
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bioeng. Biotechnol. , 11 December 2014
Sec. Computational Genomics
Volume 2 - 2014 | https://doi.org/10.3389/fbioe.2014.00065
This article is part of the Research Topic Bioinformatics of Non-Coding RNAs with Applications to Biomedicine: Recent Advances and Open Challenges View all 11 articles
The use of synthetic non-coding RNAs for post-transcriptional regulation of gene expression has not only become a standard laboratory tool for gene functional studies but it has also opened up new perspectives in the design of new and potentially promising therapeutic strategies. Bioinformatics has provided researchers with a variety of tools for the design, the analysis, and the evaluation of RNAi agents such as small-interfering RNA (siRNA), short-hairpin RNA (shRNA), artificial microRNA (a-miR), and microRNA sponges. More recently, a new system for genome engineering based on the bacterial CRISPR-Cas9 system (Clustered Regularly Interspaced Short Palindromic Repeats), was shown to have the potential to also regulate gene expression at both transcriptional and post-transcriptional level in a more specific way. In this mini review, we present RNAi and CRISPRi design principles and discuss the advantages and limitations of the current design approaches.
Natural regulatory RNAs are a heterogenous group of endogenous non-coding RNAs that modulate biological processes at many levels through different mechanisms. They have inspired the design of synthetic RNA molecules, such as riboswitches, sensors, and controllers, as key elements for programing cellular behaviors, as well as antisense-based approaches for specific gene expression regulation, which is the focus of this mini-review (Sharma et al., 2008; Culler et al., 2010; Liang et al., 2011).
RNA interference (RNAi) was discovered in 1998, when Andrew Fire and Craig C. Mello reported the capability of exogenous double-stranded RNAs (dsRNA) to silence genes in a specific manner in C. elegans (Fire et al., 1998). Central molecules in RNAi are microRNA (miRNA) and small-interfering RNA (siRNA).
miRNAs are small endogenous non-coding RNAs, typically 18–22 bp long, which derive from longer hairpin-shaped precursors called pre-miRNA (Bartel, 2004). A pre-miRNA can encode one or two different mature miRNAs, one from each arm (-5p and -3p). Pre-miRNAs come, in turn, from primary transcripts, called pri-miRNA, which are transcribed from miRNA genes. Mature miRNAs are incorporated into effector protein complexes called RISCs (RNA-induced silencing complex) and exert their regulatory function by binding specific target mRNAs through perfect or, more often, partial sequence complementarity, leading to the inhibition of their translation or promoting their degradation.
siRNAs are mostly exogenous dsRNA molecules derived from viral RNAs or artificially introduced into the cell (Chu and Rana, 2007).
The use of artificially designed siRNA has become a common and powerful strategy for the knock-down of gene expression yielding functional including therapeutic phenotypes (Gunsalus and Piano, 2005; Kim and Rossi, 2007). Several optimizations have been proposed in order to improve their efficacy and specificity (Liu et al., 2012b). Although research is focused on the development of selective delivery systems, a crucial factor is the presence of undesired off-target effects. siRNAs are designed to be perfectly complementary to their target sequences, ideally with few or no off-target genes. However, several studies have shown that a siRNA can bind mRNAs through partial complementarity, in a miRNA-like way, thus leading to undesirable and not easily predictable side effects (Birmingham et al., 2006; Jackson et al., 2006). In fact, despite the advances made in the recent past years, miRNA-target recognition has revealed itself to be a very dynamic mechanism influenced by many factors, which are only partially understood (Bartel, 2009; Thomas et al., 2010).
Along with specific gene silencing, the artificial repression of miRNAs can also provide a valuable tool for functional studies and have important therapeutic applications (Esquela-Kerscher and Slack, 2006; Garofalo et al., 2008; Croce, 2009). Two different strategies have been developed for the specific inhibition of miRNAs: antagomiRs and miRNA sponges (Krützfeldt et al., 2005; Ebert et al., 2007). The former consist of small RNAs exhibiting anti-complementarity to the miRNA to repress. The latter are longer RNA transcripts that act as attractors for miRNAs by distracting them from their original targets.
Finally, a novel methodology for artificial gene expression and miRNA regulation based on Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) has been recently proposed (Qi et al., 2013). CRISPR interference (CRISPRi) employs an engineered CRISPR/Cas system to control gene expression at the transcriptional level through a catalytically inactive Cas9 protein. Recent studies have shown that the CRISPR/Cas system can also target RNA (Hale et al., 2009).
In this mini-review, we summarize RNAi and CRISPRi design principles and discuss the advantages and limitations of the current approaches.
siRNA are usually synthesized as double-stranded RNA duplexes or as hairpin-shaped molecules called shRNA. The siRNA design process consists of the identification of a functional binding site on a target mRNA sequence, which will correspond to the sense strand of the siRNA. The anti-sense sequence is obtained as the complement to the sense strand.
Many studies have been conducted to determine the features associated to functional siRNAs and have allowed to establish siRNA design rules. Elbashir et al. (2001b) suggest to choose the 23-nt sequence motif AA(N19)TT as binding site, where N19 means any combination of 19 nucleotides (nt) and corresponds to the sense strand of the siRNA. The complement to AA(N19) corresponds to the anti-sense strand (Figures 1A–C).
