
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bioeng. Biotechnol. , 11 December 2018
Sec. Tissue Engineering and Regenerative Medicine
Volume 6 - 2018 | https://doi.org/10.3389/fbioe.2018.00194
This article is part of the Research Topic Medical and Industrial Applications of Microfluidic-based Cell/Tissue Culture and Organs-on-a-Chip View all 13 articles
Movement of skeletal-muscle fibers is generated by the coordinated action of several cells taking part within the locomotion circuit (motoneurons, sensory-neurons, Schwann cells, astrocytes, microglia, and muscle-cells). Failures in any part of this circuit could impede or hinder coordinated muscle movement and cause a neuromuscular disease (NMD) or determine its severity. Studying fragments of the circuit cannot provide a comprehensive and complete view of the pathological process. We trace the historic developments of studies focused on in-vitro modeling of the spinal-locomotion circuit and how bioengineered innovative technologies show advantages for an accurate mimicking of physiological conditions of spinal-locomotion circuit. New developments on compartmentalized microfluidic culture systems (cμFCS), the use of human induced pluripotent stem cells (hiPSCs) and 3D cell-cultures are analyzed. We finally address limitations of current study models and three main challenges on neuromuscular studies: (i) mimic the whole spinal-locomotion circuit including all cell-types involved and the evaluation of independent and interdependent roles of each one; (ii) mimic the neurodegenerative response of mature neurons in-vitro as it occurs in-vivo; and (iii) develop, tune, implement, and combine cμFCS, hiPSC, and 3D-culture technologies to ultimately create patient-specific complete, translational, and reliable NMD in-vitro model. Overcoming these challenges would significantly facilitate understanding the events taking place in NMDs and accelerate the process of finding new therapies.
From the physiological and anatomical points of view, the mechanosensory-motor circuit is complex, involving several cell-types with specific natural environments. Traditionally, it has been studied coculturing different cell-types on the same platform from animal origin in 2D (Vilmont et al., 2016; Charoensook et al., 2017; Happe et al., 2017) and 3D (Morimoto et al., 2013; Martin et al., 2015; Smith et al., 2016), or from human origin (Guo et al., 2011; Demestre et al., 2015), or mixed species (Yoshida et al., 2015; Prpar Mihevc et al., 2017). These models provide valuable information in understanding some of the mechanisms underlying the system; but to the date they have not been able to replicate the exact human complexity of physiological functional-units formed by the connection of different cell-types, arising from separated microenvironments. Compartmentalized microfluidic culture systems (cμFCS) (Bhatia and Ingber, 2014) represent an alternative to overcome those problems and, combined with 3D-culture techniques and the use of human induced pluripotent stem cells, they could help recreating neuromuscular physiology of humans in-vitro.
This review aims to: (i) provide basic insights about the locomotion circuit and neuromuscular diseases required for its in-vitro modeling; (ii) review the breakthrough of bioengineered technologies for neuromuscular-systems; (iii) discuss the limitations and challenges of current study models and future prospects.
Locomotion circuit, also known as mechanosensory-motor circuit or reflex-arc circuit, is responsible for executing voluntary, and reflex skeletal-muscle movement, alternating flexion, and extension of the muscle (McCrea, 2001; Purves et al., 2004; Kiehn and Dougherty, 2013). The coordinated-action of cells taking part within is what generates movement: (i) motor-neurons (MN) are in charge of carrying information from the central nervous system to the muscle (Kandel et al., 2013); (ii) sensory-neurons (SN) carry information from the periphery of the body (the muscle in this case) to the central nervous system (Kandel et al., 2013); (iii) interneurons innervate motoneurons and are linked to their pattern of sensory input (Côté et al., 2018); (iv) Schwann cells are small cells that form a myelin-sheath around MN and SN axons that insulates them and enhances signal conduction (Kandel et al., 2013); (v) astrocytes maintain synapses, modulate the transmission of the signal, regulate blood flow, and availability of oxygen, nutrients, and survival factors onto neurons (Rindt et al., 2015); (vi) microglia are phagocytic and immunocompetent cells within the central nervous system, able to induce MN cell-death (Sargsyan et al., 2005; Frakes et al., 2014); (vii) skeletal-muscle cells are multinucleated and elongated cells, with sarcomeric striations that form muscle-fibers distributed in fascicle fashion and are the last executors of voluntary and reflex skeletal-muscle movement (Marieb, 2015; Tortora and Derrickson, 2017).
The events that take part within the neuromuscular-circuit to guide the movement in mammals could be resumed as follows. Once the brain takes the decision of initiating a movement, the signal is transmitted from neocortical projecting neurons through the spinal-cord. Then the spinal-locomotion circuit takes part of guiding the voluntary and reflex skeletal-muscle movement (Purves et al., 2004; Kandel et al., 2013; Tortora and Derrickson, 2017): (1) somatic α-motoneurons (MNs) arising from the ventral-horns of spinal-cord, send the input to the synaptic end-bulbs, triggering calcium flows inwards, and the release of the neurotransmitter acetylcholine (ACh) in the neuromuscular junction (NMJ) between the motoneuron and the motor-end plate of extrafusal muscle-fibers; (2) ACh binds specifically to the skeletal-muscle motor-end plates' ACh-receptors (AChR), inducing contraction of sarcolemma, releasing calcium into the sarcoplasm, that binds to troponin on the thin filaments, facilitating myosin-actin binding, and triggering muscle contraction; (3) intrafusal muscle fibers, located interspersed parallel to extrafusal fibers, change in length as the whole muscle changes; (4) sensory-neurons (SN) sense muscle fiber elongation through muscle-spindle—formed by SN nerve endings wrapped around central areas of intrafusal fibers—, and contraction through Golgi tendon organ—formed by encapsulated structures of collagen fibers located at the joint between muscle fibers and tendons that compress innervating a single SN axon—propagating an impulse signal back to the spinal-cord where is modulated by local interneurons and; (5) γ-motoneurons modulate excitatory input adjusting the contractibility of the muscle-spindle by stimulating intrafusal fibers adapting them to an appropriate length; (6) the integration of both afferent signals from the muscle-spindle and the Golgi tendon organ travels through the spinal-cord to the brain to have awareness of the position of the muscle and movement (muscle extension-flexion state), coordinating movements.
Failures in any part of this circuit can hamper coordinated muscle movement and be the cause of neuromuscular diseases (NMDs) or be the consequence that defines their severity (Gogliotti et al., 2012). The term of NMD comprises several diseases with different origins and affectations (such as muscular dystrophy, amyotrophic lateral sclerosis, myasthenia gravis, or spinal muscular atrophy). The effects of NMDs are reflected in the mechanosensory-motor circuit at different cellular levels—including sensory and motor neurons (Jablonka et al., 2006; Gogliotti et al., 2012), Schwann cells (Hunter et al., 2016; Vilmont et al., 2016; Santosa et al., 2018), astrocytes (Rindt et al., 2015), microglia (Frakes et al., 2014; Cooper-Knock et al., 2017), muscle (Martínez-Hernández et al., 2014; Maimon et al., 2018)—, as well as in the connections among them—NMJ (Uzel et al., 2016b; Maimon et al., 2018; Santhanam et al., 2018), muscle spindle (Rumsey et al., 2010; Guo et al., 2017)—, or intraspinal circuits. However, they all share symptoms such as: peripheral hypotonia, muscle weakness, and orthopedic deformities, among others (Bhatt, 2016; Morrison, 2016; Mary et al., 2018). These symptoms impoverishes patient's life-quality (Mary et al., 2018). There is still no treatment for them. Current study models are far from mimicking physiology and therefore are limited on helping to find cures. The technologies here reviewed (Figure 1) aim to help on that direction.
