
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bacteriol., 20 March 2025
Sec. Antibiotic Resistance
Volume 4 - 2025 | https://doi.org/10.3389/fbrio.2025.1439865
Background: Antimicrobial resistance can vary significantly across different regions, making it crucial to understand the distribution of urinary pathogens and their susceptibility to antibiotics in specific settings. This knowledge is essential for guiding effective empirical treatment approaches. Additionally, antimicrobial resistance is an ongoing process that requires routine surveillance and monitoring studies to stay ahead of emerging resistance patterns. In Ethiopia, including southern Ethiopia, there is a lack of recent data regarding the magnitude of the problem of antimicrobial resistance in urinary tract infections (UTIs). Due to limited microbiology laboratory capabilities, routine culture and antibiotic susceptibility testing are not performed in the region. Consequently, the treatment of UTIs in Ethiopia relies on an empirical basis, with physicians prescribing different drugs without the guidance of culture and antibiotic susceptibility tests. This practice can potentially contribute to the overuse of antibiotics and the development of resistant microbial species. To address this knowledge gap and promote the rational use of existing antimicrobials, it is essential to have up-to-date information on the prevalence of uropathogens causing UTIs and their antimicrobial susceptibility patterns. This study aims to determine the most prevalent uropathogens causing UTIs and their susceptibility to antimicrobial agents among individuals suspected of having UTIs in the study area. By conducting this study, we aim to provide valuable insights into the current situation of antimicrobial resistance in UTIs in southern Ethiopia. This information will help guide clinicians in selecting appropriate antibiotics for empirical treatment and contribute to the overall efforts in combating antimicrobial resistance.
Methods and materials: A cross-sectional study was carried out from April to September 2022 at Bule Hora University Teaching Hospital. The study included a total of 279 suspected patients who visited the outpatient department. Sociodemographic data and associated factors were assessed using a pretested questionnaire. Clean catch midstream specimens were collected following standard microbiological procedures. The identification of gram-negative organisms was done using TSI, oxidase, H2S production, gas production, motility, indole production, urease production, citrate utilization, and lysine decarboxylation and deamination. Antibiotic susceptibility testing of the isolates was conducted using the Kirby–Bauer disc diffusion method. Data analysis was performed using SPSS version 25, and descriptive statistics were used to summarize the findings. The results were presented in words and tables. Binary logistic regression was used to determine the statistical association between predictors and outcome variables.
Results: A total of 279 individuals participated in the study. The overall prevalence of gram-negative uropathogens was 11.8%. The predominant bacteria isolated was Escherichia coli (57.7%) followed by Klebsiella species (n = 9, 27.3%) and Proteus species (n = 3, 9.1%).
Conclusion: Bacterial uropathogens, especially resistant strains of E. coli, pose a significant threat. Amoxicillin–clavulanic acid, trimethoprim–sulfamethoxazole, and ciprofloxacin were found to be ineffective against these strains. Regular surveillance of uropathogenic bacteria’s sensitivity to antibiotics can help clinicians make better treatment choices. More research is needed on both gram-positive and gram-negative bacteria, focusing on the molecular characterization of resistant genes.
Urinary tract infection (UTI) is characterized by the invasion and growth of microbes in any part of the urinary tract, spanning from the renal cortex to the urethral meatus. It ranks as the second most prevalent infectious condition in community medical settings, affecting individuals across all age groups, from neonates to the elderly (Baron et al., 2013).
Globally, UTIs contribute to approximately 150 million cases annually, leading to significant morbidity and mortality (Totsika et al., 2012). Additionally, UTIs can manifest asymptomatically, with reported prevalence rates of 0.37% and 0.47% in boys and girls, respectively, and a 3% prevalence of asymptomatic bacteriuria in women. Notably, studies have indicated a high recurrence rate of UTIs in children, ranging from 19% to 30% in various age groups under 16 years old (Foxman, 2014; Shaikh et al., 2020; Veauthier and Miller, 2020; Guideline, 2023).
Urinary tract infections affecting the kidneys pose a significant and grave health concern across all sex and age demographics (Murgia et al., 2018; Sabih and Leslie, 2022). Women, in particular, face a higher susceptibility to UTIs compared to men, attributed to anatomical differences and the shorter length of the female urethra (Mambatta et al., 2015). UTIs rank among the most prevalent infectious diseases, imposing a substantial financial burden on society, notably impacting young women. Moreover, the incidence of UTIs escalates with age, notably surpassing the age of 65 (Murgia et al., 2018). Furthermore, individuals undergoing urethral catheterization, urological surgeries, and manipulations; long-term elderly male patients; and those with debilitating illnesses are identified as being at a heightened risk of developing nosocomial UTIs (Iacovelli et al., 2014).
Bacterial uropathogens are the most frequently encountered causes of UTI, which can usually be demonstrated for most episodes, and the common bacterial species seen in various patient groups have now been well defined. The frequently isolated organisms include gram negatives like Escherichia coli, Proteus mirabilis, Klebsiella pneumoniae, and Enterobacter species and gram-positive bacteria such as Staphylococcus saprophyticus, Enterococcus faecalis, and Streptococcus agalactiae (group B Streptococcus) (Johansen and Naber, 2014; Wiedemann et al., 2014). Escherichia coli, the most prevalent community-acquired UTI, accounts for 70%–95% of acute and uncomplicated UTIs in adults, followed by P. mirabilis, Klebsiella spp., and S. saprophyticus accounting for 5%–10% of the cases collectively. On the other hand, secondary pathogens such as Pseudomonas are mostly seen in complicated UTIs. Additionally, a small fraction of UTIs can have a hematogenous origin with relatively uncommon microorganisms (like Staphylococcus aureus, Candida spp., Salmonella spp., and Mycobacterium tuberculosis) (Lo et al., 2013; Murgia et al., 2018). Moreover, 90% of UTI causative agents were reported from gram-negative bacteria, while the rest (10%) of the cases were from gram-positive bacteria (Seifu and Gebissa, 2018).
