
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bee Sci., 07 March 2025
Sec. Bee Genetics
Volume 3 - 2025 | https://doi.org/10.3389/frbee.2025.1507903
This article is part of the Research TopicWomen in Bee ScienceView all 3 articles
Declining social insects such as bumble bees are particularly vulnerable to loss of genetic diversity. Population delimitation is a precondition for measuring genetic diversity but usually requires extensive genetic data and comprehensive knowledge about gene flow barriers. As a first step towards a delimitation strategy that does not rely on genetic data, we compiled existing knowledge about Bombus population structures and (potential) gene flow barriers. We reviewed studies examining genetic structuring in Bombus species and assessed the impact of different ecological and environmental factors on their gene flow. Generally, we found that declining species and clearly isolated populations exhibit genetic structuring for which some underlying factors can be correlated with isolation-by-landscape approaches. For widespread species, isolation-by-environment approaches can help elucidate subtle factors impeding gene flow between populations, even though such species are capable of maintaining gene flow across large stepping stone populations. However, to better inform isolation-by landscape/environment models that could contribute to a landscape-based population delimitation strategy, more research into habitat requirements and dispersal ranges has to be conducted.
Genetic diversity is crucial to the survival of species, as it enhances their ability to adapt to environmental changes, including pressures such as climate change (Spielman et al., 2004). Small and/or isolated populations can experience inbreeding (Darvill et al., 2012), potentially perilous impacts of genetic drift (Kimura and Ohta, 1971) and restrictions to gene flow, i. e. genetic exchange, between individuals or populations, which all contribute to the loss of genetic diversity within a population. This leads to declines in a species’ adaptation ability and an increased chance of inbreeding depression which can push populations and subsequently species towards extinction (Wright, 1931; Keller, 2002; Frankham, 2005). Maintaining and monitoring genetic variation should therefore pose an important objective in the conservation of biodiversity (Laikre, 2010; Hoban et al., 2020). Recent studies suggested that conservation measures should target functional genetic variation directly affecting fitness rather than genome-wide, neutral genetic variation (Kyriazis et al., 2021; Teixeira and Huber, 2021). While targeting specific deleterious genetic variations may advance conservation in some cases (van Oosterhout, 2020; Kyriazis et al., 2021), the correlation between low genome-wide genetic diversity and the decreased fitness of populations has been firmly established both theoretically and empirically (DeWoody et al., 2021; Kardos et al., 2021). Since genetic diversity impacts the genetic health and resilience of populations, a number of metrics have been introduced to assess genetic diversity in order to inform conservation efforts on extinction risks and trends in population fitness. Commonly used measures are the number of polymorphisms or metrics of heterozygosity and nucleotide diversity on individual and population levels (Balkenhol et al., 2016). These metrics require comprehensive genetic analyses [depending on the metric: microsatellite data, single nucleotide polymorphisms (SNPs), mitochondrial DNA (mtDNA)], however, appropriate data on a global scale remains limited. Consequently, assessments concerning risk estimates or conservation goals frequently have to be made without sufficient data on genetic diversity. This has led to a need for more general guidelines that could inform conservation efforts with or without extensive genetic analyses (Laikre et al., 2020; Hoban et al., 2022, 2024). A particularly relevant parameter is the effective population size (Ne), which in essence, describes the number of adult individuals within a population that contribute to the gene pool of the subsequent generation. In other words, the Ne is defined as the number of individuals in a population that would undergo an equivalent loss of genetic diversity to that experienced by an idealized population (Wright, 1931; Frankham, 2019) (for further information see Supplementary Material). Regular assessment of the Ne can help monitor key aspects of genetic diversity, including the risk of loss of genetic diversity due to drift, the risk of inbreeding depression and potential for adaption and long-term resilience (Hoban et al., 2020). It is suggested that the long-term persistence of a population across various taxa requires an Ne larger than 500 individuals (Hoban et al., 2021). This threshold constitutes the basis of the Ne500 indicator, a headline indicator for genetic diversity monitoring within the Kunming-Montreal Global Biodiversity Framework (CBD/COP/DEC/15/4, CBD/COP/DEC/15/5, CBD/COP/15/L.26). The Ne500 indicator describes the proportion of populations within a species with an Ne greater than 500 (Hoban et al., 2020, 2024) and promises a straightforward approach to monitoring and assessing genetic diversity on previously unmatched scales, even in the absence of comprehensive genetic data (for further information see Supplementary Material). Regardless of the chosen metric and/or method to assess genetic diversity, different populations within a species need to be defined and delimited from one another. This requires a comprehensive understanding of gene flow, biological and geographical barriers. However, for many insect species with their widespread distribution and difficult to measure population sizes, this knowledge is currently lacking. Thus, population delimitation proves difficult, and it has been advised to exclude insects from the initial implementation of genetic diversity monitoring (Mastretta-Yanes et al., 2023).
Recent studies have documented worrying declines in many bumble bee species (Potts et al., 2010; Soroye et al., 2020) which are of vital importance as pollinators with significant economic and ecological value (Rao and Stephen, 2009; Bänsch et al., 2021). Bumble bees as well as other pollinating insects are facing a multitude of increasing environmental pressures. In recent years factors such as pathogens, climate change, agricultural practices and the use of pesticides have been identified as main drivers for the decline in pollinating insects (Potts et al., 2010; Hristov et al., 2020). It can be expected that declining bumble bee species face a reduction of genetic diversity (Ellis et al., 2006; Cameron et al., 2011; Darvill et al., 2012; Maebe et al., 2015). Bumble bees are particularly vulnerable to loss of genetic diversity due to small effective population sizes, as most individuals are sterile workers (Chapman and Bourke, 2001; Packer and Owen, 2001). Additionally bumble bees, like other Hymenoptera, are haplodiploid, thus all males are hemizygous at all loci, which further reduces genetic diversity compared to diploids (Wright, 1933; Hedrick, 2005). Furthermore, it has been demonstrated that low genetic diversity may lead to increased occurrences of sterile males within a population due to the haplodiploid sex determination mechanism, which consequently reduces overall population fitness (Zayed, 2004; Zayed and Packer, 2005). The actual extent to which reduced genetic diversity may affect the fitness of wild bumble bee populations remains unclear for wild populations (Goulson et al., 2008; Cameron et al., 2011). All in all, bumble bees constitute a group of insects of particular importance for genetic diversity monitoring and should thus be incorporated into respective monitoring efforts as soon as possible. At the same time, associated obstacles regarding population delimitation need to be overcome.
In regards to the Ne500 indicator, all groups of individuals of a species that are capable of exchanging at least one migrant on average per generation are considered a population because gene flow between the groups is deemed sufficient (Mastretta-Yanes et al., 2023). For some taxa, this can be estimated through considering dispersal distances and geographical barriers or it can be achieved through the assessment of genetic data, enabling the identification of current and historical patterns of genetic variation and thereby facilitating the investigation of gene flow and gene flow barriers in a variable landscape (Packer and Owen, 2001; Frankham et al., 2011; Mastretta-Yanes et al., 2023) (for further information see Supplementary Material). The analysis of genetic variation typically encompasses two key aspects: genetic diversity and genetic structure. Genetic diversity refers to the genetic variation observed within an individual or population, whereas genetic structuring provides insights into the distribution of variations among populations or geographical areas (Balkenhol et al., 2016). Identifying genetic structuring in populations is therefore essential for the assessment of gene flow barriers (Frankham et al., 2011).
The resolution of genetic structure is influenced by the methodological approach employed (Figure 1). For example, the use of a single-locus approach based on the identification of mitochondrial DNA (mtDNA), lineages and subsequent construction of haplotype networks can result in limited resolution (Figure 1A), and mtDNA is also typically slower to respond to contemporary changes in population structure (Freeland, 2019). Consequently, the resolution of mtDNA approaches may be inadequate for elucidating current connectivity. Microsatellite markers (Figure 1B) are multiallelic short nucleotide sequences distributed throughout the genome and are widely employed in genetic studies (Storfer et al., 2010). They exhibit a significantly higher mutation rate than mtDNA, thereby enabling the detection of population structure with greater precision (Freeland, 2019). A further increase in resolution can be achieved through the utilization of single-nucleotide polymorphisms (SNPs, Figure 1C). Single-nucleotide polymorphisms are bi-allelic, which results in a reduction in the amount of information per locus (Morin et al., 2004). This is offset by a much larger number of SNPs generated, which makes them increasingly important as they seem to identify genetic clusters that depict subtle genetic structuring not detected by microsatellites (Morin et al., 2004; Zimmerman et al., 2020; Heraghty et al., 2023; Skey et al., 2023) (for further information see Supplementary Material).
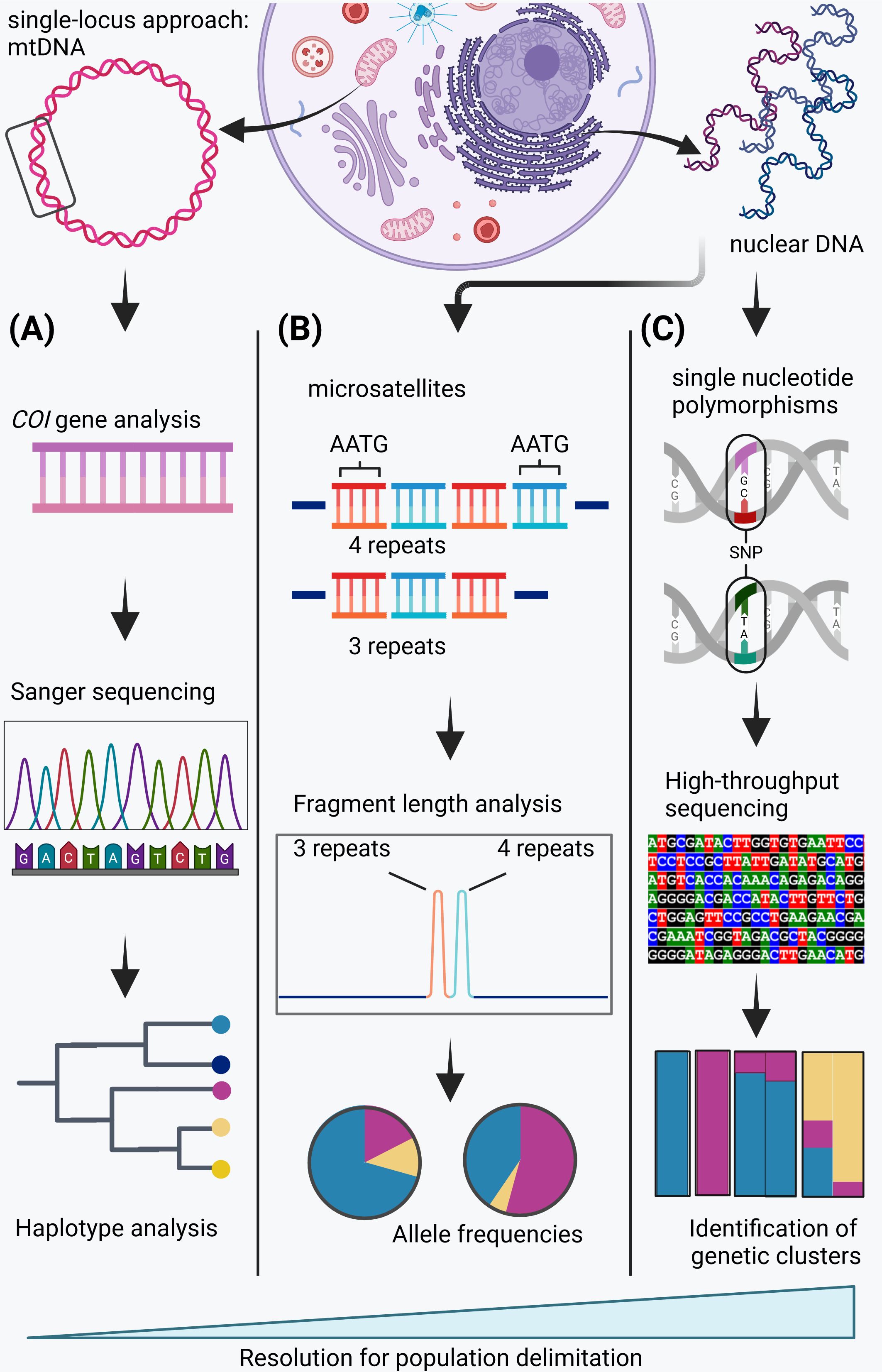
Figure 1. Summary of different genetic data used to assess genetic population structure. (A) The single-locus approach using mitochondrial DNA (mtDNA) which usually relies on the cytochrome oxidase I (COI) gene. The gene sequence is traditionally determined using Sanger sequencing. Mitochondrial DNA analysis allows the assessment of haplotypes and thereby genetic lineages, but its resolution for population delimitation is low. (B) Microsatellite analysis from nuclear DNA. Microsatellites are short oligo motifs that are repeated at different frequencies and repeat lengths are assessed using fragment length analysis, which allows the determination of allele frequencies in different populations. The resolution of microsatellite analysis for population delimitation is medium and may allow to detect genetic structure. (C) Single nucleotide polymorphisms (SNPs) from nuclear DNA. SNPs can be detected using high-throughput sequencing and allows the identification of genetic clusters and thereby even subtle genetic structuring. The resolution of SNP analysis for population delimitation is high.