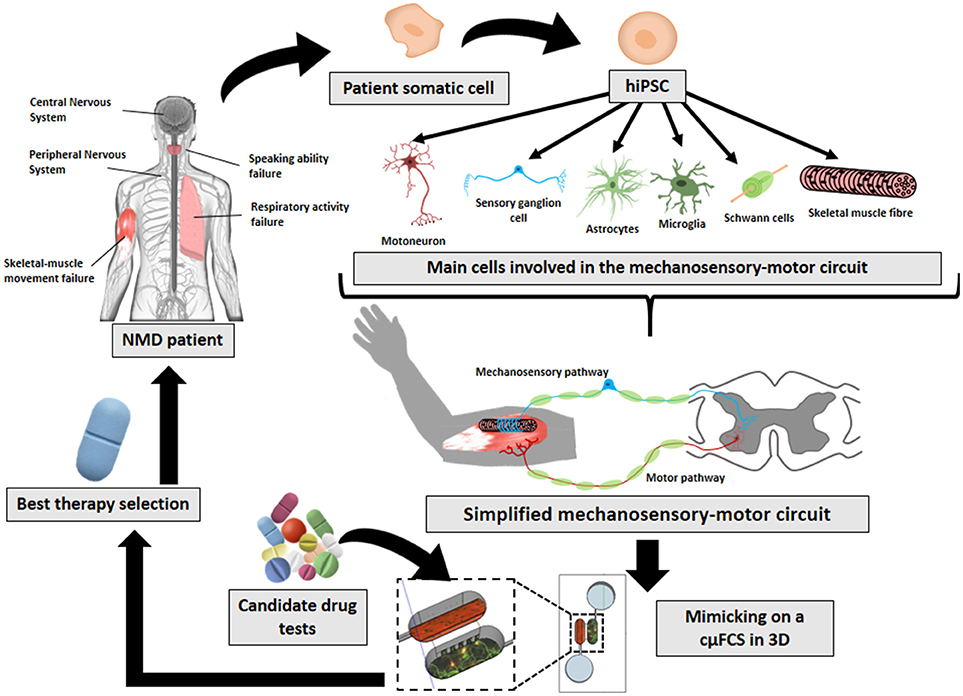
Figure 1. Future prospects on in-vitro neuromuscular disease modeling. The implementation and combination of later innovative technologies (hiPSCs, cμFCS, 3D cell-culture) would facilitate the recreation of patient's physiological conditions with his own disease-carrying cells mimicking the mechanosensory-motor circuit in 3D inside a microfluidic compartmentalized device. And finally, it would serve to find specific treatments for each NMD.
NMD and NMJ in-vitro models have gone through a long evolution history (Thomson et al., 2012): the use of primary cells, cell lines and stem cells; animal-animal cocultures, human-human cocultures, xeno-cocultures; and disease-specific studies. But as reported, most of the research has been done until now by coculturing healthy and diseased cells; primary cells or cell lines with stem cells; and mostly on rodent models or xeno-cultures (human-animal models). However, rodent models offer limited benefit translated into clinic as they do not carry human genetic background. Therefore, personalized medicine needs patient-specific isogenic disease models. In this regard, human induced pluripotent stem cells (hiPSCs) offer the possibility of obtaining different isogenic cell-types from patient's somatic cells, by overexpressing some transcription factors (Takahashi and Yamanaka, 2006), later reviewed (Amabile and Meissner, 2009). They can serve both for creating study models that mimic the physiopathology of the patient, with the further development of future therapeutic transplantation strategies (Su et al., 2013).
The use of induced pluripotent stem cells for disease-modeling, drug-screening, and regenerative therapies of NMDs has widely evolved in the last years (Selvaraj and Perlingeiro, 2018). Nevertheless, few studies have cultured hiPSC-derived motoneurons with hiPSC-derived skeletal-muscle cells in-vitro (Demestre et al., 2015; Puttonen et al., 2015; Maffioletti et al., 2018; Osaki et al., 2018). Puttonen et al. (2015) reported a method for the simultaneous differentiation of motoneurons and myotubes from patient-specific hiPSCs, obtaining neuronal differentiation, multinucleated spontaneously contracting myotubes, and functional NMJs on a 2D monolayer. In contrast, Demestre et al. used hiPSCs from healthy donors differentiated separately to motoneurons and myotubes, and subsequently cocultured in 2D. They observed AChR formation in the muscle and neurites outgrowing from motoneurons within the first weeks; but AChR aggregation, maturation of muscle cells, and NMJ formation was not detected until 3 weeks of monolayer cocultures (Demestre et al., 2015).
The advantages and disadvantages between 2D and 3D hiPSC cultures for neurodegenerative disease studies were recently reviewed (Centeno et al., 2018). Briefly, the main outstanding contributions of 3D cultures on NMDs are that: (i) it has been proved that 2D monolayers present in some cases altered gene expression whereas 3D cultures display a genotype more relevant to in-vivo (Smith et al., 2012; Centeno et al., 2018); (ii) cells cultured in 3D acquire more in-vivo like phenotype (lower proliferation rate, areas with different levels of oxygen distribution, higher cell-to-cell, and cell-extracellular matrix interactions, increased viability, proliferation, differentiation, and response to stimuli of other cells) (LaPlaca et al., 2010; Antoni et al., 2015; Centeno et al., 2018); (iii) some higher-order processes, such as angiogenesis, occur inherently in 3D (Baker and Chen, 2012; Centeno et al., 2018). But so far, only two studies have been able to mimic the NMJ in 3D using hiPSC-derived motoneurons and skeletal-muscle cells in-vitro (Maffioletti et al., 2018; Osaki et al., 2018). Maffioletti et al. (2018) used somatic cells from muscular dystrophy patients to create hiPSC-derived isogenic multilineages (skeletal-muscle cells, vascular endothelial cells, pericytes, and motoneurons), subsequently cocultured embedded in fibrin hydrogels. Osaki et al. (2018) created an ALS microphysiological 3D model culturing ALS-hiPSC-derived neural stem cells with hiPSC-derived skeletal-myoblasts embedded in collagen-Matrigel composites.
However, there are still no studies utilizing hiPSC-derived sensory-neurons and hiPSC-derived muscle-cells to mimic the sensory pathway of the spinal reflex-arc circuit. The latest advances on this respect were published by the group of J.J. Hickman using sensory-neurons derived from human neural progenitor cells and intrafusal fibers generated from human skeletal-muscle stem cells, cocultured on a 2D monolayer (Guo et al., 2017).