In a setting where there is an intake of antibiotics without prescription, antimicrobial susceptibility testing is considered a crucial technique to investigate and decide the appropriate treatment regime along with pathological identification via evaluating and measuring the halos of inhibition around disks according to the standard reference tables which create great insight into an overall antimicrobial susceptibility profile (Murgia et al., 2018; Sakeena et al., 2018).
Since antimicrobial resistance varies regionally, it is necessary to know the distribution of urinary pathogens and their susceptibility to antibiotics in a particular setting (Farrell et al., 2003) that could support the most effective empirical treatment (Farajnia et al., 2009). Moreover, antimicrobial resistance is an evolving process that needs routine surveillance and monitoring studies (Beyene et al., 2011). In Ethiopia, the approach to treating UTIs relies heavily on empirical methods due to limited access to routine culture and antibiotic susceptibility testing. Physicians in the region, including the study area, often prescribe various medications without the guidance of these tests when managing patients with suspected UTIs. This practice may contribute to the misuse of antibiotics and the emergence of resistant microbial strains, posing a significant public health concern.
Moreover, there is a notable lack of recent data, particularly in southern Ethiopia, regarding the prevalence and impact of antimicrobial resistance in UTI cases. To promote the careful use of available antimicrobials, it is imperative to have updated and comprehensive information on antimicrobial resistance patterns accessible across all healthcare levels. These data serve as a crucial tool in guiding treatment decisions and combating the growing challenge of antimicrobial resistance in the region. Therefore, this study was carried out to determine the most prevalent uropathogens that caused UTI and their antimicrobial susceptibility pattern among UTI-suspected individuals in the study area.
An institution-based cross-sectional study was undertaken among outpatient attendees at BHUTH, situated 467 km south of Addis Ababa. Bule Hora, the capital of West Guji and located at 5035′N, 38015′E/5.5830N, 38.2500E and 1,716 m above sea level, has an average annual rainfall of 648 mm. Bule Hora is home to one governmental general hospital and one health center, serving a catchment population of approximately 1,296,475 people. The hospital provides outpatient services to an annual total of 172,800 attendees. It is estimated that approximately 70 outpatient attendees per month present with clinically diagnosed UTIs.
All patients attending the OPD of the BHUTH were the source population. All patients who visited the OPD for complaints of a suspected urinary tract infection during the study period were included in the study population, while patients who had taken antibiotics 2 weeks prior to the data collection period and clients on follow-up were excluded.
The sample size was calculated using Epi Info version 7 using the single population proportion formula with the proportion of 18.8% of uropathogens from a previous study (Yismaw et al., 2012), with 95% confidence intervals and a 5% margin of error.
As a result, the initial sample size was determined to be 258. However, accounting for potential non-response rates, the final sample size was adjusted to 279. To reach this final sample size, consecutive patients who provided consent and were available during the study period were included until the intended sample size was successfully attained.
These include the bacterial profile of uropathogens and the antimicrobial susceptibility pattern.
These include sociodemographic characteristics (age, sex, residence, income, educational status, marital status) and clinical factors (presence of symptoms, history of catheterization, history of previous UTI, dysuria, frequency of micturition, blood in urine).
Information regarding sociodemographic details and selected associated factors concerning urinary tract infections was gathered through a structured questionnaire administered via interviews. The questionnaire was initially developed in English and subsequently translated into Afaan Oromo and Amharic. To ensure accuracy, the translations were then reverted back to English for validation. Data collection was facilitated by trained data collectors working within the Outpatient Department (OPD) setting. For each participant, a single clean catch/midstream urine sample was collected specifically for culture analysis, maintaining strict adherence to standardized collection protocols.
All specimens were promptly collected and transported to the microbiology laboratory in sterile containers. In cases where delays were anticipated, samples were stored in the hospital laboratory’s refrigerator. The urine specimens underwent both presumptive and further identification procedures with strict adherence to standard operational procedures (SOPs).
Upon reaching the laboratory, the specimens were processed without delay. For samples designated for culture, urine specimens were inoculated onto sterile MacConkey (Mac) agar plates within 30 min of collection. Subsequently, the plates were placed in an overnight incubation at 37°C. Identification was carried out based on colony morphology, gram staining, and biochemical tests, ensuring meticulous attention to detail in the laboratory procedures.
On the initial day, the urine specimens were plated on MacConkey agar and then placed in an incubator set at 35°C–37°C for a duration of 18–48 h. Subsequently, on the following day, standard microbiological protocols were employed to characterize the bacterial colonies that had developed on each culture medium. This involved a detailed analysis of colony morphology, gram staining, and the implementation of biochemical tests on the colonies from each plate in accordance with established procedures.
