- 1Department of Biobehavioral Sciences, Columbia University, New York, NY, United States
- 2Faculty of Health and Medicine, Lancaster University, Lancaster, United Kingdom
- 3School of Medical Imaging, Hebei Medical University, Shijiazhuang, China
- 4School of Medicine, Shanghai Jiao Tong University, Shanghai, China
- 5School of Medicine, University of Galway, Galway, Ireland
- 6College of Health, Medicine and Wellbeing, University of Newcastle, Newcastle, NSW, Australia
- 7Faculty of Medicine, University of Iceland, Reykjavík, Iceland
- 8Division of Biosciences, Faculty of Life Sciences, University College London, London, United Kingdom
- 9School of Medicine, Royal College of Surgeons in Ireland, Dublin, Ireland
Background: Neurexins, essential synaptic proteins, are linked to neurodevelopmental and neuropsychiatric disorders like autism spectrum disorder (ASD) and schizophrenia.
Objective: Through this systematic review, we aimed to shed light on the relationship between neurexin dysfunction and its implications in neurodevelopmental and neuropsychiatric manifestations. Both animal and human-induced pluripotent stem cell (hiPSC) models served as our primary investigative platforms.
Methods: Utilizing the PRISMA 2020 guidelines, our search strategy involved scouring articles from the PubMed and Google Scholar databases covering a span of two decades (2003–2023). Of the initial collection, 27 rigorously evaluated studies formed the essence of our review.
Results: Our review suggested the significant ties between neurexin anomalies and neurodevelopmental and neuropsychiatric outcomes, most notably ASD. Rodent-based investigations delineated pronounced ASD-associated behaviors, and hiPSC models derived from ASD-diagnosed patients revealed the disruptions in calcium dynamics and synaptic activities. Additionally, our review underlined the integral role of specific neurexin variants, primarily NRXN1, in the pathology of schizophrenia. It was also evident from our observation that neurexin malfunctions were implicated in a broader array of these disorders, including ADHD, intellectual challenges, and seizure disorders.
Conclusion: This review accentuates the cardinal role neurexins play in the pathological process of neurodevelopmental and neuropsychiatric disorders. The findings underscore a critical need for standardized methodologies in developing animal and hiPSC models for future studies, aiming to minimize heterogeneity. Moreover, we highlight the need to expand research into less studied neurexin variants (i.e., NRXN2 and NRXN3), broadening the scope of our understanding in this field. Our observation also projects hiPSC models as potent tools for bridging research gaps, promoting translational research, and fostering the development of patient-specific therapeutic interventions.
Introduction
Neurexins, primarily transcribed from three genes (in animals: Nrxn1, Nrxn2, Nrxn3; in humans: NRXN1, NRXN2, NRXN3) (Khoja et al., 2023), are pivotal presynaptic adhesion proteins within the nervous system. These proteins play critical roles in synapse formation and function, manifesting in two primary forms: the longer α-neurexins and the shorter β-neurexins (Reissner et al., 2013; Zhang et al., 2022; Trotter et al., 2023). α-neurexins, characterized by six large extracellular laminin/neurexin/sex hormone-binding (LNS) globulin domains and three interspersed epidermal growth factor (EGF)-like regions; along with β-neurexins, featuring only the sixth LNS domain and absent EGF-like regions, both play a pivotal role in synaptic function (Cuttler et al., 2021). These proteins, expressed at both excitatory and inhibitory synapses (Reissner et al., 2013), contribute to neurotransmission, synaptic plasticity, and neuronal development (Trotter et al., 2023). Their structures, particularly in the neurexin 1 protein, have been elucidated in various species, though human-specific structural data remains elusive (Cuttler et al., 2021).
The intricacies of neurexin functionality are underscored by their binding capability to various postsynaptic ligands, including neuroligins, cerebellins, and leucine-rich repeat transmembrane proteins (LRRTMs) (Francks, 2011; Asede et al., 2020). Such interactions underscore the multifaceted nature of neurexins in synaptic operations and their potential association with neurodevelopmental and neuropsychiatric disorders when dysfunctional (Kasem et al., 2018). Perturbations in neurexin signaling have been implicated in various neurodevelopmental and neuropsychiatric disorders, including autism spectrum disorders (ASD), schizophrenia (SCZ), bipolar disorder (BD), and attention deficit hyperactivity disorder (ADHD) (Kasem et al., 2018; Cuttler et al., 2021).
It should be noted that deletions of NRXN 1–3 and these neurodevelopmental and neuropsychiatric disorders are highly interconnected and not simple one-to-one relationships (Kasem et al., 2018). While not all individuals with NRXN deletions inevitably develop neurodevelopmental or neuropsychiatric disorders, a single NRXN deletion could potentially enhance the risk for a multitude of such disorders. Deletions of neurexin-1α, for instance, have been linked with a significantly elevated risk for SCZ, ASD, and intellectual disability (Reichelt et al., 2012). In contrast, a single disorder could also result from different deletions of neurexin genes. For example, dysfunctions across all three neurexin genes (i.e., NRXN 1–3) have been identified as contributors to a disrupted balance between excitatory and inhibitory neurotransmission in human ASD (Khoja et al., 2023). These findings further highlight the need to figure out the complex association between neurexins and these disorders.
Previous research has investigated the role of neurexins using both in vivo and in vitro approaches (Cuttler et al., 2021; Gomez et al., 2021). While genetic studies have greatly expanded our understanding of the involvement of neurexins in these disorders (Tromp et al., 2021), a significant gap remains in the functional analysis of these genetic aberrations. Understanding the precise molecular and cellular outcomes of neurexin dysfunction can pave the way for targeted therapeutic interventions. Herein lies the importance of in vitro human models, specifically induced pluripotent stem cells (iPSCs). By reprogramming somatic cells from patients into iPSCs, and subsequently differentiating these cells into neurons, researchers can create patient-specific neural models that recapitulate disease phenotypes (Jusop et al., 2023). This approach allows for the study of neurexin dysfunction in a cellular context that closely mirrors the patient’s genetic and epigenetic background (Lee et al., 2020). Meanwhile, somatic cells from healthy population could be used for comparison (Raab et al., 2014). On the other hand, animal models, particularly rodents, have traditionally served as invaluable tools for dissecting the physiological consequences of neurexin dysfunction (Bryda, 2013; Tromp et al., 2021). They offer insights into behavioral manifestations, neuronal circuitry alterations, and potential compensatory mechanisms that might arise due to the dysfunctional neurexin signaling. By employing both iPSCs and animal models, researchers could achieve a more comprehensive understanding that combines human-specific cellular insights with in vivo physiological observations (Rowe and Daley, 2019).
Yet, despite the existing plethora of studies on neurexin dysfunction’s relevance to neurodevelopmental and neuropsychiatric disorders, comprehensive systematic review encompassing both iPSC and animal model perspectives is lacking. This systematic review aims to bridge this knowledge gap by amalgamating existing research, shedding light on the molecular, cellular, physiological, and behavioral impacts of neurexin dysfunction, while highlighting both their commonalities and differences.
Materials and methods
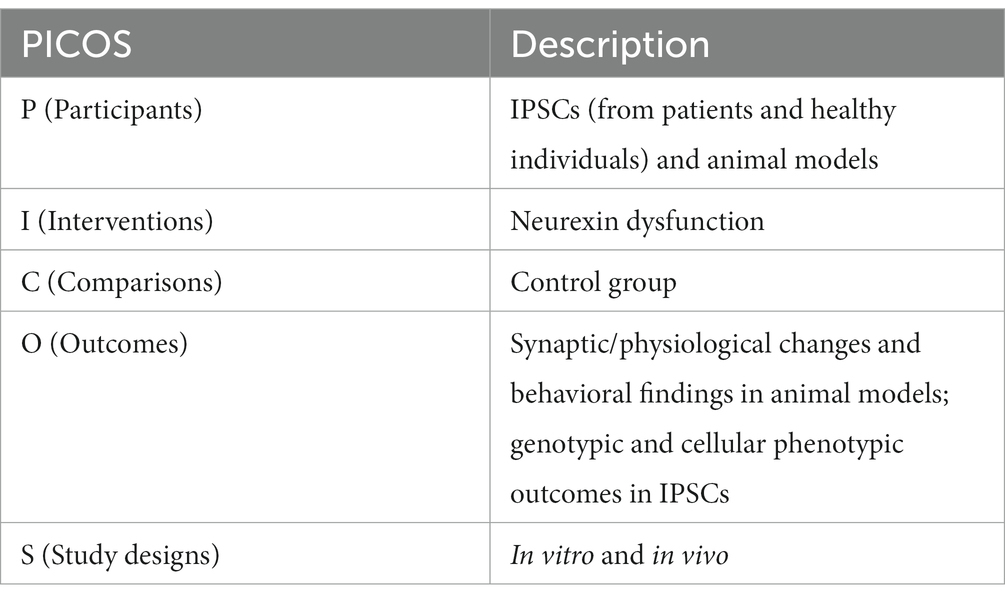
This systematic review was conducted in accordance with the 2020 guidelines and principles detailed in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement and checklist (Shan et al., 2022). The PICOS (Participants; Interventions; Comparisons; Outcomes; Study designs) are defined in Table 1.
