
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bacteriol., 06 November 2024
Sec. Molecular Bacteriology and Microbiome
Volume 3 - 2024 | https://doi.org/10.3389/fbrio.2024.1497132
This article is part of the Research TopicBiofilm Dynamics: Interdisciplinary Insights into Microbial Interactions within the Context of Microbiomes and Host HealthView all articles
Here, we view biofilm as a nested cybernetic system that cannot be studied in isolation from the dynamics in the rest of the ecosystem. We provide a systems view of B. subtilis development from the inoculation to hibernation. We propose that B. subtilis biofilm development in an aqueous environment is a temporal response to changes in the ecosystem provoked and caused by bacteria. We show that the initial bacterial growth changes physio-chemical parameters of the ecosystem, which in turn trigger the initiation of the biofilm formation and guide its development, structure, and ultimately its decay and dispersion. Formation of the biofilm is just one of the possible outputs of the bacterial cybernetic system. It is not invariably the best response that fits all environmental needs. Viewing biofilm as a cybernetic set of interrelated objects, capable of receiving, storing, processing, and exchanging information with the rest of the ecosystem, should become an integral part of biofilm studies.
Formation of biofilm is one of the many possible outcomes for a bacterial population that self-regulates and acts toward a goal of survival. The formation of the biofilm can be viewed as an emergent form of bacterial life, wherein communal life of a cell is completely different from a free-living style (Flemming et al., 2016). Not all of the bacteria in an ecosystem will form a biofilm. A substantial fraction of B. subtilis cells will remain to be planktonic in the presence of the biofilm (Lee et al., 2019; Sanchez-Vizuete et al., 2021; Krajnc et al., 2022). Some bacteria will actively disperse to different locations (Mitchell and Kogure, 2006; Koch and Subramanian, 2011; Samad et al., 2017; Krajnc et al., 2024), cells not forming biofilm may change their metabolism in an attempt to survive (Wan et al., 2018; Pisithkul et al., 2019), or change lifestyle to other non-biofilm styles (i.e. to predatory, cannibalism (Nandy et al., 2007; Rozen et al., 2009; Thiery and Kaimer, 2020; Contreras-Moreno et al., 2024). If a subpopulation of cells produces a biofilm, it may produce different biofilms (i.e. pellicles at the air-liquid interface and submerged biofilms at the solid-liquid interface) (Dergham et al., 2021; Sanchez-Vizuete et al., 2021; Krajnc et al., 2022). Thus, even in the simple model system, biofilm will describe only part of the bacterial dynamics in the ecosystem. The decision on model complexity should be based on the specific goals of the experiment. If the focus is on understanding general biofilm properties (i.e. the initiation of biofilm formation, adhesion, growth rates, morphology, thickness), a simpler, uniform model may suffice. But if the aim is to investigate more detailed phenomena like differential antibiotic resistance then models that account for microenvironments and spatial differentiation will offer more accurate and relevant insights.
Bacillus subtilis is one of the best studied model organisms and its biofilm development in simple model systems has been detailed in several studies from genomic (Hamon et al., 2004; Tosato and Bruschi, 2004; Kearns et al., 2005), transcriptomic (Xu et al., 2022; Yang et al., 2022), proteomic (Miller and Diaz-Torres, 1999; Branda et al., 2006), as well as metabolomic point of view (Pisithkul et al., 2019). The most informative studies have been conducted in a well-defined, semi-closed model systems consisting of water column in contact with air, with a defined chemical composition and no additional feeding from the beginning to the end of the process, where only a limited exchange of gasses through the air-liquid interface was allowed. Typically, no external shear stress is imposed on the model system and fluid flow is induced solely by bacterial mobility. The model system has a single bacterial species and is maintained at constant temperature and pressure (Dergham et al., 2021; Sanchez-Vizuete et al., 2021; Krajnc et al., 2022). While this is clearly an oversimplification of most natural ecosystems it allows one to communicate ideas, understand biofilm processes at a fundamental level, and make predictions about biofilms in the absence of external disturbances. After inoculation of B. subtilis in the simple model system cells grow rapidly and alter conditions in the medium so that within hours bacteria are forced to seek alternative habitats within the ecosystem. In particular, oxygen concentration may become critically low to support aerobic growth in the suspension (da Silva et al., 2013). The cells sense oxygen concentration gradient, swim in the direction up the gradient, and accumulate at the air-liquid interface (Figure 1A). Rapidly the air-liquid interface becomes the preferred environment within the ecosystem where cells have access to high oxygen concentrations from the air as well as nutrients from the medium (Sanchez-Vizuete et al., 2021; Krajnc et al., 2022). Flagellum-based mobility plays a key role in interface colonization (Kobayashi, 2007; Hölscher et al., 2015; Lee et al., 2019; Shoup and Ursell, 2023). As bacterial cells accumulate at the interface, the increased fluid density causes patches of bacterial cells to sink, initiating a form of bioconvection that begins as a gravitational Rayleigh–Taylor instability (Figure 1B) (Jánosi et al., 1998). When fully developed, bioconvection enables the downward transport of oxygen-charged water from the interface, providing a quick fix to an increased oxygen demand in the bulk of the solution (Jánosi et al., 1998). Bioconvection, however, does not allow formation of a stable structure at the air-liquid interface. Similarly, also in E. coli, vertical gradients arising at the air-liquid interface create cell-density inversions, which drive bioconvection, much like in Bacillus subtilis (Shoup and Ursell, 2023). Recent studies have shown that individual B. subtilis cells are weakly mechanically connected already in suspension (Sretenovic et al., 2017; Dogsa et al., 2023). The mechanically coupled bacterial structures enable interconnections between individual cells and coordinated motion of a group of bacteria that are up to 100 μm apart. Although such structures are generally not considered biofilms, they exhibit characteristics analogous to biofilms such as self-made weak viscoelastic extracellular matrix which strengthens with increasing bacterial density (Krajnc et al., 2022). It is likely that such assemblies can float to the surface forming a pre-biofilm structure (Lee et al., 2019; Krajnc et al., 2022). During the formation of an initially weak biofilm at the air-liquid interface a major redistribution of bacteria in the water column occurs (Krajnc et al., 2022). Cells move either to the air-liquid interface or to the bottom of the water column (Figure 1A) (Dergham et al., 2021; Sanchez-Vizuete et al., 2021; Krajnc et al., 2022). The two strategies have radically different outcome. The biofilm at the air-liquid interface enables further survival of bacteria, whereas most of the cells in the sedimented biofilm die.