For gram-negative organisms, a series of tests including TSI, oxidase, H2S production, gas production, motility, indole production, urease production, citrate utilization, and lysine decarboxylation and deamination were conducted. Antimicrobial susceptibility testing was carried out for all gram-negative isolates on Mueller–Hinton agar utilizing the standardized Kirby–Bauer diffusion technique as per the guidelines of the Clinical and Laboratory Standards Institute (CLSI). The selection and number of antibiotics used varied depending on the isolated organism, ensuring a tailored approach to antimicrobial testing.
The SOPs meticulously followed during the preparation of reagents, culture media, specimen processing, culturing, and microscopic examination adhered to the guidelines established by the Ethiopian Public Health Institute (EPHI) and relevant textbooks. Upholding the quality of the bacterial culture process, the control strains E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were employed.
In order to uphold the precision of the collected data, a pretest of the data collection tool (questionnaire) was conducted on a 5% sample of the total study population, distinct from those included in the main study. Corrections were promptly implemented based on the pretest feedback, ensuring data completeness before entry into the system.
After coding, the data were checked for completeness, inconsistency, and outliers by looking at their distribution. Incomplete and inconsistent data were excluded from the analysis. Data were analyzed using SPSS version 25. Descriptive statistics like percentage and frequency were used to describe the study sample. Binary and logistic regression was used to determine the statistical association between predictors and outcome variables. A P-value <0.05 with a 95% confidence interval was considered statistically significant.
The study included a total of 279 outpatients, achieving a 100% response rate. The mean age of the mothers involved was 30.1 years, with a standard deviation of 4.8 years. Notably, more than half of the participants (n = 160, 57.3%) fell within the age range of 25–34 years, implying a relatively youthful sample. Regarding residency, the majority of the participants (n = 187, 67.0%) were urban residents, indicating a predominant focus on an urban population. In terms of the occupational distribution of the participants, a significant proportion (n = 94, 33.7%) were housewives.
Furthermore, a large majority of the participants (n = 246, 88.1%) were married, indicating that the study primarily included individuals in committed relationships. In terms of educational background, it is noteworthy that a significant proportion (n = 109, 39.1%) of the participants did not attend formal education. This highlights the importance of considering educational disparities and their potential impact on the study findings (Table 1).
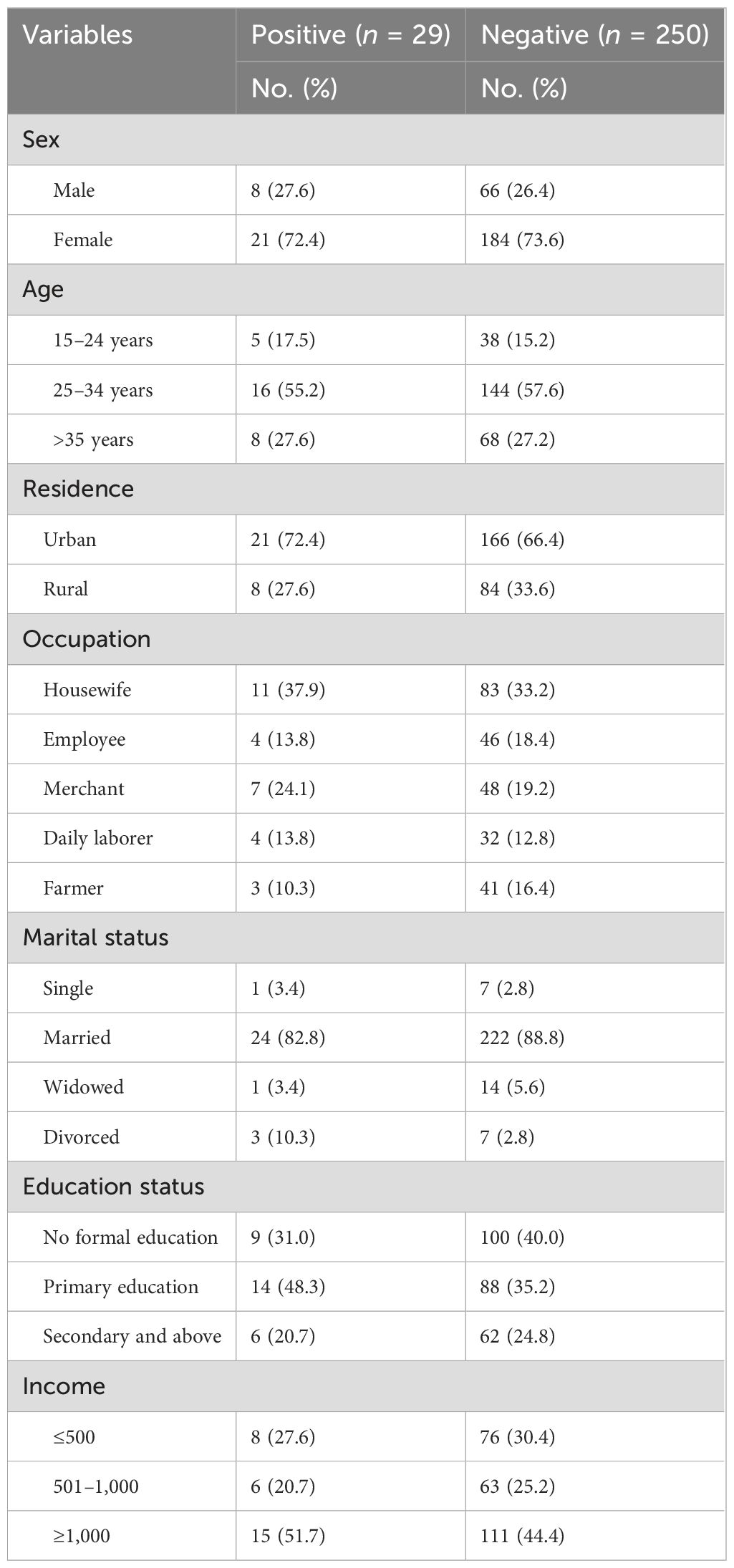
Table 1. Sociodemographic characteristics of outpatient attendants at Bule Hora University Teaching Hospital, 2022.