Once genetic structuring has been identified, environmental factors can be related to genetic differentiation in order to identify underlying patterns that shape gene flow between individuals. Spatial and environmental heterogeneity in landscapes is expected to shape genetic variation and population genetic structure (Manel et al., 2003), but not all species appear to respond to environmental and landscape conditions in the same way (Melero et al., 2022). One possible method to uncovering patterns of genetic differentiation is the isolation-by-distance (IBD) approach (Figure 2A). This approach is based on the fundamental assumption that dispersal and mating choices among populations are related to geographic distance (Wright, 1943). However, IBD assumes movement in straight lines and environmental characteristics that might influence the movement of individuals are not included (Balkenhol et al., 2016). In the past decades new fields such as landscape genetics have arisen, combining population genetics, landscape ecology and spatial analytical techniques to improve the understanding of evolutionary processes such as gene flow (Balkenhol et al., 2016). Thus, isolation-by-landscapes (IBL) approaches were developed, incorporating the influence of landscape on a species dispersal pattern (Storfer et al., 2010). One frequently employed application of IBL is isolation-by-resistance (IBR) in which landscape characteristics are converted to resistance surfaces (Figure 2B). A resistance surface is a spatial layer in which each grid cell is assigned a resistance value based on the extent to which the landscape impedes the dispersal of organisms, thereby limiting gene flow (Spear et al., 2010). Parameterization of this model may be achieved through the use of techniques such as environmental niche modeling (ENM) or habitat suitability modeling (Balkenhol et al., 2016). Typically, ENMs are based on broad-scale predictors using bioclimatic variables, which tend to capture the broad-scale features of species distributions. Habitat suitability variables, such as land cover types, are more likely to capture finer-scale contributions to species distributions (Elith and Franklin, 2013). These resistance layers must be converted into measures of population connectivity, which can be achieved through the utilization of circuit theory. In this approach, the landscape is represented as a conductance layer, not unlike an electrical node connected with resistors in electrical circuits (McRae, 2006). This approach assumes a positive correlation between genetic distance and resistance and integrates all potential pathways between populations (McRae, 2006). A more recent approach, isolation-by-environment (IBE), predicts a correlation between genetic distance and environmental dissimilarity, which is independent of geographic distance (Wang and Bradburd, 2014) (Figure 2C). This is based on the hypothesis that populations are more genetically differentiated when there is an increase in habitat dissimilarity. The underlying processes that could be responsible for this pattern include natural or sexual selection against immigrants, reduced hybrid fitness or biased dispersal (Wang and Bradburd, 2014).
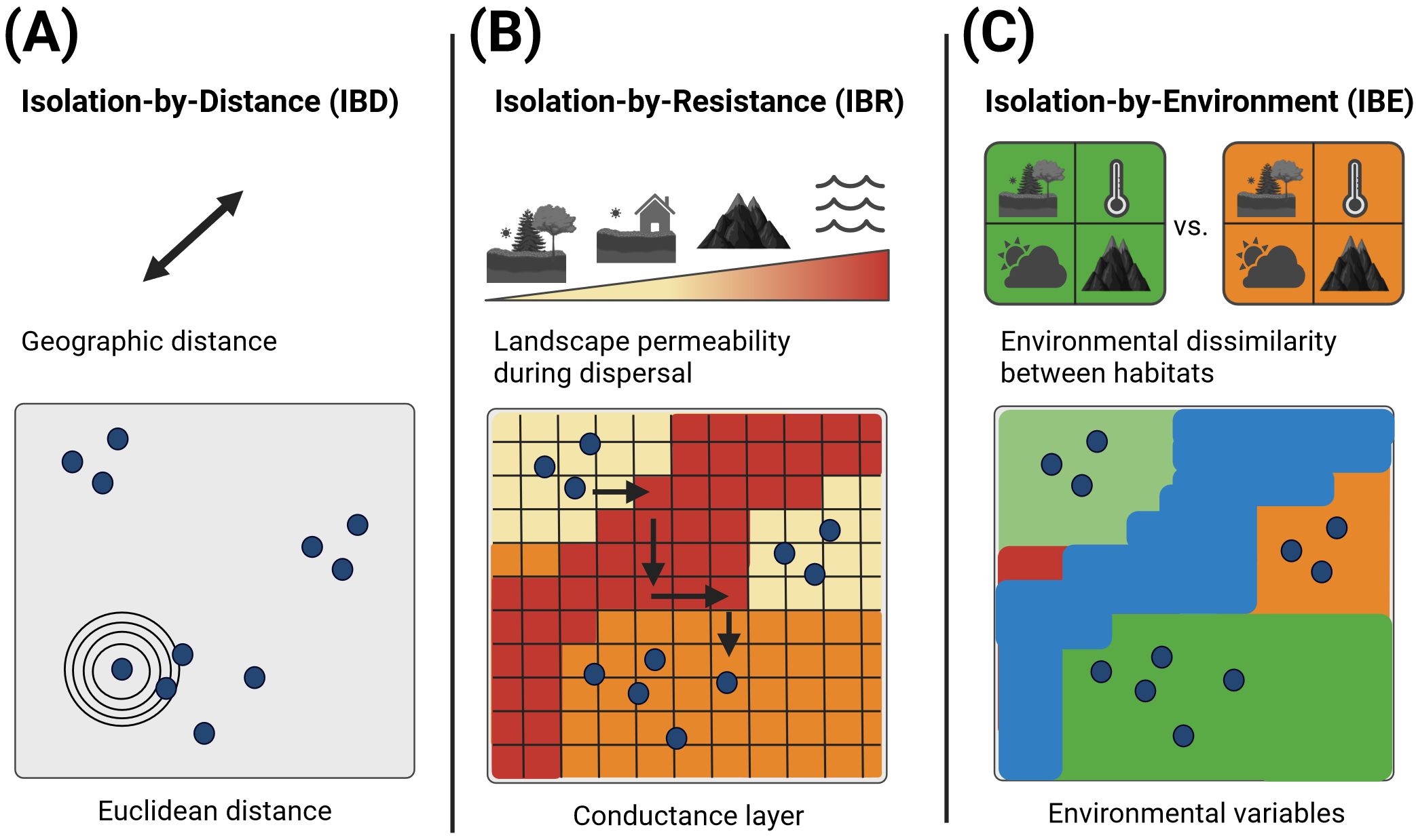
Figure 2. Summary of three main approaches correlating genetic structuring and environmental parameters. The blue dots indicate theoretical sampling points where individuals have been genetically analyzed. (A) The isolation-by-distance approach relates genetic structuring to geographic distance. (B) The isolation-by-resistance approach incorporates the influence of the landscape on species dispersal by integrating all possible pathways of dispersal between populations while considering the varying permeability of the landscape. A frequently employed approach is the conversion of landscape characteristics into a conductance layer assuming a positive correlation between genetic distance and landscape resistance to dispersal. (C) The isolation-by-environment approach relates genetic structuring to environmental dissimilarity independent of geographic distance. Matrix regression-based methods are utilized to relate environmental dissimilarities to genetic distance.
For purposes of genetic diversity monitoring, delineating populations solely on the basis of genetic data is not economically feasible, as it would require the availability of extensive genetic data sets that depict genetic structuring at regional, national or international scales with sufficient resolution. Accordingly, the objective of this review is to identify the factors and considerations that impede or facilitate gene flow, with a view to establishing population delimitation strategies that do not rely solely on genetic data. We therefore reviewed the scientific literature regarding genetic structuring much of which utilized IBL/IBE approaches to identify contributing factors affecting gene flow among bumble bee populations (Figure 3A). Additionally, we reviewed the potential of ecological factors, including resource availability, habitat requirements and fragmentation, as well as dispersal ranges, in order to ascertain their capacity to impede or facilitate gene flow between bumble bee populations so they may be incorporated into future population delimitation strategies (Figure 3B).