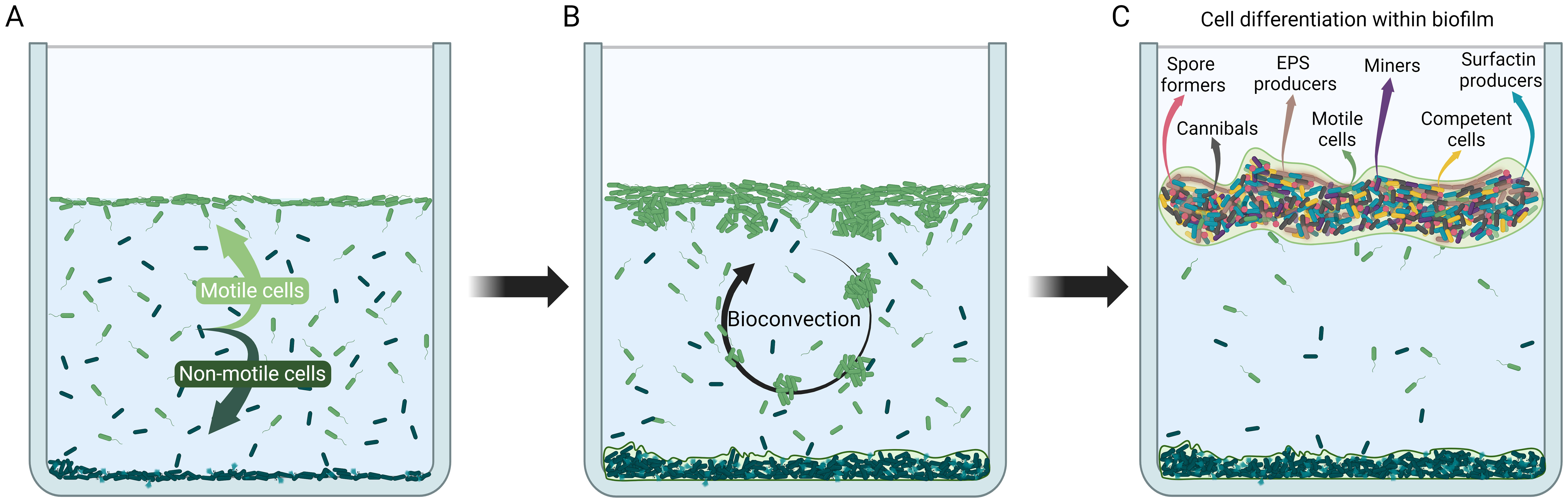
Figure 1. Schematic illustration of cybernetic system development through cell differentiation and behavior during B. subtilis biofilm formation. (A) Initial cell redistribution in the system. When growth conditions in the water column deteriorate motile cells move towards the air-liquid interface and begin to form a floating biofilm, whereas non-motile cells settle at the bottom and mostly die (Dergham et al., 2021; Sanchez-Vizuete et al., 2021; Krajnc et al., 2022; Shoup and Ursell, 2023). (B) Bioconvection. Up-swimming cells searching for a source of oxygen cause instabilities in the biofilm at the air-liquid interface which eventually cause down-welling plumes of cells starting a bioconvection cycle (Hopkins and Fauci, 2002; Lee et al., 2019; Shoup and Ursell, 2023). (C) Cell differentiation in the mature floating biofilm (Lopez et al., 2009; Lopez and Kolter, 2010). Biofilm is segregated along chemical gradients and individual cells are in different local microenvironments. Cells adapt to local microenvironments and differentiate into specialized subpopulation types (depicted by different colors and shapes) that enhance survival of the biofilm cybernetic unit. Created in BioRender. Podnar, E. (2024). BioRender.com/w33p163.
After reaching the air-liquid interface cells continue to grow, but cell division is impaired, resulting in formation of long bacterial filaments (Kobayashi, 2007; Chai et al., 2010; Krajnc et al., 2022). The entangled filament network provides a scaffold for newly synthesized extracellular matrix components (Chai et al., 2010) such as extracellular polysaccharides (EpsA-O) and proteins (TasA, TapA, SipW, BslA), the synthesis of which is upregulated during biofilm formation (Pisithkul et al., 2019). As the biofilm matures, it becomes protease-resistant (Romero et al., 2010; Diehl et al., 2018) and further bacterial growth leads to the wrinkle formation in fully mature biofilms (Trejo et al., 2013; Douarche et al., 2015).