We mainly performed a systematic search of PubMed and Google Scholar databases, targeting original peer-reviewed research articles published in the last 20 years from 10/05/2003 to 10/05/2023. The search terms included combinations of the following: [(Neurexin) OR (NRXN)) AND ((IPSC) OR (Induced Pluripotent Stem Cells) OR (Mouse) OR (Rat) OR (Animal)) AND ((Schizophrenia) OR (Autism) OR (Bipolar disorder) OR (Attention-deficit/hyperactivity disorder) OR (ADHD) OR (Neurodevelopmental disorders) OR (Neuropsychiatric Disorders)]. Only English language papers were included. Papers were initially screened by Title and Abstract Only, and excluded if deemed not relevant because the study was not on animal or iPSCs models, neurexin, or any neurodevelopmental/neuropsychiatric disorder. In a second round of screening, full-text papers were assessed for eligibility, and excluded if not relevant to our objectives. Additional search criterion was studies from the manually screening of the reference lists of extracted articles between 2003 and 2023, in order to identify additional studies that may fit the systematic review objective in the current article but were not identified by the PubMed and Google Scholar databases. In case of circular demonstration of research results combined with original empirical articles, only empirical studies were retained if a systematic or narrative review addressing the potential linkage between neurexin dysfunction and these disorders emerged (Shan et al., 2021).
During the second round of screening, we meticulously screened pertinent studies that utilized animal models or IPSCs as their primary methodologies. Emphasis was placed on retaining studies that elucidated synaptic/physiological activity and/or animal behavioral outcomes pertinent to neurodevelopmental and/or neuropsychiatric disorders as a result of neurexin dysfunction. Conversely, studies that lacked explicit mention of the potential association between neurexin dysfunction and these disorders were excluded.
For the studies employing animal models, the extracted data encompassed the targeted neurexin, type of animal used, generation strategy of the model, model viability, synaptic/physiological activities, principal behavioral findings, and the inferred neurodevelopmental or neuropsychiatric disease correlation. On the other hand, data retrieval from studies employing IPSCs comprised the targeted neurexin, donor details and sample count, IPSC model generation technique, genotypic profiles, cellular phenotypic outcomes, and the extrapolated disease correlation.
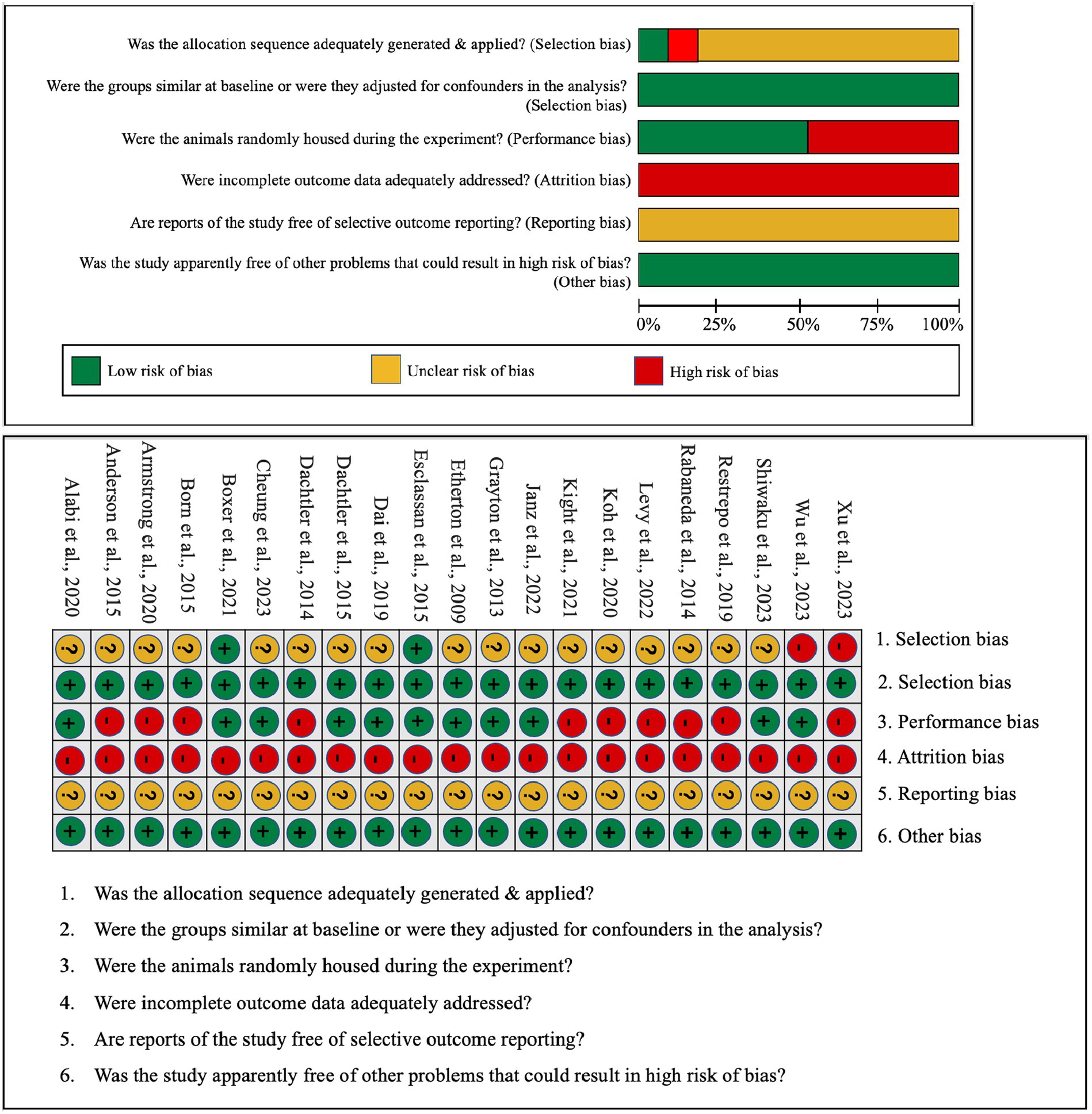
In addition, we utilized certain methods to evaluate bias risk in both in vitro and in vivo studies. The quality and risk for bias in the included in vitro (i.e., IPSC) studies were assessed by the adapted quasi-experimental studies appraisal tool by the Joanna Briggs Institute (Tufanaru et al., 2017). Each in vitro investigation was designated a risk status: low (where any detected bias would not substantively skew results), unclear (where potential bias introduces some result uncertainty), or high (indicating that bias could significantly compromise findings). For in vivo research, we employed the SYRCLE risk assessment tool, which refers to the Systematic Review Centre for Laboratory Animal Experimentation’s risk of bias tool, tailored for the included animal studies (Silveira et al., 2023). This instrument is structured around various bias categories: selection, performance, attrition, reporting, and other biases. The current systematic review involved a ten-question evaluation of the articles, with possible responses being “YES” (signifying minimal bias risk), “NO” (indicative of pronounced bias risk), and “UNCLEAR” (signifying indeterminate bias risk).
Results
From our search on the PubMed and Google Scholar databases, we procured a total of 1754 records. Subsequent exploration through a ‘citation searching’ methodology yielded an additional 8 records. Firstly, 234 were identified as duplicates and thus removed. Following a preliminary assessment of titles and abstracts, we excluded 1,477 of the 1,528 entries. Subsequently, 51 articles progressed to an advanced screening phase, from which a further 24 were eliminated due to their irrelevance to our stipulated inclusion criteria or their categorization as systematic/narrative reviews upon thorough examination of their full text. Consequently, 27 articles formed the core of our review. Referring to Figure 1 for a detailed overview of our search and screening procedures. All these studies, without exception, probed the potential link between neurexin dysfunction and neurodevelopmental and/or neuropsychiatric disorders, utilizing either animal or IPSC models as their investigative platforms. A schematic representation of neurexin-neuroligin interaction is shown in Figure 2.