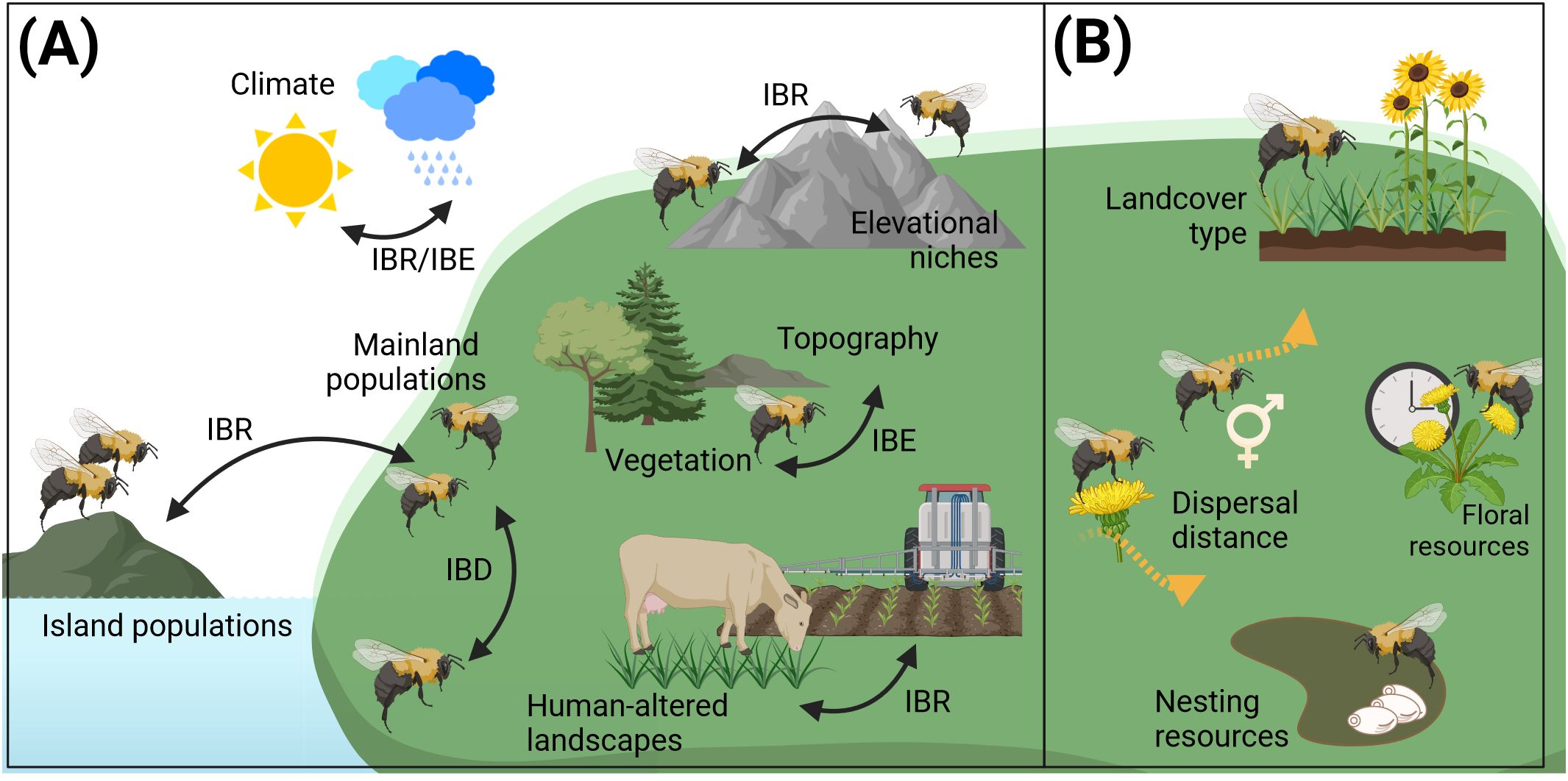
Figure 3. Summary of parameters affecting gene flow. (A) Effects identified using genetic analysis. (B) Effects that are expected to affect gene flow based on a species’ ecology. IBD, isolation-by-distance; IBE, isolation-by-environment; IBR, isolation-by-resistance.
Scientific literature was collected on 23 June 2024 via Google scholar using two search terms: (1) [population delimitation landscape distribution model dispersal model bumblebee OR bumble bee OR Bombus -parasites -disease -viruses -pathogens] and (2) [genetic structuring gene flow population bumblebee OR bumble bee OR Bombus -parasites -disease -viruses -pathogens -pollen] and on 13 January 2025 via Web of Science (WOS) using the same search terms adapted to requirements of the WOS search engine: (1) [(population delimitation OR landscape distribution OR model dispersal model) AND (bumblebee* OR bumble bee* OR Bombus) NOT parasite* NOT disease NOT virus* NOT pathogen*] and (2) [(“genetic structuring” OR “gene flow”) population AND (bumblebee* OR “bumble bee”* OR Bombus) Not parasites NOT disease NOT viruses NOT pathogens NOT pollen]. The first term was designed to primarily identify existing population delimitation strategies for bumble bees, with a particular focus on those that employed non-genetic approaches, such as landscape or dispersal models. The second search term focused on papers that provided information about gene flow, specifically genetic structuring, in bumble bees. Some research papers were found with both search terms and several papers were found through both the Google scholar and the WOS search engine. Both search engines were adjusted to sort the results by decreasing relevance. Studies returned by the Google scholar engine showed decreasing significance to our research question (assessed by study title), therefore we assessed only first 150 results. For the first search term 21 research papers and for the second search term 38 research papers were found to align with the broader topic and were thus deemed suitable for further examination. As no overall decrease in relevance was evident across the results called by the WOS search engine, all 555 results for the first search term and 98 results for the second search term were evaluated. Through the WOS search we identified four additional studies for the initial search term and another seven studies for the secondary search term, which were not identified by the Google scholar search, yet were deemed relevant. The papers were then subjected to a more detailed evaluation, during which some were dismissed on the grounds of the following aspects (1) The species studied was not of the Bombus genus, as it is not possible to assume that findings for other taxa could be transferred to the Bombus genus; (2) the focus of the study was species rather than population delimitation and genetic structuring between populations of the same species was not considered; (3) the term gene flow barrier was used as a synonym for genetic loci resisting homogenization with other genomes during the process of gene flow between diverging species (Elmer, 2005). They are distinct from environmental gene flow barriers, which are discussed in this paper; (4) general evaluation of genetic methods, as the search was directed towards reviewing what is known about genetic structuring within the Bombus genus. A further eleven studies were identified during the research process, mainly through citation networks of papers called with our search term, and included in the review. In total, 38 studies were reviewed, focusing on genetic structuring and patterns of gene flow. After reviewing these studies concerning patterns of genetic structuring, further aspects of ecology and bumble bees’ response to landscapes were investigated (see section 4).
For several bumble bee species, especially those that are widespread, no or only weak genetic structuring has been observed (Table 1). Utilizing mtDNA data, Bombus ardens, a common Bombus species in Japan and Korea, has been shown to be connected by high levels of gene flow across the Korean peninsula (Kim et al., 2009). Similarly, the following studies utilizing microsatellite data also failed to detect any significant genetic structuring. Although B. brasiliensis, a species with a wide distribution in the Brazilian Atlantic Forest, displays low levels of genetic structuring, it appears to be capable of dispersing across fragmented mosaics of anthropogenic and conserved habitats (Santos Júnior et al., 2019). No significant genetic structuring was observed in B. morio and B. pauloensis populations in Brazil (Françoso et al., 2016), as well as populations of B. lucorum in Belgium, France and Spain (Blasco-Lavilla et al., 2019). On a fine scale (<10 km) bumble bee species such as B. lapidarius, B. hortorum, B. ruderatus, B. terrestris and B. pascuorum also do not exhibit genetic structuring (Ellis et al., 2006; Dreier et al., 2014). In regards to larger scales, B. distinguendus has been demonstrated to maintain gene flow over distances of at least tens of kilometers, and potentially across the open sea, as no population structuring was evident in populations within island groups off the Scottish coast (Charman et al., 2010). The same was found for B. lapidarius and B. pascuorum across a 2100 ha estate in the United Kingdom (Carvell et al., 2012). Furthermore, evidence indicates that B. pascuorum exhibits a high degree of connectivity across Sweden (Liu et al., 2024). This phenomenon has also been observed in other Scandinavian montane bumble bee species (B. balteatus, B. hyperboreus, B. pyrrhopygus, B. alpinus, B. lapponicus and B. monticola) with considerably smaller distributions (Liu et al., 2024) and for B. vosnesenskii for North America (Lozier et al., 2011). For three species considered declining in Europe, B. ruderarius, B. soroeensis, and B. sylvarum, as well as for the more widespread B. hortorum, microsatelite data revealed no genetic structuring on national scale and across Europe between Belgium and Estonia (Maebe et al., 2019). This may indicate that these bumble bee species have maintained extensive dispersal abilities throughout Europe (Maebe et al., 2019). Surprisingly, while no genetic structuring was apparent on national scales for the widespread species in B. hypnorum, B. lapidarius and B. pascuorum, low levels of genetic structuring was present between populations in Estonia and Belgium (Maebe et al., 2019). While the aforementioned studies were based on microsatellite data, SNP data sets capable of higher resolution, revealed minimal population structure at a continental scale (>1000km) for B. vosnesenskii (Jha, 2015; Jackson et al., 2018; Heraghty et al., 2023). However, SNP data did not reveal any significant genetic structuring for B. pratorum across the UK (Huml et al., 2023).
Island systems can be used to demonstrate limitations to migration and to provide insight into the manifestation of genetic structuring. It serves as a model for the formation of genetic structuring patterns in the presence of gene flow barriers within highly fragmented landscapes (Darvill et al., 2006). Genetic structuring is frequently observed in island populations, even in species that do not exhibit such structuring in their mainland counterparts (Table 1). Previous studies revealed genetic structuring within island systems and between islands and mainland populations based on microsatellite data (Table 1). This is the case for island populations of B. pascuorum on Gotland (Liu et al., 2024), as well as for B. terrestris populations on Mediterranean islands and Tenerife (Estoup et al., 1996) and between Irish and British or continental populations (Moreira et al., 2015). Microsatellite data has also revealed significant genetic structuring in a B. ardens population located on Ulleung-do, an Island 150 km from the coast of South Korea (Jeong et al., 2023). Genetic analysis of mtDNA and nuclear DNA has revealed a significant degree of genetic differentiation between the Corsican population and mainland populations of B. terrestris, B. lucorum and B. vestalis (Lecocq et al., 2013). For island populations of B. bifarius, B. impatiens and B. pensylvanicus in the Gulf of Mexico, clear signatures of IBD in comparison to mainland populations could be inferred from the genetic structuring assessed (Lozier et al., 2011). Furthermore, clear genetic structuring correlated with IBD patterns were demonstrated for B. muscorum populations further apart than 10 km on islands in the Inner and Outer Hebrides (Scotland, UK), clearly demonstrating that gene flow is uncommon over distances further than 10 km in this island system (Darvill et al., 2006, 2010). A study on B. vosnesenskii populations on the Channel Islands (California, USA) demonstrated that incorporating the ocean as a strong dispersal limitation could better explain patterns of genetic structuring than the geographic distance (IBD) alone (Jha, 2015). Although open water may significantly impede gene flow from the mainland or other islands, it is evident that oceans and open water do not constitute absolute barriers to gene flow. In fact, populations of neighboring islands often display less differentiation from each other than those situated further away (Charman et al., 2010). The possibility of gene flow over open water has been demonstrated for widespread species such as B. terrestris (Estoup et al., 1996; Moreira et al., 2015), the south American B. morio (Francisco et al., 2016) as well as for declining species such as B. distinguendus (Charman et al., 2010) or B. muscorum, at least between island populations situated less than 10 km apart (Darvill et al., 2006; Francisco et al., 2016). Additionally, Jha (2015) suggests that B. vosnesenskii may occasionally disperse across open water bodies at distances exceeding 30 km. B. jonellus populations on the Hebrides (UK) less than 7 km apart were not significantly differentiated, and some populations even remain undifferentiated up to 104 km apart (Darvill et al., 2010). Darvill et al. (2010) suggests that potential stepping stone populations existing along the UK coast may explain the lack of genetic differentiation between some B. jonellus populations.
