
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bee Sci., 27 September 2023
Sec. Bee Physiology
Volume 1 - 2023 | https://doi.org/10.3389/frbee.2023.1251714
The European honey bee is a model organism for investigating the molecular and neural bases of the brain underlying social behaviors. Mushroom bodies (MBs) are a higher-order center of memory, learning, and sensory integration in insect brains, and honey bee MBs are a model to study adult neuronal plasticity. In the honey bee, MBs comprise three Class I Kenyon cell (KC) subtypes: large-, middle-, and small-type KCs, which are distinguished based on the size and localization of their somata, and gene expression profiles. One of the unique characteristics of honey bee MBs is that genes for ecdysone signaling are expressed in a spatially and temporarily regulated manner in the adult brain, suggesting that they play a role in the functional specialization of each KC subtype and behavioral control. A recent study reported that the transcription factor Mblk-1/E93, which functions downstream of ecdysone signaling during metamorphosis, targets genes involved in synaptic plasticity underlying memory and learning ability in the adult honey bee brain. On the other hand, the ecdysone receptor (EcR), which is expressed in small-type KCs in the MBs, was reported to target genes involved in lipid metabolism in the brain during foraging flight. The target genes for Mblk-1 and EcR in the adult brains differed from those during metamorphosis, implying that the reiterative use of some transcription factors involved in ecdysone signaling, such as EcR and Mblk-1, has contributed to the acquisition of novel MB functions in Aculeata species, including the honey bee.
The European honey bee (Apis mellifera L.) is a model organism for investigating the molecular and neural bases underlying social behaviors and advanced brain functions (Kamikouchi et al., 1998; Kucharski et al., 1998; Toma et al., 2000; Ben-Shahar et al., 2002; Whitfield et al., 2009; Suenami et al., 2018). Mushroom bodies (MBs) are a higher-order center of memory, learning, and sensory integration of the insect brain (Hammer, 1993; Heisenberg, 1998; Menzel and Giurfa, 2001), and honey bee MBs have been used as a model to study adult neuronal plasticity (Withers et al., 1993; Fahrbach et al., 1998; Groh and Rössler, 2020). In the honey bee, MBs are composed of four subtypes of interneurons, termed Kenyon cells (KCs): Class I (large-, middle-, and small-type) KCs and Class II KCs, which are distinguished based on their size and localization of their somata in the MBs, and gene expression profiles (Figure 1) (Mobbs, 1982; Strausfeld, 2002; Fahrbach, 2006; Kaneko et al., 2013; Kaneko et al., 2016; Suenami et al., 2018). Large-type KCs have been suggested to function in learning and memory based on their gene expression profiles (Kamikouchi et al., 1998; Kamikouchi et al., 2000; Takeuchi et al., 2002; Uno et al., 2012; Suenami et al., 2018), whereas small- and a part of middle-type KCs have been suggested to be related to information processing during foraging behavior based on neural activity mapping using immediate early genes (IEGs) (Kiya et al., 2007; Ugajin et al., 2013).
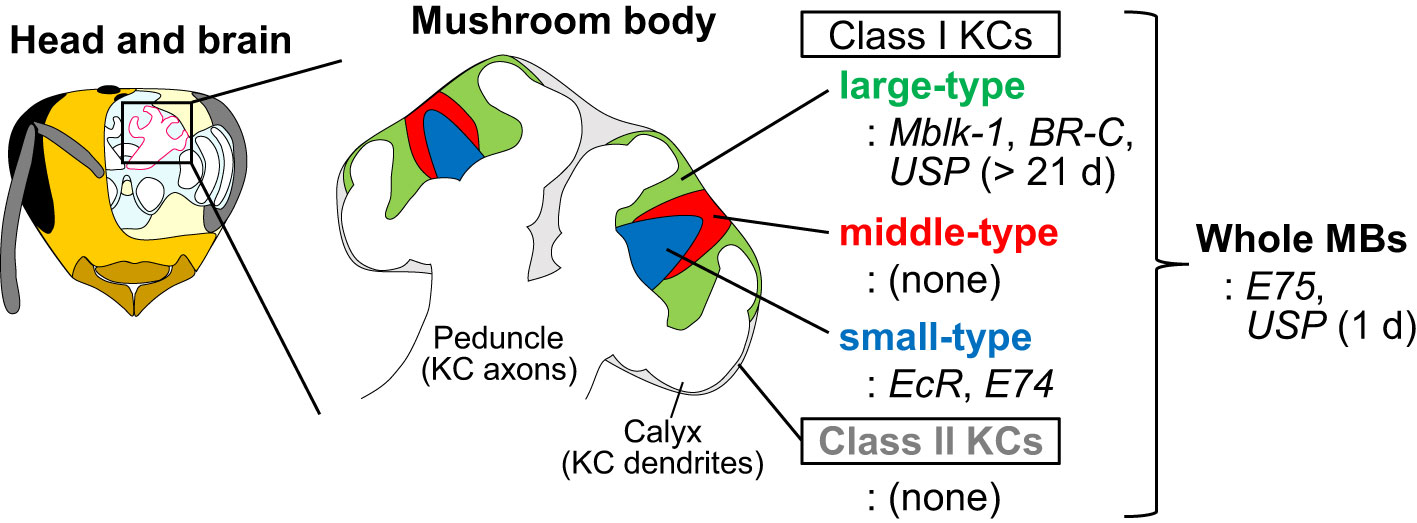
Figure 1 Schematic drawings of the head, brain, MBs, and four KC subtypes of the honey bee. Expression patterns of some MB-preferential ecdysone signaling-related genes in the MBs are shown. The figures are adapted from Matsumura et al. (2022) (Matsumura et al., 2022) with some modifications.