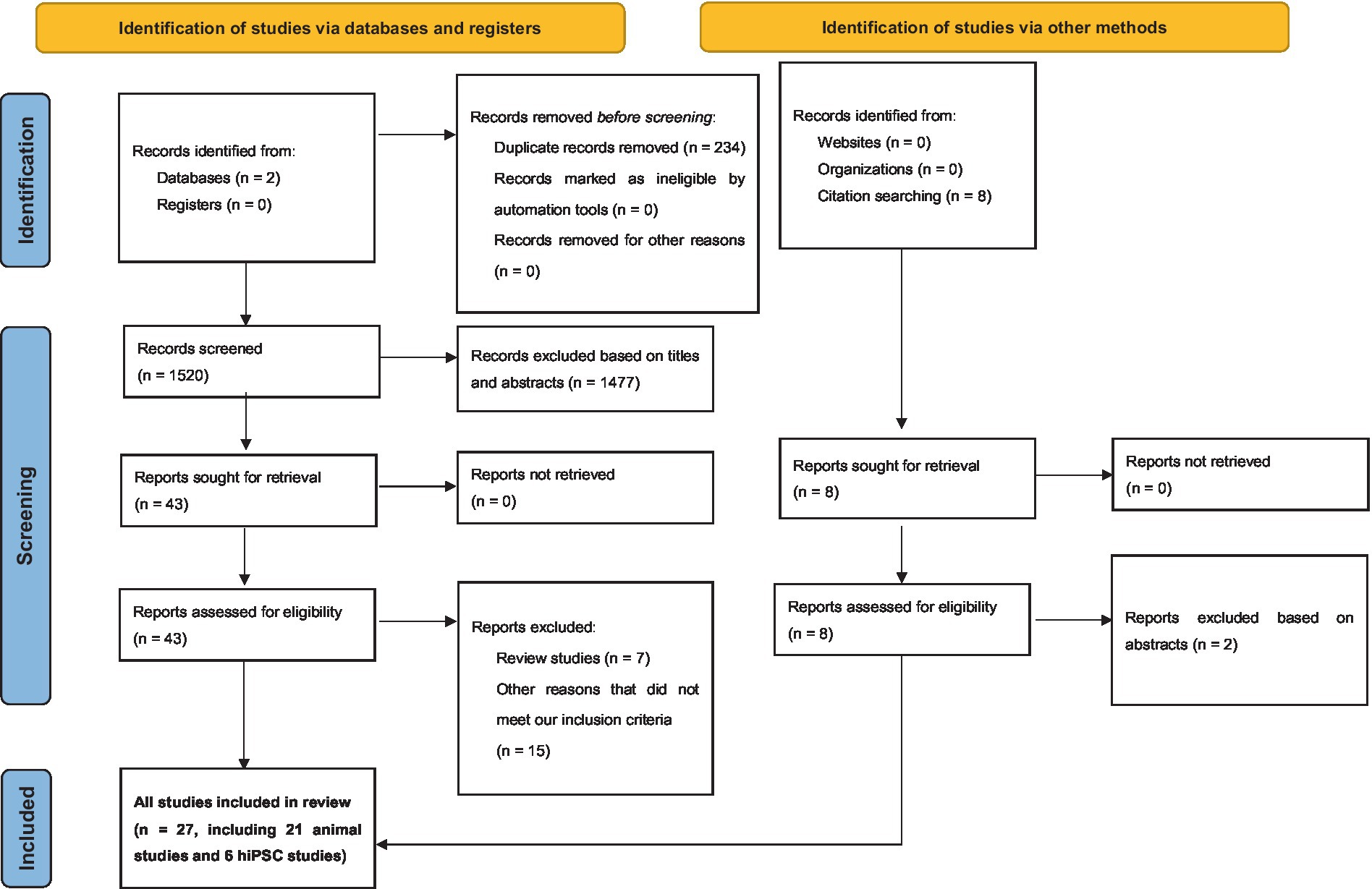
Figure 1. Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) diagram demonstrating search strategy.
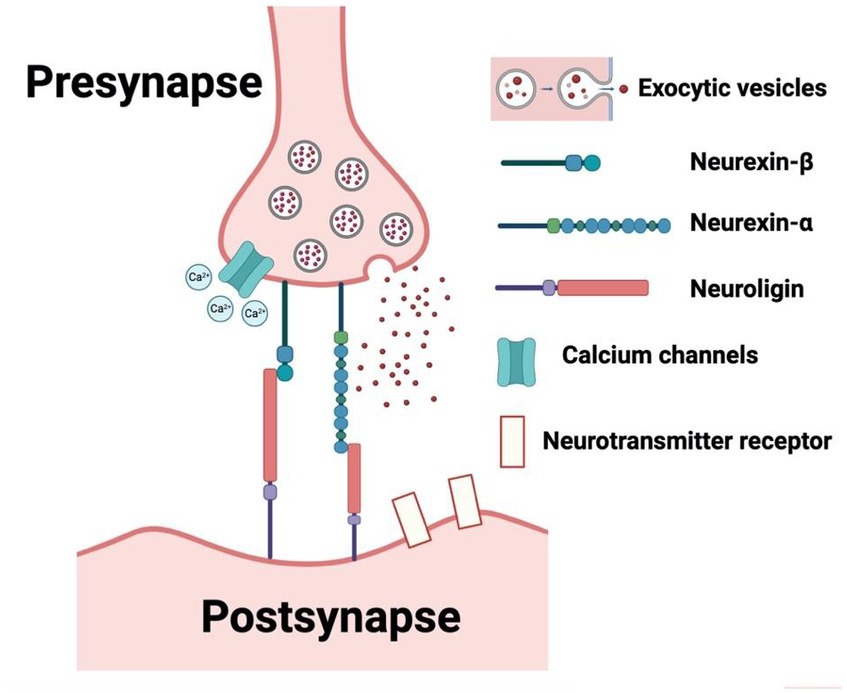
Figure 2. Schematic representation of neurexin-neuroligin interaction. (Created with BioRender.com, accessed on 28 May 2023).
In an effort to gauge the risk of bias in both in vitro and in vivo studies, we have encapsulated our quality assessment findings in Figure 3 (pertaining to IPSC studies) and Figure 4 (relevant to animal studies). A noteworthy observation is that a predominant fraction of the in vitro studies showcased minimal bias risk. However, an anomaly was discerned for the criterion addressing the frequency of outcome measurements both pre and post-intervention/exposure. Herein, a palpable bias risk was evident, attributed largely to these studies’ inclination towards contrasting different cell lines (notably control vs. NRXN-deleted) rather than employing iterative measurements on identical cells prior to and subsequent to specific interventions. As for in vivo studies, we identified salient deficiencies in certain quality components. For instance, concerning the criterion, “Was the allocation sequence generated and executed appropriately?” a mere pair of studies (Esclassan et al., 2015; Boxer et al., 2021) demonstrated a minimal bias risk owing to their explicit randomization acknowledgment. Conversely, a sizable cohort exhibited pronounced bias risks due to ambiguous animal selection during result evaluation and/or a lack of randomized housing during experimentation. Furthermore, universal bias risks were detected in the realm of incomplete outcome data reporting. In addition, the absence of accessible study protocols introduced ambiguity in reporting bias, making it challenging to ascertain if the pre-determined primary and secondary outcomes were duly reported. Lastly, when assessed against the criterion, “Was the study apparently free of other problems that could result in high risk of bias?,” all studies uniformly exhibited a minimal bias risk.