In contrast to island populations, which experience impediments to gene flow because the open sea constitutes a gene flow barrier, high elevation populations may experience isolation due to increases of habitat heterogeneity. Conditions regarding temperature, air pressure or habitat suitability can shift sharply across elevations (Cheviron and Brumfield, 2012), impeding gene flow between populations at different elevation levels. Based on microsatellite data, genetic structuring has been observed in populations of B. ignitus in the Korean Taebaek Mountains (Han et al., 2014) (Table 1). Clake et al. (2024) revealed the existence of two genetically distinct, parapatric populations of B. lapponicus sylvicola in the Canadian Rocky Mountains through the analysis of SNP data. Model predictions based on climatic variables suggest that the populations occupy areas with different climatic profiles characterized best by differences in the minimum lowest temperatures (Clake et al., 2024). B. vancouverensis exhibits local adaptations to high elevations, while populations at lower elevations do not, indicating significant population isolation (Lozier et al., 2021; Heraghty et al., 2022). For B. bifarius populations in high elevations in the United States microsatellite data demonstrated genetic structuring which could be correlated to patterns of IBD (Lozier et al., 2011). In a follow up study, the application of an IBR-model incorporating a resistance layer based on an ENM with bioclimatic variables could significantly reduce the scatter of the previous IBD analysis (Lozier et al., 2013). Similar analyses were conducted for a SNP-based study across mountain ranges in the US, which revealed weak but significant genetic structuring in mountain populations of B. vosnesenskii and B. bifarius (Jackson et al., 2018). In contrast to B. vosnesenskii populations in lower elevation habitats, IBR patterns were more pronounced in B. bifarius populations inhabiting higher elevation habitats. Similarly, discrepancies could be shown for high and low altitude populations of B. huntii, suggesting that adjacent environments constitute barriers to dispersal and gene flow (Koch et al., 2018). Furthermore, IBR analyses conducted independently of environmental variables, only testing for the effect of elevation, failed to provide evidence in support of any isolation by elevation patterns (Jha and Kremen, 2013; Jha, 2015). These findings indicate that elevation is not the primary determinant of genetic differentiation, but rather the restrictions imposed by elevational niches impede genetic connectivity, resulting in the formation of habitat islands with pronounced IBR patterns. This is consistent with other studies that have indicated that mountains do not typically represent a significant impediment to gene flow for bumble bees (Pirounakis et al., 1998; Christmas et al., 2022; Liu et al., 2024). However, high heterogeneity in habitat and, consequently, niche restrictions align with the well-documented phenomenon of “sky islands”, which describes populations of habitat specialists that are isolated in montane regions by inhospitable adjacent habitats (Brown, 1971). Conversely, IBD patterns are often more pronounced in low elevation populations because habitat heterogeneity is not as pronounced and geographic distance is the main factor contributing the patterns of genetic structuring.
While island and montane populations are constrained by natural barriers, some studies have also demonstrated how landscapes, mainly influenced by human land use change, impede gene flow. The range of B. affinis, a widespread species in North America, has contracted by approximately 70 to 90% since the late 1990s (Szymanski et al., 2016). Genetic analysis indicates a correlation between genetic structuring and geographic distance (IBD), suggesting that colonies are less abundant and potentially less well connected, at least on a larger scale (~500 km) in the remaining distribution range (Mola et al., 2024). A significant reduction in gene flow between populations has also been observed in the case of B. sylvarum across mainland landscapes in the UK, resulting in pronounced genetic structuring (Table 1). The populations of this rare and declining species are geographically isolated and the genetic structuring indicates that gene flow between them is improbable in the majority of cases (Ellis et al., 2006). Contrary to expectations, the study found no evidence for IBD patterns. This could be due to the small number of seven populations assessed compared to other studies that successfully revealed IBD (Darvill et al., 2006; Jackson et al., 2018; Mola et al., 2024). Alternatively, the basic assumption of mutation-drift equilibrium may not have been fulfilled due to recent fragmentation potentially obfuscating patterns of IBD through random genetic drift (Wright, 1943). The study posits that the populations were previously connected by a larger meta-population, which has since been degraded by recent habitat fragmentation (Ellis et al., 2006). Genetic structuring explained by IBD could be shown for B. ruderatus populations in New Zealand more than 300 km apart. However, for populations <100 km apart, significant IBR patterns could be demonstrated, suggesting that poor habitats regarding the availability of foraging resources may constrain gene flow (Bartlett et al., 2016). Subtle genetic structuring in B. ephippiatus populations in two mountain ranges in Guatemala may be the results of habitat loss at low elevations increasingly isolating the populations from each other (Landaverde-González et al., 2018). Significant patterns of genetic structuring explained by IBD were revealed by SNP data between populations of B. monticola known to have a fragmented distribution across the UK and Ireland (Huml et al., 2023). Interestingly, no significant genetic structuring was apparent between two Irish populations, less than 100 km apart and connected through habitat of high suitability, emphasizing the value of high value habitats for gene flow (Huml et al., 2023). The impact of human-altered landscapes on bumble bee gene flow can be demonstrated in the case of the widespread species B. vosnesenskii. A study based on SNP data, which included 17 sampling sites across the southwestern US coast, revealed significant genetic structuring across the mainland (Jha, 2015). IBD patterns were very weak, and patterns were better explained when incorporating oceans and human land use types. The study demonstrated that human-altered land use exerts the most significant influence on the genetic differentiation patterns of mainland populations. Similar patterns have also been revealed at regional scales (~200 km) (Jha and Kremen, 2013). Other studies, comparable in scale (~1000 km) could not find strong evidence for the influence of IBR on the dispersal of B. vosnesenskii (Lozier et al., 2011; Schenau and Jha, 2017; Jackson et al., 2018). Contrary to these studies, this study focused on the central and southern parts of the species range, which included larger human-developed areas than the northwestern part or more mountainous regions of the USA. It is also noteworthy that the sample size of individuals per population was particularly high in this study (40 vs. 20 or fewer individuals).
In comparison to isolated populations, the detection of subtle influences on the gene flow of widely distributed species that do not demonstrate clear patterns of genetic structuring is a considerably challenging endeavor. While several studies using mtDNA, microsatellites or SNP data failed to demonstrate significant genetic structuring in B. terrestris across the UK (Colgan et al., 2022) and the European continent (Estoup et al., 1996; Pirounakis et al., 1998), a study conducted on B. terrestris populations in Romania and Bulgaria revealed weak, yet significant genetic structuring inferred from a microsatellite data set (Table 1). One potential explanation for this discrepancy is the larger sample size and greater number of sampling locations included in the analysis as well as the pronounced heterogeneity of Bulgarian and Romanian landscapes (Glück et al., 2022). The study suggests that because of the vagility of B. terrestris, gene flow is not influenced by the permeability of habitats they traverse through during dispersal (IBR) nor by geographic distance (IBD), but rather by environmental factors. While IBD and IBR, based on a species distribution model, exhibited only marginal significance, IBE was found to account for over a third of genetic variations (Glück et al., 2022). In order to test for IBE, the environmental conditions were characterized with variables related to climate, vegetation and topography. The most significant predictive variables were found to be dissimilarity in temperature, Leaf Area Index (LAI) and slope inclination. The study suggests that temperature might influence emergence times of hibernating queens, isolating early colonies from late colonies. The LAI characterizes the coverage of leaves per unit of ground surface indicating the canopy cover. Bumble bees prefer open habitats for nesting therefore areas characterized by high LAI could limit the amount of nesting habitats. Similarly, the slope of a terrain could influence the suitability as a nesting habitat (Glück et al., 2022).
Genetic structuring of isolated populations or populations of rare species have been shown to correlate with IBD patterns (Darvill et al., 2006; Lozier et al., 2011; Jackson et al., 2018; Liu et al., 2024), but incorporating data about land cover encompassing specific types and extents of habitat heterogeneity improved the correlation with patterns of genetic structuring (Lozier et al., 2013). Significant variables were oceanic and human-altered impervious landcover (Jha and Kremen, 2013; Jha, 2015) (Figure 3A). While patterns of IBR can be identified in isolated or specialized species, widespread species are reported to display no or very weak genetic differentiation (Table 1) resulting in large scale IBD patterns (Mola et al., 2024). This is despite the fact that genetic data suggests that queens typically disperse no further than 10 km (Darvill et al., 2006; Charman et al., 2010; Lepais et al., 2010; Carvell et al., 2017). This indicates that a considerable number of bumble bee species are structured in metapopulations or exhibit “stepping stone” migration, whereby individual queens primarily migrate into neighboring populations that lie within the radius of their dispersal capabilities (Kimura and Weiss, 1964). Provided that these stepping stone population structures are not impeded by barriers to migration such as severe fragmentation or natural barriers such as open water, this facilitates gene flow over extensive distances. As widespread species are capable of dispersing over long distances and through a variety of land cover types, testing for IBD and IBR patterns may not be sufficient to explain subtle genetic differentiation. However, patterns of IBE could be demonstrated for B. terrestris, which was previously considered to be quasi-panmictic across Europe (Glück et al., 2022). Taking into consideration factors of bumble bee ecology, such as habitat requirements, appears to be a promising avenue for modeling potential impediments to gene flow even for species capable of dispersing in a broad spectrum of habitats. Therefore, further considerations regarding bumble bee ecology may prove beneficial in narrowing down barriers to gene flow, useful for population delimitation, on a more detailed scale.
Gene flow is contingent upon the mating of bumble bee queens and the establishment of a successful colony, thereby securing the next generation. A bumble bee queen is only able to reproduce successfully when specific habitat requirements are fulfilled, as the complex life cycle of a bumble bee requires different habitats at each specific life cycle stage (Goulson, 2010; Robinson, 2024). Additionally, these habitats have to be distributed in sufficiently close proximity in order for queens to be able to reach them (Iles et al., 2018). In order to identify gene flow barriers which could constitute separate populations, studies examining habitat requirements like nesting and flowering resources, biotope association and dispersal distances may help to narrow down aspects of landscapes or bumble bee ecology that may prohibit gene flow along certain barriers.
Generally speaking, wooded and urbanized areas, including roads, and intensively managed arable areas with sparse floral resources show the lowest bumble bee abundance and species richness but other environmental factors like recent wildfires and grazing intensity also influence abundance negatively (Grundel et al., 2010; Iserbyt et al., 2015; Kallioniemi et al., 2017) and may also indirectly affect gene flow. In landscapes shaped by agriculture, linear elements like crop verges and pastures were positively associated with abundance and species richness (Kallioniemi et al., 2017). Depending on aspects like flower morphology, some flowering crops like alfalfa, tree fruits and grapes can be beneficial for bumble bees (Rollin et al., 2013; Quinlan et al., 2021). In natural habitats, grass- and shrubland seem to be the most important landcover types associated positively with high abundance and species richness (Clake et al., 2022). Thus, the availability of flowering and nesting resources are important factors shaping bumble bee abundance and distribution in the landscape and consequently population genetic structure, although other effects like increased mortality induced by pesticides (Nicholson et al., 2024) or roads (Kallioniemi et al., 2017) may also play a role.