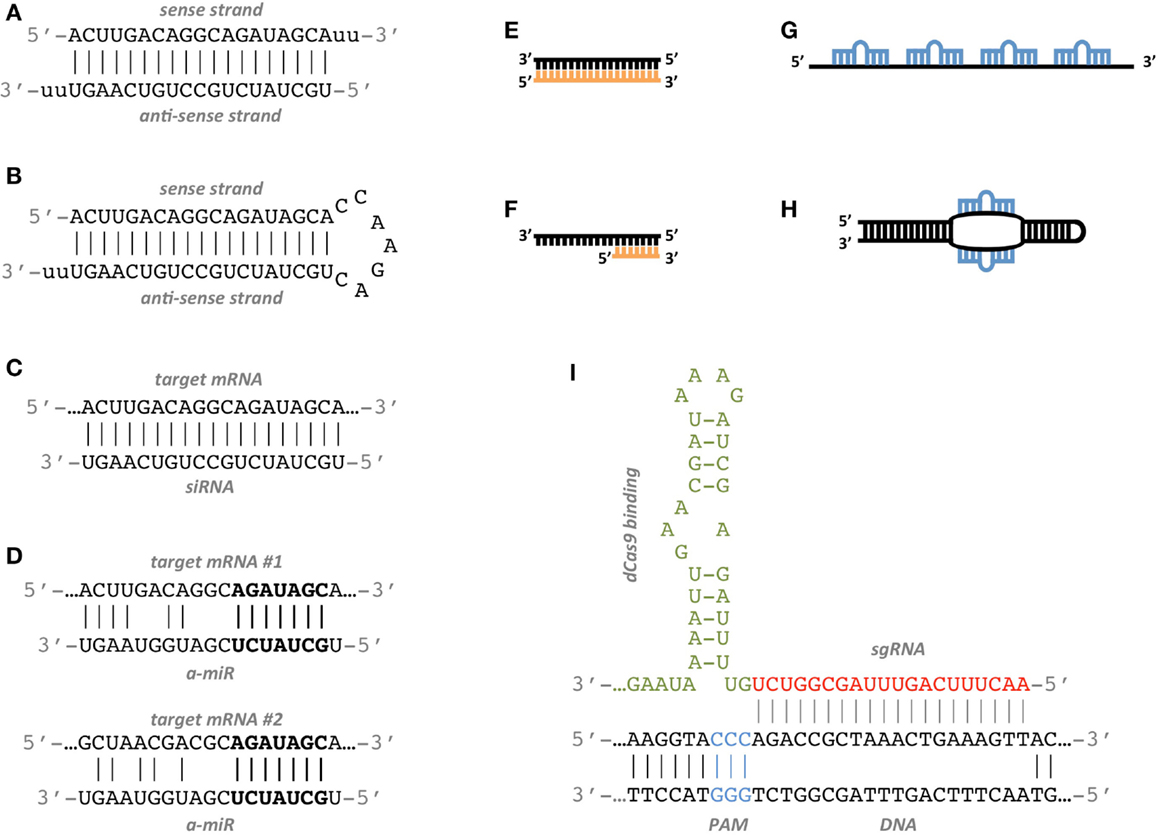
Figure 1. Artificial RNA constructs for miRNA and gene regulation. (A) Standard double strand siRNA; the anti-sense strand is the active agent which binds the target site. (B) shRNA construct; it is produced inside the target cell from a DNA construct that has been delivered to the nucleus and it expresses the anti-sense active strand. (C) The siRNA anti-sense strand binds the target mRNA with perfect complementarity. (D) Example of an a-miR sequence targeting two different sites with partial complementarity. The seed sequence of the a-miR, highlighted in bold characters, matches perfectly the target sites. (E) The antagomiR sequence (orange) perfectly matches the sequence of the target miRNA (black). (F) The Tiny LNA sequence (orange) perfectly matches the seed sequence of the target miRNA (black). (G) miRNA sponge construct with four miRNA binding sites separated by spacers. (H) Synthetic TUD construct with two exposed miRNA binding sites. (I) Model of a CRISPR sgRNA sequence binding the target DNA region. The PAM sequence (blue) is a short DNA motif juxtaposed to the DNA complementary region. The base-pairing nucleotides of the sgRNA are shown in red, while the dCas9-binding hairpin is in green.
Symmetric 3′ dTdT overhangs are added to the siRNA duplex to improve its stability and facilitate RISC loading. Although other combinations of nucleotides are acceptable, dGdG overhangs should be avoided, as they appear to be associated to decreased siRNA activity (Elbashir et al., 2001a,b; Strapps et al., 2010). siRNA duplexes often have asymmetric loading of the anti-sense versus sense strands. The strand whose 5′ end is thermodynamically less stable is preferentially incorporated into the RISC (Khvorova et al., 2003).
siRNA design rules can be classified into sequence and structure rules (See Table S1 in Supplementary Material). Sequence rules concern the position of the binding site in the target transcript and its nucleotide composition. The target region should be chosen preferably 50-100 nt downstream of the start codon and should avoid the middle of the coding sequence (Elbashir et al., 2001a; Hsieh et al., 2004). The G/C content of the binding site, and consequently of the siRNA, is relevant to the silencing activity and should be in the range of 30–55%, although values as low as 25% or as high as 79% are still associated to functional siRNAs (Reynolds et al., 2004; Liu et al., 2012a).