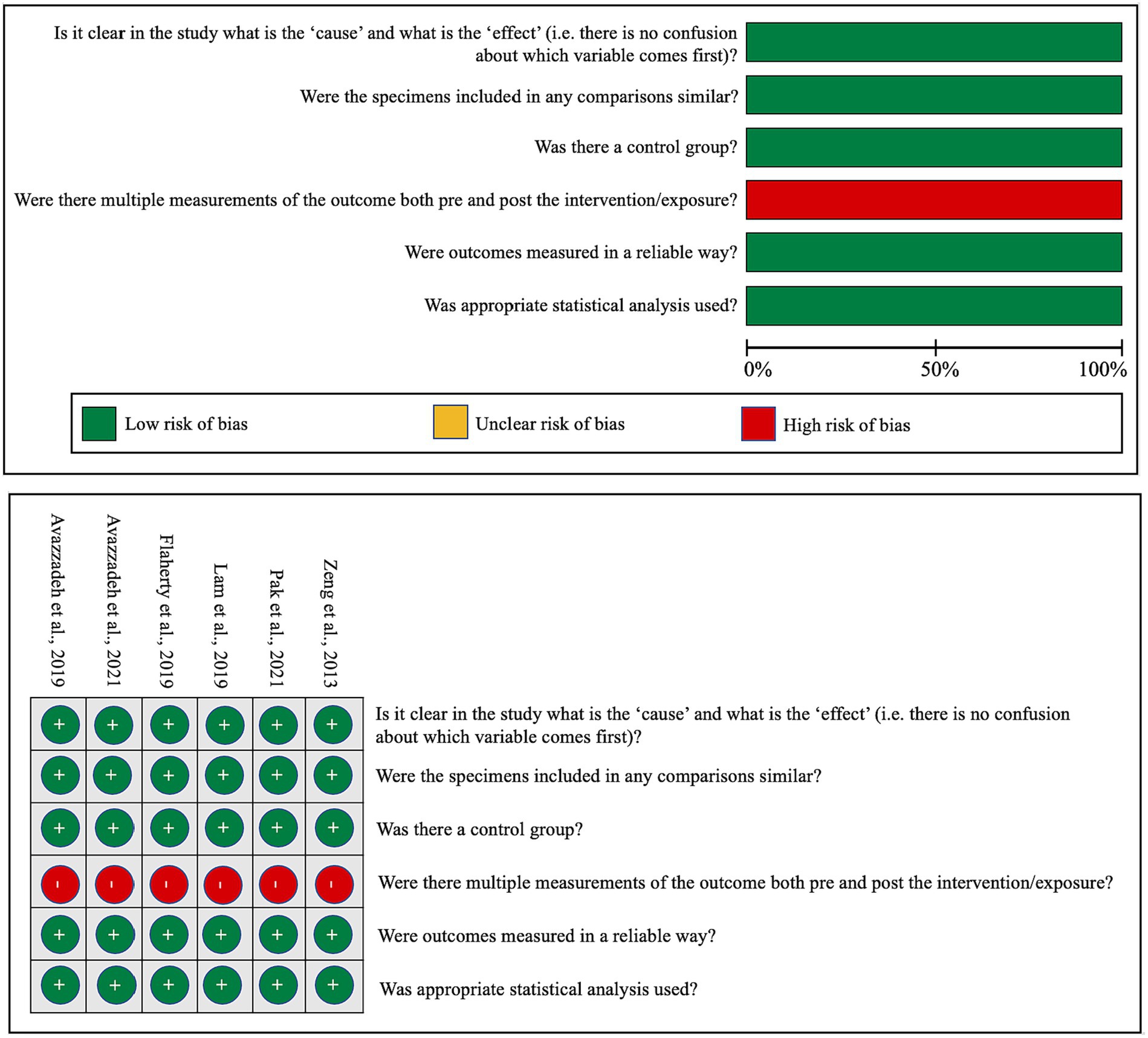
Figure 3. Qualitative analysis with adapted the quasi-experimental studies appraisal tool by the Joanna Briggs Institute.
The categorization of the 27 key studies according to their research models showed that 21 utilized animal models, while 6 employed IPSC models. An analysis of the neurexin targets within these studies revealed a distinct emphasis on nrxn1/NRXN1, especially Nrxn1α/NRXN1α. Notably, a substantial majority (more than 75%, n = 21) of these studies suggested a link to ASD as a potential associated condition, with other neurodevelopmental and neuropsychiatric disorders like schizophrenia, ADHD, and intellectual disability also being mentioned.
Within the 21 studies focusing on animal models, rodents, specifically rats and mice, were predominantly used in 19 of them, while Zebrafish and Drosophila melanogaster were each featured in one study. The research heavily favored Nrxn1α, explored in 14 studies, with less frequent examination of Nrxn2α, Nrxn3α, and Nrxnβ. Most studies adopted knockout (KO) strategies in mice, targeting specific Nrxn genes such as Nrxn1α, Nrxnβ, and Nrxn3. Variability in genetic backgrounds of these models (e.g., pure C57BL/6 J, mixed backgrounds) was noted, potentially contributing to phenotypic differences observed. For instance, while Etherton et al. (2009) and Armstrong et al. (2020) both researched Nrxn1α KO mice, their differing genetic backgrounds led to distinct behavioral outcomes, including increased aggression and impaired prepulse inhibition, respectively. Concerning model viability, the majority of studies successfully developed viable models. The earliest Nrxn loss of function mouse models were introduced by Missler et al. (2003) who generated single, double, and triple Nrxn1α/2α/3α KO mice. Tromp et al. (2021) highlighted the importance of maintaining at least two intact α-Nrxns for survival, based on variable survival rates in double and triple KO mice. In a more recent study, Cheung et al. (2023) discovered that triple Nrxn knockout in 5-HT neurons led to a significant reduction in serotonin neurons during early postnatal stages, indicating the viability of these models but with specific neural deficits. The studies consistently reported synaptic and physiological changes due to Nrxn gene disruptions. For example, Alabi et al. (2020) observed neural signaling disruptions in the striatum, and Born et al. (2015) noted a decrease in spontaneous transmitter release at excitatory synapses. Behavioral findings in these animal models displayed both similarities and variances. Commonly reported were deficits in social interactions and increased anxiety-like behavior, aligning with traits in ASD and Schizophrenia, as shown in studies like Dachtler et al. (2014) and Born et al. (2015). However, discrepancies were also apparent; Grayton et al. (2013) reported reduced locomotor activity and increased aggression, while Etherton et al. (2009) observed normal social and anxiety-like behaviors and locomotor activity. These variations suggest that the behavioral impacts of Nrxn disruptions can significantly differ due to the genetic background of the mice. The primary diseases correlated with these disruptions are predominantly ASD and schizophrenia, reflecting the strong link between Nrxn gene disruptions and these disorders. Nonetheless, the observed range of behavioral phenotypes, from social deficits to cognitive impairments, implies that the influence of these genes extends beyond these disorders, potentially affecting a wider spectrum of neurodevelopmental conditions.
In the hiPSC studies analyzed, hiPSC lines originated from diverse sources such as skin biopsies and peripheral blood mononuclear cells, collected from both healthy subjects and patients with disorders including ASD and schizophrenia. While the generation of these iPSC models consistently utilized cellular reprogramming techniques, there was a slight variation in the genetic focus across different studies. The primary attention was on NRXN1α, with investigations into the impacts of NRXN1α+/− deletions, exemplified by Avazzadeh et al. (2019, 2021), and bi-allelic NRXN1-alpha deletions, as reported by Lam et al. (2019). These studies primarily explored intragenic deletions affecting NRXN1, uncovering a spectrum of cellular phenotypes indicative of impaired neuronal function, such as the altered dynamics of calcium and sodium and modifications in neurotransmitter release, as detailed by Avazzadeh et al. (2019, 2021) and Pak et al. (2021). The cellular phenotypes identified exhibited both commonalities and differences; for instance, Avazzadeh et al. (2021) and Pak et al. (2021) observed changes in ion dynamics and neurotransmitter release, whereas Lam et al. (2019) noted a shift in cells towards a radial glia-like identity. Predominantly, the genetic and cellular alterations were linked to ASD and schizophrenia. Nevertheless, the observed range of symptoms and associated conditions, such as seizures, intellectual disability, and developmental delays, indicate that the influence of these genetic modifications might extend well beyond these disorders. This broader impact is exemplified by Avazzadeh et al. (2021), who associated NRXN1α+/− deletions with ASD and related symptoms, and Flaherty et al. (2019), who connected NRXN1 deletions to schizophrenia and bipolar disorder with psychosis.
Discussion and future directions
Our review systematically examines Nrxn knockout/knock-in (KO/KI) animal models and human-induced pluripotent stem cell (hiPSC) studies on neurexins, highlighting their complex roles in various neurodevelopmental and neuropsychiatric conditions. Utilizing diverse methods, from CRISPR/Cas9 genome editing to hiPSC-derived neuronal cultures, these studies showcase the significant effects of neurexin variations, notably in disorders like ASD and schizophrenia (Avazzadeh et al., 2019; Flaherty et al., 2019). Neurexin dysfunction is also linked to other conditions such as ADHD, intellectual disability, seizures, and developmental delays. Tables 2, 3 provide in-depth information on the specifics of each hiPSC study and Nrxn KO/KI animal investigation, including target neurexins, the animals used, model generation strategies, model viability, synaptic or physiological activities, key findings regarding animal behaviors, and related diseases.