The temporal distribution and fragmentation of flower resources are among the most significant factors influencing abundance in both arable and natural landscapes (Mayer et al., 2012; Nicholson et al., 2021). Kallioniemi et al. (2017) demonstrated that resources flowering later in the season, in contrast to those that flower earlier, have a positive effect on bumble bee abundance. With regard to fragmentation, it appears that bumble bees forage in greater numbers when they have access to a single, extensive patch rather than having to fly to multiple smaller patches (Fragoso et al., 2021; Clake et al., 2022), which would prolong the foraging distance thereby reducing the net energy intake (Goulson, 1999; Redhead et al., 2016). It can be argued that fragmented landscapes provide a greater variation of habitats with an array of different biotic and abiotic conditions (Tscharntke et al., 2012), a phenomenon that can mainly be observed in natural landscapes (e.g mountains, nature reserves) (Quinn and Harrison, 1988). Generalist bumble bee species could potentially profit from the increase in habitat diversity resulting from the fragmentation in natural landscapes, provided that different floral resources do not decrease as a result of fragmentation and persist over a longer period of time while specialized or rare species might suffer under the increased effort to reach suitable foraging habitats (Walther-Hellwig and Frankl, 2000; Tscharntke et al., 2002; Clake et al., 2022; Hemberger et al., 2023).
High diet specialization on specific plants for nectar and pollen is unusual at least for European bumble bee species (Goulson et al., 2006). A study encompassing 13 Bombus species in the UK, confirmed that widespread species are less specialized than rare species (Goulson and Darvill, 2004). This however does not lead to a particularly tight biotope association, not even in rare species (Goulson et al., 2006). That is why, in contrast to the fragmentation and temporal distribution, plant species composition seems to have less influence on bumble bee abundance and community composition. The lack of correlation may be attributed to the broad diet breadth of bumble bees (Goulson and Darvill, 2004). With regards to plant species communities, they are able to tolerate a wide range of pollinator species compositions due to the ecological redundancy of pollinators. These factors could potentially diffuse the ability to link bumble bee and plant communities clearly (Goulson and Darvill, 2004; Grundel et al., 2010).
In a study concerning (social, solitary and parasitic) wild bee species, potential nesting resources were significant predictors of wild bee distribution in general (Grundel et al., 2010). In open habitats, where dead wood is scarce, the percentage of soil nesting bees was higher and the percentage of solitary and wood nesting bees, which both utilize dead wood for nesting, were lower (Grundel et al., 2010). With regards to bumble bees, it is evident that species-specific differences in nesting habitat preference exist (Richards, 1978; Kells and Goulson, 2003). Some species appear to prefer forest boundaries, potentially benefiting from the heterogeneous ecotone (Kells and Goulson, 2003; Christman et al., 2024). For others, open terrains may provide sufficient resources to sustain them (Svensson et al., 2000; Pugesek and Crone, 2021; Diekötter et al., 2006). Furthermore, bumble bee queens appear to select the location of their nest based on net energy intake rate (Suzuki et al., 2007). This indicates that nesting sites with an abundance of floral resources in close proximity to the nest are preferred (Heinrich, 2004). A study on the species B. ardens suggests that queens will determine the location of a new nest based on their experience of resource availability in April, when nests are being established. However, colonies that experience a lack of floral resources throughout the remainder of the season will ultimately become extinct. This is why the most accurate predictions of nesting sites for B. ardens were made when data on floral resource availability and quality from the middle of May was used (Suzuki et al., 2007).
While flowering and nesting resources determine successful reproduction, dispersal abilities of bumble bees determine which habitats can be reached and in turn, how far suitable habitats can be apart and still be in reach of colony founding queens (Iles et al., 2018). It is important to differentiate between foraging ranges of workers and dispersal ranges of queens. Foraging ranges of bumble bee workers have been estimated to range between 0.5 and 1 km for B. terrestris and between 0.3 and 0.8 km for B. pascuorum, B. lapidarius and B. pratorum (Chapman et al., 2003; Darvill et al., 2004; Knight et al., 2005; Carvell et al., 2012). In contrast to this, queen dispersal ranges are much larger because queen dispersal encompasses several dispersal steps, including searching for a mate, finding a hibernation place as well as a nesting site and foraging. To our knowledge, dispersal distances for queens have only been estimated for very few species. By genetically relating queens who are full sisters with workers foraging in the previous year, a study across 14 sites in an arable landscape demonstrated that B. pascuorum and B. lapidarius queens can disperse by at least 3 and 5 km respectively (Lepais et al., 2010). A different study also in arable landscapes in the UK, estimated shorter mean queen dispersal ranges at 1.5 km for B. terrestris, 1.1 km for B. pascuorum and 0.98 km for B. lapidarius (Carvell et al., 2017). Carvell et al. (2017) found a significant positive correlation between dispersal distance and nesting habitat cover, suggesting that non-crop habitats suitable for nesting may facilitate movement of queens into the wider landscape. Therefore, differences between relative proportion and distribution of landcover types between the two study sites may produce different estimations of dispersal range (Carvell et al., 2017). Additionally, the species assessed here were all common and widespread in agricultural landscapes. To which extent dispersal ranges of rare or declining bumble bee species might differ is unclear (Darvill et al., 2006; Dreier et al., 2014; Mola et al., 2024). Overall, these results are consistent with estimations made through analyzing genetic structuring (see section 3). Although male dispersal ranges also have a direct influence on gene flow, only one study has estimated male dispersal ranges. Kraus et al. (2009) estimated the male flight radius of B. terrestris to range between 2.6 and 9.9 km. This coincides with findings regarding males of B. vosnesenskii showing high levels of relatedness up to 10 km from their natal nest (Schenau and Jha, 2017). In conclusion, male dispersal abilities seem to be similar to those of queens.
It is important to note that estimates of queen dispersal made with the genetic sibship assignment method are likely unable to detect rare long-distance movements that would take queens out of the studied area. It is therefore prudent to consider these estimations as representing a minimum, rather than a maximum, dispersal range (Lepais et al., 2010). Nevertheless, these estimates concur with the findings of genetic studies in indicating that habitats separated by more than 10 km of unsuitable matrix landscape might significantly impede gene flow (see section 3). Additionally, as demonstrated above, other factors relating to bumble bee ecology play a role in species distribution and thus gene flow. For example, the suitability of a habitat for hosting a successful colony is contingent upon the availability of adequate nesting and flowering resources. The temporal distribution of flowering resources must be sufficiently extensive to span the entire flowering season (Figure 3B). The specific composition of flowering plant species may not be a significant indicator, given the broad dietary breadth of most bumble bee species.
Despite the numerous genetic studies about bumble bees there is still no practical strategy to delimit populations based on non-genetic parameters because information about gene flow barriers from which the delimitation of population could be inferred, remains limited. Several factors make it challenging to derive information about genetic structuring and gene flow barriers from genetic data.
The majority of studies reviewed here utilized microsatellite data, of which many could not identify any or only weak genetic structuring. Consequently, a re-evaluation using SNP data could prove beneficial in identifying genetic structure that may have been undetected with microsatellites. Furthermore, SNPs can be utilized to analyze adaptive genes, thereby providing further insight into environmental factors driving diverging genetic adaptation between populations (Theodorou et al., 2018; Lozier et al., 2021; Heraghty et al., 2023).
Subtle genetic structuring may only be detected, if a sufficient number of samples from a diverse range of sampling points are included in the analysis (Glück et al., 2022). Subtle genetic structuring may not indicate severe barriers to gene flow justifying the strict delimitation of separate populations. However, identifying biotic and abiotic factors causing these patterns can inform preventive conservation measures before pressures caused by land use and climate change become severe enough to result in impervious gene flow barriers thus disrupting bumble bee populations and consequently reducing genetic diversity and ultimately fitness.
Different events at different moments in time affect patterns of genetic structuring. Additionally, there is a time lag between the disruption of gene flow and the establishment of significant genetic structuring to become detectable. For example, historic events such as climatic fluctuations and bottleneck events have had a long-term effect on genetic structuring of bumble bee populations, that is still evident today (for example: Lye et al., 2011; Françoso et al., 2016; Santos Júnior et al., 2019). At the same time, recent changes in contemporary land use have already left their mark in bumble bee populations (Jha, 2015). In studies correlating patterns of genetic structuring to past and contemporary land use at three different points in time suggests that contemporary patterns of genetic structuring were best explained with contemporary (2011) land use distribution, rather than past (2006, 2001) distributions (Jha, 2015), although other patterns may hold true for other species groups (for example: bush crickets (Holzhauer et al., 2006). Generally, studies only utilized genetic datasets representative of a single point in time. To further understand the impact of land use change on genetic structuring and the potential for putative time lags it may be necessary to analyze genetic data from several points in time (Jha, 2015).
Many studies focused on explaining observed patterns of genetic structuring with IBD and IBR approaches. However, especially for widespread species these approaches often failed to explain the observed patterns of subtle genetic structuring. Resistance layers utilized in an IBR approach can only be modeled correctly when the environmental factor constituting a dispersal constraint is present and known. Additionally, IBD and IBR approaches may be unsuitable to comprehensively explain the underlying patterns of genetic structuring of bumble bee species due to their high vagility (Glück et al., 2022). The IBE approach, which remains underutilized in studies of the Bombus genus, combines the influence of spatial heterogeneity, agents of selection and potentially many different environmental variables (Wang and Bradburd, 2014) and may thus provide a basis to re-evaluate patterns of genetic structuring that could not be explained by IBD/IBR before.
The majority of studies conducted thus far have been carried out in North America and Europe (Table 1), with considerably less studies conducted in Asia and Central America. It is evident that the degree of genetic structuring of different bumble bee species varies, not only between different species (Table 1), but also between populations of the same species in different regions (Ellis et al., 2006; Han et al., 2014; Blasco-Lavilla et al., 2019; Glück et al., 2022). Thus, findings for North American or European species and regions cannot be inferred to Asian and Central American regions or species. In light of the global scale of the endeavor to conserve genetic biodiversity, it is crucial that further studies be conducted on bumble bee species in Asia and Central America.
In the future, further findings regarding the biotic and abiotic variables that constitute gene flow barriers for a bumble bee species must be compiled. These findings may serve as a valuable basis for a population delimitation strategy that does not require extensive genetic data and may be applied to genetic diversity indicators of the Kunming-Montreal Global Biodiversity Framework (CBD/COP/DEC/15/4, CBD/COP/DEC/15/5, CBD/COP/15/L.26).
The studies reviewed here demonstrate that the capacity for bumble bees to disperse is influenced not only by distance, but also by the suitability of their habitats and the landscape matrix. In light of ever progressing human land use change (Ramankutty and Foley, 1999; Ellis et al., 2013), this knowledge not only helps to inform the design of effective conservation measures but it also emphasizes the importance of monitoring genetic diversity in important pollinators such as bumble bees.
In order to implement genetic diversity indicators, included in the Kunming-Montreal Global Biodiversity Framework (CBD/COP/DEC/15/4, CBD/COP/DEC/15/5, CBD/COP/15/L.26), for the Bombus genus, estimations of effective populations sizes (Ne) are required. However, the availability of data that could facilitate the development of a strategy for delimiting populations for the estimation of Ne based on non-genetic parameters remains limited. Studies on genetic structuring of bumble bee populations have demonstrated that the high dispersal ability of bumble bees enables the maintenance of gene flow between large meta-populations across fragmented landscapes. If habitats unsuitable for hosting successful colonies exceed the dispersal abilities, populations become isolated resulting in the establishment of genetic population structuring over time. Isolation-by-landscape/environment approaches have proven useful in identifying these patterns of gene flow barriers. We suggest that in order to delimit bumble bee populations based on gene flow barriers, the impact of land cover, dispersal ability, resource availability and habitat requirements need to be evaluated further. This knowledge could be used to refine isolation-by-environment models, which have been shown to depict genetic structuring patterns even in the widespread and abundant B. terrestris (Glück et al., 2022). Such an approach could contribute to a population delimitation strategy that does not depend on comprehensive genetic data sets thereby offering a more cost- and labor-effective alternative that is more feasible in implementing in future monitoring schemes of genetic diversity.