Numerous sequence rules regard the selection of nucleotides to prefer or avoid in specific positions of either the sense or the anti-sense strand of the duplex. For example, a higher content of A/U nucleotides in the 5′ end of the anti-sense strand of the siRNA yields higher silencing efficacy (Ui-Tei et al., 2004; Shabalina et al., 2006). Also, the 5′ half of the anti-sense strand dictates competition potency of siRNAs, which is a consequence of the RNAi machinery saturation followed by transfection of multiple siRNAs (Yoo et al., 2008).
Other relevant sequence features include the absence of internal repeats and the presence/absence of specific motifs (Reynolds et al., 2004).
Structure rules refer to the thermodynamics features of the siRNA/target duplex and are mostly expressed in terms of the nucleotide composition of the duplex itself or of the area surrounding the binding site (Chalk et al., 2004; Shabalina et al., 2006). Structure rules specify functional levels of binding energy at different positions of the duplex, and optimal energy difference between different positions of the duplex itself. Another important thermodynamic feature associated to siRNA efficacy is the structural accessibility of the target site. It has been demonstrated, in fact, that an mRNA stretch, which is not involved in a strict secondary structure exhibits a stronger binding affinity to a siRNA (or miRNA) molecule than one with a highly structured conformation (Tafer et al., 2008).
Several optimizations have been proposed to improve the activity of siRNA molecules, such as a more accurate prediction of the active strand of the duplex, design rules to avoid competition with endogenous miRNAs and vectors expressing multiple siRNAs at once (Cheng et al., 2009; Ma et al., 2014; Malefyt et al., 2014).
Many tools are available online for the design of siRNA and shRNA molecules (see Table 1).
Although siRNAs and shRNAs are designed to specifically target a single gene through perfect complementarity to the binding site, several studies show that they can partially bind to many other transcripts in a way reminiscent of the endogenous miRNAs (Birmingham et al., 2006; Jackson et al., 2006). A single miRNA can potentially regulate hundreds of different mRNAs through partial sequence complementarity. In particular, perfect base pairing of the 5′ end region of the miRNA, termed “seed,” to a binding site located in the 3′ UTR of a mRNA, is usually sufficient to yield a significant repression of the target, while other recent studies also report functional centered site-mediated interactions (Shin et al., 2010; Helwak et al., 2013; Martin et al., 2014).
This represents a relevant drawback of single-target siRNAs, especially when pools of four or five siRNA duplexes per target gene are used to achieve stronger repression but also leading to widespread off-target effects.
One approach to the off-targeting problem consists of employing pools of siRNAs, at low concentrations, that target a single gene in multiple sites (Straka and Boese, 2010). The advantage of this approach lies in the fact that such pools are both effective on that one target, while the effects of a low concentrations siRNA on other potential targets should be negligible (Arvey et al., 2010; Larsson et al., 2010). Another study showed that siRNAs with a bulge at position 2 of the anti-sense strand were able to discriminate better between perfectly matched and mismatched targets (Dua et al., 2009; Li et al., 2010).
Targeting multiple genes can also be an intended choice, as there are many biological and biomedical applications in which it is important to regulate multiple genes at once while suffering as few side effects as possible. One way to achieve this goal is to exploit the multi-targeting properties of endogenous miRNAs by employing artificially designed miRNAs, or a-miRs. Two recent papers have shown that a-miRs can successfully repress at least two targets simultaneously by binding to one or more sites in their 3′ UTRs (Figure 1D) (Arroyo et al., 2014; Lagana et al., 2014). The employment of a single multi-target a-miR in place of a pair or a pool of single-target siRNAs is likely to yield significant repression of targets with few off-target effects.
While the inhibition of over-expressed genes has been the main goal of RNAi research for years, the de-repression of down-regulated miRNA targets has increasingly gained importance over time. The inhibition of endogenous miRNAs was first introduced in 2005 by Krützfeldt et al. (2005). They employed cholesterol-conjugated oligo-ribonucleotides, which they termed “antagomiRs,” reproducing the anti-sense strand of the endogenous miRNA they inhibit. Their design is thus straightforward, as there is not much space for sequence variations (Figure 1E). Since then, a variety of chemical modifications have been proposed in order to increase binding affinity, improve nuclease resistance and facilitate in vivo delivery. They include locked nucleic acid (LNA), which possesses the highest affinity toward complementary RNA, Bifunctional oligodeoxynucleotide/antagomiR constructs, which ensure high transfection efficiency and prevention of unintended immune stimulation, and morpholino oligomers, which have been shown to be efficient inhibitors of both pri-miRNA and mature miRNA activity in zebrafish and Xenopus laevis (Summerton and Weller, 1997; Braasch and Corey, 2001; Petersen and Wengel, 2003; Ziegler et al., 2013). A further variant of antagomiRs is represented by short seed-targeting LNA oligonucleotides, called tiny LNAs. These molecules allow simultaneous inhibition of miRNAs within families sharing the same seed (Figure 1F) (Obad et al., 2011).
