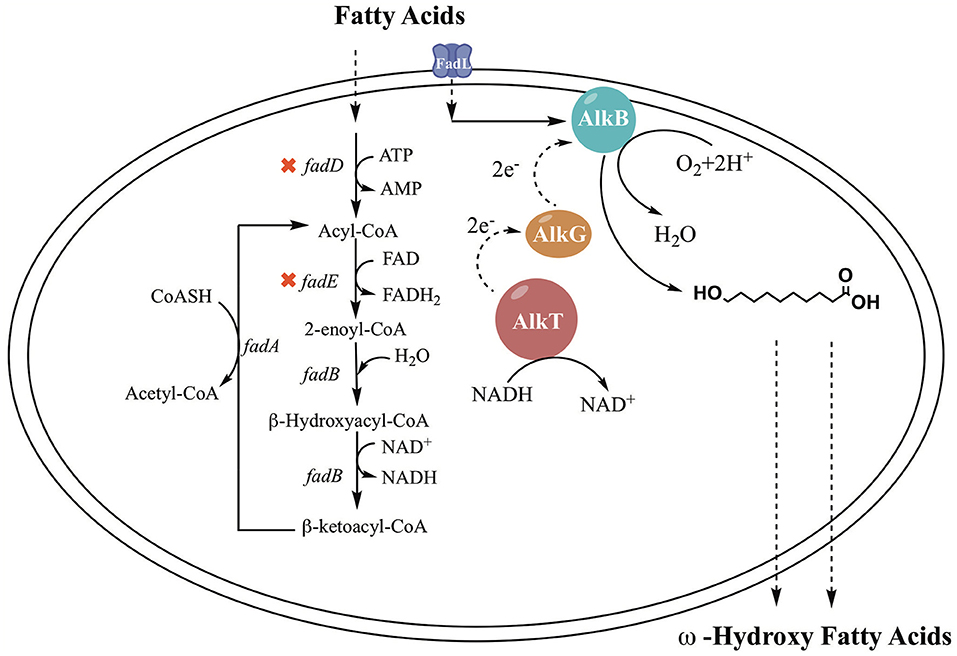
The alkane hydroxylase of Pseudomonas putida GPo1, which consists of three components (AlkB, AlkG, and AlkT), could be another notable ω-oxidation enzyme system of FAs. This system has high specificity for medium-chain fatty acid (C5-C12) oxidation at terminal positions (Grant et al., 2011; Manfred et al., 2011; Tsai et al., 2017). The mono-oxygenase encoded by alkB belongs to a large class of membrane-bound proteins and is an essential component of the alkane hydroxylase system (Grund et al., 1975; Menno et al., 1989; Beilen et al., 1992). Rubredoxin encoded by alkG and rubredoxin reductase encoded by alkT are two electron transfer proteins. The hydroxylase system in vivo operates as follows: AlkG transfers an electron from AlkT, which reduces its flavin adenine dinucleotide at the expense of NADH, then further delivers the electron to AlkB to achieve hydroxylation (Jan et al., 2002). As reported, the AlkBGT hydroxylation system has been investigated in the bioconversion of the fatty acids methyl ester and alkane (Grant et al., 2011; Manfred et al., 2011; Julsing et al., 2012; Call et al., 2016; van Nuland et al., 2016; Kadisch et al., 2017), but there are no reports on ω-HFA production.
E. coli is the ideal host to employ to exploit the hydroxylation activity of these enzymes because of its clarified metabolic background. FA metabolism in E. coli has been expounded in previous work. FA β-oxidation in E. coli is induced by the presence of FAs in the growth medium. The FA β-oxidation cycle is activated by acyl-CoA synthetase (FadD), which plays an important role in FA degradation. The catabolic oxidation cycle consists of dehydrogenation by FadE, hydration and dehydrogenation by FadB or FadJ, and thiolation with CoA by FadA or FadI (Lennen and Pfleger, 2012; Wu and San, 2014). Regulation of FA oxidation is performed by the control protein FadR, which represses the fad regulon. FadD and FadE are the critical enzymes associated with β-oxidation (Wang et al., 2015; Bule et al., 2016). An important approach is deletion of fadD and fadE, which block the breakdown of fatty acids into acyl-CoA (Steen et al., 2010; Handke et al., 2011). Cao et al. (2016) constructed a fadD knockout strain, and the engineered strain showed an enhanced ability to produce HFAs. The titer of HFAs reached 58.7 mg/L, which is 1.6-fold higher than that of the original strain. Kirtz et al. (2016) and Sung et al. (2015) observed improved production of ω-hydroxy fatty acids with a fadD knockout strain. In addition, FA transport through the cell membrane to reach intracellular enzymes is one of the hurdles in ω-HFA production. There are two approaches to overcome this hurdle: one is the addition of chemical reagents such as surfactants to enhance membrane permeabilization, and another is using a transporter protein. Many studies have been done with transporter proteins. For example, AlkL from Pseudomonas putida GPo1 was reported to transport alkanes and fatty acid methyl esters into E. coli but was not active for fatty acids (Jeon et al., 2018). In fact, the outer membrane FA transport mechanism was also clarified in E. coli. It was clearly demonstrated that FA transport is mediated by a membrane protein pump (Bonen et al., 2007). FadL is a typical transporter for FAs; it can use spontaneous conformational changes to transport hydrophobic substrates by diffusion (van den Berg, 2005). There have been many notable cases of the use of FadL to achieve increased production (Bae et al., 2014; Tan et al., 2017; Jeon et al., 2018; Wu et al., 2019).
In this study, the medium-chain alkane hydroxylation system, AlkBGT, from P. oleovorans GPo1 was introduced in E. coli strains for ω-hydroxydecanoic acid biosynthesis from decanoic acid (C10FA). Metabolic engineering strategies to reduce the fatty acid metabolism of the host strains were also investigated here. Blocking the β-oxidation of fatty acids by deletion of fadE and fadD highly increased the yield of ω-hydroxydecanoic acid. In addition, co-expression of the native FA transporter protein, FadL, enhanced the consumption rate of decanoic acid as well as the yield of ω-hydroxydecanoic acid. Octanoic acid and dodecanoic acid were also used as substrates in this hydroxylation system. The results showed that this established system has good efficiency for the synthesis of ω-hydroxyoctanoic acid and ω-hydroxydodecanoic acid. To our knowledge, the current study constitutes the first report of AlkBGT to be applied to FA hydroxylation, and it provides a generalizable framework for the production of medium-chain ω-HFAs via AlkBGT in engineered whole-cell systems. Meanwhile, this study provides a new platform for medium-chain ω-HFA production.