An important observation in terms of clinical factors is that a significant proportion of both urine culture-positive and culture-negative patients were asymptomatic. Specifically, 23 (79.3%) of the culture-positive patients and 198 (79.2%) of the culture-negative patients reported no symptoms. This suggests that the presence of bacteria in urine does not always align with the presence of symptoms.
Moreover, a majority of individuals in both the culture-positive and culture-negative groups did not have a previous history of UTI. Specifically, 22 (75.9%) of the culture-positive patients and 174 (69.6%) of the culture-negative patients had no prior history of UTI. This highlights that UTIs can manifest in individuals without a previous record of the condition.
When assessing the history of catheterization and gastrourinary abnormalities, it is intriguing to observe that nearly all culture-positive patients (n = 25, 86.2%) and culture-negative patients (n = 244, 97.6%) did not have a history of catheterization. Similarly, 237 (94.8%) of the culture-negative patients did not report any gastrourinary abnormalities. These findings suggest that these factors may not play a significant role in the presence of uropathogenic bacteria (Table 2).
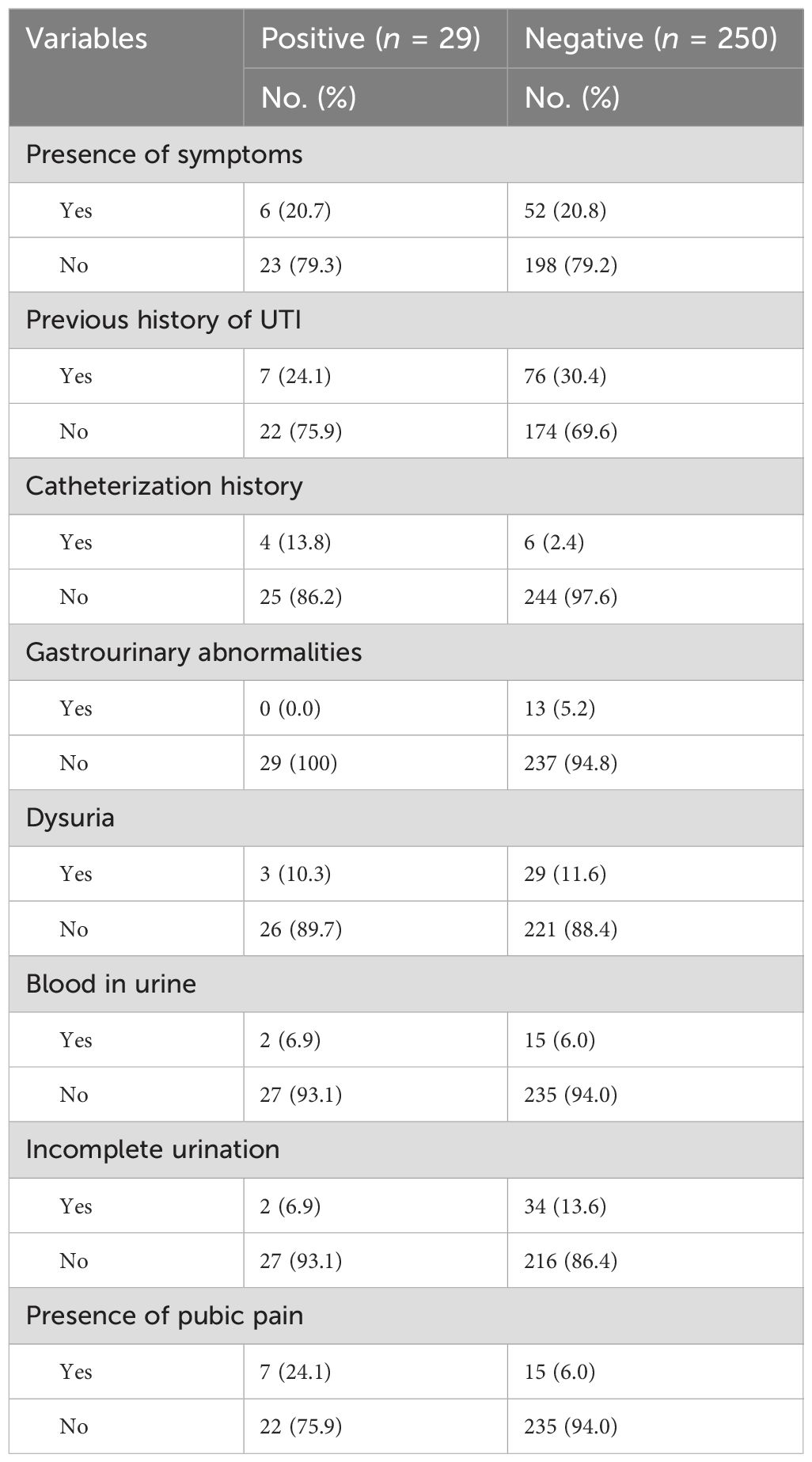
Table 2. Clinical characteristics of outpatient attendants at Bule Hora University Teaching Hospital, 2022.