AntagomiRs represent one well-established tool for miRNA functional studies, and several works have also shown successful employment of antagomiRs as therapeutic agents able to restore disease-associated pathways altered by miRNA up-regulation. Like siRNAs, AntagomiRs can also have significant off-target effects, as they act like endogenous miRNAs and may hit complementary mRNA transcripts. However, experiments have showed no detectable effect on mRNAs with perfect tiny LNA complementary sites, not even at the proteomic level (Obad et al., 2011).
miRNA sponges are an alternative to antagomiRs. They act as competitive inhibitors that distract endogenous miRNAs from their natural targets. Many sponge variants have been described, such as miRNA-target mimics, miRNA decoys, and miRNA erasers, and they all consist of RNA constructs containing multiple binding sites for the miRNA to be sponged (Figure 1G) (Carè et al., 2007; Ebert et al., 2007; Franco-Zorrilla et al., 2007; Sayed et al., 2008).
A basic sponge consists of an RNA sequence exhibiting 4–10 miRNA binding sites separated by short spacers, usually 2–4 nt long. These sites can be either bulged or perfectly complementary to the miRNAs. In the first case, a bulge at positions 9–12 of the binding site is introduced in order to prevent cleavage and degradation of the sponge. Sponges with bulged binding sites produce stronger de-repressive effects than sponges with perfect binding sites (Ebert et al., 2007). Kluiver et al. (2012) developed a methodology for the rapid generation of miRNA sponges by making use of simple constructs with up to 20 perfect or bulged miRNA binding sites.
Structural optimizations have also been proposed. TuD RNAs (tough decoy RNAs) are efficient sponges with structurally accessible and indigestible miRNA binding sites (Figure 1H) (Haraguchi et al., 2009, 2012). The optimal TuD RNA consists of a bulged stem-loop structure where both sides of the bulge are miRNA binding sites which are perfectly complementary to the miRNA sequence and which do not form any base-pairing regions longer than 9nt.
An exciting and promising advance in the field of artificial gene regulation comes from Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR). CRISPR is a natural adaptive immune system used by archaea and bacteria against phage and plasmids (Jinek et al., 2012). This system is genomically encoded by the prokaryotic chromosome and consists of a series of short repeats separated by spacer sequences that match previously encountered foreign DNA. Thus, CRISPR arrays are transcribed and processed in order to produce mature crRNAs, which are loaded onto effector protein complexes and function as a guide to target recognition and degradation.
The CRISPR/Cas system has been engineered to function with synthetic small guide RNA (sgRNA) in order to perform genome editing in eukaryotes (Mali et al., 2013) (See Table S2 in Supplementary Material). The sgRNA consists of a 20 nt crRNA sequence complementary to the target region followed by a 42 nt Cas9-binding hairpin and a 40 nt transcription terminator. The target region must be of the form N20NGG that is any 21 nucleotides followed by GG. NGG is the 3′ protospacer-adjacent motif (PAM) and is required for Cas9 binding. This particular PAM sequence is derived from Streptococcus pyogenes, but other functional PAM sequences have been characterized from other bacteria (Esvelt et al., 2013). In addition, if a U6 snRNA or T7 promoters are used to express the sgRNA, this must start with G or GG, respectively, in order to maintain transcript initiation. Thus, the target region must be of the form GN19NGG or GGN18NGG. Ultimately, the beginning of the sgRNA and the PAM sequence will depend on the specific promoters and Cas9 used (Figure 1I).
DNA breaks caused by Cas9 are repaired through either homologous recombination or non-homologous end joining (NHEJ) mechanisms, thus this system can be used to either disrupt or edit a gene (i.e., insertions and deletions). Many tools are currently available online for the design of sgRNAs (Table 1).
Besides genome-editing applications, the CRISPR/Cas9 system can be employed for gene expression regulation. The system, known as CRISPR interference (CRISPRi), is based on a catalytically dead Cas9 (dCas9) lacking endonuclease activity co-expressed with a sgRNA (Gilbert et al., 2013; Qi et al., 2013). Instead of generating DNA breaks, the recognition complex interferes with transcriptional elongation, RNA polymerase binding, or transcription factor binding, leading to efficient inhibition of gene expression. CRISPRi gene silencing is inducible and reversible and recognition of the targets depends solely on the sgRNA sequence (Qi et al., 2013).
A “seed” region has been identified as the 12nt region adjacent to the PAM site. Mismatches in the seed region can dramatically reduce the repression, while mismatches in the non-seed area can cause a mild effect. Design guidelines recommend using a length of 20–25 nt as the base-pairing region of the sgRNA (Larson et al., 2013) and provide specific design rules based on nucleotide preference for active sgRNA (Doench et al., 2014). A recent study aimed at the identification of features of effective sgRNA specific to CRISPRi, suggests that the target site should be chosen from −50 to +300 bp relative to the Transcription Start Site (TSS) of a gene (Gilbert et al., 2014). The authors observed that nucleotide homopolymers have a strongly negative effect on sgRNA activity and that the GC content of the sgRNA or the binding site is not correlated with sgRNA activity, although another study reports a decreased activity of sgRNA with low or high GC content (Doench et al., 2014). Moreover, CRISPRi activity seems to be highly sensitive to mismatches between the sgRNA and DNA sequence, thus the authors conclude that properly designed sgRNA will have minimal off-target effects. However, previous studies reported silencing activity with sgRNAs exhibiting mismatches to the target in the seed area (Cradick et al., 2013) and that off-targets might be cell type dependent and determined by various complicated factors in addition to primary DNA sequences (Duan et al., 2014). Thus, side effects still constitute a challenge, which needs to be properly addressed by further focused research.