Materials and Methods
Strains and Plasmids
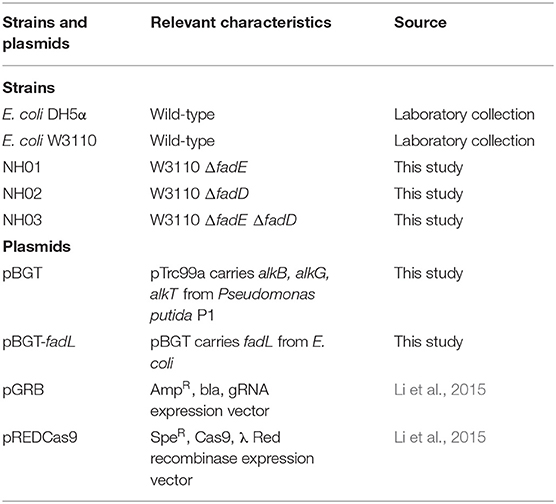
The strains and plasmids used in this study are listed in Table 1. E. coli DH5α was used for plasmid construction, and E. coli W3110 was used as the host and initial strain for further genetic manipulation. Ribosome binding sites (RBS) between alkB-alkG and alkG-alkT were calculated by online software (https://salislab.net/software/forward). The alkB, alkG, and alkT genes with optimized ribosome binding sites were cloned into pTrc99a, and the formed plasmid was named pBGT. The strains with fadD and fadE deletion were constructed using a CRISPR–Cas9 and λ Red recombination system-based genome editing system, which is composed of five elements: Cas9-expressing cassette, gRNA expression plasmid, λ Red recombination, donor template DNA, and inducible plasmid curing system for eliminating gRNA plasmid from the cells (Li et al., 2015). In brief, to construct the gRNA expression plasmid, the pGRB backbone was amplified by PCR with a pair of primers that included 20-bp spacer sequences specific for the target gene. The PCR product was then ligated via homologous recombination to obtain the desired gRNA expression plasmid. To construct donor dsDNA, a pair of 300–500-bp homologous sequences, which are upstream and downstream sequences of the target gene, were amplified by PCR separately and then fused together by overlapping PCR. The mutant strain was verified with genomic PCR after construction to ensure that the target gene had been deleted. For construction of the AlkBGT and FadL expression plasmids, the one-step cloning method was applied. The primers used in this study are listed in Table 2. Gene segments of alkB, alkG, alkT from P. putida GPo1 were amplified by PCR. Ribosome binding sites inside the trc-alkB, alkB-alkG, and alkG-alkT genes were calculated by online software version 2.0 (https://salislab.net/software/forward). The alkB, alkG, and alkT genes with optimized ribosome binding sites were cloned into pTrc99a, and the formed plasmid was named pBGT. The native fadL was further added behind alkT in pBGT with optimized ribosome binding sites, and the newly formed plasmid was named pBGT-fadL.
Media and Culture Conditions
The medium for strain construction and primary preculture was Luria-Bertani Broth (LB), containing tryptone 10 g/L, yeast extract 5 g/L, and sodium chloride 10 g/L. The modified M9 medium used for whole-cell bioconversion contained Na2HPO4·12H2O 15.12 g/L, KH2PO4 3.0 g/L, NaCl 0.5 g/L, MgSO4·7H2O 0.5 g/L, CaCl2 0.011 g/L, 1% (w/v) vitamin B1 0.2 ml/L, glucose 5 g/L, and trace elements solution 0.2 ml/L. The total amount of decanoic acid was ~500 mg/L. The trace elements solution contained (per liter): FeCl3·6H2O 5 g; MnCl2·4H2O 2 g; ZnCl2 0.684 g; CoCl2·6H2O 0.476 g, CuCl2·2H2O 0.17 g; H3BO3 0.062 g; Na2MoO4·2H2O 0.005 g.
To prepare the cell culture, the primary precultures were prepared by transferring one single colony to 3 ml of LB medium with a specific concentration of antibiotics (ampicillin 100 mg/L) at 37°C overnight. In the secondary precultures, 1 ml of the primary preculture was inoculated into 50 ml of LB medium at 37°C for 8 h. Then, a fresh 1 ml of the secondary preculture was transferred into 500-ml flasks containing 100 ml of LB medium with 2 g/L glucose at 30°C, 220 rpm. The expression of target genes was induced with IPTG when the OD600 reached 0.5–0.6, and the cells were grown for 12 h at 30°C. For whole-cell bioconversion, after 12 h of induction, the cells were harvested by centrifugation at 5,945 g and 4°C for 5 min. Then, the cells were washed twice with nitrogen-free M9 medium and resuspended in the modified M9 medium with 20 OD600, with the addition of 0.1% (vol/vol) Triton X-100 depending on the experiment; the total volume was 20 ml. The whole-cell bioconversion was processed at different temperatures (18, 25, 30, and 37°C) for 24 h. All experiments were carried out in triplicate.
Analytical Methods
Cell density was measured at 600 nm at appropriate dilutions (Bausch & Lomb Spectronic 1001). The culture was diluted to the linear range with 0.95% (W/V) NaCl.
For extraction of FA and HFA during bioconversion, 1 ml of supernatant of the cell broth was recovered by centrifuging (13,780 g, 5 min). Then, 1.5 ml of chloroform and 1.5 ml of methanol containing 15% (vol/vol) sulfuric acid were added with 500 μL of 1 g/L undecylic acid as internal standards. The mixtures were mixed by vortexing. Then, the mixtures were exposed at 100°C for 4 h for derivatization. After the heating step, the mixtures were vortexed vigorously for 20 s, stood for 3 min, and operated three times. After centrifugation for 5 min at 1,666 g, the mixture in the tube separated into two layers. The organic layer was recovered by passing through a pipette containing 1 g of anhydrous sodium sulfate for GC analysis.