Formation of the biofilm at the air-liquid interface is accompanied with a major role switch of the individual bacteria. One of the tenets of microbiology is that, when cells are homogenously mixed in an aqueous medium, cells behave identically, since they originate from a clonal ancestor and have identical genomes (Lopez et al., 2009). Although not explicitly stated, this also assumes that cells are in a uniform local environment with no chemical gradients. These assumptions can easily be challenged and questioned in the biofilm. In response to chemical gradient formation in biofilms multiple cell types are generated (Figure 1C) (Lopez et al., 2009). For example, a subpopulation of B. subtilis cells produces BslA protein at the air-biofilm interface which creates a hydrophobic barrier that prevent evaporation from the growing biofilm surface (Kobayashi and Iwano, 2012; Hobley et al., 2013). A different subpopulation become competent and is capable of taking up DNA from the environment (Dubnau, 1991; Dubnau and Provvedi, 2000), some cells can differentiate into dormant spores that are highly resistant to external stresses (Rudner and Losick, 2001; Piggot and Losick, 2014). Additionally, a subset of cells can produce an extracellular killing factor and toxin that kill (or cannibalize) cells that have not yet begun sporulation (González-Pastor et al., 2003). A fraction of the population produces extracellular matrix material that holds cells together (Vlamakis et al., 2008), and yet another subtype produces surfactin and other surface-active compounds (De Dier et al., 2015; Hölscher and Kovács, 2017) which provides antibacterial properties (Płaza et al., 2013; Horng et al., 2019; Chen X. et al., 2022). Clearly, gradients in biofilm enable several parallel feedback loops that regulate its development.
Biofilm at the air-liquid interface is now the most viable place for bacteria in the ecosystem and acts as an active barrier limiting oxygen penetration to the interior of the water column. Consequently, the number of viable bacteria in the water column below the biofilm structure significantly decreases with a concomitant increase of the dead cells in the sediment. There is, however, a limit to the growth of the biofilm at the air-liquid interface. At a critical biofilm thickness of approximately 250 μm, the transport of nutrients and water from the growth medium is impaired (Krajnc et al., 2022) and bacteria face yet another critical decision to survive. A subpopulation of cells at the forefront of the biofilm begins sporulation (Špacapan et al., 2020). During endospore formation, the mother cell undergoes lysis, which releases enzymes in the environment which will eventually terminate protease-resistant state in the biofilm and weaken its viscoelastic structure. The weakened biofilm is no longer able to support the bacterial mass. Bending of the surface increases shear force and bending moments in the plane of the biofilm, which exceeds the biofilm ability to support stress. The biofilm overturns and sediments to the bottom of the water column (Krajnc et al., 2022). The weakening of the extracellular matrix helps to release and disperse spores through air and liquid. The simple monoculture ecosystem has now entered into hibernation, a stable dormant state.
From a systems point of view, biofilm is not a permanent structure, it is better viewed as a temporary fix to the ecosystem crisis provoked and caused by bacterial activity. It is a multifaceted response induced by bacterial growth that causes population redistribution within the ecosystem, reduction of the viable habitat within the ecosystem to the air-liquid interface, it induces cell development, morphogenesis, and build-up of mechanical stress supporting structures in the biofilm (Krajnc et al., 2022). Biofilm development marks the climax of the ecosystem development, forcing bacteria to form social structures, invent division of labor, and cooperation (Claessen et al., 2014; Lyons and Kolter, 2015; Kalamara et al., 2018). However, living on a floating raft is precarious. The overexploitation of the resources exhausts the ecosystem’s capability to support bacterial growth but also at the same time bacteria sense the harsh conditions they have created and in response start the formation of endospores therefore enabling continuation of bacterial life (Nicholson et al., 2000; Ulrich et al., 2018).
Biofilm is therefore an essential but temporal cybernetic subsystem in the ecosystem that allows better bacterial survival. Cybernetic systems are those systems that correct or adjust their behavior based on feedback received form the environment towards some goal (Marinescu, 2017). Bacterial ultimate goal is long-term survival in the environment. To do so, bacteria gather information about the state of the local environment as tactile or sensory feedback and utilize genetic program to decide what to do with the information collected. For instance, B. subtilis monitors oxygen level and in response moves towards the air-liquid interface. Or it can sense the lack of nutrients, which activates one of its histidine sensor kinases and downstream genetic programs (Lazazzera, 2001; Eswaramoorthy et al., 2009; Qin et al., 2022). For instance, when environment deteriorates sensor kinase KinC is responsible for phosphorylation of master regulator Spo0A, which down-regulates transcriptional repressors AbrB and SinR (Figure 2). The downregulation of AbrB and SinR enables expression of genes for the production of extracellular matrix and formation of the biofilm while at the same time they block KinA phosphorylation and consequent entrance into sporulation (Branda et al., 2006; Vlamakis et al., 2013). An additional clue that conditions in the environment deteriorate are sensed via ppGpp or CodY-GTP cell energy status indicators and cell density monitoring via quorum sensing (Lombardía et al., 2006). For example, lactose related increase in AI-2 signal molecule production has been linked to biofilm formation (Duanis-Assaf et al., 2016). AI-2 is synthesized through LuxS pathway and is a soluble information carrier synchronizing the collective behavior of bacteria forming biofilm (Schauder et al., 2001).