Gilbert et al. also introduced the sunCas9 CRISPRa system, in which expression of a single sgRNA with one binding site is sufficient to turn on genes that are poorly expressed or that increase the expression of well-expressed genes (Gilbert et al., 2014; Tanenbaum et al., 2014).
CRISPRi can also be successfully employed to knock out miRNAs, by using a sgRNA/Cas9 complex targeting the pre-miRNA sequence (Zhao et al., 2014), and to study functional miRNA-target interactions in vivo by site-specific genome engineering (Bassett et al., 2014).
Finally, although current tools for CRISPRi are based on the DNA targeting approach described above, the discovery of other Cas proteins targeting RNA molecules, such as Cmr, suggests an alternative post-transcriptional methodology similar to RNAi (Hale et al., 2009; Zebec et al., 2014).
Both RNAi and CRISPRi represent valid approaches for artificial gene regulation and both can suffer from significant side effects which may result from factors beyond sequence match. One clear advantage of CRISPRi over RNAi is that being an exogenous system it does not compete with the endogenous machinery of miRNA processing. Nevertheless, both techniques require more work in terms of enhancing targeting efficiency and reducing side effects.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Journal/10.3389/fbioe.2014.00065/abstract
Arroyo, J. D., Gallichotte, E. N., and Tewari, M. (2014). Systematic design and functional analysis of artificial microRNAs. Nucleic Acids Res. 42, 6064–6077. doi:10.1093/nar/gku171
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arvey, A., Larsson, E., Sander, C., Leslie, C. S., and Marks, D. S. (2010). Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 6, 1–7. doi:10.1038/msb.2010.24
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi:10.1016/S0092-8674(04)00045-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi:10.1016/j.cell.2009.01.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bassett, A. R., Azzam, G., Wheatley, L., Tibbit, C., Rajakumar, T., McGowan, S., et al. (2014). Understanding functional miRNA-target interactions in vivo by site-specific genome engineering. Nat. Commun. 5, 1–11. doi:10.1038/ncomms5640
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Birmingham, A., Anderson, E. M., Reynolds, A., Ilsley-Tyree, D., Leake, D., Fedorov, Y., et al. (2006). 3’ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Meth. 3, 199–204. doi:10.1038/nmeth854
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Braasch, D. A., and Corey, D. R. (2001). Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 8, 1–7. doi:10.1016/S1074-5521(00)00058-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carè, A., Catalucci, D., Felicetti, F., Bonci, D., Addario, A., Gallo, P., et al. (2007). MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13, 613–618. doi:10.1038/nm1582
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chalk, A. M., Wahlestedt, C., and Sonnhammer, E. L. L. (2004). Improved and automated prediction of effective siRNA. Biochem. Biophys. Res. Commun. 319, 264–274. doi:10.1016/j.bbrc.2004.04.181
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, T. L., Teng, C. F., Tsai, W. H., Yeh, C. W., Wu, M. P., Hsu, H. C., et al. (2009). Multitarget therapy of malignant cancers by the head-to-tail tandem array multiple shRNAs expression system. Cancer Gene Ther. 16, 516–531. doi:10.1038/cgt.2008.102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chu, C.-Y., and Rana, T. M. (2007). Small RNAs: regulators and guardians of the genome. J. Cell. Physiol. 213, 412–419. doi:10.1002/jcp.21230
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi:10.1126/science.1231143
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cradick, T. J., Fine, E. J., Antico, C. J., and Bao, G. (2013). CRISPR/Cas9 systems targeting-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 41, 9584–9592. doi:10.1093/nar/gkt714
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Croce, C. M. (2009). Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704–714. doi:10.1038/nrg2634
Cui, W., Ning, J., Naik, U. P., and Duncan, M. K. (2004). OptiRNAi, an RNAi design tool. Comput. Methods Programs Biomed. 75, 67–73. doi:10.1016/j.cmpb.2003.09.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Culler, S. J., Hoff, K. G., and Smolke, C. D. (2010). Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 330, 1251–1255. doi:10.1126/science.1192128
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ding, Y., Chan, C. Y., and Lawrence, C. E. (2004). Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 32, W135–W141. doi:10.1093/nar/gkh449
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Doench, J. G., Hartenian, E., Graham, D. B., Tothova, Z., Hegde, M., Smith, I., et al. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. doi:10.1038/nbt.3026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dua, P., Yoo, J. W., Kim, S., and Lee, D.-K. (2009). Modified siRNA structure with a single nucleotide bulge overcomes conventional siRNA-mediated off-target silencing. Mol. Ther. 19, 1676–1687. doi:10.1038/mt.2011.109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Duan, J., Lu, G., Xie, Z., Lou, M., Luo, J., Guo, L., et al. (2014). Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Res. 24, 1009–1012. doi:10.1038/cr.2014.87
Ebert, M. S., Neilson, J. R., and Sharp, P. A. (2007). MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Meth. 4, 721–726. doi:10.1038/nmeth1079