The concentrations of FFA and HFA in each sample were quantified by the GC-FID system (GC2014, Shimadzu Co., Japan) with a flame ionization detector (FID) and a 30-m DB-5 column (30 m × 0.25 mm × 1 m, Agilent Co., Palo Alto, CA). Nitrogen was used as the carrier gas, and the flow rate was set at 1 ml/min. The injector and detector temperatures were both set at 280°C. The oven temperature was initially held at 150°C for 1 min. Thereafter, the temperature was raised with a gradient of 10°C/min until the temperature reached 200°C, then raised with a gradient of 20°C/min until the temperature reached 240°C. This temperature was held for 3 min.
Results and Discussion
Biosynthesis of ω-Hydroxydecanoic Acid From Decanoic Acid Using the AlkBGT System
In previous studies, it was demonstrated that the AlkBGT system took three steps to achieve ω-oxyfunctionalization of alkane (Jan et al., 2002). The efficiency of AlkBGT co-expression may affect the multistep reactions during ω-HFA biosynthesis. In general, ribosomal interactions with mRNA control translation initiation and the translation rate of proteins (Borujeni et al., 2014). Hence, in this experiment, the AlkBGT encoded by the alkB, alkG, and alkT genes from P. putida GPo1 was cloned with optimized RBS into vector pTrc99a (named pBGT) to generate the ω-HFA biosynthetic pathway (Figure 1).
Traditionally, the major method for performing bioconversion is to use isolated enzymes in vitro; however, this method usually ignores the structural characteristics and metabolic regulation of these enzymes during the overexpression process inside the host cells. In vivo, most oxygenases are membrane-bound proteins and have a multi-component structure (Schrewe et al., 2013). Since AlkB is membrane-bound, it seemed to be more efficient to employ whole-cell bioconversion rather than one-pot biocatalysis in vivo. To verify the hydroxylation of decanoic acid in the AlkBGT system, the whole-cell bioconversion of W3110(pBGT) was investigated first using W3110(pTrc99a) as a control. In this case, considering the solubility and dispersion status of decanoic acid in the medium, we chose DMSO as the co-solvent and TritonX-100 as the surfactant. Three conditions, specifically sodium fatty acids, fatty acids dissolved in dimethyl sulfoxide (DMSO) and fatty acids dissolved in DMSO containing 10% (vol/vol) Triton X-100, were tested. Approximately 0.5 g/L decanoic acid was added in the bioconversion medium. The experiment was performed at 30°C, and the IPTG concentration was 0.1 mM. The conversion rate and concentrations of ω-hydroxydecanoic acid from W3110(pBGT) in different conditions are shown in Figure 2. No ω-hydroxydecanoic acid accumulated in W3110(pTrc99a) (data not shown); however, the strain W3110(pBGT) expressing AlkBGT had the ability to convert decanoic acid to ω-hydroxydecanoic acid. Among these three different substrates, decanoic acid dissolved in DMSO containing 10% (vol/vol) Triton X-100 showed the best performance for ω-hydroxydecanoic acid production. The concentration and yield of ω-hydroxydecanoic acid achieved 123.15 mg/L and 0.24 mol/mol, respectively. The yield was approximately 56.6% higher than that obtained using sodium decanoic acid as the substrate (Figure 2). Hence, decanoic acid dissolved in DMSO containing 10% (vol/vol) Triton X-100 was chosen in further studies. Since AlkBGT are the key enzymes of ω-hydroxydecanoic acid production in whole-cell bioconversion, the optimized expression of AlkBGT will be critical for further improvement of ω-hydroxydecanoic acid production.
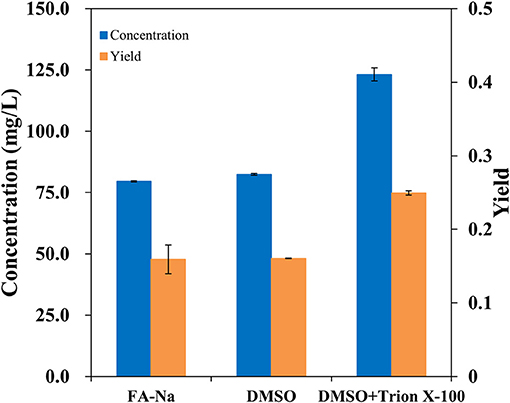
Figure 2. The effects of decanoic acid with differential status in the whole-cell bioconversion of W3110(pBGT) on the concentration (blue) and yield (orange) of ω-hydroxydecanoic acid production.
Optimization of AlkBGT Expression for ω-Hydroxydecanoic Acid Production
To make the whole-cell system more efficient, the expression level of AlkBGT in W3110(pBGT) was optimized. Different concentrations of IPTG, 0.1, 0.2, 0.4, 0.6, and 0.8 mM, were tested in the cultivation conditions. The culture without the addition of IPTG was selected as the control. Whole-cell bioconversion was performed at 25°C. The yields and concentrations of ω-hydroxydecanoic acid in W3110(pBGT) collected following cultivation with different IPTG concentrations are shown in Figure 3A. With 0 mM IPTG addition, no ω-hydroxydecanoic acid accumulated due to the low expression of AlkBGT. The ω-hydroxydecanoic acid yields of whole cells induced by 0.1, 0.4, and 0.6 mM IPTG reached 0.24, 0.27, and 0.31 mol/mol, respectively. The highest concentration of ω-hydroxydecanoic acid was produced with the addition of 0.2 mM IPTG and reached 192.22 mg/L with the highest yield of 0.41 mol/mol (Figure 3A). When the concentration of IPTG increased to 0.8 mM, the yield of ω-hydroxydecanoic acid dropped dramatically to only approximately 0.15 mol/mol, which was only 36.6% of that of 0.2 mM IPTG. Hence, this result indicated that 0.2 mM IPTG was the optimum inducing concentration for AlkBGT overexpression in W3110(pBGT), and it was applied in further whole-cell bioconversion experiments.