One of the prominent molecular characteristics of honey bee MBs is that the genes involved in ecdysone signaling are expressed preferentially in the MBs of the adult honey bee brain (Truman et al., 1994; Baehrecke, 1996; Strausfeld, 2002; Kayukawa et al., 2017; Liu et al., 2018; Suenami et al., 2018). In holometabolous insects, metamorphosis is coordinated by two insect hormones, 20-hydroxyecdysone (20E) and juvenile hormone (JH) (Baehrecke and Thummel, 1995; Buszczak and Segraves, 2000; Lee et al., 2000; Lee and Baehrecke, 2001; Truman, 2019). Although 20E induces larval-larval molting in the presence of JH, it induces larval-pupal and pulpal-adult molts in the absence of JH. 20E binds to ecdysone receptor (EcR) and its co-factor, ultraspiracle (USP) and induces a group of genes encoding transcription factors that function downstream of EcR during metamorphosis: e.g., Mushroom body large-type Kenyon cell-preferential protein-1 (Mblk-1)/E93, BR-C, E74, and E75 (Takeuchi et al., 2001; Paul et al., 2005; Paul et al., 2006; Takeuchi et al., 2007). In the honey bee, the hemolymph titer of JH increases with the division of labor of workers from nurse bees to foragers and influences the timing of the behavioral transition of workers (Robinson et al., 1991; Jassim et al., 2000). In addition, both 20E and JH modulate the expression of genes involved in ecdysone signaling in the MBs of newly emerged workers (Velarde et al., 2009). On the other hand, ecdysone signaling regulates behavior and brain function even in adult Drosophila. For example, 20E and EcR are required for the formation of long-term courtship memory (Ishimoto et al., 2009). Overexpression or knockdown of E75 in clock neurons disrupts circadian rhythms (Kumar et al., 2014). Therefore, the reiterative use of ecdysone signaling in the adult brain is not specific to honey bees. However, honey bee MBs are characteristic in that the genes for ecdysone signaling-related transcription factors are expressed in both temporally and spatially regulated (that is, KC-subtype-preferential) manner (Figure 1). Thus, it is plausible that ecdysone signaling plays an important role in the functional specialization of each KC subtype and behavioral control.
Recent studies have reported that both Mblk-1 and EcR target unique genes in the adult honey bee brain compared to those during metamorphosis (Matsumura et al., 2022; Iino et al., 2023). Matsumura et al. (2022) reported that, Mblk-1 targets two genes involved in synaptic plasticity, which underlies learning and memory abilities in the animal brain, such as Ca2+/calmodulin-dependent protein kinase II (CaMKII), besides many neural-related genes in the worker honey bee brain (Matsumura et al., 2022). On the other hand, Iino et al. (2023) reported that some EcR target genes are upregulated in the forager brain, some of which are implicated in the repression of metabolic processes (Iino et al., 2023). These findings suggest that ecdysone signaling-related transcription factors are reiteratively used to regulate brain function in the adult honey bee brain. This minireview summarizes the recent findings regarding the functions of ecdysone signaling-related transcription factors in the adult honey bee brain and discusses their possible roles in acquiring unique KC subtype functions during the evolution of nidifying Aculeata species including honey bees.
Among the genes for ecdysone-related transcription factors expressed in the MBs of the adult honey bee, Mblk-1 was originally identified as a gene that is expressed preferentially in the large-type KCs in the worker honey bee brain (Figure 1) (Takeuchi et al., 2001). Subsequent studies have shown that Mblk-1 encodes a transcription factor (Park et al., 2002; Park et al., 2003) and is constitutively and preferentially expressed in the large-type KCs in the worker brain from the pupal to adult stages (Suenami et al., 2016; Kumagai et al., 2020). Recently, Matsumura et al. (2022) conducted chromatin immunoprecipitaion-sequencing (ChIP-seq) analysis to search for candidate target genes for Mblk-1 using the MBs of adult worker brains (Matsumura et al., 2022). Among the many neural-related genes identified as Mblk-1 target gene candidates, two synaptic plasticity-related genes, CaMKII and pumilio homolog 2 (pum), were confirmed to be expressed preferentially in large-type KCs, just like Mblk-1, suggesting that Mblk-1 upregulates their expression levels in large-type KCs (Figure 2A) (Kamikouchi et al., 2000; Pasch et al., 2011; Kaneko et al., 2013). Mblk-1 binds to Mblk-1-binding elements located in introns of these synaptic plasticity-related target genes. CaMKII functions in synaptic plasticity, which is the molecular basis for learning and memory ability (Shonesy et al., 2014), and is required for long-term memory formation, even in honey bees (Matsumoto et al., 2014; Scholl et al., 2015). pum is involved in learning and memory via synaptic plasticity in Drosophila (Dubnau et al., 2003). Therefore, it is plausible that Mblk-1 functions in learning and memory via the transactivation of these synaptic plasticity-related genes in large-type KCs in the adult honey bee brain (Figure 2A).