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001a). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. doi:10.1038/35078107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Elbashir, S. M., Martinez, J., Patkaniowska, A., Lendeckel, W., and Tuschl, T. (2001b). Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20, 6877–6888. doi:10.1093/emboj/20.23.6877
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Esquela-Kerscher, A., and Slack, F. J. (2006). Oncomirs – microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269. doi:10.1038/nrc1840
Esvelt, K. M., Mali, P., Braff, J. L., Moosburner, M., Yaung, S. J., and Church, G. M. (2013). Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Meth. 10, 1116–1121. doi:10.1038/nmeth.2681
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi:10.1038/35888
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi:10.1038/ng2079
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Garofalo, M., Condorelli, G., and Croce, C. M. (2008). MicroRNAs in diseases and drug response. Curr. Opin. Pharmacol. 8, 661–667. doi:10.1016/j.coph.2008.06.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gilbert, L. A., Horlbeck, M. A., Adamson, B., Villalta, J. E., Chen, Y., Whitehead, E. H., et al. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661. doi:10.1016/j.cell.2014.09.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gilbert, L. A., Larson, M. H., Morsut, L., Liu, Z., Brar, G. A., Torres, S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. doi:10.1016/j.cell.2013.06.044
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guell, M., Yang, L., and Church, G. M. (2014). Genome editing assessment using CRISPR genome analyzer (CRISPR-GA). Bioinformatics 30, 2968–2970. doi:10.1093/bioinformatics/btu427
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gunsalus, K. C., and Piano, F. (2005). RNAi as a tool to study cell biology: building the genome-phenome bridge. Curr. Opin. Cell Biol. 17, 3–8. doi:10.1016/j.ceb.2004.12.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hale, C. R., Zhao, P., Olson, S., Duff, M. O., Graveley, B. R., Wells, L., et al. (2009). RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139, 945–956. doi:10.1016/j.cell.2009.07.040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haraguchi, T., Nakano, H., Tagawa, T., Ohki, T., Ueno, Y., Yoshida, T., et al. (2012). A potent 2′-O-methylated RNA-based microRNA inhibitor with unique secondary structures. Nucleic Acids Res. 40, e58–e58. doi:10.1093/nar/gkr1317
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haraguchi, T., Ozaki, Y., and Iba, H. (2009). Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 37, e43–e43. doi:10.1093/nar/gkp040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heigwer, F., Kerr, G., and Boutros, M. (2014). Correspondence. Nat. Meth. 11, 122–123. doi:10.1038/nmeth.2812
Helwak, A., Kudla, G., Dudnakova, T., and Tollervey, D. (2013). Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153, 654–665. doi:10.1016/j.cell.2013.03.043
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hsieh, A., Bo, R., Manola, J., Vazquez, F., Bare, O., Khvorova, A., et al. (2004). A library of siRNA duplexes targeting the phosphoinositide 3-kinase pathway: determinants of gene silencing for use in cell-based screens. Nucleic. Acids Res. 32, 893–901. doi:10.1093/nar/gkh238
Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S., Agarwala, V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. doi:10.1038/nbt.2647
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., et al. (2013). Brief communications. Nat. Biotechnol. 31, 227–229. doi:10.1038/nbt.2501
Ichihara, M., Murakumo, Y., Masuda, A., Matsuura, T., Asai, N., Jijiwa, M., et al. (2007). Thermodynamic instability of siRNA duplex is a prerequisite for dependable prediction of siRNA activities. Nucleic Acids Res. 35, e123–e123. doi:10.1093/nar/gkm699
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jackson, A. L., Burchard, J., Schelter, J., Chau, B. N., Cleary, M., Lim, L., et al. (2006). Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12, 1179–1187. doi:10.1261/rna.25706
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jiang, P., Wu, H., Da, Y., Sang, F., Wei, J., Sun, X., et al. (2007). RFRCDB-siRNA: Improved design of siRNAs by random forest regression model coupled with database searching. Comput. Methods Programs Biomed. 87, 230–238. doi:10.1016/j.cmpb.2007.06.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi:10.1126/science.1225829
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Katoh, T., and Suzuki, T. (2007). Specific residues at every third position of siRNA shape its efficient RNAi activity. Nucleic Acids Res. 35, e27–e27. doi:10.1093/nar/gkl1120
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khvorova, A., Reynolds, A., and Jayasena, S. D. (2003). Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216. doi:10.1016/S0092-8674(03)00801-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, D. H., and Rossi, J. J. (2007). Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 8, 173–184. doi:10.1038/nrg2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kluiver, J., Gibcus, J. H., Hettinga, C., Adema, A., Richter, M. K. S., Halsema, N., et al. (2012). Rapid generation of MicroRNA sponges for microRNA inhibition. PLoS ONE 7:e29275. doi:10.1371/journal.pone.0029275