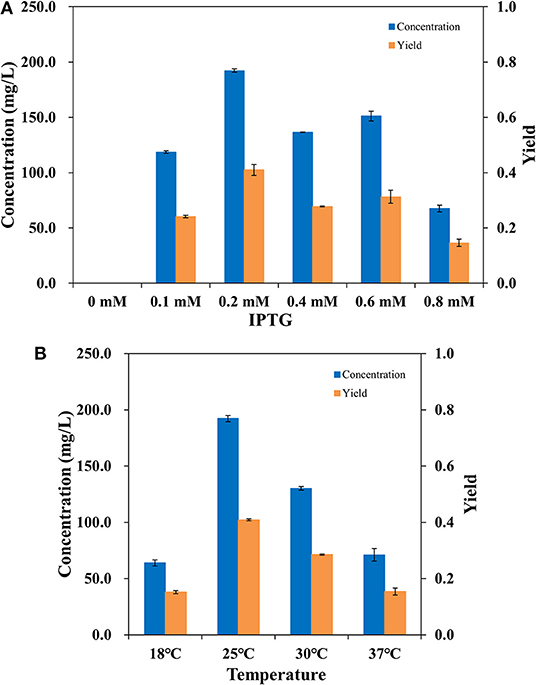
Figure 3. The concentration (blue) and yield (orange) of ω-hydroxydecanoic acid produced by whole-cell conversion of W3110(pBGT) cultured with different concentrations of IPTG (A). The concentration (blue) and yield (orange) of ω-hydroxydecanoic acid produced at different temperatures by the whole-cell conversion of W3110(pBGT) cultured with 0.2 mM IPTG (B).
Incubation temperature affects the catalysis of enzymes during the process of whole-cell bioconversion. To further improve the biocatalysis of AlkBGT in whole-cell bioconversion, four different temperatures, 18, 25, 30, and 37°C, were investigated during bioconversion. The yields and concentrations of ω-hydroxydecanoic acid of W3110(pBGT) in different temperatures are shown in Figure 3B. When whole-cell bioconversion was performed at 18 and 37°C, the concentrations of ω-hydroxydecanoic acid were similar, approximately 63.92 and 70.98 mg/L, respectively, with low yields of ω-hydroxydecanoic acid. At 30°C, the concentration and yield of ω-hydroxydecanoic acid on decanoic acid increased to 130.1 mg/L and 0.28 mol/mol, which were approximately two times higher than those at 18°C. The best result was achieved at 25°C among all of these different temperatures. The highest concentration and yield of ω-hydroxydecanoic acid on decanoic acid reached 192.22 mg/L and 0.41 mol/mol (Figure 3B). Therefore, 25°C was the optimum temperature for whole-cell bioconversion and was applied for further studies.
Further Genetic Modification of the FA Degradation Pathway
In a previous study, it was demonstrated that single deletion of fadD and fadE or double deletion showed high enhancement of FA accumulation (Steen et al., 2010; Li et al., 2012; Cao et al., 2016; Jawed et al., 2016). Hence, it seemed that deletion of fadE and fadD in E. coli might block the β-oxidation of decanoic acid, the precursor of ω-hydroxydecanoic acid, and further enhance the yield of ω-hydroxydecanoic acid. Engineered strains NH01 (ΔfadE), NH02 (ΔfadD), and NH03 (ΔfadE ΔfadD) were constructed via a CRISPR-Cas9-based approach. pBGT was transformed into NH01, NH02 and NH03 and formed NH01(pBGT), NH02(pBGT), and NH03(pBGT), respectively. The yields and concentrations of ω-hydroxydecanoic acid of these strains are shown in Figure 4. As a result, higher yields of ω-hydroxydecanoic acid were found in these three mutants than in W3110(pBGT). The yield of ω-hydroxydecanoic acid on decanoic acid in NH01(pBGT) achieved 0.47 mol/mol, which was 14.6% higher than that in W3110(pBGT); however, it was 13% lower than that in NH02(pBGT) (0.54 mol/mol). The deletion of fadD seemed to prevent decanoic acid degradation more efficiently than the deletion of fadE. This result was also consistent with a previous study (Bae et al., 2014). This result might be explained because NH01 with only fadE knockout was still able to synthesize Acyl-CoA from FA, and then Acyl-CoA might be utilized further by other metabolic pathways. The highest yield of ω-hydroxydecanoic acid was found in NH03(pBGT) with deletions of both fadE and fadD, reaching 0.70 mol/mol, which was 70.7% higher than that of W3110(pBGT). Deleting the key genes fadE and fadD related to the β-oxidation cycle showed large effects on ω-hydroxydecanoic acid accumulation during whole-cell bioconversion due to the reduction of the catabolic degradation of decanoic acid.
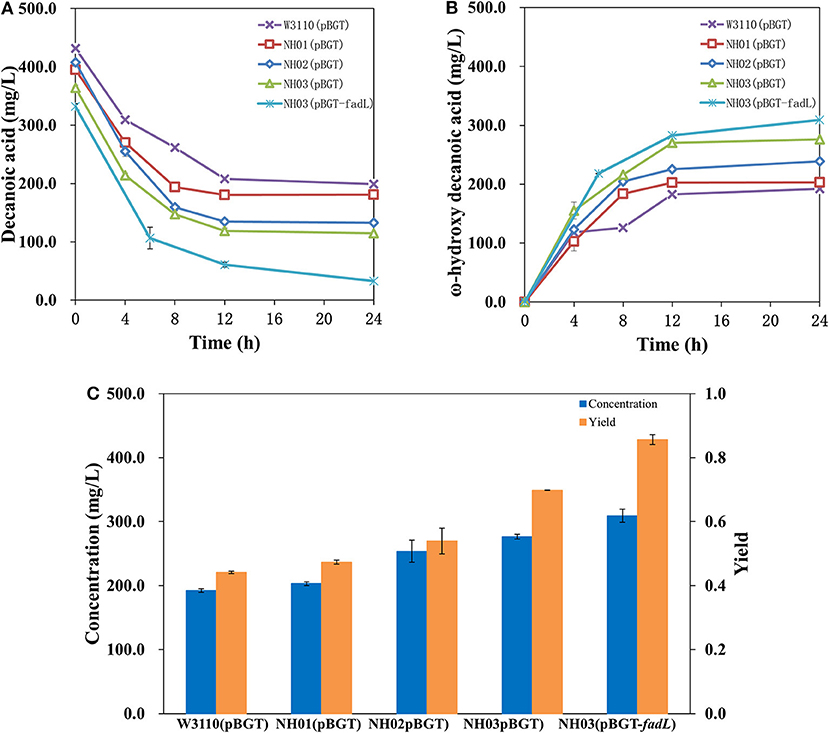
Figure 4. Profiles of decanoic acid (C10FA) consumption and ω-hydroxydecanoic acid (C10HFA) accumulation in the whole-cell bioconversion of W3110(pBGT), NH01(pBGT), NH02(pBGT), NH03(pBGT), and NH03(pBGT-fadL). (A) The concentration of decanoic acid, (B) the concentration of ω-hydroxydecanoic acid. The summary of the concentration (blue) and yield (orange) of ω-hydroxydecanoic acid at the final time point in different engineered strains (C).