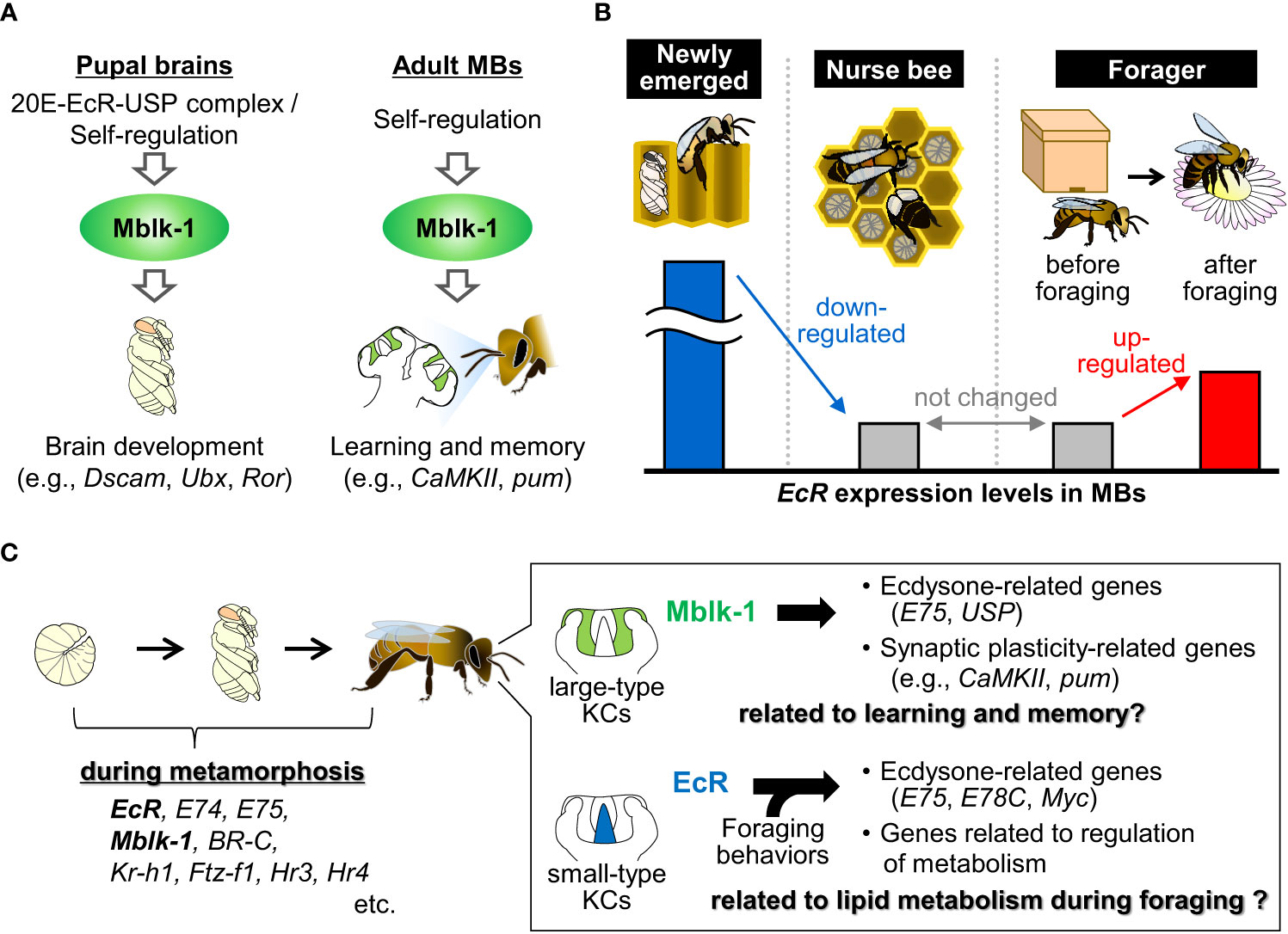
Figure 2 Summary of the expression and functions of Mblk-1 and EcR in the adult honey bee brain. (A) Proposed model for the induction and target genes/functions of Mblk-1 in the pupal brains (left) and adult MBs (right) of the honey bee, respectively. (B) EcR expression levels in the MBs of newly emerged workers, nurse bees and foragers before and after foraging. (C) Genes for ecdysone signaling-related transcription factors during metamorphosis (left), and proposed target genes and functions of Mblk-1 and EcR in the adult honey bee brain (right).
In addition to neural-related genes, two ecdysone signaling-related genes, USP and E75, were identified as Mblk-1 candidate target genes in the brains of adult workers (Matsumura et al., 2022). However, the expression patterns of USP and E75 are different from that of Mblk-1; USP is expressed in the entire MBs in 1-day-old workers but it is expressed more strongly in large-type KCs in foragers than in 1-day-old workers (Velarde et al., 2009), and E75 is expressed in the entire MBs in the brain of workers (Figure 1) (Paul et al., 2006). Therefore, it is plausible that Mblk-1 contributes to the large-type KC preferential expression of USP in foragers, at least partly, and that transcription factors other than Mblk-1 also regulate its expression in the entire MBs in 1-day-old workers, as well as the expression of E75 in adult honey bee workers. Considering that the E75 and USP expression levels in the MBs of newly emerged workers are affected by exogenous JH treatment (Velarde et al., 2009), and considering that the Mblk-1 expression levels in the brain are higher in foragers than that in nurse bees (Liu et al., 2022), it might be that the expression levels of Mblk-1, and thus its downstream genes, are also regulated by JH signaling.
Matsumura et al. (2022) also compared profiles of Mblk-1 target gene candidates in pupal and adult worker brains (Matsumura et al., 2022). Approximately half of pupal Mblk-1 target gene candidates, including some developmental genes, such as Dscam, Ubx, and Ror are specifically detected in pupal brains but not in adult worker brains. These findings suggest that Mblk-1 alters its target genes in the brain between the pupal and adult stages (Figure 2A).
Liu et al. (2022) recently reported that Mblk-1 expressed in the worker brain is related to sugar responsiveness in foragers via the regulation of the expression of a gustatory receptor gene (AmGR1) (Liu et al., 2022). However, it seems unclear at present whether Mblk-1 expression in large-type KCs is directly related to the regulation of sugar responsiveness and whether Mblk-1 directly targets AmGR1. For example, the expression of Mblk-1 in brain regions apart from MBs, such as suboesophageal zone, a primary gustatory center, might underlie this functionality.
In adult honey bees, EcR is expressed in the ovaries of queens, a reproductive caste, and involved in oogenesis but not in the abdomens of workers, a non-reproductive caste (Takeuchi et al., 2007; Uno et al., 2012). However, EcR is also expressed in the adult brains, especially strongly in small-type KCs in the MBs, of both castes (Takeuchi et al., 2007), suggesting that EcR is related not only to oogenesis but also to brain functions in adult honey bees. Velarde et al. (2009) reported that the EcR expression levels in MBs are higher in newly emerged workers than in foragers (Figure 2B) (Velarde et al., 2009). Notably, the increase in the volume of MB calyces, where KCs extend their dendrites, takes place in newly emerged workers in an experience-independent manner (Fahrbach et al., 1998), suggesting that the high EcR expression levels might be related to this enlargement. Singe et al. (2018) and Iino et al. (2022) reported that EcR is induced in the forager brain by foraging flight (Figure 2B) (Singh et al., 2018; Iino et al., 2020). Therefore, it is plausible that the elevated EcR expression levels are also associated with the foraging experience-dependent increase in the volume of MB calyces (Withers et al., 1995; Farris et al., 2001). So far, small-type KCs have been suggested to function in sensory information processing and/or cognitive processes associated with foraging such as navigation, orientation, and learning and memory because kakusei, HR38, and Egr, all of which are IEGs (Yamazaki et al., 2006; Kiya et al., 2007; Fujita et al., 2013; Ugajin et al., 2013), are induced in small-type KCs during foraging flight in worker honey bees (Yamazaki et al., 2006; Kiya et al., 2007; Kaneko et al., 2013; Ugajin et al., 2013). Therefore, it is possible that EcR also functions in the information processing and/or the regulation of physiological states during foraging in adult honey bee brains.