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Krützfeldt, J., Rajewsky, N., Braich, R., Rajeev, K. G., Tuschl, T., Manoharan, M., et al. (2005). Silencing of microRNAs in vivo with “antagomirs”. Nature 438, 685–689. doi:10.1038/nature04303
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lagana, A., Acunzo, M., Romano, G., Pulvirenti, A., Veneziano, D., Cascione, L., et al. (2014). miR-Synth: a computational resource for the design of multi-site multi-target synthetic miRNAs. Nucleic Acids Res. 42, 5416–5425. doi:10.1093/nar/gku202
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Larson, M. H., Gilbert, L. A., Wang, X., Lim, W. A., Weissman, J. S., and Qi, L. S. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180–2196. doi:10.1038/nprot.2013.132
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Larsson, E., Sander, C., and Marks, D. (2010). mRNA turnover rate limits siRNA and microRNA efficacy. Mol. Syst. Biol. 6, 1–9. doi:10.1038/msb.2010.89
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, X., Yoo, J. W., Lee, J. H., Hahn, Y., Kim, S., and Lee, D.-K. (2010). Identification of sequence features that predict competition potency of siRNAs. Biochem. Biophys. Res. Commun. 398, 92–97. doi:10.1016/j.bbrc.2010.06.041
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liang, J. C., Bloom, R. J., and Smolke, C. D. (2011). Engineering biological systems with synthetic RNA molecules. Mol. Cell 43, 915–926. doi:10.1016/j.molcel.2011.08.023
Liu, Q., Zhou, H., Cui, J., Cao, Z., and Xu, Y. (2012a). Reconsideration of in-silico siRNA design based on feature selection: a cross-platform data integration perspective. PLoS ONE 7:e37879. doi:10.1371/journal.pone.0037879
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, Q., Zhou, H., Zhu, R., Xu, Y., and Cao, Z. (2012b). Reconsideration of in silico siRNA design from a perspective of heterogeneous data integration: problems and solutions. Brief Bioinform 15, 292–305. doi:10.1093/bib/bbs073
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lu, Z. J., and Mathews, D. H. (2008). OligoWalk: an online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 36, W104–W108. doi:10.1093/nar/gkn250
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, H., Zhang, J., and Wu, H. (2014). Designing Ago2-specific siRNA/shRNA to avoid competition with endogenous miRNAs. Mol Ther Nucleic Acids 3, e176. doi:10.1038/mtna.2014.27
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, M., Ye, A. Y., Zheng, W., and Kong, L. (2013). A guide RNA sequence design platform for the CRISPR/Cas9 system for model organism genomes. Biomed Res. Int. 2013, 1–4. doi:10.1155/2013/270805
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Malefyt, A. P., Wu, M., Vocelle, D. B., Kappes, S. J., Lindeman, S. D., Chan, C., et al. (2014). Improved asymmetry prediction for short interfering RNAs. FEBS J. 281, 320–330. doi:10.1111/febs.12599
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi:10.1126/science.1232033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martin, H. C., Wani, S., Steptoe, A. L., Krishnan, K., Nones, K., Nourbakhsh, E., et al. (2014). Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol. 15, R51. doi:10.1186/gb-2014-15-3-r51
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matveeva, O., Nechipurenko, Y., Rossi, L., Moore, B., Saetrom, P., Ogurtsov, A. Y., et al. (2007). Comparison of approaches for rational siRNA design leading to a new efficient and transparent method. Nucleic Acids Res. 35, e63–e63. doi:10.1093/nar/gkm088
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Montague, T. G., Cruz, J. M., Gagnon, J. A., Church, G. M., and Valen, E. (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401–W407. doi:10.1093/nar/gku410
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Naito, Y., Ui-Tei, K., Nishikawa, T., Takebe, Y., and Saigo, K. (2006). siVirus: web-based antiviral siRNA design software for highly divergent viral sequences. Nucleic Acids Res. 34, W448–W450. doi:10.1093/nar/gkl214
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Naito, Y., Yoshimura, J., Morishita, S., and Ui-Tei, K. (2009). siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics 10:392. doi:10.1186/1471-2105-10-392
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Obad, S., Santos dos, C. O., Petri, A., Heidenblad, M., Broom, O., Ruse, C., et al. (2011). Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 43, 371–378. doi:10.1038/ng.786
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Petersen, M., and Wengel, J. (2003). LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 21, 74–81. doi:10.1016/S0167-7799(02)00038-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi:10.1016/j.cell.2013.02.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ran, F. A., Hsu, P. D., Lin, C.-Y., Gootenberg, J. S., Konermann, S., Trevino, A. E., et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389. doi:10.1016/j.cell.2013.08.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W. S., and Khvorova, A. (2004). Rational siRNA design for RNA interference. Nat. Biotechnol. 22, 326–330. doi:10.1038/nbt936
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sayed, D., Rane, S., Lypowy, J., He, M., Chen, I.-Y., Vashistha, H., et al. (2008). MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol. Biol. Cell 19, 3272–3282. doi:10.1091/mbc.E08-02-0159