Effect of Native Fadl Overexpression on Enhanced Decanoic Acid Consumption and ω-Hydroxydecanoic Acid Accumulation
Uptake of substrates, especially hydrophobic molecules, into cells through transporter proteins is critical and is one of the limiting steps in whole-cell bioconversion. It was proven by isotopic labeling that the deletion of fadD decreased FA transport levels (Weimar et al., 2002). Therefore, an important aspect of enhancing catalysis is to improve transport ability. In this study, overexpression of the transporter AlkL also showed no effect on ether decanoic acid consumption or ω-hydroxydecanoic acid accumulation (data not shown). The outer membrane protein FadL of E. coli plays an important role in importing FAs into the cell, and FadL allows the entry of small hydrophobic molecules into the outer membrane through a hydrophobic channel (Call et al., 2016). It was reported that overexpression of fadL resulted in increased conversion of long-chain fatty acids to ω-hydroxy long-chain fatty acids (Bae et al., 2014). To determine whether FadL might increase catalysis for ω-hydroxydecanoic acid production, the plasmid pBGT-fadL was constructed. The profiles of decanoic acid consumption and ω-hydroxydecanoic acid accumulation in whole-cell bioconversion of NH03(pBGT-fadL) are shown in Figure 4C. After 24 h of whole-cell bioconversion, 308.98 mg/L ω-hydroxydecanoic acid was observed, and the yield of ω-hydroxydecanoic acid on decanoic acid reached 0.86 mol/mol. The yield of ω-hydroxydecanoic acid on decanoic acid in NH03(pBGT-fadL) increased by ~22.8 and 110% relative to yields of the strains NH03(pBGT) and W3110(pBGT), respectively (Figure 4C). FadL overexpression increased the concentration and yield of ω-hydroxydecanoic acid, which might be because the expression of the transporter FadL enhanced the availability of decanoic acid for the AlkBGT hydroxylation system inside the cell. It was reported that a Pseudomonas aeruginosa outer membrane protein, ExFadLO, which belongs to the family of FadL proteins, was involved in the export of long-chain oxygenated fatty acids. According to a 3D model structure, ExFadLO had similar features and structural conformation to those described for previously crystallized FadL transporters from E. coli (Martínez et al., 2013). However, no conclusive evidence has been obtained for this hypothesis.
Biosynthesis of ω-Hydroxyoctanoic Acid and ω-Hydroxydodecanoic Acid
The study above suggested that a ω-hydroxydecanoic acid biosynthesis system with high efficiency has been established. It was demonstrated that AlkBGT is known to catalyze the oxyfunctionalization of medium-chain alkanes and fatty acids (Kusunose et al., 1964; Eggink et al., 1987). To further explore the substrate spectrum of this engineered whole-cell catalysis system of NH03 (pBGT-fadL), octanoic acid and dodecanoic acid, which are medium-chain fatty acids, were employed as substrates. The results are shown in Figures 5A,B. The bioconversion process of octanoic acid and dodecanoic acid was similar to that of decanoic acid. In the initial phase, highly increased product concentrations and decreased substrate levels were observed, whereas after 12 h of incubation, the rates of product accumulation and substrate consumption became slow. At 24 h, the accumulation of ω-hydroxyoctanoic acid reached 275.48 mg/L, and the yield of ω-hydroxyoctanoic acid on octanoic acid was 0.63 mol/mol. The accumulation of ω-hydroxydodecanoic acid reached 249.03 mg/L with a yield of 0.59 mol/mol (Figure 5C). It was noted that there are differences in efficiency among different fatty acid chain lengths. This phenomenon is consistent with a previous report, and it may be attributed to a specific amino acid position in the substrate-binding pocket of AlkB that determines the length of substrate (van Beilen et al., 2005). In the process of substrate uptake, small hydrophobic compounds can easily diffuse into the cell, but large hydrophobic molecules might be restrained by the outer membrane to some extent. However, most hydrophobic substrates are often toxic to living cells, and toxic substances that are detrimental impact bioconversion negatively, which is the major drawback in production (Scheps et al., 2013). Thereby, the lower bioconversion efficiency of octanoic acid than decanoic acid was probably caused by the variable toxicity of different chain lengths of fatty acids that accumulated in cells.
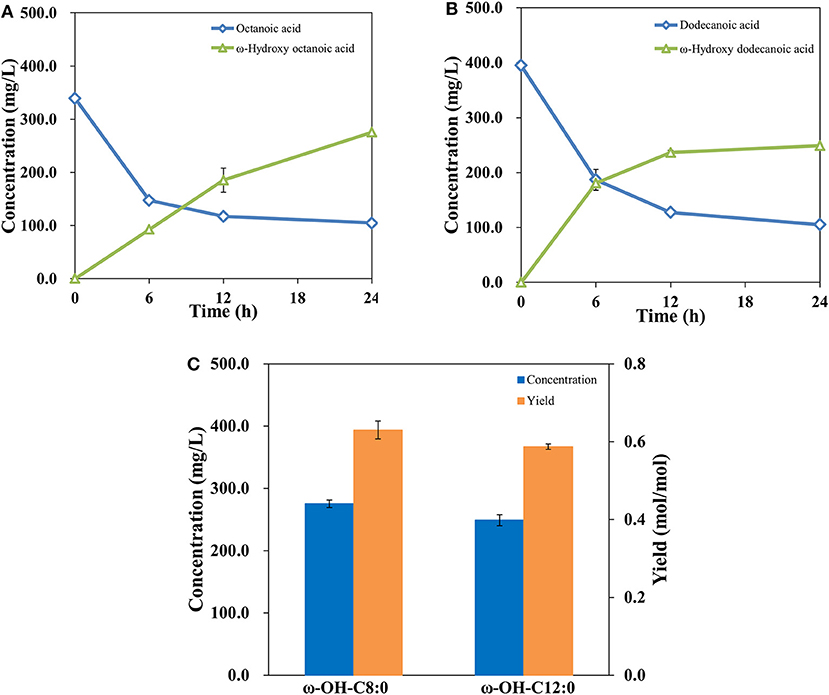
Figure 5. Profiles of octanoic acid (C8FA) consumption and ω-hydroxy octanoic acid (C8HFA) accumulation in whole-cell bioconversion of NH03(pBGT-fadL) (A). Profiles of dodecanoic acid (C12FA) consumption and ω-hydroxy dodecanoic acid (C12HFA) accumulation in whole-cell bioconversion of NH03(pBGT-fadL) (B). Summary of the concentrations (blue) and yields (orange) of ω-hydroxyoctanoic acid and ω-hydroxydodecanoic acid at the final time point in NH03(pBGT-fadL) (C).