Recently, Iino et al. (2023) conducted ChIP-seq analysis using the whole brains of nurse bees and foragers to investigate the function of EcR in the adult honey bee brain (Iino et al., 2023). Most EcR target gene candidates are common to both nurse bees and foragers and include genes for canonical ecdysone signaling, such as Hr4, E75, and Kr-h1, whereas several genes known to be targets of EcR, including Mblk-1, are not detected. GO enrichment analysis of EcR target gene candidates also detected functions related to canonical ecdysone signaling, such as post-embryonic development, oogenesis, and neurogenesis. However, genes related to the regulation of metabolism were also enriched, suggesting a novel function of EcR in adult honey bee brains. Additionally, RNA-seq analysis revealed that several EcR target gene candidates detected in the brains of foragers, including those for ecdysone signaling-related transcription factors, such as E75, E78C, and Myc, are upregulated in the brains of foragers after foraging compared with those of nurse bees and foragers before foraging (Iino et al., 2023). Some of these genes are involved in the regulation of lipid levels and metabolism in Drosophila and mice (Inagaki et al., 2009; Su and Peng, 2020; Praggastis et al., 2021). Since foragers require a large amount of energy for the vibration of flight muscles and brain neural activity during foraging flight (Stabentheiner and Kovac, 2016; Rittschof et al., 2018), by regulating the transcription of the identified target metabolism genes, honey bee EcR may reduce energy consumption in the brain and/or generate the energy needed for foraging behaviors through lipolysis. Since EcR expression is also detected in brain regions other than MBs (Iino et al., 2020), the function of EcR in the regulation of metabolism suggested by Iino et al. (2023) may be related not only to small-type KCs but also to the other brain regions (Iino et al., 2023). It is noteworthy that E75 was detected as a target gene candidate not only for Mblk-1 but also for EcR, suggesting that E75 expression is differentially regulated in each KC subtype in the adult honey bee brain (Matsumura et al., 2022; Iino et al., 2023). This may partly explain the previous finding that E75 is expressed in all KC subtypes (Paul et al., 2006). Finally, the target genes and thus the functions of EcR induced in the MBs of newly emerged workers in a foraging experience-independent manner remain unknown.
In the adult honey bee MB, the expression patterns of EcR and its conventional downstream gene Mblk-1 are different. EcR is expressed preferentially in small-type KCs, whereas Mblk-1 is expressed preferentially in large-type KCs (Figure 1). Matsumura et al. (2022) reported that Mblk-1-binding regions containing GA-rich sequences are located upstream of Mblk-1 and that Mblk-1 can transactivate a reporter gene through one of these Mblk-1 binding regions in vitro (Matsumura et al., 2022). In contrast, Mblk-1 has not been identified as a candidate EcR target gene in the adult honey bee brain (Iino et al., 2023). Therefore, the constitutive expression of Mblk-1 in large-type KCs may be accounted for by its self-regulation, independent of ecdysone and EcR, although the initial large-type KC preferential expression of Mblk-1 may be induced by ecdysone and EcR at the pupal stage (Figure 2A).
Recent studies shed light on the possible evolution of KC subtypes in the MBs in Hymenoptera. The number of KC subtypes has been proposed to have increased from one in solitary sawflies, basal hymenopteran species (Symphyta), to two in parasitic wasps, and then to three in nidifying Aculeata species associated with behavioral evolution in Hymenoptera (Oya et al., 2017). Recently, Kuwabara et al. (2023) conducted a comparative transcriptome analysis to propose a model for evolutionary dynamics, in which the number of subtypes increased from one ancestral, multifunctional KC type through functional segregation and divergence (specialization) (Kuwabara et al., 2023). They also suggested that CaMKII expression and its functions in long-term memory are located in the entire MBs in the turnip sawfly, which are composed of a single ancestral-like KC subtype, whereas they are localized to large-type KCs in honey bees. Considering that Mblk-1 expression is restricted to some KC populations in the MBs of the brains of two ant species, another Aculeate species (Sheng et al., 2020; Li et al., 2022), whereas it is not detected in the brain of turnip sawflies (Matsumura et al., 2022), large-type KC preferential Mblk-1 expression in adult brains has likely been acquired in Aculeata (Matsumura et al., 2022). Therefore, Mblk-1 may have contributed, at least in part, to the functional specialization of large-type KCs, such as advanced learning and memory abilities, by elevating CaMKII expression and upregulating other synaptic plasticity-related genes in Aculeata MBs (Figure 2C). Regarding EcR, small-type KC preferential expression and upregulation after foraging flight in the adult brain are also observed in the bumble bee Bombus ignitus, a close honey bee relative (Iino et al., 2020), suggesting a conserved function of EcR in the adult brains at least in Apidae. EcR may have contributed to the functional segregation or specialization of small-type KCs by regulating downstream gene expression, although the EcR expression patterns in the adult brains of sawflies and other Aculeata species need to be clarified in the future.
Finally, it is necessary to substantiate whether the genes identified as target gene candidates for Mblk-1 and EcR (Matsumura et al., 2022; Iino et al., 2023) are up- or down-regulated by these transcription factors, for example, by examining changes in expression levels of these target gene candidates in the adult worker bee brain in which Mblk-1/EcR is knocked down in the future research.
In this Minireview, we mainly focused on the possible functions of Mblk-1 and EcR in worker honey bee brains. Some questions have arisen regarding the mechanisms and functions of the induction of these ecdysone signaling-related transcription factors in the brains of adult honey bees and their relationship to the acquisition of behavioral traits characteristic of Aculeata species, including honey bees.
The first is the relationship between the transcriptional regulation by these ecdysone-related transcription factors and the hemolymph 20E titer in adult worker honey bees. Considering that the hemolymph ecdysteroid titer in workers transiently rises on 3-day after the emergence (Hartfelder et al., 2002; Amdam et al., 2004) and the EcR expression levels are affected by exogenous 20E treatment in newly emerged workers (Velarde et al., 2009), EcR might be activated directly by hemolymph 20E at least in newly emerged workers. However, Yamazaki et al. (2011) suggested that ecdysone is synthesized in the brains of adult workers (Yamazaki et al., 2011). Therefore, EcR might also be activated directly by 20E synthesized in the worker brain. In contrast, Matsumura et al. (2022) proposed that Mblk-1 may be induced autonomously by self-regulation independent of ecdysone (Matsumura et al., 2022). This could be testified by examining whether the knockdown of genes involved in the ecdysteroid synthesis alters the Mblk-1/E93 expression levels.