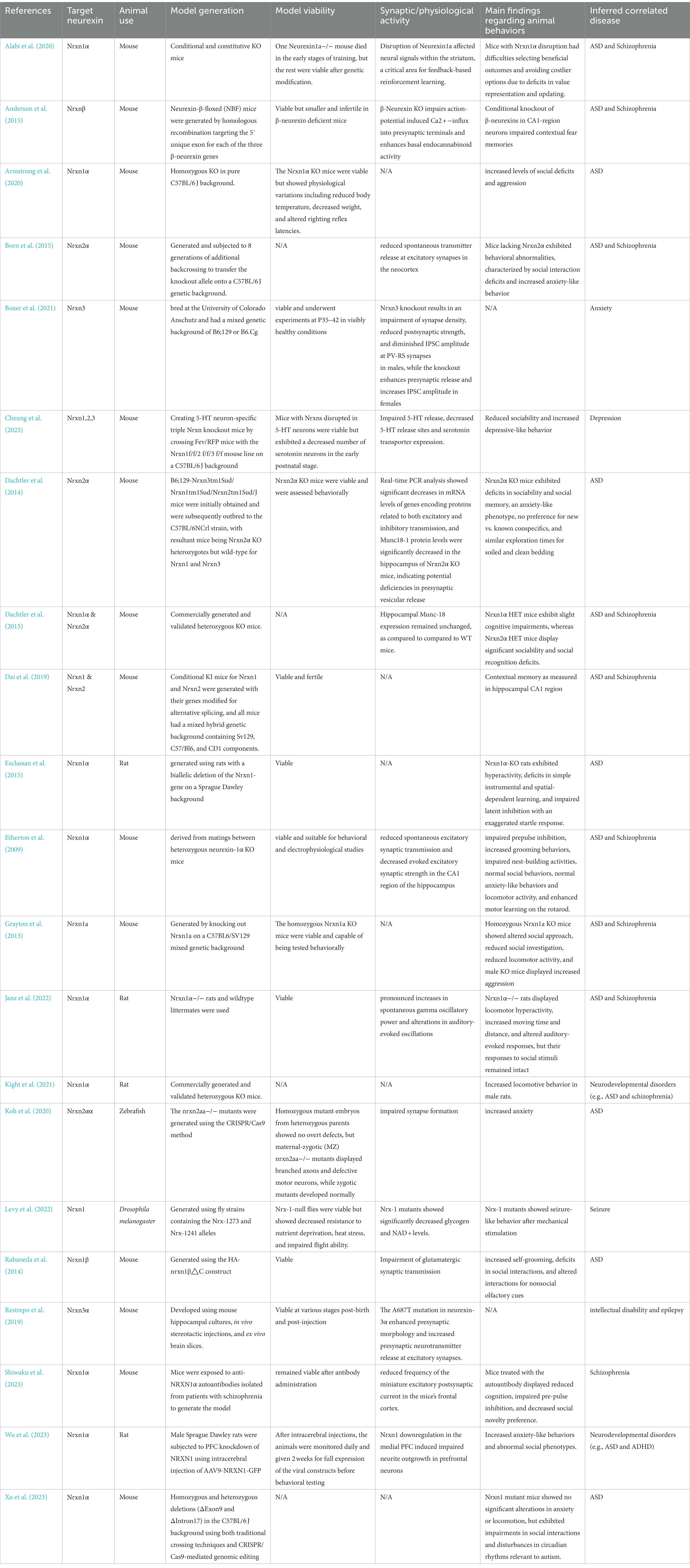
Table 2. Overview of existing Nrxn KO/KI animal models detailing target neurexin, animal use, model generation, model viability, synaptic/physiological activity, main findings regarding animal behaviors, and inferred correlated disease.
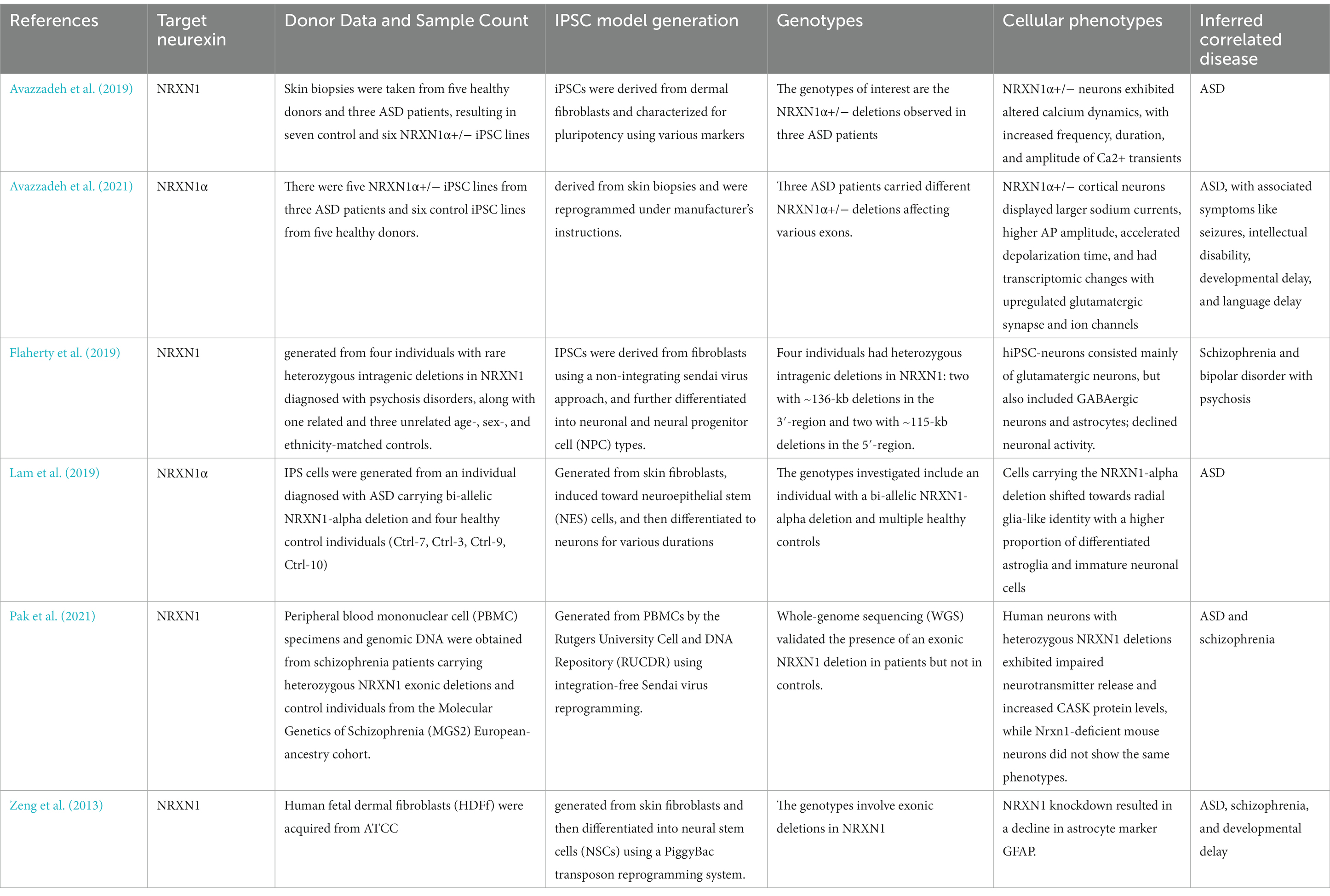
Table 3. Summary of hiPSC studies on NRXN outlining details of target neurexin, donor data and sample count, IPSC model generation, genotypes, cellular phenotypes and inferred correlated disease.
Recognizing the genetic link between neurexin variants and an increased risk for neurodevelopmental and neuropsychiatric disorders, our study seeked to understand the neuronal functions of these genes and their potential role in initiating, progressing, and exacerbating mental disorders. We have expanded upon previous reviews by generating animal models of Nrxns and integrating relevant data from hiPSC studies to better evaluate the mental conditions associated with these genes (Tromp et al., 2021).
Neurexin dysfunction in autism-spectrum disorders
ASD, as a complex neurodevelopmental disorder, presents with a spectrum of cognitive and behavioral phenotypes, including social interaction deficits and repetitive behaviors (Evans et al., 2023). The identification of rare missense mutations in exon 1 of NRXN1β in ASD patients (p.S14L and p.T40S), absent in controls, first established a genetic link with NRXNs (Feng et al., 2006). Recent research has increasingly highlighted the role of NRXN1α variants in the development of ASD (Avazzadeh et al., 2021). The distinct behavioral phenotypes observed in Nrxn1 mutant mice models, such as social deficits and altered circadian rhythms (Levy et al., 2022), resonate with the human ASD condition, suggesting a possible conserved molecular mechanism across species. The resemblance in behavioral phenotypes could be attributed to the fundamental role of NRXN1 in synaptic formation and neurotransmitter release, processes crucial for normal social and cognitive functions.
In hiPSC models derived from ASD patients, perturbations in calcium dynamics and ionic pathways (Avazzadeh et al., 2019) point towards a synaptic dysfunction at a cellular level. Such findings may explain the diverse cognitive and behavioral outcomes in ASD, aligning with theories that posit synaptic homeostasis disruptions as a core component of ASD pathophysiology (Bourgeron, 2015). The variation in cellular responses to NRXN1 mutations among different studies might be attributed to the inherent genetic diversity among individuals with ASD, reflecting the heterogeneity of the disorder. Furthermore, the impact of NRXN1 mutations extends beyond synaptic dysfunction to broader neural network alterations. The observation of a shift towards radial glia-like identity in cells with NRXN1-alpha deletion (Lam et al., 2019) suggests a profound effect on neural development. This could potentially disrupt the balance between excitatory and inhibitory circuits, a crucial aspect of ASD’s neural basis.