Traditional coculture methods do not consider: (i) the different microenvironment requirements of muscle, nerves, and neurons; (ii) the distal connections as they are physically separated in-vivo. Furthermore, finding a medium composition compatible for the long-term coculture of both cell-types could be challenging, as several medium components ideal for MN are incompatible with long-term maintenance of skeletal-muscle cells (Thomson et al., 2012; Tong et al., 2014). Hence, a coculture system between neurons and muscle offers limited benefits on mimicking the pathophysiology of NMDs.
The use of compartmentalized microfluidic culture systems (cμFCS) is increasingly growing for neurobiology studies due to the advantages offered compared to classical coculture systems, reviewed in Box 1.
Box 1. Top 10 advantages of compartmentalized microfluidic culture systems (cμFCS) compared to traditional coculture systems for mimicking spinal-locomotion circuit in-vitro.
1. Fluidic control. Compartmentalization enables to control de fluidic environment and provide each cell-type required nutrients to efficiently mature, facilitating survival, functionality, and long-term coculture (Park et al., 2006; Tong et al., 2014).
2. Experiment feasibility. Ease to study, enhance, control, and monitor some processes (cell proliferation, differentiation, directional growth, migration, and media diffusion from one compartment to the other) (Kamm and Bashir, 2014; Esch et al., 2015).
3. Microenvironment spatiotemporal control and monitoring. Independent manipulation of each compartment cells or extracellular matrix hydrogels could be a critical point to assess some processes. Compartmentalized platforms make possible to dissect molecular and cellular events occurring in somal vs. axonal compartments (effect of compounds, drug-sensitivitiy test, etc.) (Yang et al., 2009; Hur et al., 2011; Zahavi et al., 2015); to track individual axons through microchannels (Hosmane et al., 2011); and to assess axon-specific molecules through immunostaining, protein lisate isolation, and mRNA isolation (Saal et al., 2014; Zahavi et al., 2015).
4. Customisability of cμFCS designs. Control over microchannel geometry and dimensions (and therefore sifting of cells or compounds that can pass from one compartment to the other), number of compartments (and therefore number or different microenvironments, if required), compartment division tool (micochannels, microgrooves, membranes), distance between compartments, possibility to include reservoirs, covered, or opened compartments, scalability, and device size (Park et al., 2009; Yang et al., 2009; Hosmane et al., 2010; Uzel et al., 2016b).
5. Customisable engineering features. Possibility to integrate and take control over parameters: shear-stress flows (Joanne Wang et al., 2008; Shin, 2009); mechanical (Hosmane et al., 2011), optical (Renault et al., 2015; Jang et al., 2016), and electrical stimuli (Hallfors et al., 2013); topographical cues or micropatterns (Hoffman-Kim et al., 2010); chemical gradients (Uzel et al., 2016a); and sensory systems (Shen et al., 2004; Jeong et al., 2018).
6. Cost-effectiveness. Low volume of cells and reagents are required (Millet et al., 2007).
7. Control over the polarity of neural development. Culturing somas on one compartment makes possible to take control over axon and dendrite polarity during the development, and hence, to mimic axon injuries and study post-injury regeneration easily (Peyrin et al., 2011; Tong et al., 2015; Renault et al., 2016).
8. Possibility to mimic neural distal connections. Neurons growing on one compartment can extend their axons to interact with the other compartment cells, mimicking distal connections (Yang et al., 2009; Zahavi et al., 2015; Maimon et al., 2018).
9. Possibility to integrate control over the polarity of myocyte differentiation. Skeletal-muscle cells usually adopt randomized distribution in-vitro, whereas the presence of aligned micropatterns for 2D culture, cantilevers for 3D culture, or some stimuli methods (mechanical, electrical, or optical) integrated on the cμFCS can enhance its appropriate differentiation and functionality (Tourovskaia et al., 2008; Hume et al., 2012; Uzel et al., 2014, 2016b).
10. Better mimicking of physiological conditions and possibility to connect with other microfluidic platforms. This facilitates the study interactions between different physiological functional-units (Maschmeyer et al., 2015), as well as to integrate blood-flow effects (Maoz et al., 2018), or in a future, to mimic a full human-on-a-chip (Williamson et al., 2013; Kamm and Bashir, 2014), enhancing the development of therapies or diagnostic tools (Esch et al., 2014, 2015; Kamm and Bashir, 2014).
Most relevantly, they enable independent culture conditions for neurons and muscle cells, each supplied by its own microenvironment requirements in different interconnected but fluidically isolated compartments, whilst axons can still go through microgroves connecting cells of both compartments.
First compartmentalized microfluidic culture system (cμFCS) for neurobiology studies on neurotrophic effects of dorsal rood ganglion cells (Campenot, 1977) used Teflon-made open compartments, silicone-glue, and microchannels shattered in glass. Since then, and after soft-lithography fabrication improvements, cμFCS have widely evolved in the last 10 years for spinal-locomotion circuit studies (Supplementary Table 1). Most compartmentalized cocultures are nowadays performed onto polydimethylsiloxane (PDMS)-based platforms with two cellular compartments separated through microchannels or, as described in the only two publications found using a 3D cell coculture, through a gel region (Uzel et al., 2016b; Osaki et al., 2018). Different cell sources are used for MN (predominantly mouse embryonic primary cells) and muscle-cells (using equally rodent hind limb primary skeletal-muscle cells and C2C12 cell-line).
Besides, there are progressively more commercial cμFCS on the market (Zhang and Radisic, 2017), available for neuromuscular and other neurobiology studies in 2D or 3D, connecting two compartments through microchannels, microposts, or membranes, from the following companies: (i) Xona neuron devices (Xona Microfluidics, California), used in some neuromuscular studies (Southam et al., 2013; Blizzard et al., 2015); (ii) ANANDA Neuro-Device and Coculture-Device (Advanced Nano Design Applications Devices, Canada) employed by Magdesian et al. (2016) combined with AFM measurements to study neuronal growth; (iii) OrganoPlates for 3D culture (Mimetas BV, Netherlands) used to differentiate stem cells into neurons in 3D (Moreno et al., 2015) or for high-throughput evaluation of compounds in glia and neuronal 3D culture (Wevers et al., 2016); (iv) AIM 3D culture chips (AimBiotech, Singapore), employed mostly in cancer-research (Jenkins et al., 2018), although with a great potential to be used in neuromuscular studies, as performed by Uzel et al. (2016b) with a similar custom made device; (v) Neural Diode (MicroBrain Biotech, France) to reconstruct oriented neural network monolayer cultures (Peyrin et al., 2011; Deleglise et al., 2014); (vi) Idealized coculture chips (Synvivo, USA), with different options of radial slits or pillars utilized in many cases to mimic the BBB (Prabhakarpandian et al., 2013), or linear slits for compartmentalization purposes.