Generally, in the study, a total of 33 gram-negative organisms were isolated, where E. coli is the most prevalent (n = 19, 57.6%) and the rest includes Klebsiella species (n = 9, 27.3%), Proteus species (n = 3, 9.1%), and Pseudomonas species (n = 2, 6.0%). None of those organisms were resistant to antibiotics used in the study. Finally, the majority of E. coli isolates were resistant to amoxicillin–clavulanic acid (54%), trimethoprim–sulfamethoxazole (63.4%), and ciprofloxacin (53.0%) (Table 3).
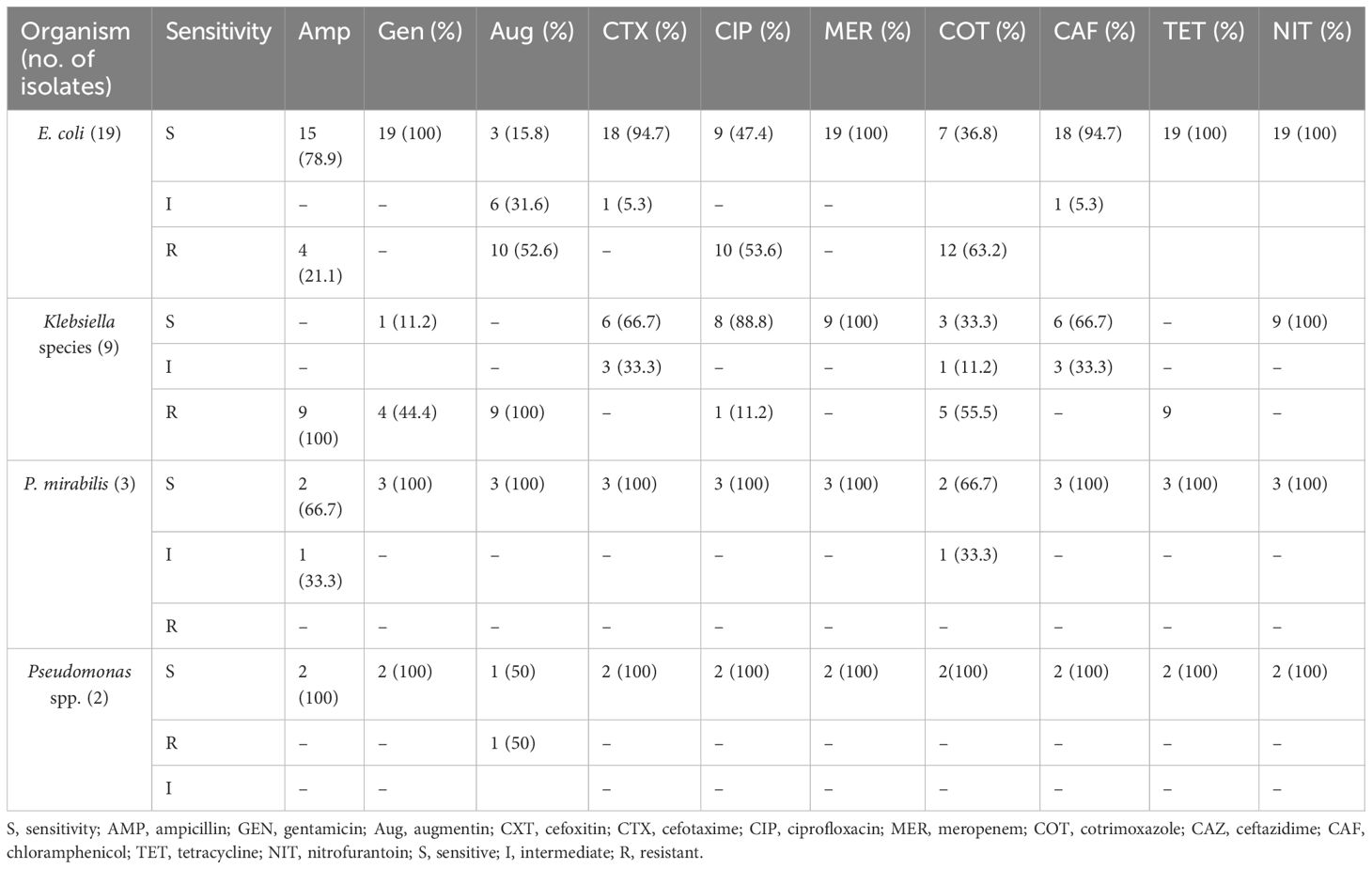
Table 3. Antimicrobial resistance pattern of bacterial isolates from outpatient attendants at Bule Hora University Teaching Hospital, 2022.