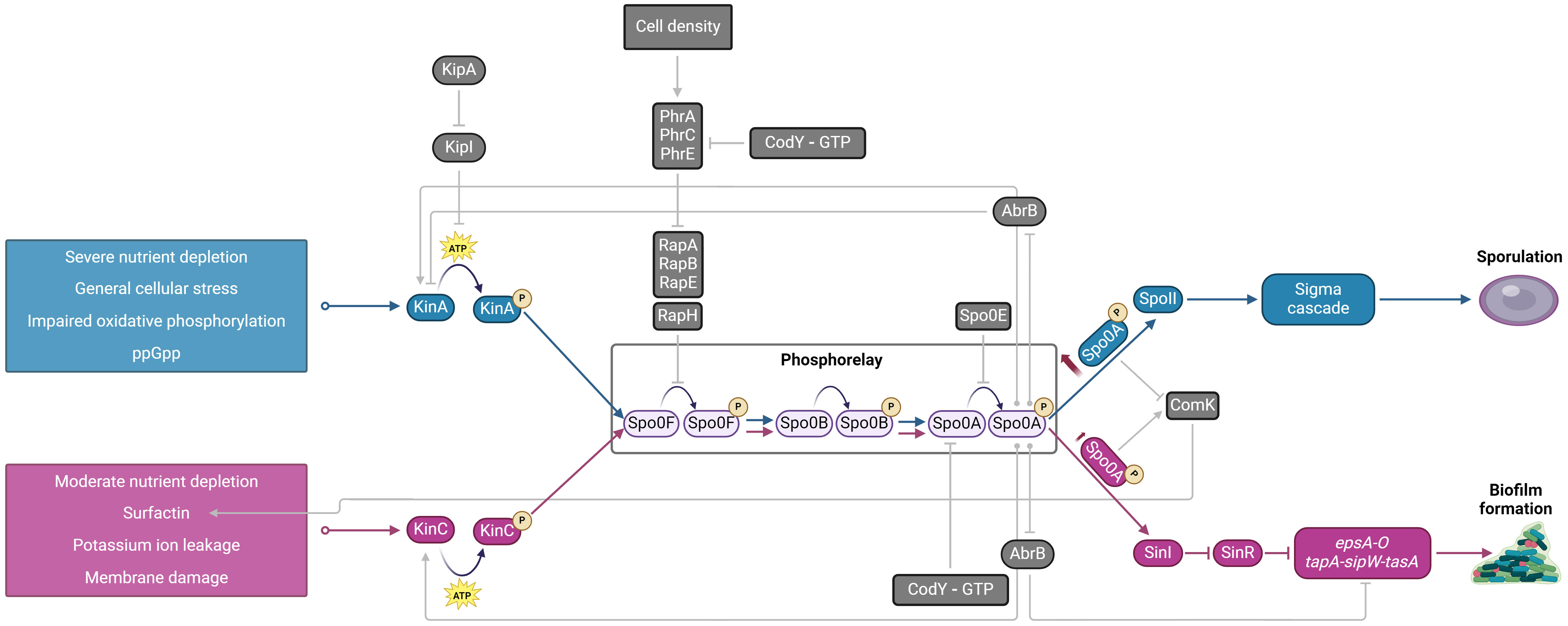
Figure 2. Schematic diagram of the cybernetic subsystem in Bacillus subtilis regulated via KinA (blue) and KinC (pink) sensor kinases that results in either sporulation or biofilm formation. Both biofilm matrix production and sporulation have a common phosphorelay pathway that activates Spo0A, a master transcriptional regulator. The two signaling pathways result in differential activation of Spo0A. High levels of Spo0A~P via KinA phosphorelay result in sporulation, whereas low levels of Spo0A~P via KinC result in biofilm formation. Additional external and internal signals that modulate this cybernetic subsystem are indicated in gray. KinA, sensor kinase; KinC, sensor kinase; KipI, inhibitor of KinA; KipA, inhibitor of KipI; Spo0F, Spo0B, Spo0A, phosphotransferases of the sporulation initiation phosphorelay; RapA, RapB, RapE, RapH, aspartate phosphatases, that dephosphorylate Spo0F; PhrA, PhrC, PhrE, aspartate phosphatase regulators; Spo0E, phosphatase; AbrB, transcriptional regulator, repressor of KinA, epsA-O and tapA-sipW-tasA operons; ComK, competence transcription factor, affects the production of surfactin, SinR, transcriptional regulator, epsA-O and tapA-sipW-tasA operons repressor, SinI, antagonist of SinR; epsA-O, extracellular polysaccharide synthesis; tapA-sipW-tasA, extracellular proteins synthesis, CodY, GTP-binding protein. Created in BioRender. Podnar, E. (2024) BioRender.com/b91s087.
Cells in the biofilm must make further decisions that bring population closer to survival. One such decision is to divide labor and to differentiate into various cell subpopulations (Shank and Kolter, 2011; Qin et al., 2022). Several physiological subpopulations were documented in the B. subtilis biofilms (i.e. EPS producers, surfactin producers, competence subpopulation, cannibals, epidermis builders, miners) (Lopez and Kolter, 2010). The differentiation into subpopulations is based on bistable gene switches (Lopez et al., 2009). The basis of the bistable response is a nonlinear induction of gene expression. Specifically, induction of a given gene leads to a stronger induction of the gene and once cells reach a certain threshold of gene expression, a hypersensitive response in the induction of the gene occurs (Dubnau and Losick, 2006). For instance, the cascade of genes required for a subpopulation of cells that are competent is induced by the regulator ComK. The expression of comK is self-induced by the ComK regulator, and when levels of ComK reach a threshold the expression of its own gene comK increases nonlinearly (Maamar and Dubnau, 2005; Smits et al., 2005). Increased concentration of ComX is sensed by membrane kinase ComP that phosphorylates ComA, which in a feedforward loop positively regulates surfactin biosynthesis. Increased surfactin concentration regulates activation of KinC which through Spo0 phosphorelay positively regulates more extracellular matrix production and consequently more biofilm growth. Through such cybernetic regulatory actions’ cells become firmly locked in a biofilm mode which ensures better survival in deteriorated environment.
The basic algorithm that governs the bacterial cybernetic actions can therefore be summarized as: take the inputs from the environment through sensors, make decision through signaling cascade and gene transcription, output an action that will bring bacteria closer to the goal of survival (i.e. produce EPS and form biofilm), loop through this process to further refine the outcome (differentiate into different subpopulations) until maximal reproduction and survival of the system is achieved. This cyclical process ensures that bacteria continuously adapt to the environment they are shaping, thereby optimizing their chances for survival and reproduction.