However, and despite the advantages offered by both hiPSC and cμFCS technologies, at the moment only two studies have attempted to mimic the spinal-locomotion circuit combining both technologies (Osaki et al., 2018; Santhanam et al., 2018). Santhanam et al. (2018) seed healthy donor's hiPSC-derived MN and human skeletal-muscle fibers in 2D, for dose-evaluation study of toxins affecting the NMJ. Osaki et al. create an ALS microphysiological 3D in-vitro study-model and compare it with a healthy model (muscle contraction, recovery, and response to drugs administered via endothelial cell barrier).
Uzel et al. (2016b) combine cμFCS with 3D cell-culture techniques for neuromuscular studies. Additionally, Santhanam et al. (2018) have attempted to mimic the spinal-locomotion circuit combining hiPSCs and cμFCS technologies. And Maffioletti et al. (2018) perform 3D culture of hiPSCs for NMD studies. But thus far, there is one single study combining 3D culture, hiPSCs and cμFCS as NMD models (Osaki et al., 2018). The three innovative technologies offer advantages (Figure 1), but the novelty itself comes with the challenge of developing, tuning, and implementing them together in a unique and biologically reproducible functional platform.
The co-culture of main cell-types participating in the spinal-locomotion circuit is mandatory to provide a native microenvironment, including the inherent release of growth-factors, as well as to support the viability and maturation of both muscle and neurons, and axon elongation of MNs (Gingras et al., 2008). For instance, spinal MNs cannot achieve proper maturation even after long-term maintenance, unless cultured with muscle-cells, and Schwann cells, as previously reviewed (Bucchia et al., 2018).
Besides, both SN and MN could be altered in particular NMDs (Jablonka et al., 2006; Rumsey et al., 2010; Guo et al., 2017), but not being many available studies focused on the muscle spindle (Taylor et al., 2005; Dagberg and Alstermark, 2006; Rumsey et al., 2010; Bewick and Banks, 2015; Matthews, 2015; Guo et al., 2017) challenges the task of mimicking and characterizing the mechanosensory spinal-locomotion circuit. Additionally, glial cells are also affected and involved in several neuromuscular pathologies (Lobsiger and Cleveland, 2007; Vilmont et al., 2016; Bucchia et al., 2018). Yet, most publications do not consider them.
Most in-vitro NMD studies are focused on the α-MN-muscle connection (Supplementary Table 1), very few on the γ-MN-muscle connection (Colón et al., 2017), and some on the SN-muscle connection (Taylor et al., 2005; Dagberg and Alstermark, 2006; Rumsey et al., 2010; Bewick and Banks, 2015; Matthews, 2015; Guo et al., 2017; Levin et al., 2017) and fewer on the internal SN-MN connection (Schwab and Ebert, 2014). But there is still very little known about what happens beyond those connections, to what extent cell-strategies for synaptic-specificity contribute on the formation of a functional connection (Fukuhara et al., 2013; Maimon et al., 2018).
Studying parts of the circuit cannot provide a comprehensive and complete view of the pathological process, and functional alterations occurring within it. The big challenge that remains out there on NMD studies is the modeling of the whole spinal-locomotion circuit with all cell-types involved, and the evaluation of independent and interdependent roles of each part of the circuit on the development of a particular NMD.
In a broad sense, neurodegeneration is a process characterized by the progressive functional loss of a population of neurons by intrinsic cell-death or the loss of support cells (i.e., oligodendrocytes or astrocytes). Most NMDs are characterized by this devastating phenomenon (i.e., motoneuron diseases: amyotrophic lateral sclerosis, or spinal muscular atrophy, etc.).
Indeed, studying only functional changes in MNs cannot give a comprehensive and complete picture of the process as it is also regulated by non-neuronal cells (Lobsiger and Cleveland, 2007; Bucchia et al., 2018; Maimon et al., 2018). For instance, a recent study performed by Maimon et al. (2018) demonstrated that axon degeneration only occurred with both MN and muscle cells carrying the genetic mutation indicative of the disorder. Furthermore, the morphology of spinal MN axons in-vitro differs from the one presented in-vivo: contrary to in-vivo, axonal terminals in-vitro manifest growth-cones, and are prone to regenerate and lengthen in response to neurotrophic factors required for the in-vitro maintenance of the culture (Bucchia et al., 2018). On top of that, another caveat is the fact that extrapolating effects of short-term studies to longer-term disease processes is often not correlated. Therefore, mimicking neurodegeneration response in-vitro as it occurs in-vivo endures to this day as a challenge. And consequently, there is no effective treatment for neurodegenerative NMDs to promote axonal regeneration yet.
Finding causes and treatment for NMD requires an accurate modeling of the microphysiological conditions that the patient is suffering. But reproducing the complete spinal-locomotion (reflex-arc) circuit in-vitro is very complex. Later progresses in neuromuscular-mimicking in-vitro systems, have been achieved incorporating increasingly evolving technologies of hiPSCs, cμFCS, and 3D cell-culture techniques here reviewed. The combination of novel technologies in the proper manner has proved to result in the acquisition of more reliable results (Uzel et al., 2016b; Maffioletti et al., 2018; Osaki et al., 2018; Santhanam et al., 2018). But there is still room for improvement. Future studies should focus on addressing unsolved questions related to: mimicking the whole spinal-locomotion circuit (including all cell-types involved, as well as evaluating the independent and interdependent roles of each one), defining the specific role of the factors that determine the NMD and their severity; mimicking neurodegeneration processes; and above all, finding treatments for NMD.
MB-M wrote the manuscript with contributions from AH, JdR, and JS.