Studies have demonstrated variations in the prevalence of urinary tract infections across different population groups, ranging from 9.16% to 43.69% (Kashef et al., 2010; Beyene and Tsegaye, 2011; Eshwarappa et al., 2011; Muvunyi et al., 2011; Alemu et al., 2012; Yismaw et al., 2012; Ayoade et al., 2013; Kabew et al., 2013; Irenge et al., 2014; Can et al., 2015; Guyomard-Rabenirina et al., 2016; Lehrasab et al., 2016). These studies have identified various causal organisms and risk factors associated with urinary tract infections. The current study reveals a prevalence of gram-negative organisms at 10.4%. This finding aligns with similar studies conducted in different regions of Ethiopia, such as 11.6% in Addis Ababa (Assefa et al., 2008), 10.4% in Gondar (Alemu et al., 2012), and 9.5% in Bahir Dar (Emiru et al., 2013). Moreover, these results are consistent with the findings from studies conducted in India, Iran, and other regions of Ethiopia (Beyene and Tsegaye, 2011; Eshwarappa et al., 2011; Yismaw et al., 2012; Ayoade et al., 2013).
The study findings indicate that E. coli is the most prevalent pathogen at 57.6%, followed by Klebsiella species at 27.3%. These results are consistent with the studies conducted in India, Pakistan, South Africa, and Addis Ababa (Eshwarappa et al., 2011; Demilie et al., 2014; Guyomard-Rabenirina et al., 2016; Lehrasab et al., 2016). The high prevalence of E. coli in these findings may be attributed to its significant presence in the rectal area, leading to contamination of the genitalia and migration to the urinary tract, thus causing UTIs. Moreover, E. coli’s possession of specialized virulence factors such as P-fimbriae and S-fimbriae adherence factors enables them to colonize and invade the urinary epithelium effectively. This enhances their capacity to adhere to both vaginal and uroepithelial cells (Kot, 2017).
The majority of E. coli isolates demonstrate resistance to key antibiotics, with rates of resistance at 54% for amoxicillin–clavulanic acid, 63.4% for trimethoprim–sulfamethoxazole, and 53.0% for ciprofloxacin. These findings are consistent with a study conducted at the University of Gondar Comprehensive Specialized Hospital in northwest Ethiopia, which revealed that over 75% of gram-negative isolates exhibited resistance to amoxicillin–clavulanate (80.1%) and trimethoprim–sulfamethoxazole (78.3%).
Escherichia coli isolates exhibited significant resistance rates to trimethoprim–sulfamethoxazole (76.5%) and amoxicillin–clavulanate (74%) (Kasew et al., 2022). Similar resistance rates were observed in Sudan, at 90% for amoxicillin–clavulanate and at 88% for trimethoprim–sulfamethoxazole (Mechal et al., 2021). Additionally, ciprofloxacin resistance was noted at 28.8% in another study (Belete and Saravanan, 2020).
These findings underscore that certain antibiotics may not be reliable for empirical therapy due to prolonged high levels of reported resistance. This study, along with several of our previous works (Gezmu et al., 2016; Mama et al., 2019), aligns with this long-standing trend of antibiotic resistance, highlighting the need for caution when selecting treatment options.
The study’s scope did not encompass gram-positive, anaerobic organisms and lacked molecular characterization of resistant genes. To achieve a thorough comprehension of uropathogenic bacteria and their resistance mechanisms, it is vital to consider the entire spectrum of organisms involved, including gram-positive and anaerobic species. Serological identification can provide valuable insights into the specific strains and variants present in the samples.
Moreover, the molecular characterization of resistant genes is crucial to unravel the genetic foundations of antibiotic resistance. Identifying the specific genes and mechanisms at play can inform the development of targeted interventions and more effective treatment strategies.
Future research endeavors should address these limitations by incorporating a broader array of organisms, conducting serological identification, and performing molecular characterization of resistant genes. This holistic approach will advance our understanding of uropathogenic bacteria and facilitate the formulation of enhanced treatment protocols.
The escalating presence of antibiotic-resistant bacterial uropathogens, particularly E. coli, underscores the urgency of the situation. Alarmingly, commonly used antibiotics such as amoxicillin–clavulanic acid, trimethoprim–sulfamethoxazole, and ciprofloxacin are showing reduced efficacy against these pathogens.
To tackle this pressing issue, it is imperative to conduct regular surveillance on the susceptibility of uropathogenic bacteria. This proactive measure will furnish clinicians with essential data to make well-informed decisions regarding the most suitable antibiotics for treatment. Furthermore, it is advisable to undertake further research focusing on both gram-positive and gram-negative bacteria, with a specific emphasis on molecular profiling of resistant genes.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethical clearance was obtained from the Bule Hora University Research and Publication Directorate. After the purpose of the study was explained to the participants, informed consent with a written signature was obtained. They were informed to withdraw at any time and/or to refrain from responding to questions. Study participants were also informed that all data obtained from them could be kept confidential using code instead of any personal identifiers. Furthermore, the research procedures were conducted in accordance with the principles expressed in the World Medical Association’s Declaration of Helsinki.
AE: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. DT: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. AA: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. BJ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research and/or publication of this article.
The authors would like to thank the Institute of Health, Bule Hora University and the individuals who provided comments and suggestions. We also extend our great appreciation to the data collectors for their selfless provision of continuous support and facilitation in the overall data collection processes.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
CA-UTI, community-acquired urinary tract infection; UTI, urinary tract infection; BHUTH, Bule Hora University Hospital; SOP, standard operational procedures; CLED, cystine lactose electrolyte deficient; Mac, MacConkey; CONS, coagulase-negative Staphylococcus; EPHI, Ethiopian Public Health Institute; CLSI, Clinical Laboratory Standard Institute.