Conclusion
In this study, several strategies were applied to enhance the whole-cell bioconversion of strains, including optimized overexpression of alkBGT from P. putida GPo1, deletion of the FA β-oxidation-associated key enzymes FadE and FadD, and overexpression of the native FadL. The concentrations of ω-hydroxyoctanoic acid, ω-hydroxydecanoic acid and ω-hydroxydodecanoic acid in NH03(pBGT-fadL) reached 275.48, 308.98, and 249.03 mg/L, respectively. The yields reached 0.63, 0.86, and 0.56 mol/mol, respectively. This study demonstrated the great potential of these engineered strains for the production of ω-hydroxy medium-chain fatty acids from medium-chain fatty acids by whole-cell bioconversion.
Data Availability Statement
All datasets generated for this study are included in the manuscript/supplementary files.
Author Contributions
HW, GB, and K-YS designed the experiments. QH performed the research experiments. QH, GB, K-YS, and HW analysis the data. QH and HW wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (Grant No. 2017YFB0309302), the National Natural Science Foundation of China (Grant No. 21776083), the Fok Ying-Tong Education Foundation, China (Grant No. 161017), the Fundamental Research Funds for the Central Universities (Grant No. 22221818014), and the 111 Project (B18022), the Science and Technology Commission of Shanghai Municipality (Grant No. 17JC1404800). This study was partially supported by the Open Funding Project of the State Key Laboratory of Bioreactor Engineering.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahsan, M., Patil, M., Jeon, H., Sung, S., Chung, T., and Yun, H. (2018). Biosynthesis of nylon 12 monomer, ω-aminododecanoic acid using artificial self-sufficient P450, AlkJ and ω-TA. Catalysts 8:400. doi: 10.3390/catal8090400
Bae, J.H., Park, B.G., Jung, E., Lee, P.G., and Kim, B.G. (2014). fadD deletion and fadL overexpression in Escherichia coli increase hydroxy long-chain fatty acid productivity. Appl. Microbiol. Biotechnol. 98, 8917–8925. doi: 10.1007/s00253-014-5974-2
Beilen, V. J. B., Penninga, D., and Witholt, B. (1992). Topology of the membrane-bound alkane hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 267:17.
Biermann, U., Bornscheuer, U., Meier, M.A., Metzger, J.O., and Schäfer, H.J. (2011). Oils and fats as renewable raw materials in chemistry. Angew. Chem. Int. Ed. 50, 3854–3871. doi: 10.1002/anie.201002767
Bonen, A., Chabowski, A., Luiken, J. J., and Glatz, J.F. (2007). Is membrane transport of FFA mediated by lipid, protein, or both? Mechanisms and regulation of protein-mediated cellular fatty acid uptake: molecular, biochemical, and physiological evidence. Physiology 22, 15–29. doi: 10.1152/physiologyonline.2007.22.1.15
Bordeaux, M., Galarneau, A., and Drone, J. (2012). Catalytic, mild, and selective oxyfunctionalization of linear alkanes: current challenges. Angew. Chem. Int. Ed. 51, 10712–10723. doi: 10.1002/anie.201203280
Borujeni, E.A., Channarasappa, S.A., and Salis, M.H. (2014). Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 42, 2646–2659. doi: 10.1093/nar/gkt1139
Bowen, C. H., Bonin, J., Kogler, A., Barba-Ostria, C., and Zhang, F. (2016). Engineering Escherichia coli for conversion of glucose to medium-chain ω-hydroxy fatty acids and α,ω-dicarboxylic acids. ACS Synth. Biol. 18, 200–206. doi: 10.1021/acssynbio.5b00201
Bule, M., Luo, Y., Bennett, G., Karanjikar, M., Rooney, W., and San, K.-Y. (2016). Direct bioconversion of sorghum extract sugars to free fatty acids using metabolically engineered Escherichia coli strains: value addition to the sorghum bioenergy crop. Biomass Bioenergy 93, 217–226. doi: 10.1016/j.biombioe.2016.07.020
Call, T.P., Akhtar, M.K., Baganz, F., and Grant, C. (2016). Modulating the import of medium-chain alkanes in E. coli through tuned expression of FadL. J. Biol. Eng. 10:5. doi: 10.1186/s13036-016-0026-3
Cao, Y., Cheng, T., Zhao, G., Niu, W., Guo, J., Xian, M., et al. (2016). Metabolic engineering of Escherichia coli for the production of hydroxy fatty acids from glucose. BMC Biotechnol. 16:26. doi: 10.1186/s12896-016-0257-x
Cao, Y., and Zhang, X. (2013). Production of long-chain hydroxy fatty acids by microbial conversion. Appl. Microbiol. Biotechnol. 97, 3323–3331. doi: 10.1007/s00253-013-4815-z
Cheng, L., Miao, M., Zhang, T., Jiang, B., and Mu, W.M. (2010). Advances on research of antifungal lactic acid bacteria as a biopreservative in food industry:a review. Food Fermentat. Ind. 36, 129–133.
Craft, D.L., Madduri, K.M., Eshoo, M., and Wilson, C.R. (2003). Identification and characterization of the CYP52 family of Candida tropicalis ATCC 20336, important for the conversion of fatty acids and alkanes to α, ω-dicarboxylic acids. Appl. Environ. Microbiol. 69, 5983–5991. doi: 10.1128/aem.69.10.5983-5991.2003
Cross, K.C., Lionel, H., Opie, and Radda, A.G.K. (1995). Is lactate-induced myocardial ischaemic injury mediated by decreased pH or increased intracellular lactate? J. Mol. Cell Cardiol. 27, 1369–1381. doi: 10.1006/jmcc.1995.0130
Eggink, G., Lageveen, R.G., Altenburg, B., and Witholt, B. (1987). Controlled and functional expression of the Pseudomonas oleovorans alkane utilizing system in Pseudomonas putida and Escherichia coli. J. Biol. Chem. 262, 11712–11718.