This work was supported by Networking Biomedical Research Center (CIBER), Spain. CIBER is an initiative funded by the VI National R&D Plan 2008–2011, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions, and the Instituto de Salud Carlos III, with the support of the European Regional Development Fund. This work was funded by the CERCA Programme and by the Commission for Universities and Research of the Department of Innovation, Universities, and Enterprise of the Generalitat de Catalunya (2017 SGR 1079). This work was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) through the projects MINDS (Proyectos I+D Excelencia + FEDER): TEC2015-70104-P and BIOBOT (Programa Explora Ciencia/Tecnología): TEC2015-72718-EXP. This research was supported by grants from the Spanish Ministry of Economy, Industry and Competitiveness (MEICO / FEDER) (BFU2015-67777-R), the Spanish Prion Network (Prionet Spain, AGL2015-71764-REDT and AGL2017-90665-REDT), the Generalitat de Catalunya (SGR2017-648), CIBERNED (PRY-2016-2, MFDEND), CERCA Programme/Generalitat de Catalunya and La Marató de TV3 to JADR. AH was supported by a fellowship from the Juan de la Cierva program from MINECO.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2018.00194/full#supplementary-material
Amabile, G., and Meissner, A. (2009). Induced pluripotent stem cells: current progress and potential for regenerative medicine. Trends Mol. Med. 15, 59–68. doi: 10.1016/j.molmed.2008.12.003
Antoni, D., Burckel, H., Josset, E., and Noel, G. (2015). Three-dimensional cell culture: a breakthrough in vivo. Int. J. Mol. Sci. 16, 5517–5527. doi: 10.3390/ijms16035517
Baker, B. M., and Chen, C. S. (2012). Deconstructing the third dimension – how 3D culture microenvironments alter cellular cues. J. Cell Sci. 125, 3015–3024. doi: 10.1242/jcs.079509
Bewick, G. S., and Banks, R. W. (2015). Mechanotransduction in the muscle spindle. Pflügers Arch. Eur. J. Physiol. 467, 175–190. doi: 10.1007/s00424-014-1536-9
Bhatia, S. N., and Ingber, D. E. (2014). Microfluidic organs-on-chips. Nat. Biotechnol. 32, 760–772. doi: 10.1038/nbt.2989
Bhatt, J. M. (2016). The epidemiology of neuromuscular diseases. Neurol. Clin. 34, 999–1021. doi: 10.1016/j.ncl.2016.06.017
Blizzard, C. A., Southam, K. A., Dawkins, E., Lewis, K. E., King, A. E., Clark, J. A., et al. (2015). Identifying the primary site of pathogenesis in amyotrophic lateral sclerosis - vulnerability of lower motor neurons to proximal excitotoxicity. Dis. Model. Mech. 8, 215–224. doi: 10.1242/dmm.018606
Bucchia, M., Merwin, S. J., Re, D. B., and Kariya, S. (2018). Limitations and challenges in modeling diseases involving spinal motor neuron degeneration in vitro. Front. Cell. Neurosci. 12:61. doi: 10.3389/fncel.2018.00061
Côté, M. P., Murray, L. M., and Knikou, M. (2018). Spinal control of locomotion: individual neurons, their circuits and functions. Front. Physiol. 9:784. doi: 10.3389/fphys.2018.00784
Campenot, R. B. (1977). Local control of neurite development by nerve growth factor (chemotaxis/culture methods/retrograde transport/sympathetic ganglia). Cell Biol. 74, 4516–4519.
Centeno, E. G. Z., Cimarosti, H., and Bithell, A. (2018). 2D versus 3D human induced pluripotent stem cell-derived cultures for neurodegenerative disease modelling. Mol. Neurodegener. 13:27. doi: 10.1186/s13024-018-0258-4
Charoensook, S. N., Williams, D. J., Chakraborty, S., Leong, K. W., and Vunjak-Novakovic, G. (2017). Bioreactor model of neuromuscular junction with electrical stimulation for pharmacological potency testing. Integr. Biol. 9, 956–967. doi: 10.1039/C7IB00144D
Colón, A., Guo, X., Akanda, N., Cai, Y., and Hickman, J. J. (2017). Functional analysis of human intrafusal fiber innervation by human γ-motoneurons. Sci. Rep. 7:17202. doi: 10.1038/s41598-017-17382-2
Cooper-Knock, J., Green, C., Altschuler, G., Wei, W., Bury, J. J., Heath, P. R., et al. (2017). A data-driven approach links microglia to pathology and prognosis in amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 5:23. doi: 10.1186/s40478-017-0424-x
Dagberg, B., and Alstermark, B. (2006). Improved organotypic cell culture model for analysis of the neuronal circuit involved in the monosynaptic stretch reflex. J. Neurosci. Res. 84, 460–469. doi: 10.1002/jnr.20888
Deleglise, B., Magnifico, S., Duplus, E., Vaur, P., Soubeyre, V., Belle, M., et al. (2014). β-amyloid induces a dying-back process and remote trans-synaptic alterations in a microfluidic-based reconstructed neuronal network. Acta Neuropathol. Commun. 2:145. doi: 10.1186/s40478-014-0145-3
Demestre, M., Orth, M., Föhr, K. J., Achberger, K., Ludolph, A. C., Liebau, S., et al. (2015). Formation and characterisation of neuromuscular junctions between hiPSC derived motoneurons and myotubes. Stem Cell Res. 15, 328–336. doi: 10.1016/j.scr.2015.07.005
Esch, E. W., Bahinski, A., and Huh, D. (2015). Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 14, 248–260. doi: 10.1038/nrd4539
Esch, M. B., Smith, A. S., Prot, J. M., Oleaga, C., Hickman, J. J., and Shuler, M. L. (2014). How multi-organ microdevices can help foster drug development. Adv. Drug Deliv. Rev. 69–70, 158–169. doi: 10.1016/j.addr.2013.12.003
Frakes, A. E., Ferraiuolo, L., Haidet-Phillips, A. M., Schmelzer, L., Braun, L., Miranda, C. J., et al. (2014). Microglia induce motor neuron death via the classical NF-κB pathway in amyotrophic lateral sclerosis. Neuron 81, 1009–1023. doi: 10.1016/j.neuron.2014.01.013
Fukuhara, K., Imai, F., Ladle, D. R., Katayama, K. I., Leslie, J. R., Arber, S., et al. (2013). Specificity of monosynaptic sensory-motor connections imposed by repellent Sema3E-PlexinD1 signaling. Cell Rep. 5, 748–758. doi: 10.1016/j.celrep.2013.10.005
Gingras, M., Beaulieu, M. M., Gagnon, V., Durham, H. D., and Berthod, F. (2008). In vitro study of axonal migration and myelination of motor neurons in a three-dimensional tissue-engineered model. Glia 56, 354–364. doi: 10.1002/glia.20617
Gogliotti, R. G., Quinlan, K. A., Barlow, C. B., Heier, C. R., Heckman, C. J., and DiDonato, C. J. (2012). Motor neuron rescue in spinal muscular atrophy mice demonstrates that sensory-motor defects are a consequence, not a cause, of motor neuron dysfunction. J. Neurosci. 32, 3818–3829. doi: 10.1523/JNEUROSCI.5775-11.2012
Guo, X., Colon, A., Akanda, N., Spradling, S., Stancescu, M., Martin, C., et al. (2017). Tissue engineering the mechanosensory circuit of the stretch reflex arc with human stem cells: sensory neuron innervation of intrafusal muscle fibers. Biomaterials 122, 179–187. doi: 10.1016/j.biomaterials.2017.01.005
Guo, X., Gonzalez, M., Stancescu, M., Vandenburgh, H. H., and Hickman, J. J. (2011). Neuromuscular junction formation between human stem cell-derived motoneurons and human skeletal muscle in a defined system. Biomaterials 32, 9602–9611. doi: 10.1016/j.biomaterials.2011.09.014
Hallfors, N., Khan, A., Dickey, M. D., and Taylor, A. M. (2013). Integration of pre-aligned liquid metal electrodes for neural stimulation within a user-friendly microfluidic platform. Lab Chip 13, 522–526. doi: 10.1039/C2LC40954B
Happe, C. L., Tenerelli, K. P., Gromova, A. K., Kolb, F., and Engler, A. J. (2017). Mechanically patterned neuromuscular junctions-in-a-dish have improved functional maturation. Mol. Biol. Cell 28, 1950–1958. doi: 10.1091/mbc.e17-01-0046
Hoffman-Kim, D., Mitchel, J. A., and Bellamkonda, R. V. (2010). Topography, cell response, and nerve regeneration. Annu. Rev. Biomed. Eng. 12, 203–231. doi: 10.1146/annurev-bioeng-070909-105351
Hosmane, S., Fournier, A., Wright, R., Rajbhandari, L., Siddique, R., Yang, I. H., et al. (2011). Valve-based microfluidic compression platform: single axon injury and regrowth. Lab Chip 11, 3888–3895. doi: 10.1039/c1lc20549h
Hosmane, S., Yang, I. H., Ruffin, A., Thakor, N., and Venkatesan, A. (2010). Circular compartmentalized microfluidic platform: study of axon–glia interactions. Lab Chip 10, 741–747. doi: 10.1039/b918640a
Hume, S. L., Hoyt, S. M., Walker, J. S., Sridhar, B. V., Ashley, J. F., Bowman, C. N., et al. (2012). Alignment of multi-layered muscle cells within three-dimensional hydrogel macrochannels. Acta Biomater. 8, 2193–2202. doi: 10.1016/j.actbio.2012.02.001
Hunter, G., Powis, R. A., Jones, R. A., Groen, E. J., Shorrock, H. K., Lane, F. M., et al. (2016). Restoration of SMN in schwann cells reverses myelination defects and improves neuromuscular function in spinal muscular atrophy. Hum. Mol. Genet. 25, 2853–2861. doi: 10.1093/hmg/ddw141
Hur, E. M., Yang, I. H., Kim, D. H., Byun, J., Saijilafu, X.U. W. L., et al. (2011). Engineering neuronal growth cones to promote axon regeneration over inhibitory molecules. Proc. Natl. Acad. Sci.U.S.A. 108, 5057–5062. doi: 10.1073/pnas.1011258108
Hyun Sung, P, Su, L., McDonald, J., Thakor, N., and In Hong, Y (2013). “Neuromuscular junction in a microfluidic device,” in 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (IEEE), 2833–2835.