Inevitable, bacterial actions in a closed environment will exhaust the environmental resources. In case of severe nutrient depletion yet another output of the cybernetic system is triggered and bacteria enter the survival mode (formation of endospores). The entry into sporulation is a very well-studied bacterial cybernetic response that does not occur homogenously in the bacterial population, but rather occurs in a subpopulation of cells and is regulated by a series of feedback and feed-forward loops (Tan and Ramamurthi, 2014). As shown in Figure 2 the main trigger for endospore formation is severe nutrient deprivation that is sensed and feedforward through KinA sensor and Spo0A master transcriptional regulator. Spo0A is under the control of a positive feedback loop through activation of itself, positive control of KinA and KinC sensors as well as by a modified double-repression system (Lopez et al., 2009). In the latter case, the Spo0A acts as a repressor which represses abrB, which represses sigH, which is an activator of spo0A. Therefore, until a threshold of Spo0A is reached, AbrB will repress sigH, keeping Spo0A levels low. Low levels of Spo0A are conductive for biofilm growth. On the other hand, an increase of Spo0A expression will alleviate the AbrB-mediated repression of SigH and the balance will be shifted to induce further spo0A expression (Strauch et al., 1990; Predich et al., 1992) which will trigger a subpopulation of cells to switch to a sporulation pathway.
The decision to go to the biofilm or to the endospore pathway is made based on the environmental input. The environmental inputs are mainly but not exclusively channeled through KinA and KinC sensors (Figure 2). KinA is activated in response to severe nutrient depletion (Tojo et al., 2013), impaired oxidative phosphorylation (Kolodkin-Gal et al., 2013), and general cellular stress cues such as ppGpp (Tojo et al., 2013). This ensures that irreversible commitment to sporulation is initiated only when cells experience severe environmental stress. KinA drives the accumulation of Spo0A~P to levels that trigger the activation of genes essential for sporulation (Veening et al., 2004, 2005). At high concentrations, Spo0A~P also represses the expression of SinI, thereby preventing biofilm formation saving the energy and reinforcing the cell’s commitment to sporulation (Chen et al., 2023). On the other hand, when environmental conditions are not as severe and there is a moderate nutrient depletion, KinC kinases acts on a common phosphorelay system and produce lower levels of Spo0A~P (Chen Z. et al., 2022). Lower levels of Spo0A~P promotes the expression of SinI (Fujita et al., 2005), which sequesters and inactivates SinR (Bai et al., 1993; Chai et al., 2008), a transcriptional repressor of biofilm-related genes. The inactivation of SinR results in the derepression of the epsA-O and tapA-sipW-tasA operons, leading to the production of the extracellular matrix necessary for biofilm formation (Kearns et al., 2005; Chu et al., 2006). In addition, as shown in Figure 2, there are other external and internal signals (i.e. PhrA, PhrC, PhrE, GTP energy status, ComK) that fine tune and complement KinA and KinC system.
As demonstrated for B. subtilis cybernetic system, there is no single solution to a given goal of survival. At any time, different phenotypes are present and ready to act, which explains why in bacteria bet hedging strategy is so successful. In suspension planktonic life style is the best solution when resources are plentiful and bacterial numbers are low. When environment deteriorates swimmers are better suited and dominate the population, only to be superseded by air-liquid dwellers where surfactin producers may get an upper hand, which in turn will give way to extracellular matrix producers and biofilm formers. In starvation cannibals may be best adapted, but finally when conditions are not compatible with vegetative growth endospore formers will ensure long-term survival of the population. However, even endospore is not a permanent solution for survival as its viability will eventually decay and spore will need a periodical rejuvenation. In conclusion, biofilm is just one of the possible outputs of the bacterial cybernetic system to the ever changing environment. It is not invariably the best response. Biofilm is a rather complex cybernetic system that is composed of different subpopulations that are themselves composed of individual bacteria which are the fundamental cybernetic units in the ecosystem able to respond to the changing environment ensuring the long-term survival.
MB: Conceptualization, Writing – review & editing, Investigation, Writing – original draft. DS: Conceptualization, Writing – review & editing, Funding acquisition, Supervision.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The work was supported by the Slovenian Research Agency (ARIS), research project L7-3186, L2-50060, as well as national program grant P4-0116.
We sincerely thank Dr. Eli Podnar for her help with BioRender.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bai U., Mandic-Mulec I., Smith I. (1993). SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 7, 139–148. doi: 10.1101/GAD.7.1.139
Branda S. S., Chu F., Kearns D. B., Losick R., Kolter R. (2006). A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59, 1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x
Chai Y., Chu F., Kolter R., Losick R. (2008). Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 67, 254–263. doi: 10.1111/j.1365-2958.2007.06040.x
Chai Y., Kolter R., Losick R. (2010). Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability. Mol. Microbiol. 78, 218–229. doi: 10.1111/J.1365-2958.2010.07335.X
Chen X., Lu Y., Shan M., Zhao H., Lu Z., Lu Y. (2022). A mini-review: mechanism of antimicrobial action and application of surfactin. World J. Microbiol. Biotechnol. 38, 1–10. doi: 10.1007/S11274-022-03323-3/FIGURES/2
Chen Z., Srivastava P., Zarazúa-Osorio B., Marathe A., Fujita M., Igoshin O. A. (2022). Bacillus subtilis histidine kinase KinC activates biofilm formation by controlling heterogeneity of single-cell responses. MBio 13, 1–19. doi: 10.1128/MBIO.01694-21
Chen Z., Zarazúa-Osorio B., Srivastava P., Fujita M., Igoshin O. A. (2023). The slowdown of growth rate controls the single-cell distribution of biofilm matrix production via an SinI-SinR-SlrR network. mSystems 8, 1–19. doi: 10.1128/MSYSTEMS.00622-
Chu F., Kearns D. B., Branda S. S., Kolter R., Losick R. (2006). Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol. Microbiol. 59, 1216–1228. doi: 10.1111/J.1365-2958.2005.05019.X
Claessen D., Rozen D. E., Kuipers O. P., Søgaard-Andersen L., Van Wezel G. P. (2014). Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 12, 115–124. doi: 10.1038/nrmicro3178
Contreras-Moreno F. J., Pérez J., Muñoz-Dorado J., Moraleda-Muñoz A., Marcos-Torres F. J. (2024). Myxococcus xanthus predation: an updated overview. Front. Microbiol. 15. doi: 10.3389/FMICB.2024.1339696
da Silva S. B., Cantarelli V. V., Ayub M. A. Z. (2013). Production and optimization of poly-γ-glutamic acid by Bacillus subtilis BL53 isolated from the Amazonian environment. Bioprocess Biosyst. Eng. 37, 469–479. doi: 10.1007/S00449-013-1016-1
De Dier R., Fauvart M., Michiels J., Vermant J. (2015). “The role of biosurfactants in bacterial systems,” in The physical basis of bacterial quorum communication, ed. Hagen S. (New York, NY: Springer), 189–204.