LG: Conceptualization, Data curation, Formal Analysis, Investigation, Writing – original draft, Writing – review & editing. JD: Conceptualization, Resources, Supervision, Writing – review & editing, Funding acquisition. WS: Conceptualization, Resources, Supervision, Writing – review & editing, Methodology, Project administration, Validation, Visualization, Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The presented study is part of the MonViA project. The joint project Monitoring of biodiversity in agricultural landscapes (MonViA) has been funded by the German Federal Ministry of Food and Agriculture.
We thank the following colleagues for valuable input during the conceptual phase of this study: Felix Kirsch, Lasse Krüger, Leonie Lakemann, Brandon Seah, Johanna Stahl, Demetra Rakosy, Frank Sommerlandt, Toni Kasiske (all Thünen Institute of Biodiversity), Sophie Ogan (Coordination Unit Climate, Soil, Biodiversity, Thünen Institute) and Niels Hellwig (Anhalt University of Applied Sciences).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frbee.2025.1507903/full#supplementary-material
ENM, environmental niche modeling; IBD, isolation-by-distance; IBE, isolation-by-environment; IBL, isolation-by-landscape; IBR, isolation-by-resistance; LAI, Leaf Area Index; mtDNA, mitochondrial DNA; SNPs, single nucleotide polymorphisms.
Balkenhol N., Cushman S., Storfer A., Waits L. (2016). Landscape genetics: Concepts, methods, applications (Chichester: Wiley-Blackwell).
Bänsch S., Tscharntke T., Gabriel D., Westphal C. (2021). Crop pollination services: Complementary resource use by social vs solitary bees facing crops with contrasting flower supply. J. Appl. Ecol. 58, 476–485. doi: 10.1111/1365-2664.13777
Bartlett M., Hale R., Hale M. (2016). Habitat quality limits gene flow between populations of Bombus ruderatus in the South Island, New Zealand. Conserv. Genet. 17, 703–713. doi: 10.1007/s10592-016-0816-7
Blasco-Lavilla N., Ornosa C., Michez D., La Rúa P. (2019). Contrasting patterns of genetic and morphological diversity in the bumblebee Bombus lucorum (Hymenoptera: Apidae: Bombus) along a European gradient. J. Insect Conserv. 23, 933–943. doi: 10.1007/s10841-019-00178-2
Brown J. H. (1971). Mammals on mountaintops: Nonequilibrium insular biogeography. Am. Nat. 105, 467–478. doi: 10.1086/282738
Cameron S. A., Lozier J. D., Strange J. P., Koch J. B., Cordes N., Solter L. F., et al. (2011). Patterns of widespread decline in North American bumble bees. Proc. Natl. Acad. Sci. U.S.A. 108, 662–667. doi: 10.1073/pnas.1014743108
Carvell C., Bourke A. F. G., Dreier S., Freeman S. N., Hulmes S., Jordan W. C., et al. (2017). Bumblebee family lineage survival is enhanced in high-quality landscapes. Nature 543, 547–549. doi: 10.1038/nature21709
Carvell C., Jordan W. C., Bourke A. F. G., Pickles R., Redhead J. W., Heard M. S., et al. (2012). Molecular and spatial analyses reveal links between colony-specific foraging distance and landscape-level resource availability in two bumblebee species. Oikos 121, 734–742. doi: 10.1111/j.1600-0706.2011.19832.x
Chapman R. E., Bourke A. F. G. (2001). The influence of sociality on the conservation biology of social insects. Ecol. Lett. 4, 650–662. doi: 10.1046/j.1461-0248.2001.00253.x
Chapman R. E., Wang J., Bourke A. F. G. (2003). Genetic analysis of spatial foraging patterns and resource sharing in bumble bee pollinators. Mol. Ecol. 12, 2801–2808. doi: 10.1046/j.1365-294X.2003.01957.x
Charman T. G., Sears J., Green R. E., Bourke A. F. G. (2010). Conservation genetics, foraging distance and nest density of the scarce Great Yellow Bumblebee (Bombus distinguendus). Mol. Ecol. 19, 2661–2674. doi: 10.1111/j.1365-294X.2010.04697.x
Cheviron Z. A., Brumfield R. T. (2012). Genomic insights into adaptation to high-altitude environments. Heredity 108, 354–361. doi: 10.1038/hdy.2011.85
Christman M. E., Spears L. R., Burchfield E. K., Pearse W. D., Strange J. P., Ramirez R. A. (2024). Bumble bee responses to climate and landscapes: Investigating habitat associations and species assemblages across geographic regions in the United States of America. Glob. Change Bio. 30, e17380. doi: 10.1111/gcb.17380
Christmas M. J., Jones J. C., Olsson A., Wallerman O., Bunikis I., Kierczak M., et al. (2022). A genomic and morphometric analysis of alpine bumblebees: Ongoing reductions in tongue length but no clear genetic component. Mol. Ecol. 31, 1111–1127. doi: 10.1111/mec.16291
Clake D. J., Rogers S. M., Galpern P. (2022). Landscape complementation is a driver of bumble bee (Bombus sp.) abundance in the Canadian Rocky Mountains. Landsc. Ecol. 37, 713–728. doi: 10.1007/s10980-021-01389-2
Clake D. J., Rogers S. M., Galpern P. (2024). Cryptic genotypic and phenotypic diversity in parapatric bumble bee populations associated with minimum cold temperatures. Biodivers. Conserv. 33, 485–507. doi: 10.1007/s10531-023-02753-1
Colgan T. J., Arce A. N., Gill R. J., Ramos Rodrigues A., Kanteh A., Duncan E. J., et al. (2022). Genomic signatures of recent adaptation in a wild bumblebee. Mol. Biol. Evol. 39, msab366. doi: 10.1093/molbev/msab366
Darvill B., Ellis J. S., Lye G. C., Goulson D. (2006). Population structure and inbreeding in a rare and declining bumblebee, Bombus muscorum (Hymenoptera: Apidae). Mol. Ecol. 15, 601–611. doi: 10.1111/j.1365-294X.2006.02797.x
Darvill B., Knight M. E., Goulson D. (2004). Use of genetic markers to quantify bumblebee foraging range and nest density. Oikos 107, 471–478. doi: 10.1111/j.0030-1299.2004.13510.x
Darvill B., Lepais O., Woodall L. C., Goulson D. (2012). Triploid bumblebees indicate a direct cost of inbreeding in fragmented populations. Mol. Ecol. 21, 3988–3995. doi: 10.1111/j.1365-294X.2012.05679.x
Darvill B., O’Connor S., Lye G. C., Waters J., Lepais O., Goulson D. (2010). Cryptic differences in dispersal lead to differential sensitivity to habitat fragmentation in two bumblebee species. Mol. Ecol. 19, 53–63. doi: 10.1111/j.1365-294X.2009.04423.x
DeWoody J. A., Harder A. M., Mathur S., Willoughby J. R. (2021). The long-standing significance of genetic diversity in conservation. Mol. Ecol. 30, 4147–4154. doi: 10.1111/mec.16051
Diekötter T., Walther-Hellwig K., Conradi M., Suter M., Frankl R. (2006). Effects of landscape elements on the distribution of the rare bumblebee species Bombus muscorum in an agricultural landscape. Biodivers. Conserv. 15, 57–68. doi: 10.1007/s10531-004-2932-9
Dreier S., Redhead J. W., Warren I. A., Bourke A. F. G., Heard M. S., Jordan W. C., et al. (2014). Fine-scale spatial genetic structure of common and declining bumble bees across an agricultural landscape. Mol. Ecol. 23, 3384–3395. doi: 10.1111/mec.12823
Elith J., Franklin J. (2013). “Species distribution modeling,” in Encyclopedia of biodiversity. Ed. Levin S. A. (Academic Press, Amsterdam), 692–705.
Ellis E. C., Kaplan J. O., Fuller D. Q., Vavrus S., Klein Goldewijk K., Verburg P. H. (2013). Used planet: a global history. Proc. Natl. Acad. Sci. U.S.A. 110, 7978–7985. doi: 10.1073/pnas.1217241110
Ellis J. S., Knight M. E., Darvill B., Goulson D. (2006). Extremely low effective population sizes, genetic structuring and reduced genetic diversity in a threatened bumblebee species, Bombus sylvarum (Hymenoptera: Apidae). Mol. Ecol. 15, 4375–4386. doi: 10.1111/j.1365-294X.2006.03121.x
Elmer K. R. (2005). “Barrier loci and evolution,” in Encyclopedia of life sciences (Wiley, Chichester), 1–7.
Estoup A., Solignac M., Cornuet J. M., Goudet J., Scholl A. (1996). Genetic differentiation of continental and island populations of Bombus terrestris (Hymenoptera: Apidae) in Europe. Mol. Ecol. 5, 19–31. doi: 10.1111/j.1365-294X.1996.tb00288.x
Fragoso F. P., Jiang Q., Clayton M. K., Brunet J. (2021). Patch selection by bumble bees navigating discontinuous landscapes. Sci. Rep. 11, 8986. doi: 10.1038/s41598-021-88394-2
Francisco F. O., Santiago L. R., Mizusawa Y. M., Oldroyd B. P., Arias M. C. (2016). Genetic structure of island and mainland populations of a Neotropical bumble bee species. J. Insect Conserv. 20, 383–394. doi: 10.1007/s10841-016-9872-z
Françoso E., Zuntini A. R., Carnaval A. C., Arias M. C. (2016). Comparative phylogeography in the Atlantic forest and Brazilian savannas: pleistocene fluctuations and dispersal shape spatial patterns in two bumblebees. BMC Evol. Biol. 16, 267. doi: 10.1186/s12862-016-0803-0
Frankham R. (2005). Genetics and extinction. Biol. Conserv. 126, 131–140. doi: 10.1016/j.biocon.2005.05.002
Frankham R. (2019). “Conservation genetics,” in Encyclopedia of ecology. Ed. Fath B. D. (Elsevier B.V, Amsterdam, Oxford, Cambridge), 382–390.
Frankham R., Ballou J. D., Briscoe D. A. (2011). Introduction to conservation genetics (Cambridge: Cambridge University Press).
Glück M., Geue J. C., Thomassen H. A. (2022). Environmental differences explain subtle yet detectable genetic structure in a widespread pollinator. BMC Ecol. Evo. 22, 8. doi: 10.1186/s12862-022-01963-5
Goulson D. (1999). Foraging strategies of insects for gathering nectar and pollen, and implications for plant ecology and evolution. Perspect. Plant Ecol. Evol. Syst. 2, 185–209. doi: 10.1078/1433-8319-00070
Goulson D. (2010). Bumblebees: Behaviour, ecology, and conservation (Oxford: Oxford University Press).