Then, why are ecdysone signaling-related transcription factors reiteratively used in the worker honey bee brain, among many other transcription factors? Mblk-1/E93 acts as an “adult specifier” and BR-C as a “pupal specifier” in the holometabolous insect (Zhou and Riddiford, 2002; Konopova and Jindra, 2008; Ureña et al., 2014). EcR, Mblk-1, and BR-C, all of which are key factors in metamorphosis, are preferentially expressed in honey bee MBs. These ecdysone-related transcription factors regulate the expression of many downstream genes in a tissue-dependent manner during metamorphosis (Lee et al., 2000; Liu et al., 2015; Uyehara and McKay, 2019). Therefore, the reiterative use of ecdysone-related transcription factors might have caused drastic changes in gene expression and contributed to conferring the unique characteristic to each KC subtype during the evolution of the hymenopteran insect.
Finally, what are the possible roles of ecdysone signaling in the acquisition of behavioral traits of honey bees? Both learning and memory abilities, in which Mblk-1 is proposed to be involved, and the metabolic regulation in brains, in which EcR is proposed to be involved, seem to be behavioral and physiological traits especially important for nidifying Aculeate species, including honey bees (Figure 2C). After foraging, adult Aculeata species need to return to their nest to feed their brood (Capaldi et al., 2000; Goulson and Stout, 2001; Gathmann and Tscharntke, 2002; Klein et al., 2004; Woodgate et al., 2016); therefore, they are assumed to have excellent foraging ability and highly advanced learning and memory abilities (Pyke, 1978; Dyer, 1996; Menzel and Giurfa, 2001; Hendricsma et al., 2019). The reiterative use of ecdysone-related transcription factors may have contributed to increasing the functional complexities of MBs by conferring unique gene expression profiles to each KC subtype, leading to the acquisition of behavioral and physiological regulatory mechanisms specific to Aculeata, including honey bees (Figure 2C). It seems that the analysis of the functional diversification of ecdysone-related transcription factors in hymenopteran species remains to be an attractive subject in the search for molecular mechanisms underlying the honey bee behaviors and the driving force behind their evolution. Several other transcription factors are involved in ecdysone signaling that are reiteratively used in the adult honey bee brain besides Mblk-1 and EcR. Therefore, by comparing the expression patterns of these transcription factors and identifying their target genes, it may be possible to test the hypothesis that ecdysone signaling reiteratively used in the adult honey bee brain may be involved in behavioral regulation and the acquisition of brain function specific to Aculeata, including honey bees.
All authors contributed to the article and approved the submitted version. YM and HK created the figures.
This work was supported by JSPS KAKENHI (Grant-in-Aid for Challenging Exploratory Research) Grant Number 23K18148.
The authors are grateful to Dr. Shiori Iino for her critical reading of the manuscript. The authors also thank the reviewers for their helpful comments that improved the manuscript. Finally, the authors are grateful to Dr. Susan Fahrbach, Editor of the Bee Physiology Section of Frontiers in Bee Science, for her kind encouragement to submit our manuscript to this journal's Editor's Showcase.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amdam G. V., Hartfelder K., Norberg K., Hagen A., Omholt S. W. (2004). Altered physiology in worker honey bees (Hymenoptera : Apidae) ingested with the mite Varroa destructor (Acari : Varroidae): a factor in colony loss during overwintering? J. Econ Entomol 97, 741–747. doi: 10.1603/0022-0493(2004)097[0741:APIWHB]2.0.CO;2
Baehrecke E. H. (1996). Ecdysone signaling cascade and regulation of Drosophila metamorphosis. Arch. Insect Biochem. Physiol. 33, 231–244. doi: 10.1002/(SICI)1520-6327(1996)33:3/4<231::AID-ARCH5>3.0.CO;2-V
Baehrecke E. H., Thummel C. S. (1995). The Drosophila E93 gene from the 93F early puff displays stage- and tissue-specific regulation by 20-hydroxyecdysone. Dev. Biol. 171, 85–97. doi: 10.1006/dbio.1995.1262
Ben-Shahar Y., Robichon A., Sokolowski M. B., Robinson G. E. (2002). Influence of gene action across different time scales on behavior. Science 296, 741–744. doi: 10.1126/science.1069911
Buszczak M., Segraves W. A. (2000). Insect metamorphosis: out with the old, in with the new. Curr. Biol. 10, R830–R833. doi: 10.1016/S0960-9822(00)00792-2
Capaldi E. A., Smith A. D., Osborne J. L., Fahrbach S. E., Farris S. M., Reynolds D. R., et al. (2000). Ontogeny of orientation flight in the honeybee revealed by harmonic radar. Nature 403, 537–540. doi: 10.1038/35000564
Dubnau J., Chiang A. S., Grady L., Barditch J., Gossweiler S., McNeil, et al. (2003). The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 13, 286–296. doi: 10.1016/S0960-9822(03)00064-2
Dyer F. C. (1996). Spatial memory and navigation by honeybees on the scale of the foraging range. J. Exp. Biol. 199, 147–154. doi: 10.1242/jeb.199.1.147
Fahrbach S. E. (2006). Structure of the mushroom bodies of the insect brain. Annu. Rev. Entomol 51, 209–232. doi: 10.1146/annurev.ento.51.110104.150954
Fahrbach S. E., Moore D., Capaldi E. A., Farris S. M., Robinson G. E. (1998). Experience-expectant plasticity in the mushroom bodies of the honeybee. Lean Mem 5, 115–123. doi: 10.1101/lm.5.1.115