Eschenfeldt, W.H., Zhang, Y., Samaha, H., Stols, L., Eirich, L.D., Wilson, C.R., et al. (2003). Transformation of fatty acids catalyzed by cytochrome P450 monooxygenase enzymes of Candida tropicalis. Appl. Environ. Microbiol. 69, 5992–5999. doi: 10.1128/aem.69.10.5992-5999.2003
Fer, M., Corcos, L., Dréano, Y., Plée-Gautier, E., Salaun, J.P., Berthou, F., et al. (2008). Cytochromes P450 from family 4 are the main omega hydroxylating enzymes in humans: CYP4F3B is the prominent player in PUFA metabolism. J. Lipid Res. 49, 2379–2389. doi: 10.1194/jlr.m800199-jlr200
Grant, C., Woodley, J.M., and Baganz, F. (2011). Whole-cell bio-oxidation of n-dodecane using the alkane hydroxylase system of P. putida GPo1 expressed in E. coli. Enzyme Microb. Technol. 48, 480–486. doi: 10.1016/j.enzmictec.2011.01.008
Grund, A., Shapori, J., Fennewald, M., Bacha, P., Leahy, J., Markbreiter, K., et al. (1975). Regulation of alkane oxidation in Pseudomonas putida. J. Bacteriol. 123, 546–556.
Handke, P., Lynch, S.A., and Gill, R.T. (2011). Application and engineering of fatty acid biosynthesis in Escherichia coli for advanced fuels and chemicals. Metab. Eng. 13, 28–37. doi: 10.1016/j.ymben.2010.10.007
Jan, B.N. M., Smits, T.H.M., Roth, C., Balada, S. B., and Witholt, B. (2002). Rubredoxins involved in alkane oxidation. J. Bacteriol. 184, 1722–1732. doi: 10.1128/jb.184.6.1722-1732.2002
Jawed, K., Mattam, A.J., Fatma, Z., Wajid, S., Abdin, M.Z., and Yazdani, S.S. (2016). Engineered production of short chain fatty acid in Escherichia coli using fatty acid synthesis pathway. PLoS ONE 11:e0160035. doi: 10.1371/journal.pone.0160035
Jeon, E.Y., Song, J.W., Cha, H.J., Lee, S.M., Lee, J., and Park, J.B. (2018). Intracellular transformation rates of fatty acids are influenced by expression of the fatty acid transporter FadL in Escherichia coli cell membrane. J. Biotechnol. 281, 161–167. doi: 10.1016/j.jbiotec.2018.07.019
Joo, S.-Y., Yoo, H.-W., Sarak, S., Kim, B.-G., and Yun, H. (2019). Enzymatic synthesis of ω-hydroxydodecanoic acid by employing a cytochrome P450 from Limnobacter sp. 105 MED. Catalysts 9:54. doi: 10.3390/catal9010054
Julsing, M.K., Schrewe, M., Cornelissen, S., Hermann, I., Schmid, A., and Buhler, B. (2012). Outer membrane protein AlkL boosts biocatalytic oxyfunctionalization of hydrophobic substrates in Escherichia coli. Appl. Environ. Microbiol. 78, 5724–5733. doi: 10.1128/aem.00949-12
Kadisch, M., Julsing, M. K., Schrewe, M., Jehmlich, N., Scheer, B., von Bergen, M., et al. (2017). Maximization of cell viability rather than biocatalyst activity improves whole-cell ω-oxyfunctionalization performance. Biotechnol. Bioeng. 114, 874–884. doi: 10.1002/bit.26213
Kirtz, M., Klebensberger, J., Otte, K.B., Richter, S.M., and Hauer, B. (2016). Production of -hydroxy octanoic acid with Escherichia coli. J. Biotechnol. 20, 30–33. doi: 10.1016/j.jbiotec.2016.05.017
Koay, G.F.L., Chuah, T.-G., Zainal-Abidin, S., Ahmad, S., and Choong, T.S.Y. (2011). Development, characterization and commercial application of palm based dihydroxystearic acid and its derivatives: an overview. J. Oleo Sci. 60, 237–265. doi: 10.5650/jos.60.237
Kusunose, M., Kusunose, E., and Coon, J. M. (1964). Enzymatic ω-oxidation of fatty acids. J. Biol. Chem. 239, 1374–1380. doi: 10.1201/b18138-5
Lennen, R.M., and Pfleger, B.F. (2012). Engineering Escherichia coli to synthesize free fatty acids. Trends Biotechnol. 30, 659–667. doi: 10.1016/j.tibtech.2012.09.006
Li, H., Pinot, F., Sauveplane, V., Werck-Reichhart, D., Diehl, P., Schreiber, L., et al. (2010). Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant. Cell 22, 173–190. doi: 10.1105/tpc.109.070326
Li, M., Zhang, X., Agrawal, A., and San, K.Y. (2012). Effect of acetate formation pathway and long chain fatty acid CoA-ligase on the free fatty acid production in E. coli expressing acy-ACP thioesterase from Ricinus communis. Metab. Eng. 14, 380–387. doi: 10.1016/j.ymben.2012.03.007
Li, Y., Lin, Z., Huang, C., Zhang, Y., Wang, Z., Tang, Y.J., et al. (2015). Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab. Eng. 31, 13–21. doi: 10.1016/j.ymben.2015.06.006
Lin, B., and Tao, Y. (2017). Whole-cell biocatalysts by design. Microb. Cell Fact. 16:106. doi: 10.1186/s12934-017-0724-7
Liu, C., Liu, F., Cai, J., Xie, W., Long, T.E., Turner, S.R., et al. (2011). Polymers from fatty acids: poly(ω-hydroxyl tetradecanoic acid) synthesis and physico-mechanical studies. Biomacromolecules 12, 3291–3298. doi: 10.1021/bk-2012-1105.ch009
Manfred, S., Magnusson, O.A., Christian, W., Bruno, B., and Andreas, S. (2011). Kinetic analysis of terminal and unactivated C-H bond oxyfunctionalization in fatty acid methyl esters by monooxygenase-based whole-cell biocatalysis. Adv. Synth. Catal. 353, 3485–3495. doi: 10.1002/adsc.201100440
Martínez, E., Estupiñán, M., Pastor, F. I., Busquets, M., Díaz, P., and Manresa, A. (2013). Functional characterization of ExFadLO, an outer membrane protein required for exporting oxygenated long-chain fatty acids in Pseudomonas aeruginosa. Biochimie 95, 290–298. doi: 10.1016/j.biochi.2012.09.032
Menno, K.R., Mark, P.G., Van, D.L., Philip, R., Jaap, K., Philip, H.V.L., et al. (1989). The Pseudomonas oleovorans alkane hydroxylase gene. Sequence and expression. J. Biol. Chem. 264, 5534–5541.