While most ASD research has focused on NRXN1 deletions, studies also implicate NRXN2 and NRXN3. While some research, such as Gauthier et al.’s, identified specific truncating mutations in NRXN2α associated with ASD (Gauthier et al., 2011), other studies reported broader de novo deletions in NRXN2 linked to autistic behaviors and developmental delays (Mohrmann et al., 2011; Boyle et al., 2015). Additionally, a study by Vaags et al. (2012) found NRXN3 mutations in both individuals with ASD and their unaffected siblings, indicating reduced penetrance and suggesting a complex genetic interaction rather than a straightforward cause-effect relationship. In contrast, a study by Yuan et al. reported a co-segregating NRXN3α deletion in a child with ASD (Yuan et al., 2018). In contrast, a study by Yuan et al. reported a co-segregating NRXN3α deletion in a child with ASD. This evidence indicates an inconsistent role for NRXN2 and NRXN3 in ASD, and further research is needed to clarify their specific contributions to the disorder.
Neurexin dysfunction in schizophrenia
Neurexin variations, particularly those in NRXN1, are consistently implicated in schizophrenia (Flaherty et al., 2019; Pak et al., 2021), a severe mental disorder characterized by delusions, hallucinations, and cognitive deficits (Iqbal et al., 2023). Studies have established a strong link between NRXN1 and schizophrenia. For example, rodent models exposed to anti-NRXN1α autoantibodies from schizophrenia patients exhibited cognitive impairments typical of the disorder (Restrepo et al., 2019; Shiwaku et al., 2023). Concurrently, hiPSC studies have shown reduced neuronal activity and neurotransmitter release in neurons from schizophrenia patients with NRXN1 deletions (Flaherty et al., 2019; Pak et al., 2021). This aligns with the known role of neurotransmitters like dopamine, glutamate, and serotonin in the pathophysiology of schizophrenia, underscoring neurexins’ relevance in this context (Beeraka et al., 2022).
As of now, there has been no research confirming a connection between NRXN2 and schizophrenia. However, a significant study on the NRXN3 gene and schizophrenia was conducted with a Chinese Han population, involving 1,214 schizophrenia patients and 1,517 control subjects. This study identified three specific Single Nucleotide Polymorphisms (SNPs) within NRXN3 that were associated with schizophrenia, located in the first and second introns of the gene (Hu et al., 2013). These findings suggest a potential link between NRXN3 and schizophrenia, though further research is needed for a more comprehensive understanding.
Neurexin dysfunction in other neurodevelopmental/neuropsychiatric disorders
The impact of neurexins extends beyond ASD and schizophrenia, affecting a variety of other conditions. Mutations in Nrxn3α, for example, are linked to intellectual disabilities and epilepsy (Shiwaku et al., 2023). Research has also shown that Nrxn1 influenced seizure-like behaviors in flies (Avazzadeh et al., 2019). Additionally, studies indicated that individuals with epilepsy were more likely to develop certain neuropsychiatric conditions (Tolchin et al., 2020). Further, it has been found that reduced Nrxn1 expression in rats could lead to symptoms associated with various neurodevelopmental disorders, including ADHD (Wu et al., 2023). These varied symptoms demonstrate the wide-ranging role of neurexins in synaptic functions, suggesting that disruptions in neurexin activity can trigger a range of neurodevelopmental and neuropsychiatric symptoms, influenced by other genetic and environmental factors (Gupta et al., 2023). The overlapping symptoms across different disorders point to potential common molecular mechanisms that warrant further investigation.
Advantages and limitations
Our review’s expansive scope, encompassing both animal and human-derived model systems from fruit flies to humans, provides a holistic perspective on neurexin’s neurobiological significance. While animal models shed light on multifaceted behaviors, hiPSC models offer unparalleled cellular-level details, bridging the translational gap (Xu et al., 2023).
Nevertheless, challenges persist. A significant limitation across the studies is the diversity in methodologies, ranging from different techniques in model generation, such as traditional crossing vs. CRISPR/Cas9 genomic editing, to various animal models and iPSC sources (study methodologies vary, introducing potential inconsistencies). The pronounced focus on ASD and schizophrenia might inadvertently minimize neurexins’ role in other neurodevelopmental or neuropsychiatric disorders. While animal models are informative, translating these findings directly to humans can be uncertain. Additionally, hiPSC models, despite their sophistication, may not fully capture the intricacies of human brain development in vivo (Dixon and Muotri, 2023).
Our review primarily concentrates on certain models and does not include others, such as human pluripotent stem cell-derived forebrain organoid models. These models, as discussed in the study by Sebastian et al. (2023), provided essential insights developmental-timing- and cell-type-specific vulnerabilities associated with NRXN1 deletions in the context of schizophrenia. Omitting these advanced models might lead to a gap in our understanding of neuronal development and the pathology related to NRXN1 gene changes. Furthermore, the current research landscape is heavily focused on NRXN1, with less attention given to the roles of NRXN2 and NRXN3.
Future recommendations
To the best of our knowledge, this is the first systematic review study to present potential associations between neurexin dysfunction and a spectrum of neurodevelopmental and neuropsychiatric disorders. Moving forward, several key research considerations must be addressed. First, we believe there is an undeniable urgency for establishing standardized protocols in generating animal and hiPSC models, in order to provide a consistent framework for comparison and replication. Second, while the current literature predominantly focuses on NRXN1, future endeavors should cast a wider net to thoroughly investigate NRXN2 and NRXN3. Thirdly, future studies would benefit from incorporating data from organoid models, as discussed earlier, to gain a more complete understanding of NRXN-related disorders. Lastly, there is a notable imbalance between studies using animal models and those employing hiPSCs. Given hiPSCs’ ability to provide unique insights into human-specific cellular processes, increasing the number of hiPSC studies with varied sample sizes is essential. These studies not only enhance our understanding of human cellular and molecular dynamics but also pave the way for personalized medicine through the use of patient-derived cells.
Conclusion
This systematic review comprehensively explores the connections between neurexin dysfunction and a range of neurodevelopmental and neuropsychiatric disorders, employing a dual approach of animal models and hiPSC studies. The review highlights the significant implications of neurexin dysregulation, particularly in ASD and schizophrenia, while also acknowledging the broad spectrum of other neurodevelopmental disorders influenced by neurexin anomalies. The findings emphasize the need for more standardized methodologies in future research, particularly in the development of animal and hiPSC models. Additionally, the review underscores the importance of broadening our investigative focus beyond the primarily studied NRXN1 to include other less explored neurexin variants. This approach is crucial for deepening our understanding of these complex disorders and advancing towards personalized therapeutic interventions. The potential of hiPSC models as powerful tools in this research domain is particularly noted, promising to bridge existing gaps and propel translational research forward.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
DS: Conceptualization, Methodology, Resources, Validation, Writing – original draft, Writing – review & editing. YS: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. YZ: Conceptualization, Methodology, Data curation, Writing – original draft, Writing – review & editing. CH: Investigation, Methodology, Validation, Visualization, Writing – review & editing. WX: Formal analysis, Methodology, Validation, Visualization, Writing – review & editing. ZL: Investigation, Validation, Visualization, Writing – review & editing. FG: Conceptualization, Investigation, Visualization, Writing – review & editing. QO: Formal analysis, Investigation, Visualization, Writing – review & editing. ZiD: Data curation, Investigation, Methodology, Writing – review & editing. ZhD: Conceptualization, Investigation, Methodology, Resources, Validation, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alabi, O. O., Davatolhagh, M. F., Robinson, M., Fortunato, M. P., Vargas Cifuentes, L., Kable, J. W., et al. (2020). Disruption of Nrxn1α within excitatory forebrain circuits drives value-based dysfunction. elife 9:e54838. doi: 10.7554/eLife.54838
Anderson, G. R., Aoto, J., Tabuchi, K., Földy, C., Covy, J., Yee, A. X., et al. (2015). β-Neurexins control neural circuits by regulating synaptic endocannabinoid signaling. Cell 162, 593–606. doi: 10.1016/j.cell.2015.06.056
Armstrong, E. C., Caruso, A., Servadio, M., Andreae, L. C., Trezza, V., Scattoni, M. L., et al. (2020). Assessing the developmental trajectory of mouse models of neurodevelopmental disorders: social and communication deficits in mice with Neurexin 1α deletion. Genes Brain Behav. 19:e12630. doi: 10.1111/gbb.12630
Asede, D., Joseph, A., and Bolton, M. M. (2020). Deletion of NRXN1α impairs long-range and local connectivity in amygdala fear circuit. Transl. Psychiatry 10:242. doi: 10.1038/s41398-020-00926-y
Avazzadeh, S., McDonagh, K., Reilly, J., Wang, Y., Boomkamp, S. D., McInerney, V., et al. (2019). Increased Ca2+ signaling in NRXN1α+/− neurons derived from ASD induced pluripotent stem cells. Mol. Autism. 10:52. doi: 10.1186/s13229-019-0303-3
Avazzadeh, S., Quinlan, L. R., Reilly, J., McDonagh, K., Jalali, A., Wang, Y., et al. (2021). NRXN1α+/− is associated with increased excitability in ASD iPSC-derived neurons. BMC Neurosci. 22:56. doi: 10.1186/s12868-021-00661-0
Beeraka, N. M., Avila-Rodriguez, M. F., and Aliev, G. (2022). Recent reports on redox stress-induced mitochondrial DNA variations, neuroglial interactions, and NMDA receptor system in pathophysiology of schizophrenia. Mol. Neurobiol. 59, 2472–2496. doi: 10.1007/s12035-021-02703-4
Born, G., Grayton, H. M., Langhorst, H., Dudanova, I., Rohlmann, A., Woodward, B. W., et al. (2015). Genetic targeting of NRXN2 in mice unveils role in excitatory cortical synapse function and social behaviors. Front. Synaptic Neurosci. 7:3. doi: 10.3389/fnsyn.2015.00003
Bourgeron, T. (2015). From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat. Rev. Neurosci. 16, 551–563. doi: 10.1038/nrn3992
Boxer, E. E., Seng, C., Lukacsovich, D., Kim, J., Schwartz, S., Kennedy, M. J., et al. (2021). Neurexin-3 defines synapse- and sex-dependent diversity of GABAergic inhibition in ventral subiculum. Cell Rep. 37:110098. doi: 10.1016/j.celrep.2021.110098
Boyle, M. I., Jespersgaard, C., Nazaryan, L., Ravn, K., Brøndum-Nielsen, K., Bisgaard, A. M., et al. (2015). Deletion of 11q12.3-11q13.1 in a patient with intellectual disability and childhood facial features resembling Cornelia de Lange syndrome. Gene 572, 130–134. doi: 10.1016/j.gene.2015.07.016
Bryda, E. C. (2013). The mighty mouse: the impact of rodents on advances in biomedical research. Mo. Med. 110, 207–211.