Dergham Y., Sanchez-Vizuete P., Le Coq D., Deschamps J., Bridier A., Hamze K., et al. (2021). Comparison of the genetic features involved in Bacillus subtilis biofilm formation using multi-culturing approaches. Microorganisms 9, 1–17. doi: 10.3390/microorganisms9030633
Diehl A., Roske Y., Ball L., Chowdhury A., Hiller M., Molière N., et al. (2018). Structural changes of TasA in biofilm formation of Bacillus subtilis. Proc. Natl. Acad. Sci. 115, 3237–3242. doi: 10.1073/PNAS.1718102115
Dogsa I., Kostanjšek R., Stopar D. (2023). eDNA provides a scaffold for autoaggregation of B. subtilis in bacterioplankton suspension. Microorganisms 11, 322. doi: 10.3390/microorganisms11020332
Douarche C., Allain J. M., Raspaud E. (2015). Bacillus subtilis bacteria generate an internal mechanical force within a biofilm. Biophys. J. 109, 2195. doi: 10.1016/J.BPJ.2015.10.004
Duanis-Assaf D., Steinberg D., Chai Y., Shemesh M. (2016). The LuxS based quorum sensing governs lactose induced biofilm formation by Bacillus subtilis. Front. Microbiol. 6. doi: 10.3389/FMICB.2015.01517/BIBTEX
Dubnau D. (1991). Genetic competence in Bacillus subtilis. Microbiol. Rev. 55, 395–424. doi: 10.1128/MR.55.3.395-424.1991
Dubnau D., Losick R. (2006). Bistability in bacteria. Mol. Microbiol. 61, 564–572. doi: 10.1111/J.1365-2958.2006.05249.X
Dubnau D., Provvedi R. (2000). Internalizing DNA. Res. Microbiol. 151, 475–480. doi: 10.1016/S0923-2508(00)00166-2
Eswaramoorthy P., Guo T., Fujita M. (2009). In vivo domain-based functional analysis of the major sporulation sensor kinase, KinA, in Bacillus subtilis. J. Bacteriol. 191, 5358. doi: 10.1128/JB.00503-09
Flemming H. C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Fujita M., González-Pastor J. E., Losick R. (2005). High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187, 1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005/SUPPL_FILE/TABLE_S.DOC
González-Pastor J. E., Hobbs E. C., Losick R. (2003). Cannibalism by sporulating bacteria. Science 301, 510–513. doi: 10.1126/SCIENCE.1086462
Hamon M. A., Stanley N. R., Britton R. A., Grossman A. D., Lazazzera B. A. (2004). Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52, 847–860. doi: 10.1111/J.1365-2958.2004.04023.X
Hobley L., Ostrowski A., Rao F. V., Bromley K. M., Porter M., Prescott A. R., et al. (2013). BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc. Natl. Acad. Sci. U.S.A. 110, 13600–13605. doi: 10.1073/PNAS.1306390110/-/DCSUPPLEMENTAL/PNAS.201306390SI.PDF
Hölscher T., Bartels B., Lin Y., Gallegos-Monterrosa R., Price-Whelan A., Kolter R., et al. (2015). Motility, chemotaxis and aerotaxis contribute to competitiveness during bacterial pellicle biofilm development. J. Mol. Biol. 427, 3695–3708. doi: 10.1016/J.JMB.2015.06.014
Hölscher T., Kovács Á.T. (2017). Sliding on the surface: bacterial spreading without an active motor. Environ. Microbiol. 19, 2537–2545. doi: 10.1111/1462-2920.13741
Hopkins M. M., Fauci L. J. (2002). A computational model of the collective fluid dynamics of motile micro-organisms. J. Fluid Mech. 455, 149–174. doi: 10.1017/S0022112001007339
Horng Y. B., Yu Y. H., Dybus A., Hsiao F. S. H., Cheng Y. H. (2019). Antibacterial activity of Bacillus species-derived surfactin on Brachyspira hyodysenteriae and Clostridium perfringens. AMB Express 9, 1–9. doi: 10.1186/S13568-019-0914-2/TABLES/4
Jánosi I. M., Kessler J. O., Horváth V. K. (1998). Onset of bioconvection in suspensions of Bacillus subtilis. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics 58, 4793–4800. doi: 10.1103/PhysRevE.58.4793
Kalamara M., Spacapan M., Mandic-Mulec I., Stanley-Wall N. R. (2018). Social behaviours by Bacillus subtilis: quorum sensing, kin discrimination and beyond. Mol. Microbiol. 110, 863–878. doi: 10.1111/MMI.14127
Kearns D. B., Chu F., Branda S. S., Kolter R., Losick R. (2005). A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55, 739–749. doi: 10.1111/J.1365-2958.2004.04440.X
Kobayashi K. (2007). Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J. Bacteriol. 189, 4920–4931. doi: 10.1128/JB.00157-07
Kobayashi K., Iwano M. (2012). BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol. Microbiol. 85, 51–66. doi: 10.1111/J.1365-2958.2012.08094.X
Koch D. L., Subramanian G. (2011). Collective hydrodynamics of swimming microorganisms: Living fluids. Annu. Rev. Fluid Mech. 43, 637–659. doi: 10.1146/ANNUREV-FLUID-121108-145434/CITE/REFWORKS
Kolodkin-Gal I., Elsholz A. K. W., Muth C., Girguis P. R., Kolter R., Losick R. (2013). Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase. Genes Dev. 27, 887. doi: 10.1101/GAD.215244.113