Ionescu, A., Zahavi, E. E., Gradus, T., Ben-Yaakov, K., and Perlson, E. (2016). Compartmental microfluidic system for studying muscle–neuron communication and neuromuscular junction maintenance. Eur. J. Cell Biol. 95, 69–88. doi: 10.1016/j.ejcb.2015.11.004
Jablonka, S., Karle, K., Sandner, B., Andreassi, C., von Au, K., and Sendtner, M. (2006). Distinct and overlapping alterations in motor and sensory neurons in a mouse model of spinal muscular atrophy. Hum. Mol. Genet. 15, 511–518. doi: 10.1093/hmg/ddi467
Jang, J. M., Lee, J., Kim, H., Jeon, N. L., and Jung, W. (2016). One-photon and two-photon stimulation of neurons in a microfluidic culture system. Lab Chip 16, 1684–1690. doi: 10.1039/C6LC00065G
Jenkins, R. W., Aref, A. R., Lizotte, P. H., Ivanova, E., Stinson, S., Zhou, C. W., et al. (2018). Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 8, 196–215. doi: 10.1158/2159-8290.CD-17-0833
Jeong, S., Kim, S., Buonocore, J., Park, J., Welsh, C. J., Li, J., et al. (2018). A three-dimensional arrayed microfluidic blood–brain barrier model with integrated electrical sensor array. IEEE Trans. Biomed. Eng. 65, 431–439. doi: 10.1109/TBME.2017.2773463
Joanne Wang, C., Li, X., Lin, B., Shim, S., Ming, G., and Levchenko, A. (2008). A microfluidics-based turning assay reveals complex growth cone responses to integrated gradients of substrate-bound ECM molecules and diffusible guidance cues. Lab Chip 8, 227–237. doi: 10.1039/b713945d
Kamm, R. D., and Bashir, R. (2014). Creating living cellular machines. Ann. Biomed. Eng. 42, 445–459. doi: 10.1007/s10439-013-0902-7
Kandel, E. R., Schwartz, J. H., Jessel, T. M., Siegelbaum, S. A., and Hudspeth, A. J. (eds.). (2013). Principles of Neural Science, 5th Edn. New York, NY: Mc Graw Hill Medical.
Kiehn, O., and Dougherty, K. (2013). “Locomotion: circuits and physiology,” in Neuroscience in the 21st Century, eds D. W. Pfaff (New York, NY: Springer), 1209–1236. doi: 10.1007/978-1-4614-1997-6_42
LaPlaca, M. C., Vernekar, V. N., Shoemaker, J. T., and Cullen, D. K. (2010). “Three-Dimensional Neuronal Cultures,” in Methods in Bioengineering: 3D Tissue Engineering, eds F. Berthiaume and J. R. Morgan (Norwood, OH: Artech House), 187–204.
Levin, E., Andreadaki, A., Gobrecht, P., Bosse, F., and Fischer, D. (2017). Nociceptive DRG neurons express muscle lim protein upon axonal injury. Sci. Rep. 7:643. doi: 10.1038/s41598-017-00590-1
Lobsiger, C. S., and Cleveland, D. W. (2007). Glial cells as intrinsic components of non-cell-autonomous neurodegenerative disease. Nat. Neurosci. 10, 1355–1360. doi: 10.1038/nn1988
Maffioletti, S. M., Sarcar, S., Henderson, A. B. H., Mannhardt, I., Pinton, L., Moyle, L. A., et al. (2018). Three-dimensional human iPSC-derived artificial skeletal muscles model muscular dystrophies and enable multilineage tissue engineering. Cell Rep. 23, 899–908. doi: 10.1016/j.celrep.2018.03.091
Magdesian, M. H., Lopez-Ayon, G. M., Mori, M., Boudreau, D., Goulet-Hanssens, A., Sanz, R., et al. (2016). Rapid mechanically controlled rewiring of neuronal circuits. J. Neurosci. 36, 979–987. doi: 10.1523/JNEUROSCI.1667-15.2016
Maimon, R., Ionescu, A., Bonnie, A., Sweetat, S., Wald-Altman, S., Inbar, S., et al. (2018). miR126-5p down-regulation facilitates axon degeneration and NMJ disruption via a non-cell-autonomous mechanism in ALS. J. Neurosci. 38, 5478–5494. doi: 10.1523/JNEUROSCI.3037-17.2018
Maoz, B. M., Herland, A., FitzGerald, E. A., Grevesse, T., Vidoudez, C., Pacheco, A. R., et al. (2018). A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat. Biotechnol. 36, 865–874. doi: 10.1038/nbt.4226
Marieb, E. N. (ed.). (2015). Essentials of Human Anatomy & Physiology, 11th Edn. Harlow: Pearson Education.