Alemu A., Moges F., Shiferaw Y., Tafess K., Kassu A., Anagaw B., et al. (2012). Bacterial profile and drug susceptibility pattern of urinary tract infection in pregnant women at University of Gondar Teaching Hospital, Northwest Ethiopia. BMC Res. Notes 5, 1–7. doi: 10.1186/1756-0500-5-197
Assefa A., Asrat D., Woldeamanuel Y., G/Hiwot Y., Abdella A., Melesse T. (2008). Bacterial profile and drug susceptibility pattern of urinary tract infection in pregnant women at Tikur Anbessa Specialized Hospital Addis Ababa, Ethiopia. Ethiopian Med. J. 46, 227–235.
Ayoade F., Osho A., Fayemi S. O., Oyejide N. E., Ibikunle A. A (2013). Unusually high prevalence of asymptomatic bacteriuria among male university students on Redemption Camp, Ogun State, Nigeria. Afr. J. Clin. Exp. Microbiol. 14, 19–22.
Baron E. J., Miller J. M., Weinstein M. P., Richter S. S., Gilligan P. H., Thomson R. B., et al. (2013). A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM) a. Clin. Infect. Dis. 57, e22–e121.
Belete M. A., Saravanan M. (2020). A systematic review on drug resistant urinary tract infection among pregnant women in developing countries in Africa and Asia; 2005–2016. Infect. Drug Resist. 18, 1465–1477. doi: 10.2147/IDR.S250654
Beyene W., Jira C., Sudhakar M. (2011). Assessment of quality of health care in Jimma zone, southwest Ethiopia. Ethiopian J. Health Sci.
Beyene G., Tsegaye W. (2011). Bacterial uropathogens in urinary tract infection and antibiotic susceptibility pattern in Jimma University specialized hospital, southwest Ethiopia. Ethiopian J. Health Sci. 21 (2), 141–146. doi: 10.4314/ejhs.v21i2.69055
Can F., Azap O. K., Seref C., Ispir P., Arslan H., Ergonul O. (2015). Emerging Escherichia coli O25b/ST131 clone predicts treatment failure in urinary tract infections. Clin. Infect. Dis. 60, 523–527. doi: 10.1093/cid/ciu864
Demilie T., Beyene G., Melaku S., Tsegaye W. (2014). Diagnostic accuracy of rapid urine dipstick test to predict urinary tract infection among pregnant women in Felege Hiwot Referral Hospital, Bahir Dar, North West Ethiopia. BMC Res. Notes 7, 1–5. doi: 10.1186/1756-0500-7-481
Emiru T., Beyene G., Tsegaye W., Melaku S. (2013). Associated risk factors of urinary tract infection among pregnant women at Felege Hiwot Referral Hospital, Bahir Dar, North West Ethiopia. BMC Res. Notes 6, 1–6. doi: 10.1186/1756-0500-6-292
Eshwarappa M., Dosegowda R., Vrithmani A. I., Khan M. W., Kumar P. S., Kempegowda P. P. (2011). Clinico-microbiological profile of urinary tract infection in south India. Indian J. Nephrol. 21, 30. doi: 10.4103/0971-4065.75226
Farajnia S., Alikhani M. Y., Ghotaslou R., Naghili B., Nakhlband A. (2009). Causative agents and antimicrobial susceptibilities of urinary tract infections in the northwest of Iran. Int. J. Infect. Dis. 13, 140–144. doi: 10.1016/j.ijid.2008.04.014
Farrell D., Morrissey I., De Rubeis D., Robbins M., Felmingham D. (2003). A UK multicentre study of the antimicrobial susceptibility of bacterial pathogens causing urinary tract infection. J. infect. 46, 94–100. doi: 10.1053/jinf.2002.1091
Foxman B. (2014). Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect. Dis. Clinics 28, 1–13.
Gezmu T., Regasa B., Manilal A., Mama M., Hailu T., Merdekios B. (2016). Prevalence, diversity and antimicrobial resistance of bacteria isolated from the UTI patients of Arba Minch Province, southern Ethiopia. Transl. BioMed. 7, 81. doi: 10.21767/2172-0479.100081
Guyomard-Rabenirina S., Malespine J., Ducat C., Sadikalay S., Falord M., Harrois D., et al. (2016). Temporal trends and risks factors for antimicrobial resistant Enterobacteriaceae urinary isolates from outpatients in Guadeloupe. BMC Microbiol. 16, 1–8. doi: 10.1186/s12866-016-0749-9
Iacovelli V., Gaziev G., Topazio L., Bove P., Vespasiani G., Finazzi Agrò E. (2014). Nosocomial urinary tract infections: A review. Urol. J. 81, 222–227. doi: 10.5301/uro.5000092
Irenge L. M., Kabego L., Vandenberg O., Chirimwami R. B., Gala J.-L. (2014). Antimicrobial resistance in urinary isolates from inpatients and outpatients at a tertiary care hospital in South-Kivu Province (Democratic Republic of Congo). BMC Res. Notes 7, 1–6. doi: 10.1186/1756-0500-7-374
Kabew G., Abebe T., Miheret A. (2013). A retrospective study on prevalence and antimicrobial susceptibility patterns of bacterial isolates from urinary tract infections in Tikur Anbessa Specialized Teaching Hospital Addis Ababa, Ethiopia, 2011. Ethiopian J. Health Dev. 27, 111–117.