Krajnc M., Fei C., Košmrlj A., Kalin M., Stopar D. (2024). Mechanical constraints to unbound expansion of B. subtilis on semi-solid surfaces. Microbiol. Spectr. 12, 1–16. doi: 10.1128/SPECTRUM.02740-23/SUPPL_FILE/REVIEWER-COMMENTS.PDF
Krajnc M., Stefanic P., Kostanjšek R., Mandic-Mulec I., Dogsa I., Stopar D. (2022). Systems view of Bacillus subtilis pellicle development. NPJ Biofilms Microb. 8, 1–11. doi: 10.1038/s41522-022-00293-0
Lazazzera B. A. (2001). The intracellular function of extracellular signaling peptides. Peptides 22, 1519–1527. doi: 10.1016/S0196-9781(01)00488-0
Lee L. M., Rosenberg G., Rubinstein S. M. (2019). A sequence of developmental events occurs underneath growing Bacillus subtilis pellicles. Front. Microbiol. 10. doi: 10.3389/FMICB.2019.00842/BIBTEX
Lombardía E., Rovetto A. J., Arabolaza A. L., Grau R. R. (2006). A LuxS-dependent cell-to-cell language regulates social behavior and development in Bacillus subtilis. J. Bacteriol. 188, 4442–4452. doi: 10.1128/jb.00165-06/asset/41a986ff-8dc8-4e6c-a490-a9e914bbc4db/assets/graphic/zjb0120658220007.jpeg
Lopez D., Kolter R. (2010). Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol. Rev. 34, 134–149. doi: 10.1111/j.1574-6976.2009.00199.x
Lopez D., Vlamakis H., Kolter R. (2009). Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 33, 152–163. doi: 10.1111/J.1574-6976.2008.00148.X
Lyons N. A., Kolter R. (2015). On the evolution of bacterial multicellularity. Curr. Opin. Microbiol. 24, 21–28. doi: 10.1016/J.MIB.2014.12.007
Maamar H., Dubnau D. (2005). Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56, 615–624. doi: 10.1111/J.1365-2958.2005.04592.X
Marinescu D. C. (2017). “Complex systems”, in Complex Systems and Clouds, ed. Marinescu D. C. (Amsterdam: Elsevier), 1–32.
Miller B. S., Diaz-Torres M. R. (1999). Proteome analysis of biofilms: Growth of Bacillus subtilis on solid medium as model. Methods Enzymol. 310, 433–441. doi: 10.1016/S0076-6879(99)10034-X
Mitchell J. G., Kogure K. (2006). Bacterial motility: links to the environment and a driving force for microbial physics. FEMS Microbiol. Ecol. 55, 3–16. doi: 10.1111/J.1574-6941.2005.00003.X
Nandy S. K., Bapat P. M., Venkatesh K. V. (2007). Sporulating bacteria prefers predation to cannibalism in mixed cultures. FEBS Lett. 581, 151–156. doi: 10.1016/J.FEBSLET.2006.12.011
Nicholson W. L., Munakata N., Horneck G., Melosh H. J., Setlow P. (2000). Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64, 548. doi: 10.1128/MMBR.64.3.548-572.2000
Piggot P. J., Losick R. (2002). “Sporulation genes and intercompartmental regulation” in Bacillus subtilis and its closest relatives: From genes to cells, ed. Sonenshein A. L., Hoch J. A., Losick R. (Washington: American Society for Microbiology), 483–518.
Pisithkul T., Schroeder J. W., Trujillo E. A., Yeesin P., Stevenson D. M., Chaiamarit T., et al. (2019). Metabolic remodeling during biofilm development of Bacillus subtilis. mBio 10, 1–32. doi: 10.1128/mBio.00623-19
Płaza G. A., Turek A., Król E., Szczygłowska R. (2013). Antifungal and antibacterial properties of surfactin isolated from Bacillus subtilis growing on molasses. Afr. J. Microbiol. Res. 7, 3165–3170. doi: 10.5897/AJMR2013.5565
Predich M., Nair G., Smith I. (1992). Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing sigma H. J. Bacteriol. 174, 2771–2778. doi: 10.1128/JB.174.9.2771-2778.1992
Qin Y., Angelini L. L., Chai Y. (2022). Bacillus subtilis cell differentiation, biofilm formation and environmental prevalence. Microorganisms 10, 1–10. doi: 10.3390/MICROORGANISMS10061108
Romero D., Aguilar C., Losick R., Kolter R. (2010). Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 107, 2230–2234. doi: 10.1073/pnas.0910560107
Rozen D. E., Philippe N., Arjan De Visser J., Lenski R. E., Schneider D. (2009). Death and cannibalism in a seasonal environment facilitate bacterial coexistence. Ecol. Lett. 12, 34–44. doi: 10.1111/J.1461-0248.2008.01257.X
Rudner D. Z., Losick R. (2001). Morphological coupling in development: lessons from prokaryotes. Dev. Cell 1, 733–742. doi: 10.1016/S1534-5807(01)00094-6
Samad T., Billings N., Birjiniuk A., Crouzier T., Doyle P. S., Ribbeck K. (2017). Swimming bacteria promote dispersal of non-motile Staphylococcal species. ISME J. 11, 1933–1937. doi: 10.1038/ISMEJ.2017.23
Sanchez-Vizuete P., Dergham Y., Bridier A., Deschamps J., Dervyn E., Hamze K., et al. (2021). The coordinated population redistribution between Bacillus subtilis submerged biofilm and liquid-air pellicle. Biofilm 4, 1–8. doi: 10.1016/J.BIOFLM.2021.100065