Martin, N. R., Passey, S. L., Player, D. J., Mudera, V., Baar, K., Greensmith, L., et al. (2015). Neuromuscular junction formation in tissue-engineered skeletal muscle augments contractile function and improves cytoskeletal organization. Tissue Eng. Part A 21, 2595–2604. doi: 10.1089/ten.tea.2015.0146
Martínez-Hernández, R., Bernal, S., Alias, L., and Tizzano, E. F. (2014). Abnormalities in early markers of muscle involvement support a delay in myogenesis in spinal muscular atrophy. J. Neuropathol. Exp. Neurol. 73, 559–567. doi: 10.1097/NEN.0000000000000078
Mary, P., Servais, L., and Vialle, R. (2018). Neuromuscular diseases: diagnosis and management. Orthop. Traumatol. Surg. Res. 104, S89–S95. doi: 10.1016/j.otsr.2017.04.019
Maschmeyer, I., Lorenz, A. K., Schimek, K., Hasenberg, T., Ramme, A. P., Hübner, J., et al. (2015). A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 15, 2688–2699. doi: 10.1039/C5LC00392J
Matthews, P. B. (2015). Where anatomy led, physiology followed: a survey of our developing understanding of the muscle spindle, what it does and how it works. J. Anat. 227, 104–114. doi: 10.1111/joa.12345
McCrea, D. A. (2001). Spinal circuitry of sensorimotor control of locomotion. J. Physiol. 533, 41–50. doi: 10.1111/j.1469-7793.2001.0041b.x
Millet, L. J., Stewart, M. E., Sweedler, J. V., Nuzzo, R. G., and Gillette, M. U. (2007). Microfluidic devices for culturing primary mammalian neurons at low densities. Lab Chip 7, 987–994. doi: 10.1039/b705266a
Moreno, E. L., Hachi, S., Hemmer, K., Trietsch, S. J., Baumuratov, A. S., Hankemeier, T., et al. (2015). Differentiation of neuroepithelial stem cells into functional dopaminergic neurons in 3D microfluidic cell culture. Lab Chip 15, 2419–2428. doi: 10.1039/C5LC00180C
Morimoto, Y., Kato-Negishi, M., Onoe, H., and Takeuchi, S. (2013). Three-dimensional neuron–muscle constructs with neuromuscular junctions. Biomaterials 34, 9413–9419. doi: 10.1016/j.biomaterials.2013.08.062
Morrison, B. (2016). Neuromuscular diseases. Semin. Neurol. 36, 409–418. doi: 10.1055/s-0036-1586263
Osaki, T., Uzel, S. G. M., and Kamm, R. D. (2018). Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. Sci. Adv. 4:eaat5847. doi: 10.1126/sciadv.aat5847
Park, J., Koito, H., Li, J., and Han, A. (2009). Microfluidic compartmentalized co-culture platform for CNS axon myelination research. Biomed. Microdev. 11, 1145–1153. doi: 10.1007/s10544-009-9331-7
Park, J. W., Vahidi, B., Taylor, A. M., Rhee, S. W., and Jeon, N. L. (2006). Microfluidic culture platform for neuroscience research. Nat. Protoc. 1, 2128–2136. doi: 10.1038/nprot.2006.316
Peyrin, J. M., Deleglise, B., Saias, L., Vignes, M., Gougis, P., Magnifico, S., et al. (2011). Axon diodes for the reconstruction of oriented neuronal networks in microfluidic chambers. Lab Chip 11, 3663–3673. doi: 10.1039/c1lc20014c
Prabhakarpandian, B., Shen, M. C., Nichols, J. B., Mills, I. R., Sidoryk-Wegrzynowicz, M., Aschner, M., et al. (2013). SyM-BBB: a microfluidic blood brain barrier model. Lab Chip 13, 1093–1101. doi: 10.1039/c2lc41208j
Prpar Mihevc, S., Pavlin, M., Darovic, S., Živin, M., Podbregar, M., Rogelj, B., et al. (2017). Modelling FUS mislocalisation in an in vitro model of innervated human muscle. J. Mol. Neurosci. 62, 318–328. doi: 10.1007/s12031-017-0940-y
Purves, D., Augustine, G. J., Hall, W. C., LaMantia, A. S., McNamara, J. O., and Williams, S. M. (eds) (2004). Neuroscience, 3rd Edn. Sunderland: Sinauer Associates, Inc.
Puttonen, K. A., Ruponen, M., Naumenko, N., Hovatta, O. H., Tavi, P., and Koistinaho, J. (2015). Generation of functional neuromuscular junctions from human pluripotent stem cell lines. Front. Cell. Neurosci. 9:473. doi: 10.3389/fncel.2015.00473
Renault, R., Durand, J. B., Viovy, J. L., and Villard, C. (2016). Asymmetric axonal edge guidance: a new paradigm for building oriented neuronal networks. Lab Chip 16, 2188–2191. doi: 10.1039/C6LC00479B
Renault, R., Sukenik, N., Descroix, S., Malaquin, L., Viovy, J. L., Peyrin, J. M., et al. (2015). Combining microfluidics, optogenetics and calcium imaging to study neuronal communication in vitro. PLoS ONE 10:e0120680. doi: 10.1371/journal.pone.0120680
Rindt, H., Feng, Z., Mazzasette, C., Glascock, J. J., Valdivia, D., Pyles, N., et al. (2015). Astrocytes influence the severity of spinal muscular atrophy. Hum. Mol. Genet. 24, 4094–4102. doi: 10.1093/hmg/ddv148
Rumsey, J. W., Das, M., Bhalkikar, A., Stancescu, M., and Hickman, J. J. (2010). Tissue engineering the mechanosensory circuit of the stretch reflex arc: sensory neuron innervation of intrafusal muscle fibers. Biomaterials 31, 8218–8227. doi: 10.1016/j.biomaterials.2010.07.027
Saal, L., Briese, M., Kneitz, S., Glinka, M., and Sendtner, M. (2014). Subcellular transcriptome alterations in a cell culture model of spinal muscular atrophy point to widespread defects in axonal growth and presynaptic differentiation. RNA 20, 1789–1802. doi: 10.1261/rna.047373.114
Santhanam, N., Kumanchik, L., Guo, X., Sommerhage, F., Cai, Y., Jackson, M., et al. (2018). Stem cell derived phenotypic human neuromuscular junction model for dose response evaluation of therapeutics. Biomaterials 166, 64–78. doi: 10.1016/j.biomaterials.2018.02.047
Santosa, K. B., Keane, A. M., Jablonka-Shariff, A., Vannucci, B., and Snyder-Warwick, A. K. (2018). Clinical relevance of terminal Schwann cells: an overlooked component of the neuromuscular junction. J. Neurosci. Res. 96, 1125–1135. doi: 10.1002/jnr.24231
Sargsyan, S. A., Monk, P. N., and Shaw, P. J. (2005). Microglia as potential contributors to motor neuron injury in amyotrophic lateral sclerosis. Glia 51, 241–253. doi: 10.1002/glia.20210
Schwab, A. J., and Ebert, A. D. (2014). Sensory neurons do not induce motor neuron loss in a human stem cell model of spinal muscular atrophy. PLoS ONE 9:e103112. doi: 10.1371/journal.pone.0103112