Cheung, A., Konno, K., Imamura, Y., Matsui, A., Abe, M., Sakimura, K., et al. (2023). Neurexins in serotonergic neurons regulate neuronal survival, serotonin transmission, and complex mouse behaviors. elife 12:e85058. doi: 10.7554/eLife.85058
Cuttler, K., Hassan, M., Carr, J., Cloete, R., and Bardien, S. (2021). Emerging evidence implicating a role for neurexins in neurodegenerative and neuropsychiatric disorders. Open Biol. 11:210091. doi: 10.1098/rsob.210091
Dachtler, J., Glasper, J., Cohen, R. N., Ivorra, J. L., Swiffen, D. J., Jackson, A. J., et al. (2014). Deletion of α-neurexin II results in autism-related behaviors in mice. Transl. Psychiatry 4:e484. doi: 10.1038/tp.2014.123
Dachtler, J., Ivorra, J. L., Rowland, T. E., Lever, C., Rodgers, R. J., and Clapcote, S. J. (2015). Heterozygous deletion of α-neurexin I or α-neurexin II results in behaviors relevant to autism and schizophrenia. Behav. Neurosci. 129, 765–776. doi: 10.1037/bne0000108
Dai, J., Aoto, J., and Südhof, T. C. (2019). Alternative splicing of presynaptic Neurexins differentially controls postsynaptic NMDA and AMPA receptor responses. Neuron 102, 993–1008.e5. doi: 10.1016/j.neuron.2019.03.032
Dixon, T. A., and Muotri, A. R. (2023). Advancing preclinical models of psychiatric disorders with human brain organoid cultures. Mol. Psychiatry 28, 83–95. doi: 10.1038/s41380-022-01708-2
Esclassan, F., Francois, J., Phillips, K. G., Loomis, S., and Gilmour, G. (2015). Phenotypic characterization of nonsocial behavioral impairment in neurexin 1α knockout rats. Behav. Neurosci. 129, 74–85. doi: 10.1037/bne0000024
Etherton, M. R., Blaiss, C. A., Powell, C. M., and Südhof, T. C. (2009). Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc. Natl. Acad. Sci. U. S. A. 106, 17998–18003. doi: 10.1073/pnas.0910297106
Evans, M. M., Kim, J., Abel, T., Nickl-Jockschat, T., and Stevens, H. E. (2023). Developmental disruptions of the dorsal striatum in autism spectrum disorder. Biol. Psychiatry 95, 102–111. Advance online publication. doi: 10.1016/j.biopsych.2023.08.015
Feng, J., Schroer, R., Yan, J., Song, W., Yang, C., Bockholt, A., et al. (2006). High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci. Lett. 409, 10–13. doi: 10.1016/j.neulet.2006.08.017
Flaherty, E., Zhu, S., Barretto, N., Cheng, E., Deans, P. J. M., Fernando, M. B., et al. (2019). Neuronal impact of patient-specific aberrant NRXN1α splicing. Nat. Genet. 51, 1679–1690. doi: 10.1038/s41588-019-0539-z
Francks, C. (2011). Leucine-rich repeat genes and the fine-tuning of synapses. Biol. Psychiatry 69, 820–821. doi: 10.1016/j.biopsych.2010.12.018
Gauthier, J., Siddiqui, T. J., Huashan, P., Yokomaku, D., Hamdan, F. F., Champagne, N., et al. (2011). Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum. Genet. 130, 563–573. doi: 10.1007/s00439-011-0975-z
Gomez, A. M., Traunmüller, L., and Scheiffele, P. (2021). Neurexins: molecular codes for shaping neuronal synapses. Nat. Rev. Neurosci. 22, 137–151. doi: 10.1038/s41583-020-00415-7
Grayton, H. M., Missler, M., Collier, D. A., and Fernandes, C. (2013). Altered social behaviours in neurexin 1α knockout mice resemble core symptoms in neurodevelopmental disorders. PLoS One 8:e67114. doi: 10.1371/journal.pone.0067114
Gupta, R., Advani, D., Yadav, D., Ambasta, R. K., and Kumar, P. (2023). Dissecting the relationship between neuropsychiatric and neurodegenerative disorders. Mol. Neurobiol. 60, 6476–6529. doi: 10.1007/s12035-023-03502-9. Advance online publication. doi:doi: 10.1007/s12035-023-03502-9
Hu, X., Zhang, J., Jin, C., Mi, W., Wang, F., Ma, W., et al. (2013). Association study of NRXN3 polymorphisms with schizophrenia and risperidone-induced bodyweight gain in Chinese Han population. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 43, 197–202. doi: 10.1016/j.pnpbp.2012.12.007
Iqbal, S., Han, J., Matorin, A., Husnain, A., Ullah, U., Naeem, R., et al. (2023). Schizophrenia and chronic psychotic disorders: update on treatment options and new trends. Psychiatr. Ann. 53, 155–159. doi: 10.3928/00485713-20230313-02
Janz, P., Bainier, M., Marashli, S., Schoenenberger, P., Valencia, M., and Redondo, R. L. (2022). Neurexin1α knockout rats display oscillatory abnormalities and sensory processing deficits back-translating key endophenotypes of psychiatric disorders. Transl. Psychiatry 12:455. doi: 10.1038/s41398-022-02224-1
Jusop, A. S., Thanaskody, K., Tye, G. J., Dass, S. A., Wan Kamarul Zaman, W. S., and Nordin, F. (2023). Development of brain organoid technology derived from iPSC for the neurodegenerative disease modelling: a glance through. Front. Mol. Neurosci. 16:1173433. doi: 10.3389/fnmol.2023.1173433
Kasem, E., Kurihara, T., and Tabuchi, K. (2018). Neurexins and neuropsychiatric disorders. Neurosci. Res. 127, 53–60. doi: 10.1016/j.neures.2017.10.012
Khoja, S., Haile, M. T., and Chen, L. Y. (2023). Advances in neurexin studies and the emerging role of neurexin-2 in autism spectrum disorder. Front. Mol. Neurosci. 16:1125087. doi: 10.3389/fnmol.2023.1125087
Kight, K. E., Argue, K. J., Bumgardner, J. G., Bardhi, K., Waddell, J., and McCarthy, M. M. (2021). Social behavior in prepubertal neurexin 1α deficient rats: a model of neurodevelopmental disorders. Behav. Neurosci. 135, 782–803. doi: 10.1037/bne0000482
Koh, A., Tao, S., Jing Goh, Y., Chaganty, V., See, K., Purushothaman, K., et al. (2020). A Neurexin2aa deficiency results in axon pathfinding defects and increased anxiety in zebrafish. Hum. Mol. Genet. 29, 3765–3780. doi: 10.1093/hmg/ddaa260
Lam, M., Moslem, M., Bryois, J., Pronk, R. J., Uhlin, E., Ellström, I. D., et al. (2019). Single cell analysis of autism patient with bi-allelic NRXN1-alpha deletion reveals skewed fate choice in neural progenitors and impaired neuronal functionality. Exp. Cell Res. 383:111469. doi: 10.1016/j.yexcr.2019.06.014
Lee, K. M., Hawi, Z. H., Parkington, H. C., Parish, C. L., Kumar, P. V., Polo, J. M., et al. (2020). The application of human pluripotent stem cells to model the neuronal and glial components of neurodevelopmental disorders. Mol. Psychiatry 25, 368–378. doi: 10.1038/s41380-019-0495-0
Levy, K. A., Weisz, E. D., and Jongens, T. A. (2022). Loss of neurexin-1 in Drosophila melanogaster results in altered energy metabolism and increased seizure susceptibility. Hum. Mol. Genet. 31, 3422–3438. doi: 10.1093/hmg/ddac115