Goulson D., Darvill B. (2004). Niche overlap and diet breadth in bumblebees; are rare species more specialized in their choice of flowers? Apidologie 35, 55–63. doi: 10.1051/apido:2003062
Goulson D., Hanley M. E., Darvill B., Ellis J. S. (2006). Biotope associations and the decline of bumblebees (Bombus spp.). J. Insect. Conserv. 10, 95–103. doi: 10.1007/s10841-006-6286-3
Goulson D., Lye G. C., Darvill B. (2008). Decline and conservation of bumble bees. Annu. Rev. Entomol. 53, 191–208. doi: 10.1146/annurev.ento.53.103106.093454
Grundel R., Jean R. P., Frohnapple K. J., Glowacki G. A., Scott P. E., Pavlovic N. B. (2010). Floral and nesting resources, habitat structure, and fire influence bee distribution across an open-forest gradient. Ecol. Appl. 20, 1678–1692. doi: 10.1890/08-1792.1
Han T., Park H., Park I. G., Yoon H. J., Kim K.-H., Lee H. J. (2014). Genetic structure of Korean populations of bumblebees Bombus ignitus (Hymenoptera: Apidae) as revealed by microsatellite markers. Entomol. Res. 44, 262–270. doi: 10.1111/1748-5967.12077
Hedrick P. W. (2005). A standardized genetic differentiation measure. Evol 59, 1633–1638. doi: 10.1111/j.0014-3820.2005.tb01814.x
Heinrich B. (2004). Bumblebee economics: With a new preface. Cambridge, Massachusetts, London, England: Harvard University Press.
Hemberger J., Bernauer O. M., Gaines-Day H. R., Gratton C. (2023). Landscape-scale floral resource discontinuity decreases bumble bee occurrence and alters community composition. Ecol. Appl. 33, e2907. doi: 10.1002/eap.2907
Heraghty S. D., Jackson J. M., Lozier J. D. (2023). Whole genome analyses reveal weak signatures of population structure and environmentally associated local adaptation in an important North American pollinator, the bumble bee Bombus vosnesenskii. Mol. Ecol. 32, 5479–5497. doi: 10.1111/mec.17125
Heraghty S. D., Rahman S. R., Jackson J. M., Lozier J. D. (2022). Whole genome sequencing reveals the structure of environment-associated divergence in a broadly distributed montane bumble bee, Bombus vancouverensis. Insect Syst. Divers. 6. doi: 10.1093/isd/ixac025
Hoban S., Archer F. I., Bertola L. D., Bragg J. G., Breed M. F., Bruford M. W., et al. (2022). Global genetic diversity status and trends: towards a suite of Essential Biodiversity Variables (EBVs) for genetic composition. Biol. Rev. 97, 1511–1538. doi: 10.1111/brv.12852
Hoban S., Bruford M., D’Urban Jackson J., Lopes-Fernandes M., Heuertz M., Hohenlohe P. A., et al. (2020). Genetic diversity targets and indicators in the CBD post-2020 Global Biodiversity Framework must be improved. Biol. Conserv. 248, 108654. doi: 10.1016/j.biocon.2020.108654
Hoban S., Da Silva J. M., Hughes A., Hunter M. E., Kalamujić Stroil B., Laikre L., et al. (2024). Too simple, too complex, or just right? Advantages, challenges, and guidance for indicators of genetic diversity. Bio. Sci. 74, 269–280. doi: 10.1093/biosci/biae006
Hoban S., Paz-Vinas I., Aitken S., Bertola L. D., Breed M. F., Bruford M. W., et al. (2021). Effective population size remains a suitable, pragmatic indicator of genetic diversity for all species, including forest trees. Biol. Conserv. 253, 108906. doi: 10.1016/j.biocon.2020.108906
Holzhauer S. I. J., Ekschmitt K., Sander A.-C., Dauber J., Wolters V. (2006). Effect of historic landscape change on the genetic structure of the bush-cricket Metrioptera roeseli. Landsc. Ecol. 21, 891–899. doi: 10.1007/s10980-005-0438-9
Hristov P., Shumkova R., Palova N., Neov B. (2020). Factors associated with honey bee colony losses: A mini-review. Vet. Sci. 7, 166. doi: 10.3390/vetsci7040166
Huml J. V., Ellis J. S., Rustage S., Brown M. J. F., Billington R., Knight M. E. (2023). The tragedy of the common? A comparative population genomic study of two bumblebee species. Insect Conserv. Divers. 16, 335–354. doi: 10.1111/icad.12626
Iles D. T., Williams N. M., Crone E. E. (2018). Source-sink dynamics of bumblebees in rapidly changing landscapes. J. Appl. Ecol. 55, 2802–2811. doi: 10.1111/1365-2664.13175
Iserbyt S., Vray S., Dendoncker N., Viart S., Rasmont P. (2015). High-resolution distribution of bumblebees (Bombus spp.) in a mountain area marked by agricultural decline. Ann. Soc Entomol. Fr. 51, 375–391. doi: 10.1080/00379271.2016.1141664
Jackson J. M., Pimsler M. L., Oyen K. J., Koch-Uhuad J. B., Herndon J. D., Strange J. P., et al. (2018). Distance, elevation and environment as drivers of diversity and divergence in bumble bees across latitude and altitude. Mol. Ecol. 27, 2926–2942. doi: 10.1111/mec.14735
Jeong S. Y., Yoon H. J., Park J. S., Kim M. J., Kim I. (2023). Population genetic characteristics of the bumble bee Bombus ardens (Hymenoptera: Apidae) in South Korea using novel microsatellite markers. J. Asia-Pac. Entomol. 26, 102071. doi: 10.1016/j.aspen.2023.102071
Jha S. (2015). Contemporary human-altered landscapes and oceanic barriers reduce bumble bee gene flow. Mol. Ecol. 24, 993–1006. doi: 10.1111/mec.13090
Jha S., Kremen C. (2013). Urban land use limits regional bumble bee gene flow. Mol. Ecol. 22, 2483–2495. doi: 10.1111/mec.12275
Kallioniemi E., Åström J., Rusch G. M., Dahle S., Åström S., Gjershaug J. O. (2017). Local resources, linear elements and mass-flowering crops determine bumblebee occurrences in moderately intensified farmlands. Agric. Ecosyst. Environ. 239, 90–100. doi: 10.1016/j.agee.2016.12.039
Kardos M., Armstrong E. E., Fitzpatrick S. W., Hauser S., Hedrick P. W., Miller J. M., et al. (2021). The crucial role of genome-wide genetic variation in conservation. Proc. Natl. Acad. Sci. U.S.A. 118, e2104642118. doi: 10.1073/pnas.2104642118
Keller L. (2002). Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241. doi: 10.1016/S0169-5347(02)02489-8
Kells A. R., Goulson D. (2003). Preferred nesting sites of bumblebee queens (Hymenoptera: Apidae) in agroecosystems in the UK. Biol. Conserv. 109, 165–174. doi: 10.1016/S0006-3207(02)00131-3
Kim M. J., Yoon H. J., Im H. H., Jeong H. U., Kim M. I., Kim S. R., et al. (2009). Mitochondrial DNA sequence variation of the bumblebee, Bombus ardens (Hymenoptera: Apidae). J. Asia. Pac. Entomol. 12, 133–139. doi: 10.1016/j.aspen.2009.02.003
Kimura M., Ohta T. (1971). Theoretical aspects of population genetics (Princeton, NJ: Princeton Univ. Press).
Kimura M., Weiss G. H. (1964). The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics 49, 561–576. doi: 10.1093/genetics/49.4.561
Knight M. E., Martin A. P., Bishop S., Osborne J. L., Hale R. J., Sanderson R. A., et al. (2005). An interspecific comparison of foraging range and nest density of four bumblebee (Bombus) species. Mol. Ecol. 14, 1811–1820. doi: 10.1111/j.1365-294X.2005.02540.x
Koch J. B. U., Vandame R., Mérida-Rivas J., Sagot P., Strange J. (2018). Quaternary climate instability is correlated with patterns of population genetic variability in Bombus huntii. Ecol. Evol. 8, 7849–7864. doi: 10.1002/ece3.4294
Kraus F. B., Wolf S., Moritz R. F. A. (2009). Male flight distance and population substructure in the bumblebee Bombus terrestris. J. Anim. Ecol. 78, 247–252. doi: 10.1111/j.1365-2656.2008.01479.x
Kyriazis C. C., Wayne R. K., Lohmueller K. E. (2021). Strongly deleterious mutations are a primary determinant of extinction risk due to inbreeding depression. Evol. Lett. 5, 33–47. doi: 10.1002/evl3.209
Laikre L. (2010). Genetic diversity is overlooked in international conservation policy implementation. Conserv. Genet. 11, 349–354. doi: 10.1007/s10592-009-0037-4
Laikre L., Hoban S., Bruford M. W., Segelbacher G., Allendorf F. W., Gajardo G., et al. (2020). Post-2020 goals overlook genetic diversity. Sci. Adv. 367, 1083–1085. doi: 10.1126/science.abb2748
Landaverde-González P., Baltz L. M., Escobedo-Kenefic N., Mérida J., Paxton R. J., Husemann M. (2018). Recent low levels of differentiation in the native Bombus ephippiatus (Hymenoptera: Apidae) along two Neotropical mountain-ranges in Guatemala. Biodivers. Conserv. 27, 3513–3531. doi: 10.1007/s10531-018-1612-0
Lecocq T., Vereecken N. J., Michez D., Dellicour S., Lhomme P., Valterová I., et al. (2013). Patterns of genetic and reproductive traits differentiation in Mainland vs. Corsican populations of bumblebees. PloS One 8, e65642. doi: 10.1371/journal.pone.0065642
Lepais O., Darvill B., O’Connor S., Osborne J. L., Sanderson R. A., Cussans J., et al. (2010). Estimation of bumblebee queen dispersal distances using sibship reconstruction method. Mol. Ecol. 19, 819–831. doi: 10.1111/j.1365-294X.2009.04500.x
Liu Y., Olsson A., Larva T., Cantwell-Jones A., Gill R. J., Cederberg B., et al. (2024). Genomic variation in montane bumblebees in Scandinavia: High levels of intraspecific diversity despite population vulnerability. Mol. Ecol. 33, e17251. doi: 10.1111/mec.17251
Lozier J. D., Parsons Z. M., Rachoki L., Jackson J. M., Pimsler M. L., Oyen K. J., et al. (2021). Divergence in body mass, wing loading, and population structure reveals species-specific and potentially adaptive trait variation across elevations in montane bumble bees. Insect Syst. Divers. 5. doi: 10.1093/isd/ixab012
Lozier J. D., Strange J. P., Koch J. B. U. (2013). Landscape heterogeneity predicts gene flow in a widespread polymorphic bumble bee, Bombus bifarius (Hymenoptera: Apidae). Conserv. Genet. 14, 1099–1110. doi: 10.1007/s10592-013-0498-3
Lozier J. D., Strange J. P., Stewart I. J., Cameron S. A. (2011). Patterns of range-wide genetic variation in six North American bumble bee (Apidae: Bombus) species. Mol. Ecol. 20, 4870–4888. doi: 10.1111/j.1365-294X.2011.05314.x
Lye G. C., Lepais O., Goulson D. (2011). Reconstructing demographic events from population genetic data: the introduction of bumblebees to New Zealand. Mol. Ecol. 20, 2888–2900. doi: 10.1111/j.1365-294X.2011.05139.x
Maebe K., Karise R., Meeus I., Mänd M., Smagghe G. (2019). Pattern of population structuring between Belgian and Estonian bumblebees. Sci. Rep. (Scientific Reports) 9, 9651. doi: 10.1038/s41598-019-46188-7
Maebe K., Meeus I., Ganne M., Meulemeester T., Biesmeijer K., Smagghe G. (2015). Microsatellite analysis of museum specimens reveals historical differences in genetic diversity between declining and more stable bombus species. PloS One 10, e0127870. doi: 10.1371/journal.pone.0127870
Manel S., Schwartz M. K., Luikart G., Taberlet P. (2003). Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 18, 189–197. doi: 10.1016/S0169-5347(03)00008-9
Mastretta-Yanes A., Ishihama F., Castillo L. (2023).Resources for the paper multinational evaluation of genetic diversity indicators for the Kunming-Montreal Global Biodiversity Monitoring framework: Guidance document. Available online at: https://github.com/AliciaMstt/GeneticIndicators (Accessed July 29, 2024).