Farris S. M., Robinson G. E., Fahrbach S. E. (2001). Experience- and age-related outgrowth of intrinsic neurons in the mushroom bodies of the adult worker honeybee. J. Nerurosci 21, 6395–6404. doi: 10.1523/JNEUROSCI.21-16-06395.2001
Fujita N., Nagata Y., Nishiuchi T., Sato M., Iwami M., Kiya T. (2013). Visualization of neural activity in insect brains using a conserved immediate early gene, hr38. Curr. Biol. 23, 2063–2070. doi: 10.1016/j.cub.2013.08.051
Gathmann A., Tscharntke T. (2002). Foraging ranges of solitary bees. J. Anim. Ecol. 71, 757–764. doi: 10.1046/j.1365-2656.2002.00641.x
Goulson D., Stout J. C. (2001). Homing ability of the bumblebee Bombus terrestris (Hymenoptera: Apidae). Apidologie 32, 105–111. doi: 10.1051/apido:2001115
Groh C., Rössler W. (2020). Analysis of synaptic microcircuits in the mushroom bodies of the honeybee. Insects 11, 43. doi: 10.3390/insects11010043
Hammer M. (1993). An identified neuron mediates the unconditioned stimulus in associative olfactory learning in honeybees. Nature 366, 59–63. doi: 10.1038/366059a0
Hartfelder K., Bitondi M. M. G., Santana W. C., Simões Z. L. P. (2002). Ecdysteroid titer and reproduction in queens and workers of the honey bee and of a stingless bee: loss of ecdysteroid function at increasing levels of sociality? Insect Biochem. Mol. Biol. 32, 211–216. doi: 10.1016/S0965-1748(01)00100-X
Heisenberg M. (1998). What do the mushroom bodies do for the insect brain? An introduction. Learn Mem 5, 1–10. doi: 10.1101/lm.5.1.1
Hendricsma H. P., Toth A. L., Shafir S. (2019). Individual and colony level foraging decisions of bumble bees and honey bees in relation to balancing of nutrient needs. Front. Ecol. Evol. 7, 177. doi: 10.3389/fevo.2019.00177
Iino S., Oya S., Kakutani T., Kohno H., Kubo T. (2023). Identification of ecdysone receptor target genes in the worker honey bee brains during foraging behavior. Sci. Rep. 13, 10491. doi: 10.1038/s41598-023-37001-7
Iino S., Shiota Y., Nishimura M., Asada S., Ono M., Kubo T. (2020). Neural activity mapping of bumble bee (Bombus ignitus) brains during foraging flight using immediate early genes. Sci. Rep. 10, 7887. doi: 10.1038/s41598-020-64701-1
Inagaki T., Tachibana M., Magoori K., Kudo H., Tanaka T., Okamura M., et al. (2009). Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells 14, 991–1001. doi: 10.1111/j.1365-2443.2009.01326.x
Ishimoto H., Sakai T., Kitamoto T. (2009). Ecdysone signaling regulates the formation of long-term courtship memory in adult Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 106, 6381–6386. doi: 10.1073/pnas.0810213106
Jassim O., Huang Z. Y., Robinson G. E. (2000). Juvenile hormone profiles of worker honey bees, Apis mellifera, during normal and accelerated behavioural development. J. Insect Physiol. 46, 243–249. doi: 10.1016/S0022-1910(99)00176-6
Kamikouchi A., Takeuchi H., Sawata M., Natori S., Kubo T. (2000). Concentrated expression of Ca2+/calmodulin-dependent protein kinase II and protein kinase C in the mushroom bodies of the brain of the honeybee Apis mellifera L. J. Comp. Neurol. 417, 501–510. doi: 10.1002/(SICI)1096-9861(20000221)417:4<501::AID-CNE8>3.0.CO;2-4
Kamikouchi A., Takeuchi H., Sawata M., Ohashi K., Natori S., Kubo T. (1998). Preferential expression of the gene for a putative inositol 1,4,5-trisphosphate receptor homolog in the mushroom bodies of the brain of the worker honeybee Apis mellifera L. Biochem. Biophys. Res. Commun. 242, 181–186. doi: 10.1006/bbrc.1997.7870
Kaneko K., Ikeda T., Nagai M., Hori S., Umatani C., Tadano H., et al. (2013). Novel middle-type Kenyon cells in the honeybee brain revealed by area-preferential gene expression analysis. PloS One 8, e71732. doi: 10.1371/annotation/1fa31a02-1b58-4361-98eb-5c213e5d5336
Kaneko K., Suenami S., Kubo T. (2016). Gene expression profiles and neural activities of Kenyon cell subtypes in the honeybee brain: identification of novel ‘middle-type’ Kenyon cells. Zool Lett. 2, 14. doi: 10.1186/s40851-016-0051-6
Kayukawa T., Jyuraku A., Ito Y., Shinoda T. (2017). Molecular mechanism underlying juvenile hormone-mediated repression of precocious larval–adult metamorphosis. Proc. Natl. Acad. Sci. U.S.A. 114, 1057–1062. doi: 10.1073/pnas.1615423114
Kiya T., Kunieda T., Kubo T. (2007). Increased neural activity of a mushroom body neuron subtype in the brains of forager honeybees. PloS One 2, e371. doi: 10.1371/journal.pone.0000371
Klein A. M., Steffan-Dewenter I., Tscharntke T. (2004). Foraging trip duration and density of megachilid bees, eumenid wasps and pompilid wasps in tropical agroforestry systems. J. Anim. Ecol. 73, 517–525. doi: 10.1111/j.0021-8790.2004.00826.x
Konopova B., Jindra M. (2008). Broad-Complex acts downstream of Met in juvenile hormone signaling to coordinate primitive holometabolan metamorphosis. Development 135, 559–568. doi: 10.1242/dev.016097
Kucharski R., Maleszka R., Hayward D. C., Ball E. E. (1998). A royal jelly protein is expressed in a subset of Kenyon cells in the mushroom bodies of the honey bee brain. Naturwissenschaften 85, 343–346. doi: 10.1007/s001140050512
Kumagai H., Kunieda T., Nakamura K., Matsumura Y., Namiki M., Kohno H., et al. (2020). Developmental stage-specific distribution and phosphorylation of Mblk-1, a transcription factor involved in ecdysteroid-signaling in the honey bee brain. Sci. Rep. 10, 8735. doi: 10.1038/s41598-020-65327-z
Kumar S., Chen D., Jang C., Nall A., Zheng X., Sehgal A. (2014). An ecdysone-responsive nuclear receptor regulates circadian rhythms in Drosophila. Nat. Commun. 5, 5697. doi: 10.1038/ncomms6697