Missler, M., Zhang, W., Rohlmann, A., Kattenstroth, G., Hammer, R. E., Gottmann, K., et al. (2003). Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature 423, 939–948. doi: 10.1038/nature01755
Mohrmann, I., Gillessen-Kaesbach, G., Siebert, R., Caliebe, A., and Hellenbroich, Y. (2011). A de novo 0.57 Mb microdeletion in chromosome 11q13.1 in a patient with speech problems, autistic traits, dysmorphic features and multiple endocrine neoplasia type 1. Eur. J. Med. Genet. 54, e461–e464. doi: 10.1016/j.ejmg.2011.04.006
Pak, C., Danko, T., Mirabella, V. R., Wang, J., Liu, Y., Vangipuram, M., et al. (2021). Cross-platform validation of neurotransmitter release impairments in schizophrenia patient-derived NRXN1-mutant neurons. Proc. Natl. Acad. Sci. U. S. A. 118:e2025598118. doi: 10.1073/pnas.2025598118
Raab, S., Klingenstein, M., Liebau, S., and Linta, L. (2014). A comparative view on human somatic cell sources for iPSC generation. Stem Cells Int. 2014:768391, 1–12. doi: 10.1155/2014/768391
Rabaneda, L. G., Robles-Lanuza, E., Nieto-González, J. L., and Scholl, F. G. (2014). Neurexin dysfunction in adult neurons results in autistic-like behavior in mice. Cell Rep. 8, 338–346. doi: 10.1016/j.celrep.2014.06.022
Reichelt, A. C., Rodgers, R. J., and Clapcote, S. J. (2012). The role of neurexins in schizophrenia and autistic spectrum disorder. Neuropharmacology 62, 1519–1526. doi: 10.1016/j.neuropharm.2011.01.024
Reissner, C., Runkel, F., and Missler, M. (2013). Neurexins. Genome Biol. 14:213. doi: 10.1186/gb-2013-14-9-213
Restrepo, S., Langer, N. J., Nelson, K. A., and Aoto, J. (2019). Modeling a Neurexin-3α human mutation in mouse neurons identifies a novel role in the regulation of Transsynaptic signaling and neurotransmitter release at excitatory synapses. J. Neurosci. 39, 9065–9082. doi: 10.1523/JNEUROSCI.1261-19.2019
Rowe, R. G., and Daley, G. Q. (2019). Induced pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Genet. 20, 377–388. doi: 10.1038/s41576-019-0100-z
Sebastian, R., Jin, K., Pavon, N., Bansal, R., Potter, A., Song, Y., et al. (2023). Schizophrenia-associated NRXN1 deletions induce developmental-timing- and cell-type-specific vulnerabilities in human brain organoids. Nat. Commun. 14:3770. doi: 10.1038/s41467-023-39420-6
Shan, D., Li, S., Xu, R., Nie, G., Xie, Y., Han, J., et al. (2022). Post-COVID-19 human memory impairment: a PRISMA-based systematic review of evidence from brain imaging studies. Front. Aging Neurosci. 14:1077384. doi: 10.3389/fnagi.2022.1077384
Shan, D., Zheng, Y., and Froud, K. (2021). Brain-derived neurotrophic factor as a clinical biomarker in predicting the development of post-stroke depression: a review of evidence. Cureus 13:e15662. doi: 10.7759/cureus.15662
Shiwaku, H., Katayama, S., Gao, M., Kondo, K., Nakano, Y., Motokawa, Y., et al. (2023). Analyzing schizophrenia-related phenotypes in mice caused by autoantibodies against NRXN1α in schizophrenia. Brain Behav. Immun. 111, 32–45. doi: 10.1016/j.bbi.2023.03.028
Silveira, S. R., Sahm, B. D., Kreve, S., and dos Reis, A. C. (2023). Osseointegration, antimicrobial capacity and cytotoxicity of implant materials coated with graphene compounds: a systematic review. Jpn. Dent. Sci. Rev. 59, 303–311. doi: 10.1016/j.jdsr.2023.08.005
Tolchin, B., Hirsch, L. J., and LaFrance, W. C. Jr. (2020). Neuropsychiatric aspects of epilepsy. Psychiatr. Clin. North Am. 43, 275–290. doi: 10.1016/j.psc.2020.02.002
Tromp, A., Mowry, B., and Giacomotto, J. (2021). Neurexins in autism and schizophrenia-a review of patient mutations, mouse models and potential future directions. Mol. Psychiatry 26, 747–760. doi: 10.1038/s41380-020-00944-8
Trotter, J. H., Wang, C. Y., Zhou, P., Nakahara, G., and Südhof, T. C. (2023). A combinatorial code of neurexin-3 alternative splicing controls inhibitory synapses via a trans-synaptic dystroglycan signaling loop. Nat. Commun. 14:1771. doi: 10.1038/s41467-023-36872-8
Tufanaru, C., Munn, Z., Aromataris, E., Campbell, J., and Hopp, L. (2017). “Systematic reviews of effectiveness” in Joanna Briggs Institute Reviewer’s Manual. eds. E. Aromataris and Z. Munn (Adel: Joanna Briggs Inst)
Vaags, A. K., Lionel, A. C., Sato, D., Goodenberger, M., Stein, Q. P., Curran, S., et al. (2012). Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am. J. Hum. Genet. 90, 133–141. doi: 10.1016/j.ajhg.2011.11.025
Wu, D., Zhu, J., You, L., Wang, J., Zhang, S., Liu, Z., et al. (2023). NRXN1 depletion in the medial prefrontal cortex induces anxiety-like behaviors and abnormal social phenotypes along with impaired neurite outgrowth in rat. J. Neurodev. Disord. 15:6. doi: 10.1186/s11689-022-09471-9
Xu, B., Ho, Y., Fasolino, M., Medina, J., O'Brien, W. T., Lamonica, J. M., et al. (2023). Allelic contribution of Nrxn1α to autism-relevant behavioral phenotypes in mice. PLoS Genet. 19:e1010659. doi: 10.1371/journal.pgen.1010659
Yuan, H., Wang, Q., Liu, Y., Yang, W., He, Y., Gusella, J. F., et al. (2018). A rare exonic NRXN3 deletion segregating with neurodevelopmental and neuropsychiatric conditions in a three-generation Chinese family. American journal of medical genetics. Part B, neuropsychiatric genetics: the official publication of the international society of. Psychiatr. Genet. 177, 589–595. doi: 10.1002/ajmg.b.32673
Zeng, L., Zhang, P., Shi, L., Yamamoto, V., Lu, W., and Wang, K. (2013). Functional impacts of NRXN1 knockdown on neurodevelopment in stem cell models. PLoS One 8:e59685. doi: 10.1371/journal.pone.0059685
Keywords: animal models, human induced pluripotent stem cells, disease modeling, neurexins, neuropsychiatric diseases
Citation: Shan D, Song Y, Zhang Y, Ho CW, Xia W, Li Z, Ge F, Ou Q, Dai Z and Dai Z (2024) Neurexin dysfunction in neurodevelopmental and neuropsychiatric disorders: a PRIMSA-based systematic review through iPSC and animal models. Front. Behav. Neurosci. 18:1297374. doi: 10.3389/fnbeh.2024.1297374
Edited by:
Thomas Nicholas Ferraro, Cooper Medical School of Rowan University, United StatesReviewed by:
João Peça, University of Coimbra, PortugalLuye Qin, University of South Dakota, United States
Copyright © 2024 Shan, Song, Zhang, Ho, Xia, Li, Ge, Ou, Dai and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan Shan, ZC5zaGFuQGxhbmNhc3Rlci5hYy51aw==; Zhihao Dai, emhpaGFvZGFpMjBAcmNzaS5jb20=
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work
 Dan Shan
Dan Shan Yuming Song3†
Yuming Song3† Zhihao Dai
Zhihao Dai