Kasew D., Desalegn B., Aynalem M., Tila S., Diriba D., Afework B., et al. (2022). Antimicrobial resistance trend of bacterial uropathogens at the university of Gondar comprehensive specialized hospital, northwest Ethiopia: A 10 years retrospective study. PloS One 17, e0266878. doi: 10.1371/journal.pone.0266878
Kashef N., Djavid G. E., Shahbazi S. (2010). Antimicrobial susceptibility patterns of community-acquired uropathogens in Tehran, Iran. J. Infect. Develop. Countries 4, 202–206. doi: 10.3855/jidc.540
Kot B. (2017). Virulence factors and innovative strategies for the treatment and control of uropathogenic Escherichia coli. Escherichia coli-recent advances on physiology, pathogenesis and biotechnological applications.
Lehrasab W., Aziz T., Naheem Ahmed I. A. (2016). Causative organisms of urinary tract infection and their sensitivity pattern in children. Ann. PIMS ISSN 1815, 2287.
Lo D. S., Shieh H. H., Ragazzi S. L. B., Koch V. H. K., Martinez M. B., Gilio A. E. (2013). Community-acquired urinary tract infection: age and gender-dependent etiology. Braz. J. Nephrol. 35, 93–98. doi: 10.5935/0101-2800.20130016
Mama M., Manilal A., Gezmu T., Kidanewold A., Gosa F., Gebresilasie A. (2019). Prevalence and associated factors of urinary tract infections among diabetic patients in Arba Minch Hospital, Arba Minch province, South Ethiopia. Turkish J. Urol. 45, 56. doi: 10.5152/tud.2018.32855
Mambatta A. K., Jayarajan J., Rashme V. L., Harini S., Menon S., Kuppusamy J. (2015). Reliability of dipstick assay in predicting urinary tract infection. J. Family Med. primary Care 4, 265. doi: 10.4103/2249-4863.154672
Mechal T., Hussen S., Desta M. (2021). Bacterial profile, antibiotic susceptibility pattern and associated factors among patients attending adult OPD at Hawassa University Comprehensive Specialized Hospital, Hawassa, Ethiopia. Infect. Drug Resist., 99–110. doi: 10.2147/IDR.S287374
Murgia L., Stalio O., Arienzo A., Ferrante V., Cellitti V., Di Somma S., et al. (2018). Management of urinary tract infections: problems and possible solutions. Urinary tract infection—The result of the strength of the pathogen, or the weakness of the host.
Muvunyi C. M., Masaisa F., Bayingana C., Mutesa D. L., Musemakweri A., Muhirwa G., et al. (2011). Decreased susceptibility to commonly used antimicrobial agents in bacterial pathogens isolated from urinary tract infections in Rwanda: need for new antimicrobial guidelines. Am. J. Trop. Med. hygiene 84, 923. doi: 10.4269/ajtmh.2011.11-0057
Sabih A., Leslie S. W. (2022). “Complicated urinary tract infections,” in StatPearls (Europe PMC: StatPearls Publishing).
Sakeena M., Bennett A. A., McLachlan A. J. (2018). Non-prescription sales of antimicrobial agents at community pharmacies in developing countries: a systematic review. Int. J. antimicrobial Agents 52, 771–782. doi: 10.1016/j.ijantimicag.2018.09.022
Seifu W. D., Gebissa A. D. (2018). Prevalence and antibiotic susceptibility of Uropathogens from cases of urinary tract infections (UTI) in Shashemene referral hospital, Ethiopia. BMC Infect. Dis. 18, 30. doi: 10.1186/s12879-017-2911-x.2018
Shaikh N., Osio V. A., Wessel C. B., Jeong J. H. (2020). Prevalence of asymptomatic bacteriuria in children: a meta-analysis. J. Pediatr. 217, 110–117. e4. doi: 10.1016/j.jpeds.2019.10.019
Johansen T.E.B., Naber K.G. (2014). Urinary tract infections. Antibiotics 3, 375–377. doi: 10.3390/antibiotics3030375
Totsika M., Moriel D. G., Idris A., Rogers B. A., Wurpel D. J., Phan M. D. (2012). Uropathogenic Escherichia coli mediated urinary tract infection. Curr. Drug Targets 13, 1386–1399. doi: 10.2174/138945012803530206
Veauthier B., Miller M. V. (2020). Urinary tract infections in young children and infants: common questions and answers. Am. Family Phys. 102, 278–285.
Wiedemann B., Heisig A., Heisig P. (2014). Uncomplicated urinary tract infections and antibiotic resistance—epidemiological and mechanistic aspects. Antibiotics 3, 341–352. doi: 10.3390/antibiotics3030341
Keywords: antimicrobial, bacterial, Ethiopia, profile, susceptibility, uropathogens
Citation: Edin A, Tilahun D, Jara BA and Ayele A (2025) Bacterial profile and antimicrobial susceptibility pattern of uropathogens among suspected patients attending Bule Hora University Teaching Hospital, southern Ethiopia. Front. Bacteriol. 4:1439865. doi: 10.3389/fbrio.2025.1439865
Received: 28 May 2024; Accepted: 24 February 2025;
Published: 20 March 2025.
Edited by:
Aseer Manilal, Komar University of Science and Technology, IraqReviewed by:
Santosh Dulal, Management Sciences for Health, United StatesCopyright © 2025 Edin, Tilahun, Jara and Ayele. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alo Edin, amlibG9zYTFAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.