Kuwabara T., Kohno H., Hatakeyama M., Kubo T. (2023). Evolutionary dynamics of mushroom body Kenyon cell types in hymenopteran brains from multifunctional type to functionally specialized types. Sci. Adv. 9, eadd4201. doi: 10.1126/sciadv.add4201
Lee C. Y., Baehrecke E. H. (2001). Steroid regulation of autophagic programmed cell death during development. Development 128, 1443–1455. doi: 10.1242/dev.128.8.1443
Lee C. Y., Wendel D. P., Reid P., Lam G., Thummel C. S., Baehrecke E. H. (2000). E93 directs steroid-triggered programmed cell death in Drosophila. Mol. Cell 6, 433–443. doi: 10.1016/S1097-2765(00)00042-3
Li Q., Wang M., Zhang P., Liu Y., Guo Q., Zhu Y., et al. (2022). A single-cell transcriptomic atlas tracking the neural basis of division of labour in an ant superorganism. Nat. Ecol. Evol. 6, 1191–1204. doi: 10.1038/s41559-022-01784-1
Liu X., Dai F., Guo E., Li K., Ma L., Tian L., et al. (2015). 20-Hydroxyecdysone (20E) Primary response gene E93 modulates 20E signaling to promote Bombyx larval-pupal metamorphosis. J. Biol. Chem. 290, 27370–27383. doi: 10.1074/jbc.M115.687293
Liu S., Li K., Gao Y., Liu X., Chen W., Ge W., et al. (2018). Antagonistic actions of juvenile hormone and20-hydroxyecdysone within the ring gland determine developmental transitions in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 115, 139–144. doi: 10.1073/pnas.1716897115
Liu F., Wu L., Zhang Y., Li Q., Li L., Huang Z. Y., et al. (2022). Mblk-1 regulates sugar responsiveness in honey bee (Apis mellifera) foragers. Insect Sci. 29, 683–690. doi: 10.1111/1744-7917.12971
Matsumoto Y., Sandoz J. C., Devaud J. M., Lormant F., Mizunami M., Giurfa M. (2014). cGMP-gated channels, calmodulin, AC, and CamKII required for late, but not early, long-term memory formation in the honeybee. Learn Mem 21, 272–286. doi: 10.1101/lm.032037.113
Matsumura Y., To T. K., Kunieda T., Kohno H., Kakutani T., Kubo T. (2022). Mblk-1/E93, an ecdysone related-transcription factor, targets synaptic plasticity-related genes in the honey bee mushroom bodies. Sci. Rep. 12, 21367. doi: 10.1038/s41598-022-23329-z
Menzel R., Giurfa M. (2001). Cognitive architecture of a mini-brain: the honeybee. Trends Cognit. Sci. 5, 62–71. doi: 10.1016/S1364-6613(00)01601-6
Mobbs P. G. (1982). The brain of the honeybee Apis mellifera. I. The connections and spatial organization of the mushroom bodies. Phil Trans. R Soc. London B 298, 309–354. doi: 10.1098/rstb.1982.0086
Oya S., Kohno H., Kainoh Y., Ono M., Kubo T. (2017). Increased complexity of mushroom body Kenyon cell subtypes in the brain is associated with behavioral evolution in hymenopteran insects. Sci. Rep. 7, 13785. doi: 10.1038/s41598-017-14174-6
Park J.-M., Kunieda T., Kubo T. (2003). The activity of Mblk-1, a mushroom body-selective transcription factor from the honeybee, is modulated by the Ras/MAPK pathway. J. Biol. Chem. 278, 18689–18694. doi: 10.1074/jbc.M300486200
Park J.-M., Kunieda T., Takeuchi H., Kubo T. (2002). DNA binding properties of Mblk-1, a putative transcription factor from the honeybee. Biochem. Biophys. Res. Commun. 291, 23–28. doi: 10.1006/bbrc.2002.6397
Pasch E., Muenz T. S., Rössler W. (2011). CaMKII is differentially localized in synaptic regions of Kenyon cells within the mushroom bodies of the honeybee brain. J. Comp. Neurol. 519, 3700–3712. doi: 10.1002/cne.22683
Paul R. K., Takeuchi H., Kubo T. (2006). Expression of two ecdysteroid-regulated genes, Broad-Complex and E75, in the brain and ovary of the honeybee (Apis mellifera L. ). Zoolog Sci. 23, 1085–1092. doi: 10.2108/zsj.23.1085
Paul R. K., Takeuchi H., Matsuo Y., Kubo T. (2005). Gene expression of ecdysteroid-regulated gene E74 of the honeybee in ovary and brain. Insect Mol. Biol. 14, 9–15. doi: 10.1111/j.1365-2583.2004.00524.x
Praggastis S. A., Lam G., Horner M. A., Nam H. J., Thummel C. S. (2021). The Drosophila E78 nuclear receptor regulates dietary triglyceride uptake and systemic lipid levels. Dev. Dyn 250, 640–651. doi: 10.1002/dvdy.287
Pyke G. H. (1978). Optimal foraging: Movement patterns of bumble bees between inflorescences. Theor. Popul Biol. 13, 72–98. doi: 10.1016/0040-5809(78)90036-9
Rittschof C. C., Vekaria H. J., Palmer J. H., Sullivan P. G. (2018). Brain mitochondrial bioenergetics change with rapid and prolonged shifts in aggression in the honey bee, Apis mellifera. J. Exp. Biol. 221, jeb176917. doi: 10.1242/jeb.176917
Robinson G. E., Strambi C., Strambi A., Feldlaufer M. F. (1991). Comparison of juvenile hormone and ecdysteroid haemolymph titres in adult worker and queen honey bees (Apis mellifera). J. Insect Physiol. 37, 929–935. doi: 10.1016/0022-1910(91)90008-N
Scholl C., Kübert N., Muenz T. S., Rössler W. (2015). CaMKII knockdown affects both early and late phases of olfactory long-term memory in the honeybee. J. Exp. Biol. 218, 3788–3796. doi: 10.1242/jeb.124859
Sheng L., Shields E. J., Gospocic J., Glastad K. M., Ratchasanmuang P., Berger S. L., et al. Social reprogramming in ants induces longevity-associated glia remodeling. Sci. Adv.20206, eaba9869. doi: 10.1126/sciadv.aba9869
Shonesy B. C., Jalan-Sakrikar N., Cavener V. S., Colbran R. J. (2014). CaMKII: A molecular substrate for synaptic plasticity and memory. Prog. Mol. Biol. Transl. Sci. 122, 61–87. doi: 10.1016/B978-0-12-420170-5.00003-9
Singh A. S., Shah A., Brockmann A. (2018). Honey bee foraging induces upregulation of early growth response protein 1, hormone receptor 38 and candidate downstream genes of the ecdysteroid signalling pathway. Insect Mol. Biol. 27, 90–98. doi: 10.1111/imb.12350
Stabentheiner A., Kovac H. (2016). Honeybee economics: optimisation of foraging in a variable world. Sci. Rep. 6, 28339. doi: 10.1038/srep28339