Selvaraj, S., and Perlingeiro, R. C. R. (2018). “Induced pluripotent stem cells for neuromuscular diseases: potential for disease modeling, drug screening, and regenerative medicine,” in Reference Module in Biomedical Sciences, eds C. A. McQueen, W. F. Boron, E. L. Boulpaep, A. Bradshaw, Ralph, D. B. Bylund, S. J. Enna, et al. (Elsevier). doi: 10.1016/B978-0-12-801238-3.65504-6
Shen, N. Y., Liu, Z., Jacquot, B. C., Minch, B. A., and Kan, E. C. (2004). Integration of chemical sensing and electrowetting actuation on chemoreceptive neuron MOS (CνMOS) transistors. Sens. Actuators B Chem. 102, 35–43. doi: 10.1016/j.snb.2003.10.013
Shin, H. S. (2009). Shear stress effect on transfection of neurons cultured in microfluidic devices. J. Nanosci. Nanotechnol. 9, 7330–7335. doi: 10.1166/jnn.2009.1769
Smith, A. S., Passey, S. L., Martin, N. R. W., Player, D. J., Mudera, V., Greensmith, L., et al. (2016). Creating interactions between tissue-engineered skeletal muscle and the peripheral nervous system. Cells. Tissues. Organs 202, 143–158. doi: 10.1159/000443634
Smith, S. J., Wilson, M., Ward, J. H., Rahman, C. V., Peet, A. C., Macarthur, D. C., et al. (2012). Recapitulation of tumor heterogeneity and molecular signatures in a 3D brain cancer model with decreased sensitivity to histone deacetylase inhibition. PLoS ONE 7:e52335. doi: 10.1371/journal.pone.0052335
Southam, K. A., King, A. E., Blizzard, C. A., McCormack, G. H., and Dickson, T. C. (2013). Microfluidic primary culture model of the lower motor neuron–neuromuscular junction circuit. J. Neurosci. Methods 218, 164–169. doi: 10.1016/j.jneumeth.2013.06.002
Su, H., Wang, L., Cai, J., Yuan, Q., Yang, X., Yao, X., et al. (2013). Transplanted motoneurons derived from human induced pluripotent stem cells form functional connections with target muscle. Stem Cell Res. 11, 529–539. doi: 10.1016/j.scr.2013.02.007
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Taylor, M. D., Holdeman, A. S., Weltmer, S. G., Ryals, J. M., and Wright, D. E. (2005). Modulation of muscle spindle innervation by neurotrophin-3 following nerve injury. Exp. Neurol. 191, 211–222. doi: 10.1016/j.expneurol.2004.09.015
Thomson, S. R., Wishart, T. M., Patani, R., Chandran, S., and Gillingwater, T. H. (2012). Using induced pluripotent stem cells (iPSC) to model human neuromuscular connectivity: promise or reality? J. Anat. 220, 122–130. doi: 10.1111/j.1469-7580.2011.01459.x
Tong, Z., Segura-Feliu, M., Seira, O., Homs-Corbera, A., Del Río, J. A., and Samitier, J. (2015). A microfluidic neuronal platform for neuron axotomy and controlled regenerative studies. RSC Adv. 5, 73457–73466. doi: 10.1039/C5RA11522A
Tong, Z., Seira, O., Casas, C., Reginensi, D., Homs-Corbera, A., Samitier, J., et al. (2014). Engineering a functional neuro-muscular junction model in a chip. RSC Adv. 4, 54788–54797. doi: 10.1039/C4RA10219C
Tortora, G. J., and Derrickson, B. (eds.). (2017). Principles of Anatomy and Physiology, 15th Edn. Danvers, MA: John Wiley & Sons, Inc.
Tourovskaia, A., Li, N., and Folch, A. (2008). Localized acetylcholine receptor clustering dynamics in response to microfluidic focal stimulation with agrin. Biophys. J. 95, 3009–3016. doi: 10.1529/biophysj.107.128173
Uzel, S. G., Platt, R. J., Subramanian, V., Pearl, T. M., Rowlands, C. J., Chan, V., et al. (2016b). Microfluidic device for the formation of optically excitable, three-dimensional, compartmentalized motor units. Sci. Adv. 2:e1501429–e1501429. doi: 10.1126/sciadv.1501429
Uzel, S. G. M., Amadi, O. C., Pearl, T. M., Lee, R. T., So, P. T. C., and Kamm, R. D. (2016a). Simultaneous or sequential orthogonal gradient formation in a 3D cell culture microfluidic platform. Small 12, 612–622. doi: 10.1002/smll.201501905
Uzel, S. G. M., Pavesi, A., and Kamm, R. D. (2014). Microfabrication and microfluidics for muscle tissue models. Prog. Biophys. Mol. Biol. 115, 279–293. doi: 10.1016/j.pbiomolbio.2014.08.013
Vilmont, V., Cadot, B., Ouanounou, G., and Gomes, E. R. (2016). A system for studying mechanisms of neuromuscular junction development and maintenance. Development 143, 2464–2477. doi: 10.1242/dev.130278
Wevers, N. R., van Vught, R., Wilschut, K. J., Nicolas, A., Chiang, C., Lanz, H. L., et al. (2016). High-throughput compound evaluation on 3D networks of neurons and glia in a microfluidic platform. Sci. Rep. 6:38856. doi: 10.1038/srep38856
Williamson, A., Singh, S., Fernekorn, U., and Schober, A. (2013). The future of the patient-specific body-on-a-chip. Lab Chip 13, 3471–3480. doi: 10.1039/c3lc50237f
Yang, I. H., Siddique, R., Hosmane, S., Thakor, N., and Höke, A. (2009). Compartmentalized microfluidic culture platform to study mechanism of paclitaxel-induced axonal degeneration. Exp. Neurol. 218, 124–128. doi: 10.1016/j.expneurol.2009.04.017
Yoshida, M., Kitaoka, S., Egawa, N., Yamane, M., Ikeda, R., Tsukita, K., et al. (2015). Modeling the early phenotype at the neuromuscular junction of spinal muscular atrophy using patient-derived iPSCs. Stem Cell Rep. 4, 561–568. doi: 10.1016/j.stemcr.2015.02.010
Zahavi, E. E., Ionescu, A., Gluska, S., Gradus, T., Ben-Yaakov, K., and Perlson, E. (2015). A compartmentalized microfluidic neuromuscular co-culture system reveals spatial aspects of GDNF functions. J. Cell Sci. 128, 1241–1252. doi: 10.1242/jcs.167544
Keywords: neuromuscular circuit, compartmentalized microfluidic culture systems (cμFCS), hiPSC, 3D-culture, in-vitro models
Citation: Badiola-Mateos M, Hervera A, del Río JA and Samitier J (2018) Challenges and Future Prospects on 3D in-vitro Modeling of the Neuromuscular Circuit. Front. Bioeng. Biotechnol. 6:194. doi: 10.3389/fbioe.2018.00194
Received: 31 August 2018; Accepted: 27 November 2018;
Published: 11 December 2018.
Edited by:
Massimo Alberti, Agency for Science, Technology and Research (A*STAR), SingaporeReviewed by:
Michela Pozzobon, Università degli Studi di Padova, ItalyCopyright © 2018 Badiola-Mateos, Hervera, del Río and Samitier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Josep Samitier, anNhbWl0aWVyQGliZWNiYXJjZWxvbmEuZXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.