Picataggio, S., Rohrer, T., Deanda, K., Lanning, D., Reynolds, R., Mielenz, J., et al. (1992). Metabolic engineering of Candida tropicalis for the production of long-chain dicarboxylic acids. Nature 10:5. doi: 10.1038/nbt0892-894
Scheps, D., Honda Malca, S., Richter, S.M., Marisch, K., Nestl, B.M., and Hauer, B. (2013). Synthesis of ω-hydroxy dodecanoic acid based on an engineered CYP153A fusion construct. Microb. Biotechnol. 6, 694–707. doi: 10.1111/1751-7915.12073
Schrewe, M., Julsing, M.K., Buhler, B., and Schmid, A. (2013). Whole-cell biocatalysis for selective and productive C-O functional group introduction and modification. Chem. Soc. Rev. 42, 6346–6377. doi: 10.1039/c3cs60011d
Steen, E.J., Kang, Y., Bokinsky, G., Hu, Z., Schirmer, A., Mcclure, A., et al. (2010). Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–562. doi: 10.1038/nature08721
Sugiharto, Y.E.C., Lee, H., Fitriana, A.D., Lee, H., Jeon, W., Park, K., et al. (2018). Effect of decanoic acid and 10-hydroxydecanoic acid on the biotransformation of methyl decanoate to sebacic acid. AMB Express. 8:75. doi: 10.1186/s13568-018-0605-4
Sung, C., Jung, E., Choi, K. Y., Bae, J. H., Kim, M., Kim, J., et al. (2015). The production of ω-hydroxy palmitic acid using fatty acid metabolism and cofactor optimization in Escherichia coli. Appl. Microbiol. Biotechnol. 99, 6667–6676. doi: 10.1007/s00253-015-6630-1
Tan, Z., Black, W., Yoon, J. M., Shanks, J. V., and Jarboe, L. R. (2017). Improving Escherichia coli membrane integrity and fatty acid production by expression tuning of FadL and OmpF. Microb. Cell Fact. 16:38. doi: 10.1186/s12934-017-0650-8
Tsai, Y. F., Luo, W. I., Chang, J. L., Chang, C. W., Chuang, H. C., Ramu, R., et al. (2017). Electrochemical hydroxylation of C3-C12 n-alkanes by recombinant alkane hydroxylase (AlkB) and rubredoxin-2 (AlkG) from Pseudomonas putida GPo1. Sci. Rep. 7:8369. doi: 10.1038/s41598-017-08610-w
USDA (2018). World Agricultural Supply and Demand Estimates. Washington, DC: United States Department of Agriculture.
van Beilen, J. B., Smits, T. H., Roos, F. F., Brunner, T., Balada, S. B., Röthlisberger, M., et al. (2005). Identification of an amino acid position that determines the substrate range of integral membrane alkane hydroxylases. J. Bacteriol. 187, 85–91. doi: 10.1128/JB.187.1.85-91.2005
van den Berg, B. (2005). The FadL family: unusual transporters for unusual substrates. Curr. Opin. Struct. Biol. 15, 401–407. doi: 10.1016/j.sbi.2005.06.003
van Nuland, Y. M., Eggink, G., and Weusthuis, R. A. (2016). Application of AlkBGT and AlkL from Pseudomonas putida GPo1 for selective alkyl ester ω-oxyfunctionalization in Escherichia coli. Appl. Environ. Microbiol. 82, 3801–3807. doi: 10.1128/aem.00822-16
Wang, D., Wu, H., Thakker, C., Beyersdorf, J., Bennett, G.N., and San, K.-Y. (2015). Efficient free fatty acid production in engineered Escherichia coli strains using soybean oligosaccharides as feedstock. Biotechnol. Prog. 31, 686–694. doi: 10.1002/btpr.2092
Weimar, D. J., DiRusso, C.C., Delio, R., and Black, N.P. (2002). Functional role of fatty acyl-coenzyme A synthetase in the transmembrane movement and activation of exogenous long-chain fatty acids. J. Biol. Chem. 277, 29369–29376. doi: 10.1074/jbc.m107022200
Weng, P.F., and Wu, Z.F. (2008). Advances of hydroxyl fatty acids production by bioconversion of fatty acids with microbe. J. Chin. Cereals Oil Assoc. 23, 203–206.
Wu, H., and San, K.-Y. (2014). Efficient odd straight medium chain free fatty acid production by metabolically engineered Escherichia coli. Biotechnol. Bioeng. 111, 2209–2219. doi: 10.1002/bit.25296
Wu, J., Wang, Z., Zhang, X., Zhou, P., Xia, X., and Dong, M. (2019). Improving medium chain fatty acid production in Escherichia coli by multiple transporter engineering. Food Chem. 30, 628–634. doi: 10.1016/j.foodchem.2018.08.102
Keywords: Escherichia coli, AlkBGT, medium-chain fatty acids, ω-hydroxy fatty acids, FadL
Citation: He Q, Bennett GN, San K-Y and Wu H (2019) Biosynthesis of Medium-Chain ω-Hydroxy Fatty Acids by AlkBGT of Pseudomonas putida GPo1 With Native FadL in Engineered Escherichia coli. Front. Bioeng. Biotechnol. 7:273. doi: 10.3389/fbioe.2019.00273
Received: 10 July 2019; Accepted: 01 October 2019;
Published: 17 October 2019.
Edited by:
Zhi-Qiang Liu, Zhejiang University of Technology, ChinaReviewed by:
Lidan Ye, Zhejiang University, ChinaSimon, Congqiang Zhang, Agency for Science, Technology and Research (A*STAR), Singapore
Laura R. Jarboe, Iowa State University, United States
Copyright © 2019 He, Bennett, San and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Wu, aHd1QGVjdXN0LmVkdS5jbg==
 Qiaofei He1
Qiaofei He1 George N. Bennett
George N. Bennett Ka-Yiu San
Ka-Yiu San Hui Wu
Hui Wu