Schauder S., Shokat K., Surette M. G., Bassler B. L. (2001). The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41, 463–476. doi: 10.1046/J.1365-2958.2001.02532.X
Shank E. A., Kolter R. (2011). Extracellular signaling and multicellularity in Bacillus subtilis. Curr. Opin. Microbiol. 14, 741–747. doi: 10.1016/J.MIB.2011.09.016
Shoup D., Ursell T. (2023). Bacterial bioconvection confers context-dependent growth benefits and is robust under varying metabolic and genetic conditions. J. Bacteriol. 205, e0023223. doi: 10.1128/jb.00232-23
Smits W. K., Eschevins C. C., Susanna K. A., Bron S., Kuipers O. P., Hamoen L. W. (2005). Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol. Microbiol. 56, 604–614. doi: 10.1111/J.1365-2958.2005.04488.X
Špacapan M., Danevčič T., Štefanic P., Porter M., Stanley-Wall N. R., Mandic-Mulec I. (2020). The ComX quorum sensing peptide of Bacillus subtilis affects biofilm formation negatively and sporulation positively. Microorganisms 8, 1131. doi: 10.3390/MICROORGANISMS8081131
Sretenovic S., Stojković B., Dogsa I., Kostanjšek R., Poberaj I., Stopar D. (2017). An early mechanical coupling of planktonic bacteria in dilute suspensions. Nat. Commun. 8, 1–10. doi: 10.1038/s41467-017-00295-z
Strauch M., Webb V., Spiegelman G., Hoch J. A. (1990). The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. U.S.A. 87, 1801–1805. doi: 10.1073/PNAS.87.5.1801
Tan I. S., Ramamurthi K. S. (2014). Spore formation in Bacillus subtilis. Environ. Microbiol. Rep. 6, 212. doi: 10.1111/1758-2229.12130
Thiery S., Kaimer C. (2020). The predation strategy of myxococcus xanthus. Front. Microbiol. 11. doi: 10.3389/FMICB.2020.00002/BIBTEX
Tojo S., Hirooka K., Fujita Y. (2013). Expression of kinA and kinB of Bacillus subtilis, necessary for sporulation initiation, is under positive stringent transcription control. J. Bacteriol. 195, 1656–1665. doi: 10.1128/JB.02131-12
Tosato V., Bruschi C. V. (2004). Knowledge of the Bacillus subtilis genome: Impacts on fundamental science and biotechnology. Appl. Microbiol. Biotechnol. 64, 1–6. doi: 10.1007/S00253-003-1513-2/METRICS
Trejo M., Douarche C., Bailleux V., Poulard C., Mariot S., Regeard C., et al. (2013). Elasticity and wrinkled morphology of Bacillus subtilis pellicles. Proc. Natl. Acad. Sci. U.S.A. 110, 2011–2016. doi: 10.1073/pnas.1217178110
Ulrich N., Nagler K., Laue M., Cockell C. S., Setlow P., Moeller R. (2018). Experimental studies addressing the longevity of Bacillus subtilis spores – The first data from a 500-year experiment. PloS One 13, 1–14. doi: 10.1371/JOURNAL.PONE.0208425
Veening J. W., Hamoen L. W., Kuipers O. P. (2005). Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis. Mol. Microbiol. 56, 1481–1494. doi: 10.1111/J.1365-2958.2005.04659.X
Veening J. W., Smits W. K., Hamoen L. W., Jongbloed J. D. H., Kuipers O. P. (2004). Visualization of differential gene expression by improved cyan fluorescent protein and yellow fluorescent protein production in Bacillus subtilis. Appl. Environ. Microbiol. 70, 6809–6815. doi: 10.1128/AEM.70.11.6809-6815.2004/ASSET/40421C5C-A35D-46BA-B3A6-38E245443DC4/ASSETS/GRAPHIC/ZAM0110449180006.JPEG
Vlamakis H., Aguilar C., Losick R., Kolter R. (2008). Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 22, 945–953. doi: 10.1101/GAD.1645008
Vlamakis H., Chai Y., Beauregard P. B., Losick R., Kolter R. (2013). Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168. doi: 10.1038/NRMICRO2960
Wan N., Wang H., Ng C. K., Mukherjee M., Ren D., Cao B., et al. (2018). Bacterial metabolism during biofilm growth investigated by13C tracing. Front. Microbiol. 9. doi: 10.3389/FMICB.2018.02657/FULL
Xu S., Cao Q., Liu Z., Chen J., Yan P., Li B., et al. (2022). Transcriptomic analysis reveals the role of tmRNA on biofilm formation in Bacillus subtilis. Microorganisms 10, 1–22. doi: 10.3390/MICROORGANISMS10071338/S1
Keywords: Bacillus subtilis, cybernetic system, systems view, regulation, biofilms
Citation: Blaznik M and Stopar D (2024) B. subtilis biofilm as a cybernetic system. Front. Bacteriol. 3:1497132. doi: 10.3389/fbrio.2024.1497132
Received: 16 September 2024; Accepted: 21 October 2024;
Published: 06 November 2024.
Edited by:
Luisa Jordao, National Health Institute Doutor Ricardo Jorge (INSA), PortugalReviewed by:
Brandon W. Peterson, University Medical Center Groningen, NetherlandsCopyright © 2024 Blaznik and Stopar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Stopar, ZGF2aWQuc3RvcGFyQGJmLnVuaS1sai5zaQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.