Mayer C., Michez D., Chyzy A., Brédat E., Jacquemart A.-L. (2012). The abundance and pollen foraging behaviour of bumble bees in relation to population size of whortleberry (Vaccinium uliginosum). PloS One 7, e50353. doi: 10.1371/journal.pone.0050353
McRae B. H. (2006). Isolation by resistance. Evolution 60, 1551–1561. doi: 10.1111/j.0014-3820.2006.tb00500.x
Melero Y., Evans L. C., Kuussaari M., Schmucki R., Stefanescu C., Roy D. B., et al. (2022). Local adaptation to climate anomalies relates to species phylogeny. Commun. Biol. 5, 143. doi: 10.1038/s42003-022-03088-3
Mola J. M., Pearse I. S., Boone M. L., Evans E., Hepner M. J., Jean R. P., et al. (2024). Range-wide genetic analysis of an endangered bumble bee (Bombus affinis, Hymenoptera: Apidae) reveals population structure, isolation by distance, and low colony abundance. J. Insect Sci. 24. doi: 10.1093/jisesa/ieae041
Moreira A. S., Horgan F. G., Murray T. E., Kakouli-Duarte T. (2015). Population genetic structure of Bombus terrestris in Europe: Isolation and genetic differentiation of Irish and British populations. Mol. Ecol. 24, 3257–3268. doi: 10.1111/mec.13235
Morin P. A., Luikart G., Wayne R. K. (2004). SNPs in ecology, evolution and conservation. Trends Ecol. Evol. 19, 208–216. doi: 10.1016/j.tree.2004.01.009
Nicholson C. C., Hayes J.-M., Connolly S., Ricketts T. H. (2021). Corridors through time: Does resource continuity impact pollinator communities, populations, and individuals? Ecol. Appl. 31, e02260. doi: 10.1002/eap.2260
Nicholson C. C., Knapp J., Kiljanek T., Albrecht M., Chauzat M.-P., Costa C., et al. (2024). Pesticide use negatively affects bumble bees across European landscapes. Nature 628, 355–358. doi: 10.1038/s41586-023-06773-3
Packer L., Owen R. (2001). Population genetic aspects of pollinator decline. Curr. Landscape Ecol. Rep. 5. doi: 10.5751/ES-00267-050104
Pirounakis K., Koulianos S., Schmid-Hempel P. (1998). Genetic variation among European populations of Bombus pascuorum (Hymenoptera: Apidae) from mitochondrial DNA sequence data. Eur. J. Entomol. 95, 27–33.
Potts S. G., Biesmeijer J. C., Kremen C., Neumann P., Schweiger O., Kunin W. E. (2010). Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25, 345–353. doi: 10.1016/j.tree.2010.01.007
Pugesek G., Crone E. E. (2021). Contrasting effects of land cover on nesting habitat use and reproductive output for bumble bees. Ecosphere 12, e03642. doi: 10.1002/ecs2.3642
Quinlan G. M., Milbrath M. O., Otto C. R. V., Isaacs R. (2021). Honey bee (Apis mellifera) colonies benefit from grassland/pasture while bumble bee (Bombus impatiens) colonies in the same landscapes benefit from non-corn/soybean cropland. PloS One 16, e0257701. doi: 10.1371/journal.pone.0257701
Quinn J. F., Harrison S. P. (1988). Effects of habitat fragmentation and isolation on species richness: evidence from biogeographic patterns. Oecologia 75, 132–140. doi: 10.1007/BF00378826
Ramankutty N., Foley J. A. (1999). Estimating historical changes in global land cover: Croplands from 1700 to 1992. Glob. Biogeochem. Cycles 13, 997–1027. doi: 10.1029/1999GB900046
Rao S., Stephen W. P. (2009). Bumble bee pollinators in red clover seed production. Crop Sci. 49, 2207–2214. doi: 10.2135/cropsci2009.01.0003
Redhead J. W., Dreier S., Bourke A. F. G., Heard M. S., Jordan W. C., Sumner S., et al. (2016). Effects of habitat composition and landscape structure on worker foraging distances of five bumble bee species. Ecol. Appl. 26, 726–739. doi: 10.1890/15-0546
Richards K. W. (1978). Nest site selection by bumble bees (Hymenoptera: Apidae) in southern Alberta. Can. Entomol. 110, 301–318. doi: 10.4039/Ent110301-3
Robinson J. L. (2024). Project-specific bumble bee habitat quality assessment. MethodsX 12, 102571. doi: 10.1016/j.mex.2024.102571
Rollin O., Bretagnolle V., Decourtye A., Aptel J., Michel N., Vaissière B. E., et al. (2013). Differences of floral resource use between honey bees and wild bees in an intensive farming system. Agric. Ecosyst. Environ. 179, 78–86. doi: 10.1016/j.agee.2013.07.007
Santos Júnior J. E., Silveira F. A., Oliveira U., Dias C. A. R., Santos F. R. (2019). Conservation and historical distribution of two bumblebee species from the Atlantic Forest. Syst. Biodivers. 17, 22–38. doi: 10.1080/14772000.2018.1530313
Schenau E., Jha S. (2017). High levels of male diploidy but low levels of genetic structure characterize Bombus vosnesenskii populations across the Western US. Conserv. Genet. 18, 597–605. doi: 10.1007/s10592-016-0900-z
Skey E. D., Ottewell K. M., Spencer P. B., Shaw R. E. (2023). Empirical landscape genetic comparison of single nucleotide polymorphisms and microsatellites in three arid-zone mammals with high dispersal capacity. Ecol. Evol. 13, e10037. doi: 10.1002/ece3.10037
Soroye P., Newbold T., Kerr J. (2020). Climate change contributes to widespread declines among bumble bees across continents. Science 367, 685–688. doi: 10.1126/science.aax8591
Spear S. F., Balkenhol N., Fortin M.-J., McRae B. H., Scribner K. (2010). Use of resistance surfaces for landscape genetic studies: considerations for parameterization and analysis. Mol. Ecol. 19, 3576–3591. doi: 10.1111/j.1365-294X.2010.04657.x
Spielman D., Brook B. W., Frankham R. (2004). Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. U. S. A. 101, 15261–15264. doi: 10.1073/pnas.0403809101
Storfer A., Murphy M. A., Spear S. F., Holderegger R., Waits L. P. (2010). Landscape genetics: where are we now? Mol. Ecol. 19, 3496–3514. doi: 10.1111/j.1365-294X.2010.04691.x
Suzuki Y., Kawaguchi L. G., Toquenaga Y. (2007). Estimating nest locations of bumblebee Bombus ardens from flower quality and distribution. Ecol Res. 22, 220–227. doi: 10.1007/s11284-006-0010-3
Svensson B., Lagerlöf J., G. Svensson B. (2000). Habitat preferences of nest-seeking bumble bees (Hymenoptera: Apidae) in an agricultural landscape. Agric. Ecosyst. Environ. 77, 247–255. doi: 10.1016/S0167-8809(99)00106-1
Szymanski J., Smith T., Horton A., Parkin M., Ragan L., Masson G., et al. (2016). Rusty Patched Bumble Bee (Bombus affinis) species status assessment final report (Bloomington, MN: U.S. Fish and Wildlife Service).
Teixeira J. C., Huber C. D. (2021). The inflated significance of neutral genetic diversity in conservation genetics. Proc. Natl. Acad. Sci. U.S.A. 118, e2015096118. doi: 10.1073/pnas.2015096118
Theodorou P., Radzevičiūtė R., Kahnt B., Soro A., Grosse I., Paxton R. J. (2018). Genome-wide single nucleotide polymorphism scan suggests adaptation to urbanization in an important pollinator, the red-tailed bumblebee (Bombus lapidarius L.). Proc. Biol. Sci. 285, 20172806. doi: 10.1098/rspb.2017.2806
Tscharntke T., Steffan-Dewenter I., Kruess A., Thies C. (2002). Contribution of small habitat fragments to conservation of insect communities of grassland-cropland landscapes. Ecol. Appl. 12, 354. doi: 10.2307/3060947
Tscharntke T., Tylianakis J. M., Rand T. A., Didham R. K., Fahrig L., Batáry P., et al. (2012). Landscape moderation of biodiversity patterns and processes - eight hypotheses. Biol. Rev. Camb. Philos. Soc. 87, 661–685. doi: 10.1111/j.1469-185X.2011.00216.x
van Oosterhout C. (2020). Mutation load is the spectre of species conservation. Nat. Ecol. Evol. 4, 1004–1006. doi: 10.1038/s41559-020-1204-8
Walther-Hellwig K., Frankl R. (2000). Foraging habitats and foraging distances of bumblebees, Bombus spp. (Hym., Apidae), in an agricultural landscape. J. Appl. Entomol. 124, 299–306. doi: 10.1046/j.1439-0418.2000.00484.x
Wang I. J., Bradburd G. S. (2014). Isolation by environment. Mol. Ecol. 23, 5649–5662. doi: 10.1111/mec.12938
Wright S. (1931). Evolution in Mendelian populations. Genetics 16, 97–159. doi: 10.1093/genetics/16.2.97
Wright S. (1933). Inbreeding and homozygosis. Proc. Natl. Acad. Sci. U.S.A. 19, 411–420. doi: 10.1073/pnas.19.4.411
Zayed A. (2004). Effective population size in Hymenoptera with complementary sex determination. Heredity 93, 627–630. doi: 10.1038/sj.hdy.6800588
Zayed A., Packer L. (2005). Complementary sex determination substantially increases extinction proneness of haplodiploid populations. Proc. Natl. Acad. Sci. U.S.A. 102, 10742–10746. doi: 10.1073/pnas.0502271102
Keywords: Bombus, gene flow, genetic diversity, isolation-by-landscape, landscape genetics, monitoring, pollinators, population ecology
Citation: Gornall L, Dauber J and Sickel W (2025) Population delimitation in bumble bees - strategies and research gaps. Front. Bee Sci. 3:1507903. doi: 10.3389/frbee.2025.1507903
Received: 08 October 2024; Accepted: 19 February 2025;
Published: 07 March 2025.
Edited by:
Helen Wallace, Queensland University of Technology, AustraliaReviewed by:
Jonathan Shaw Ellis, University of Plymouth, United KingdomCopyright © 2025 Gornall, Dauber and Sickel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wiebke Sickel, d2llYmtlLnNpY2tlbEB0aHVlbmVuLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.