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schramm, G., and Ramey, R. (2005). siRNA design including secondary structure target site prediction. Nat. Meth. 2. doi:10.1038/nmeth780
Shabalina, S. A., Spiridonov, A. N., and Ogurtsov, A. Y. (2006). Computational models with thermodynamic and composition features improve siRNA design. BMC Bioinformatics 7:65. doi:10.1186/1471-2105-7-65
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sharma, V., Nomura, Y., and Yokobayashi, Y. (2008). Engineering complex Riboswitch regulation by dual genetic selection. J. Am. Chem. Soc. 130, 16310–16315. doi:10.1021/ja805203w
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shin, C., Nam, J.-W., Farh, K. K.-H., Chiang, H. R., Shkumatava, A., and Bartel, D. P. (2010). Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell 38, 789–802. doi:10.1016/j.molcel.2010.06.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Straka, M., and Boese, Q. (2010). Current topics in RNAi: why rational pooling of siRNAs is SMART. Thermo Fisher Scientific Inc
Strapps, W. R., Pickering, V., Muiru, G. T., Rice, J., Orsborn, S., Polisky, B. A., et al. (2010). The siRNA sequence and guide strand overhangs are determinants of in vivo duration of silencing. Nucleic Acids Res. 38, 4788–4797. doi:10.1093/nar/gkq206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Summerton, J., and Weller, D. (1997). Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 7, 187–195. doi:10.1089/oli.1.1997.7.187
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tafer, H., Ameres, S. L., Obernosterer, G., Gebeshuber, C. A., Schroeder, R., Martinez, J., et al. (2008). The impact of target site accessibility on the design of effective siRNAs. Nat. Biotechnol. 26, 578–583. doi:10.1038/nbt1404
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S., and Vale, R. D. (2014). A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646. doi:10.1016/j.cell.2014.09.039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thomas, M., Lieberman, J., and Lal, A. (2010). Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 17, 1169–1174. doi:10.1038/nsmb.1921
Ui-Tei, K., Naito, Y., Takahashi, F., Haraguchi, T., Ohki-Hamazaki, H., Juni, A., et al. (2004). Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 32, 936–948. doi:10.1093/nar/gkh247
Upadhyay, S. K., and Sharma, S. (2014). SSFinder: high throughput CRISPR-Cas target sites prediction tool. Biomed Res. Int. 2014, 1–4. doi:10.1155/2014/742482
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vert, J. P., Foveau, N., Lajaunie, C., and Vandenbrouck, Y. (2006). An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics 7:520. doi:10.1186/1471-2105-7-520
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xiao, A., Cheng, Z., Kong, L., Zhu, Z., Lin, S., Gao, G., et al. (2014). CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30, 1180–1182. doi:10.1093/bioinformatics/btt764
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xie, S., Shen, B., Zhang, C., Huang, X., and Zhang, Y. (2014). sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS ONE 9:e100448. doi:10.1371/journal.pone.0100448
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, P., Zhang, Y., Kang, L., Roossinck, M. J., and Mysore, K. S. (2006). Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol. 142, 429–440. doi:10.1104/pp.106.083295
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yoo, J. W., Kim, S., and Lee, D.-K. (2008). Competition potency of siRNA is specified by the 5’-half sequence of the guide strand. Biochem. Biophys. Res. Commun. 367, 78–83. doi:10.1016/j.bbrc.2007.12.099
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zebec, Z., Manica, A., Zhang, J., White, M. F., and Schleper, C. (2014). CRISPR-mediated targeted mRNA degradation in the Archaeon Sulfolobus solfataricus. Nucleic Acids Res. 42, 5280–5288. doi:10.1093/nar/gku161
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, Y., Dai, Z., Liang, Y., Yin, M., Ma, K., He, M., et al. (2014). Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci. Rep. 4:3943. doi:10.1038/srep03943
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhu, L. J., Holmes, B. R., Aronin, N., and Brodsky, M. H. (2014). CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PLoS ONE 9:e108424. doi:10.1371/journal.pone.0108424
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ziegler, S., Eberle, M. E., Wölfle, S. J., Heeg, K., and Bekeredjian-Ding, I. (2013). Bifunctional oligodeoxynucleotide/antagomir constructs: evaluation of a new tool for microRNA silencing. Nucleic Acid Therapeut. 23, 427–434. doi:10.1089/nat.2013.0447
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: RNAi, siRNA, miRNA, a-miR, AntagomiR, Sponge, CRISPRi
Citation: Laganà A, Shasha D and Croce CM (2014) Synthetic RNAs for gene regulation: design principles and computational tools. Front. Bioeng. Biotechnol. 2:65. doi: 10.3389/fbioe.2014.00065
Received: 30 September 2014; Accepted: 23 November 2014;
Published online: 11 December 2014.
Edited by:
Christian M. Zmasek, Sanford-Burnham Medical Research Institute, USAReviewed by:
Rui Alves, Universitat de Lleida, SpainCopyright: © 2014 Laganà, Shasha and Croce. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alessandro Laganà, Department of Molecular Virology, Immunology and Medical Genetics, Comprehensive Cancer Center, The Ohio State University, 460 West 12th Avenue, Room 0995, Columbus, OH 43210, USA e-mail:YWxlc3NhbmRyby5sYWdhbmFAb3N1bWMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.