Strausfeld N. J. (2002). Organization of the honey bee mushroom body: representation of the calyx within the vertical and gamma lobes. J. Comp. Neurol. 450, 4–33. doi: 10.1002/cne.10285
Su X., Peng D. (2020). New insight into sortilin in controlling lipid metabolism and the risk of atherogenesis. Biol. Rev. Camb Philos. Soc. 95, 232–243. doi: 10.1111/brv.12561
Suenami S., Oya S., Kohno H., Kubo T. (2018). Kenyon cell subtypes/populations in the honey bee mushroom bodies: possible function based on their gene expression profiles, differentiation, possible evolution, and application of genome editing. Front. Psychol. 9, 1717. doi: 10.3389/fpsyg.2018.01717
Suenami S., Paul R. K., Takeuchi H., Okude G., Fujiyuki T., Shirai K., et al. (2016). Analysis of the differentiation of Kenyon cell subtypes using three mushroom body-preferential genes during metamorphosis in the honeybee (Apis mellifera L.). PloS One 11, e0157841. doi: 10.1371/journal.pone.0157841
Takeuchi H., Fujiyuki T., Shirai K., Matsuo Y., Kamikouchi A., Fujinawa Y., et al. (2002). Identification of genes expressed preferentially in the honeybee mushroom bodies by combination of differential display and cDNA microarray. FEBS Lett. 513, 230–234. doi: 10.1016/S0014-5793(02)02319-0
Takeuchi H., Kage E., Sawata M., Kamikouchi A., Ohashi K., Ohara M., et al. (2001). Identification of a novel gene, Mblk-1, that encodes a putative transcription factor expressed preferentially in the large-type Kenyon cells of the honeybee brain. Insect Mol. Biol. 10, 487–494. doi: 10.1046/j.0962-1075.2001.00288.x
Takeuchi H., Paul R. K., Matsuzaka E., Kubo T. (2007). EcR-A expression in the brain and ovary of the honeybee (Apis mellifera L.). Zool Sci. 24, 596–603. doi: 10.2108/zsj.24.596
Toma D. P., Bloch G., Moore D., Robinson G. E. (2000). Changes in period mRNA levels in the brain and division of labor in honey bee colonies. Proc. Natl. Acad. Sci. U.S.A. 97, 6914–6919. doi: 10.1073/pnas.97.12.6914
Truman J. W. (2019). The evolution of insect metamorphosis. Curr. Biol. 29, R1252–R1268. doi: 10.1016/j.cub.2019.10.009
Truman J. W., Talbot W. S., Fahrbach S. E., Hogness D. S. (1994). Ecdysone receptor expression in the CNS correlates with stage-specific responses to ecdysteroids during Drosophila and Manduca development. Development 120, 219–234. doi: 10.1242/dev.120.1.219
Ugajin A., Kunieda T., Kubo T. (2013). Identification and characterization of an Egr ortholog as a neural immediate early gene in the European honeybee (Apis mellifera L.). FEBS Lett. 587, 3224–3230. doi: 10.1016/j.febslet.2013.08.014
Uno Y., Fujiyuki T., Morioka M., Kubo T. (2012). Mushroom body-preferential expression of proteins/genes involved in endoplasmic reticulum Ca2+-transport in the worker honeybee (Apis mellifera L. ) brain. Insect Mol. Biol. 22, 52–61. doi: 10.1111/imb.12002
Ureña E., Manjón C., Franch-Marro X., Martín D. (2014). Transcription factor E93 specifies adult metamorphosis in hemimetabolous and holometabolous insects. Proc. Natl. Acad. Sci. U.S.A. 111, 7024–7029. doi: 10.1073/pnas.1401478111
Uyehara C. M., McKay D. J. (2019). Direct and widespread role for the nuclear receptor EcR in mediating the response to ecdysone in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 116, 9893–9902. doi: 10.1073/pnas.1900343116
Velarde R. A., Robinson G. E., Fahrbach S. E. (2009). Coordinated responses to developmental hormones in the Kenyon cells of the adult worker honey bee brain (Apis mellifera L. ). J. Insect Physiol. 55, 59–69. doi: 10.1016/j.jinsphys.2008.10.006
Whitfield C. W., Cziko A.-M., Robinson G. E. (2009). Gene expression profiles in the brain predict behavior in individual honey bees. Science 302, 296–299. doi: 10.1126/science.1086807
Withers G. S., Fahrbach S. E., Robinson G. E. (1993). Selective neuroanatomical plasticity and division of labour in the honeybee. Nature 364, 238–240. doi: 10.1038/364238a0
Withers G. S., Fahrbach S. E., Robinson G. E. (1995). Effects of experience and juvenile hormone on the organization of the mushroom bodies of honey bees. J. Neurobiol. 26, 130–144. doi: 10.1002/neu.480260111
Woodgate J. L., Makinson J. C., Lim K. S., Reynolds A. M., Chittka L. (2016). Life-long radar tracking of bumblebees. PloS One 11, e0160333. doi: 10.1371/journal.pone.0160333
Yamazaki Y., Kiuchi M., Takeuchi H., Kubo T. (2011). Ecdysteroid biosynthesis in workers of the European honeybee Apis mellifera L. Insect Biochem. Mol. Biol. 41, 283–293. doi: 10.1016/j.ibmb.2011.01.005
Yamazaki Y., Shirai K., Paul R. K., Fujiyuki T., Wakamoto A., Takeuchi H., et al. (2006). Differential expression of HR38 in the mushroom bodies of the honeybee brain depends on the caste and division of labor. FEBS Lett. 580, 2667–2670. doi: 10.1016/j.febslet.2006.04.016
Keywords: honey bee, hymenoptera, mushroom body, Kenyon cell, ecdysone signaling, Mblk-1/E93, ecdysone receptor, chromatin immunoprecipitation-sequencing
Citation: Matsumura Y, Kohno H and Kubo T (2023) Possible functions of ecdysone signaling reiteratively used in the adult honey bee brain. Front. Bee Sci. 1:1251714. doi: 10.3389/frbee.2023.1251714
Received: 02 July 2023; Accepted: 07 September 2023;
Published: 27 September 2023.
Edited by:
Susan Elizabeth Fahrbach, University of Manitoba, CanadaReviewed by:
Eirik Søvik, Volda University College, NorwayCopyright © 2023 Matsumura, Kohno and Kubo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takeo Kubo, c3RrdWJvQGJzLnMudS10b2t5by5hYy5qcA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.